Abstract
Frogs are ideal organisms for studying sex chromosome evolution because of their diversity in sex chromosome differentiation and sex-determination systems. We review 222 anuran frogs, spanning ~220 Myr of divergence, with characterized sex chromosomes, and discuss their evolution, phylogenetic distribution and transitions between homomorphic and heteromorphic states, as well as between sex-determination systems. Most (~75%) anurans have homomorphic sex chromosomes, with XY systems being three times more common than ZW systems. Most remaining anurans (~25%) have heteromorphic sex chromosomes, with XY and ZW systems almost equally represented. There are Y-autosome fusions in 11 species, and no W-/Z-/X-autosome fusions are known. The phylogeny represents at least 19 transitions between sex-determination systems and at least 16 cases of independent evolution of heteromorphic sex chromosomes from homomorphy, the likely ancestral state. Five lineages mostly have heteromorphic sex chromosomes, which might have evolved due to demographic and sexual selection attributes of those lineages. Males do not recombine over most of their genome, regardless of which is the heterogametic sex. Nevertheless, telomere-restricted recombination between ZW chromosomes has evolved at least once. More comparative genomic studies are needed to understand the evolutionary trajectories of sex chromosomes among frog lineages, especially in the ZW systems.
Keywords: anurans, sex chromosome diversity, homomorphy, heteromorphy, sex determination, fountain of youth, sexually antagonistic selection, sex-determination turnover, telomere-restricted recombination
1. Sex Chromosome Evolution
Across the tree of life, species determine sex either by using environmental cues or with sex chromosomes, which are the subject of this review. The asymmetrical inheritance of sex chromosomes with respect to sex makes them a genomic hotspot, compared to autosomes, for sex-specific selection and sexual conflict [1,2,3,4]. Sex chromosomes have independently evolved multiple times and show varied levels of divergence from each other in the heterogametic sex (in XY males or ZW females; [5,6,7,8,9,10]). The Y/W chromosomes in mammals, most birds and insects, are highly differentiated, and many functional genes have degenerated or were lost completely due to the long-term arrest of recombination with the X/Z. In sharp contrast, sex chromosomes are usually homomorphic (indistinguishable under the microscope) and rich in gene content in many reptiles, fish, amphibians and dioecious flowering plants and are thought to represent early stages of sex chromosome evolution [7,10,11,12,13]. The reason why the trajectory of sex chromosome evolution differs so dramatically across the tree of life is an unresolved question in evolutionary biology.
The canonical model of sex chromosome evolution suggests that sexually antagonistic (SA) genes play a key role in the process of sex chromosome degeneration [5,14,15,16,17,18]. Sex chromosomes are thought to evolve from a pair of autosomes starting with a sex-determining mutation. Male beneficial mutations are favored to accumulate in the vicinity of the sex-determining locus on the sex-limited Y chromosome (or female-beneficial mutations on the W), which selects for the arrest of recombination in the region because it contains the fittest possible haplotype for the heterogametic sex. The arrival of new mutations with sex-specific beneficial effects selects for an extension of the region of recombination arrest to a longer haplotype. Over evolutionary time, the recombination arrest leads to progressive differentiation and degeneration of sex-limited chromosomes through mechanisms, such as Muller’s ratchet, genetic hitchhiking, reduced purifying selection, and stronger genetic drift [5,14,15,16,17,18]. This model has been widely accepted to account for the recombination suppression and degeneration in the highly differentiated sex chromosomes in mammals, most birds and insects [7,10].
In the past decade, with the advancement and reduced price of genomic sequencing, studies of many non-model organisms have revealed a remarkable diversity in the rate of sex chromosome differentiation, as well as in the dynamics of birth and death of sex chromosomes (i.e., sex chromosome turnovers) in many fishes, amphibians and reptiles (reviewed by [19]). In many cases, there is very limited differentiation between the sex chromosomes in the heterogametic sex despite their old age [20,21,22,23,24,25]. In other words, they do not follow the degeneration path predicted by the canonical model of sex chromosome evolution. Furthermore, there has been little empirical support to demonstrate the role of sexually antagonistic (SA) genes in sex chromosome recombination suppression, because the phenotypic effects of SA genes are difficult to demonstrate and because it is hard to show the causal effects of SA genes (since SA genes precede recombination arrest, both recombining and arrest status are needed in closely related species to test this; reviewed by [4]). In some frog lineages, studies have shown that recombination suppression between XY chromosomes is likely due to genome-wide male-specific reduced recombination, because it is restricted to the telomeric regions in males (females recombine across the full length of their chromosomes), which challenges the universal role of sexually antagonistic (SA) genes in driving sex chromosome recombination arrest [26,27,28,29]. Studies on a variety of stages of sex chromosome evolution from homomorphic to highly degenerated, particularly the less studied early stage, are still needed to fully understand sex chromosome evolution.
2. Sex Chromosome Diversity in Frogs
Frogs offer an ideal system for advancing our understanding of sex chromosome diversity and evolution; this is because they harbor various stages of sex chromosome differentiation and diverse sex-determination systems between different species and, sometimes, across and within populations of the same species [26,30,31,32,33]. Earlier studies showed that the majority (~95%) of studied frog species had homomorphic sex chromosomes [30,33]. Recent studies have revealed a diversity of sex chromosome systems in frogs, including heteromorphic and multiple sex chromosomes, especially in species distributed in neotropical regions (i.e., Central and South America) [12,34,35,36]. Heteromorphic sex chromosomes show signs of degeneration, such as extensive accumulation of transposable elements and other repeats, resulting in an enlargement of the sex-limited chromosomes (Y or W), increased heterochromatinization, or a diminishment of their size and gene loss. Both are consequences of the long-term recombination suppression between the sex chromosomes [13,37].
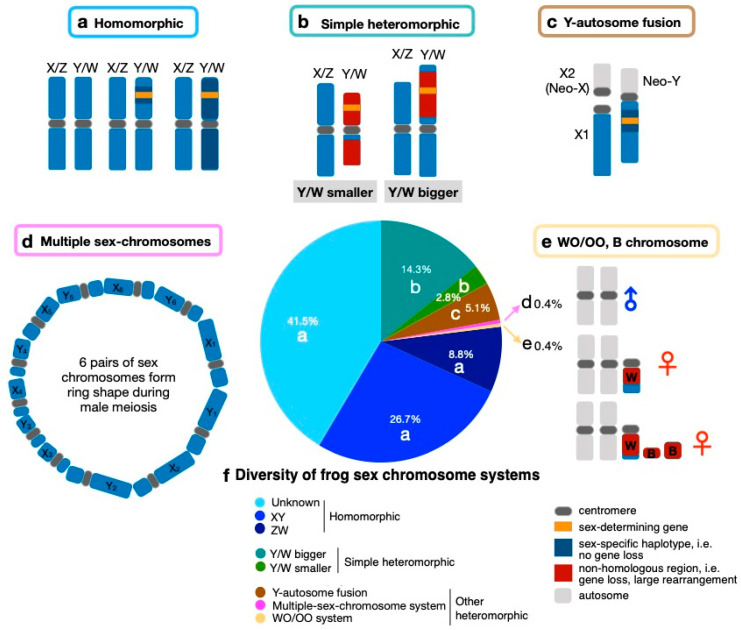
In this review, we have compiled a list of 222 Anuran species, from 23 different families, with known sex chromosome systems from the literature, spanning ~220 Myr (Supplementary Materials Table S1). We discuss the pattern, diversity and evolution of their sex chromosomes. The majority (~75%) have homomorphic sex chromosomes (Figure 1a, Table S1), with more male heterogametic (26.7%, XX/XY) than female heterogametic (8.8%, ZW/ZZ) systems, and 41.5% with an unknown system (Figure 1f). The DNA sequence difference between two sex chromosomes varies from most of their length to being restricted to the sex-determining region, or no differentiation, as detected in populations of the common frog Rana temporaria (Figure 1a, [38,39]). Less commonly, overall, ~25% of frog species have heteromorphic sex chromosomes (Figure 1f). Among the simple heteromorphic sex chromosome system (composed of a single pair), there are slightly more ZW (Nspecies = 23–24) than XY (Nspecies = 19–20, Glandirana rugosa has both XY and ZW) systems (Figure 1b). The second commonest heteromorphic sex chromosome system involves Y-autosome fusions (no fusion with a W is known), which occur in ~5.1% of all surveyed species (Figure 1c). One interesting sex chromosome system was discovered in the Brazilian smoky jungle frog (Leptodactylus pentadactylus) which has six pairs of sex chromosomes and five autosomal pairs [40] (Figure 1d). Another unique sex chromosome system involves the coexistence of female-specific W (WO/OO system) and supernumerary (or B) chromosomes of various sizes among populations in the New Zealand frog Leiophlma hochstetteri [41,42] (Figure 1e).
Figure 1.
Diversity of frog sex chromosome systems. (a) Homomorphic sex chromosomes—sex-specific haplotype can vary largely from very small to the full length of the homologous X/Z, within or across populations within species. (b) Simple heteromorphic sex chromosomes—the Y/W may be bigger or smaller than X/Z. (c) Y-autosome fusion generating Neo-X (X2) and Neo-Y. (d) Multiple sex chromosomes in Leptodactylus pentadactylus, where 6 pairs of sex chromosomes form a ring shape during meiosis. (e) WO/OO and supernumerary (B) chromosomes in Leiopelma hochstetteri, with population-specific W and various forms of B chromosomes, which are never observed in males. (f) Pie chart shows the proportion of sex chromosome systems in Anuran frogs. All the non-homomorphic sex chromosome types, such as b, c, d and e systems, are heteromorphic.
