Abstract
Background
Sequencing of 16S rRNA genes has become a powerful technique to study microbial communities and their responses towards changing environmental conditions in various ecosystems. Several tools have been developed for the prediction of functional profiles from 16S rRNA gene sequencing data, because numerous questions in ecosystem ecology require knowledge of community functions in addition to taxonomic composition. However, the accuracy of these tools relies on functional information derived from genomes available in public databases, which are often not representative of the microorganisms present in the studied ecosystem. In addition, there is also a lack of tools to predict functional gene redundancy in microbial communities.
Results
To address these challenges, we developed Tax4Fun2, an R package for the prediction of functional profiles and functional gene redundancies of prokaryotic communities from 16S rRNA gene sequences. We demonstrate that functional profiles predicted by Tax4Fun2 are highly correlated to functional profiles derived from metagenomes of the same samples. We further show that Tax4Fun2 has higher accuracies than PICRUSt and Tax4Fun. By incorporating user-defined, habitat-specific genomic information, the accuracy and robustness of predicted functional profiles is substantially enhanced. In addition, functional gene redundancies predicted with Tax4Fun2 are highly correlated to functional gene redundancies determined for simulated microbial communities.
Conclusions
Tax4Fun2 provides researchers with a unique tool to predict and investigate functional profiles of prokaryotic communities based on 16S rRNA gene sequencing data. It is easy-to-use, platform-independent and highly memory-efficient, thus enabling researchers without extensive bioinformatics knowledge or access to high-performance clusters to predict functional profiles. Another unique feature of Tax4Fun2 is that it allows researchers to calculate the redundancy of specific functions, which is a potentially important measure of how resilient a community will be to environmental perturbation. Tax4Fun2 is implemented in R and freely available at https://github.com/bwemheu/Tax4Fun2.
Keywords: Metagenomics, Functional predictions, 16S rRNA gene, Bioinformatics, Microbiome, Multifunctional redundancy, Ecosystem functioning
Background
Microorganisms are key players in ecosystem functioning [1, 2]. For example, host-associated microorganisms significantly contribute to the health of their host organism, e.g., by providing essential nutrients or by enhancing the host’s resilience to pathogens or pests [3–5]. High-throughput sequencing of 16S rRNA genes is a powerful and widely used tool to study the taxonomic or phylogenetic composition of microbial communities in a variety of marine [6–8], terrestrial [9, 10] and host-associated [11–13] environments. However, numerous questions in biogeochemistry and ecology require knowledge of microbial community function, rather than, or in addition to, the taxonomic or phylogenetic composition [14]. Investigating the composition of microbial communities and their functional capabilities is of particular importance when the effect of changing environmental conditions or anthropogenic perturbations on ecosystem services is being assessed [15].
Many ecosystems are threatened by environmental perturbations. A key question in microbial ecology is whether, and to what degree, microbial communities contain functionally redundant members that may provide stability to ecosystem processes in the face of these perturbations [8, 16–18]. It has been proposed that the evaluation of multiple functions provide a more comprehensive picture on the role of biodiversity for maintaining ecosystem functions [19]. However, the simultaneousness assessment of multiple functions is time-consuming [20]. Some researchers have started to address this question by developing metatranscriptome-based [20] or metagenome-based [21] approaches for quantifying the multifunctional redundancy of microbial communities [20, 21]. To the best of our knowledge, no tools are currently available to provide a standardized method for the simultaneous calculation of functional redundancy for multiple functions.
In past years, several freely available tools including PICRUSt [22], Tax4Fun [23], Piphillin [24], Faprotax [25] and paprica [26] have been developed for the prediction of functional profiles inferred from 16S rRNA gene sequence data. Although these tools cannot replace the functional assessment via metagenomic shotgun sequencing, they have provided unique insights into functional capabilities of prokaryotic communities in diverse habitats, such as microbial mats [27], soil [28–31], marine seawater [25, 26, 32, 33], sediment [8, 34], rumen [35, 36] and the plant endosphere [37, 38].
The predictive power of these tools relies on functional information derived from genomes available in public databases. This information is used to predict functional profiles even if no close reference genomes are present in these databases. However, available genomes do not necessarily represent the functional diversity present in the ecosystem investigated. This problem has motivated the development of predictive tools specific for the rumen microbiome [35] or marine microorganisms [25]. Given the rapidly increasing number of available genomes, in particular through metagenome-assisted genome binning [39], and that many research groups have access to unpublished, habitat-specific genomic information, the incorporation of such data is likely to enhance the accuracy of functional inferences.
Here, we introduce Tax4Fun2, a new and improved version of Tax4Fun [23]. Tax4Fun2 is platform-independent, user-friendly and highly memory-efficient. It can incorporate habitat-specific and user-defined data. Although Tax4Fun2 focuses on prokaryotic data, eukaryotic data can also be incorporated. We show that the incorporation of habitat-specific data improves the practical utility of Tax4Fun2 for microbiome datasets from a wide range of ecosystems. Comparative analysis further shows that Tax4Fun2 has a higher accuracy than PICRUSt [22] and Tax4Fun [23]. Another unique feature of Tax4Fun2 is that it enables researchers to calculate the functional redundancy of multiple functions, which is critical for the prediction of how likely a specific ecosystem function is to be lost during environmental perturbation. This information might be important in ecosystem biomonitoring and the prioritisation of environmental management actions.
