Abstract
Mutations in RPGRORF15 are associated with rod-cone or cone/cone-rod dystrophy, the latter associated with mutations at the distal end. We describe the phenotype associated with a novel variant in the terminal codon of the RPGRORF15 c.3457T>A (Ter1153Lysext*38), which results in a C-terminal extension. Three male patients from two families were recruited, aged 31, 35, and 38 years. Genetic testing was performed by whole exome sequencing. Filtered variants were analysed according to the population frequency, ClinVar database, the variant’s putative impact, and predicted pathogenicity; and were classified according to the ACMG guidelines. Examination included visual acuity (Snellen), colour vision (Ishihara), visual field, fundus autofluorescence (FAF), optical coherence tomography (OCT), and electrophysiology. All patients were myopic, and had central scotoma and reduced colour vision. Visual acuities on better eyes were counting fingers, 0.3 and 0.05. Electrophysiology showed severely reduced cone-specific responses and macular dysfunction, while the rod-specific response was normal. FAF showed hyperautofluorescent ring centred at the fovea encompassing an area of photoreceptor loss approximately two optic discs in diameter (3462–6342 μm). Follow up after 2–11 years showed enlargement of the diameter (avg. 100 μm/year). The novel c.3457T>A (Ter1153Lysext*38) mutation in the terminal RPGRORF15 codon is associated with cone dystrophy, which corresponds to the previously described phenotypes associated with mutations in the distal end of the RPGRORF15. Minimal progression during follow-up years suggests a relatively stable disease after the initial loss of the central cones.
Keywords: RPGR, ORF15, cone-dystrophy
1. Introduction
1.1. Molecular Genetics of RPGR
RPGR (retinitis pigmentosa GTPase regulator) is a gene located on the X chromosome with several different protein coding transcripts, expressed in various tissues. The two major RPGR isoforms are a constitutive RPGRex1–19, derived from exons 1–19, encoding a protein of 815 amino acids; and RPGRORF15, which shares exons 1–14 with the constitutive isoform and contains a large ORF15 as its terminal exon, encoding a protein of 1152 amino acids [1] (illustrated in Figure 1). Both are expressed in the retina, with the RPGRORF15 being the predominant one [2].
Figure 1.
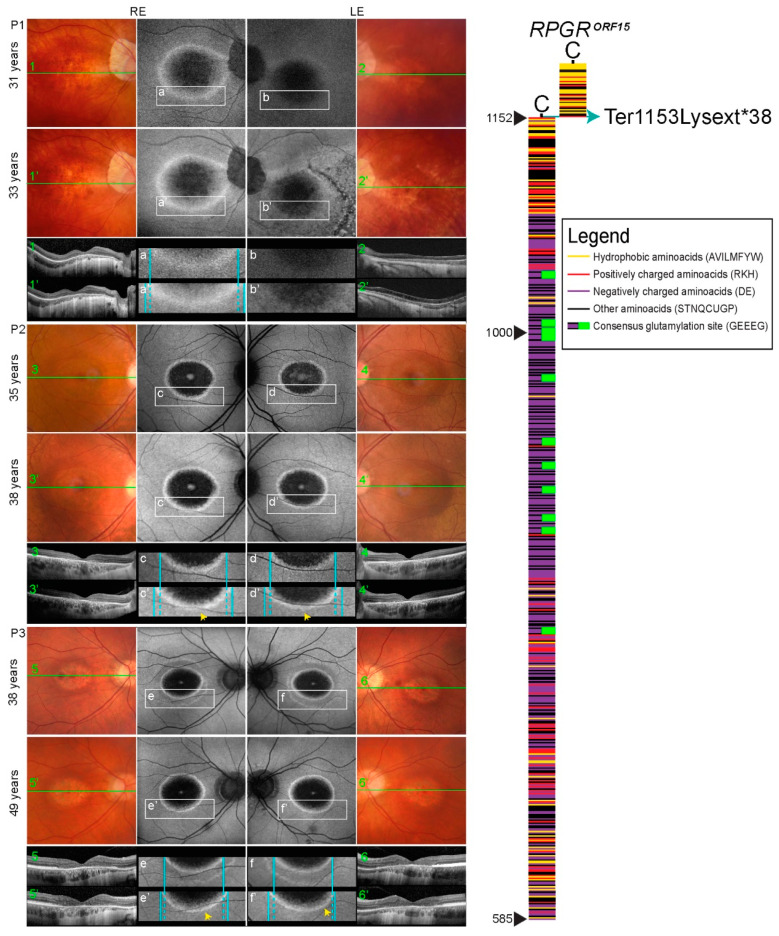
Retinal imaging (colour fundus, FAF, OCT) and RPGRORF15 scheme. Colour fundus images (1–6, 1′–6′ represent follow-up), corresponding FAF (a–f, a’–f’ represent follow-up) and ring enlargement demonstration (a–f, a’–f’ represent follow-up), corresponding OCT (1–6, 1′-6′ represent follow-up) of all patients included in the study, from their first and last examination. Colour fundus images in BE of all the patients show Bull’s eye appearance of the macula, optic pallor, and attenuated vessels; Patient 1 in his LE (2 and 2′) also presents bone spicules in the central and peripheral retina due to retinal detachment. FAF in BE of all the patients show the hyperautofluorescent ring; Patient 1 in his LE (b and b’) also presents RPE mottling in the inferior temporal retina (after retinal detachment). All the patients showed the hyperautofluorescent ring enlargement displayed in Figure 4. OCT in BE of all the patients shows absent RPE, Ise, and ELM in the central macula; Patient 2 in his LE (d and d’) and Patient 3 in his BE (e and e’, f and f’) present remnants of ELM in the foveola. In the right part, schematic representation of the amino acid structure of the RPGRORF15 and the predicted effect of the mutation c.3457T>A (Ter1153Lysext*38) elongating the protein by 38 aminoacids, is being presented.
1.2. Function of the RPGR Protein
The function of the RPGR protein has not yet been fully elucidated. The RPGRex1–19 isoform is widely expressed in tissues [2,3,4] and is found within cells at the transition zone of primary and motile cilia [5,6], or at centrosomes and their constituent centrioles in dividing cells [7]. In the retina, the RPGRex1–19 isoform localises to the developing and mature photoreceptor connecting cilium (CC), and shows a slightly different developmental expression pattern and affinity for the axonemal fraction compared with the other major RPGR isoform RPGRORF15, suggesting overlapping, but also distinct, functions [8]. The RPGRex1–19 transcript encodes a predicted 90kDa protein. The C-terminus of the RPGRex1–19 has an isoprenylation signal, suggesting that this isoform is membrane bound, consistent with its reported attachment to endoplasmic reticulum membranes in addition to its presence at the connecting cilium [9]. There has also been suggested a localization of RPGRex1–19 isoform to the Golgi [10] and other compartments in the inner segment that may indicate a possible involvement of RPGR in post-Golgi sorting of cargo-containing vesicles, phototransduction components, and other outer segment proteins towards the connecting cilium [7,9,11]. The RPGRORF15 isoform is predicted to be 127 kDa and exon ORF15 includes an acidic, repetitive, glutamic acid/glycine-rich domain and a basic C-terminal domain [12]. The repetitive domain length is varying considerably among species [2,13]. In contrast, the basic C-terminal domain is highly conserved among vertebrates [14]. In dividing cells, RPGRORF15 is present in centrosomes, while in nondividing cells containing primary cilia it is found in the transition zone of the ciliary axoneme, as mentioned, the equivalent structure to the photoreceptor connecting cilium [5,14,15]. Some studies suggest that RPGRORF15 binds to the basal body and the axoneme [7,16]. Other studies suggest that RPGRORF15 is present in the photoreceptor outer segment (OS), however the studies are inconclusive, possibly due to differences between laboratories because of the use of different antibodies, tissue processing procedures, or species differences in the OS structure [17,18]. The exons 1–14 of both isoforms encode a structure similar to regulator of chromosome condensation 1 (RCC1) at the N-terminus [3,19]. RCC1 is a well characterised protein that functions as a guanine nucleotide exchange factor for Ran (a Ras-related nuclear protein) and is thought to play an important role in nucleocytoplasmic transport and regulation of cell division as well as the predominant ciliary localization [1,7,20]. RanGTP serves as an energy source for molecular motors that move cargo through the nuclear pore complex [11]. The resultant high concentration of RanGTP in the connecting cilium as generated by RPGR could enable a putative RanGTP-dependent process that drives unidirectional movement of opsins and other cargo across the connecting cilium to the outer segment [21,22]. RPGR-interacting protein (RPGRIP1) localises to the connecting cilia and is thought to hold RPGR in this location because it consists of an N-terminal region predicted to form coiled coil structure linked to a C-terminal tail that binds RPGR [23]. RPGR interacts with several other ciliary proteins, some of which have also been associated with cone/cone-rod dystrophy (reviewed in Discussion).
1.3. Phenotypes Associated with Mutations in RPGR
To date, over 300 disease-causing variants have been identified in RPGR [2,24]. All were found in the shared exons of both isoforms and the exon ORF15, while none were reported in the exons 16–19 [25]. The ORF15 exon contains an unusual repetitive sequence encoding 567 amino acids rich in glycine and glutamic acid residues that is considered to be a ‘mutational hot spot’ [2].
