Abstract
Background
Postmenopausal osteoporosis, a common disease among elderly women, is linked to estrogen deficiency, mechanical loading, and genotype. Circular RNAs (circRNAs) are formed through reverse splicing of the splice donor at the 3′ end and the splice accepter at the 5′ end in pre-mRNA and have been shown to be involved in the development of multiple diseases. Based on their high sequence conservation and stability, circRNAs may be useful biomarkers in different diseases. However, the roles of circRNAs in postmenopausal osteoporosis remain incompletely understood.
Material/Methods
Fifty-three postmenopausal women were assigned to either the postmenopausal osteoporosis group (n=28) or the control group (n=25). Reverse-transcriptase quantitative polymerase chain reaction (RT-qPCR) analysis was performed to determine the differential expression of circRNAs between the 2 groups. Receiver-operating characteristic (ROC) curve analysis was conducted to evaluate the clinical diagnostic value of circRNA. Prediction of the binding sites between circRNA and miRNAs was conducted using miRanda and RNAhybrid. The function of the circRNA in osteoclastogenesis was determined by circRNA overexpression followed by tartrate-resistant acid phosphatase staining and RT-qPCR analysis.
Results
Among 4 circRNAs previously identified by RNA-sequencing analysis as differentially expressed in patients with postmenopausal osteoporosis, only hsa_circ_0021739 showed a significant difference in expression between the groups and was downregulated in patients with postmenopausal osteoporosis. The hsa_circ_0021739 expression level was determined to be correlated with the lumbar vertebra, femur, and forearm T-scores. Overexpression of hsa_circ_0021739 decreased the level of hsa-miR-502-5p and inhibited the differentiation of osteoclasts.
Conclusions
The circRNA hsa_circ_0021739 is a potential blood biomarker for postmenopausal osteoporosis. In addition, hsa-miR-502-5p is a likely target of hsa_circ_0021739, which acts to regulate the differentiation of osteoclasts.
Keywords: Genetic Association Studies; Osteoporosis; Osteoporosis, Postmenopausal; RNA, Small Untranslated
Background
Osteoporosis is a bone metabolic disorder characterized by a decline in bone quality and strength that leads to a high incidence of fracture due to increased bone fragility [1]. Postmenopausal osteoporosis (PMOP) usually occurs within 5–10 years after menopause in women. The decrease in estrogen that occurs after menopause promotes osteoclast formation and bone resorption, leading to PMOP [2]. In China, more than 2.69 million osteoporosis-related fractures occurred in 2015, with women experiencing approximately 3 times more fractures than men. The total number of osteoporosis-related fractures is expected to increase to 4.83 million in 2035 [3]. With continued aging of a large proportion of the population, osteoporosis has become a major public health challenge. Therefore, strategies for the early identification and prevention of osteoporosis are of great clinical significance [4].
Circular RNAs (circRNAs), a type of noncoding RNA, are formed through reverse splicing of the splice donor at the 3′ end and the splice accepter at the 5′ end in pre-mRNA. The covalently closed annular structure of circRNAs is different from the linear structure of mRNAs due to high sequence conservation and stability [5]. In the cytoplasm, most circRNAs are composed of 1 or more exons that originate from protein-coding genes. Some circRNAs have both intron and exon forms and are known as exon-intron circRNAs (EIciRNAs), while others remain as introns due to accidental splicing and are known as circular intronic RNAs (ciRNAs). Both EIciRNAs and ciRNAs are typically located in the nucleus [6]. The widespread application of RNA sequencing (RNA-seq) technology has enabled researchers to begin elucidating the biological functions of circRNAs. An important feature of most circRNAs is their function as miRNA sponges. For example, research has suggested that ciRs-7 (CDR1as) possesses more than 70 binding sites that allow it to function as an inhibitor of miR-7, which influences the expression of miR-7-regulated target genes in cancer [7]. CircIGSF11 contributes to the differentiation of osteoblasts by sponging miR-199b-5p [8]. In addition, circRNAs participate in the regulation of parental gene transcription. For instance, EIciRNAs combine with U1 small nuclear ribonucleoproteins to regulate the activity of RNA polymerase II and promote the expression of parental genes [9]. Researchers have also found that circRNAs act as protein sponges that interact with RNA-binding proteins. CircRNAs with special sites, such as AUG sites and internal ribosome entry sites, may be translated under certain conditions to produce unique circRNA peptides [6,10].