Implementation
Tax4Fun2 is provided as an R [40] package with a current default reference dataset build from 12,377 archaeal and bacterial genomes available through the NCBI RefSeq database. The current version is 1.1.5. Tax4Fun2 is platform-independent and enables researchers without extensive bioinformatics knowledge to predict functional profiles in an efficient and user-friendly manner. In the following sections, we provide further details in the generation of the reference data and the technical implementation of Tax4Fun2.
Tax4Fun2 reference data
Tax4Fun2 is supplied with two reference datasets (Ref99NR and Ref100NR) referring to the similarity threshold used during clustering of the 16S rRNA gene sequences. Each dataset consists of an association matrix with 16S rRNA gene reference sequences associated with functional reference profiles (number of entries in the full association matrix: 4584 and 18,479 for Ref99NR and Ref100NR, respectively). Due to its smaller size, the Ref99NR database is less hardware-demanding and predictions are generated faster compared to the Ref100NR. In order to obtain the default reference data, we included the function buildReferenceData, which will automatically download and build the reference data.
The reference datasets were generated as follows: we downloaded all complete genomes and all genomes with the status ‘chromosome’ from NCBI RefSeq (assessed on 18 August 2018), resulting in 275 archaeal and 12,102 bacterial genomes. Barrnap version 0.9 (https://github.com/tseemann/barrnap) was used to identify and extract all 16S rRNA gene sequences. All rRNA gene sequences were subsequently concatenated into a single file, sorted by decreasing length and clustered using the UCLUST algorithm implemented in USEARCH version 10.240 [41] at 99 and 100% sequence similarity, respectively. The longest sequence of each cluster serves as the 16S rRNA reference sequence. Functional profiles for each genome were generated as follows: open-reading-frames were predicted with prodigal version 2.6.3 [42]. Functional profiles were calculated based on obtained protein sequences with UProC version 1.2.0 [43] using the KEGG Orthology (KO) database for prokaryotes (July 2018 release; [44]) as reference. To account for differences in rRNA copy numbers, functional profiles were normalized by the number of 16S rRNA genes identified in each genome. Due to the heterogeneity of 16S rRNA genes within a genome, the functional reference profile for each 16S rRNA reference sequence was generated based on the 16S rRNA clustering results; a single functional reference profile is the average normalized functional profile of each genome with at least one 16S rRNA gene affiliated to the cluster. If more than one 16S rRNA gene sequence of a genome was assigned to a cluster, the normalized profile of the genome was multiplied by the number of 16S rRNA genes affiliated to the cluster before calculating the mean profile. To calculate phylogenetic distances, two phylogenetic trees (one tree for each reference dataset) are included in the reference data. These were generated as follows: all 16S rRNA reference sequences were aligned with SINA version 1.2.11 [45] against the latest Silva ARB release (SILVA_132_SSUREF_NR99) [46]. The phylogenetic tree was subsequently calculated using RaxML version 8.2.11 [47] under a GTRGAMMA model and a random seed of 12,345.
Extending the reference data of Tax4Fun2 using specific genomes
The predictive power of available tools, such as PICRUSt or Tax4Fun, is limited by the number of genomes available in public databases that also need frequent updates. Moreover, available genomes do not necessarily represent the functional diversity present in the ecosystem investigated. In Tax4Fun2, we address these issues by allowing users to build their own habitat-specific reference data sets (Fig. 1). In order to build such a dataset, the user needs to provide a set of genomes. To assist with the extraction of 16S rRNA sequences and the functional annotation of these genomes, we implemented two functions in the Tax4Fun2 package: extractSSU and assignFunctions. Ribosomal RNA sequences (16S rRNA or 18S rRNA) are identified by BLAST search using the SILVA SSURef database version 132 [46] (preclustered at 90% with uclust). Functional profiles are generated by BLASTp with diamond against the KEGG KO database [48]. Protein sequences are predicted prior to functional annotation using prodigal version 2.6.3 [42]. Currently, the functional annotation is only available for prokaryotes but will be extended to eukaryotes in later versions. The extracted rRNA sequences and functional profiles are subsequently used to build reference data sets using the addUserDataByClustering or addUserData functions (Fig. 1). vsearch is required to use the first function [49]. vsearch is freely available at https://github.com/torognes/vsearch and included in Tax4Fun2 as part of the reference data. The latter function bypasses the clustering step with vsearch and is recommended if only a small number of distinct genomes shall be used as reference data.
Fig. 1.