Two distinctly different phenotypes have been recognised in RPGR patients, the more frequent primarily affecting rods (retinitis pigmentosa, RP), and the other primarily affecting cones (cone/cone-rod dystrophy) [26,27]. Myopia is a common feature in both [27,28]. RPGR-associated RP is one of the most frequent (over 70% of X-linked RP [26]) and most severe forms of retinitis pigmentosa [29]. It is characterized with presentation in childhood, with first reported symptoms being nyctalopia and peripheral visual loss [27]. RPGR is the causative gene in approximately 1% cases of cone/cone-rod dystrophy [30,31]. It is characterised by central visual loss (reduced acuity, colour vision, and central scotoma), and in some patients photophobia [29]. The patients with rod system dysfunction also report night blindness and may exhibit peripheral visual field loss [29]. Most patients have myopia, with 50–72% having a refractive error of greater than −6 dioptres [27,28]. ERG in cone dystrophy typically shows delayed and reduced light adapted (LA) ERGs and, in cone-rod dystrophy also abnormal dark adapted (DA) ERGs [32]. There is typically early and severe macular involvement in cone/cone-rod dystrophy cases, characterised by pattern ERG reduction, although pattern ERGs might be relatively high in young or mild cases, with relatively small rings of increased parafoveolar FAF [32]. Due to the X-linked recessive inheritance of RPGR, the disease predominantly affects males, however, a retinal phenotype can also be seen in female carriers, caused by the inactivation of the normal X chromosome, the severity depending on the degree of normal X chromosome inactivation [33].
Fundus autofluorescence imaging (FAF) in RPGR retinopathy often reveals parafoveal rings of increased autofluorescence, delineating the border between the affected and unaffected retina [27,30,32,34,35] and can be used to follow the disease progression. In RP patients, the area within the ring corresponds to the preserved part of the retina [34,36]. This is common to RP of different genetic background and many studies have shown the progressive constriction of the ring with time [37,38,39,40]. On the contrary, the rings in cone/cone-rod dystrophy encompass the area of degenerating retina [27,30,32]. Studies have shown increasing of the ring diameter with time in RPGR cone dystrophy cases [31,35,41].
It is not known why the RPGR mutations result in two contrasting disorders. It seems that the phenotype depends on the location of the mutation: Mutations in the exons 1–14 and the proximal part of the ORF15 exon usually result in retinitis pigmentosa, while the mutations in the distal end of the ORF15 exon cause cone/cone-rod dystrophy [11,29,42,43,44]. There is however a watershed zone of approximately 100 aminoacids between the two regions, where mutations can result in either phenotype, even within the same family [29], the reasons for which are not understood. It is also not clear whether the cone and cone-rod subtypes also depend on the mutation location.
We present the clinical characteristics of three patients harbouring a novel C-terminal extension variant resulting in cone dystrophy due to the loss of the terminal codon of the RPGRORF15, c.3457T>A (Ter1153Lysext*38).
2. Materials and Methods
2.1. Patients
The study included three male patients from two families aged 31, 35, and 38 years, ascertained from the Eye Hospital University Medical Centre Ljubljana, Slovenia. The study was conducted in agreement with the Declaration of Helsinki. Informed written consent was obtained from the patients.
2.2. Genetic and Bioinformatic Analysis
Genetic analysis was performed in a proband from each family (Patient 1 and Patient 2). Genomic DNA was extracted from blood samples according to the standard procedure. Whole exome sequencing was performed. Sequencing of the defined clinical target was performed using next-generation sequencing on the isolated DNA sample. Briefly, the fragmentation and enrichment of the isolated DNA sample were performed according to the Illumina Nextera Coding Exome capture protocol, with subsequent sequencing on Illumina NextSeq 550 in 2 × 100 cycles (Illumina, San Diego, CA, USA). After duplicates were removed, the reads were aligned to the UCSC hg19 reference assembly using the BWA algorithm (v0.6.3) and variants were called using the GATK framework (v2.8). Only variants exceeding the quality score of 30.0 and depth of 5 were used for down-stream analyses. Variant annotation was performed using ANNOVAR and snpEff algorithms, with pathogenicity predictions in dbNSFPv2 database. Reference gene models and transcript sequences are based on the RefSeq database. Structural variants were assessed using CONIFER v0.2.2 algorithm. Variants with population frequency exceeding 1% in gnomAD, synonymous variants, intronic variants and variants outside the clinical target were filtered out during analyses. An in-house pipeline was used for bioinformatic analyses of exome sequencing data, in accordance with GATK best practice recommendations [45]. The interpretation of sequence variants was based on ACMG/AMP standards and guidelines [46]. Sequencing the DNA sample, we reached median coverage of 67× and covered over 99.9% targeted regions with minimum 10× depth of coverage [47]. The presence of the mutation in the population was examined in the gnomAD database (gnomad.broadinstitute.org, accessed on 06.01.2021). Single-nucleotide polymorphism (SNP) analysis comparing X-chromosomes of probands from each family was performed in order to inspect the possibility of a founder effect.
2.3. Clinical Examination
An accurate family history was recorded, and all patients underwent a complete ophthalmic examination, which included best-corrected visual acuity (Snellen), slit lamp biomicroscopy, and dilated fundus examination. Retinal fundus photographs were obtained by conventional 35° fundus colour photographs (Topcon, Tokyo, Japan). Fundus autofluorescence imaging (FAF) (30° and 55° of the central retina) and optical coherence tomography (OCT) extending 8 mm of the macula was performed with a confocal scanning laser ophthalmoscope (Spectralis; Heidelberg Engineering, Heidelberg, Germany). The horizontal diameters and areas of the hyperautofluorescent rings on FAF were measured using an automated viewing module available in the Spectralis software. The outer border of the ring was used for the measurement. The integrity of the photoreceptors was determined by qualitatively assessing the inner segment ellipsoid (ISe) band of the photoreceptors on the OCT. Full-field electroretinography (ffERG) and pattern electroretinography (PERG) were performed to incorporate the International Society for Clinical Electrophysiology of Vision Standards [48,49].
3. Results
3.1. Genetic Findings
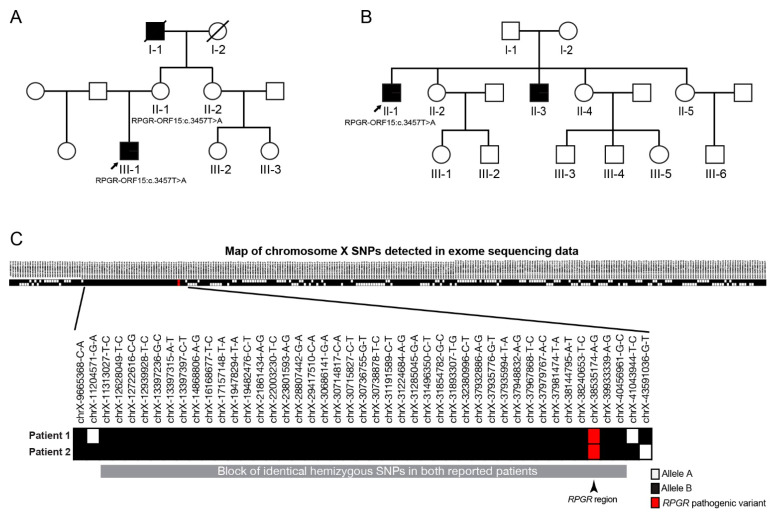
Genetic testing in the probands from two independent Slovenian families identified a novel variant c.3457T>A (Ter1153Lysext*38) in the RPGRORF15. The variant is predicted to disrupt the terminal codon of the RPGRORF15, resulting in a C-terminal extension of the protein by 38 aminoacids ending with a new stop codon. The variant has not been reported in the biomedical literature and is not present in the GnomAD population database (gnomad.broadinstitute.org, accessed on 06.01.2021). Single-nucleotide polymorphism analysis comparing X-chromosomes of the probands is shown in Figure 2C. On the X-chromosome we identified a large block of hemizygous SNPs shared by both probands with an estimated size of 29 megabases, which suggests that observation of an identical variant in two independent families may be due to a founder effect.
Figure 2.
Pedigrees of two families harbouring the RPGRORF15 c.3457T>A mutation and single-nucleotide polymorphism (SNP) analysis. (A)—family of Patient 1 (III-1), (B)—family of Patients 2 (II-1) and 3 (III-3). Probands from each family are marked with an arrow. Other living relatives were not affected; the mother of Patient 1 had mild myopia. (C)—Single-nucleotide polymorphism analysis comparing X-chromosome variants of Patient 1 (Family 1) and Patient 2 (Family 2) has been performed in order to inspect the possibility of a founder effect. The X-chromosomes differed in the majority of the segments, except in one where they involved the same hemizygous variants (enlarged segment at the bottom of the Figure). The findings suggest that the studied variant is being transmitted on the same haplotype and our patients could have inherited the variant from a shared ancestor.
