Abstract
Cross-species transmission of emerging viruses happens occasionally due to epidemiological, biological, and ecological factors, and it has caused more concern recently. Covert mortality nodavirus (CMNV) was revealed to be a unique shrimp virus that could cross species barrier to infect vertebrate fish. In the present study, CMNV reverse transcription-nested PCR (RT-nPCR)-positive samples were identified from farmed sea cucumber (Apostichopus japonicas) in the CMNV host range investigation. The amplicons of RT-nPCR from sea cucumber were sequenced, and its sequences showed 100% identity with the RNA-dependent RNA polymerase gene of the original CMNV isolate. Histopathological analysis revealed pathologic changes, including karyopyknosis and vacuolation of the epithelial cells, in the sea cucumber intestinal tissue. The extensive positive hybridization signals with CMNV probe were shown in the damaged epithelial cells in the in situ hybridization assay. Meanwhile, transmission electron microscopy analysis revealed CMNV-like virus particles in the intestine epithelium. All the results indicated that the sea cucumber, an Echinodermata, is a new host of CMNV. This study supplied further evidence of the wide host range of CMNV and also reminded us to pay close attention to its potential risk to threaten different aquaculture animal species.
Keywords: covert mortality nodavirus (CMNV), natural infection, sea cucumber (Apostichopus japonicas), in situ hybridization, TEM
1. Introduction
For most viruses, the virus–receptor interactions determine viral host range and therefore constitute the interspecies barrier of viral infection, eventually leading to the strong host specificity of a virus [1,2]. However, the cross-species transmission of emerging viruses happens occasionally due to a variety of epidemiological, biological, and ecological factors [3,4,5]. Furthermore, RNA viruses more easily cross species boundaries on account of the lack of exonuclease proofreading activity and easy variation [6], such as coronaviruses [7], the avian influenza virus [8], and the rabies virus [9].
Nodaviridae is composed of two genera, Alphanodavirus and Betanodavirus [10]. Alphanodaviruses were mostly isolated from insects and their host range also appears to be restricted to insects [11], except for Nodamura virus (NoV) and Flock House virus (FHV) [12,13]. NoV was originally isolated from mosquitoes (Culex tritaeniorhynchus) [14]; however, it could also lethally infect mammals, including suckling mice and suckling hamsters [10,15]. FHV, another alphanodavirus isolated from Costelytra zealandica (Coleoptera: Scarabaeidae), is capable of replicating in many species of plants, including chenopodium Chenopodium hybridum, barley Hordeum vulgare, and tobacco Nicotiana tabacum [12,16]. Therefore, the hosts of NoV and FHV are not only confined to insects, and they also possess the capacity of cross-species transmission as well. However, Betanodavirus mainly infects larvae, juvenile or adult marine fish, and is different from Alphanodavirus [17,18,19].
Covert mortality nodavirus (CMNV) is an alphanodavirus first isolated from shrimp with viral covert mortality disease (VCMD) in China and later also found in Thailand and Ecuador [20,21,22,23]. It can also infect various major farmed shrimp, including Penaeus vannamei, Penaeus chinensis, Marsupenaeus japonicus, and Penaeus monodon, and VCMD outbreaks in shrimp farms represent a significant threat to the shrimp culture industry [21]. Additionally, other crustaceans (including the hyperiid amphipod Parathemisto gaudichaud, amphipod Corophium sinense Zhang, and the ghost crab Ocypode cordimandus) and several fish species (including gobiid fish Mugilogobius abei, Japanese flounder Paralichthys olivaceus, and goldfish Carassius auratus) are also natural hosts of CMNV. And the host range spanning crustaceans and fish illustrates the capacity of CMNV to spread crossing the species barrier [24,25,26].
Sea cucumber (Holothuians) is a common marine invertebrate [27], and it is the generic term for Holothuroidea, which belongs to the invertebrate Echinodermata. Apostichopus japonicus is the sea cucumber species with the largest social demand, and its aquaculture has become an emerging marine industry [28,29,30]. In a systematic investigation of CMNV natural hosts and vectors, CMNV reverse transcription-nested PCR (RT-nPCR)-positive sea cucumber individuals were accidentally found in shrimp farming ponds. In this study, we describe here the outcome of the detection of CMNV in sea cucumber by RT-nPCR, in situ hybridization (ISH), histopathology, and transmission electron microscopy (TEM). Our study provides significant novel insights into the new natural host discovery of CMNV.