Peripheral blood mononuclear cells (PBMCs) are crucial during the differentiation of osteoclasts, given that they are the precursors of osteoclasts and secret osteoclastogenesis-related cytokines [11]. Osteoclasts are mainly derived from peripheral blood circulatory monocytes migrating to bone [12]. Researchers have demonstrated that PBMCs are related to the bone resorptive activity [13] and that reactive oxygen species signaling in PBMCs can act as therapeutic target in prednisolone-induced osteoporosis [14].
We previously identified 260 circRNAs that are differentially expressed in the PBMCs of PMOP patients, including 154 downregulated circRNAs and 106 upregulated circRNAs [15]. In the present study, we chose the top 4 differentially expressed circRNAs to verify the results of our previous RNA-seq analysis by quantitative polymerase chain reaction (qPCR) analysis. We found that hsa_circ_0021739 was significantly downregulated in PMOP patients and that hsa_circ_0021739 overexpression influenced the differentiation of monocyte-derived osteoclasts.
Material and Methods
Patients and Samples
Fifty-three postmenopausal female patients hospitalized at the Second Affiliated Hospital of Harbin Medical University between September 2018 and August 2019 provided blood samples from which the human PBMCs were collected. According to the diagnostic criteria for PMOP, dual-energy X-ray absorptiometry was used to determine the bone mineral density (BMD) of the femur and lumbar vertebra. The patients were assigned to the control group or PMOP group based on the lumbar or femur BMD T-scores, with cutoff scores of ≥-1.0 and ≤-2.5, respectively, for assignment [16]. None of the patients had diseases such as diabetes, thyroid diseases, or pituitary diseases that could have influenced bone metabolism. Patient age, weight, height, and other clinical data were also collected in this study.
This study received approval from the Second Affiliated Hospital of Harbin Medical University’s Medical Ethics Committee (Number KY2016-187). Informed consent for participation was provided by all patients enrolled.
Isolation of PBMCs
Patient blood samples were processed within 6 h after collection. The density gradient centrifugation technique was applied to isolate mononuclear cells using a Histopaque-1077 (Sigma-Aldrich, USA), as recommended by the manufacturer. The mononuclear cells were immediately used for total RNA extraction or stored in 1 mL of TRIzol reagent at −80°C until use.
Real-Time Reverse-Transcriptase qPCR
circRNAs, miRNAs, and other types of RNA were extracted using TRIzol reagent (Invitrogen, USA). A Transcriptor First-Strand cDNA Synthesis Kit (Roche, Switzerland) was used for reverse transcribing circRNAs into cDNA and a FastStart Universal SYBR Green Master (ROX) Kit (Roche, Switzerland) was used for cDNA amplification. The reverse-transcription reaction system volume for circRNA was 20 μL with random primers, and the reaction procedure was 25°C for 10 min, 55°C for 30 min, 85°C for 5 min, and 4°C for 5 min. The reverse-transcriptase (RT)-qPCR system volume for circRNA was 20 μL, and the amplification procedure was 95°C for 5 min with 1 cycle. The cycle reaction procedure was 95°C for 15 s, 60°C for 15 s, and 72°C for 32 s, for 40 cycles. The melting curve procedure was 95°C for 15 s, 60°C for 60 s, and 95°C for 15 s, for 1 cycle. A miRcute Plus miRNA First-Strand cDNA Kit (TIANGEN, China) was used for miRNA reverse transcription, while a miRcute Plus miRNA qPCR (SYBR Green) Kit (TIANGEN, China) was used to perform RT-PCR. The reverse-transcription reaction system volume for miRNA was 20 μL, and the reaction procedure was 42°C for 60 min, 95°C for 3 min, and 4°C for 5 min. The RT-qPCR system volume for miRNA was 20 μL, and the amplification procedure was 95°C for 5 min, for 1 cycle. The cycle reaction procedure was 95°C for 20 s and 60°C for 34 s, for 40 cycles. The melting curve procedure was 95°C for 15 s, 60°C for 60 s, and 95°C for 15 s, for 1 cycle. The 2−ΔΔCT method was used to determine the fold change in gene expression. The primers for the circRNAs were designed by GENESEED (Guangzhou, China).
Target Prediction
The potential miRNA targets of hsa_circ_0021739 were predicted using Circbank (http://www.circbank.cn/), starBase (http://starbase.sysu.edu.cn/), and Circular RNA Interactome (https://circinteractome.nia.nih.gov/). Binding sites between miRNA and hsa_circ_0021739 were predicted using software based on miRanda (Memorial Sloan Kettering Cancer Center, USA) and RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/).