3.2. Clinical Presentation
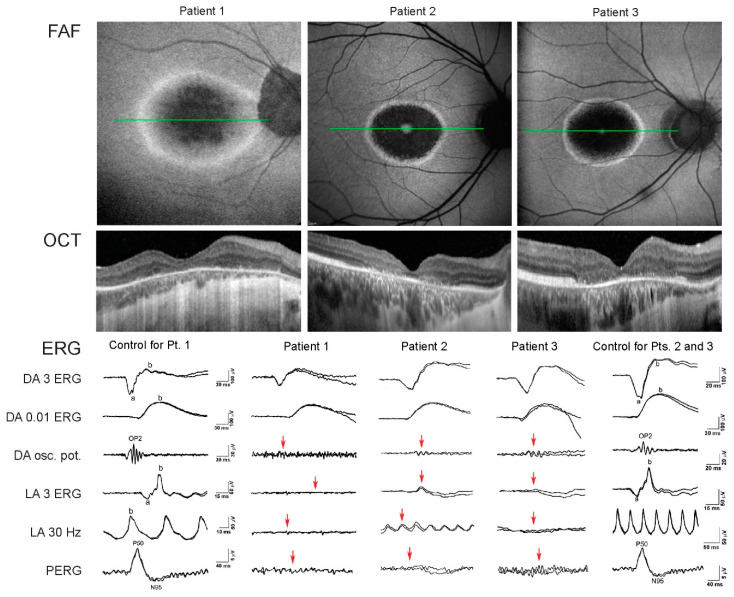
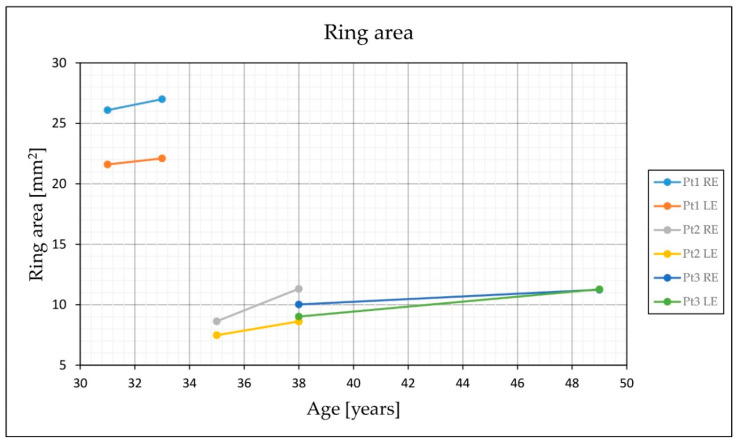
The family pedigrees and clinical characteristics of the 3 male patients are shown in Table 1 and Figure 2, respectively. All had myopia from childhood and adult-onset cone dystrophy. The visual loss appeared in the early 30s in all three, consisting of loss of the central vision (N = 3), photophobia (N = 2), and/or loss of colour discrimination (N = 2). One patient (Patient 2) also reported night vision difficulties. Patient 1 additionally suffered from recurring rhegmatogenous retinal detachment on the left eye at 16 and 31 years of age. At the first exam at the clinic for retinal dystrophies at the median age of 35 years (range, 31–38 years) the median best-corrected visual acuity on the better eye was 1.5 logMAR (range 0.2–1.8; decimal Snellen 0.03; range 0.015–0.6). All had central scotoma while fundus examination revealed mottled pigment in the macula, arteriolar attenuation and optic disc pallor. Pigment formation in the macula and periphery was observed in the left eye of Patient 1 as a remnant of the retinal detachment. Fundus autofluorescence in all eyes showed hyperautofluorescent rings centred at the fovea, encompassing an area of reduced autofluorescence approximately two optic discs in diameter (3462–6342 μm). The rings delineated the loss of the outer retina in the fovea on OCT (Figure 1). Electrophysiology showed significantly reduced to undetectable PERG and/or mfERGs, corresponding to the loss of macular function, normal to borderline dark-adapted ERG, and significantly reduced to undetectable light-adapted ERG (Figure 3). The patients were followed for a median time of 3 years (range, 2–8 years). At their latest exam at the median age of 38 years (range 33–49 years), the median BCVA was 1.5 logMAR (range 0.5–1.5; Snellen 0.03; range 0.03–0.3). FAF showed enlargement of the hyperautofluorescent ring diameters of about 100 μm per year (range, 31–194 μm); while the enlargement of the ring area was calculated to be about 0.33 mm2 per year (range, 0.15–0.63 mm2) (Figure 4).
Table 1.
Clinical characteristics of the included patients. Data from the first and last exam are stated when they differed.
| Patient ID | Age at the First and Last Examination (years) | Age at Onset | Ishihara | Refraction (Dioptre) | BCVA, logMAR (Snellen Decimal) | Visual Field | Fundus Features | Fundus Autofluorescence | OCT | Ring Area [mm2] (Ring Diameter [μm]) |
Electroretinography | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BE | RE | LE | RE | LE | RE | LE | ||||||||
| 1 | 31; 33 |
Childhood: Refraction error (myopia), early 30s: loss of central vision, photophobia | 1/15 | −17.00–3.00/75° | −2.25–0.75/11° (pseudophakic eye after vitrectomy) |
1.5 (0.03); 1.6 (0.02) |
2.1 (0.0075); 1.5 (0.03) |
Central scotoma |
BE: Bull’s eye appearance of macula, optic pallor, attenuated vessels. LE: Bone spicules in the central and peripheral retina (after retinal detachment) | BE: Hyperautoflourescent ring; LE: RPE mottling in the inferior temporal retina (after retinal detachment) |
BE: Absent RPE, Ise, and ELM in the central macula | 26.1 (6342); 27.0 (6635) |
21.6 (4267); 22.1 (4280) |
BE: Undetectable PERG, normal DA ERG, undetectable LA ERG, significantly reduced mfERG |
| 2 * | 35; 38 |
Childhood: refraction error (myopia), early 30s: Loss of central vision, difficulties in colour discrimination and night blindness | 1/15 | −2.25–1.0/34° | −2.0–0.5/139° | 0.3 (0.5); 0.7 (0.2) |
0.2 (0.6); 0.5 (0.3) |
Central scotoma |
BE: Bull’s eye appearance of macula, optic pallor, attenuated vessels | BE: Hyperautofluorescent ring |
BE: Absent RPE, Ise, and ELM in the central macula LE: Remnants of ELM in the foveola |
8.6 (3618); 11.3 (4284) | 7.5 (3462); 8.6 (3623) |
BE: Significantly reduced PERG, normal DA ERG, significantly reduced and delayed LA ERG, reduced mfERG |
| 3 * | 38; 49 |
Childhood refraction error (myopia), early 30s: Photophobia, difficulties in colour discrimination | 1/15 | −12.0–2.0/80° | −14.0–4.0/90° | 2.1 (0.0075); 1.5 (0.03) | 1.8 (0.015); 1.5 (0.03) |
Central scotoma |
BE: Bull’s eye appearance of macula, optic pallor, attenuated vessels | BE: Hyperautofluorescent ring |
BE: Absent RPE, Ise, and ELM in the central macula with remnants of the ELM in the foveola | 10 (3917); 11.2 (4100) | 9.0 (3778); 11.3 (4081) |
BE: Undetectable PERG; normal DA ERG, significantly reduced to undetectable LA ERG |
Abbreviation explanation: LE—left eye, RE—right eye, BE—both eyes, BCVA—best corrected visual acuity, CC—cum correctione, P CC—partial cum correctione, OCT—optical coherence tomography, ERG—electroretinography, mfERG—multifocal ERG, PERG—pattern ERG, LA—light adapted, DA—dark adapted. * Brothers.
Figure 3.
FAF, OCT, and ERG of the three studied patients. Note that although the brothers (Patients 2 and 3) had similar FAF and OCT phenotypes, Patient 2 had better LA ERG and residual PERG response coinciding with better visual acuity.
Figure 4.
Enlargement of the hyperautofluorescent ring area with time. Enlargement of the hyperautofluorescent ring was observed in all patients (see also Figure 1). Ring area has been observed and measured between first and last examinations in all patients. Exact measurements are given in Table 1.
4. Discussion
4.1. Phenotype Associated with the Terminal Mutation in RPGRORF15
The novel C-terminal extension variant c.3457T>A (Ter1153Lysext*38) due to the loss of the terminal RPGRORF15 codon was associated with myopia and adult onset cone dystrophy in all three patients.
It has been previously shown that mutations in the proximal part of the RPGRORF15 result in RP while mutations in the distal end result in cone and cone-rod dystrophy [42,44,50,51,52]. It has not yet been elucidated however whether the latter two phenotypic subtypes occur interchangeably or are also location specific. Considering that the mutation in the last codon resulted in cone-dystrophy in all three patients we hypothesize that the involvement of rods diminishes downstream of the ORF15 exon. It is possible that the cone involvement increases concomitantly with the decrease of rod involvement or else is present at the same level in all phenotypes and becomes more prominent with diminished rod involvement. Sandberg et al. reported faster deterioration of visual acuity in patients with RPGR-associated RP in comparison to RP caused by variants in other genes, at around 4–5% per year (in comparison to RHO patients with decline of 1.6% per year), which has been attributed to a greater rate of foveal thinning and outer nuclear layer loss [53]. This may suggest an additional direct cone involvement already in the RPGR-RP phenotype. Studies have however shown a faster rate of visual acuity decline in RPGR-cone-rod patients in comparison to the RPGR-RP patients with approximately half of the cone-rod patients reaching legal blindness at the age of 40–50 [27,28], whereas patients with RP were reported to reach legal blindness at a median age of 77 [53]. This is comparable to our observation, as among our three patients, two were legally blind before the age of 40 (at the ages 31 and 38 years) whereas Patient 2 had still some functional central vision at the age of 38 years.