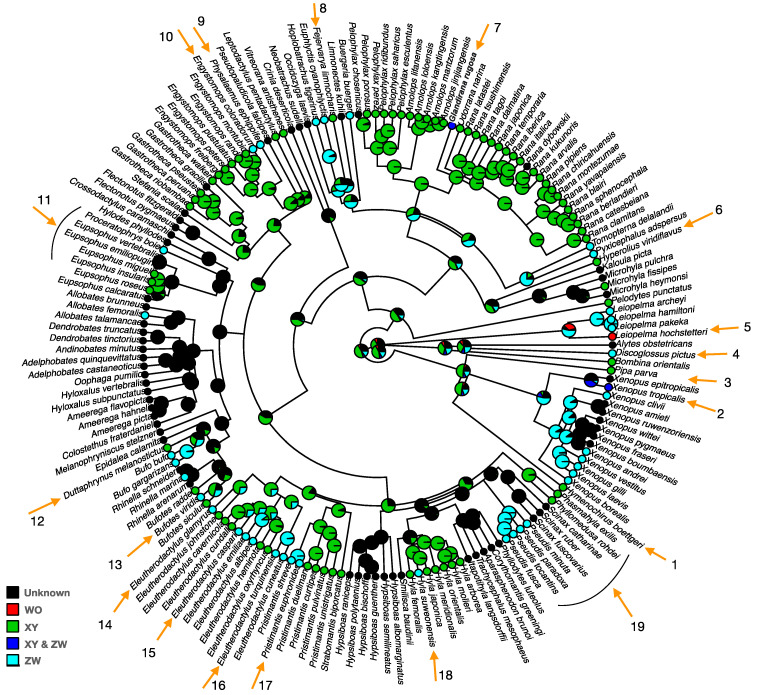
There are at least 19 turnovers between sex-determination systems (including XY to ZW, ZW to XY, and ZW to WO) during anuran evolution spanning ~220 Myr (Figure 2, Figure S1). It was not possible to reconstruct the ancestral status due to many transitions along the phylogeny (Figure 2), but the stochastic mapping approach using the ARD model estimated XY had the highest probability among the known sex-determination systems. The sex-determination system is almost exclusively XY in Ranidae frogs (with the exception of Glandirana rugosa which has both XY and ZW systems) and in Hyla tree frogs (except H. suweonensis which has ZW). All Pseudis frogs have a ZW system. The number of sex-chromosome transitions is expected to be higher than the number estimated for sex-determination system transitions (Figure 2), because turnovers between chromosomes within the same sex chromosome system are common. There are at least 13 turnovers within almost exclusively the XY system in 28 Ranidae frogs [43]. Frequent sex-determination system turnovers also occur in geckos (17–25 transitions [44]), stickleback fishes (2–3 transitions, [45]) and salmonid fishes (3 transitions, [46]). Sex chromosome turnover within a given sex-determination system has also been documented in Oryzias medaka fishes (five transitions, [47]).
Figure 2.
Distribution of various sex-determination systems along Anurans. The phylogenetic tree is obtained from http://www.timetree.org/, accessed on 5 February 2021 and the pattern of sex chromosome diversity and the ancestral status reconstruction were analyzed and visualized with the “Phytools” package in R (version 3.6.3), using a stochastic mapping approach with the ARD (all rates different matrix) transition rate model for discrete character traits [48,49]. Pie charts display the posterior probability of sex-determination systems on each node of the phylogenetic tree. Numbered arrows indicate transitions between sex-determination systems.
3. Sex Determination in Frogs
Sex in most amphibians is genetically determined, although non-genetic sex determination has also been reported [50]. Many genes with a known association with gonadal differentiation have been mapped to the sex chromosomes of various frog species. Genes with feminization effects include Dm-w, Cyp19, Sf1, Foxl2, Sox3, and genes with masculinization effects include Dmrt1, Amh, Ar, Cyp17 (reviewed in [32,51]). Little is known about the molecular mechanisms underlying sex determination in frogs. The only confirmed sex-determining gene is Dm-w, located on the W chromosome of the African clawed frog Xenopus laevis, where it has a crucial role in primary ovary formation [52,53]. Dm-w shares high protein sequence identity (~89%) with the DNA-binding domain of its paralog Dmrt1, but does not contain a domain with homology to the transactivation domain of Dmrt1 [54]. It has been hypothesized that Dm-w binds the target gene Dmrt1 in the gonads of ZW females during sex determination, which prevents Dmrt1 from interacting with its binding site, thus inducing ovary formation [52,53]. A recent study has found that the Dm-w gene is not always associated with female development in many Pipidae species (except in Xenopus laevis, X. gilli) and has further identified sex chromosomes in three additional pipid frogs (see Figure 3, [55]).
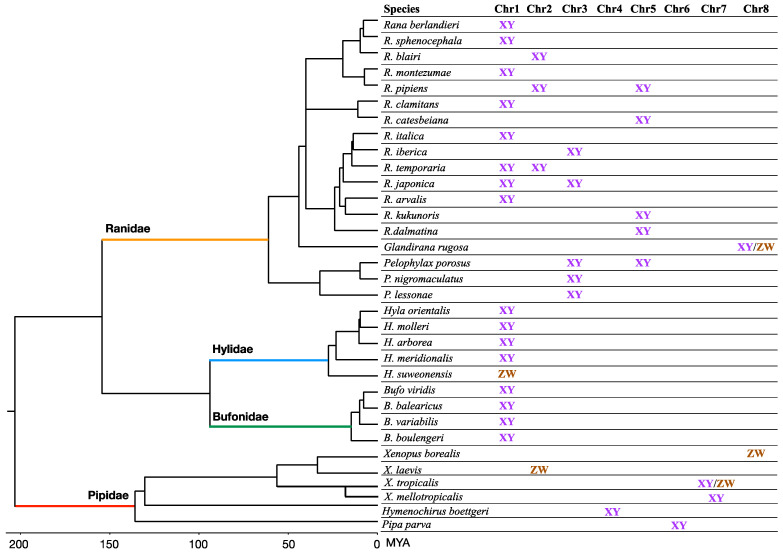
Figure 3.
Overview of known sex-determination systems among anurans across four families (branch in distinct colors) spanning ~200 Mya divergence, due to the well-conserved karyotype and genome-wide synteny revealed by comparative genomics. The chromosome number is based on the genome of Xenopus tropicalis. The chromosomes (Chr9 and Chr10) that are never used as sex chromosomes are omitted. Sex chromosomes were identified from various publications [43,55,59,60,62,63,64,65,66,67]. The tree is obtained from http://www.timetree.org/, accessed on 5 February 2021 and visualized in Figtree (http://tree.bio.ed.ac.uk/software/figtree/, accessed on 5 February 2021).
Frog genomes exhibit considerable synteny [56,57,58], despite long periods of independent evolution (e.g., approximately 266 Myr of divergence between Narorana parkeri and Xenopus tropicalis or 204 Myr between Rana temporaria and Xenopus tropicalis). Genome-wide syntenic block analysis shows that amphibians have fewer inter-chromosomal rearrangements than mammals but have a comparable rate of intrachromosomal rearrangements [56]. It is therefore possible to refer to the chromosome numbers of Xenopus when comparing frog genomes (Figure 3).
A study in three divergent groups of anurans (Bufo siculus, Hyla arborea and Rana temporaria) found that various sex-linked genes, including FGA, SMARCB1, Dmrt1, map to Xenopus tropicalis chromosome 1 (Chr1) and show a strong association with sex (Figure 3, [58]). Other studies using microsatellites found sex-linked haplotypes on Chr1, in addition, in at least four Hyla tree frog species and four Bufo species [59,60]. Dmrt1 is known to play a key role in sex differentiation across many animal lineages. Advances in RAD (restriction site associated DNA) sequencing allow us to rapidly expand the identification of sex chromosomes in many more species. A recent study identified five chromosomes (Chr1, Chr2, Chr3, Chr5, Chr8) in 28 Ranidae to be associated with sex determination (Figure 3) [43]. The non-random co-option of these five chromosomes as sex chromosomes was probably because genes on these chromosomes are involved in the sex-determination cascade in amphibians. Such candidate genes include Dmrt1 and Amh on Chr1, Fgf9, Amhr2 and Rspo1 on Chr2, Cyp19 on Chr3, Foxl2 on Chr5, Sox9 on Chr7, Ar and Sox3 on Chr8 [43,51,61]. Furthermore, one study has further extended the sex chromosome list in pipid frogs (Chr2, Chr4, Chr6, Chr7, Chr8) [55], which leaves homologs of only X. tropicalis Chr9 and Chr10 as not being associated with sex in frogs.