The Tax4Fun workflow. 16S rRNA gene sequences are initially aligned against the reference sequences provided with Tax4Fun2 to identify the nearest neighbour. If user-defined data is supplied, the 16S rRNA gene sequences are additionally aligned against the sequences added by the user. Nearest neighbours in the user data will be preferred if both search attempts result in significant hits. The OTU abundances for each sample are summarized based on the results from the nearest neighbour search. An association matrix (AM) containing the functional profiles of those references identified in the 16S rRNA search is generated. The summarized abundances and the functional profiles stored in the AM are merged and a metagenome is predicted for each sample. FTU and FSU values are provided as a log file. To include user data and generate a habitat-specific data set, Tax4Fun2 provides functions to functionally annotate prokaryotic genomes and to extract 16S rRNA gene sequences. User-defined reference data sets are subsequently generated and can be included in the prediction. If large amounts of genomes are provided, extracted 16S rRNA gene sequences can be optionally clustered using the uclust algorithm
Predicting functional profiles with Tax4Fun2
In the initial step of Tax4Fun2, user-supplied 16S rRNA gene sequences (operational taxonomic units or exact sequence variants [50], also known as zero-radius OTUs, but here simply referred to as OTUs) are searched against the 16S rRNA reference sequences by BLAST using the runRefBlast function (Fig. 1). Other tools, such as paprica [26], use algorithms to place query sequences into phylogenetic trees. These algorithms usually provide a very accurate phylogenetic placement. However, placing sequences is very hardware intensive and most algorithms are restricted to Unix or Unix-like operating systems. Due to these limitations, we decided to use a BLAST-based approach, because we only need to identify the closest match in the database. If user data is supplied, the next-neighbour search is repeated using the user-generated reference data. Following the assumption that users provide habitat or site-specific data, user-generated data is preferred to the default reference data. Specifically, if the next neighbour search for one OTU against both the default reference database and the user-generated database resulted in significant hits, the user data is incorporated in the functional prediction.
Functional predictions are subsequently calculated using the makeFunctionalPrediction function (Fig. 1). During this step, the OTU table supplied by the user is summarized based on the results of the next neighbour search. A specific association matrix in the summarized table is generated containing only the functional reference profiles of the next neighbours. For each sample, the abundance information from the OTU table and the functional information stored in the specific association matrix are converted into a sample-specific functional profile. Predicted profiles are later summarized based on KEGG pathways. Only OTUs passing a defined similarity threshold (default = 97%) are considered in the functional prediction. The fraction of OTUs having no close hit in the reference data and hence are unused in the subsequent prediction (fraction of taxonomic units unused = FTU) as well as the amount of sequences assigned to these unused taxonomic units (fraction of sequences unused = FSU) is recorded. FTU and FSU values may serve as an additional quality indicator for the predicted metagenomes as high FTU and/or FSU values indicate that predictions were made only for a small fraction of the total microbial community.
Calculation and validation of the functional redundancy index (FRI)
To date, there is no tool available to predict functional redundancies based on 16S rRNA data. Here, we introduce the functional redundancy index (FRI), which describes the (multi-) functional redundancy of a prokaryotic community, i.e., the redundancy of multiple functions in the investigated community. The FRI incorporates the phylogenetic distribution (distance) of community members harbouring a specific function and their proportion in the community. In Tax4Fun2, the functional redundancy index is calculated using the function calculateFunctionalRedundancy (Fig. 2).
Fig. 2.

The FRI workflow. 16S rRNA gene sequences are initially aligned against the reference sequences provided with Tax4Fun2 to identify the nearest neighbour. The OTU abundances for each sample are converted to presence-absence data and subsequently summarized based on the results from the nearest neighbour search. An association matrix (AM) containing the normalized functional profiles of those references identified in the 16S rRNA search is generated. The normalized functional profile in the AM is multiplied by the summarized OTU abundance. We assume that a function is present if the abundance of a function is greater than 50%. For instance, if 5 OTUs were affiliated to one 16S rRNA reference seqeunce, we would assume that every function with a minimum abundance of 10% in the normalized reference profile would be present in the AM. The FRI is finally calculated based on those community members possessing the function and their phylogenetic distribution. The amount of sequences/OTUs unused in the prediction (FSU/FTU) is provided in a log file 16S rRNA gene sequences are initially aligned against the reference sequences provided with Tax4Fun2 to identify the nearest neighbour. The presence of each OTU are summarized based on the results from the nearest neighbour search. An association matrix (AM) containing the functional profiles of those references identified in the 16S rRNA search is generated. The summarized abundances and the functional profiles stored in the AM are multiplied. Functions being present in all genomes are assumed to be present in the entire cluster. The functional redundancy is subsequently predicted from the average phylogenetic distance of all community members potentially possessing the function. This distance is subsequently normalized either by the average phylogenetic distance of the total reference tree (absolute) or by the distance found in the surveyed community (relative). FTU and FSU values are provided as a log file. The functions performing each step are marked with numbers
Following next neighbour search, the OTU table is summarized based on the results of the next neighbour search and converted to binary data (presence/absence). The normalized functional profile associated with each reference sequence is also converted to a binary data (presence/absence) using a cut-off of 100% meaning that a function is considered to be present if it was observed in all genomes assigned to the reference profile. If the next neighbour search identifies the same neighbour for several OTUs, then the normalized functional profile associated with the 16S rRNA reference sequence is multiplied by the number of OTUs before it is being converted to binary data. This is based on the assumption that the probability a specific function present in a community is higher the more distinct the phylotypes associated to a single reference are present. The redundancy of any function present in the community is subsequently calculated by multiplying the average phylogenetic distance of all community members possessing the function with their proportion in the community. The product is normalized by the average phylogenetic distance of the total community. To account for differences in average phylogenetic distances between different communities, we provide the possibility to calculate the absolute and the relative FRI (aFRI and rFRI). To calculate the aFRI, the average phylogenetic distance of all species in the full 16S rRNA reference tree is used for normalization, whereas the rFRI is normalized by the average phylogenetic distance of those species in the 16S rRNA reference tree identified as being present in the surveyed samples during next neighbour search. The rFRI can be used to compare samples within one survey, whereas the aFRI allows the comparison of functional redundancy indices across different ecosystems. The latter is important as multifunctional redundancy comparisons between or among different environments generate a more robust depiction of (regional) variation in the resilience/vulnerability of microbial communities [21].
