Abstract
Background
Anaerobic digestion (AD) of protein-rich grass silage was performed in experimental two-stage two-phase biogas reactor systems at low vs. increased organic loading rates (OLRs) under mesophilic (37 °C) and thermophilic (55 °C) temperatures. To follow the adaptive response of the biomass-attached cellulolytic/hydrolytic biofilms at increasing ammonium/ammonia contents, genome-centered metagenomics and transcriptional profiling based on metagenome assembled genomes (MAGs) were conducted.
Results
In total, 78 bacterial and archaeal MAGs representing the most abundant members of the communities, and featuring defined quality criteria were selected and characterized in detail. Determination of MAG abundances under the tested conditions by mapping of the obtained metagenome sequence reads to the MAGs revealed that MAG abundance profiles were mainly shaped by the temperature but also by the OLR. However, the OLR effect was more pronounced for the mesophilic systems as compared to the thermophilic ones. In contrast, metatranscriptome mapping to MAGs subsequently normalized to MAG abundances showed that under thermophilic conditions, MAGs respond to increased OLRs by shifting their transcriptional activities mainly without adjusting their proliferation rates. This is a clear difference compared to the behavior of the microbiome under mesophilic conditions. Here, the response to increased OLRs involved adjusting of proliferation rates and corresponding transcriptional activities. The analysis led to the identification of MAGs positively responding to increased OLRs. The most outstanding MAGs in this regard, obviously well adapted to higher OLRs and/or associated conditions, were assigned to the order Clostridiales (Acetivibrio sp.) for the mesophilic biofilm and the orders Bacteroidales (Prevotella sp. and an unknown species), Lachnospirales (Herbinix sp. and Kineothrix sp.) and Clostridiales (Clostridium sp.) for the thermophilic biofilm. Genome-based metabolic reconstruction and transcriptional profiling revealed that positively responding MAGs mainly are involved in hydrolysis of grass silage, acidogenesis and / or acetogenesis.
Conclusions
An integrated -omics approach enabled the identification of new AD biofilm keystone species featuring outstanding performance under stress conditions such as increased OLRs. Genome-based knowledge on the metabolic potential and transcriptional activity of responsive microbiome members will contribute to the development of improved microbiological AD management strategies for biomethanation of renewable biomass.
Keywords: Metagenome assembled genomes, Integrated -omics, Polyomics, Anaerobic digestion, Biogas, Bioconversion, Microbial community structure, Methane, Metabolic activity
Background
Important part of bioeconomical strategies for sustainable and carbon dioxide (CO2) - neutral energy production is the anaerobic digestion (AD) and biomethanation of renewable raw materials. Beside manure and slurries from agricultural husbandry, also agriculturally produced biomass such as maize silage (‘energy crops’) or material from landscape management were utilized [1, 2].
Crop biomass is rich in long-chained carbohydrates such as cellulose, hemicellulose, and xylan and additionally contains considerable amounts of proteins depending on the particular crop. To digest crop biomass as sole substrate and at high organic loading rates (OLRs), specially designed biogas reactors are advantageous such as staged degradation step (‘phase’) separated reactor systems consisting of a cellulolysis / hydrolysis fermenter and a downstream methanogenesis reactor. Compared to single phase reactors, these two-stage two-phase reactors possess several advantages, for example, the more stable operation (under particular process conditions) combined with higher bioenergy yields [2–6].
The degradation of high-molecular compounds requires the direct access of microbial specialists. Accordingly, crop biomass is colonized by a cellulolytic biofilm which composition varies with the abiotic environmental conditions, as example, the process temperature, but also relies on the physico-chemical characteristics of crop material and process liquids [7]. Once the biofilm is established, it functions as a cooperative consortium leading to enhanced biomass degradation and, in consequence, to biogas production [8, 9].
Comprehensive microbiome analyses by means of metagenome sequencing revealed that many biogas microbiome members could not be classified down to the species level and hence are currently unknown (‘microbial dark matter’) [10, 11]. As example published previously, in the metagenome dataset determined for a biogas plant operated under thermophilic temperature regime, only 18% of the included 16S rRNA gene sequences were assignable to a taxonomically established genus [10]. In addition, information on the metabolic activity of biogas biofilm microbial communities is only rudimentarily available. Most of the corresponding studies focus on the methanogenic sub-community, while the metabolic potential of the entire biogas biofilm microbiome remains poorly characterized [12, 13]. For this propose, the integration of different -omics approaches (integrated or poly-omics), as example, the combination of metagenome with metatranscriptome datasets, is indispensable to distinguish between metabolically active and less active microbial species. Genome-centered biogas microbiome analyses already disclosed and prospectively will disclose further functionalities and interactions of keystone microbiome members [14–21].
The degradation of plant biomass by hydrolytic enzymes of surface attached cellulolytic and hydrolytic bacteria and the subsequent secondary fermentation still represent bottlenecks in the engineered optimization of biogas processes. For economically optimal AD process operation, cellulolytic / hydrolytic biofilms adapted to maximal OLRs and, with special respect to the fermentation of substrates with high contents of nitrogen-containing compounds such as proteins and peptides (e.g., grass silage), microbiomes featuring tolerance to high ammonium / ammonia contents are essentially required. It was hypothesized that such biofilm members, in particular functional keystone species, will become recognizable by their increasing abundances and transcriptional activities under demanding process conditions.
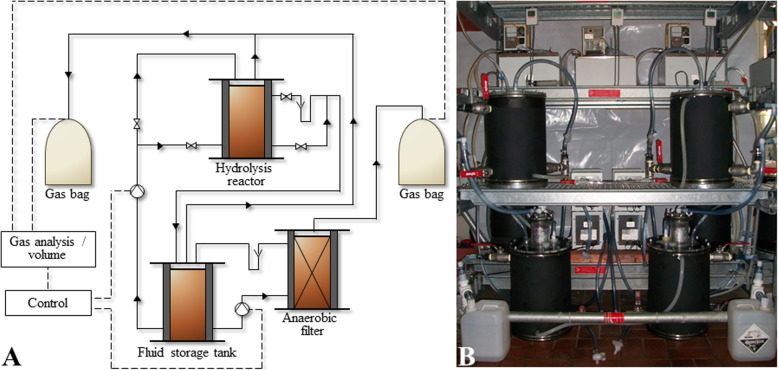
To unravel the structure, functionality, and metabolic activity of such cellulolytic / hydrolytic biofilm microbiomes, in this study, an integrated -omics approach was applied consisting of parallel microbial metagenome and metatranscriptome analyses. To obtain direct access to these microbiomes, biofilm samples grown on the surface of ryegrass silage digested in the hydrolysis reactors (HR) of two-stage two-phase biogas reactor systems (Fig. 1) operated under mesophilic (37 °C) and thermophilic (55 °C) temperature regime and at two (low and increased) OLRs were analyzed. Metabolic reconstruction for candidate metagenome-assembled genomes (MAGs) and corresponding genome-centered transcriptome analyses provided insights into life-styles and activities of adapted species.
Fig. 1.
Flow scheme (a) and picture (b) of the two-stage two-phase biogas reactor system
Results
Biogas reactor performance and long-term microbial dynamics
Four biogas reactor systems were operated in parallel over a period of longer than 750 days resulting in two (biological) replicates for mesophilic (M1, M2) and two replicates for thermophilic conditions (T1, T2) (Fig. 1). Sampling of substrate surface attached biofilms was conducted for two different OLRs, i.e., 500 g (low OLR) and 1500 g (increased OLR) of perennial ryegrass silage.
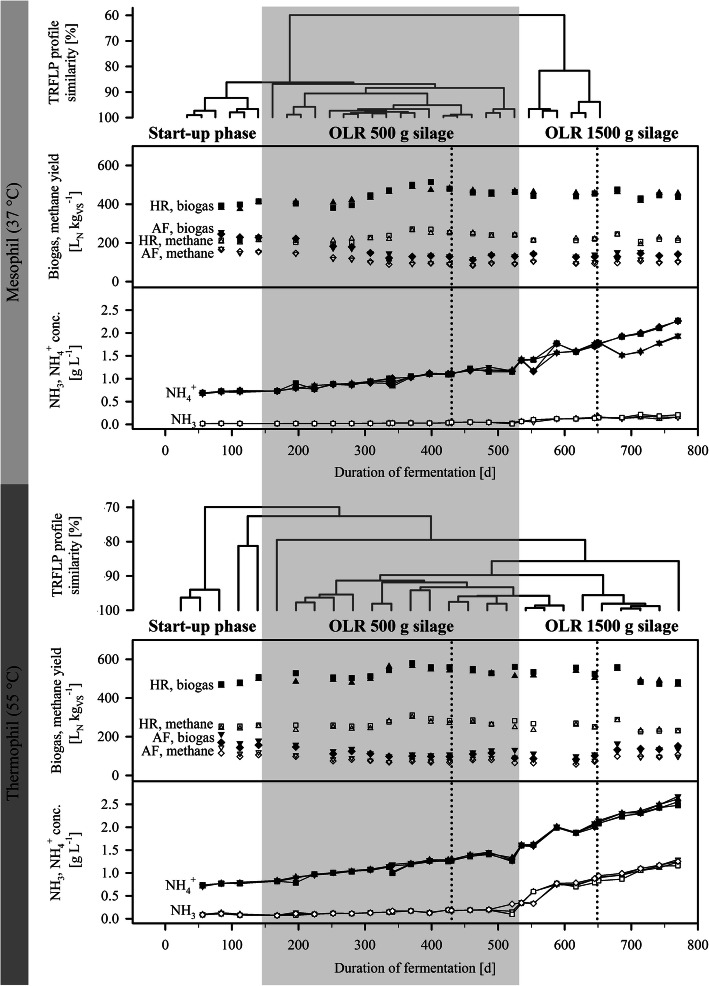
The summarized average biogas yields from hydrolytic reactors (HR) and anaerobic filters (AF) were constantly 601 ± 18 l normalized for standard temperature and pressure (LN) per kg volatile substances (VS) (M1) and 599 ± 26 LN kgVS− 1 (M2) for the reactor systems operated under mesophilic temperature regime with an average methane content of 56 ± 2% (v/v) (Fig. 2). Under the thermophilic temperature regime, the summarized systems’ biogas yields were slightly higher with average values of 645 ± 27 LN kgVS− 1 (T1) and 644 ± 19 LN kgVS− 1 (T2) with a slightly lower average methane content of 54 ± 2% (v/v) each. The average methane yields ranged from 337 ± 20 LN kgVS− 1 (M2) to 348 ± 19 LN kgVS− 1 (T2) which is less than 5% lower than the reference value of, on average, 353 LN kgVS− 1 as determined by standard batch fermentation tests according to the German technical standard VDI 4630. Even if, in general, the methane content in the biogas was higher in the AF than in the HR (mesophilic, 71 ± 3% vs. 51 ± 2%; thermophilic, 70 ± 2% vs. 50 ± 2%), most of the biogas and methane was produced in the HR (mesophilic, 74 ± 6% and 67 ± 6% on average; thermophilic, 79 ± 4% to 81 ± 4% and 73 ± 4% to 76 ± 4% depending on the respective process status).
Fig. 2.