Cell Culture and Transfection
PBMCs of the healthy postmenopausal female patients were separated from blood samples using a Histopaque-1077 (Sigma-Aldrich, USA). Alpha-minimum essential medium (α-MEM) containing 10% fetal bovine serum was used to culture the mononuclear cells (2.5×106/well in 24-well cluster plates) in a 5% CO2 humidified atmosphere at 37°C. Anchorage-independent lymphocytes were removed during cultivation, and the remaining monocytes were collected for use during transfection. The construction and verification of hsa_circ_0021739 expression in pLO5-ciR were done by GENESEED (Guangzhou, China). The monocytes were transfected using a hsa_circ_0021739-overexpressing plasmid or control plasmid with Lipofectamine 2000 and Opti-MEM (Invitrogen, USA).
Monocyte differentiation into osteoclasts was induced by exchanging the medium with medium supplemented with 25 ng/mL human macrophage colony-stimulating factor (M-CSF, PeproTech, USA) and 40 ng/mL human receptor activator of nuclear factor (NF)-κB ligand (RANKL, PeproTech, USA). Staining with tartrate-resistant acid phosphatase (TRAP) was conducted to observe osteoclast growth.
Statistical Analysis
SPSS version 22.0 and GraphPad Prism version 7.0 were used for all statistical analyses. Variables with a nonnormal distribution are presented as median±quartile and were analyzed using the rank sum test. Normally distributed continuous variables are presented as mean±standard deviation (SD) and were analyzed using t test. The Shapiro-Wilk test and P-P test were used to check for normal distribution. Relative expression levels are presented as ΔCT (ΔCT=target gene CT – internal reference gene CT). Pearson correlation analysis was performed to identify correlations between clinical characteristics and hsa_circ_0021739 levels. Evaluation of the diagnostic value of the circRNAs was carried out by receiver-operating characteristic (ROC) curve analysis. A P value of <0.05 was considered to be statistically significant. According to the sample size estimation calculated using PASS 11 software, we needed at least 23 patients in the PMOP group and 23 in the control group (α=0.05, tolerance error=0.3).
Results
Characteristics of the Participants
The 53 included participants, aged 49–65 years, were assigned to either the PMOP group (n=28) or the control group (n=25). No significant differences in body mass index, height, weight, age, lymphocyte count, and monocyte count were found between the PMOP and control groups (all P>0.05; Table 1). However, significant differences in the T-scores and BMD measurements for the lumbar vertebra, femur, and forearm were observed between the 2 groups (all P<0.001).
Table 1.
Clinical characteristics of the participants.
| Characteristic | Control group (n=25) | PMOP group (n=28) | P value |
|---|---|---|---|
| Age, years | 57.00±5.19 | 59.50±4.37 | 0.063 |
| Height, cm | 161.80±5.02 | 159.27±4.21 | 0.052 |
| Weight, kg | 60.64±9.08 | 56.39±6.74 | 0.057 |
| BMI, kg/m2 | 23.11±2.99 | 22.22±2.37 | 0.233 |
| Lymphocyte count, 109/L | 2.09±0.61 | 1.70±0.62 | 0.137 |
| Monocyte count, 109/L | 0.32±0.18 | 0.26±0.12 | 0.103 |
| T-score | |||
| Lumbar vertebra | −0.31±0.61 | −3.10±0.66 | <0.001* |
| Femur | −0.24±0.60 | −2.74±0.52 | <0.001* |
| Forearm | −0.47±0.84 | −3.18±1.37 | <0.001* |
| BMD, g/cm2 | |||
| Lumbar vertebra | 1.003±0.066 | 0.709±0.064 | <0.001* |
| Femur | 0.813±0.076 | 0.556±0.066 | <0.001* |
| Forearm | 0.655±0.055 | 0.504±0.076 | <0.001* |
Data are presented as mean±SD or median±quartile.
P<0.05.