Electrophysiology can be used to distinguish between cone and cone-rod dystrophy. Light-adapted ERGs are typically abnormal in both, whereas in cone-rod dystrophy, there is an additional abnormality of the dark-adapted ERGs. There is typically early and severe macular involvement in both, which is characterised by pattern ERG and multifocal ERG reduction, where pattern ERG responses might be relatively high in young or mild cases, with relatively small rings of increased parafoveal FAF [32]. All three patients studied had abnormal PERG and/or mfERG corresponding to the photoreceptor loss in the macula. All three also had severely abnormal light-adapted ERGs, indicating severe generalised cone dysfunction. Patient 2, who had the best visual acuity and lowest refraction error, also had better LA ERG and residual PERG response (Figure 3). Their rod-specific responses were normal. However, the dark-adapted bright flash ERGs exhibit borderline amplitudes in 2 patients. Due to a combined contribution of both cone and rod system activity to this response, its borderline abnormality was most probably a consequence of severe cone dysfunction. This was previously shown in stationary cone dysfunction syndromes, namely in complete achromatopsia [54], although it is not yet clear whether a lack of functional cones might additionally affect rods or their neural pathways [55]. Additionally, high myopia, which was also present in our patients, can cause the reduction of ERG amplitudes [56,57,58].
A ring of increased autofluorescence is often seen in RPGR-cone/cone-rod dystrophy, delineating the border between affected and unaffected retina [27,30,32]. As has been observed previously [31,35,41], the rings in the studied patients enlarged with time, however, the ring enlargement was minimal (approx. 100 μm per year in diameter) suggesting a relatively stable disease after the initial loss of the foveal cones. Interestingly, the patient who had suffered from retinal detachment in the past appeared to have a smaller ring in the vitrectomized eye (Figure 1), however, the low quality of the FAF image on that eye could have had an influence on the measurement. In a recent study, Lima et al. reported three patients with cone-rod dystrophy, one of them harbouring RPGR mutation, and progressive expansion of the hyperautofluorescence ring area in 24-months follow-up [41]. The progression of the ring area in their study was faster (mean 9.2% per year) in comparison to ours (calculated at mean 2.9% per year). Their patients were younger (mean 21 years; range 18–25 years) and had smaller initial ring sizes (mean 1965 μm) in comparison to ours (mean 4231 μm). The difference in enlargement rate may reflect faster ring enlargement in early stages.
4.2. Female Carriers
Female carriers of the RPGR mutation are usually either asymptomatic or have delayed onset of the symptoms and show a milder phenotype [59]. Skewed inactivation of the X chromosome is thought to be responsible for manifesting more severe phenotype [44,60], however, other modifying genetic factors have also been suggested [61]. RPGR carriers appear to have four main patterns of fundus appearance: Normal or near normal pattern, a tapetal reflex, focal or patchy pigmentary retinopathy limited to a quadrant or hemisphere, and three or more quadrants of bone spicule pigmentation or atrophy [62]. Comander et al. reported 101 females from a family with RPGR mutation, showing that 40% of carriers had at least one abnormal test assessing visual acuity, visual field, or dark adaptation, with those carrying variants in RPGRORF15 having lower 30 Hz amplitudes on ERG compared to carriers with variants in exons 1–14 [62]. Recently, Talib et al. described the phenotypic spectrum of 125 female carriers of RPGR mutations from 49 pedigrees of RP and cone/cone-rod dystrophy (COD/CORD) [63]. The authors report a frequent (70%) occurrence of signs and/or symptoms in both, RP and COD/CORD carrier groups, with complete expression of RP or CORD in 29 heterozygotes (23%), carriers of ORF15 mutations [63]. Both studies reported worse visual function in carriers of RPGRORF15 compared to carriers of the RPGR exon 1–14 mutations [62,63]. In our case, there was no family history of retinal disease in female carriers except for mild myopia in the mother of Patient 1. Nevertheless, the female carriers were not examined thoroughly at our institution, therefore a mild phenotype cannot be excluded. Considering the frequent occurrence of retinal disease in female carriers, it is important that they undergo regular ophthalmic follow-up because of the possible late-onset of the disease.
4.3. Haplotype Analysis
The results of the SNP analysis of probands from each family (Figure 2C) suggest that the RPGR variant is present on a common haplotype and may therefore originate from a common ancestor (founder effect).
4.4. RPGR C-Terminal Extension Variants’ Pathogenicity
RPGR variants resulting in the loss of the stop codon have not been conclusively established as a pathogenic mechanism in this gene. A single variant affecting the stop codon has been reported to date (c.3458A>C; Ter1153Serext*38 [30]), however, no convincing segregation or functional evidence has been provided to support its pathogenicity. The identification of two individuals with an overlapping phenotype and presence of the variant affecting the RPGR stop codon contributes novel evidence in support of the pathogenicity and further establishes this type of variants as pathogenic mechanism in this gene. According to the literature, it is still not clear whether all the loss of function RPGR variants result in retinal degeneration through the loss of protein function or gain a new one. Hong et al. suggested that some variants may cause a severe phenotype due to a dominant gain of function mechanism, resulting from the accumulation of truncated products [64]. Our stop loss variant extends the protein sequence with additional 38 residues, which may lead to a gain of a novel toxic or disruptive function. Therefore, our study also adds to the evidence for the pathogenic role of gain of function variants in RPGR.
4.5. Review of RPGR-Interacting Proteins Associated with Cone or Cone-Rod Dystrophy
The pathogenesis behind the RPGR-retinopathy is not clear [28,65]. The RPGR isoforms are localized in the connecting cilium of both cone and rod photoreceptors [28] and contain RCC1-like domains which facilitate interaction with other proteins [4,7,64,66]. Mutations within these domains may affect protein interactions [4] and with that transport between inner and outer segments [67]. The RPGR protein interacts with various ciliary proteins, such as the δ subunit of rod cyclic guanosine monophosphate phosphodiesterase (PDE δ), structural maintenance of chromosomes 1 and 3 (SMC1 and SMC3), GTPase Rab8A, whirlin, gelsolin, ARL3, INPP5E, RPGRIP1L (RPGRIP1 like), NPHP1, NPHP4, NPHP5 (IQCB1), NPHP6 (CEP290), and TTLL5 [12,16,68,69,70,71,72,73,74,75,76,77,78,79] in the mammalian retina. These protein interactions are known to differ among rods and cones, and certain mutations may have more effects on cone interactions than those of rods and vice versa [28]. Some insight into the pathogenesis behind RPGR-cone/cone-rod dystrophy may be gained by studying the proteins that interact with RPGR and also associate with the phenotype above. One of those is RPGRIP1, known to associate with LCA (Leber congenital amaurosis) [80,81,82] as well as childhood-onset cone-rod dystrophy [83]. Recently, cone/cone-rod dystrophy was also described in patients harbouring mutations in TTLL5 (tubulin tyrosine ligase like-5) and ARL3 (Arf-like protein 3), both interacting partners of RPGR [76,84]. TTLL5 localizes to the ciliary base and is important for the glutamylation of the ORF15 [85]. Glutamylation of the RPGRORF15 as posttranslational modification is critical for its function in photoreceptors [76]. ARL3 (ADP-ribosylation factor-like protein 3), a small G protein, is mainly situated on the connecting cilium myoid region of the inner segments of cone photoreceptors and acts as an allosteric factor for the release of lipidated proteins bound to PDE6δ (δ unit of cGMP phosphodiesterase) [86]. Loss of ARL3 function may impair the trafficking of the lipidated outer segment proteins, leading to outer segment shortening and slow retinal degeneration [87]. Previously, missense variants in ARL3 were reported to cause Joubert syndrome, characterized by hypoplasia of the cerebellar vermis, developmental delay, renal anomalies, and rod-cone dystrophy [88]. However, in a recent study, a novel missense variant p.(Arg99Ile) in ARL3 has been described, resulting in a cone-rod dystrophy [86]. CEP290 (NPHP6) is primarily involved in syndromic and non-syndromic LCA [89], however, a rare form of cone-dominated retinal dystrophy associated with mutations in CEP290 has recently been described in two siblings [90]. They were firstly diagnosed with oligocone trichromacy (OT), which is thought to be a stationary condition, but then converted into a progressive degenerative disease [90]. It has been hypothesized that differing phenotypes indicate different functions of CEP290 in rods and cones [91] or cones being more vulnerable compared to rods due to their higher metabolism [92]. Similar reasons could be behind the phenotypic spectrum of RPGR-retinopathy. A novel insight into RPGR function has recently been reported [93]. By studying the cilia in patient’s fibroblasts, the researchers showed that ORF15 mutation resulted in cilia length defects, mutant mRNA instability, and in some cases a significant increase in the RPGR 1–19/RPGR ORF15 ratio. The authors proposed that the relative levels of both RPGR isoforms are critical for optimal cilia growth as overexpression of the RPGRex1–19 resulted in longer cilia, while that of RPGRORF15 resulted in shorter cilia [93]. These observations could be of importance in the development of gene therapy, which is currently underway [93,94]. Further clinical and in vitro studies are needed to fully elucidate the complexity of RPGR function in the retina.