Biogas and methane yields, and NH4+- and NH3-contents in the biogas reactor effluents. Bacterial 16S rRNA gene targeting TRFLP analyses results are shown exemplary for different time points at different process conditions. Dotted lines indicate time points of sampling for NGS. ▲,△, hydrolysis reactor (HR) of reactor system M1 (mesophil) resp. T1 (thermophil);▼, ▽, downstream AF of reactor system M1 (mesophil) resp. T1 (thermophil); ■, □, HR of reactor system M2 (mesophil) resp. T2 (thermophil); ◆, ◇, AF of reactor system M2 (mesophil) resp. T2 (thermophil); LN, liters normalized to 0 °C and 1013 hPa; VS, volatile substances; OLR, organic loading rate
The main product of bacterial fermentation in the HR was acetic acid. Highest concentrations were determined for fermentation day 2 with values ranging from 0.80 g L− 1 (start-up phase) to 5.30 g L− 1 (OLR 1500 g silage) and under the thermophilic conditions, from 0.91 g L− 1 (start-up phase) to 4.21 g L− 1 (OLR 1500 g silage). Propionic acid was produced only in minor amounts of up to 0.58 g L− 1 (mesophilic) and 0.43 g L− 1 (thermophilic). The volatile fatty acids (VFA) accumulation throughout the ongoing fermentation process was not observed.
Due to the nitrogen content of the silage of 7.6 g kgFM− 1, a NH4+ accumulation of up to 2.3 g L− 1 (mesophilic) and 2.7 g L− 1 (thermophilic) was observed in the process fluids during the fermentation and within the entire biogas reactor system (Fig. 2). In contrast to the fermentations at mesophilic temperatures, under thermophilic temperature regime also an accumulation of cytotoxic NH3 of up to 1.3 g L− 1 occurred.
As revealed by bacterial 16S rRNA gene targeting TRFLP analysis, the bacterial community structure had adapted during the ongoing biomethanation experiment (Fig. 2). It is assumed that adaptation of the community primarily occurred as response to the increase in OLR. Likewise, also the increase in NH4+ concentrations and, in particular during the thermophilic fermentations, the increase in NH3 concentrations may had affected the community composition.
Cellulolytic / hydrolytic biofilm community structure
To characterize the structure of the bacterial biofilms established on the surface of ryegrass silage digestate in the mesophilic and thermophilic HRs, next generation sequencing (NGS) of the 16S rRNA gene was performed. Due to the used primer sets, primarily 16S rRNA genes for the domain Bacteria were detected. 16S rRNA gene sequences for methanogenic Archaea were only detected for mesophilic biofilm samples (in maximum, with an abundance of 1.2% in case of the HR biofilm sample M1OLR1500) and assigned to genus Methanosaeta, i.e., Methanothrix.
Overall, 66% of the 16S rRNA gene sequences were classified into operational taxonomic units (OTUs) that could be assigned to known genera. 34% remained unassigned at genus rank (Additional file 1). In general, the thermophilic biofilms exhibited a slightly lower diversity than the mesophilic biofilms (Shannon indices in average 2.9 vs. 3.2).
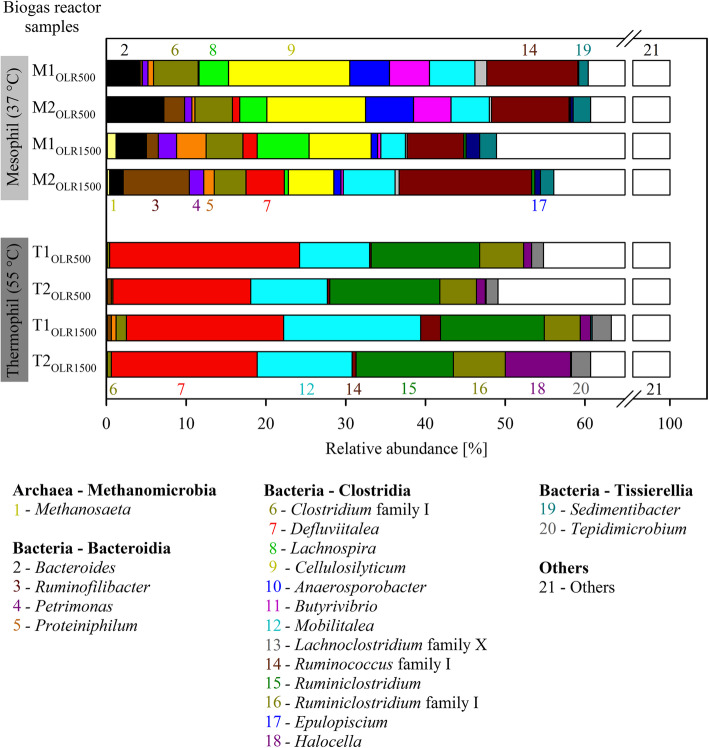
In the mesophilic biofilms, a number of genera were exclusively detected belonging to the classes Clostridia (i.e., Cellulosilyticum, Lachnospira, Anaerosporobacter, Butyrivibrio, and Epulopiscium), Bacteroidia (i.e., Bacteroides, and Petrimonas), Tissierellia (i.e., Sedimentibacter) (Fig. 3, Additional file 2). In general, the abundance of particular OTUs varied between the biological replicates and OLRs indicating varying microbiome structure.
Fig. 3.
Taxonomic profiling of the hydrolysis reactor (HR) biofilm microbiome. Relative abundances are shown for the most abundant genera of microbial sub-communities as determined by 16S rRNA gene amplicon NGS. M1, M2, mesophilic replicates; T1, T2, thermophilic replicates; OLR500, OLR1500, organic loading rates of 500 or 1500 g ryegrass silage. For further details on sample denomination, refer to Fig. 2
In the thermophilic biofilms, members of the genera Ruminiclostridium and Halocella (phylum Firmicutes, class Clostridia), and Tepidimicrobium (class Tissierellia) were found, which were not or only detected in minor amounts in the mesophilic biofilms. Also in these biofilms, the abundance of OTUs varied between biological replicates and OLRs. Some genera were present in all biofilms, such as Ruminococcus, Defluviitalea, and Mobilitalea (class Clostridia), but all showing substantial differences in abundance in mesophilic and thermophilic biofilms (Fig. 3, Additional file 2).
Under mesophilic temperature regime, the increase in OLR had a positive effect on the abundance of some members, e.g., of the genera Proteiniphilum (OLR 500 g: 0.7 resp. 0.4% vs. OLR 1500 g: 3.7 resp. 1.3%) and Defluviitalea (OLR 500 g: 0.1 resp. 0.9% vs. OLR 1500 g: 1.8 resp. 4.8%). Other biofilm members decreased in abundance after increase of OLR, such as members of genera Anaerosporobacter (OLR 500 g: 5.0 resp. 6.0% vs. OLR 1500 g: 0.8 resp. 0.9%) and Cellulosilyticum (OLR 500 g: 15.2 resp. 12.4% vs. OLR 1500 g: 7.8 resp. 5.7%) (Additional file 2).
At thermophilic temperatures, only the genus Mobilitalea benefitted from the increased OLR (OLR 500 g: 8.8 resp. 9.6% vs. OLR 1500 g: 17.2 resp. 11.9%) (Additional file 2). All other genera remained at more or less similar abundances. Thus, it can be assumed that Mobilitalea species are more tolerant towards increased NH3 concentrations compared to other species.
MAG abundance in the cellulolytic/hydrolytic biofilms depending on the temperature and the OLR
To determine and characterize differential abundances of species represented by metagenomically assembled genomes (MAGs) in the cellulolytic/hydrolytic biofilms established on the surface of ryegrass silage, microbial metagenome sequencing followed by a combined assembly of sequence data and genome binning were applied (Additional file 3). In total, 157 MAGs were compiled and taxonomically classified (Additional file 4). 74 MAGs were assigned to the domain Bacteria, five MAGs to Archaea, and 78 remained unclassified at domain level. At the phylum level, the MAGs were allocated to the Firmicutes (55), Bacteroidota (12), Euryarchaeota (5), Spirochaetota (2), Fibrobacterota (1), and Cloacimonadota (1). At the family level, only 53 MAGs were classifiable among others to the Lachnospiraceae (19), Bacteroidaceae (4), Ruminoclostridiaceae (3), Clostridiaceae (3), indicating occurrence of so far unknown microbial species and/or insufficient representation of adequate reference genomes in databases that are available for comparative analyses.
To uncover the relative abundances of the compiled MAGs in the HRs analyzed, metagenome sequences obtained from the mesophilic and thermophilic microbial communities were mapped onto the MAG sequences. Only 78 MAGs featuring contamination rates below 10% were considered for this approach. Deeper metagenome sequencing would certainly have improved the completeness and number of compiled MAGs. However, sequencing depth always is a trade-off between incurred costs and expected results.
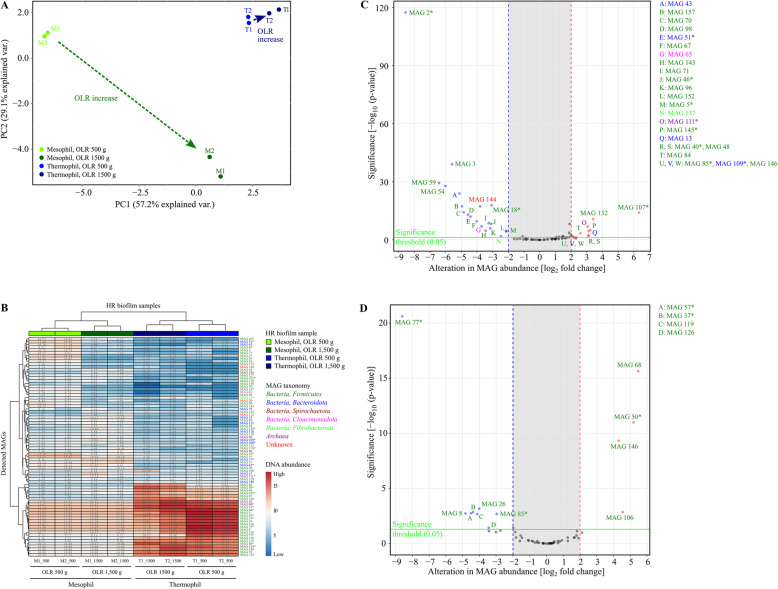
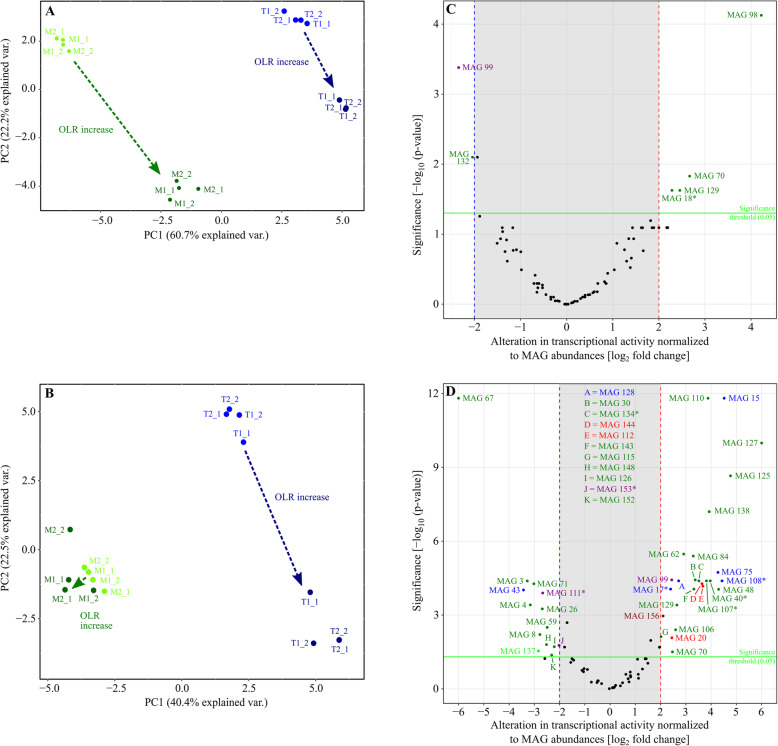
Principal component (PCA) analysis of MAG abundance profiles revealed close clustering of replicates confirming reproducibility of the treatments carried out (Fig. 4a). Moreover, temperatures (mesophilic vs. thermophilic) led to a clear separation of MAG abundance profiles in the PC plots. Likewise, the OLRs (OLR500 vs. OLR1500) differentiate MAG abundances. However, this effect is far more pronounced for the mesophilic systems as compared to the thermophilic ones (Fig. 4a).