2. Materials and Methods
2.1. Sample Collection
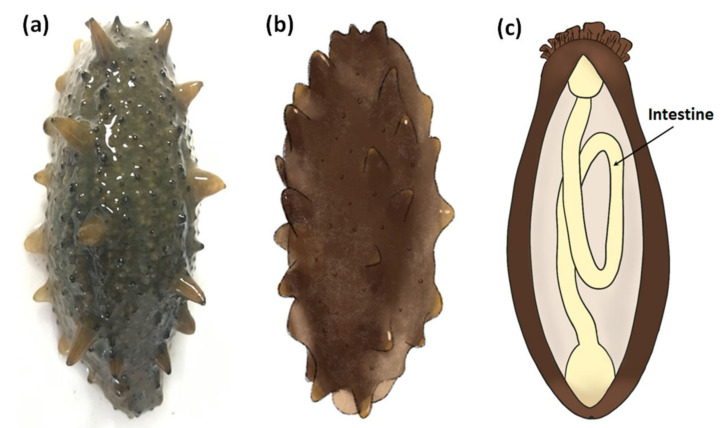
In June 2018, we found that the farming P. vannamei collected from polyculture ponds of shrimp and sea cucumber was infected with CMNV. Considering the capacity of CMNV cross-species infection and the needs of further exploration of its host range, four sea cucumber individuals (length 7–8 cm, co-inhabiting with the P. vannamei in the same polyculture pond) (Figure 1a,b) were randomly collected for CMNV detection. The collected sea cucumber individuals looked normal, but the body was not compacted enough (somewhat soft) compared with healthy individuals. These sea cucumbers were primarily examined after dissecting the body along the longitudinal axis, and the thinning intestinal tissue (Figure 1c) was selected to be divided into three parts and preserved. One part was preserved in 4% paraformaldehyde solution in PBS (PFA-PBS) (Sinopharm, Beijing, China) for ISH detection and histopathological analysis. Another part was fixed in 2.5% glutaraldehyde solution (Solarbio, Beijing, China) for electron microscopic examinations. Residual intestinal tissues were preserved in RNAstore solution (Tiangen, Beijing, China) for molecular biological analysis.
Figure 1.
The topography and schematic diagram of the intestine of sea cucumber (Apostichopus japonicas) in this study. (a) Topography of sea cucumber collected from the polyculture ponds infected with covert mortality nodavirus (CMNV). (b) Schematic diagram of sea cucumber. (c) Schematic diagram of the longitudinal section of sea cucumber. Intestinal tissue is pointed by the black arrow.
2.2. Total RNA Extraction
The total RNA of intestinal tissues was extracted from four sea cucumber individuals using RNAiso Plus Reagent (Takara, Dalian, China) according to the manufacturer’s instructions. The tissues samples were first homogenized in RNAiso Plus, and then trichloromethane was added into the homogenate for protein degeneration. Finally, isopropanol was used to get total RNA from liquid supernatant obtained by centrifugation. The concentration and purity of purified RNA were measured by Nanodrop 2000 (Thermo Scientific, Waltham, MA, USA).
2.3. Reverse Transcription-Nested PCR (RT-nPCR)
The total RNA from the intestinal tissue of sea cucumber was used as a template for RT-PCR analysis. First, the first-step PCR amplification was conducted by using the template of 1 μL total RNA (concentration of the template was 100–200 ng/μL) and a PrimeScript One Step RT-PCR Kit (TaKaRa, Dalian, China) with the primer sets of CMNV-F1/R1 (CMNV-F1: 5′-AAATACGGCGATGACG-3′, CMNV-R1: 5′-ACGAAGTGCCCA-CAGAC-3′) according to the recommended procedures, and the annealing temperature was 52 °C. Using the first-step RT-PCR products as templates, the second-step PCR was carried out using a TaKaRa Ex Taq Kit (TaKaRa, Dalian, China) with the primer sets of CMNV-D-F1/R1 (CMNV-D-F1: 5′-TCGCGTATTCGTGGAT-3′, CMNV-D-R1: 5′-TAGGGTCAAAAGGTGTAGT-3′), and the annealing temperature was 52 °C. The expected CMNV target fragments of the first and second rounds of the PCR amplifications were 619 bp and 413 bp amplicons from the CMNV RNA-dependent RNA polymerase (RdRp) gene, respectively. Then, the amplicons were resolved by 2% agarose gel electrophoresis for 0.5 h.
2.4. Sequence Alignment and Phylogenetic Tree Analysis
The amplicons (413 bp) from the second step of the RT-nPCR of sea cucumber were sent for commercial sequencing to Sangon Biological Engineering (Shanghai, China) Co. Ltd. The obtained 413 bp RdRp gene fragment sequences were subjected to multiple sequence alignment by the online software of BLASTn (https://www.ncbi.nlm.nih.gov/, accessed on 4 February 2021). Then, the phylogenetic tree, based on 25 relevant RdRp protein sequences retrieved from the GenBank database (Table 1) and the deduced amino acids sequence of the 413 bp gene fragment, was constructed by using the software MEGA 6.0 [31]. The tree was finally optimized through the online tool of iTOL (https://itol.embl.de/, accessed on 4 February 2021).
Table 1.
Names and abbreviations for viral species of Nodaviridae.