To test the FRI accuracy, we simulated 1000 prokaryotic communities each consisting of 100 genomes randomly selected from the 12,377 genomes used to generate the reference data. To assess the phylogenetic distance between the genomes, we extracted 63 marker protein sequences based on hmm profiles derived from PFAM version 31 [51] and TIGRFAM version 15 [52]. The 63 marker proteins were selected because their corresponding genes were present in 90% of all 12,377 genomes and, if present, were single-copy genes in 99% of them. These criteria were applied to archaea and bacteria independently. The extracted protein sequences of each hmm profile were aligned using mafft version 7.3.11 [53]. Afterwards, aligned protein sequences for each genome were concatenated. The phylogenomic tree was calculated using FastTree version 2.1.10 [54]. Functional profiles for each genome were converted to presence-absence data and the FRI was calculated for each function using the genome tree and the presence-absence data. The 16S rRNA gene sequences present in each genome subset were separately clustered in operational taxonomic units (OTUs) at 97% similarity with UCLUST implemented in USEARCH [41]. An OTU table was generated based on the clustering. Each OTU was represented by its longest sequence.
Results and discussion
Tax4Fun2 evaluation
We first applied Tax4Fun2 in comparison to Tax4Fun [23] and PICRUSt [22] using the same paired samples (16S rRNA gene and metagenome data), which were used to validate both tools, i.e. samples derived from the human microbiome, mammalian guts, soil and from a hypersaline microbial mat (Table 1), in addition to ten marine seawater samples taken in the North Sea [7] and 90 kelp-associated samples collected within the Marine Microbes Framework Data Initiative (http://www.bioplatforms.com/marine-microbes).
Table 1.
Accession numbers of samples/studies used to validate Tax4Fun2
| Origin | Sample number | Accession numbers |
|---|---|---|
| Human Microbiome | 41 | SRS011271, SRS011452, SRS011529, SRS011584, SRS011586, SRS013234, SRS013252, SRS013258, SRS013506, SRS013687, SRS013711, SRS013723, SRS014235, SRS014287, SRS014343, SRS014613, SRS014629, SRS014923, SRS015133, SRS015190, SRS015425, SRS015450, SRS015574, SRS015578, SRS015762, SRS015782, SRS015854, SRS015960, SRS016002, SRS016018, SRS016095, SRS016111, SRS016203, SRS016225, SRS016331, SRS016335, SRS016349, SRS016434, SRS016533, SRS016553, SRS016559 |
| Mammalian Gut | 56 | 4,461,284–301, 4,461,341–55, 4,461,357–58, 4,461,360–80, 4,461,383 (MG-RAST) |
| Microbial Mat | 10 | 4,440,963–71 (MG-RAST) |
| Soil | 14 | 4,477,803–5, 4,477,807, 4,477,872–7, 4,477,899, 4,477,902–4 (MG-RAST) |
| Water | 10 | SRA060677 |
| Kelp | 90 | 57,884–936, 57,938–56, 87,958–74, 58,019–20 (https://data.bioplatforms.com/organization/about/australian-microbiome).) |
Functional profiles were predicted using the default workflows. For PICRUSt, processed sequences were clustered using QIIME version 1.8 [55] by closed-reference picking against the Greengenes database (version 13_5; [56]) and normalized prior to functional prediction. For Tax4Fun, OTU sequences were taxonomically classified by BLAST search [57] against the SILVA database (SILVA_123_SSURef_Nr99) [46].
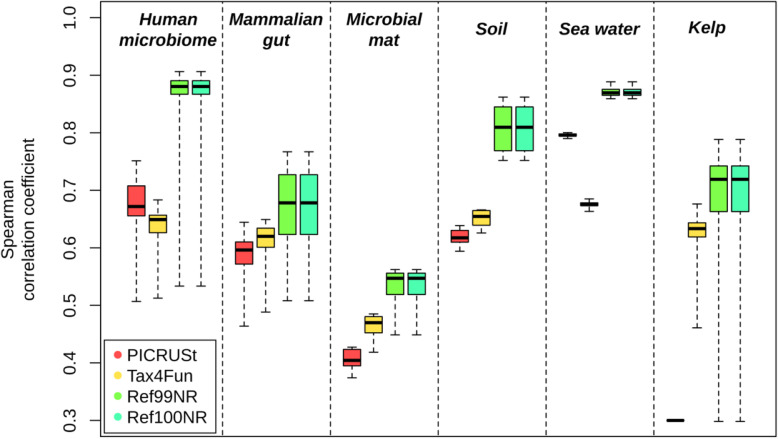
We evaluated the predictive power of each tool by comparing the functional profiles predicted from the 16S rRNA data to functional profiles generated directly from the metagenomes using Spearman rank correlations. Comparing the profiles predicted with PICRUSt, Tax4Fun and Tax4Fun2 with metagenome-derived profiles clearly showed that Tax4Fun2 outperforms PICRUSt and Tax4Fun across all six tested datasets (Fig. 3). In addition, Tax4Fun2 was more than 20 times faster than Tax4Fun due to the smaller reference database.
Fig. 3.
