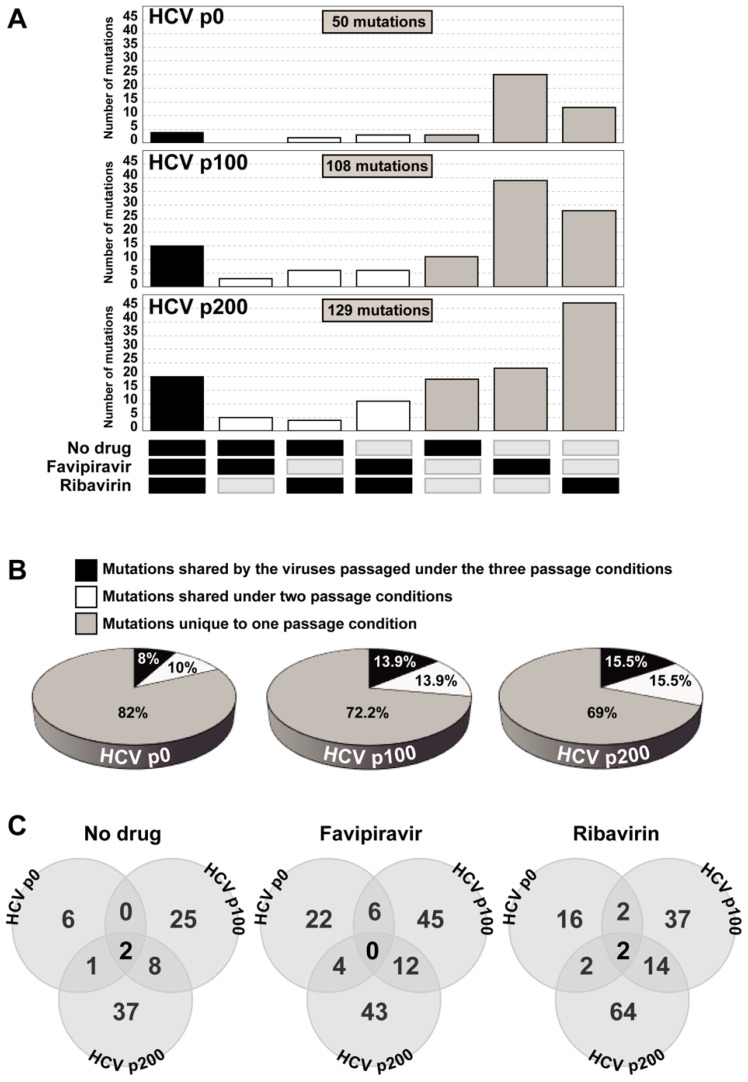
Figure 3.
Shared and unique mutations in the hepatitis C virus populations passaged in absence or presence of inhibitors. (A) number of mutations (ordinate) that participated in mutational waves in populations derived from HCV p0, HCV p100, and HCV p200 (excluding mutations that appear only in the initial populations), which were shared among conditions of the same virus and also among viruses passaged in the absence or presence of inhibitors; the passage conditions in which mutations were found is indicated by the filled rectangles drawn below the abscissa; note that the majority of mutations was unique to one condition (grey bars). (B) summary of the percentage of unique and shared mutations in the populations derived from HCV p0, HCV p100, and HCV p200. The complete list of mutations and deduced amino acid substitutions is given in Tables S3–S5 of [19]. (C) Venn diagram of mutations that are shared among HCV p0, HCV p100 and HCV p200 in the three experimental conditions (no drug, favipiravir, and ribavirin). The experimental procedures are described in the text.