4. Homomorphic Sex Chromosomes in Frogs
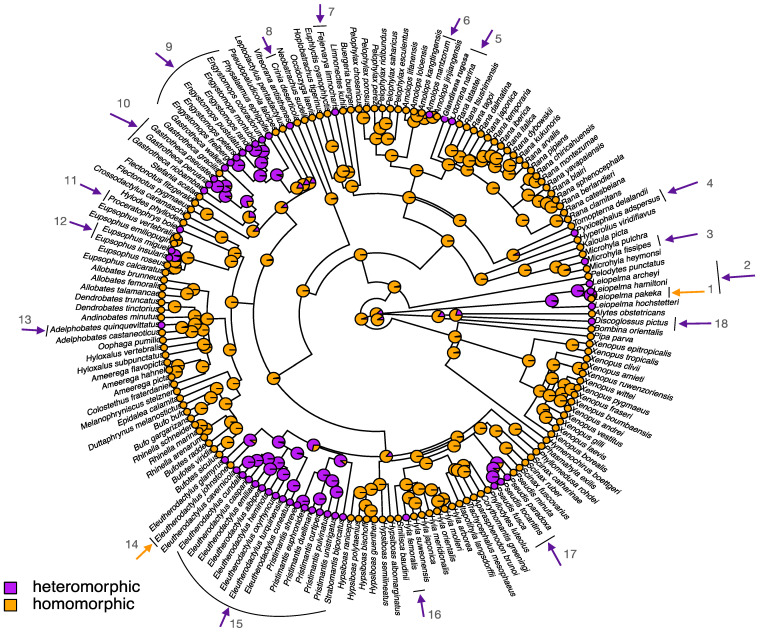
The majority (~75%) of studied anurans (20/23 studied families) have homomorphic sex chromosomes (Figure 1, Table S1). Most species with homomorphic sex chromosomes occur in Bufonidae (100% of the family), Pipidae (100%), Dendrobatidae (91%), Ranidae (88%) and Hylidae (88%) (Figure 4). Among species with homomorphic sex chromosomes, there are three times more species (Nspecies = 57–58) with male heterogamety (XX/XY), than those (Nspecies = 18–19) with female heterogamety (ZZ/ZW). The remaining 54% of species with homomorphic sex chromosomes have an unknown (NA) sex chromosome system due to limited cytogenetic analysis (Table S1).
Figure 4.
Distribution of homomorphic and heteromorphic sex chromosomes along the phylogeny of studied anurans; see more details in Table S1. Homomorphic sex chromosomes are indicated in orange, and heteromorphic in purple. The tree is obtained from http://www.timetree.org/, accessed on 5 February 2021 and the ancestral state reconstruction of sex chromosome morphology is analyzed and visualized with the “Phytools” package in R (version 3.6.3), using a stochastic mapping approach with the ER (equal rate) transition rate model, suitable for binary character traits [48,49]. Pie charts display the posterior probabilities of sex chromosome types on each node of the phylogenetic tree. Numbered arrows indicate transitions in sex chromosome morphology, purple arrows indicate from homomorphy to heteromorphy, and orange arrows from heteromorphy to homomorphy.
One of the best studied homomorphic sex chromosome systems is in the European common frog Rana temporaria. This species is distributed in Europe as far north as Scandinavia and as far east as the Urals, but not in most of Iberia, southern Italy and the southern Balkans [68,69,70]. Consistent with earlier karyotype studies [71], genetic and genomic studies have confirmed the species to have a homomorphic male heterogametic XX/XY system [38,72,73]. Variation in the Dmrt1 gene is consistent with a sex-determination role across populations in this species [38]. There is an interesting polymorphism in Y-chromosome differentiation (Chr1), which was identified using sex-linked markers, including the Dmrt1 region. The Y-chromosome may be fully differentiated (Y-specific haplotypes as long as Chr1), proto-Y (Y-specific haplotype is restricted to the region surrounding Dmrt1), and undifferentiated Y (no Y-specific haplotype is identified) (Figure 1a, [38,39,73]). Multiple haplotypes of each Y-chromosome differentiation pattern have been found [39,74]. The three levels of Y-chromosome differentiation follow a latitudinal cline within Sweden, where populations in the south have the majority (>81%) of males with proto-Y. This proportion decreases as latitude increases so that northern Swedish populations exclusively have a fully differentiated Y-chromosome haplotype [73]. The association with temperature, implied by this latitudinal cline, is not repeated in Swiss populations where temperature varies with altitude. Instead, Y-chromosome haplotypes are better associated with the phylogeographic signal, similar to the distribution pattern of mitochondrial haplotypes [74,75]. Remarkably, Y-chromosome haplotype polymorphisms involving all three differentiation levels were found within single Swiss populations [39,72], and the haplotypes appear selectively neutral [29]. The forces maintaining the polymorphism within populations remain unclear [29,39].
Intraspecific polymorphisms of homomorphic sex chromosomes have been documented in many frog species (Figure 3). Chr1 is the sex chromosome in R. temporaria throughout its distribution range, but Chr2 has also been shown to co-segregate with sex (along with Chr1) in one northern Swedish population [66,67]. Pelophylax porosus uses Chr5 in the Okayama form in western Japan and Chr3 in the Nagoya form in eastern Japan [76]. Two chromosomes in Rana pipiens, Chr2 and Chr5, have been described as sex-linked in various populations between lineages of eastern and western USA [43,77]. R. japonica uses Chr1 to determine the sex in western Japan but Chr3 in eastern Japan [78]. However, no association was found between sex and any of these two chromosomes in the Akita (northern Honshu) population, suggesting another undetected intraspecific sex chromosome polymorphism in R. japonica [78]. Chr8 is probably involved in the intraspecific sex-determination system turnover in Glandirana rugosa, from an XY to ZW sex-determination system [61]. Finally, Xenopus tropicalis Chr7 is associated with multiple sex-determination system turnovers, where W, Z, and Y chromosomes occur in the same population in Ghana, and the degenerate W and Y probably evolved from the Z chromosomes [62].
4.1. Mechanism/Forces to Maintain Homomorphic Sex Chromosomes
Two mutually nonexclusive hypotheses have been proposed to explain the maintenance of homomorphic sex chromosomes in frogs. The “fountain of youth” model states that deleterious mutations could be purged from the sex-limited chromosomes (Y or W), and long-term differentiation could be prevented by occasional sex-chromosome recombination [79]. Supporting evidence has been found in European tree frogs, which inherited the same pair of sex chromosomes from a common ancestor approximately 5 Mya ago. Their sex chromosomes remained homomorphic despite the lack of XY recombination in males, and haplotypes at sex-linked markers further cluster by species and not by gametologs, suggesting a history of recurrent XY recombination [20,22,80]. Using an Approximate Bayesian Computation approach, Guerrero et al. [21] showed the rate of XY recombination in this tree frog group was significantly different from zero, while being ∼10−6 lower than that of XX recombination. A possible mechanism allowing XY recombination is the occasional sex reversal of XY individuals (i.e., sex-reversed XY females), resulting from incomplete genetic control over sex determination [79]. If the arrest of XY recombination in males is a property of male meiosis and not the male genotype, then Y chromosomes would recombine with X chromosomes in the occasional sex-reversed XY females, preventing Y chromosomes from progressive differentiation and degeneration. Sex-reversal experiments in a series of taxa (crested newts, medaka fish, housefly) have confirmed that sex differences in recombination largely depend on phenotypic sex, not genotypic sex [81,82,83]. Additional support for this “fountain-of-youth” model comes from genetic mapping in wild populations of the common frog R. temporaria, which showed that XY recombination only depends on phenotypic sex. Wild XX males showed recombination restriction similar to XY males, while wild sex-reversed XY females recombined as much as XX females [84].
The second hypothesis for maintenance of homomorphic sex chromosomes states that sex chromosome turnovers are frequent enough that sex chromosomes are replaced by another autosome pair before they have time to degenerate [85,86]. Van Doorn and Kirkpatrick [87,88] proposed a role for sexually antagonistic (SA) genes in driving sex chromosome turnover, where a male-benefiting mutation accumulating on an autosome favors the evolution of a masculinizing mutation in its vicinity. This mechanism is well-illustrated in Cichlidae fishes but with a female-beneficial mutation, which allowed a new ZW system to invade an XY system via a mutation on the proto-W chromosome [89]. Blaser et al. [90] proposed a role for the mutation load that accumulates on sex chromosomes to drive the sex chromosome turnovers. Later, Blaser et al. [91] proposed the “hot-potato model”, which combined SA and deleterious effects. They showed that SA alleles located on a chromosome, after it has been co-opted for sex, induce the recombination arrest, and the ensuing accumulation of deleterious mutations would generate pressure for a new sex-chromosome transition. This “hot-potato model” makes two predictions, (1) that the type of heterogamety would be conserved during transitions, and (2) that autosomes are not recruited randomly, with some autosomes being more likely to be co-opted for sex. Both predictions were supported in a study of 28 Ranidea, which showed at least 13 sex chromosome turnovers while preserving (with one exception) the XY system, although genetic drift could not be excluded to account for fast sex chromosome turnovers. Similarly, the model is supported by the frequent sex chromosome turnovers within Oryzias medaka fishes, where five turnovers occur within the XY system and two in the ZW system [47].
5. Heteromorphic Sex Chromosomes in Frogs
Recent frog studies have brought up the proportion of frog species with heteromorphic sex chromosomes to ~25% among these karyotyped frogs (Table S1). Among the simple heteromorphic sex chromosomes, female heterogametic (ZW) species are slightly more common than those with male heterogamety (XY, Nspecies = 23 vs. 19, Glandirana rugosa has both XY and ZW systems). Interestingly, 68% of these species with XY systems and 83% with ZW systems have larger Ys/Ws chromosomes than their homologous Xs/Zs chromosomes (Table S1, Figure 1b), suggesting most of the simple heteromorphic sex chromosome systems in frogs are at a relatively early stage of sex chromosome evolution.