5. Conclusions
The present study describes the clinical phenotype of three patients harbouring a mutation in the terminal codon of RPGRORF15. The phenotype of cone dystrophy in all three patients is consistent with previous observation that mutations in the distal end result in cone/cone-rod phenotypes. Furthermore, the fact that proximal mutations in ORF15 exon cause RP, the terminal mutation Ter1153Lysext*38 cone-dystrophy and mutations in between cone or cone-rod dystrophy, we propose that the involvement of rods diminishes downstream of the ORF15 exon.
Acknowledgments
We would like to thank Barbara Klemenc for the preparation of the figures.
Abbreviations
| RPGR | retinitis pigmentosa GTPase regulator |
| FAF | fundus autofluorescence |
| OCT | optical coherence tomography |
| RCC1 | regulator of chromosome condensation 1 |
| CC | connecting cilium |
| OS | outer segment |
| RP | retinitis pigmentosa |
| ERG | electroretinography |
| LE | left eye |
| RE | right eye |
| BE | both eyes |
| BCVA | best corrected visual acuity |
| mfERG | multifocal ERG |
| PERG | pattern EGR |
| ffERG | full field ERG |
| LA | light adapted |
| DA | dark adapted |
| RPE | retinal pigment epithelium |
| ELM | external limiting membrane |
| Ise band | inner segment ellipsoid band |
| LCA | Leber congenital amaurosis |
| SNP | single-nucleotide polymorphism |
| COD | cone dystrophy |
| CORD | cone-rod dystrophy |
Author Contributions
Conceptualization: V.H., A.F. and M.Š.; methodology: V.H., A.F., M.Š., M.V., B.P. and A.M.; formal analysis: V.H., A.F., M.J.-V., M.H., J.S., M.Š., investigation: V.H., A.F., M.H., M.Š., M.J.-V., J.S., B.P., M.V., A.M.; resources: A.F., M.V., M.J.-V., J.S., B.P., M.H., A.M.; data curation: A.F., M.Š., V.H., writing—original draft preparation: V.H., A.F., M.Š., writing—review and editing: V.H., A.F., M.Š., J.S., M.H., B.P., M.V., A.M., M.J.-V., visualization: V.H., A.F., M.Š., supervision: A.F., M.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Slovenian research agency ARRS J3-1750.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki. Ethical review and approval were waived for this study as all examinations were done as a part of routine diagnostic procedures. The patients provided informed consent for the genetic analysis and publication of their medical information.
Informed Consent Statement
Informed consent was obtained from the subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to personal data protection.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hadjebi O., Casas-Terradellas E., Garcia-Gonzalo F.R., Rosa J.L. The RCC1 superfamily: From genes, to function, to disease. Biochim. Biophys. Acta (BBA) Bioenerg. 2008;1783:1467–1479. doi: 10.1016/j.bbamcr.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 2.Vervoort R., Lennon A., Bird A.C., Tulloch B., Axton R., Miano M.G., Meindl A., Meitinger T., Ciccodicola A., Wright A.F. Mutational hot spot within a new RPGR exon in X-linked retinitis pigmentosa. Nat. Genet. 2000;25:462–466. doi: 10.1038/78182. [DOI] [PubMed] [Google Scholar]
- 3.Meindl A., Dry K., Herrmann K., Manson E., Ciccodicola A., Edgar A., Carvalho M., Achatz H., Hellebrand H., Lennon A., et al. A gene (RPGR) with homology to the RCC1 guanine nucleotide exchange factor is mutated in X–linked retinitis pigmentosa (RP3) Nat. Genet. 1996;13:35–42. doi: 10.1038/ng0596-35. [DOI] [PubMed] [Google Scholar]
- 4.Roepman R., Van Duijnhoven G., Rosenberg T., Pinckers A.J.L.G., Bleeker-Wagemakers L.M., Bergen A.A.B., Post J., Beck A., Reinhardt R., Ropers H.-H., et al. Positional cloning of the gene for X-linked retinitis pigmentosa 3: Homology with the guanine-nucleotide-exchange factor RCC. Hum. Mol. Genet. 1996;5:1035–1041. doi: 10.1093/hmg/5.7.1035. [DOI] [PubMed] [Google Scholar]
- 5.Hong D.-H., Pawlyk B., Sokolov M., Strissel K.J., Yang J., Tulloch B., Wright A.F., Arshavsky V.Y., Li T. RPGR Isoforms in Photoreceptor Connecting Cilia and the Transitional Zone of Motile Cilia. Investig. Ophthalmol. Vis. Sci. 2003;44:2413–2421. doi: 10.1167/iovs.02-1206. [DOI] [PubMed] [Google Scholar]
- 6.Iannaccone A., Breuer D.K., Wang X.F., Kuo S.F., Normando E.M., Filippova E., Baldi A., Hiriyanna S., Macdonald C.B., Baldi F., et al. Clinical and immunohistochemical evidence for an X linked retinitis pigmentosa syndrome with recurrent infections and hearing loss in association with an RPGR mutation. J. Med. Genet. 2003;40:118. doi: 10.1136/jmg.40.11.e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He S., Parapuram S.K., Hurd T.W., Behnam B., Margolis B., Swaroop A., Khanna H. Retinitis Pigmentosa GTPase Regulator (RPGR) protein isoforms in mammalian retina: Insights into X-linked Retinitis Pigmentosa and associated ciliopathies. Vis. Res. 2008;48:366–376. doi: 10.1016/j.visres.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright R.N., Hong D.-H., Perkins B. Misexpression of the Constitutive Rpgrex1-19Variant Leads to Severe Photoreceptor Degeneration. Investig. Ophthalmol. Vis. Sci. 2011;52:5189–5201. doi: 10.1167/iovs.11-7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patil H., Guruju M.R., Cho K.-I., Yi H., Orry A., Kim H., Ferreira P.A. Structural and functional plasticity of subcellular tethering, targeting and processing of RPGRIP1 by RPGR isoforms. Biol. Open. 2011;1:140–160. doi: 10.1242/bio.2011489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan D., Swain P.K., Breuer D., Tucker R.M., Wu W., Fujita R., Rehemtulla A., Burke D., Swaroop A. Biochemical Characterization and Subcellular Localization of the Mouse Retinitis Pigmentosa GTPase Regulator (mRpgr) J. Biol. Chem. 1998;273:19656–19663. doi: 10.1074/jbc.273.31.19656. [DOI] [PubMed] [Google Scholar]
- 11.Tee J.J.L., Smith A.J., Hardcastle A.J., Michaelides M. RPGR-associated retinopathy: Clinical features, molecular genetics, animal models and therapeutic options. Br. J. Ophthalmol. 2016;100:1022–1027. doi: 10.1136/bjophthalmol-2015-307698. [DOI] [PubMed] [Google Scholar]
- 12.Megaw R.D., Soares D.C., Wright A.F. RPGR: Its role in photoreceptor physiology, human disease, and future therapies. Exp. Eye Res. 2015;138:32–41. doi: 10.1016/j.exer.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong D.-H., Pawlyk B.S., Adamian M., Sandberg M.A., Li T. A Single, Abbreviated RPGR-ORF15 Variant Reconstitutes RPGR Function In Vivo. Investig. Ophthalmol. Vis. Sci. 2005;46:435–441. doi: 10.1167/iovs.04-1065. [DOI] [PubMed] [Google Scholar]
- 14.Shu X., Fry A.M., Tulloch B., Manson F.D.C., Crabb J.W., Khanna H., Faragher A.J., Lennon A., He S., Trojan P., et al. RPGR ORF15 isoform co-localizes with RPGRIP1 at centrioles and basal bodies and interacts with nucleophosmin. Hum. Mol. Genet. 2005;14:1183–1197. doi: 10.1093/hmg/ddi129. [DOI] [PubMed] [Google Scholar]
- 15.Gakovic M., Shu X., Kasioulis I., Carpanini S., Moraga I., Wright A.F. The role of RPGR in cilia formation and actin stability. Hum. Mol. Genet. 2011;20:4840–4850. doi: 10.1093/hmg/ddr423. [DOI] [PubMed] [Google Scholar]
- 16.Khanna H., Hurd T.W., Lillo C., Shu X., Parapuram S.K., He S., Akimoto M., Wright A.F., Margolis B., Williams D.S., et al. RPGR-ORF15, Which Is Mutated in Retinitis Pigmentosa, Associates with SMC1, SMC3, and Microtubule Transport Proteins. J. Biol. Chem. 2005;280:33580–33587. doi: 10.1074/jbc.M505827200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mavlyutov T.A., Zhao H., Ferreira P.A. Species-specific subcellular localization of RPGR and RPGRIP isoforms: Implications for the phenotypic variability of congenital retinopathies among species. Hum. Mol. Genet. 2002;11:1899–1907. doi: 10.1093/hmg/11.16.1899. [DOI] [PubMed] [Google Scholar]