| Virus | Abbreviation | GenBank No. * |
|---|---|---|
| Covert mortality nodavirus | CMNV | AIL48199.1 |
| Flock House virus | FHV | AEQ39075.1 |
| Gungahlin Chrysomya noda-like virus | GCNV | QIJ70031.1 |
| Newington virus | NeV | AMO03244.1 |
| Drosophila melanogaster American nodavirus (ANV) strain SW-2009a | DmANV-SW-2009a | ACU32794.1 |
| Black beetle virus | BBV | YP_053043.1 |
| Wenzhou noda-like virus 6 strain | Wenzhou NLV-6 | APG76600.1 |
| Hubei noda-like virus 21 strain | Hubei NLV-21 | APG76486.1 |
| Hubei noda-like virus 22 strain | Hubei NLV-22 | APG76466.1 |
| Boolarra virus | BoV | NP_689439.1 |
| Nodamura virus | NoV | NP_077730.1 |
| Shuangao insect virus 11 strain | SIV-11 | YP_009337806.1 |
| Wenzhou noda-like virus 7 strain | Wenzhou NLV-7 | APG76642.1 |
| Beihai noda-like virus 25 strain | Beihai NLV-25 | APG76164.1 |
| Beihai mantis shrimp virus 6 strain | BMSV-6 | YP_009333376.1 |
| Macrobrachium rosenbergii nodavirus | MrNV | AAQ83832.1 |
| Beihai noda-like virus 30 strain | Beihai NLV-30 | APG76125.1 |
| Striped jack nervous necrosis virus | SJNNV | NP_599247 |
| Golden pompano nervous necrosis virus | GPNNV | ACX54065 |
| Redspotted grouper nervous necrosis virus | RGNNV | ACX69744 |
| Dragon grouper nervous necrosis virus | DGNNV | AAU85148 |
| Tiger puffer nervous necrosis virus | TPNNV | YP_00328875 |
| Atlantic cod nodavirus | ACNV | ABR23192 |
| Atlantic halibut nodavirus | AHNV | AAY34458 |
| Barfin flounder nervous necrosis virus | BFNNV | YP_003288756 |
GenBank No.* indicate the GenBank accession numbers of the amino acid sequence of RNA-dependent RNA polymerase used in this study.
2.5. In Situ Hybridization (ISH) and Histopathological Analysis
Fixation, dehydration and paraffin embedding of the tissue samples were conducted following the histological method reported by Bell and Lightner [32]. Two paraffin-embedded sections (3 µm) were prepared. One of the sections was subjected to CMNV ISH analysis according to the published papers [21,25], and the other section was stained with routine hematoxylin and eosin-phloxine (H&E) according to previously described procedures [33]. The ISH sections were counterstained using the Nuclear Fast Red solution (Solarbio, Beijing, China) [34]. Finally, the sections of ISH detection and H&E staining were analyzed under the Nikon Eclipse E80i microscope (Nikon Co., Tokyo, Japan), and the image acquisition was accomplished through the slide scanning system of Pannoramic MIDI (3DHISTECH Ltd., Budapest, Hungary).
2.6. Transmission Electron Microscopy Analysis
To detect the presence of CMNV particles in sea cucumber, the intestinal tissues (approximately 1 mm3) were first preserved in 2.5% glutaraldehyde for 24 h at 4 °C, and then further fixed in 1% osmium tetroxide for 2 h. Finally, the tissues were embedded in plastic resin [35,36]. An ultramicrotome (Leica EM UC7) was used to prepare ultrathin sections (50 nm) of a resin block, and the obtained sections were stained with uranyl acetate and lead citrate [37,38]. Eventually, all the sections were examined using a JEOL JEM-1200 electron microscope (equipped with a field emission gun).
3. Results
3.1. Detection of CMNV in Sea Cucumber by RT-nPCR
Two out of the four RNA samples from the sea cucumbers produced the expected amplicons (413 bp) in the RT-nPCR assay (Figure 2a). In addition, the 413 bp amplicons of the second-step PCR were then sequenced to confirm the exact sequence information of RdRp-targeted fragments.
Figure 2.
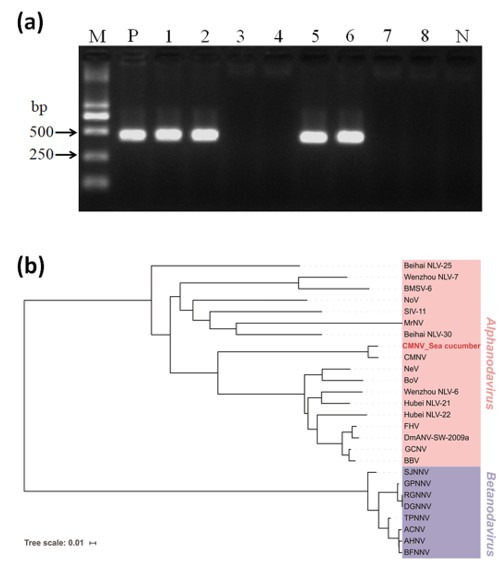
Electrophoretogram of amplicons from the CMNV RT-nPCR assay and phylogenetic analysis. (a) Electrophoretogram of amplicons from the second-step PCR of the CMNV RT-nPCR assay. M: DL2000 molecular weight marker. Lanes 1–8: the PCR products of four RNA samples from sea cucumbers’ intestines (each sample was done with two replicates; samples 1, 2, 3, and 4 were shown in lanes 1 and 2, 3 and 4, 5 and 6, 7 and 8, respectively). P: positive control; N: negative control. (b) Analysis of the phylogenetic tree based on the deduced amino acid sequences of the RdRp gene from the CMNV-positive sea cucumber sample and other nodaviruses (abbreviations of other viruses shown in Table 1). The CMNV isolate of the sea cucumber sampled from farming ponds is highlighted in red. Viral species of Alphanodavirus genus and Betanodavirus genus are shown in pink and lavender background, respectively. The phylogeny tree was derived using the neighbor-joining method by the MEGA 6.0 program. The scale bar is 0.01.