circRNA Expression in the PMOP and Control Groups
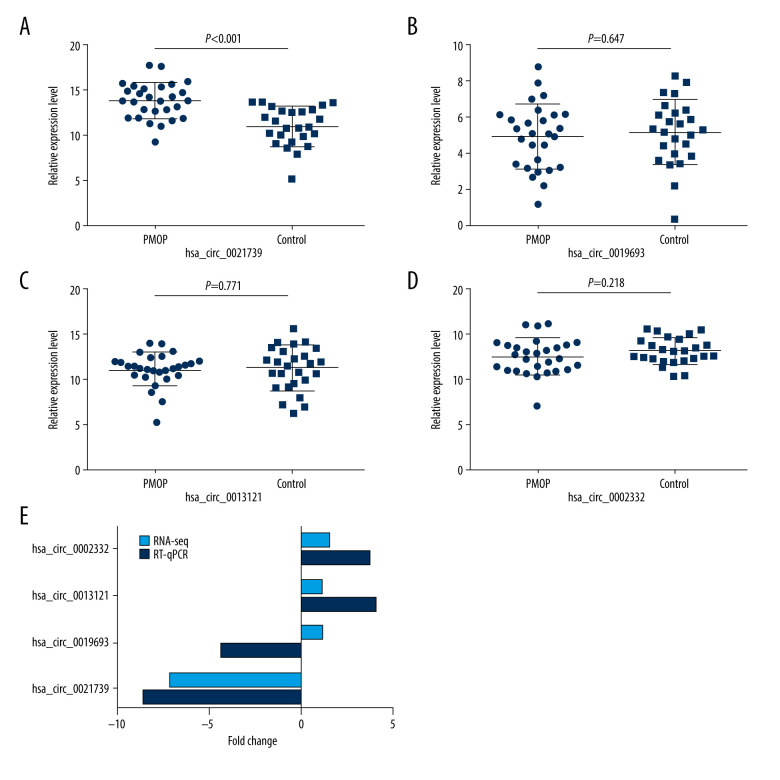
We chose 4 circRNAs, 2 downregulated and 2 upregulated, from among those identified as differentially expression in PMOP patients by RNA-seq analysis in our previous study (Table 2) for measurement by RT-qPCR in the PMOP and control groups. The primers are listed in Table 3. The resulting relative expression levels (∆CT) are shown in Figure 1A–1D. The trends in the RT-qPCR results were identical to those of the RNA-seq results, except for hsa_circ_0019693 (Figure 1E). Moreover, only the results for hsa_circ_0021739 expression showed statistical significance. The actual hsa_circ_0021739 expression level in the PBMCs was determined to be significantly lower for PMOP patients than for controls (P<0.001). No significant difference in expression level was found between the 2 groups for the other 3 circRNAs. Thus, we chose hsa_circ_0021739 for further study.
Table 2.
Differential expression of circRNAs on RNA-seq.
| CircRNA | Regulation | Chromosome | Fold change | P value |
|---|---|---|---|---|
| hsa_circ_0021739 | Down | Chr6 | −8.51 | <0.001 |
| hsa_circ_0019693 | Down | Chr5 | −4.29 | 0.001 |
| hsa_circ_0013121 | Up | Chr20 | 4.08 | 0.003 |
| hsa_circ_0002332 | Up | Chr11 | 3.69 | 0.008 |
Table 3.
Primers for circRNAs and miRNAs.
| RNA | Primer |
|---|---|
| hsa_circ_0021739 | F: 5′-CAATGACAACGCAGCAGAGT-3′ R: 5′-GAGCTGAGGCTGTGCTTGAT-3′ |
| hsa_circ_0019693 | F: 5′-CATCATCCCCATTCCGAAGG-3′ R: 5′-GTGCTGCTCGAGCTTGGCAT-3′ |
| hsa_circ_0013121 | F: 5′-AGTCTCCTGTCGCCGTGGTT-3′ R: 5′-GCCGTGCACTTCCTGGACCA-3′ |
| hsa_circ_0002332 | F: 5′-GTGAGCGTTTGTTGAACAAG-3′ R: 5′-CTATGTCTGGCTCTCCAGCG-3′ |
| GAPDH | F: 5′-CGGATTTGGTCGTATTGGG-3′ R: 5′-CCTGGAAGATGGTGATGGG-3′ |
| hsa-miR-502-5p | F: 5′-ATCCTTGCTATCTGGGTGCTA-3′ |
| U6 | F: 5′-CTCGCTTCGGCAGCACA-3′ |
Figure 1.
The relative expression levels of circRNAs. (A–D) The relative expression levels (ΔCT) of hsa_circ_0021739, hsa_circ_0019693, hsa_circ_0013121, and hsa_circ_0002332 in the postmenopausal osteoporosis group (PMOP; n=28) and healthy postmenopausal group (control; n = 25). (E) Differences (fold change) in RNA-sequencing (RNA-seq) and reverse-transcriptase quantitative polymerase chain reaction (RT-qPCR) results (2−ΔΔCT).