- 18.Shu X., Zeng Z., Eckmiller M.S., Gautier P., Vlachantoni D., Manson F.D.C., Tulloch B., Sharpe C., Górecki D.C., Wright A.F. Developmental and Tissue Expression ofXenopus laevis RPGR. Investig. Ophthalmol. Vis. Sci. 2006;47:348–356. doi: 10.1167/iovs.05-0858. [DOI] [PubMed] [Google Scholar]
- 19.Shu X., Black G.C., Rice J.M., Hart-Holden N., Jones A., O’Grady A., Ramsden S.C., Wright A.F. RPGRmutation analysis and disease: An update. Hum. Mutat. 2007;28:322–328. doi: 10.1002/humu.20461. [DOI] [PubMed] [Google Scholar]
- 20.Bischoff F.R., Ponstingl H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC. Nat. Cell Biol. 1991;354:80–82. doi: 10.1038/354080a0. [DOI] [PubMed] [Google Scholar]
- 21.Ferreira P.A., Nakayama T.A., Pak W.L., Travis G.H. Cyclophilin-related protein RanBP2 acts as chaperone for red/green opsin. Nat. Cell Biol. 1996;383:637–640. doi: 10.1038/383637a0. [DOI] [PubMed] [Google Scholar]
- 22.Hong D.-H., Pawlyk B.S., Shang J., Sandberg M.A., Berson E.L., Li T. A retinitis pigmentosa GTPase regulator (RPGR)- deficient mouse model for X-linked retinitis pigmentosa (RP3) Proc. Natl. Acad. Sci. USA. 2000;97:3649–3654. doi: 10.1073/pnas.97.7.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong D.-H., Yue G., Adamian M., Li T. Retinitis Pigmentosa GTPase Regulator (RPGR)-interacting Protein Is Stably Associated with the Photoreceptor Ciliary Axoneme and Anchors RPGR to the Connecting Cilium. J. Biol. Chem. 2001;276:12091–12099. doi: 10.1074/jbc.M009351200. [DOI] [PubMed] [Google Scholar]
- 24.Shu X., McDowall E., Brown A.F., Wright A.F. The human retinitis pigmentosa GTPase regulator gene variant database. Hum. Mutat. 2008;29:605–608. doi: 10.1002/humu.20733. [DOI] [PubMed] [Google Scholar]
- 25.Sharon D., Bruns G.A., McGee T.L., Sandberg M.A., Berson E.L., Dryja T.P. X-linked retinitis pigmentosa: Mutation spectrum of the RPGR and RP2 genes and correlation with visual function. Investig. Ophthalmol. Vis. Sci. 2000;41:2712–2721. [PubMed] [Google Scholar]
- 26.Sharon D., Sandberg M.A., Rabe V.W., Stillberger M., Dryja T.P., Berson E.L. RP2 and RPGR Mutations and Clinical Correlations in Patients with X-Linked Retinitis Pigmentosa. Am. J. Hum. Genet. 2003;73:1131–1146. doi: 10.1086/379379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Talib M., Van Schooneveld M.J., Thiadens A.A., Fiocco M., Wijnholds J., Florijn R.J., Schalij-Delfos N.E., Van Genderen M.M., Putter H., Cremers F.P.M., et al. Clinical and Genetic Characteristics of Male Patients with Rpgr-Associated Retinal Dystrophies. Retina. 2019;39:1186–1199. doi: 10.1097/IAE.0000000000002125. [DOI] [PubMed] [Google Scholar]
- 28.Thiadens A.A.H.J., Soerjoesing G.G., Florijn R.J., Tjiam A.G., Hollander A.I.D., Born L.I.V.D., Riemslag F.C., Bergen A.A.B., Klaver C.C.W. Clinical course of cone dystrophy caused by mutations in the RPGR gene. Graefe’s Arch. Clin. Exp. Ophthalmol. 2011;249:1527–1535. doi: 10.1007/s00417-011-1789-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Silva S.R., Arno G., Robson A.G., Fakin A., Pontikos N., Mohamed M.D., Bird A.C., Moore A.T., Michaelides M., Webster A.R., et al. The X-linked retinopathies: Physiological insights, pathogenic mechanisms, phenotypic features and novel therapies. Prog. Retin. Eye Res. 2020:100898. doi: 10.1016/j.preteyeres.2020.100898. [DOI] [PubMed] [Google Scholar]
- 30.Birtel J., Eisenberger T., Gliem M., Müller P.L., Herrmann P., Betz C., Zahnleiter D., Neuhaus C., Lenzner S., Holz F.G., et al. Clinical and genetic characteristics of 251 consecutive patients with macular and cone/cone-rod dystrophy. Sci. Rep. 2018;8:4824. doi: 10.1038/s41598-018-22096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gill J.S., Georgiou M., Kalitzeos A., Moore A.T., Michaelides M. Progressive cone and cone-rod dystrophies: Clinical features, molecular genetics and prospects for therapy. Br. J. Ophthalmol. 2019;103:711–720. doi: 10.1136/bjophthalmol-2018-313278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robson A.G., Michaelides M., Saihan Z., Bird A.C., Webster A.R., Moore A.T., Fitzke F.W., Holder G.E. Functional characteristics of patients with retinal dystrophy that manifest abnormal parafoveal annuli of high density fundus autofluorescence; a review and update. Doc. Ophthalmol. 2007;116:79–89. doi: 10.1007/s10633-007-9087-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fahim A.T., Daiger S.P. The Role of X-Chromosome Inactivation in Retinal Development and Disease. Adv. Exp. Med. Biol. 2016;854:325–331. doi: 10.1007/978-3-319-17121-0_43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robson A.G., El-Amir A., Bailey C., Egan C.A., Fitzke F.W., Webster A.R., Bird A.C., Holder G.E. Pattern ERG Correlates of Abnormal Fundus Autofluorescence in Patients with Retinitis Pigmentosa and Normal Visual Acuity. Investig. Ophthalmol. Vis. Sci. 2003;44:3544–3550. doi: 10.1167/iovs.02-1278. [DOI] [PubMed] [Google Scholar]
- 35.Robson A.G., Michaelides M., Luong V.A., E Holder G., Bird A.C., Webster A.R., Moore A.T., Fitzke F.W. Functional correlates of fundus autofluorescence abnormalities in patients with RPGR or RIMS1 mutations causing cone or cone rod dystrophy. Br. J. Ophthalmol. 2007;92:95–102. doi: 10.1136/bjo.2007.124008. [DOI] [PubMed] [Google Scholar]
- 36.Robson A.G., Tufail A., Fitzke F., Bird A.C., Moore A.T., E Holder G., Webster A.R. Serial Imaging and Structure-Function Correlates of High-Density Rings of Fundus Autofluorescence in Retinitis Pigmentosa. Retina. 2011;31:1670–1679. doi: 10.1097/IAE.0b013e318206d155. [DOI] [PubMed] [Google Scholar]
- 37.Aizawa S., Mitamura Y., Baba T., Hagiwara A., Ogata K., Yamamoto S. Correlation between visual function and photoreceptor inner/outer segment junction in patients with retinitis pigmentosa. Eye. 2008;23:304–308. doi: 10.1038/sj.eye.6703076. [DOI] [PubMed] [Google Scholar]
- 38.Fakin A., Jarc-Vidmar M., Glavač D., Bonnet C., Petit C., Hawlina M. Fundus autofluorescence and optical coherence tomography in relation to visual function in Usher syndrome type 1 and 2. Vis. Res. 2012;75:60–70. doi: 10.1016/j.visres.2012.08.017. [DOI] [PubMed] [Google Scholar]
- 39.Robson A.G., Saihan Z., A Jenkins S., Fitzke F.W., Bird A.C., Webster A.R., Holder G.E. Functional characterisation and serial imaging of abnormal fundus autofluorescence in patients with retinitis pigmentosa and normal visual acuity. Br. J. Ophthalmol. 2006;90:472–479. doi: 10.1136/bjo.2005.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wakabayashi T., Sawa M., Gomi F., Tsujikawa M. Correlation of fundus autofluorescence with photoreceptor morphology and functional changes in eyes with retinitis pigmentosa. Acta Ophthalmol. 2009;88:e177–e183. doi: 10.1111/j.1755-3768.2010.01926.x. [DOI] [PubMed] [Google Scholar]
- 41.Lima L.H., Zett C., Kniggendorf V., Marianelli B., De Carvalho R.A.P., Farah M.E., Sallum J.M.F. Progressive expansion of the hyperautofluorescent ring in cone-rod dystrophy patients. Ophthalmic Genet. 2018;39:492–499. doi: 10.1080/13816810.2018.1461911. [DOI] [PubMed] [Google Scholar]
- 42.Ebenezer N.D., Michaelides M., Jenkins S.A., Audo I., Webster A.R., Cheetham M.E., Stockman A., Maher E.R., Ainsworth J.R., Yates J.R., et al. Identification of NovelRPGRORF15 Mutations in X-linked Progressive Cone-Rod Dystrophy (XLCORD) Families. Investig. Ophthalmol. Vis. Sci. 2005;46:1891–1898. doi: 10.1167/iovs.04-1482. [DOI] [PubMed] [Google Scholar]