3.2. Phylogenetic Analyses
The result of multiple sequence alignment based on the 413 bp amplicons (413 bp, GenBank No. MW678771) from the second-step PCR showed that the CMNV sequences of sea cucumber shared 100% identity with the known CMNV RdRp gene (GenBank no. KM112247). Phylogenetic analysis revealed that the deduced amino acid sequences of CMNV RdRp from sea cucumber samples were clustered closely into the embranchment of the primordial CMNV isolate (Figure 2b). Additionally, the phylogenetic tree showed that the CMNV RdRp fragment sequences from sea cucumber were clustered into Alphanodavirus, and that members from Betanodavirus were clustered into the other independent branch (Table 1).
3.3. Detection of CMNV in Sea Cucumber by ISH and Histopathological Analysis
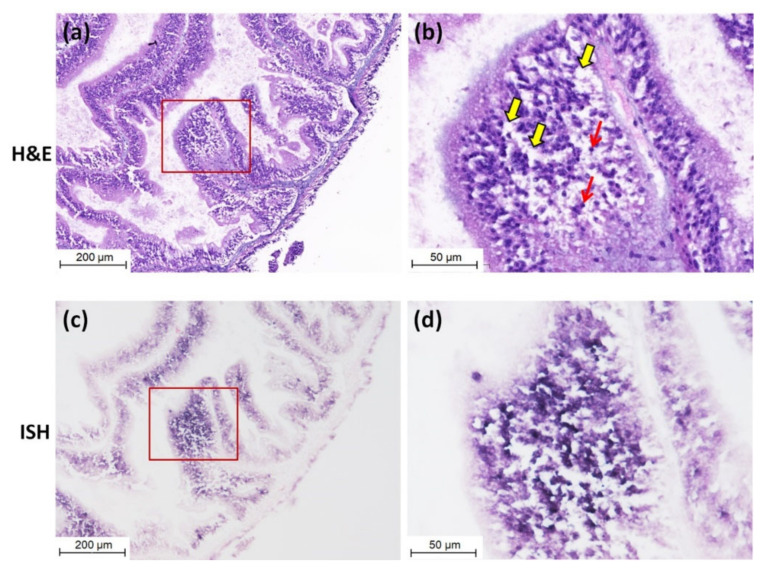
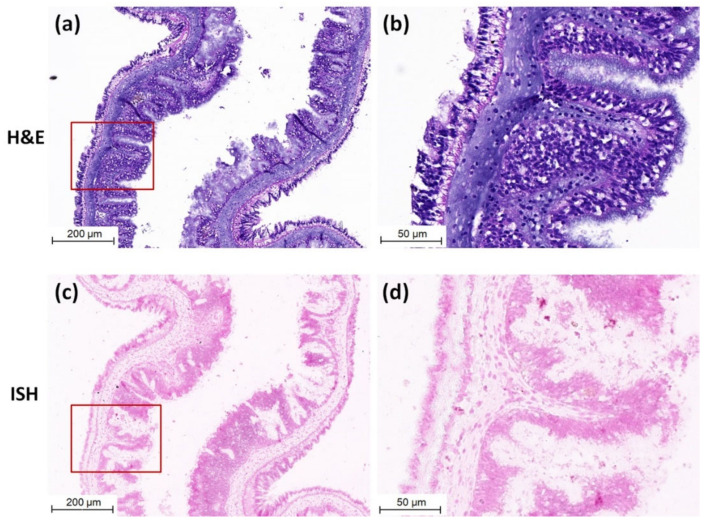
Purple positive hybridization signals of CMNV probe were widely present in the intestine of CMNV RT-nPCR-positive sea cucumber (Figure 3c), and especially more pronounced in intestinal epithelial cells (Figure 3d). Meanwhile, histological examination revealed obvious histopathological lesions in the same sites, such as villi decreasing, villus epithelium cells becoming uncompact and acidophilic (comparing with the normal villus epithelium cells), karyopyknosis (red arrows) and extensive vacuolation (yellow arrows) in the intestinal epithelial cells (Figure 3a,b). The result of ISH and histopathological analysis of the intestine of sea cucumber of negative control is shown in Figure 4a–d. No obvious histopathological changes and CMNV-positive hybridization signals were observed in the intestinal tissue (Figure 4c,d).
Figure 3.
Micrographs of hematoxylin and eosin-phloxine (H&E) staining and in situ hybridization (ISH) of the intestine of sea cucumber naturally infected with CMNV. (a) Micrographs of H&E staining of the intestine. (b) Magnified micrographs from the red-framed areas of (a). Karyopyknosis (red arrows) and extensive vacuolation (yellow arrows) were observed in the intestinal epithelial cells. (c) Micrographs of ISH of the intestine. (d) Magnified micrographs from the red-framed areas of (c). Intense CMNV positive hybridization signals (colored deep-purple) were detected in the intestinal epithelial cells. Scale bars: (a,c) 200 µm, (b,d) 50 µm.
Figure 4.
Micrographs of H&E staining and ISH of the intestinal tissue of sea cucumber of negative control. (a) Micrographs of H&E staining of negative control intestinal tissue. (b) Magnified micrographs from the red-framed areas of (a). No obvious histopathological change was observed in the intestinal tissue. (c) Micrographs of ISH of negative control intestinal tissue. (d) Magnified micrographs from the red-framed areas of (c). No CMNV positive hybridization signal was detected in the intestinal tissue. Scale bars: (a,c) 200 µm, (b,d) 50 µm.