Correlations between functional profiles obtained from metagenomic datasets and those predicted from 16 s rRNA data. Predictions were made with PICRUSt, Tax4Fun, and Tax4Fun2. Predictions with Tax4Fun2 were made using both supplied default reference datasets (Ref99NR and Ref100NR). Note that PICRUSt did not generate any prediction for the kelp data
It should be noted that a direct comparison of functional profiles predicted with all three tools is difficult due to several changes in the KEGG Orthology since PICRUSt and Tax4Fun were developed (deprecated and new functional orthologs). Hence, predicted functional profiles as well as those obtained by metagenomic shotgun sequencing were converted to relative abundances prior to comparison. Only functions present in the metagenomic profile and in the predicted profile were considered in each comparison. On average, more than 95% of all functions in the human microbiome as well as the marine, soil and kelp-derived metagenomes were affiliated to these shared KOs. In the microbial mat and the mammalian gut samples, more than 74 and 78% of all predicted functions were affiliated to these KOs, respectively.
Using user-defined data increased the accuracy and reduced the FTU
Following the assumption that any additional genomic information specific for the investigated habitat further enhances the predictive power of Tax4Fun2, we used 68 metagenome-assembled genomes (MAGs) generated from the 90 kelp-associated metagenomes to build a kelp-specific reference dataset. These genomes were selected because at least one 16S rRNA gene sequence was identified in each genome.
Using the default data, the median Spearman correlation coefficient was 0.72. Incorporating the kelp-specific data substantially increased the power of the functional prediction (median Spearman correlation coefficient = 0.86) and reduced the fraction of the sequences not used in the prediction (Fig. 4), showing that a lack of suitable reference genomes did initially limit Tax4Fun2’s performance. Moreover, using the kelp-specific dataset enabled us to predict functional profiles for samples, which failed when using only the default reference data, because next neighbour search resulted in no close matches. These results demonstrate the benefits of incorporating user-defined, habitat-specific reference databases, which distinguishes Tax4Fun2 from all other published tools.
Fig. 4.
Correlations between functional profiles retrieved from 90 kelp metagenomes and those predicted with Tax4Fun2 without and with user data added and the fraction of zOTUs and sequences unused in the prediction
Functional redundancy index
The simultaneousness assessment of multiple functions is usually very time-consuming [20]. Previous studies have focused on a limited number of community functions (e.g., [58–60]). However, the degree of functional redundancy in any given system depends on the functions considered [14]. In addition, it is difficult to draw conclusions about functional redundancy based on a single environmental situation, as species that are functionally redundant in one environment might be pivotal in another [61]. Hence, the extent of functional redundancy change as the ecological contribution of a species might change between different environments. Nonetheless, a contemporary concern for the conservation of biodiversity and the development of management strategies is that decision makers require quantitative measures as part of science-based negotiations and communications. In order to provide those measures when assessing natural or human-induced impacts on an ecosystem, we introduced the FRI with respect to multiple functions in Tax4Fun2. A high FRI indicates that a specific function is almost ubiquitous in all community members, whereas a low FRI suggests that the function is present in a few closely related species. A FRI of 0 indicates that a function has been detected in only one community member or is not present at all. Consequently, the lower the FRI the higher the probability that a function gets lost after community shifts or perturbations.
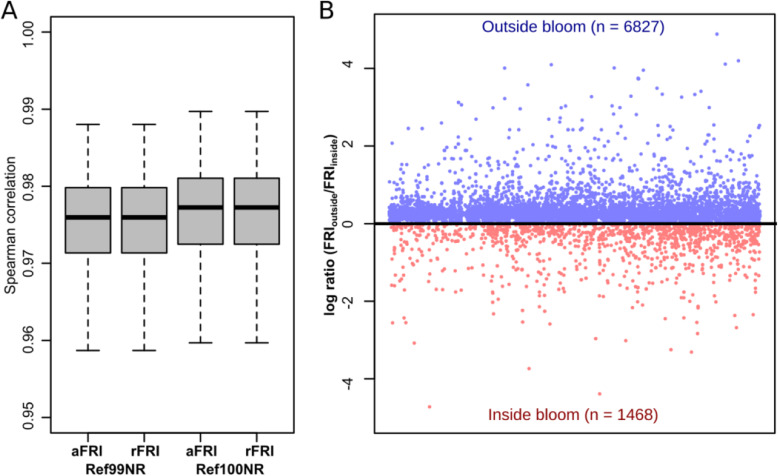
To test the accuracy, we simulated 1000 microbial communities and calculated FRI values based on 16S rRNA gene data using Tax4Fun2. The FRIs calculated for each function were subsequently compared to the FRIs calculated directly from the genomes of each simulation by Spearman rank correlation. The comparison revealed that Tax4Fun2 provides a good estimate of the functional redundancy present in the microbial community (Spearman rank correlation > 90%) (Fig. 5a).
Fig. 5.