- 43.Nguyen X.-T.-A., Talib M., Van Schooneveld M.J., Brinks J., Brink J.T., Florijn R.J., Wijnholds J., Verdijk R.M., Bergen A.A., Boon C.J. RPGR-Associated Dystrophies: Clinical, Genetic, and Histopathological Features. Int. J. Mol. Sci. 2020;21:835. doi: 10.3390/ijms21030835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang L., Yin X., Feng L., You D., Wu L., Chen N., Li A., Li G., Ma Z. Novel Mutations of RPGR in Chinese Retinitis Pigmentosa Patients and the Genotype-Phenotype Correlation. PLoS ONE. 2014;9:e85752. doi: 10.1371/journal.pone.0085752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Depristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., Del Angel G., Rivas M.A., Hanna M., et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–423. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meynert A.M., Bicknell L.S., E Hurles M., Jackson A.P., Taylor M.S. Quantifying single nucleotide variant detection sensitivity in exome sequencing. BMC Bioinform. 2013;14:195. doi: 10.1186/1471-2105-14-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bach M., Brigell M.G., Hawlina M., Holder G.E., Johnson M.A., McCulloch D.L., Meigen T., Viswanathan S. ISCEV standard for clinical pattern electroretinography (PERG): 2012 update. Doc. Ophthalmol. 2012;126:1–7. doi: 10.1007/s10633-012-9353-y. [DOI] [PubMed] [Google Scholar]
- 49.McCulloch D.L., Marmor M.F., Brigell M.G., Hamilton R., Holder G.E., Tzekov R., Bach M. ISCEV Standard for full-field clinical electroretinography (2015 update) Doc. Ophthalmol. 2015;130:1–12. doi: 10.1007/s10633-014-9473-7. [DOI] [PubMed] [Google Scholar]
- 50.Branham K.E., Othman M., Brumm M., Karoukis A.J., Atmaca-Sonmez P., Yashar B.M., Schwartz S.B., Stover N.B., Trzupek K.M., Wheaton D.K.H., et al. Mutations inRPGRandRP2Account for 15% of Males with Simplex Retinal Degenerative Disease. Investig. Ophthalmol. Vis. Sci. 2012;53:8232–8237. doi: 10.1167/iovs.12-11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Demirci F.Y.K., Rigatti B.W., Wen G., Radak A.L., Mah T.S., Baic C.L., Traboulsi E.I., Alitalo T., Ramser J., Gorin M.B. X-Linked Cone-Rod Dystrophy (Locus COD1): Identification of Mutations in RPGR Exon ORF. Am. J. Hum. Genet. 2002;70:1049–1053. doi: 10.1086/339620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Michaelides M., Hardcastle A.J., Hunt D.M., Moore A.T. Progressive Cone and Cone-Rod Dystrophies: Phenotypes and Underlying Molecular Genetic Basis. Surv. Ophthalmol. 2006;51:232–258. doi: 10.1016/j.survophthal.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 53.Sandberg M.A., Rosner B., Weigel-DiFranco C., Dryja T.P., Berson E.L. Disease Course of Patients with X-linked Retinitis Pigmentosa due toRPGRGene Mutations. Investig. Ophthalmol. Vis. Sci. 2007;48:1298–1304. doi: 10.1167/iovs.06-0971. [DOI] [PubMed] [Google Scholar]
- 54.Verdon W.A. A Wave Analysis of The Electro-Retinogram (ERG) in Congenital Achromatopsia: 2:40 pm (VS-330) Optom. Vis. Sci. 1995;72:187. doi: 10.1097/00006324-199512001-00296. [DOI] [Google Scholar]
- 55.Michaelides M., Hunt D.M., Moore A.T. The cone dysfunction syndromes. Br. J. Ophthalmol. 2004;88:291–297. doi: 10.1136/bjo.2003.027102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perlman I., Meyer E., Haim T., Zonis S. Retinal function in high refractive error assessed electroretinographically. Br. J. Ophthalmol. 1984;68:79–84. doi: 10.1136/bjo.68.2.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Westall C.A., Dhaliwal H.S., Panton C.M., Sigesmund D., Levin A.V., Nischal K.K., Héon E. Values of electroretinogram responses according to axial length. Doc. Ophthalmol. 2001;102:115–130. doi: 10.1023/A:1017535207481. [DOI] [PubMed] [Google Scholar]
- 58.Yamamoto S., Nitta K., Kamiyama M. Cone electroretinogram to chromatic stimuli in myopic eyes. Vis. Res. 1997;37:2157–2159. doi: 10.1016/S0042-6989(96)00303-3. [DOI] [PubMed] [Google Scholar]
- 59.Rozet J.-M. Dominant X linked retinitis pigmentosa is frequently accounted for by truncating mutations in exon ORF15 of the RPGR gene. J. Med. Genet. 2002;39:284–285. doi: 10.1136/jmg.39.4.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fahim A.T., Sullivan L.S., Bowne S.J., Jones K.D., Wheaton D.K., Khan N.W., Heckenlively J.R., Jayasundera K.T., Branham K.H., Andrews C.A., et al. X-Chromosome Inactivation Is a Biomarker of Clinical Severity in Female Carriers of RPGR-Associated X-Linked Retinitis Pigmentosa. Ophthalmol. Retin. 2020;4:510–520. doi: 10.1016/j.oret.2019.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pelletier V., Jambou M., Delphin N., Zinovieva E., Stum M., Gigarel N., Dollfus H., Hamel C., Toutain A., Dufier J.-L., et al. Comprehensive survey of mutations in RP2 and RPGR in patients affected with distinct retinal dystrophies: Genotype–phenotype correlations and impact on genetic counseling. Hum. Mutat. 2006;28:81–91. doi: 10.1002/humu.20417. [DOI] [PubMed] [Google Scholar]
- 62.Comander J., Weigel-DiFranco C., Sandberg M.A., Berson E.L. Visual Function in Carriers of X-Linked Retinitis Pigmentosa. Ophthalmol. 2015;122:1899–1906. doi: 10.1016/j.ophtha.2015.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Talib M., Van Schooneveld M.J., Van Cauwenbergh C., Wijnholds J., Brink J.B.T., Florijn R.J., Schalij-Delfos N.E., Dagnelie G., Van Genderen M.M., De Baere E., et al. The Spectrum of Structural and Functional Abnormalities in Female Carriers of Pathogenic Variants in theRPGRGene. Investig. Ophthalmol. Vis. Sci. 2018;59:4123–4133. doi: 10.1167/iovs.17-23453. [DOI] [PubMed] [Google Scholar]
- 64.Hong D.-H., Pawlyk B.S., Adamian M., Li T. Dominant, gain-of-function mutant produced by truncation of RPGR. Investig. Ophthalmol. Vis. Sci. 2004;45:36–41. doi: 10.1167/iovs.03-0787. [DOI] [PubMed] [Google Scholar]
- 65.Demirci F.Y.K., Gupta N., Radak A.L., Rigatti B.W., Mah T.S., Milam A.H., Gorin M.B. Histopathologic study of X-linked cone-rod dystrophy (CORDX1) caused by a mutation in the RPGR exon ORF. Am. J. Ophthalmol. 2005;139:386–388. doi: 10.1016/j.ajo.2004.08.041. [DOI] [PubMed] [Google Scholar]
- 66.Roepman R., Bernoud-Hubac N., Schick D.E., Maugeri A., Berger W., Ropers H.-H., Cremers F.P.M., Ferreira P.A. The retinitis pigmentosa GTPase regulator (RPGR) interacts with novel transport-like proteins in the outer segments of rod photoreceptors. Hum. Mol. Genet. 2000;9:2095–2105. doi: 10.1093/hmg/9.14.2095. [DOI] [PubMed] [Google Scholar]
- 67.Zhao Y., Hong D.-H., Pawlyk B., Yue G., Adamian M., Grynberg M., Godzik A., Li T. The retinitis pigmentosa GTPase regulator (RPGR)- interacting protein: Subserving RPGR function and participating in disk morphogenesis. Proc. Natl. Acad. Sci. USA. 2003;100:3965–3970. doi: 10.1073/pnas.0637349100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khanna H., Davis E.E., Murga-Zamalloa C.A., Estrada-Cuzcano A., Lopez I., Hollander A.I.D., Zonneveld M.N., Othman M.I., Waseem N., Chakarova C.F., et al. A common allele in RPGRIP1L is a modifier of retinal degeneration in ciliopathies. Nat. Genet. 2009;41:739–745. doi: 10.1038/ng.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Linari M., Ueffing M., Manson F., Wright A., Meitinger T., Becker J. The retinitis pigmentosa GTPase regulator, RPGR, interacts with the delta subunit of rod cyclic GMP phosphodiesterase. Proc. Natl. Acad. Sci. USA. 1999;96:1315–1320. doi: 10.1073/pnas.96.4.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Megaw R., Abu-Arafeh H., Jungnickel M., Mellough C., Gurniak C., Witke W., Zhang W., Khanna H., Mill P., Dhillon B., et al. Gelsolin dysfunction causes photoreceptor loss in induced pluripotent cell and animal retinitis pigmentosa models. Nat. Commun. 2017;8:271. doi: 10.1038/s41467-017-00111-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Murga-Zamalloa C.A., Atkins S.J., Peranen J., Swaroop A., Khanna H. Interaction of retinitis pigmentosa GTPase regulator (RPGR) with RAB8A GTPase: Implications for cilia dysfunction and photoreceptor degeneration. Hum. Mol. Genet. 2010;19:3591–3598. doi: 10.1093/hmg/ddq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Murga-Zamalloa C., Swaroop A., Khanna H. Multiprotein Complexes of Retinitis Pigmentosa GTPase Regulator (RPGR), a Ciliary Protein Mutated in X-Linked Retinitis Pigmentosa (XLRP) Adv. Exp. Med. Biol. 2009;664:105–114. doi: 10.1007/978-1-4419-1399-9_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Otto E.A., Loeys B., Khanna H., Hellemans J., Sudbrak R., Fan S., Muerb U., O’Toole J.F., Helou J., Attanasio M., et al. Nephrocystin-5, a ciliary IQ domain protein, is mutated in Senior-Loken syndrome and interacts with RPGR and calmodulin. Nat. Genet. 2005;37:282–288. doi: 10.1038/ng1520. [DOI] [PubMed] [Google Scholar]