3.4. Detection of CMNV in Sea Cucumber by TEM Analysis
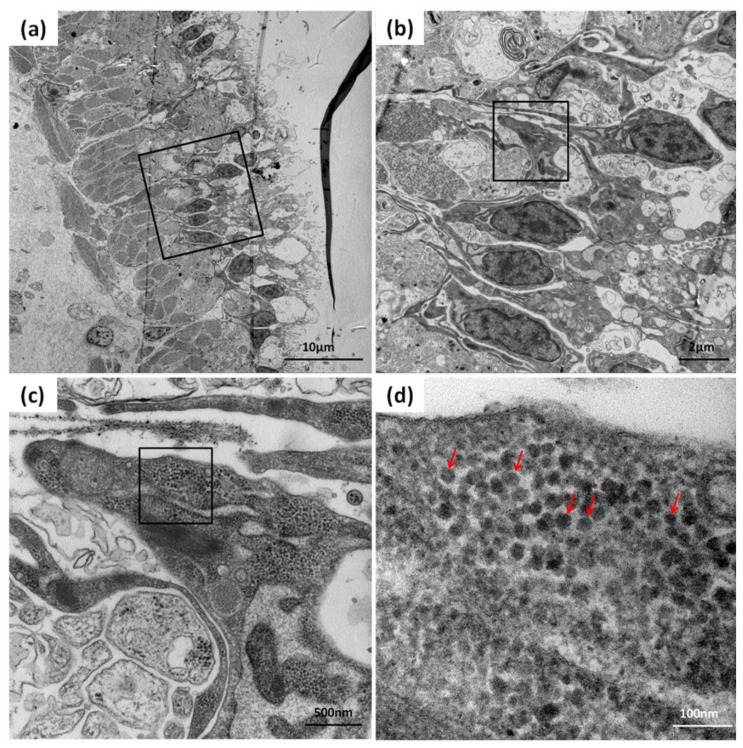
TEM analysis was used to further confirm the CMNV infection in the sea cucumber. TEM micrographs of the ultrathin sections of the intestinal tissue of sea cucumber revealed the presence of massive CMNV-like virus particles of about 28–32 nm in the intestinal villus epithelium cells (Figure 5a–d).
Figure 5.
TEM micrographs of an ultrathin section of the intestinal epithelial cells of sea cucumber from the farm infected with CMNV. (b–d) show magnified micrographs in the black-framed areas of (a–c), respectively. Note the scattering of CMNV-like virus particles (red arrows) at the intestinal villus epithelium cells. Scale bars: (a) 10 µm, (b) 2 µm, (c) 500 nm, (d) 100 nm.
4. Discussion
Recently, the phenomenon of cross-species transmission of a few viruses has negatively affected areas such as human life, animal husbandry, and aquaculture and, meanwhile, also piqued the concern of scholars [25,39,40]. Notably, CMNV, an RNA virus isolated from invertebrate shrimp, can cross the host barrier to infect vertebrate fish, including the marine fishes gobiid fish Mugilogobius abei and Japanese flounder Paralichthys olivaceus [24,25], as well as the freshwater fish goldfish Carassius auratus [25]. CMNV infection in fish reminds that we should pay attention to the potential ability of CMNV to cross more species barriers. Hence, it makes sense to strengthen the investigation of the host range of CMNV. In this study, we proved that sea cucumber is a new natural host of CMNV—something that, to the best of our knowledge, has not been reported before in echinoderms.
CMNV-positive individuals were found in the farming sea cucumber by RT-nPCR. Thus, a further investigation was performed to confirm the infection of CMNV in sea cucumber. The histopathological analysis revealed that obvious pathological lesions, including karyopyknosis and vacuolation of the epithelial cells, occur in the sea cucumber intestinal tissue, and these lesions are similar to the intestinal lesions of gobiid fish Mugilogobius abei infected with CMNV [24]. Consistently, extensive positive hybridization signals of the CMNV probe were also revealed in the damaged epithelial cells in the ISH assay. Meanwhile, the presence of CMNV-like particles in lesion sites of the intestine was also confirmed by TEM analysis. In addition, it was found that the amplicons of RT-nPCR from sea cucumber were highly identical to the CMNV original RdRp gene. Therefore, the above results demonstrated that the sea cucumber could be infected by CMNV and turned into a new natural host of CMNV. In addition, sea cucumber has been continually attacked by disease in recent years [41,42,43], and whether CMNV is one of the important pathogens should be of concern.
During the past decade, the rapid and disorderly expansion of sea cucumber aquaculture was companied by various diseases that caused high mortality of sea cucumbers, resulting in grievous economic losses to the culture industry of sea cucumber [44,45]. Frequent outbreaks of disease has obviously hindered the sustainable development of sea cucumber culture industry. The studies on infectious agents of sea cucumber have been frequently reported in the past 10 years [46]. For many of these disease events, most research has focused on bacterial pathogens such as Vibrio harveyi [47], Vibrio splendidus [48], and Lactococcus garviaeae [49], while studies on viral pathogens have been relatively rare. Some scholars successfully isolated DNA viruses from diseased sea cucumbers, reproduced the disease symptoms, and speculated that the virus might be one of the pathogens causing diseases [46,50,51]. However, to date, no virus has been definitely identified and named. The discovery of CMNV in sea cucumber might provide more possibility and theoretical basis for the research of viral pathogens in diseased sea cucumber.