Heteromorphic sex chromosomes in frogs have been relatively understudied, and the forces allowing their escape from the mechanisms maintaining homomorphy in most frog species remain largely unknown. Phylogenetic analysis suggests the likely ancestral status to be homomorphic sex chromosomes in frogs (88% probability; Figure 4). Heteromorphic sex chromosomes span several lineages and also occur within closely related species in genera with primarily homomorphic sex chromosomes, suggesting that heteromorphy has independently evolved multiple times (at least 16; Figure 4). Interestingly, the inverse transition (from heteromorphy to homomorphy) is rare (two times in our compiled dataset; Figure 4). Five frog genera have primarily heteromorphic sex chromosomes (Eleutherodactylus, Gastrotheca, Pristimantis, Engystomops and Pseudis; Figure 4). The stability of the sex-determination system varies between the lineages, with Pseudis being exclusively ZW, Pristimantis and Gastrotheca having mostly a XY system (60–80% of species), and Eleutherodactylus has an equal proportion of XY and ZW systems (Figure 2).
The canonical sex chromosome evolution model predicts that the Y and W chromosomes eventually become small because they degenerate and that most of their genetic content is lost due to the absence of recombination with the X or Z, respectively. However, the canonical model does not explicitly predict an expansion stage of sex-limited Y/W chromosomes during their evolution [5,14,15,16,17,18]. Such an expansion has been observed in plants [13] and fishes, amphibians, reptiles and some birds, many species of which have larger Ys and Ws than their Xs and Zs counterparts, suggesting that the accumulation of transposons and expansion of repetitive sequences can increase their size during their early differentiation [37,92]. This enlargement can be a rapid and effective mechanism to make the emerging Ys and Ws different from the other chromosomes and further prevent crossovers between the sex chromosomes. The shrinking of the Y and W chromosome to their familiar small size in mammals and birds is proposed to only occur at a later phase of their degeneration [37]. The enlarged Y/W chromosomes of some species are thus considered “younger” than the shrunk Y/W chromosomes of other species. Phylogenetic analysis based on current datasets supports this as the enlarged Y/W are “younger” than the shrunk ones in Gastrotheca frogs (Figure S2). However, the overall pattern in the whole phylogeny could also be explained by lineage-specific selection on the rate of sex chromosome differentiation (Section 5.1), and more data on such heteromorphic sex chromosome systems in the future will help resolve this issue. As large Ys/Ws lose genetic material to turn to small, they are expected to pass through a stage where they are identical in size to the homologous Xs/Zs chromosomes [13,37]. This hypothesis has gained support from genomic analysis of fish species with various stages of early evolution of their sex chromosomes [93]. Whether repetitive sequences are mainly responsible for the enlargement of Ys/Ws in anurans, as well as whether homomorphic sex chromosomes have passed through an enlarged stage of Ys/Ws in their past evolutionary history, is not clear for most frog species and would require comparative genomic analysis.
5.1. What Forces Result in Heteromorphic Sex Chromosomes in Frog Lineages?
The reasons why ~25% of studied frogs have escaped the mechanisms that seem to maintain homomorphic sex chromosomes in other species (“fountain of youth” and/or “hot-potato model”) remain unclear. Homomorphy has been maintained for over 100 Myr in many frog species (Figure S3). The old age of heteromorphic sex chromosome systems is also not a good explanation because phylogenetic analysis shows them to be as old as ~150 Mya and as recent as ~10 Mya (Figure S3).
Of particular interest are the frog lineages Eleutherodactylus, Gastrotheca, Pristimantis, Engystomops, which have primarily heteromorphic sex chromosomes (Figure S3, Table S1). They are all distributed in highly restricted ranges in the neotropics (central and south America) and many of the species occur in single Caribbean islands (sometimes more than one species co-occur in one island), or in small populations [12,30]. Their small population sizes might result in lineage-specific fast degeneration as an outcome of the fixation of deleterious genetic variation. Another possibility is lineage-specific selection on the rate of sex chromosome differentiation, a situation found in palaeognathous and neognathous birds in which limited divergence between sex chromosomes (i.e., sex chromosome are homomorphic and largely recombining) were detected spanning >90 Myr, suggesting slow rates of sex chromosome degeneration compared to the rest of bird lineages with highly degenerated sex chromosomes [24,94,95,96]. Unlike these primarily non-flying birds, these frog lineages might show faster degeneration, as observed in the heteromorphic sex chromosomes of the salamander genus Aneides, which are restricted to isolated populations [30]. Most frogs with homomorphic sex chromosomes are widely distributed in the mainland, such as in the lineages of Rana, Hyla, and Bufo frogs [30]. In addition, the frog lineages with primarily heteromorphic sex chromosomes (Eleutherodactylus, Gastrotheca, Pristimantis, Engystomops) have unique life-history traits that might contribute to fast sex chromosome degeneration. For instance, all Eleutherodactylus and Pristimantis frogs have direct development, i.e., they lack free-living aquatic tadpoles and instead hatch from terrestrial eggs as miniature adults [97,98,99,100]. They experience a reduced developmental time to reach adulthood, which leads to shorter generation times and could possibly accelerate sex chromosome evolution. These frogs, together with Gastrotheca frogs, also have parental care, and some show pronounced sexual dimorphism in body color and size [100]. Engystomops frogs have a foam-nest behavior, where the egg jelly is beaten into white foam by the male during amplexus, which functions as parental care. The developing eggs in the foam nest are removed from the aquatic environment and are protected from desiccation and predators [100]. These life history traits might lead to pronounced sexual selection compared to other frog lineages, which could generate strong selection for sexually antagonistic genes, making them more central in sex chromosome evolution, including their degeneration, in these lineages.
Other frogs with heteromorphic sex chromosomes occur in linages with a majority of species with homomorphic sex chromosomes (Figure 4). In these cases, it is more difficult to argue for demographic and life history explanations that allow the escape from the forces normally maintaining homomorphic sex chromosomes. One possibility that can allow the canonical model of sex chromosome evolution to hold despite male-specific absence of recombination (see Section 7) is the occurrence of a chromosomal inversion involving the sex-determining region, so that recombination arrest depends on genotypic, not phenotypic sex. For example, XY sex-reversed individuals would not recombine if the Y was fixed for an inversion. This is particularly relevant for W chromosome evolution. Since female-specific telomere-specific recombination does not seem to occur in frogs, the most likely mechanism of recombination restriction is similar to the mechanisms familiar from the canonical model of sex chromosome evolution. One case of recombination restriction is known (in Buergeria buergeri) where the ZW bivalent is ring-shaped, although the mechanism is unclear [101]. More comparative studies of sexual dimorphism and genomic composition across species with both homomorphic and heteromorphic sex chromosomes are needed to understand the different evolutionary trajectories of their sex chromosomes.
6. Sex Chromosome–Autosome Fusion
Fusions between the sex chromosomes and autosomes have been described for 11 frog species (Figure 1, Table S1). All are Y-autosome fusions in species with male heterogametic (XX/XY) sex chromosome systems. The higher fusion rate in the XX/XY system (11/94, compared to 0/45 in ZZ/ZW species) in frogs should be considered preliminary and taken with caution because the sample size is small. Nevertheless, it is consistent with the higher rate of fusions of Y chromosomes with autosomes, compared to X, Z or W chromosomes, which were observed in fishes and squamate reptiles [102]. Population genetic models suggest that direct selection acting on fusions or sexually antagonistic selection cannot alone account for the predominance of Y-autosome fusions. Instead, the most plausible explanations are that fusions are slightly deleterious, and the mutation rate is probably male-biased, or the reproductive sex ratio is female-biased [102].
7. Extremely Sexual Dimorphic Recombination Pattern
Many species show heterochiasmy, in which the location and rate of recombination differ between the sexes [103,104,105,106]. The most extreme case is achiasmy (the absence of recombination in one sex), which always occurs in the heterogametic sex (e.g., XY males in Drosophila; ZW females in butterflies) and has evolved at least 29–34 times independently in animals [103]. Sexual dimorphism in recombination may be a byproduct of mechanistic differences between meiosis in males and females, or it may be adaptive and selected to promote tight linkage of beneficial alleles on the Y or W [104,106,107,108]. Neither explanation is adequate for all species [106]. Furthermore, sex differences in recombination can vary in degree and direction even between closely related species [106,107,108].