- 74.Patnaik S.R., Raghupathy R.K., Zhang X., Mansfield D., Shu X. The Role of RPGR and Its Interacting Proteins in Ciliopathies. J. Ophthalmol. 2015;2015:414781. doi: 10.1155/2015/414781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rao K.N., Zhang W., Li L., Ronquillo C., Baehr W., Khanna H. Ciliopathy-associated protein CEP290 modifies the severity of retinal degeneration due to loss of RPGR. Hum. Mol. Genet. 2016;25:2005–2012. doi: 10.1093/hmg/ddw075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun X., Park J.H., Gumerson J., Wu Z., Swaroop A., Qian H., Roll-Mecak A., Li T. Loss of RPGR glutamylation underlies the pathogenic mechanism of retinal dystrophy caused by TTLL5 mutations. Proc. Natl. Acad. Sci. USA. 2016;113:E2925–E2934. doi: 10.1073/pnas.1523201113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wright R.N., Hong D.-H., Perkins B. RpgrORF15Connects to the Usher Protein Network through Direct Interactions with Multiple Whirlin Isoforms. Investig. Ophthalmol. Vis. Sci. 2012;53:1519–1529. doi: 10.1167/iovs.11-8845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Q., Giacalone J.C., Searby C., Stone E.M., Tucker B.A., Sheffield V.C. Disruption of RPGR protein interaction network is the common feature of RPGR missense variations that cause XLRP. Proc. Natl. Acad. Sci. USA. 2019;116:1353–1360. doi: 10.1073/pnas.1817639116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roepman R., Wolfrum U. Protein Networks and Complexes in Photoreceptor Cilia. Alzheimer’s Dis. 2007;43:209–235. doi: 10.1007/978-1-4020-5943-8_10. [DOI] [PubMed] [Google Scholar]
- 80.Abouzeid H., Othman I.S., Schorderet D.F. A Novel Recessive RPGRIP1 Mutation Causing Leber Congenital Amaurosis. Klin. Mon. Augenheilkd. 2016;233:456–459. doi: 10.1055/s-0041-111815. [DOI] [PubMed] [Google Scholar]
- 81.Dryja T.P., Adams S.M., Grimsby J.L., McGee T.L., Hong D.-H., Li T., Andréasson S., Berson E.L. Null RPGRIP1 Alleles in Patients with Leber Congenital Amaurosis. Am. J. Hum. Genet. 2001;68:1295–1298. doi: 10.1086/320113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gerber S., Perrault I., Hanein S., Barbet F., Ducroq D., Ghazi I., Martin-Coignard D., Leowski C., Homfray T., Dufier J.-L., et al. Complete exon-intron structure of the RPGR-interacting protein (RPGRIP1) gene allows the identification of mutations underlying Leber congenital amaurosis. Eur. J. Hum. Genet. 2001;9:561–571. doi: 10.1038/sj.ejhg.5200689. [DOI] [PubMed] [Google Scholar]
- 83.Hameed A., Abid A., Aziz A., Ismail M., Mehdi S.Q., Khaliq S. Evidence of RPGRIP1 gene mutations associated with recessive cone-rod dystrophy. J. Med. Genet. 2003;40:616–619. doi: 10.1136/jmg.40.8.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wätzlich D., Vetter I., Gotthardt K., Miertzschke M., Chen Y.-X., Wittinghofer A., Ismail S. The interplay between RPGR, PDEδ and Arl2/3 regulate the ciliary targeting of farnesylated cargo. EMBO Rep. 2013;14:465–472. doi: 10.1038/embor.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sergouniotis P.I., Chakarova C., Murphy C., Becker M., Lenassi E., Arno G., Lek M., MacArthur D.G., Bhattacharya S.S., Moore A.T., et al. Biallelic Variants in TTLL5, Encoding a Tubulin Glutamylase, Cause Retinal Dystrophy. Expanding the Spectrum of BAF-Related Disorders: De Novo Variants in SMARCC2 Cause a Syndrome with Intellectual Disability and Developmental Delay. Am. J. Hum. Genet. 2014;94:760–769. doi: 10.1016/j.ajhg.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sheikh S.A., Sisk R.A., Schiavon C.R., Waryah Y.M., Usmani M.A., Steel D.H., Sayer J.A., Narsani A.K., Hufnagel R.B., Riazuddin S., et al. Homozygous Variant inARL3Causes Autosomal Recessive Cone Rod Dystrophy. Investig. Ophthalmol. Vis. Sci. 2019;60:4811–4819. doi: 10.1167/iovs.19-27263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hanke-Gogokhia C., Wu Z., Gerstner C.D., Frederick J.M., Zhang H., Baehr W. Arf-like Protein 3 (ARL3) Regulates Protein Trafficking and Ciliogenesis in Mouse Photoreceptors. J. Biol. Chem. 2016;291:7142–7155. doi: 10.1074/jbc.M115.710954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Alkanderi S., Molinari E., Shaheen R., Elmaghloob Y., Stephen L.A., Sammut V., Ramsbottom S.A., Srivastava S., Cairns G., Edwards N., et al. ARL3 Mutations Cause Joubert Syndrome by Disrupting Ciliary Protein Composition. Am. J. Hum. Genet. 2018;103:612–620. doi: 10.1016/j.ajhg.2018.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Den Hollander A.I., Koenekoop R.K., Yzer S., Lopez I., Arends M.L., Voesenek K.E.J., Zonneveld M.N., Strom T.M., Meitinger T., Brunner H.G., et al. Mutations in the CEP290 (NPHP6) Gene Are a Frequent Cause of Leber Congenital Amaurosis. Am. J. Hum. Genet. 2006;79:556–561. doi: 10.1086/507318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Roosing S., Cremers F.P.M., Riemslag F.C.C., Zonneveld-Vrieling M.N., Talsma H.E., Klessens-Godfroy F.J.M., Hollander A.I.D., Born L.I.V.D. A Rare Form of Retinal Dystrophy Caused by Hypomorphic Nonsense Mutations in CEP. Genes. 2017;8:208. doi: 10.3390/genes8080208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cideciyan A.V., Aleman T.S., Jacobson S.G., Khanna H., Sumaroka A., Aguirre G.K., Schwartz S.B., Windsor E.A.M., He S., Chang B., et al. Centrosomal-ciliary geneCEP290/NPHP6 mutations result in blindness with unexpected sparing of photoreceptors and visual brain: Implications for therapy of Leber congenital amaurosis. Hum. Mutat. 2007;28:1074–1083. doi: 10.1002/humu.20565. [DOI] [PubMed] [Google Scholar]
- 92.Miyazono S., Shimauchi-Matsukawa Y., Tachibanaki S., Kawamura S. Highly efficient retinal metabolism in cones. Proc. Natl. Acad. Sci. USA. 2008;105:16051–16056. doi: 10.1073/pnas.0806593105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Moreno-Leon L., West E.L., O’Hara-Wright M., Li L., Nair R., He J., Anand M., Sahu B., Chavali V.R.M., Smith A.J., et al. RPGR isoform imbalance causes ciliary defects due to exon ORF15 mutations in X-linked retinitis pigmentosa (XLRP) Hum. Mol. Genet. 2021;29:3706–3716. doi: 10.1093/hmg/ddaa269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cehajic-Kapetanovic J., Xue K., De La Camara C.M.-F., Nanda A., Davies A., Wood L.J., Salvetti A.P., Fischer M.D., Aylward J.W., Barnard A.R., et al. Initial results from a first-in-human gene therapy trial on X-linked retinitis pigmentosa caused by mutations in RPGR. Nat. Med. 2020;26:354–359. doi: 10.1038/s41591-020-0763-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to personal data protection.