Correlation between predicted and genome-based functional redundancy indices (a) and functional redundancy indices inside and outside a phytoplankton bloom (b). A log ratio greater than 0 indicates that a function is more redundant outside the bloom. All predictions were made using a 97% similarity cut off
We further calculated FRIs using the marine seawater samples. Six of these samples were taken inside a phytoplankton bloom and three samples were taken outside the bloom [7]. Nearly 7000 functions displayed a higher functional redundancy index outside the bloom, whereas only 1468 functions had higher redundancies inside the bloom (Fig. 5b). This indicates that the functional redundancy greatly shifts during the phytoplankton bloom. Phytoplankton blooms are usually characterized by a substrate-controlled succession, i.e. distinct bacterial clades dominate the bacterioplankton community at different stages during and shortly after the bloom [62]. Consequently, community members involved in the turnover of certain substrates at a specific stage are predominant. For instance, the SAR92 clade, the Roseobacter RCA cluster and the genus Polaribacter were more abundant in bloom samples [3]. Because we did not observe significant differences in the phylogenetic diversity of bacterioplankton communities derived from bloom and reference samples, functions predominantly associated with dominant community members are more redundant in the bloom whereas all other functions display higher redundancies in the reference samples.
Conclusion
With Tax4Fun2, we provide an easy-to-use, platform-independent R package, which enables researchers to predict and investigate functional profiles of prokaryotic communities based on 16S rRNA gene data. We demonstrate the high predictive power of Tax4Fun2, providing superior results to any other established tool. The key strength of Tax4Fun2 is the incorporation of user-defined and habitat-specific data, which further enhances the accuracy of the predictions. Another unique feature of Tax4Fun2 is that it enables researchers to calculate functionial redundancies, which is a relevant parameter for ecosystem monitoring and the development of management strategies to safeguard optimal ecosystem functionality. Nonetheless, functional profiles and functional redundancies are predictions only and should be treated with caution.
Availability and requirements
Project name: Tax4Fun2.
Project homepage: https://github.com/bwemheu/Tax4Fun2
Operating system(s): Platform-independent.
Programming language: R.
Other requirements: BLAST+ 2.7.1 or later, R packages ape and seqinr.
License: GNU General Public License v3.0.
Any restrictions to use by non-academics: no.
Acknowledgements
Not applicable.
Abbreviations
- BLAST
Basic Local Alignment Search Tool
- NCBI
National Center for Biotechnology Information
- FRI
Functional redundancy index
Authors’ contributions
BW led the project. BW, PM and FW designed and implemented the final Tax4Fun2 algorithms, to which TT and RD made critical contributions. JAT collected and analysed the kelp dataset. BW, PM and RD coordinated the online implementation. FW and BW wrote the manuscript, with feedback from all other authors. All authors approved the final version of the manuscript.
Funding
B.W. and R.D. were supported by the German Research Foundation (DFG). T.T. and J.A.T. were supported by Bioplatforms Australia. F.W. and E.L.J. were supported by the Australian Research Council (ARC Linkage Project SHRP021212).
Availability of data and materials
The dataset(s) supporting the conclusions of this article is available at https://zenodo.org/records/10035668.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
2/20/2024
Following publication of this article, the URL in the 'Availability of data and materials' declaration has been updated (to https://zenodo.org/records/10035668) because the server in which the data referred to in this declaration was previously stored has been shut down.
References
- 1.Falkowski PG, Fenchel T, Delong EF. The microbial engines that drive Earth’s biogeochemical cycles. Science. 2008;320(5879):1034–1039. doi: 10.1126/science.1153213. [DOI] [PubMed] [Google Scholar]
- 2.Azam F, Malfatti F. Microbial structuring of marine ecosystems. Nat Rev Microbiol. 2007;5(10):782–791. doi: 10.1038/nrmicro1747. [DOI] [PubMed] [Google Scholar]
- 3.Heintz-Buschart A, Wilmes P. Human Gut Microbiome: Function Matters. Trends Microbiol. 2018;26(7):563–574. doi: 10.1016/j.tim.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Vandenkoornhuyse P, Quaiser A, Duhamel M, Le Van A, Dufresne A. The importance of the microbiome of the plant holobiont. New Phytol. 2015;206(4):1196–1206. doi: 10.1111/nph.13312. [DOI] [PubMed] [Google Scholar]
- 5.Egan S, Harder T, Burke C, Steinberg P, Kjelleberg S, Thomas T. The seaweed holobiont: understanding seaweed–bacteria interactions. FEMS Microbiol Rev. 2013;37(3):462–476. doi: 10.1111/1574-6976.12011. [DOI] [PubMed] [Google Scholar]
- 6.Sun MY, Dafforn KA, Johnston EL, Brown MV. Core sediment bacteria drive community response to anthropogenic contamination over multiple environmental gradients. Environ Microbiol. 2013;15(9):2517–2531. doi: 10.1111/1462-2920.12133. [DOI] [PubMed] [Google Scholar]
- 7.Wemheuer B, Wemheuer F, Hollensteiner J, Meyer F-D, Voget S, Daniel R. The green impact: bacterioplankton response towards a phytoplankton spring bloom in the southern North Sea assessed by comparative metagenomic and metatranscriptomic approaches. Front Microbiol. 2015;6:805. doi: 10.3389/fmicb.2015.00805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polónia ARM, Cleary DFR, Freitas R, Gomes NCM, de Voogd NJ. Archaeal and bacterial communities of Xestospongia testudinaria and sediment differ in diversity, composition and predicted function in an Indonesian coral reef environment. J Sea Res. 2017;119:37–53. doi: 10.1016/j.seares.2016.10.007. [DOI] [Google Scholar]