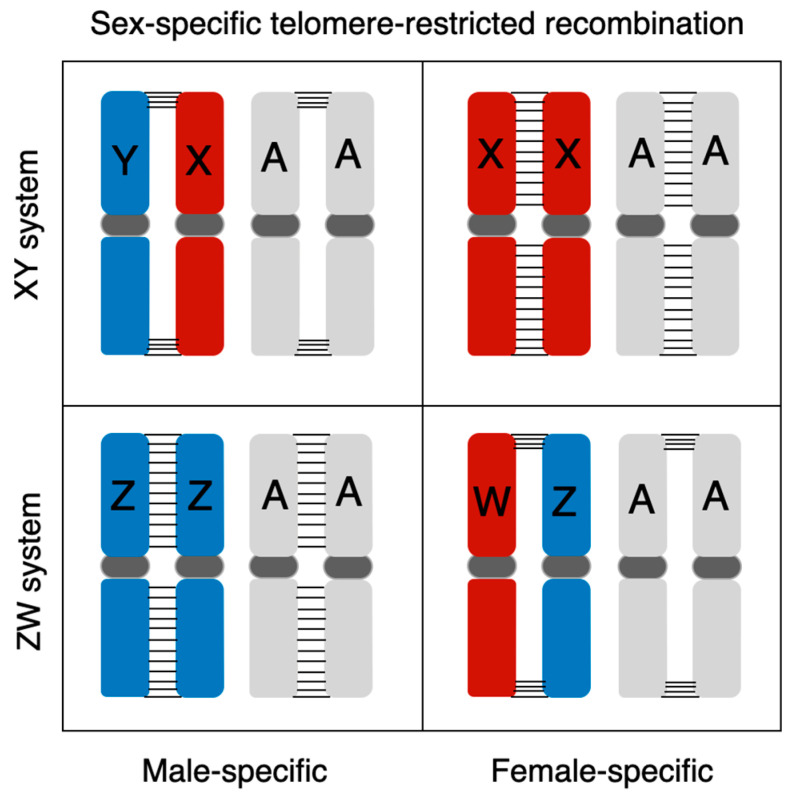
Cytogenetic studies of frogs show that during male meiosis, ring-shaped bivalents form in most frog lineages with male heterogametic (XX/XY) sex chromosomes (male-specific telomere-restricted recombination), suggesting that most recombination occurs in the telomere regions in males but across the chromosome length in females (Figure 5, [12,30]). Extreme heterochiasmy has been confirmed by directly estimating sex-specific recombination with genetic mapping in many frogs and some poecilid fishes with XY systems [20,43,109,110], as well as in a few ZW systems in frogs [25,111], where males always show reduced recombination. However, all of these studies involve frogs with homomorphic sex chromosomes. Cytogenetic analysis of female meiosis in the Kajika frog (Buergeria buergeri), which has a homomorphic ZW system, showed the ZW bivalent to be ring-shaped, while the autosomes showed recombination across their length, suggesting that female-specific telomere-restricted recombination has not evolved in this case (Figure 5, [30,107]). Little is known about female-specific telomere-restricted recombination in other female heterogametic (ZZ/ZW) frog species, especially in those with heteromorphic sex chromosomes, which has the potential to affect evolution of the W chromosome in the same way that male-specific telomere-restricted recombination influences Y evolution in most studied frogs. The possible evolution of female-specific telomere-restricted recombination (Figure 5), or chromosome-specific (i.e., ZW) recombination suppression, can both affect the evolution of ZW systems, and future comparative studies of homomorphic and heteromorphic ZW systems are needed to understand their relative importance in sex-chromosome evolution.
Figure 5.
Schematic drawing of sex-specific telomere-restricted recombination in male heterogametic (XX/XY) and female heterogametic (ZZ/ZW) sex chromosome systems. “A” refers to all autosomes, and horizontal bar “-“ indicates recombination between homologous chromosomes. Note that female-specific telomere-restricted recombination has not been described in studied anurans. The only case of telomere-specific ZW recombination (in Buergeria buergeri) does not affect the autosomes.
Acknowledgments
We would like to thank Nicolas Perrin lab members for sex chromosome work in various Ranidae frogs and related discussions. We also thank three anonymous reviewers for their constructive comments to improve this manuscript.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/genes12040483/s1, Table S1. Characterized sex chromosome systems in Anura frogs from published literature. The columns show species, chromosome number, sex chromosome morphology, sex-determination system, references and other relevant notes. Figure S1. Distribution of various sex-determination systems along Anurans. The phylogenetic tree with estimated divergence time is obtained from http://www.timetree.org/, accessed on 5 February 2021. Pie charts display the posterior probability of sex-determination systems on each node of the phylogenetic tree. Figure S2. Distribution of homomorphic and various heteromorphic sex chromosome systems (i.e., Y/W bigger than X/Z, Y/W smaller than X/Z, other forms) along the phylogenetic relationship in the studied anurans. The phylogenetic tree with estimated divergence time is obtained from http://www.timetree.org/, accessed on 5 February 2021. Pie charts display the posterior probability of sex chromosome morphology on each node of the phylogenetic tree. Figure S3. Distribution of homomorphic and heteromorphic sex chromosomes along the phylogenetic relationship in the studied anurans. Homomorphic sex chromosomes are indicated in orange, and heteromorphic in purple. The phylogenetic tree with estimated divergence time is obtained from http://www.timetree.org/, accessed on 5 February 2021. Pie charts display the posterior probability of sex chromosome morphology on each node of the phylogenetic tree.
Author Contributions
W.-J.M., P.V. designed the study. W.-J.M. collected data from literature and performed analyses. W.-J.M. and P.V. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work is supported by the National Science Foundation (NSF) grant, Division of Molecular and Cellular Bioscience 2047052.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All datasets in this review have been provided in Supplementary Table S1.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mullon C., Wright A.E., Reuter M., Pomiankowski A., Mank J.E. Evolution of Dosage Compensation under Sexual Selection Differs between X and Z Chromosomes. Nat. Commun. 2015;6:7720. doi: 10.1038/ncomms8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mank J.E., Hosken D.J., Wedell N. Conflict on the Sex Chromosomes: Cause, Effect, and Complexity. Cold Spring Harb. Perspect. Biol. 2014;6:1–14. doi: 10.1101/cshperspect.a017715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachtrog D. Y-chromosome Evolution: Emerging Insights into Processes of Y-chromosome Degeneration. Nat. Rev. Genet. 2013;14:113–124. doi: 10.1038/nrg3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wright A.E., Dean R., Zimmer F., Mank J.E. How to Make a Sex Chromosome. Nat. Commun. 2016;7:12087. doi: 10.1038/ncomms12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bull J.J.J. Evolution of Sex Determining Mechanisms. Benjamin/Cummings Pub. Co.; Menlo Park, CA, USA: 1983. [Google Scholar]
- 6.Bachtrog D., Kirkpatrick M., Mank J.E., McDaniel S.F., Pires J.C., Rice W.R., Valenzuela N. Are All Sex Chromosomes Created Equal? Trends Genet. 2011;27:350–357. doi: 10.1016/j.tig.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 7.Beukeboom L.W., Perrin N. Evolution of Sex Determination. Oxford University Press; Oxford, UK: 2014. [Google Scholar]
- 8.Bachtrog D. The Temporal Dynamics of Processes Underlying Y Chromosome Degeneration. Genetics. 2008;179:1513–1525. doi: 10.1534/genetics.107.084012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charlesworth B., Charlesworth D. The Degeneration of Y Chromosomes. Philos. Trans. R. Soc. B Biol. Sci. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bachtrog D., Mank J.E., Peichel C.L., Kirkpatrick M., Otto S.P., Ashman T.L., Hahn M.W., Kitano J., Mayrose I., Ming R., et al. Sex Determination: Why so Many Ways of Doing It? PLoS Biol. 2014;12:e1001899. doi: 10.1371/journal.pbio.1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devlin R.H., Nagahama Y. Sex Determination and Sex Differentiation in Fish: An Overview of Genetic, Physiological, and Environmental Influences. Aquaculture. 2002;208:191–364. doi: 10.1016/S0044-8486(02)00057-1. [DOI] [Google Scholar]
- 12.Schmid M., Steinlein C., Bogart J.P., Feichtinger W., León P., La Marca E., Díaz L.M., Sanz A., Chen S.H., Hedges S.B. The Chromosomes of Terraranan Frogs: Insights into Vertebrate Cytogenetics. Cytogenet. Genome Res. 2010;130–131:1–568. doi: 10.1159/000301339. [DOI] [PubMed] [Google Scholar]
- 13.Ming R., Bendahmane A., Renner S.S. Sex Chromosomes in Land Plants. Annu. Rev. Plant Biol. 2011;62:485–514. doi: 10.1146/annurev-arplant-042110-103914. [DOI] [PubMed] [Google Scholar]
- 14.Fisher R.A. The Evolution of Dominance. Biol. Rev. Biol. Proc. Camb. Philos. Soc. 1931;6:345–368. doi: 10.1111/j.1469-185X.1931.tb01030.x. [DOI] [Google Scholar]
- 15.Charlesworth B., Crow J.F. Model for Evolution of Y Chromosomes and Dosage Compensation. Proc. Natl. Acad. Sci. USA. 1978;75:5618–5622. doi: 10.1073/pnas.75.11.5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charlesworth D., Charlesworth B., Marais G. Steps in the Evolution of Heteromorphic Sex Chromosomes. Heredity. 2005;95:118–128. doi: 10.1038/sj.hdy.6800697. [DOI] [PubMed] [Google Scholar]