It is known that sea cucumber, a scavenger, is the pivotal component of the marine environment [52], playing an important role in the recycling of marine ecosystems [53]. In addition, sea cucumber is considered an important aquaculture species in East Asian countries because it has high nutritional value and is rich in multiple nutrients and biologically active substances [54,55,56,57]. Because of increased market demand, sea cucumber culture has become one of the pillar industries of China’s aquaculture industry [58,59]. The natural infection of CMNV in sea cucumber reveals the potential risk of CMNV to the maintaining of sea cucumber natural resources in marine ecosystems, as well as to the aquaculture of the increasingly important species.
5. Conclusions
Overall, the results of this study demonstrated that CMNV is a broad host range virus that deserves close attention to its potential risk to infect more aquaculture animal species. Considering the key ecological function of sea cucumber in the marine environment and the high economic value of this aquatic species, the potential negative impact of the spread and prevalence of CMNV in sea cucumber on the marine ecosystem and social economy should be of concern. The present study contributes to further investigation of the viral pathogen of diseased sea cucumber.
Author Contributions
C.W., C.L. and Q.Z. designed this experiment. W.W. drew the diagram of sea cucumber. C.W. completed the experiments and organized the data. Q.Z., L.Y., S.S. and J.H. collected the samples used in this study. Q.Z. conducted the transmission electron microscopy analysis. C.W. wrote the manuscript, and C.L. and Q.Z. revised it. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National R&D Program of China (2017YFC1404503), National Natural Science Foundation of China (32073016), Central Public-interest Scientific Institution Basal Research Fund, CAFS (No. 2020TD39), Project of Species Conservation from the MARA-Marine Fisheries Resources Collection and Preservation, and Central Public-interest Scientific Institution Basal Research Fund, YSFRI, CAFS (No. 20603022020005).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Schneider-Schaulies J. Cellular receptors for viruses: Links to tropism and pathogenesis. J. Gen. Virol. 2000;81:1413–1429. doi: 10.1099/0022-1317-81-6-1413. [DOI] [PubMed] [Google Scholar]
- 2.Kuiken T., Holmes E.C., McCauley J., Rimmelzwaan G.F., Williams C.S., Grenfell B.T. Host Species Barriers to Influenza Virus Infections. Science. 2006;312:394–397. doi: 10.1126/science.1122818. [DOI] [PubMed] [Google Scholar]
- 3.Leopardi S., Holmes E.C., Gastaldelli M., Tassoni L., Priori P., Scaravelli D., Zamperin G., De Benedictis P. Interplay between co-divergence and cross-species transmission in the evolutionary history of bat coronaviruses. Infect. Genet. Evol. 2018;58:279–289. doi: 10.1016/j.meegid.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parrish C.R., Holmes E.C., Morens D.M., Park E.-C., Burke D.S., Calisher C.H., Laughlin C.A., Saif L.J., Daszak P. Cross-Species Virus Transmission and the Emergence of New Epidemic Diseases. Microbiol. Mol. Biol. Rev. 2008;72:457–470. doi: 10.1128/MMBR.00004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson C.K., Hitchens P.L., Evans T.S., Goldstein T., Thomas K., Clements A., Joly D.O., Wolfe N.D., Daszak P., Karesh W.B., et al. Spillover and pandemic properties of zoonotic viruses with high host plasticity. Sci. Rep. 2015;5:14830. doi: 10.1038/srep14830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holmes E.C. The Evolution and Emergence of RNA Viruses. Oxford University Press; Oxford, UK: 2009. [Google Scholar]
- 7.Wang L.-F., Anderson D.E. Viruses in bats and potential spillover to animals and humans. Curr. Opin. Virol. 2019;34:79–89. doi: 10.1016/j.coviro.2018.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bourret V., Lyall J., Frost S.D.W., Teillaud A., Smith C.A., LeClaire S., Fu J., Gandon S., Guérin J.-L., Tiley L.S. Adaptation of avian influenza virus to a swine host. Virus Evol. 2017;3:vex007. doi: 10.1093/ve/vex007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding N.-Z., Xu D.-S., Sun Y.-Y., He H.-B., He C.-Q. A permanent host shift of rabies virus from Chiroptera to Carnivora associated with recombination. Sci. Rep. 2017;7:1–9. doi: 10.1038/s41598-017-00395-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Owens F., Di Serio F., Li S., Pallás V., Randles J., Sano V. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Academic Press; San Diego, CA, USA: 2012. pp. 1221–1234. [Google Scholar]
- 11.Johnson K.L., Price B.D., Ball L.A. Recovery of Infectivity from cDNA Clones of Nodamura Virus and Identification of Small Nonstructural Proteins. Virology. 2003;305:436–451. doi: 10.1006/viro.2002.1769. [DOI] [PubMed] [Google Scholar]