- 9.Yu H, Ling N, Wang T, Zhu C, Wang Y, Wang S, et al. Responses of soil biological traits and bacterial communities to nitrogen fertilization mediate maize yields across three soil types. Soil Tillage Res. 2019;185:61–69. doi: 10.1016/j.still.2018.08.017. [DOI] [Google Scholar]
- 10.Tripathi BM, Kim M, Kim Y, Byun E, Yang J-W, Ahn J, et al. Variations in bacterial and archaeal communities along depth profiles of Alaskan soil cores. Sci Rep. 2018;8(1):504. doi: 10.1038/s41598-017-18777-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zozaya-Valdés E, Roth-Schulze AJ, Egan S, Thomas T. Microbial community function in the bleaching disease of the marine macroalgae Delisea pulchra. Environ Microbiol. 2017;19(8):3012–3024. doi: 10.1111/1462-2920.13758. [DOI] [PubMed] [Google Scholar]
- 12.Granzow S, Kaiser K, Wemheuer B, Pfeiffer B, Daniel R, Vidal S, et al. The effects of cropping regimes on fungal and bacterial communities of wheat and faba bean in a greenhouse pot experiment differ between plant species and compartment. Front Microbiol. 2017;8(902). 10.3389/fmicb.2017.00902. [DOI] [PMC free article] [PubMed]
- 13.Hartman K, van der Heijden MGA, Roussely-Provent V, Walser J-C, Schlaeppi K. Deciphering composition and function of the root microbiome of a legume plant. Microbiome. 2017;5(1):2. doi: 10.1186/s40168-016-0220-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Louca S, Polz MF, Mazel F, Albright MB, Huber JA, O’Connor MI, et al. Function and functional redundancy in microbial systems. Nature Ecology Evolution. 2018;2(6):936–943. doi: 10.1038/s41559-018-0519-1. [DOI] [PubMed] [Google Scholar]
- 15.Birrer SC, Dafforn KA, Sun MY, Williams RBH, Potts J, Scanes P, et al. Using meta-omics of contaminated sediments to monitor changes in pathways relevant to climate regulation. Environ Microbiol. 2019;21(1):389–401. doi: 10.1111/1462-2920.14470. [DOI] [PubMed] [Google Scholar]
- 16.Allison SD, Martiny JBH. Resistance, resilience, and redundancy in microbial communities. Proc National Acad Sci. 2008;105(Supplement 1):11512–11519. doi: 10.1073/pnas.0801925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jurburg SD, Salles JF. Functional redundancy and ecosystem function—the soil microbiota as a case study. In: Lo YH, Blanco JA, Roy S, editors. Biodiversity in Ecosystems-Linking Structure and Function: InTech. 2015. pp. 29–49. [Google Scholar]
- 18.Moya A, Ferrer M. Functional redundancy-induced stability of gut microbiota subjected to disturbance. Trends Microbiol. 2016;24(5):402–413. doi: 10.1016/j.tim.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 19.Gamfeldt L, Hillebrand H, Jonsson PR. Multiple functions increase the importance of biodiversity for overall ecosystem functioning. Ecology. 2008;89(5):1223–1231. doi: 10.1890/06-2091.1. [DOI] [PubMed]
- 20.Beier S, Shen D, Schott T, Jürgens K. Metatranscriptomic data reveal the effect of different community properties on multifunctional redundancy. Mol Ecol. 2017;26(24):6813–6826. doi: 10.1111/mec.14409. [DOI] [PubMed] [Google Scholar]
- 21.Miki T, Yokokawa T, Matsui K. Biodiversity and multifunctionality in a microbial community: a novel theoretical approach to quantify functional redundancy. Proc R Soc B Biol Sci. 2014;281(1776):20132498. doi: 10.1098/rspb.2013.2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotech. 2013;31(9):814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aßhauer KP, Wemheuer B, Daniel R, Meinicke P. Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics. 2015;31(17):2882–2884. doi: 10.1093/bioinformatics/btv287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwai S, Weinmaier T, Schmidt BL, Albertson DG, Poloso NJ, Dabbagh K, et al. Piphillin: improved prediction of metagenomic content by direct inference from human microbiomes. PLoS One. 2016;11(11):e0166104. doi: 10.1371/journal.pone.0166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Louca S, Parfrey LW, Doebeli M. Decoupling function and taxonomy in the global ocean microbiome. Science. 2016;353(6305):1272–1277. doi: 10.1126/science.aaf4507. [DOI] [PubMed] [Google Scholar]
- 26.Bowman JS, Ducklow HW. Microbial communities can be described by metabolic structure: a general framework and application to a seasonally variable, depth-stratified microbial community from the coastal West Antarctic peninsula. PLoS One. 2015;10(8):e0135868. doi: 10.1371/journal.pone.0135868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koo H, Hakim JA, Morrow CD, Eipers PG, Davila A, Andersen DT, et al. Comparison of two bioinformatics tools used to characterize the microbial diversity and predictive functional attributes of microbial mats from Lake Obersee, Antarctica. J Microbiol Methods. 2017;140:15–22. doi: 10.1016/j.mimet.2017.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaiser K, Wemheuer B, Korolkow V, Wemheuer F, Nacke H, Schöning I, et al. Driving forces of soil bacterial community structure, diversity, and function in temperate grasslands and forests. Sci Rep. 2016;6:33696. doi: 10.1038/srep33696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herzog S, Wemheuer F, Wemheuer B, Daniel R. Effects of fertilization and sampling time on composition and diversity of entire and active bacterial communities in German grassland soils. PLoS One. 2015;10(12):e0145575. doi: 10.1371/journal.pone.0145575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berkelmann D, Schneider D, Engelhaupt M, Heinemann M, Christel S, Wijayanti M, et al. How rainforest conversion to agricultural systems in Sumatra (Indonesia) affects active soil bacterial communities. Front Microbiol. 2018;9(2381). 10.3389/fmicb.2018.02381. [DOI] [PMC free article] [PubMed]