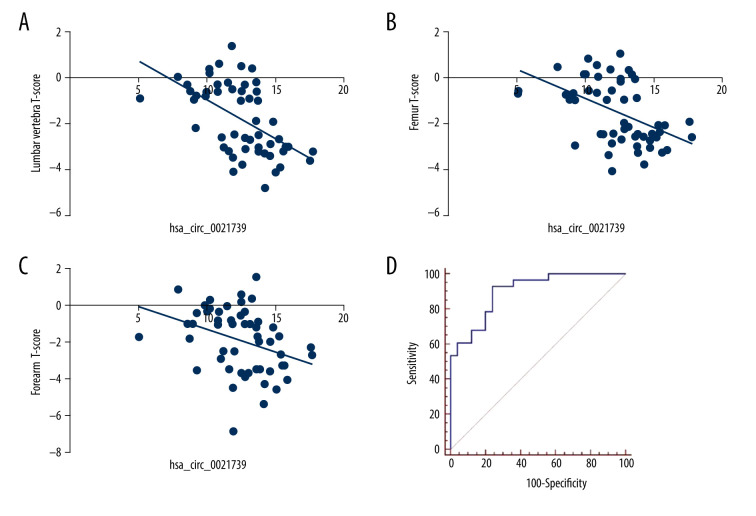
Correlation Between Clinical Characteristics and hsa_circ_0021739
We analyzed the correlations between hsa_circ_0021739 expression and PMOP patients’ clinical characteristics. The hsa_circ_0021739 expression level was determined to be correlated with the lumbar vertebra, femur, and forearm T-scores (P<0.05, Figure 2A–2C). However, no correlation was found between hsa_circ_0021739 expression and other clinical factors, such as age, height, and weight (Table 4).
Figure 2.
Correlation between hsa_circ_0021739 expression and clinical characteristics of patients with postmenopausal osteoporosis (PMOP). (A–C) Correlation between hsa_circ_0021739 expression and T-scores for the lumbar vertebra, femur, and forearm. (D) Receiver-operating characteristic (ROC) curve for hsa_circ_0021739 expression in peripheral blood mononuclear cells (PBMCs) of PMOP patients (area under the curve [AUC]: 0.849, confidence interval [CI]: 0.724–0.932, P<0.001).
Table 4.
Pearson analysis of correlations between hsa_circ_0021739 and clinical characteristics of PMOP patients.
| Characteristic | r | P value |
|---|---|---|
| Age, years | 0.157 | 0.261 |
| Height, cm | −0.264 | 0.057 |
| Weight, kg | −0.219 | 0.116 |
| BMI, kg/m2 | −0.118 | 0.399 |
| Lymphocyte count, 109/L | −0.260 | 0.060 |
| Monocyte count, 109/L | −0.017 | 0.090 |
| T-score | ||
| Lumbar vertebra | −0.542 | <0.001* |
| Femur | −0.459 | <0.001* |
| Forearm | −0.349 | 0.011* |
ROC Curve Analysis of the Diagnostic Value of hsa_circ_0021739 Expression
To determine the diagnostic value of hsa_circ_0021739 expression for PMOP, ROC curve analysis was performed. For the ability of hsa_circ_0021739 expression to detect PMOP, the area under the curve (AUC) was 0.849 (confidence interval [CI] 0.724–0.932, P<0.001, cutoff level 12.51), with a sensitivity of 100% and specificity of 42.9% (Figure 2D).
Hsa_circ_0021739 Regulates Osteoclastogenesis of PBMCs
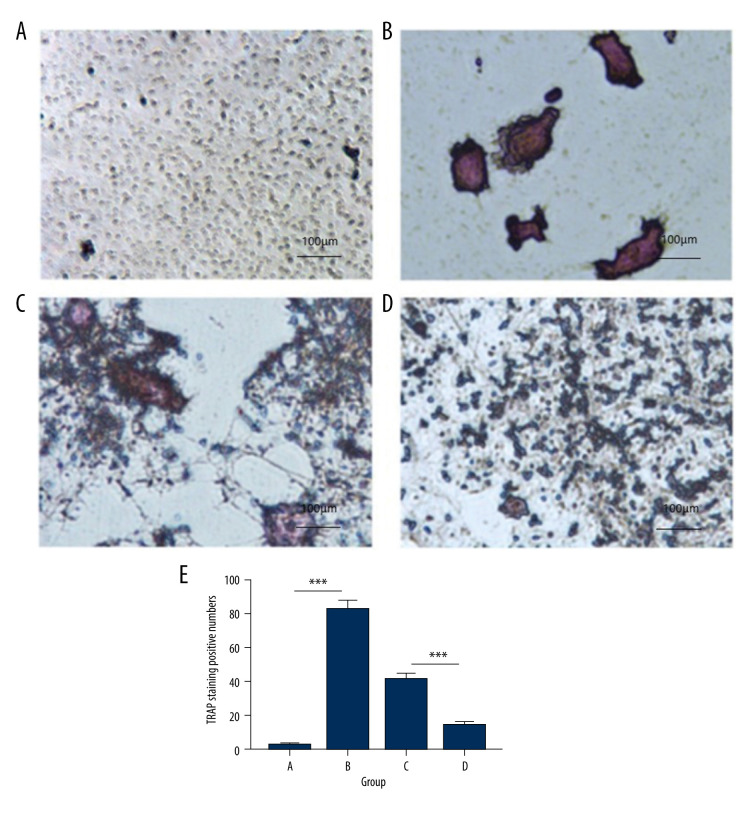
To investigate the role of hsa_circ_0021739 in osteoclast differentiation, monocytes from the patients’ PBMCs were transfected with an hsa_circ_0021739 overexpression vector or an empty vector. After 8 days of treatment with M-CSF and RANKL, the number of TRAP-positive cells (osteoclasts) differed significantly between the PBS control group, the empty vector group, and the hsa_circ_0021739 overexpression group (Figure 3). The number of osteoclasts was significantly decreased in the hsa_circ_0021739 overexpression group.