Fig. 4.
Alteration of abundance of 157 metagenome-assembled genomes (MAGs) detected in HR biofilms at mesophilic (M) and thermophilic (T) process temperature in response to the increase of organic loading rate (OLR) from 500 to 1500 g ryegrass silage as deduced from metagenome data. PCA plots are shown for principle component analyses of scaled and centered rlog transformed metagenome read counts mapped on MAGs (a). The hierarchical clustering of rlog transformed abundance values for 78 (contamination below 10%) selected MAGs detected in HR biofilms is visualized in (b). Green, mesophilic HR biofilms; blue, thermophilic HR biofilms. For further details on sample denomination, refer to Figs. 2 and 3. Alteration in abundance values of 78 MAGs selected in response of the increase of OLR from 500 to 1500 g ryegrass silage at mesophilic (c) and thermophilic (d) process temperature. Colors of the MAGs indicate taxonomic affiliation (green, Firmicutes; blue, Bacteroidota; violet, Archaea; pink, Cloacimonadota; light green, Fibrobacterota; dark red, Spirochaetota; red, unknown affiliation). * indicate MAGs with completeness above 50% and contamination rate less than 10% as listed in Table 1 and in Additional file 4
MAG abundance profiles are visualized for all conditions tested (two temperatures and two OLRs in replicates) in heat-maps for the 78 MAGs tested (Fig. 4b). Cluster analysis revealed that the temperature is the most important factor that drives shaping of the community followed by the OLR. Replicates are very similar to each other regarding MAG abundance profiles under the conditions tested.
Relative abundances of the following MAGs significantly increased (log2 fold-change of > 2 and –log10(p-value) of > 0.05) under mesophilic conditions when the OLR was raised to 1500 g: MAG 13, 40, 48, 84, 85, 107, 109, 111, 132, 145 and 146 as shown by volcano plot analysis (Fig. 4c). Relative abundances of several more MAGs decreased under high OLRs. Under thermophilic conditions, the MAGs 50, 68, 106 and 146 increased in abundance upon OLR raise (Fig. 4d). Responsive MAGs featuring completeness value of more than 50% and contamination less than 10% were further analyzed regarding their genetic potential and transcriptional activity. These are MAGs 40, 50, 85, 107, 109, 111 and 145 (see below).
Functional potential of MAGs positively responding to increased OLR
To gain insights into the functional potential of MAGs positively responding to increased OLRs, genetic determinants for utilization of carbohydrates (Fig. 5) as well as the key enzymes of AD pathways were analyzed (Additional file 5). The genetic determinants were categorized according to the four stages of the AD process, namely hydrolysis, acidogenesis, acetogenesis and methanogenesis as described previously [23].
Fig. 5.
Genes encoding carbohydrate-active enzymes predicted for bacterial metagenome-assembled genomes (MAGs) most abundant in hydrolysis reactor (HR) biofilms. * These MAGs showed increased transcriptional activity in response to the organic loading rate and were therefore additionally analyzed
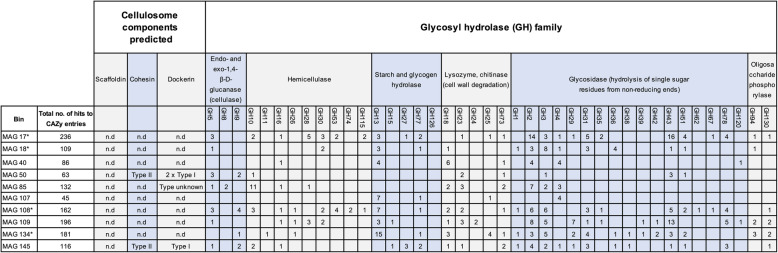
Regarding the functional context ‘hydrolysis’, the genetic potential for utilization of carbohydrates was characterized in the selected bacterial MAGs. Genes encoding carbohydrate-active enzymes were determined by applying the HMM-based Carbohydrate-active enzyme annotation database dbCAN v7 [24] (Fig. 5). Between 45 and 236 genes encoding cellulosomal proteins or enzymes with predicted activities on carbohydrates were identified in each of the bacterial MAGs analyzed.
Obtained results subdivided the analyzed MAGs into two groups. Group I members were predicted to encode cellulosome structures required for efficient degradation of cellulose, comprising dockerin-containing glycosyl hydrolases (GHs), corresponding cohesin-containing scaffoldins, and enzymes acting on large carbohydrate molecules. Some of the identified enzymes contain carbohydrate-binding motifs. MAG 50 (family Defluviitaleaceae), MAG 85 (phylum Firmicutes) and 145 (affiliated to the genus Jeotgalibaca) represent group I members. In general, for cellulolytic/hydrolytic biofilms, the MAGs belonging to group I are of great importance, since they represent bacterial candidates featuring the potential for efficient decomposition of complex carbohydrates such as cellulose, hemicellulose and xylan.
The remaining MAGs were classified to group II representing secondary fermentative bacteria mainly utilizing mono-, di- and oligosaccharides for energy production as supplied by group I bacteria. Group II comprises MAGs lacking genes for proteins involved in cellulosome formation and assembly. However, due to the incompleteness of these MAGs (Table 1), it cannot be excluded that cellulosome genes were missed during the assembly and binning processes.
Table 1.
Taxonomic affiliations and characteristics of selected metagenome-assembled genomes (MAGs) most abundant in HR biofilms
| MAG | Taxonomic affiliation1 | MAG features | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phylum | Class | Order | Family | Genus | Comleteness2 [%] | Contamination2 [%] | Size [bp] | GC content [%] | No. of genes | No. of rrn genes | No. of tRNA genes | |
| Bacteria | ||||||||||||
| 13 | Firmicutes | Unknown | Unknown | Unknown | Unknown | 29.74 | 3.43 | 1,239,774 | 44.81 | 1159 | 3 | 31 |
| 17* | Bacteroidetes | Bacteroidia | Bacteroidales | Bacteroidaceae | Prevotella sp. | 68.02 | 8.98 | 2,689,597 | 48.90 | 2294 | 2 | 39 |
| 18* | Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | Acetivibrio sp. | 68.50 | 3.65 | 2,855,089 | 39.03 | 2917 | n/d3 | 15 |
| 40* | Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | Herbinix sp. | 68.42 | 7.66 | 1,849,940 | 36.65 | 1755 | n/d | 8 |
| 48 | Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | Herbinix sp. | 26.48 | 2.63 | 770,644 | 37.4 | 614 | n/d | 3 |
| 50 | Firmicutes | Clostridia | Lachnospirales | Defluviitaleaceae | Unknown | 51.74 | 6.14 | 1,725,349 | 31.02 | 1775 | n/d | 31 |
| 68 | Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | Herbinix sp. | 52.17 | 4.77 | 2,358,488 | 46.61 | 2310 | 2 | 27 |
| 84 | Firmicutes | Clostridia | Lachnospirales | Unknown | Unknown | 35.22 | 5.14 | 1,970,476 | 38.81 | 1672 | 2 | 17 |
| 85 | Firmicutes | Unknown | Unknown | Unknown | Unknown | 80.11 | 3.08 | 2,011,311 | 47.98 | 2076 | n/d | 33 |
| 106 | Firmicutes | Clostridia | Clostridiales | Clostridiaceae | Clostridium sp. | 33.09 | 4.39 | 883,787 | 31.79 | 812 | n/d | 17 |
| 107* | Firmicutes | Clostridia | Clostridiales | Clostridiaceae | Clostridium sp. | 50.06 | 6.97 | 1,539,556 | 29.37 | 1648 | n/d | 17 |
| 108* | Bacteroidetes | Bacteroidia | Bacteroidales | Unknown | Unknown | 79.01 | 8.21 | 2,032,215 | 47.60 | 2025 | 1 | 29 |
| 109 | Bacteroidetes | Bacteroidia | Bacteroidales | Dysgonomonadaceae | Proteiniphilum sp. | 63.52 | 7.19 | 2196,928 | 45.07 | 2196 | 1 | 27 |
| 132 | Firmicutes | Clostridia | Unknown | Unknown | Unknown | 26.26 | 2.37 | 1,284,977 | 37.03 | 1286 | n/d | 16 |
| 134* | Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | Kineothrix sp. | 78.46 | 9.27 | 2,788,291 | 40.86 | 2946 | n/d | 31 |
| 145 | Firmicutes | Bacilli | Lactobacillales | Aerococcaceae | Jeotgalibaca sp. | 63.37 | 9.48 | 2,346,037 | 46.04 | 2647 | n/d | 14 |
| 146 | Firmicutes | Bacilli | Unknown | Unknown | Unknown | 33.22 | 0.00 | 861,844 | 38.49 | 705 | 1 | 13 |
| Archaea | ||||||||||||
| 111 | Euryarchaeota | Methanobacteria | Methanobacteriales | Methanobacteriaceae | Methanobacterium sp. | 73.74 | 8.57 | 2,134,563 | 37.98 | 2406 | 2 | 51 |
Furthermore, genetic determinants encoding key enzymes required for utilization of different organic molecules such as pyruvate, lactate, ethanol, acetate, propionate and butyrate representing important metabolites of the acidogenesis and acetogenesis were analyzed in the MAGs selected (Additional file 5). As examples, the bacterial MAGs 109 (Proteiniphilum sp.) and 145 (Jeotgalibaca sp.) encode high numbers of key genes featuring predicted functions in pyruvate metabolism (between 14 and 21), also representing the KEGG (map 00620) modules for utilization of lactate (between 5 and 7) and acetate (8) only in case of the MAG 145. In MAG 145, essential genes encoding enzymes of the Wood-Ljungdahl pathway (8) were identified, which plays an important role in carbon fixation and acetate utilization.
During acetogenesis, several bacterial species utilize propionic acid employing the methylmalonyl-CoA or the acrylyl-CoA pathway of the propanoate metabolism. The bacterial MAGs 109 (Proteiniphilum sp.) and 145 (Jeotgalibaca sp.) possess several genes (between 5 and 9) for enzymes that were assigned to the methylmalonyl-CoA branch of the propanoate metabolism. Furthermore, MAGs 109 and 145 additionally encode the methylmalonyl-CoA mutase Mut (EC 5.4.99.2) and the methylmalonyl-CoA/ethylmalonyl-CoA epimerase (EC 5.1.99.1) representing the key enzymes of this metabolism as described by Sikore et al. [23]. Therefore, these MAGs were predicted to be involved in the propionic acid metabolism in the analyzed cellulolytic/hydrolytic biofilms. Moreover, MAGs 50, 85, 109 and 145 most probably are involved in butanoate metabolism since they possess between 2 and 6 of key genes classified to the butanoate pathway (KEGG map00650). The butanoate pathway is not completely encoded in the MAGs analyzed. However, genes encoding relevant key enzymes described by Sikora et al. [23] were identified in the genomes of these Bacteria. The MAG 109 harbors a gene encoding PFL-pyruvate formate lyase (EC 2.3.1.54), whereas MAG 145 possesses the gene encoding butyrate kinase (EC 2.7.2.7) indicating the importance of these bacteria for AD at mesophilic temperatures.