- 17.Rice W.R. The Accumulation of Sexually Antagonistic Genes as a Selective Agent Promoting the Evolution of Reduced Recombination between Primitive Sex Chromosomes. Evolution. 1987;41:911–914. doi: 10.1111/j.1558-5646.1987.tb05864.x. [DOI] [PubMed] [Google Scholar]
- 18.Bachtrog D. A Dynamic View of Sex Chromosome Evolution. Curr. Opin. Genet. Dev. 2006;16:578–585. doi: 10.1016/j.gde.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 19.Furman B.L.S., Metzger D.C.H., Darolti I., Wright A.E., Sandkam B.A., Almeida P., Shu J.J., Mank J.E. Sex Chromosome Evolution: So Many Exceptions to the Rules. Genome Biol. Evol. 2020;12:750–763. doi: 10.1093/gbe/evaa081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stöck M., Horn A., Grossen C., Lindtke D., Sermier R., Betto-Colliard C., Dufresnes C., Bonjour E., Dumas Z., Luquet E., et al. Ever-young Sex Chromosomes in European Tree Frogs. PLoS Biol. 2011;9:e1001062. doi: 10.1371/journal.pbio.1001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guerrero R.F., Kirkpatrick M., Perrin N. Cryptic Recombination in the Ever-young Sex Chromosomes of Hylid Frogs. J. Evol. Biol. 2012;25:1947–1954. doi: 10.1111/j.1420-9101.2012.02591.x. [DOI] [PubMed] [Google Scholar]
- 22.Stöck M., Savary R., Betto-Colliard C., Biollay S., Jourdan-Pineau H., Perrin N. Low Rates of X-Y Recombination, not Turnovers, Account for Homomorphic Sex Chromosomes in Several Diploid Species of Palearctic Green Toads (Bufo viridis Subgroup) J. Evol. Biol. 2013;26:674–682. doi: 10.1111/jeb.12086. [DOI] [PubMed] [Google Scholar]
- 23.Yazdi H.P., Silva W.T.A.F., Suh A. Why do Some Sex Chromosomes Degenerate More Slowly Than Others? The Odd Case of Ratite Sex Chromosomes. Genes. 2020;11:1153. doi: 10.3390/genes11101153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yazdi H.P., Ellegren H. Old but Not (So) Degenerated-slow Evolution of Largely Homomorphic Sex Chromosomes in Ratites. Mol. Biol. Evol. 2014;31:1444–1453. doi: 10.1093/molbev/msu101. [DOI] [PubMed] [Google Scholar]
- 25.Furman B.L.S., Evans B.J. Divergent Evolutionary Trajectories of Two Young, Homomorphic, and Closely Related Sex Chromosome Systems. Genome Biol. Evol. 2018;10:742–755. doi: 10.1093/gbe/evy045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perrin N. Sex-chromosome Evolution in Frogs: What Role for Sex-antagonistic Genes? Phil. Trans. R. Soc. B. 2020 doi: 10.1098/rstb.2020.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma W.-J., Veltsos P., Toups M.A., Rodrigues N., Sermier R., Jeffries D.L., Perrin N. Tissue Specificity and Dynamics of Sex-biased Gene Expression in a Common Frog Population with Differentiated, yet Homomorphic, Sex Chromosomes. Genes. 2018;9:294. doi: 10.3390/genes9060294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma W.-J., Veltsos P., Sermier R., Parker D.J., Perrin N. Evolutionary and Developmental Dynamics of Sex-biased Gene Expression in Common Frogs with Proto-Y chromosomes. Genome Biol. 2018;19:156. doi: 10.1186/s13059-018-1548-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Veltsos P., Rodrigues N., Studer T., Ma W.-J., Sermier R., Leuenberger J., Perrin N. No Evidence That Y-chromosome Differentiation Affects Male Fitness in a Swiss Population of Common Frogs. J. Evol. Biol. 2020;33:401–409. doi: 10.1111/jeb.13573. [DOI] [PubMed] [Google Scholar]
- 30.Schmid M., Nanda I., Steinlein C., Kausch K., Haaf T., Epplen J.T. Sex-Determining Mechanisms and Sex Chromosomes in Amphibia. In: Green D.M., Sessions S.K., editors. Amphibian Cytogenetics and Evolution. Academic Press; San Diego, CA, USA: 1991. [Google Scholar]
- 31.Malcom J.W., Kudra R.S., Malone J.H. The Sex Chromosomes of Frogs: Variability and Tolerance Offer Clues to Genome Evolution and Function. J. Genom. 2014;2 doi: 10.7150/jgen.8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura M. Sex Determination in Amphibians. Semin. Cell Dev. Biol. 2009;20:271–282. doi: 10.1016/j.semcdb.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 33.Ggert C.E. Sex Determination: The Amphibian Models. Reprod. Nutr. Dev. 2004;4:539–549. doi: 10.1051/rnd. [DOI] [PubMed] [Google Scholar]
- 34.Gatto K.P., Busin C.S., Lourenço L.B. Unraveling the Sex Chromosome Heteromorphism of the Paradoxical Frog Pseudis tocantins. PLoS ONE. 2016;11:e0156176. doi: 10.1371/journal.pone.0156176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmid M., Steinlein C. Chromosome Banding in Amphibia. XXXVI. Multimorphic Sex Chromosomes and an Enigmatic Sex Determination in Eleutherodactylus johnstonei (Anura, Eleutherodactylidae) Cytogenet. Genome Res. 2018;154:86–98. doi: 10.1159/000487660. [DOI] [PubMed] [Google Scholar]
- 36.Schmid M., Feichtinger W., Steinlein C., Rupprecht A., Haaf T., Kaiser H. Chromosome Banding in Amphibia: XXIII. Giant W Sex Chromosomes and Extremely Small Genomes in Eleutherodactylus euphronides and Eleutherodactylus shrevei (Anura, Leptodactylidae) Cytogenet. Genome Res. 2002;97:81–94. doi: 10.1159/000064055. [DOI] [PubMed] [Google Scholar]
- 37.Schartl M., Schmid M., Nanda I. Dynamics of Vertebrate Sex Chromosome Evolution: From Equal Size to Giants and Dwarfs. Chromosoma. 2016;125:553–571. doi: 10.1007/s00412-015-0569-y. [DOI] [PubMed] [Google Scholar]
- 38.Ma W.-J., Rodrigues N., Sermier R., Brelsford A., Perrin N. Dmrt1 Polymorphism Covaries with Sex-determination Patterns in Rana temporaria. Ecol. Evol. 2016;6:5017–5117. doi: 10.1002/ece3.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodrigues N., Studer T., Dufresnes C., Ma W., Veltsos P., Perrin N. Dmrt1 Polymorphism and Sex-chromosome Differentiation in Rana temporaria. Mol. Ecol. 2017;26:4897–4905. doi: 10.1111/mec.14222. [DOI] [PubMed] [Google Scholar]
- 40.Gazoni T., Haddad C.F.B., Narimatsu H., Cabral-de-Mello D.C., Lyra M.L., Parise-Maltempi P.P. More Sex Chromosomes than Autosomes in the Amazonian Frog Leptodactylus pentadactylus. Chromosoma. 2018;127:269–278. doi: 10.1007/s00412-018-0663-z. [DOI] [PubMed] [Google Scholar]
- 41.Green D.M. Structure and Evolution of B Chromosomes in Amphibians. Cytogenet. Genome Res. 2004;106:235–242. doi: 10.1159/000079293. [DOI] [PubMed] [Google Scholar]
- 42.Green D.M., Zeyl C.W., Sharbel T.F. The Evolution of Hypervariable Sex and Supernumerary (B) Chromosomes in the Relict New Zealand Frog, Leiopelma hochstetteri. J. Evol. Biol. 1993;6:417–441. doi: 10.1046/j.1420-9101.1993.6030417.x. [DOI] [Google Scholar]
- 43.Jeffries D.L., Lavanchy G., Sermier R., Sredl M.J., Miura I., Borzée A., Barrow L.N., Canestrelli D., Crochet P.A., Dufresnes C., et al. A Rapid Rate of Sex-chromosome Turnover and Non-random Transitions in True Frogs. Nat. Commun. 2018;9:4088. doi: 10.1038/s41467-018-06517-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gamble T., Coryell J., Ezaz T., Lynch J., Scantlebury D.P., Zarkower D. Restriction site-associated DNA Sequencing (RAD-seq) Reveals an Extraordinary Number of Transitions among Gecko Sex-determining Systems. Mol. Biol. Evol. 2015;32:1296–1309. doi: 10.1093/molbev/msv023. [DOI] [PubMed] [Google Scholar]
- 45.Ross J.A., Urton J.R., Boland J., Shapiro M.D., Peichel C.L. Turnover of Sex Chromosomes in the Stickleback Fishes (Gasterosteidae) PLoS Genet. 2009;5:e1000391. doi: 10.1371/journal.pgen.1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Phillips R.B. Evolution of the Sex Chromosomes in Salmonid Fishes. Cytogenet. Genome Res. 2013;141:177–185. doi: 10.1159/000355149. [DOI] [PubMed] [Google Scholar]
- 47.Kikuchi K., Hamaguchi S. Novel Sex-determining Genes in Fish and Sex Chromosome Evolution. Dev. Dyn. 2013;242:339–353. doi: 10.1002/dvdy.23927. [DOI] [PubMed] [Google Scholar]
- 48.Revell M.L.J. Phytools: An R Package for Phylogenetic Comparative Biology (and Other Things) Methods Ecol. Evol. 2012;3:217–223. doi: 10.1111/j.2041-210X.2011.00169.x. [DOI] [Google Scholar]