Figure 3.
Tartrate-resistant acid phosphatase (TRAP) staining of osteoclasts in different groups (magnification, ×400). (A) Control group exposed to phosphate-buffered saline (PBS). (B) Cells exposed to receptor activator of nuclear factor (NF)-κB ligand (RANKL) and macrophage colony-stimulating factor (M-CSF) for osteoclast induction. (C) Empty vector group exposed to RANKL and M-CSF. (D) hsa_circ_0021739 overexpression group exposed to RANKL and M-CSF. (E) Microscopic images showing cell morphology and numbers of TRAP-positive cells. *** P<0.0001.
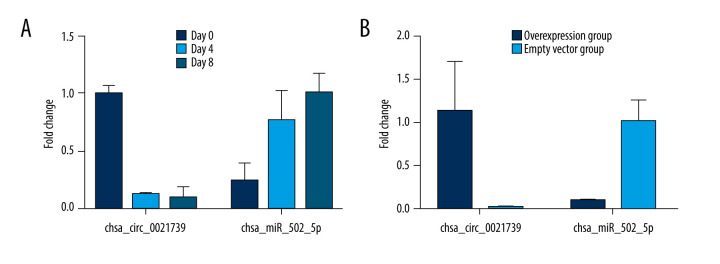
Research has suggested that circRNAs perform a microRNA-absorbing function. To determine whether hsa_circ_0021739 can sponge microRNAs, we used starBase, Circular RNA Interactome, and Circbank to predict potential miRNA targets of hsa_circ_0021739 and miRanda to determine the binding sites of the miRNAs. We found that hsa-miR-502-5p had a higher score (score 158, P=0.0396) and more binding sites than other circRNAs (Figure 4). The hsa-miR-502-5p and hsa_circ_0021739 expression levels were determined during osteoclastogenesis (days 0, 4, and 8) and after transfection using RT-qPCR (Figure 5). The results showed that hsa-miR-502-5p expression increased during osteoclast differentiation, while hsa_circ_0021739 expression visibly decreased. After transfection with the hsa_circ_0021739 overexpression vector, the level of hsa-miR-502-5p was found to be remarkably downregulated. Therefore, hsa-miR-502-5p may be a miRNA target of hsa_circ_0021739.
Figure 4.

Potential binding sites between circRNAs and miRNAs. The possible binding sites between hsa_circ_0021739 and hsa-miR-502-5p were predicted using software based on miRanda and RNAhybrid.
Figure 5.
Expression levels of hsa_circ_0021739 and hsa-miR-502-5p during osteoclastogenesis. (A) Expression levels of hsa_circ_0021739 and hsa-miR-502-5p determined during osteoclastogenesis on the days 0, 4, and 8 using reverse-transcriptase quantitative polymerase chain reaction (RT-qPCR). (B) the expression levels of hsa_circ_0021739 and hsa-miR-502-5p determined during transfection using RT-qPCR.
Discussion
As an important type of noncoding RNA in vivo, circRNAs play a variety of roles in certain biological processes [17]. Along with in-depth studies of circRNAs, relationships between circRNAs and multiple diseases, including cardiovascular disease, cancer, neurodegenerative disorders, and autoimmune disease, have been revealed [17–20]. Some studies have suggested that circRNAs can be used as novel biomarkers for disease prediction. However, the function of circRNAs in the PBMCs of PMOP patients remains unclear. In the present study, 4 differentially expressed circRNAs were selected from our previous study [15], and the relationship between circRNA expression levels and PMOP were verified. In PBMCs of PMOP patients, significant downregulation of hsa_circ_0021739 was detected, and its expression level was positively correlated with the femur, forearm, and lumbar vertebra T-scores. ROC curve analysis indicated that hsa_circ_0021739 expression showed high diagnostic value for PMOP (AUC=0.849). Similarly, previous studies have shown that hsa_circ_0001275 and hsa_circ_0016624 may function as potential new diagnostic biomarkers for PMOP [21,22]. Imbalance between osteoclast and osteoblast numbers can cause bone metabolism disorders, such as osteoporosis and bone metastases [23]. We found that the overexpression of hsa_circ_0021739 inhibited osteoclast differentiation, indicating that hsa_circ_0021739 may perform a molecular function during osteoclastogenesis. At present, no studies have investigated the functions of hsa_circ_0021739.