The formation of methane, the last step of AD, is performed by members of the phylum Euryarchaeota. In the analyzed biofilms, MAG 111 (genus Methanobacterium sp.) was detected as most abundant archaeon positively responding to the increase in OLRs. Twenty-two genes encoding key enzymes of the hydrogenotrophic methanogenesis pathway were identified in this MAG illustrating the importance of this pathway under increased OLRs.
Transcriptional activity of MAGs in response to temperature and OLR
To determine the transcriptional activities of the compiled MAGs, the metatranscriptomes from the different reactor systems were sequenced capturing the prevailing conditions (temperature: mesophilic vs. thermophilic, OLRs of 500 g vs. 1500 g). Mapping of the transcriptome sequences for each dataset to the assembled contigs allowed determination of the MAG’s transcriptional activities under the conditions tested.
Principal component (PCA) analysis revealed clear separation of the MAG’s transcriptional activity patterns in relation to the temperature (mesophilic and thermophilic) and the OLRs 500 g and 1500 g (Fig. 6a). However, compared to the corresponding analysis based on MAG abundance profiles (see Fig. 4a), the thermophilic groups representing OLR500 and OLR1500 are more distinct suggesting transcriptional differences between these groups leading to the observed separation of the respective patterns. Clustering of transcriptional activity patterns yielded two main clusters representing the mesophilic and the thermophilic temperature regime and further sub-clusters representing the different OLRs (OLR500 and OLR1500) for each reactor system (Additional file 6). The transcriptional activities of the MAGs 40 (order Lachnospirales, Herbinix sp.), 48 (order Lachnospirales, Herbinix sp.), 107 (order Clostridiales, Clostridium sp.), 111 (phylum Euryarchaeota, Methanobacterium sp.) and 132 (class Clostridia, unknown species) increased in response to the OLR1500 in the mesophilic reactors. These MAGs also showed a positive response upon OLR increase in the abundance analysis (Fig. 4b). The response of further MAGs displayed in Additional file 6 is significant but not as pronounced as compared to the aforementioned ones. Likewise, MAGs 50 (class Clostridia, unknown species), 68 (class Clostridia, Herbinix sp.), 106 (class Clostridia, Clostridium sp.) and 146 (class Bacilli, unknown species) respond to the increased OLR with higher transcriptional activities in the thermophilic reactors (Additional file 6). These observations may be explained by the obvious connection between transcriptional activity increase and proliferation enhancement. However, for the thermophilic reactor systems, there are 14 more MAGs significantly responding to the higher OLR1500 by increasing their transcriptional activities (Additional file 6). However, is should be noted that these MAGs were not apparent in the Volcano plot based on MAG abundance profiles (Fig. 4c-d).
Fig. 6.
Alteration in transcriptional activity of 78 selected metagenome-assembled genomes (MAGs) detected in HR biofilms at mesophilic (M) and thermophilic (T) process temperature in response of the increase of organic loading rate (OLR) from 500 to 1500 g ryegrass silage. PCA plots are shown for principle component analyses of scaled and centered rlog transformed metatrabscriptome read counts mapped on MAGs (a). Transcripts were normalized on the total number of sequencing reads, assuming equal MAG abundance, and were averaged for biological replicates (b). Green, mesophilic HR biofilms; blue, thermophilic HR biofilms. For further details on sample denomination, refer to Figs. 2 and 3. Alteration in transcript abundance of MAGs selected in response of the increase of OLR from 500 to 1500 g ryegrass silage at mesophilic (c) and thermophilic (d) process temperature. Colors of the MAGs indicate taxonomic affiliation (green, Firmicutes; blue, Bacteroidota; violet, Archaea; pink, Cloacimonadota; light green, Fibrobacterota; dark red, Spirochaetota; red, unknown affiliation). * indicate MAGs with completeness above 50% and contamination rate less than 10% as listed in Table 1
Transcriptional activity of MAGs normalized to MAG abundances
In order to investigate the transcriptional activity of MAGs regardless of their abundances, transcriptome read counts for MAGs were also normalized according to MAG abundances (for details refer Material and Methods). Examination of resulting normalized transcriptional rates by applying PCA revealed well separated groups representing the different OLRs (OLR500 and OLR1500) only for the thermophilic reactor systems (Fig. 6b). This observation indicates that under thermophilic conditions, particular MAGs are able to respond to the OLR by shifting their transcriptional activity without simultaneously adjusting their replication rates. Under mesophilic conditions, this behavior only is of minor importance. MAGs responding to the higher OLR with increased normalized transcription rates are depicted in Fig. 6c-d. These are the MAG 18 (order Lachnospirales, Acetivibrio sp.) originating from the mesophilic biofilm as well as MAGs 17 (order Bacteriodales, Prevotella sp.), 40 (order Lachnospirales, Herbinix sp.), 107 (order Clostridiales, Clostridium sp.), 108 (order Bacteroidales, unknown species) and 134 (order Lachnospirales, Kineothrix sp.) obtained from the thermophilic HRs.
In summary, under thermophilic conditions, several MAGs feature an additional transcriptional enhancement that is decoupled from their replication rates which may be explained by increased metabolic activity at higher (thermophilic) temperatures.
Transcriptional profiling of most active cellulolytic / hydrolytic biofilm MAGs
To study the transcriptional activities related to OLRs in more detail, MAG 18 for the mesophilic biofilm as well as MAGs 17, 40, 107, 108 and 134 for the thermophilic biofilm were selected, since they showed the highest transcriptional increases in response to OLR increase (Fig. 6c-d, Table 1). Transcriptional rates represented by TPM values were calculated for all genes in each MAG. Subsequently, lists of the 100 most highly transcribed genes based on their TPM values were compiled for each MAG, additionally specifying the encoded gene products and their predicted functions. Transcripts with TPM values of ≤0.5 were not considered further due to their low significance. Transcripts encoding proteins involved in mandatory functions (basic house-keeping functions) such as transcription and translation were not further considered. The top five transcripts of each MAG as well as remaining genes that are of importance for assessing the metabolism of the organism were further analyzed (see Table 2).
Table 2.
Most actively transcribed genes for enzymes involved in carbohydrate or protein utilization of selected HR biofilm MAGs
| Metagenome assembled genome (MAG)1 | Assumed taxonomic affiliation | Analyzed HR2 biofilm sample | Gene transcripts involved in carbohydrate or protein utilization | Predicted functional role in AD4 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GenID | Gene length | Putative gene product | Gene | EC number | Transcripts mapped | Normalized number of transcripts [TPM]3 | ||||
| 17 | Bacteroidales (Prevotella sp.) | T1OLR1500 | Bin_17_01477 | 366 | Deoxyguanosinetriphosphate triphosphohydrolase | dgt | 3.1.5.1 | 141 | 3.9 | n/s |
| Bin_17_01805 | 171 | Hypothetical protein | n/s | n/s | 28 | 3.4 | ||||
| Bin_17_00602 | 177 | 50S ribosomal protein | rpmD | n/s | 10 | 1.5 | ||||
| Bin_17_00172 | 651 | 75 | 0.8 | |||||||
| Bin_17_00603 | 519 | 50S ribosomal protein | rpsE | n/s | 53 | 0.8 | ||||
| Bin_17_00862 | 1041 | L-3,5-diaminohexanoate dehydrogenase | kdd | 1.4.1.11 | 92 | 0.6 | ||||
| Bin_17_00014 | 579 | N-acetylhexosamine 1-kinase | nahK | 2.7.1.162 | 38 | 0.5 | ||||
| 18 | Clostridiales (Acetivibrio sp.) | M1OLR1500 | Bin_18_02374 | 207 | Small; acid-soluble spore protein C2 | sspC | 4876 | 619 | Hydrolysis | |
| Bin_18_01750 | 144 | Hypothetical protein | n/s | n/s | 462 | 145 | ||||
| Bin_18_01164 | 192 | Hypothetical protein | n/s | n/s | 384 | 58 | ||||
| Bin_18_00189 | 786 | Sulfurtransferase | tusA | 2.8.1.- | 3938 | 53 | ||||
| Bin_18_01827 | 108 | Hypothetical protein | n/s | n/s | 74 | 46 | ||||
| Bin_18_02918 | 711 | Glycerol uptake facilitator protein | glpF | n/s | 573 | 9 | ||||
| Bin_18_01829 | 912 | Lactose transport system permease protein | lacF | n/s | 760 | 9 | ||||
| Bin_18_01831 | 1377 | Beta-glucosidase A | bglA | 3.2.1.21 | 798 | 6 | ||||
| Bin_18_01301 | 1272 | Peptidoglycan binding domain protein | n/s | 678 | 5 | |||||
| Bin_18_01830 | 858 | L-arabinose transport system permease protein | araQ | n/s | 413 | 5 | ||||
| Bin_18_01016 | 1119 | D-galactose-binding periplasmic protein precursor | mglB | n/s | 309 | 3 | ||||
| 40 | Lachnospirales (Herbinix sp.) | M1OLR1500 | Bin_40_01523 | 225 | Acid-soluble spore protein C2 | sspC_1 | n/s | 42,076 | 2942 | Hydrolysis |
| Bin_40_01017 | 114 | Manganese containing catalase | n/s | n/s | 5156 | 1711 | ||||
| Bin_40_00454 | 147 | Hypothetical protein | n/s | n/s | 5508 | 1017 | ||||
| Bin_40_00073 | 246 | Hypothetical protein | n/s | n/s | 17,299 | 1006 | ||||
| Bin_40_01138 | 228 | Acid-soluble spore protein C2 | sspC_2 | n/s | 14,627 | 994 | ||||
| Bin_40_00033 | 1770 | Oligopeptide-binding protein | oppA | n/s | 14,445 | 50 | ||||
| Bin_40_00081 | 819 | Maritimacin | n/s | n/s | 4915 | 42 | ||||
| Bin_40_00131 | 681 | Thiamine transporter | thiT | n/s | 3522 | 38 | ||||
| Bin_40_00519 | 1116 | Trehalose import ATP-binding protein | sugC | n/s | 6385 | 37 | ||||
| Bin_40_00505 | 1008 | Glyceraldehyde-3-phosphate dehydrogenase | gapN | 1.2.1.12 | 4163 | 27 | ||||
| Bin_40_01590 | 1329 | Maltose/maltodextrin-binding protein precursor | malX | n/s | 5638 | 27 | ||||
| 107 | Clostridiales (Clostridium sp.) | T2OLR1500 | Bin_107_01262 | 183 | Small; acid-soluble spore protein C1 | sspC1_1 | n/s | 29,669 | 3262 | Hydrolysis |
| Bin_107_01261 | 180 | Small; acid-soluble spore protein C2 | sspC2_2 | n/s | 24,756 | 2829 | ||||
| Bin_107_01260 | 171 | Small; acid-soluble spore protein C2 | sspC2_1 | n/s | 16,600 | 2136 | ||||
| Bin_107_00122 | 186 | Small; acid-soluble spore protein C1 | sspC1_2 | n/s | 9026 | 956 | ||||
| Bin_107_00144 | 234 | Glutaredoxin-3 | grxC | n/s | 7124 | 459 | ||||
| Bin_107_01233 | 258 | Peptidase domain protein | n/s | n/s | 1245 | 65 | ||||
| Bin_107_00711 | 99 | Enolase | eno | 4.2.1.11 | 87 | 37 | ||||
| Bin_107_01338 | 1530 | 4-alpha-glucanotransferase | malQ | 2.4.1.2 | 4863 | 20 | ||||
| Bin_107_00018 | 114 | L-cystine import ATP-binding protein | tcyN | n/s | 58 | 19 | ||||
| Bin_107_01344 | 678 | Trehalose import ATP-binding protein | sugC | 3.6.3.- | 1732 | 19 | ||||
| Bin_107_01340 | 894 | Maltose transport system permease protein | malG | n/s | 2415 | 19 | ||||
| Bin_107_01038 | 531 | PTS system, maltose-specific, EIICB component | malP | n/s | 1017 | 16 | ||||
| Bin_107_01656 | 1002 | Glyceraldehyde-3-phosphate dehydrogenaseGlyceraldehyde-3-phosphate dehydrogenase | gapN | 1.2.1.12 | 2090 | 14 | ||||
| Bin_107_01134 | 627 | Lysozym | acm | 3.2.1.17 | 1086 | 13 | ||||
| Bin_107_00091 | 1881 | Amylopullulanase | apu | 3.2.1.41 | 3989 | 13 | ||||