- 12.Selling B.H., Allison R.F., Kaesberg P. Genomic RNA of an insect virus directs synthesis of infectious virions in plants. Proc. Natl. Acad. Sci. USA. 1990;87:434–438. doi: 10.1073/pnas.87.1.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurd H. Review of “Insect Pathogens: Molecular Approaches and Techniques” by S. P. Stock, J. Vandenberg, I. Glazer and N. Boemare. Parasites Vectors. 2009;2:28. doi: 10.1186/1756-3305-2-28. [DOI] [Google Scholar]
- 14.Scherer W.F., Verna J.E., Richter G.W. Nodamura Virus, an Ether- and Chloroform-Resistant Arbovirus from Japan. Am. J. Trop. Med. Hyg. 1968;17:120–128. doi: 10.4269/ajtmh.1968.17.120. [DOI] [PubMed] [Google Scholar]
- 15.Ball L.A., Johnson K.L. Reverse Genetics of Nodaviruses. Adv. Appl. Microbiol. 1999;53:229–244. doi: 10.1016/s0065-3527(08)60350-4. [DOI] [PubMed] [Google Scholar]
- 16.Scotti P.D., Dearing S., Mossop D.W. Flock house virus: A Nodavirus isolated fromCostelytra zealandica (White) (Coleoptera: Scarabaeida) Arch. Virol. 1983;75:181–189. doi: 10.1007/BF01315272. [DOI] [PubMed] [Google Scholar]
- 17.Iwamoto T., Okinaka Y., Mise K., Mori K.-I., Arimoto M., Okuno T., Nakai T. Identification of Host-Specificity Determinants in Betanodaviruses by Using Reassortants between Striped Jack Nervous Necrosis Virus and Sevenband Grouper Nervous Necrosis Virus. J. Virol. 2004;78:1256–1262. doi: 10.1128/JVI.78.3.1256-1262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furusawa R., Okinaka Y., Uematsu K., Nakai T. Screening of freshwater fish species for their susceptibility to a betanodavirus. Dis. Aquat. Org. 2007;77:119–125. doi: 10.3354/dao01841. [DOI] [PubMed] [Google Scholar]
- 19.Souto S., Lopez-Jimena B., Alonso M., García-Rosado E., Bandín I. Experimental susceptibility of European sea bass and Senegalese sole to different betanodavirus isolates. Veter. Microbiol. 2015;177:53–61. doi: 10.1016/j.vetmic.2015.02.030. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Q., Liu Q., Liu S., Yang H., Liu S., Zhu L., Yang B., Jin J., Ding L., Wang X., et al. A new nodavirus is associated with covert mortality disease of shrimp. J. Gen. Virol. 2014;95:2700–2709. doi: 10.1099/vir.0.070078-0. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Q., Xu T., Wan X., Liu S., Wang X., Li X., Dong X., Yang B., Huang J. Prevalence and distribution of covert mortality nodavirus (CMNV) in cultured crustacean. Virus Res. 2017;233:113–119. doi: 10.1016/j.virusres.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 22.Thitamadee S., Prachumwat A., Srisala J., Jaroenlak P., Salachan P.V., Sritunyalucksana K., Flegel T.W., Itsathitphaisarn O. Review of current disease threats for cultivated penaeid shrimp in Asia. Aquaculture. 2016;452:69–87. doi: 10.1016/j.aquaculture.2015.10.028. [DOI] [Google Scholar]
- 23.Pooljun C., Direkbusarakom S., Chotipuntu P., Hirono I., Wuthisuthimethavee S. Development of a TaqMan real-time RT-PCR assay for detection of covert mortality nodavirus (CMNV) in penaeid shrimp. Aquaculture. 2016;464:445–450. doi: 10.1016/j.aquaculture.2016.06.044. [DOI] [Google Scholar]
- 24.Liu S., Wang X., Xu T., Li X., Du L., Zhang Q. Vectors and reservoir hosts of covert mortality nodavirus (CMNV) in shrimp ponds. J. Invertebr. Pathol. 2018;154:29–36. doi: 10.1016/j.jip.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 25.Wang C., Wang X.H., Liu S., Sang S.W., Zhang Q.L. Preliminary study on the natural infection of Carassius auratus with covert mortality nodavirus (CMNV) Prog. Fish. Sci. 2019;40:25–32. [Google Scholar]
- 26.Zhang Q.L., Liu S., Li J., Xu T.T., Wang X.H., Fu G.M., Li X.P., Sang S.W., Bian X.D., Hao J.W. Evidence for Cross-Species Transmission of Covert Mortality Nodavirus to New Host of Mugilogobius abei. Front. Microbiol. 2018;9:1447. doi: 10.3389/fmicb.2018.01447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Byrne M., Rowe F., Uthicke S. Molecular taxonomy, phylogeny and evolution in the family Stichopodidae (Aspidochirotida: Holothuroidea) based on COI and 16S mitochondrial DNA. Mol. Phylogenetics Evol. 2010;56:1068–1081. doi: 10.1016/j.ympev.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 28.Li J.Y. Ph.D. Thesis. Ocean University of China; Qingdao, China: 2007. The Immunological Characters and Pathogenic Study of Cultured Apostichopus japonicus. [Google Scholar]
- 29.McElroy S. Beche-de-mer species of commercial value—An update. SPC Beche-De-Mer Info. Bull. 1990;2:2–7. [Google Scholar]
- 30.Han H., Yi Y.H., Li L., Liu B.S., Pan M.X., Yan B., Wang X.H. Triterpene glycosides from sea cucumber Holothuria leucospilota. Chiu J. Nat. Med. 2009;7:346–350. doi: 10.3724/SP.J.1009.2009.00346. [DOI] [Google Scholar]
- 31.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bell T.A., Lightner D.V. A Handbook of Normal Penaeid Shrimp Histology. World Aquaculture Society; Baton Rouge, LA, USA: 1988. [Google Scholar]