- 31.Semedo M, Song B, Sparrer T, Phillips RL. Antibiotic effects on microbial communities responsible for Denitrification and N(2) O production in grassland soils. Front Microbiol. 2018;9:2121. doi: 10.3389/fmicb.2018.02121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wemheuer B, Wemheuer F, Meier D, Billerbeck S, Giebel H-A, Simon M, et al. Linking compositional and functional predictions to decipher the biogeochemical significance in DFAA turnover of abundant bacterioplankton lineages in the North Sea. Microorganisms. 2017;5(4). 10.3390/microorganisms5040068. [DOI] [PMC free article] [PubMed]
- 33.Galand PE, Pereira O, Hochart C, Auguet JC, Debroas D. A strong link between marine microbial community composition and function challenges the idea of functional redundancy. The ISME J. 2018;12(10):2470–2478. doi: 10.1038/s41396-018-0158-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su Z, Dai T, Tang Y, Tao Y, Huang B, Mu Q, et al. Sediment bacterial community structures and their predicted functions implied the impacts from natural processes and anthropogenic activities in coastal area. Mar Pollut Bull. 2018;131:481–495. doi: 10.1016/j.marpolbul.2018.04.052. [DOI] [PubMed] [Google Scholar]
- 35.Wilkinson TJ, Huws SA, Edwards JE, Kingston-Smith AH, Siu-Ting K, Hughes M, et al. CowPI: a rumen microbiome Focussed version of the PICRUSt functional inference software. Front Microbiol. 2018;9:1095. doi: 10.3389/fmicb.2018.01095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biscarini F, Palazzo F, Castellani F, Masetti G, Grotta L, Cichelli A, et al. Rumen microbiome in dairy calves fed copper and grape-pomace dietary supplementations: composition and predicted functional profile. PLoS One. 2018;13(11):e0205670. doi: 10.1371/journal.pone.0205670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wemheuer F, Kaiser K, Karlovsky P, Daniel R, Vidal S, Wemheuer B. Bacterial endophyte communities of three agricultural important grass species differ in their response towards management regimes. Sci Rep. 2017;7:40914. doi: 10.1038/srep40914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang R, Liu P, Ye W. Illumina-based analysis of endophytic bacterial diversity of tree peony (Paeonia Sect. Moutan) roots and leaves. Brazilian J Microbiol. 2017;48(4):695–705. doi: 10.1016/j.bjm.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parks DH, Rinke C, Chuvochina M, Chaumeil P-A, Woodcroft BJ, Evans PN, et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol. 2017;2(11):1533–1542. doi: 10.1038/s41564-017-0012-7. [DOI] [PubMed] [Google Scholar]
- 40.Team RC. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2018. [Google Scholar]
- 41.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 42.Hyatt D, Chen G, LoCascio P, Land M, Larimer F, Hauser L. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meinicke P. UProC: tools for ultra-fast protein domain classification. Bioinformatics. 2015;31(9):1382–1388. doi: 10.1093/bioinformatics/btu843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pruesse E, Peplies J, Glöckner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28(14):1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41(D1):D590–D5D6. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2014;12:59. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 49.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Callahan BJ, McMurdie PJ, Holmes SP. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. The ISME J. 2017;11(12):2639. doi: 10.1038/ismej.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44(D1):D279–DD85. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haft DH, Selengut JD, White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31(1):371–373. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Katoh T. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 54.Price MN, Dehal PS, Arkin AP. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5(3):e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–5072. doi: 10.1128/aem.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peter H, Ylla I, Gudasz C, Romaní AM, Sabater S, Tranvik LJ. Multifunctionality and diversity in bacterial biofilms. PLoS One. 2011;6(8):e23225. doi: 10.1371/journal.pone.0023225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mori AS, Isbell F, Fujii S, Makoto K, Matsuoka S, Osono T. Low multifunctional redundancy of soil fungal diversity at multiple scales. Ecol Lett. 2016;19(3):249–259. doi: 10.1111/ele.12560. [DOI] [PubMed] [Google Scholar]
- 60.Delgado-Baquerizo M, Giaramida L, Reich PB, Khachane AN, Hamonts K, Edwards C, et al. Lack of functional redundancy in the relationship between microbial diversity and ecosystem functioning. J Ecol. 2016;104(4):936–946. doi: 10.1111/1365-2745.12585. [DOI] [Google Scholar]
- 61.Fetzer I, Johst K, Schäwe R, Banitz T, Harms H, Chatzinotas A. The extent of functional redundancy changes as species’ roles shift in different environments. Proc National Acad Sci United States of America. 2015;112(48):14888–14893. doi: 10.1073/pnas.1505587112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM, et al. Substrate-controlled succession of marine Bacterioplankton populations induced by a phytoplankton bloom. Science. 2012;336(6081):608–611. doi: 10.1126/science.1218344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset(s) supporting the conclusions of this article is available at https://zenodo.org/records/10035668.