| Bin_107_01040 | 1335 | 6-phospho-alpha-glucosidase | pagL | 3.2.1.- | 2015 | 10 | ||||
| Bin_107_00188 | 624 | Thiamine transporter | thiT | n/s | 692 | 9 | ||||
| Bin_107_01547 | 132 | NADP-dependent glyceraldehyde-3-phosphate dehydrogenase | gap | 1.2.1.9 | 158 | 8 | ||||
| 108 | Bacteroidales (unclassified genus) | T2OLR1500 | Bin_108_01400 | 213 | Hypothetical protein | n/s | n/s | 18 | 1.4 | n/s |
| Bin_108_00987 | 726 | Alkyl hydroperoxide reductase | ahpC | 1.11.1.15 | 85 | 0.8 | ||||
| Bin_108_00986 | 183 | Nitrite reductase [NAD(P)H] | nasD | 1.7.1.4 | 4 | 0.4 | ||||
| Bin_108_00047 | 1140 | Hypothetical protein | n/s | n/s | 60 | 0.3 | ||||
| Bin_108_01184 | 441 | 50S ribosomal protein L15 | rplO | n/s | 14 | 0.2 | ||||
| 134 | Lachnospirales (Kineothrix sp.) | T2OLR1500 | Bin_134_02838 | 177 | Transition state regulatory protein | abrB | n/s | 1527 | 181 | Hydrtolysis |
| Bin_134_00038 | 555 | Hypothetical protein | n/s | n/s | 491 | 7 | ||||
| Bin_134_00185 | 291 | ATP-dependent Clp protease, proteolytic subunit | clpP | n/s | 81 | 3 | ||||
| Bin_134_00184 | 1278 | ATP-dependent Clp protease, ATP-binding subunit | clpX | n/s | 622 | 3 | ||||
| Bin_134_00305 | 906 | HTH-type transcriptional regulator | yofA | n/s | 324 | 2 | ||||
| Bin_134_02093 | 1110 | Trehalose import ATP-binding protein | acm | 3.2.1.17 | 234 | 1 | ||||
| Bin_134_00012 | 570 | Amylopullulanase | pulA | 3.2.1.41 | 74 | 1 | ||||
| Bin_134_00958 | 3558 | Pyruvate-flavodoxin oxidoreductase | ydbK | 1.2.7.1 | 623 | 1 | ||||
1 The first five transcripts of each MAG represent the top transcripts of the corresponding MAG
2 Hydrolysis reactor
3 Transcripts per million averaged for datasets from two technical replicates
4 Anaerobic digestion
n/s Not specified
For the mesophilic process regime, MAG 18 revealed an increased transcriptional activity depending on the apparent OLR. Genes encoding different carbohydrate transport proteins were found among the most highly transcribed genes. MAG 18 also transcribed the gene bglA encoding an beta-glucosidase (EC 3.2.1.21) acting on terminal, non-reducing beta-D-glucosyl residues resulting in release of beta-D-glucose. Regarding its genome (Fig. 5 and Additional file 5) and transcriptome characteristics, a hydrolytic lifestyle is proposed for MAG 18 which is of particular interest for AD of protein-rich grass silage.
Another thermophilic bacterium showing an increased transcriptional activity in response to the apparent OLR is MAG 40. In particular, MAG 40 potentially is involved in decomposition of di- and monosaccharides trehalose, maltose and glucose. Further highly transcribed genes encode oligopeptide transport system proteins suggesting a hydrolytic lifestyle of this taxon.
MAG 107 is of particular interest since genes for a trehalose import ATB-binding protein of the proposed MalEFG transporter feature high TPM values (see Table 2). In addition, MAG 107 participated in clevage of α-1,6 and α-1,4 glycosidic bonds in branched and linear polysaccharides, producing glucose, maltose and maltotriose as degradation products, since the gene encoding an amylopullulanase PulA (EC 3.2.1.41) was identified among the highly transcribed genes. Its hydrolytic metabolism as well as the high transcriptional activity under elevated OLR and ammonia contents makes MAG 107 (presumably related to Clostridium sp.) appear as a potential candidate for further biotechnological improvement of AD of protein-rich silages under thermophilic conditions.
The highest transcribed gene in MAG 134 are the ATP-dependent Clp proteases gene for a component of an oligopeptide membrane transport system, eventually required for protein degradation. Furthermore, within the list of highly transcribed genes, the amylopullulanase (EC 3.2.1.41) gene pulA was also identified, whose gene product is involved in cleavage of glycosidic bonds in polysaccharides as described above. Based on these findings, MAG 134 might be involved in oligopeptide and amino acid degradation, and therefore, plays a crucial role in the acidogenesis / acetogenesis process of the biofilm community.
The bacterial MAGs 17 (order Bacteroidales, Prevotella sp.) and 108 (order Bacteroidales, unknown species) were moderately abundant in thermophilic HR biofilm metagenome datasets but showed significantly increased transcriptional activity when the OLR was raised from 500 to 1500 g (Fig. 6d). However, the transcripts specified by MAG 17 and 108 only contribute to the total transcriptome of the analyzed microbiome to a small extent and therefore these MAGs seem to be of minor importance or only play a secondary role. Accordingly, prediction of the functionality of these MAGs in the hydrolysis of crop biomass remains difficult.
Discussion
In this study, the cellulolytic/hydrolytic biofilm communities grown on the surface of ryegrass silage digested in mesophilic (37 °C) and thermophilic (55 °C) hydrolysis reactors under low (500 g) and increased (1500 g) OLR conditions were studied. The community profiles were determined by 16S rRNA gene amplicon analysis whereas metagenome sequencing was done to compile metagenomically assembled genomes (MAGs) for abundance estimations and reconstruction of their metabolism. In parallel, the transcriptional activities of MAGs were unraveled by metatranscriptome sequence analysis and genome-centered transcriptome mappings. As result of this study, thermophilic and mesophilic bacterial and archaeal candidate MAGs adapted to high ammonium/ammonia concentrations caused by high substrate protein contents were identified.
The reconstruction of genomes from metagenome datasets using an assembly and binning strategy enabled cultivation-independent recovery of 157 bacterial and archaeal MAGs. These MAGs represent abundant, and therefore important cellulolytic/hydrolytic biofilm community members, with 78 out of 157 MAGs featuring defined quality criteria, necessary for adequate genome sequence analyses.
A global view on the relative abundances of the compiled MAGs in the analyzed HRs revealed clear clustering of MAG abundance profiles in dependence of the temperatures and OLRs (OLR500 vs. OLR1500). Although the temperature is an important factor shaping anaerobic digestion microbiomes, some degree of overlap in taxonomic profiles was observed regarding the mesophilic and thermophilic communities analyzed in the present study. This result is in agreement with previous research since it has been shown that certain taxa are prevalent in mesophilic as well as thermophilic AD microbiomes [25, 26]. This also applies to the methanogenic archaeal sub-community [26, 27]. Therefore, presence of MAG 82 assigned to the Euryachaeota in the mesophilic and thermophilic reactor systems of this study is not unusual. The same applies e.g. also to MAG 107 and MAG 119 assigned to the domain Bacteria. Certain bacterial species were previously identified under mesophilic as well as thermophilic conditions [28, 29].
The OLR-effect is more pronounced for the mesophilic systems as compared to the thermophilic ones suggesting that under mesophilic conditions, MAGs respond to changes of the OLR by adapting their proliferation rates whereas for the thermophilic MAGs corresponding responses are of minor importance. These analyses allowed identification of mesophilic and thermophilic MAGs positively responding to increased OLRs.
Subsequent analysis of MAG’s transcriptional activities in response to the temperature and OLRs also led to a clear separation of the MAG’s transcriptional activity patterns in response to the conditions applied. As expected, particular MAGs responding to the OLR-increase by proliferation also showed higher transcriptional activities. Interestingly, normalization of transcriptional activity patterns to MAG abundances revealed that under thermophilic conditions, the patterns are clearly separated which is not the case for the mesophilic temperature regime. This result suggests that under thermophilic conditions, specific MAGs mainly respond to the OLR-increase by enhancing or reducing their transcriptional activities without changing their proliferation rates. This is different to the behavior of responsive MAGs under mesophilic conditions (see above). The most outstanding MAGs in this regard were assigned to the order Lachnospirales (Acetivibrio sp.) for the mesophilic biofilm and the orders Bacteroidales (Prevotella sp. and an unknown species), Lachnospirales (Herbinix sp. and Kineothrix sp.) and Clostridiales (Clostridium sp.) for the thermophilic biofilm.
The effects of OLRs on biofilm formation during AD of organic waste were studied previously [9, 30, 31]. However, most of the studies only focused on reactor performance and biofilm dynamics, rather than microbial community characterization. Much more frequently, the planktonic biogas fermenter microbiome and the impact of OLRs on community structures were studied. In mesophilic AD systems, an increase in OLR supported proliferation of fermentative bacteria of the phyla Firmicutes, Bacteroidota and Actinobacteria. These results are in agreement with the results of the microbial community characterization presented in this study. Apart from fermentative bacteria, the order Thermoanaerobacteriales, including several known syntrophic acetate-oxidizing bacteria e.g., Caldanaerobacter and Alkaliphilius, was shown to increase [32]. The methanogenic archaeal community mainly consisted of members of the genera Methanosarcina [33–35]. The Methanobacteria, Methanomicrobiales, and/or Methanomassiliicoccaceae members were also observed in certain AD-processes featuring a high organic load, depending on other prevailing conditions [36].
In thermophilic AD systems, an accumulation of the phyla Thermotogae, followed by Firmicutes and Bacteroidota [34] was shown in reactor systems featuring high OLRs. From the phylum Thermotogae, the genus Defluviitoga [20, 34, 37] was distinctly abundant. The genera Ruminiclostridium, Herbinix (both assigned to the Firmicutes) and Caproiciproducens (novel genus within Clostridium Cluster IV) were predominant. As methanogens, species of the genera Methanosarcina and Methanothermobacter were observed to increase in response to higher OLRs.
Despite the differences in all these biogas community analyses, the response of Firmicutes species to changes in OLRs has been shown several times [33, 38]. Members of the phylum Bacteroidota were detected in higher abundances in reactors with increased OLRs, much more often only in mesophilic AD [31, 33]. In the HR biofilms analyzed in this study, thermophilic candidates assigned to the order Bacteroidales, namely the MAGs 17 (Prevotella sp.) and 108 (unknown species), were detected as moderately abundant as well as transcriptionally active in response to the increase in OLRs. Both MAGs are assumed to participate in hydrolysis; however, they were predicted to play a secondary role in the biogas biofilm microbiome. Both MAGs showed significantly increased transcriptional activities when the OLR was raised from 500 to 1500 g. However, in the context of the transcriptional activity of the entire microbial community, the contribution of MAGs 17 and 108 only is of minor importance.