Sponging of miRNAs is a major function of circRNAs [24]. The binding sites between hsa_circ_0021739 and miRNAs were predicted using software based on miRanda and RNAhybrid to identify the potential function of hsa_circ_0021739. We chose hsa-miR-502-5p for further experiments because it received the highest score. The hsa-miR-502-5p expression levels decreased with increasing hsa_circ_0021739 levels, indicating that hsa-miR-502-5p may be a target gene of hsa_circ_0021739. Huang et al [25] found that hsa-miR-502-5p is a potential novel diagnostic biomarker for osteosarcoma. Several studies have indicated that miRNAs in bone can regulate the activity and differentiation of osteoclasts. For example, miR-27a was found to play a significant role via targeting adenomatous polyposis coli and peroxisome proliferator-activated receptor γ during estrogen-inhibited osteoclast formation [26]. In ovariectomized mice, miR-145 overexpression was found to inhibit osteoclastogenesis by regulating Smad3 expression in RANKL-induced bone marrow-derived macrophages [27]. Medical examination combined with determination of miR-194-5p levels may be a novel approach that could be used to diagnose PMOP [28]. Therefore, the function of hsa-miR-502-5p warrants further investigation.
Conclusions
In conclusion, downregulation of hsa_circ_0021739 was detected in PBMCs of PMOP patients. hsa_circ_0021739 may be able to regulate osteoclastogenesis by targeting hsa-miR-502-5p. Our study determined that hsa_circ_0021739 is a potential biomarker for PMOP. This finding provides new avenues for the development of early prevention and diagnosis methods for osteoporosis.
Footnotes
Conflict of Interests
None.
Source of support: The Financial Scheme for Application Technology Research and Development Project of the Harbin Science and Technology Bureau (2017AB9BS037) supported this study
References
- 1.NIH Consensus Development Panel on Osteoporosis Prevention Diagnosis and Therapy. Osteoporosis prevention, diagnosis, and therapy. JAMA. 2001;285:785–95. doi: 10.1001/jama.285.6.785. [DOI] [PubMed] [Google Scholar]
- 2.Feng J, Liu S, Ma S, et al. Protective effects of resveratrol on postmenopausal osteoporosis: Regulation of SIRT1-NF-kappaB signaling pathway. Acta Biochim Biophys Sin (Shanghai) 2014;46:1024–33. doi: 10.1093/abbs/gmu103. [DOI] [PubMed] [Google Scholar]
- 3.Si L, Winzenberg TM, Jiang Q, et al. Projection of osteoporosis-related fractures and costs in China: 2010–2050. Osteoporos Int. 2015;26:1929–37. doi: 10.1007/s00198-015-3093-2. [DOI] [PubMed] [Google Scholar]
- 4.Anthamatten A, Parish A. Clinical update on osteoporosis. J Midwifery Womens Health. 2019;64:265–75. doi: 10.1111/jmwh.12954. [DOI] [PubMed] [Google Scholar]
- 5.Qu S, Yang X, Li X, et al. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–48. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–91. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 7.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–12. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 8.Zhang M, Jia L, Zheng Y. circRNA expression profiles in human bone marrow stem cells undergoing osteoblast differentiation. Stem Cell Rev Rep. 2019;15:126–38. doi: 10.1007/s12015-018-9841-x. [DOI] [PubMed] [Google Scholar]
- 9.Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–64. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 10.Luo J, Liu H, Luan S, Li Z. Guidance of circular RNAs to proteins’ behavior as binding partners. Cell Mol Life Sci. 2019;76:4233–43. doi: 10.1007/s00018-019-03216-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salamanna F, Maglio M, Borsari V, et al. Peripheral blood mononuclear cells spontaneous osteoclastogenesis: Mechanisms driving the process and clinical relevance in skeletal disease. J Cell Physiol. 2016;231:521–30. doi: 10.1002/jcp.25134. [DOI] [PubMed] [Google Scholar]