- 49.Paradis E., Schliep K. Ape 5.0: An Environment for Modern Phylogenetics and Evolutionary Analyses in R. Bioinformatics. 2019;35:526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- 50.Brelsford A., Rodrigues N., Perrin N. High-density Linkage Maps Fail to Detect Any Genetic Component to Sex Determination in a Rana temporaria Family. J. Evol. Biol. 2016;29:220–225. doi: 10.1111/jeb.12747. [DOI] [PubMed] [Google Scholar]
- 51.Miura I. Sex Determination and Sex Chromosomes in Amphibia. Sex. Dev. 2017;11:298–306. doi: 10.1159/000485270. [DOI] [PubMed] [Google Scholar]
- 52.Yoshimoto S., Okada E., Umemoto H., Tamura K., Uno Y., Nishida-umehara C., Matsuda Y., Takamatsu N., Shiba T., Ito M. A W-linked DM-domain Gene, DM-W, Participates in Primary Ovary Development in Xenopus laevis. Proc. Natl. Acad. Sci. USA. 2008;105:2469–2474. doi: 10.1073/pnas.0712244105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshimoto S., Ikeda N., Izutsu Y., Shiba T., Takamatsu N., Ito M. Opposite Roles of DMRT1 and Its W-linked Paralogue, DM-W, in Sexual Dimorphism of Xenopus laevis: Implications of a ZZ/ZW-type Sex-determining System. Development. 2010;137:2519–2526. doi: 10.1242/dev.048751. [DOI] [PubMed] [Google Scholar]
- 54.Mawaribuchi S., Yoshimoto S., Ohashi S., Takamatsu N., Ito M. Molecular Evolution of Vertebrate Sex-determining Genes. Chromosom. Res. 2012;20:139–151. doi: 10.1007/s10577-011-9265-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cauret C.M.S., Gansauge M.T., Tupper A.S., Furman B.L.S., Knytl M., Song X.Y., Greenbaum E., Meyer M., Evans B.J. Developmental Systems Drift and the Drivers of Sex Chromosome Evolution. Mol. Biol. Evol. 2020;37:799–810. doi: 10.1093/molbev/msz268. [DOI] [PubMed] [Google Scholar]
- 56.Sun Y., Xiong Z., Xiang X., Liu S., Zhou W., Tu X., Zhong L. Whole-genome Sequence of the Tibetan Frog Nanorana parkeri and the Comparative Evolution of Tetrapod Genomes. Proc. Natl. Acad. Sci. USA. 2015;112:E1257–E1262. doi: 10.1073/pnas.1501764112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brelsford A., Stöck M., Betto-Colliard C., Dubey S., Dufresnes C., Jourdan-Pineau H., Rodrigues N., Savary R., Sermier R., Perrin N. Homologous Sex Chromosomes in Three Deeply Divergent Anuran Species. Evolution. 2013;67:2434–2440. doi: 10.1111/evo.12151. [DOI] [PubMed] [Google Scholar]
- 58.Palomar G., Ahmad F., Vasemägi A., Matsuba C., Nicieza A.G., Cano J.M. Comparative High-density Linkage Mapping Reveals Conserved Genome Structure but Variation in Levels of Heterochiasmy and Location of Recombination Cold Spots in the Common Frog. G3 Genes Genomes Genet. 2017;7:637–645. doi: 10.1534/g3.116.036459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dufresnes C., Borzee A., Horn A., Stock M., Ostini M., Sermier R., Wassef J., Litvinchuck S.N., Kosch T.A., Waldman B., et al. Sex-chromosome Homomorphy in Palearctic Tree Frogs Results from Both Turnovers and X-Y Recombination. Mol. Biol. Evol. 2015;32:2328–2337. doi: 10.1093/molbev/msv113. [DOI] [PubMed] [Google Scholar]
- 60.Stöck M., Croll D., Dumas Z., Biollay S., Wang J., Perrin N. A Cryptic Heterogametic Transition Revealed by Sex-linked DNA Markers in Palearctic Green Toads. J. Evol. Biol. 2011;24:1064–1070. doi: 10.1111/j.1420-9101.2011.02239.x. [DOI] [PubMed] [Google Scholar]
- 61.Miura I. An Evolutionary Witness: The Frog Rana rugosa Underwent Change of Heterogametic Sex from XY Male to ZW Female. Sex. Dev. 2008;1:323–331. doi: 10.1159/000111764. [DOI] [PubMed] [Google Scholar]
- 62.Furman B.L.S., Cauret C.M.S., Knytl M., Song X.Y., Premachandra T., Ofori-Boateng C., Jordan D.C., Horb M.E., Evans B.J. A Frog with Three Sex Chromosomes That Co-mingle Together in Nature: Xenopus tropicalis Has a Degenerate W and a Y That Evolved from a Z Chromosome. PLoS Genet. 2020;16:e1009121. doi: 10.1371/journal.pgen.1009121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roco Á.S., Olmstead A.W., Degitz S.J., Amano T., Zimmerman L.B., Bullejos M. Coexistence of Y, W, and Z Sex Chromosomes in Xenopus tropicalis. Proc. Natl. Acad. Sci. USA. 2015;112:E4752–E4761. doi: 10.1073/pnas.1505291112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bewick A.J., Anderson D.W., Evans B.J. Evolution of the Closely Related, Sex-related Genes Dm-w and Dmrt1 in African Clawed Frogs (Xenopus) Evolution. 2011;65:698–712. doi: 10.1111/j.1558-5646.2010.01163.x. [DOI] [PubMed] [Google Scholar]
- 65.Uno Y., Nishida C., Takagi C., Igawa T., Ueno N., Sumida M., Matsuda Y. Extraordinary Diversity in the Origins of Sex Chromosomes in Anurans Inferred from Comparative Gene Mapping. Cytogenet. Genome Res. 2015;145:218–229. doi: 10.1159/000431211. [DOI] [PubMed] [Google Scholar]
- 66.Rodrigues N., Vuille Y., Brelsford A., Merilä J., Perrin N. The Genetic Contribution to Sex Determination and Number of Sex Chromosomes Vary among Populations of Common Frogs (Rana temporaria) Heredity. 2016;117:25–32. doi: 10.1038/hdy.2016.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Toups M.A., Rodrigues N., Perrin N., Kirkpatrick M. A Reciprocal Translocation Radically Reshapes Sex-linked Inheritance in the Common Frog. Mol. Ecol. 2019;28:1877–1889. doi: 10.1111/mec.14990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Witschi E. Studies on Sex Differentiation and Sex Determination in Amphibians. IV. The Geographical Distribution of the Sex Races of the European Grass Frog (Rana temporaria, L.). A Contribution to the Problem of the Evolution of Sex. J. Exp. Zool. 1930;56:149–165. doi: 10.1002/jez.1400560202. [DOI] [Google Scholar]
- 69.Teacher A.G.F., Garner T.W.J., Nichols R.A. European Phylogeography of the Common Frog (Rana temporaria): Routes of Postglacial Colonization into the British Isles, and Evidence for an Irish Glacial Refugium. Heredity. 2009;102:490–496. doi: 10.1038/hdy.2008.133. [DOI] [PubMed] [Google Scholar]
- 70.Veith M., Baumgart A., Dubois A., Ohler A., Galán P., Vieites D.R., Nieto-Román S., Vences M. Discordant Patterns of Nuclear and Mitochondrial Introgression in Iberian Populations of the European Common Frog (Rana temporaria) J. Hered. 2012;103:240–249. doi: 10.1093/jhered/esr136. [DOI] [PubMed] [Google Scholar]
- 71.Spasić-Bošković O., Tanić N., Blagojević J., Vujošević M. Comparative Cytogenetic Analysis of European Brown Frogs: Rana temporaria, R. dalmatina and R. graeca. Caryologia. 1997;50:139–149. doi: 10.1080/00087114.1997.10797393. [DOI] [Google Scholar]
- 72.Rodrigues N., Betto-Colliard C., Jourdan-Pineau H., Perrin N. Within-population Polymorphism of Sex-determination Systems in the Common Frog (Rana temporaria) J. Evol. Biol. 2013;26:1569–1577. doi: 10.1111/jeb.12163. [DOI] [PubMed] [Google Scholar]
- 73.Rodrigues N., Vuille Y., Loman J., Perrin N., Konsult R., Rodrigues N. Sex-chromosome Differentiation and ‘Sex Races’ in the Common Frog (Rana temporaria) Proc. R. Soc. B Biol. Sci. 2015;282:20142726. doi: 10.1098/rspb.2014.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Phillips B.C., Rodrigues N., van Rensburg A.J., Perrin N. Phylogeography, More Than Elevation, Accounts for Sex-chromosome Differentiation in Swiss Populations of the Common Frog. Evolution. 2020:644–654. doi: 10.1111/evo.13860. [DOI] [PubMed] [Google Scholar]
- 75.Hartmann F.E., Ma W.-J. Digest: Climate Plays Marginal Role for Homomorphic Sex Chromosome Differentiation in Common Frogs†. Evolution. 2020;74:690–693. doi: 10.1111/evo.13936. [DOI] [PubMed] [Google Scholar]
- 76.Nagai Y., Doi T., Ito K., Yuasa Y., Fujitani T., Naito J.I., Ogata M., Miura I. The Distributions and Boundary of Two Distinct, Local Forms of Japanese Pond Frog, Pelophylax porosus Brevipodus, Inferred from Sequences of Mitochondrial DNA. Front. Genet. 2018;9:5–10. doi: 10.3389/fgene.2018.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wright D.A., Richards C.M. Two Sex-linked Loci in the Leopard Frog, Rana pipiens. Genetics. 1983;103:249–261. doi: 10.1093/genetics/103.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sumida M., Nishioka M. Geographic Variability of Sex-linked Loci in the Japanese Brown Frog, Rana japonica. Sci. Rep. Lab. Amphib. Biol. Hiroshima Univ. 1994;13:173–195. [Google Scholar]