Currently, this is the first study describing the impact of temperature and OLR on the composition of biomass-attached cellulolytic/hydrolytic biofilms and corresponding transcriptional responses of Metagenomically Assembled Genomes (MAGs). The observation, that thermophilic and mesophilic community members behave differently in response to increased OLRs regarding proliferation and transcriptional activity has not been described before and therefore should be further investigated in detail.
Conclusions
Beside methanogenesis, the ‘bottleneck‘ of biomass AD is the primary hydrolysis of high-molecular carbohydrates such as cellulose, xylan, and other polysaccharides decomposed to volatile fatty acids, short-chained alcohols, and CO2 / H2 as essential substrates for methanogenesis. High-throughput metagenome analyses enabled the collection of microbiome data to a large scale without the need of time-consuming cultivation [39]. Detailed information on microbiome structure and dynamics in response to process parameters and conditions is accessible by application of corresponding methods. Based on this information, phylogenetic and functional molecular ecological networks (pMEN resp. fMEN) can be developed resulting in the definition of core microbiomes comprising key species for AD [36, 40–42]. Alternatively, a network of metabolic functionality independent from microbial community structure can be established [43].
Based on a combined analysis of metagenome and metatranscriptome datasets, species featuring outstanding performance under increased OLRs were identified. This offers opportunities for biotechnological application of corresponding strains in heavy-duty biogas processes. However, corresponding strategies will require the availability of appropriate isolates. Therefore, high-throughput culturomics approaches as recently developed for human intestinal microbiome analyses [44] should be established also for engineered AD systems yielding isolates featuring the potential to serve as inoculation and / or amendment strains. In addition, isolate-derived genome datasets will support the bioinformatical analysis of metabiome-derived metaomics datasets.
The applied approach is promising for identification of metabolically active AD community members possessing specific, advantageous properties under stress and / or disturbed process conditions without the need for prior cultivation. Insights into the metabolic potential and activity of resilient, robust, and competitive AD species provides the basis for a rational design of their management to counteract process disturbances and to increase methane production rates in challenging fermentation processes utilizing renewable primary products.
Methods
Reactor set-up and sampling
Two-stage two-phase biogas reactor systems consisting each of one batch downflow hydrolysis reactor (HR, vol. 10 L), one process fluid storage tank (vol. 10 L), and one downstream upflow AF reactor (vol. 10 L), were operated at mesophilic (M, 37 °C) and thermophilic (T, 55 °C) temperatures and over a period of > 750 d (Fig. 1, Fig. 2). For each reactor system and for each process temperature, two replicates were conducted in parallel, denominated further as biological replicates. Further process details were as previously published by [3]. Start-up of all fermenters were performed using liquid fermenter material from a biogas plant converting cattle manure in co-digestion with grass and maize silage and other biomass at varying concentrations and at mesophilic temperatures.
Silage of perennial ryegrass (Lolium perenne L.) was digested as sole substrate in batches of varying amounts with retention times of 28 d (storage of bale silage at − 20 °C, cutting length 3 cm), volatile substances (VS) 32% of fresh mass (FM), total Kjeldahl nitrogen 7.6 g kgFM− 1, NH4+-N 0.7 g kgFM− 1, acetic acid 2.6 g kgFM− 1, propionic acid < 0.04 g kgFM− 1, lactic acid 2.6 g kgFM− 1, ethanol 2.2 g kgFM− 1, C/N ratio 19.3, chemical oxygen demand (COD) 357.7 g kgFM− 1, analysis of chemical properties according to [6]. The total Kjeldahl nitrogen is an approximation of a protein content in the sample. The average pH had a value between 7 (at the beginning of every experiment) and 8 (at the end of every experiment after 28 days). No spoilage was observed in the silage. Biogas yields were calculated as liters normalized to 0 °C and 1013 hPa (LN) per kilogram volatile substances (kgVS). For chemical analysis, samples were taken from the effluents of HR and AF.
For sequencing of 16S rRNA gene amplicon libraries, microbial metagenomes, and microbial metatranscriptomes, samples were taken from the silage digestate in the HR digested for 2 d. At this time point, high AD rates were detected as indicated by the fast increase of volatile fatty acids (VFA), e.g., acetic acid. Sampling was performed at two different organic loading rates (OLRs), i.e., batch-fermentation of 500 g (denominated as “low OLR”, samples MOLR500 and TOLR500) and 1500 g silage (denominated as “increased OLR”, samples MOLR1500 and TOLR1500) (Fig. 2). Excess air was removed and the bottle with approximately 200 mg of silage digestate was tightly closed with a screw cap. The biofilm on plant material surfaces originating from the digestate sample was detached using a sterile scalpel in an anaerobic chamber. Further details were described by [12]. All samples were stored at − 20 °C until further analysis except samples for RNA isolation, which were processed immediately after sampling.
Extraction of total microbial genomic DNA
Total microbial community DNA was extracted from surface attached biofilms by using the FastDNA™ Spin Kit for Soil (MP Biomedicals, USA) with Lysing Matrix E Tubes according to the manufacturer’s instructions. Mechanical cell disruption was performed using the FastPrep®-24 Instrument (MP Biomedicals, USA) for two times at 6500 rpm (speed 5) for 20 s. Further, two washing steps using SEWS-M were accomplished. Finally, the DNA was eluted in 100 μl DES. Two independent technical replicates for each HR were prepared, and subsequently pooled in equimolar amounts together to collect sufficient DNA material for sequencing purposes (Additional file 7). Quality and quantity of extracted DNA were evaluated by gel electrophoresis and photometric analysis (NanoPhotometer, Implen). All DNA samples were stored at − 20 °C until further processing.
Terminal restriction fragment length polymorphism (TRFLP) fingerprinting
The microbial community dynamics during the operation of the biogas reactor systems were monitored by DNA-based TRFLP analysis targeting the bacterial 16S rRNA gene according to the protocol previously published by [12] with the modifications as published by [45]. TRFLP profiles were determined in triplicates for microbial DNA samples purified from the HR effluent after 28 d batch fermentation. TRFLP fingerprint processing and subsequent analysis were performed according to [46] using BioNumerics 7.1 software (Applied Maths, Belgium). Similarities of fingerprint profiles were calculated using Pearson correlation with 0.5% optimization, cluster analysis was performed applying the unweighted pair group method with arithmetic mean (UPGMA) algorithm.
Next-generation-sequencing (NGS) of 16S rRNA gene amplicon libraries
The microbial community structures were taxonomically characterized by high-throughput next generation sequencing (NGS) of 16S rRNA gene amplicon libraries as described previously [47]. The libraries were constructed using the primers 515F (5′ - CTACGGGNGGCWGCAG - 3′) and 806R (5′ - GACTACHVGGGTATCTAATCC - 3′) amplifying the V3 and V4 regions of the bacterial 16S rRNA gene [48]. Two biological replicates per sample were analyzed (Additional file 7). Obtained sequence reads were used for iterative read pair merging applying the FLASH software [49]. Subsequently, the QIIME NGS analysis pipeline was applied for amplicon data processing as described previously [29]. Obtained OTUs were clustered at the 97% sequence identity level applying the QIIME NGS analysis pipeline.
Microbial metagenome library preparation, NGS, and assembly of genomes from metagenome datasets
For library preparation, microbial DNA samples were purified using the Genomic DNA Clean & Concentrator Kit (Zymo Research, USA). For each sample, DNA from two subsamples (i.e., technical replicates) were extracted. For sequencing, 1 μg of total DNA was sheared to approximately 430 bp fragments using a focused-ultrasonicator (Covaris M220, Woburn, MA, USA). Finally, the Illumina TruSeq® DNA PCR-free sample preparation kit (Illumina, Eindhoven, Netherlands) was used to construct the sequencing libraries, which were sequenced on the Illumina HiSeq 1500 sequencer using the Illumina HiSeq Rapid SBS Kit v2 (Illumina, Eindhoven, Netherlands), following a 2 × 250 indexed high output run protocol.
Furthermore, Megahit tool (v1.0.2) [50] (command line settings: --presets meta --min-contig-len 1000) was used for assembly of the pooled sequencing data of all samples applying a k-mer sizes of 21, 41, 61, 81 and 99 (iterative assembly). Paired-end metagenome reads from individual datasets were mapped versus all assembled metagenome contigs with Bowtie 2 (v2.2.4) [51] in end-to-end mode applying the option ‘sensitive’. To convert SAM to BAM, sort the alignment file and calculate read mapping statistics SAMtools (v1.0) [52] was used. Furthermore, to predict genes on assembled contigs larger than 1 kb, the gene prediction tool Prodigal v.2.6.0 [53] was applied. Predicted protein sequences were compared to NCBI’s database using the BLASTP mode of DIAMOND [54]. The resulting output file was loaded into MEGAN5 [55] for taxonomic classification of each gene sequence. In the following binning step, the abundance profile and the tetranucleotide frequencies were used to bin contigs into metagenome-assembled genomes (MAGs) with MetaBAT (v0.21.3) [56]. Mapping of the reads was always performed on all contigs, including the contigs of the MAGs and the contigs which were not binned. Subsequently, completeness, contamination, and strain heterogeneity of the MAGs were estimated with CheckM (v1.0.4) [22], using sets of clade-specific single-copy marker genes.
Microbial metatranscriptome library preparation and NGS
The total microbial RNA from two sub-samples (from the same silage biofilm as for the DNA extraction) was extracted applying the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s guidelines. In total, two technical replicates were processed for each of two parallel hydrolytic reactor communities yielding eight samples for the mesophilic system and eight samples for the thermophilic system (Additional file 7).
Subsequently, the RNA was purified using the Ambion® Turbo DNA-free Kit (ThermoFisher, Germany). Ribosomal RNA was depleted using the Ribo-Zero™ rRNA Removal Kit for Bacteria (Illumina, Madison, USA) according to the manufacturer’s instructions. The remaining mRNA transcripts were fragmented to approximately 550 bp fragments using a focused-ultrasonicator (Covaris M220, Woburn, USA). cDNA libraries for Illumina sequencing were constructed using the TruSeq RNA Library Prep Kit v2 (Illumina, Eindhoven, Netherlands). The resulting cDNA libraries were sequenced on the Illumina HiSeq 1500 machine using the Illumina HiSeq Rapid SBS Kit v2 (Illumina, Eindhoven, Netherlands) to generate 2 × 100 bp paired-end reads.
Metagenome and metatranscriptome sequence analysis
Paired end metagenome and metatranscriptome reads were mapped versus all assembled metagenome contigs with Bowtie2 [51] in end-to-end mode with option sensitive. After read mapping SAMtools [52] was used to filter the resulting BAM files for uniquely mapped reads. Reads were classified as uniquely mapped reads with a unique genomic location if and only if they could not be aligned to another location with a higher or same mapping quality. Metagenome as well as metatranscriptome reads that could be aligned to assembled MAG’s were quantified with the HTSeq-count program [57] to get an estimate of MAG abundance and overall MAG expression, respectively. To account for different MAG abundance, raw metatranscriptome read counts for each MAG plus one (pseudocount) were divided by the MAG’s raw metagenome read count plus one and rounded to integer values. The resulting counts for each MAG served as input for DESeq2 [58] for pairwise detection and quantification of differentially abundant and transcriptionally active MAGs, respectively. For DESeq2 parametrization, a beta prior and disabled Cook distance cutoff filtering was used. All other parameters remained unchanged. Fold change estimates, p-values, and regularized log-transformed (rlog) counts OF (1) metagenome read counts and (2) metatranscriptome read counts normalized for different MAG abundances as described above were emplyed to assess proper sample resp. replicate clustering with (PCA, multidimensional scaling (MDS)), and hierarchical clustering. In addition, volcano plots were used to identify significantly differentially expressed MAGs. Similar comparative analyses were performed assuming equal MAG abundances.