- 12.Parfitt AM. The bone remodeling compartment: a circulatory function for bone lining cells. J Bone Miner Res. 2001;16:1583–85. doi: 10.1359/jbmr.2001.16.9.1583. [DOI] [PubMed] [Google Scholar]
- 13.Manrique E, Castillo LM, Lazala O, et al. Bone resorptive activity of human peripheral blood mononuclear cells after fusion with polyethylene glycol. J Bone Miner Metab. 2017;35:127–41. doi: 10.1007/s00774-016-0744-0. [DOI] [PubMed] [Google Scholar]
- 14.Shymanskyy IO, Lisakovska OO, Mazanova AO, et al. Prednisolone and vitamin D(3) modulate oxidative metabolism and cell death pathways in blood and bone marrow mononuclear cells. Ukr Biochem J. 2016;88:38–47. doi: 10.15407/ubj88.05.038. [DOI] [PubMed] [Google Scholar]
- 15.Jin D, Wu X, Yu H, et al. Systematic analysis of lncRNAs, mRNAs, circRNAs and miRNAs in patients with postmenopausal osteoporosis. Am J Transl Res. 2018;10:1498–510. [PMC free article] [PubMed] [Google Scholar]
- 16.Chinese Society of Osteoporosis and Bone Mineral Research and Chinese Medical Association. A Guide to Diagnosis and Treatment of Primary Osteoporosis (2017 Edition) Chin J Osteoporosis Bone Miner Res. 2017;10:413–43. [Google Scholar]
- 17.Liu J, Li D, Luo H, Zhu X. Circular RNAs: The star molecules in cancer. Mol Aspects Med. 2019;70:141–52. doi: 10.1016/j.mam.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 18.D’Ambra E, Capauto D, Morlando M. Exploring the regulatory role of circular RNAs in neurodegenerative disorders. Int J Mol Sci. 2019;20(21):5477. doi: 10.3390/ijms20215477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li JJ, Wang W, Wang XQ, et al. A novel strategy of identifying circRNA biomarkers in cardiovascular disease by meta-analysis. J Cell Physiol. 2019;234:21601–12. doi: 10.1002/jcp.28817. [DOI] [PubMed] [Google Scholar]
- 20.Nemtsova MV, Zaletaev DV, Bure IV, et al. Epigenetic changes in the pathogenesis of rheumatoid arthritis. Front Genet. 2019;10:570. doi: 10.3389/fgene.2019.00570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao K, Zhao Q, Guo Z, et al. Hsa_Circ_0001275: a potential novel diagnostic biomarker for postmenopausal osteoporosis. Cell Physiol Biochem. 2018;46:2508–16. doi: 10.1159/000489657. [DOI] [PubMed] [Google Scholar]
- 22.Yu L, Liu Y. circRNA_0016624 could sponge miR-98 to regulate BMP2 expression in postmenopausal osteoporosis. Biochem Biophys Res Commun. 2019;516:546–50. doi: 10.1016/j.bbrc.2019.06.087. [DOI] [PubMed] [Google Scholar]
- 23.Suva LJ, Washam C, Nicholas RW, Griffin RJ. Bone metastasis: mechanisms and therapeutic opportunities. Nat Rev Endocrinol. 2011;7:208–18. doi: 10.1038/nrendo.2010.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205–11. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 25.Huang C, Wang Q, Ma S, et al. A four serum-miRNA panel serves as a potential diagnostic biomarker of osteosarcoma. Int J Clin Oncol. 2019;24:976–82. doi: 10.1007/s10147-019-01433-x. [DOI] [PubMed] [Google Scholar]
- 26.Guo L, Chen K, Yuan J, et al. Estrogen inhibits osteoclasts formation and bone resorption via microRNA-27a targeting PPARgamma and APC. J Cell Physiol. 2018;234:581–94. doi: 10.1002/jcp.26788. [DOI] [PubMed] [Google Scholar]
- 27.Yu FY, Xie CQ, Sun JT, et al. Overexpressed miR-145 inhibits osteoclastogenesis in RANKL-induced bone marrow-derived macrophages and ovariectomized mice by regulation of Smad3. Life Sci. 2018;202:11–20. doi: 10.1016/j.lfs.2018.03.042. [DOI] [PubMed] [Google Scholar]
- 28.Ding H, Meng J, Zhang W, et al. Medical examination powers miR-194-5p as a biomarker for postmenopausal osteoporosis. Sci Rep. 2017;7:16726. doi: 10.1038/s41598-017-17075-w. [DOI] [PMC free article] [PubMed] [Google Scholar]