- 79.Perrin N. Sex Reversal: A Fountain of Youth for Sex Chromosomes? Evolution. 2009;63:3043–3049. doi: 10.1111/j.1558-5646.2009.00837.x. [DOI] [PubMed] [Google Scholar]
- 80.Brelsford A., Dufresnes C., Perrin N. Trans-species Variation in Dmrt1 is Associated with Sex Determination in Four European Tree-frog Species. Evolution. 2016;70:840–847. doi: 10.1111/evo.12891. [DOI] [PubMed] [Google Scholar]
- 81.Wallace H., Wallace B.M.N., Badawy G.M.I. Lampbrush Chromosomes and Chiasmata of Sex-reversed Crested Newts. Chromosoma. 1997;106:526–533. doi: 10.1007/s004120050275. [DOI] [PubMed] [Google Scholar]
- 82.Kondo M., Nagao E., Mitani H., Shima A. Differences in Recombination Frequencies during Female and Male Meioses of the Sex Chromosomes of the Medaka, Oryzias latipes. Genet. Res. 2001;78:23–30. doi: 10.1017/S0016672301005109. [DOI] [PubMed] [Google Scholar]
- 83.Inoue H., Fukumori Y., Hiboyoshi T. Mapping of Autosomal Male-determining Factors of the Housefly, Musca domestica L., by Means of Sex-reversal. Jpn. J. Genet. 1983;58:451–461. doi: 10.1266/jjg.58.451. [DOI] [Google Scholar]
- 84.Rodrigues N., Studer T., Dufresnes C., Perrin N. Sex-chromosome Recombination in Common Frogs Brings Water to the Fountain-of-youth. Mol. Biol. Evol. 2018;35:942–948. doi: 10.1093/molbev/msy008. [DOI] [PubMed] [Google Scholar]
- 85.Schartl M. Sex Chromosome Evolution in Non-mammalian Vertebrates. Curr. Opin. Genet. Dev. 2004;14:634–641. doi: 10.1016/j.gde.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 86.Volff J.N., Nanda I., Schmid M., Schartl M. Governing Sex Determination in Fish: Regulatory Putsches and Ephemeral Dictators. Sex. Dev. 2007;1:85–99. doi: 10.1159/000100030. [DOI] [PubMed] [Google Scholar]
- 87.Van Doorn G.S., Kirkpatrick M. Turnover of Sex Chromosomes Induced by Sexual Conflict. Nature. 2007;449:909–912. doi: 10.1038/nature06178. [DOI] [PubMed] [Google Scholar]
- 88.Van Doorn G.S., Kirkpatrick M. Transitions between Male and Female Heterogamety Caused by Sex-antagonistic Selection. Genetics. 2010;186:629–645. doi: 10.1534/genetics.110.118596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gammerdinger W.J., Conte M.A., Sandkam B.A., Ziegelbecker A., Koblmüller S., Kocher T.D. Novel Sex Chromosomes in 3 Cichlid Fishes from Lake Tanganyika. J. Hered. 2018;109:489–500. doi: 10.1093/jhered/esy003. [DOI] [PubMed] [Google Scholar]
- 90.Blaser O., Grossen C., Neuenschwander S., Perrin N. Sex-chromosome Turnovers Induced by Deleterious Mutation Load. Evolution. 2013;67:635–645. doi: 10.1111/j.1558-5646.2012.01810.x. [DOI] [PubMed] [Google Scholar]
- 91.Blaser O., Neuenschwander S., Perrin N. Sex-chromosome Turnovers: The Hot-Potato Model. Am. Nat. 2014;183:140–146. doi: 10.1086/674026. [DOI] [PubMed] [Google Scholar]
- 92.Gunski R.J., Kretschmer R., Santos De Souza M., De Oliveira Furo I., Barcellos S.A., Costa A.L., Cioffi M.B., De Oliveira E.H.C., Del Valle Garnero A. Evolution of Bird Sex Chromosomes Narrated by Repetitive Sequences: Unusual W Chromosome Enlargement in Gallinula melanops (Aves: Gruiformes: Rallidae) Cytogenet. Genome Res. 2019 doi: 10.1159/000501381. [DOI] [PubMed] [Google Scholar]
- 93.Chalopin D., Volff J., Galiana D., Anderson J.L., Schartl M. Transposable Elements and Early Evolution of Sex Chromosomes in Fish. Chromosom. Res. 2015;23:545–560. doi: 10.1007/s10577-015-9490-8. [DOI] [PubMed] [Google Scholar]
- 94.Xu L., Wa Sin S.Y., Grayson P., Edwards S.V., Sackton T.B., Mank J. Evolutionary Dynamics of Sex Chromosomes of Paleognathous Birds. Genome Biol. Evol. 2019;11:2376–2390. doi: 10.1093/gbe/evz154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang Z., Zhang J., Xu X., Witt C., Deng Y., Chen G., Meng G., Feng S., Szekely T., Zhang G., et al. Phylogeny, Transposable Element and Sex Chromosome Evolution of the Basal Lineage of Birds. bioRxiv. 2019 doi: 10.1101/750109. [DOI] [Google Scholar]
- 96.Wang Z.-J., Chen G.-J., Zhang G.-J., Zhou Q. Dynamic Evolution of Transposable Elements, Demographic History, and Gene Content of Paleognathous Birds. Zool. Res. 2021;42:51–61. doi: 10.24272/j.issn.2095-8137.2020.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Townsend D.S., Stewart M.M. Direct Development in Eleutherodactylus coqui (Anura: Leptodactylidae): A Staging Table. Copeia. 1985;2:423–436. doi: 10.2307/1444854. [DOI] [Google Scholar]
- 98.García-R J.C., Crawford A.J., Mendoza Á.M., Ospina O., Cardenas H., Castro F. Comparative Phylogeography of Direct-developing Frogs (Anura: Craugastoridae: Pristimantis) in the Southern Andes of Colombia. PLoS ONE. 2012;7:e0046077. doi: 10.1371/journal.pone.0046077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Callery E.M., Fang H., Elinson R.P. Frogs without Polliwogs: Evolution of Anuran Direct Development. BioEssays. 2001;23:233–241. doi: 10.1002/1521-1878(200103)23:3<233::AID-BIES1033>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 100.Elinson R.P., del Pino E.M. Developmental Diversity of Amphibians. Wiley Interdiscip. Rev. Dev. Biol. 2012;1:345–369. doi: 10.1002/wdev.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ohta S. Sex Determination Mechanism in Buergeria buergeri (Schlegel) I. Heterozygosity of Chromosome Pair No.7 in the Female. Sci. Rep. Lab. Amphib. Biol. Hiroshima Univ. 1986;8:29–43. [Google Scholar]
- 102.Pennell M.W., Kirkpatrick M., Otto S.P., Vamosi J.C. Y Fuse? Sex Chromosome Fusions in Fishes and Reptiles. PLoS Genet. 2015;11:e1005237. doi: 10.1371/journal.pgen.1005237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Burt A., Bell G., Harvey P.H. Sex Differences in Recombination. J. Evol. Biol. 1991;4:259–277. doi: 10.1046/j.1420-9101.1991.4020259.x. [DOI] [Google Scholar]
- 104.Lenormand T., Dutheil J. Recombination Difference between Sexes: A Role for Haploid Selection. PLoS Biol. 2005;3:e63. doi: 10.1371/journal.pbio.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lenormand T., Engelstädter J., Johnston S.E., Wijnker E., Haag C.R. Evolutionary Mysteries in Meiosis. Philos. Trans. R. Soc. B Biol. Sci. 2016;371 doi: 10.1098/rstb.2016.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sardell J.M., Kirkpatrick M. Sex Differences in the Recombination Landscape. Am. Nat. 2020;195:361–379. doi: 10.1086/704943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Haldane J.B.S. Sex Ratio and Unisexual Sterility in Hybrid Animals. J. Genet. 1922;12:101–109. doi: 10.1007/BF02983075. [DOI] [Google Scholar]
- 108.Huxley J.S. Sexual Difference of Linkage in Gammarus chevreuyxi. J. Genet. 1928;20:145–156. doi: 10.1007/BF02983136. [DOI] [Google Scholar]
- 109.Brelsford A., Dufresnes C., Perrin N. High-density Sex-specific Linkage Maps of a European Tree Frog (Hyla arborea) Identify the Sex Chromosome without Information on Offspring Sex. Heredity. 2016;116:177–181. doi: 10.1038/hdy.2015.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bergero R., Gardner J., Bader B., Yong L., Charlesworth D. Exaggerated Heterochiasmy in a Fish with Sex-linked Male Coloration Polymorphisms. Proc. Natl. Acad. Sci. USA. 2019;116:6924–6931. doi: 10.1073/pnas.1818486116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dufresnes C., Brelsford A., Baier F., Perrin N. When Sex Chromosomes Recombine only in the Heterogametic Sex: Heterochiasmy and Heterogamety in Hyla Tree Frogs. Mol. Biol. Evol. 2021;38:192–200. doi: 10.1093/molbev/msaa201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets in this review have been provided in Supplementary Table S1.