Phylogenetic and functional analysis of the metagenome-assembled genomes
The Genome Taxonomy Database toolkit [59] was used to assign objective taxonomic classifications to bacterial and archaeal genomes. Each MAG was defined as single operational taxonomic unit (OTU). For MAGs with completeness values of more than 50% and a contamination rate less than 10% (Additional file 4), analysis of the genetic potential was performed using the EMGB [60] annotation system including KEGG pathway mapping and DIAMOND tool [54]. MAGs that meet the criteria mentioned above were subsequently analyzed regarding their transcriptional activities using the EMGB annotation system again.
To predict genes encoding carbohydrate-active enzymes, the carbohydrate-active enzyme database (CAZy) annotation web server dbCAN v7 [24] was used. The genes encoding enzymes acting direct on cellulose were identified by the presence of coding regions for type one or two dockerin or cohesin modules, among others.
Supplementary information
Additional file 1. Statistics of 16S rRNA gene sequence analysis.
Additional file 2. Relative abundances of most abundant 16S rRNA gene sequences.
Additional file 3. Statistics of the obtained and processed metagenome and metatranscriptome sequences.
Additional file 4. Taxonomic affiliations of metagenome-assembled genomes (MAGs) of this study originating from HR biofilms.
Additional file 5. Analysis of the key enzymes of AD pathways in metagenome-assembled genomes (MAGs). The genetic determinants were categorized according to the four stages of the AD process, namely hydrolysis, acidogenesis, acetogenesis and methanogenesis as described previously [23].
Additional file 6. Hierarchical clustering of abundance values for 78 selected metagenome-assembled genomes (MAGs) detected in HR biofilms at mesophilic and thermophilic process temperature at organic loading rate (OLR) of 500 g resp. 1500 g ryegrass silage as deduced from transcriptome data.
Additional file 7. Experimental set up and sampling scheme.
Acknowledgements
The authors gratefully acknowledge the highly valuable technical support of M. Felgentreu, M. Jäkel, K. Mundt, and G. Rehde. The bioinformatics support of the BMBF-funded project ‘Bielefeld-Gießen Center for Microbial Bioinformatics - BiGi (grant no. 031A533)’ within the German Network for Bioinformatics Infrastructure (de.NBI) is also gratefully acknowledged. ASz and A.S. acknowledge funding from the European Union’s Horizon 2020 research and innovation program under grant agreement No. 818431 (SIMBA). This output reflects only the author’s view and the Research Executive Agency (REA) cannot be held responsible for any use that may be made of the information contained therein.
Abbreviations
- AD
Anaerobic digestion
- AF
Anaerobic filter
- ANI
Average nucleotide sequence identity
- CAZymes
Carbohydrate-active enzymes
- CO2
Carbon dioxide
- COD
Chemical oxygen demand
- d
day
- FM
Fresh mass
- GH
Glycosyl hydrolase
- HR
Hydrolysis reactor
- HT
High-throughput
- kg
kilogram
- kgVS
kilograms volatile substances
- L
Liter
- LN
Liter normalized
- M
Mesophilic
- MAG
Metagenome assembled genomes
- MG
Microbial metagenome dataset
- MT
Microbial metatranscriptome dataset
- OLR
Organic loading rates
- OTU
Operational taxonomic unit
- SAOB
Syntrophic acetate-oxidizing bacteria
- T
Thermophilic
- TPM
Transcripts per million
- TRFLP
Terminal restriction fragment length polymorophism
- VFA
Volatile fatty acids
- VS
Volatile substances
Authors’ contributions
IM prepared the 16S rRNA gene amplicon libraries for sequencing, analyzed the taxonomic profiles of biofilm communities, analyzed and interpreted metagenome and metatranscriptome data, performed the comparative MAG analyses, coordinated drafting and drafted corresponding parts of the manuscript. MK participated in data analysis and visualization, and drafted parts of the introduction, results, discussion, and conclusions sections. JD conducted all AD experiments, the sampling of digestates and process liquids, DNA and RNA extraction from biofilm samples, and TRFLP analysis. JD contributed also to AD data analysis and revised the manuscript. YS also performed DNA and RNA extraction from biofilm samples, prepared the metagenome and metatranscriprome sequencing libraries, and revised the manuscript. DW and JB determined the phylogenetic relationship between the MAGs, contributed to the results and discussion section, and revised the manuscript. MB performed the comparative metagenome and metatranscriptome analysis, contributed to the material and methods section, and revised the manuscript. CJ participated in AD experiments and corresponding data analysis, and revised the manuscript. KW participated in metatranscriptome data analysis, and revised the manuscript. MR carried out the taxonomic classification of the microbial communities, contributed to the results section, and revised the manuscript. AR participated in AD experiments and TRFLP analysis and corresponding data analysis, and revised the manuscript. AP participated in the design of this study, contributed to the results and discussion sections, and revised the manuscript. ASz and CH carried out the taxonomic classification of the microbial communities based on metagenome and metatranscriptome data, performed the metagenome assembly and binning, participated in the comparative metagenome and metatranscriptome analysis, contributed to the results and discussion section, and revised the manuscript. ASz, AS, and MK conceived the study, participated in manuscript coordination, organized third-party funding, supervised all biological and bioinformatic data analyses, contributed to drafting of the manuscript and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was part of the joint project BIOGAS-MARKER supported by the German Federal Ministry of Education and Research (BMBF), grant nos. 03SF0440A and 03SF0440C.
Availability of data and materials
Sequence datasets were deposited in the European Nucleotide Archive (ENA) under the Bioproject accession numbers PRJEB27769 (metagenome datasets), E-MTAB-7533 (metatranscriptome datasets), PRJEB30260 (16S rRNA gene amplicon sequences).
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
I.M. and M.K. contributed equally to this work.
A.Scz. and A.Schl. contributed equally to this work.
Contributor Information
Irena Maus, Email: irena.maus@cebitec.uni-bielefeld.de.
Michael Klocke, Email: mklocke@atb-potsdam.de.
Jaqueline Derenkó, Email: jaqueline.derenko@gmail.com.
Yvonne Stolze, Email: ystolze@cebitec.uni-bielefeld.de.
Michael Beckstette, Email: michael.beckstette@helmholtz-hzi.de.
Carsten Jost, Email: cjost@atb-potsdam.de.
Daniel Wibberg, Email: dwibberg@cebitec.uni-bielefeld.de.
Jochen Blom, Email: jochen.blom@computational.bio.uni-giessen.de.
Christian Henke, Email: christian@cebitec.uni-bielefeld.de.
Katharina Willenbücher, Email: kwillenbuecher@atb-potsdam.de.
Madis Rumming, Email: mrumming@uni-bielefeld.de.
Antje Rademacher, Email: a-rademacher@gmx.net.
Alfred Pühler, Email: puehler@cebitec.uni-bielefeld.de.
Alexander Sczyrba, Email: asczyrba@cebitec.uni-bielefeld.de.
Andreas Schlüter, Email: aschluet@cebitec.uni-bielefeld.de.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s40793-020-00354-x.
References
- 1.Weiland P. Biogas production: current state and perspectives. Appl Microbiol Biotechnol. 2010;85:849–860. doi: 10.1007/s00253-009-2246-7. [DOI] [PubMed] [Google Scholar]
- 2.Nizami AS, Korres NE, Murphy JD. Review of the integrated process for the production of grass biomethane. Environ Sci Technol. 2009;43:8496–8508. doi: 10.1021/es901533j. [DOI] [PubMed] [Google Scholar]
- 3.Schönberg M, Linke B. The influence of the temperature regime on the formation of methane in a two-phase anaerobic digestion process. Eng Life Sci. 2012;12:279–286. doi: 10.1002/elsc.201100062. [DOI] [Google Scholar]
- 4.Pohl M, Heeg K, Mumme J. Anaerobic digestion of wheat straw – performance of continuous solid-state digestion. Bioresour Technol. 2013;146:408–415. doi: 10.1016/j.biortech.2013.07.101. [DOI] [PubMed] [Google Scholar]
- 5.Shen F, Yuan H, Pang Y, Chen S, Zhu B, Zou D, et al. Performances of anaerobic co-digestion of fruit & vegetable waste (FVW) and food waste (FW): single-phase vs. two-phase. Bioresour Technol. 2013;144:80–85. doi: 10.1016/j.biortech.2013.06.099. [DOI] [PubMed] [Google Scholar]
- 6.Linke B, Rodríguez-Abalde Á, Jost C, Krieg A. Performance of a novel two-phase continuously fed leach bed reactor for demand-based biogas production from maize silage. Bioresour Technol. 2015;177:34–40. doi: 10.1016/j.biortech.2014.11.070. [DOI] [PubMed] [Google Scholar]
- 7.Habouzit F, Hamelin J, Santa-Catalina G, Steyer J-P, Bernet N. Biofilm development during the start-up period of anaerobic biofilm reactors: the biofilm Archaea community is highly dependent on the support material. Microb Biotechnol. 2014;7:257–264. doi: 10.1111/1751-7915.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zakrzewski M, Goesmann A, Jaenicke S, Jünemann S, Eikmeyer F, Szczepanowski R, et al. Profiling of the metabolically active community from a production-scale biogas plant by means of high-throughput metatranscriptome sequencing. J Biotechnol. 2012;158:248–258. doi: 10.1016/j.jbiotec.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 9.Langer S, Schropp D, Bengelsdorf FR, Othman M, Kazda M. Dynamics of biofilm formation during anaerobic digestion of organic waste. Anaerobe. 2014;29:44–51. doi: 10.1016/j.anaerobe.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 10.Maus I, Koeck DE, Cibis KG, Hahnke S, Kim YS, Langer T, et al. Unraveling the microbiome of a thermophilic biogas plant by metagenome and metatranscriptome analysis complemented by characterization of bacterial and archaeal isolates. Biotechnol Biofuels. 2016;9:171. doi: 10.1186/s13068-016-0581-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maus I, Rumming M, Bergmann I, Heeg K, Pohl M, Nettmann E, et al. Characterization of Bathyarchaeota genomes assembled from metagenomes of biofilms residing in mesophilic and thermophilic biogas reactors. Biotechnol Biofuels. 2018;11:167. doi: 10.1186/s13068-018-1162-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rademacher A, Nolte C, Schönberg M, Klocke M. Temperature increases from 55 to 75 °C in a two-phase biogas reactor result in fundamental alterations within the bacterial and archaeal community structure. Appl Microbiol Biotechnol. 2012;96:565–576. doi: 10.1007/s00253-012-4348-x. [DOI] [PubMed] [Google Scholar]
- 13.Kouzuma A, Tsutsumi M, Ishii S, Ueno Y, Abe T, Watanabe K. Non-autotrophic methanogens dominate in anaerobic digesters. Sci Rep. 2017;7:1510. doi: 10.1038/s41598-017-01752-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bremges A, Maus I, Belmann P, Eikmeyer F, Winkler A, Albersmeier A, et al. Deeply sequenced metagenome and metatranscriptome of a biogas-producing microbial community from an agricultural production-scale biogas plant. Gigascience. 2015;4:33. doi: 10.1186/s13742-015-0073-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hassa J, Maus I, Off S, Pühler A, Scherer P, Klocke M, et al. Metagenome, metatranscriptome, and metaproteome approaches unraveled compositions and functional relationships of microbial communities residing in biogas plants. Appl Microbiol Biotechnol. 2018;102:5045–5063. doi: 10.1007/s00253-018-8976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heyer R, Kohrs F, Reichl U, Benndorf D. Metaproteomics of complex microbial communities in biogas plants. Microb Biotechnol. 2015;8:749–763. doi: 10.1111/1751-7915.12276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campanaro S, Treu L, Kougias PG, De Francisci D, Valle G, Angelidaki I. Metagenomic analysis and functional characterization of the biogas microbiome using high throughput shotgun sequencing and a novel binning strategy. Biotechnol Biofuels. 2016;9:26. doi: 10.1186/s13068-016-0441-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kougias PG, Campanaro S, Treu L, Zhu X, Angelidaki I. A novel archaeal species belonging to Methanoculleus genus identified via de-novo assembly and metagenomic binning process in biogas reactors. Anaerobe. 2017;46:23–32. doi: 10.1016/j.anaerobe.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 19.Maus I, Bremges A, Stolze Y, Hahnke S, Cibis KG, Koeck DE, et al. Genomics and prevalence of bacterial and archaeal isolates from biogas-producing microbiomes. Biotechnol Biofuels. 2017;10:264. doi: 10.1186/s13068-017-0947-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maus I, Cibis KG, Bremges A, Stolze Y, Wibberg D, Tomazetto G, et al. Genomic characterization of Defluviitoga tunisiensis L3, a key hydrolytic bacterium in a thermophilic biogas plant and its abundance as determined by metagenome fragment recruitment. J Biotechnol. 2016;232:50–60. doi: 10.1016/j.jbiotec.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 21.Campanaro S, Treu L, Rodriguez-R L, Kovalöovszki A, Ziels R, Maus I, et al. The anaerobic digestion microbiome: a collection of 1600 metagenome-assembled genomes shows high species diversity related to methane production. Biotechnol Biofuels. 2019; 10.1101/680553.
- 22.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sikora Anna, Detman Anna, Mielecki Damian, Chojnacka Aleksandra, Błaszczyk Mieczysław. Anaerobic Digestion. 2019. Searching for Metabolic Pathways of Anaerobic Digestion: A Useful List of the Key Enzymes. [Google Scholar]
- 24.Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40:W445–W451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sundberg C, Al-Soud WA, Larsson M, Alm E, Yekta SE, Svensson BH, et al. 454 pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbial Ecol. 2013;85:612–626. doi: 10.1111/1574-6941.12148. [DOI] [PubMed] [Google Scholar]
- 26.Lv Z, Wu X, Zhou B, Wang Y, Sun Y, Wang Y, et al. Effect of one step temperature increment from mesophilic to thermophilic anaerobic digestion on the linked pattern between bacterial and methanogenic communities. Bioresour Technol. 2019;292:121968. doi: 10.1016/j.biortech.2019.121968. [DOI] [PubMed] [Google Scholar]
- 27.Munk B, Guebitz GM, Lebuhn M. Influence of nitrogen-rich substrates on biogas production and on the methanogenic community under mesophilic and thermophilic conditions. Anaerobe. 2017;46:146–154. doi: 10.1016/j.anaerobe.2017.02.015. [DOI] [PubMed] [Google Scholar]
- 28.Shin SG, Han G, Lee J, Shin J, Hwang S. A snapshot of microbial community structures in 20 different field-scale anaerobic bioreactors treating food waste. J Environ Manag. 2019;248:109297. doi: 10.1016/j.jenvman.2019.109297. [DOI] [PubMed] [Google Scholar]
- 29.Stolze Y, Bremges A, Rumming M, Henke C, Maus I, Pühler A, et al. Identification and genome reconstruction of abundant distinct taxa in microbiomes from one thermophilic and three mesophilic production-scale biogas plants. Biotechnol Biofuels. 2016;9:156. doi: 10.1186/s13068-016-0565-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bassini JP, Diass N, Caoe SMS, Senra E, Laranjeira Y, Dezotti M. Effect of increasing organic loading rates on the performance of moving-bed biofilm reactors filled with different support media: assessing the activity of suspended and attached biomass fractions. Proc Saf Environ Protect. 2016;100:131–141. doi: 10.1016/j.psep.2016.01.007. [DOI] [Google Scholar]
- 31.Liu C, Wang W, Anwar N, Ma Z, Liu G, Zhang R. Effect of organic loading rate on anaerobic digestion of food waste under mesophilic and thermophilic conditions. Energy Fuel. 2017;31:2976–2984. doi: 10.1021/acs.energyfuels.7b00018. [DOI] [Google Scholar]
- 32.Kovács E, Wirth R, Maróti G, Bagi Z, Rákhely G, Kovács KL. Biogas production from protein-rich biomass: fed-batch anaerobic fermentation of casein and of pig blood and associated changes in microbial community composition. PLoS One. 2013;16:e77265. doi: 10.1371/journal.pone.0077265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Westerholm Maria, Schnürer Anna. Anaerobic Digestion. 2019. Microbial Responses to Different Operating Practices for Biogas Production Systems. [Google Scholar]
- 34.Zhang L, Ban Q, Li J. Microbial community dynamics at high organic loading rates revealed by pyrosequencing during sugar refinery wastewater treatment in a UASB reactor. Front Environ Sci Eng. 2018;12:4. doi: 10.1007/s11783-018-1045-8. [DOI] [Google Scholar]
- 35.Leite AF, Janke L, Lv Z, Harms H, Richnow HH, Nikolausz M. Improved monitoring of semi-continuous anaerobic digestion of sugarcane waste: effects of increasing organic loading rate on methanogenic community dynamics. Int J Mol Sci. 2015;25:23210–23226. doi: 10.3390/ijms161023210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu R, Yang ZH, Zheng Y, Liu JB, Xiong WP, Zhang YR, et al. Organic loading rate and hydraulic retention time shape distinct ecological networks of anaerobic digestion related microbiome. Bioresour Technol. 2018;262:184–193. doi: 10.1016/j.biortech.2018.04.083. [DOI] [PubMed] [Google Scholar]
- 37.Ben Hania W, Godbane R, Postec A, Hamdi M, Ollivier B, Fardeau ML. Defluviitoga tunisiensis gen. Nov., sp. nov., a thermophilic bacterium isolated from a mesothermic and anaerobic whey digester. Int J Syst Evol Microbiol. 2012;62:1377–1382. doi: 10.1099/ijs.0.033720-0. [DOI] [PubMed] [Google Scholar]
- 38.Zhang L, Loh KC, Sarvanantharajah S, Tong YW, Wang CH, Dai Y. Mesophilic and thermophilic anaerobic digestion of soybean curd residue for methane production: characterizing bacterial and methanogen communities and their correlations with organic loading rate and operating temperature. Bioresour Technol. 2018;288:121597. doi: 10.1016/j.biortech.2019.121597. [DOI] [PubMed] [Google Scholar]
- 39.Treu L, Kougias PG, Campanaro S, Bassani I, Angelidaki I. Deeper insight into the structure of the anaerobic digestion microbial community; the biogas microbiome database is expanded with 157 new genomes. Bioresour Technol. 2016;2016:260266. doi: 10.1016/j.biortech.2016.05.081. [DOI] [PubMed] [Google Scholar]
- 40.Zhou J, Deng Y, Luo F, He Z, Tu Q, Zhi X. Functional molecular ecological networks. MBio. 2010;2010;1. [DOI] [PMC free article] [PubMed]
- 41.Zhou J, Deng Y, Luo F, He Z, Yang Y. Phylogenetic molecular ecological network of soil microbial communities in response to elevated CO2. mBio. 2011;2:e00122–e00111. doi: 10.1128/mBio.00122-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang J, Li G, Qian Y, Yang Y, Zhang F. Microbial functional gene patterns related to soil greenhouse gas emissions in oil contaminated areas. Sci Total Environ. 2018;628–629:94–102. doi: 10.1016/j.scitotenv.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 43.Louca S, Polz MF, Mazel F, Albright MBN, Huber JA, O’Connor MI, et al. Function and functional redundancy in microbial systems. Nature Ecol Evol. 2018;2:936. doi: 10.1038/s41559-018-0519-1. [DOI] [PubMed] [Google Scholar]
- 44.Bilen M, Dufour J-C, Lagier J-C, Cadoret F, Daoud Z, Dubourg G, et al. The contribution of culturomics to the repertoire of isolated human bacterial and archaeal species. Microbiome. 2018;6:94. doi: 10.1186/s40168-018-0485-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Theuerl S, Kohrs F, Benndorf D, Maus I, Wibberg D, Schlüter A, et al. Community shifts in a well-operating agricultural biogas plant: how process variations are handled by the microbiome. Appl Microbiol Biotechnol. 2015;99:7791–7803. doi: 10.1007/s00253-015-6627-9. [DOI] [PubMed] [Google Scholar]
- 46.Klang J, Theuerl S, Szewzyk U, Huth M, Tölle R, Klocke M. Dynamic variation of the microbial community structure during the long-time mono-fermentation of maize and sugar beet silage. Microb Biotechnol. 2015;8:764–775. doi: 10.1111/1751-7915.12263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maus I, Kim YS, Wibberg D, Stolze Y, Off S, Antonczyk S, et al. Biphasic study to characterize agricultural biogas plants by high-throughput 16S rRNA gene amplicon sequencing and microscopic analysis. J Microbiol Biotechnol. 2017;27:321–334. doi: 10.4014/jmb.1605.05083. [DOI] [PubMed] [Google Scholar]
- 48.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li D, Liu CM, Luo R, Sadakane K, Lam TW. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 51.Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hyatt D, LoCascio PF, Hauser LJ, Uberbacher EC. Gene and translation initiation site prediction in metagenomic sequences. Bioinformatics. 2012;28:2223–2230. doi: 10.1093/bioinformatics/bts429. [DOI] [PubMed] [Google Scholar]
- 54.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 55.Huson DH, Mitra S, Ruscheweyh H-J, Weber N, Schuster SC. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011;21:1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kang DD, Froula J, Egan R, Wang Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ. 2015;3:e1165. doi: 10.7717/peerj.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anders S, Pyl P, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;15:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.A toolkit for assigning objective taxonomic classifications to bacterial and archaeal genomes.: Ecogenomics/GTDBTk. Python. Australian Centre for Ecogenomics; 2018. https://github.com/Ecogenomics/GTDBTk. Accessed 6 Nov 2018.
- 60.Jünemann S, Kleinbölting N, Jaenicke S, Henke C, Hassa J, Nelkner J, et al. Bioinformatics for NGS-based metagenomics and the application to biogas research. J Biotechnol. 2017;261:10–23. doi: 10.1016/j.jbiotec.2017.08.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Statistics of 16S rRNA gene sequence analysis.
Additional file 2. Relative abundances of most abundant 16S rRNA gene sequences.
Additional file 3. Statistics of the obtained and processed metagenome and metatranscriptome sequences.
Additional file 4. Taxonomic affiliations of metagenome-assembled genomes (MAGs) of this study originating from HR biofilms.
Additional file 5. Analysis of the key enzymes of AD pathways in metagenome-assembled genomes (MAGs). The genetic determinants were categorized according to the four stages of the AD process, namely hydrolysis, acidogenesis, acetogenesis and methanogenesis as described previously [23].
Additional file 6. Hierarchical clustering of abundance values for 78 selected metagenome-assembled genomes (MAGs) detected in HR biofilms at mesophilic and thermophilic process temperature at organic loading rate (OLR) of 500 g resp. 1500 g ryegrass silage as deduced from transcriptome data.
Additional file 7. Experimental set up and sampling scheme.
Data Availability Statement
Sequence datasets were deposited in the European Nucleotide Archive (ENA) under the Bioproject accession numbers PRJEB27769 (metagenome datasets), E-MTAB-7533 (metatranscriptome datasets), PRJEB30260 (16S rRNA gene amplicon sequences).