- 33.Lightner D.V. A Handbook of Shrimp Pathology and Diagnostic Procedures for Diseases of Cultured Penaeid Shrimp. World Aquaculture Society; Baton Rouge, LA, USA: 1996. [Google Scholar]
- 34.Nuovo G.J., Plaia T.W., Belinsky S.A., Baylin S.B., Herman J.G. In situ detection of the hypermethylation-induced inactivation of the p16 gene as an early event in oncogenesis. Proc. Natl. Acad. Sci. USA. 1999;96:12754–12759. doi: 10.1073/pnas.96.22.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fortunato T.M., Beltrami C., Emanueli C., De Bank P.A., Pula G. Platelet lysate gel and endothelial progenitors stimulate microvascular network formation in vitro: Tissue engineering implications. Sci. Rep. 2016;6:25326. doi: 10.1038/srep25326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zechmann B., Zellnig G. Rapid diagnosis of plant virus diseases by transmission electron microscopy. J. Virol. Methods. 2009;162:163–169. doi: 10.1016/j.jviromet.2009.07.032. [DOI] [PubMed] [Google Scholar]
- 37.Graham L., Orenstein J.M. Processing tissue and cells for transmission electron microscopy in diagnostic pathology and research. Nat. Protoc. 2007;2:2439–2450. doi: 10.1038/nprot.2007.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Panphut W., Senapin S., Sriurairatana S., Withyachumnarnkul B., Flegel T.W. A novel integrase-containing element may interact with Laem-Singh virus (LSNV) to cause slow growth in giant tiger shrimp. BMC Veter. Res. 2011;7:18. doi: 10.1186/1746-6148-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pauly M., Snoeck C.J., Phoutana V., Keosengthong A., Sausy A., Khenkha L., Nouanthong P., Samountry B., Jutavijittum P., Vilivong K., et al. Cross-species transmission of poultry pathogens in backyard farms: Ducks as carriers of chicken viruses. Avian Pathol. 2019;48:503–511. doi: 10.1080/03079457.2019.1628919. [DOI] [PubMed] [Google Scholar]
- 40.Li X., Zai J., Zhao Q., Nie Q., Li Y., Foley B.T., Chaillon A. Evolutionary history, potential intermediate animal host, and cross-species analyses of SARS-CoV-2. J. Med Virol. 2020;92:602–611. doi: 10.1002/jmv.25731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao Y., Guo M., Lv Z., Zhang W., Shao Y., Zhao X., Li C. Fas-associated death domain (FADD) in sea cucumber (Apostichopus japonicus): Molecular cloning, characterization and pro-apoptotic function analysis. Dev. Comp. Immunol. 2020;108:103673. doi: 10.1016/j.dci.2020.103673. [DOI] [PubMed] [Google Scholar]
- 42.Li H., Qiao G., Gu J., Zhou W., Li Q., Woo S., Xu D., Park S. Phenotypic and genetic characterization of bacteria isolated from diseased cultured sea cucumber Apostichopus japonicus in northeastern China. Dis. Aquat. Org. 2010;91:223–235. doi: 10.3354/dao02254. [DOI] [PubMed] [Google Scholar]
- 43.Shao Y., Li C., Ou C., Zhang P., Lu Y., Su X., Li Y., Li T. Divergent Metabolic Responses of Apostichopus japonicus Suffered from Skin Ulceration Syndrome and Pathogen Challenge. J. Agric. Food Chem. 2013;61:10766–10771. doi: 10.1021/jf4038776. [DOI] [PubMed] [Google Scholar]
- 44.Yang G., Tian X., Dong S., Peng M., Wang D., Zhang K. Effects of dietary rhubarb, Bacillus cereus, yeast polysaccharide, and florfenicol supplementation on growth, intestinal morphology, and immune responses of sea cucumber (Apostichopus japonicus) Aquac. Int. 2015;24:675–690. doi: 10.1007/s10499-015-9957-9. [DOI] [Google Scholar]
- 45.Song Z.R. Pathology of Aquatic Animal. Xiamen University Press; Xiamen, China: 2009. [Google Scholar]
- 46.Deng H., Zhou Z.-C., Wang N.-B., Liu C. The syndrome of sea cucumber (Apostichopus japonicus) infected by virus and bacteria. Virol. Sin. 2008;23:63–67. doi: 10.1007/s12250-008-2863-9. [DOI] [Google Scholar]
- 47.Morgan A. The effect of food availability on early growth, development and survival of the sea cucumber Holothuria scabra (Echinodermata: Holothuroidea) SPC Beche-De-Mer Inf. Bull. 2001;14:6–12. [Google Scholar]
- 48.Zhang C.Y., Wang Y.G., Rong X.J. Isolation and identification of causative pathogen for skin ulcerative syndrome in Apostichopus japonicus. J. Fish. China. 2006;30:118–123. [Google Scholar]
- 49.Zhang Z., Xing R., Lv Z., Shao Y., Zhang W., Zhao X., Li C. Analysis of gut microbiota revealed Lactococcus garviaeae could be an indicative of skin ulceration syndrome in farmed sea cucumber Apostichopus japonicus. Fish Shellfish Immunol. 2018;80:148–154. doi: 10.1016/j.fsi.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 50.Wang P., Chang Y., Yu J., Li C., Xu G. Acute peristome edema disease in juvenile and adult sea cucumbers Apostichopus japonicus (Selenka) reared in North China. J. Invertebr. Pathol. 2007;96:11–17. doi: 10.1016/j.jip.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang G.X., Yuan J.L., Zhao Y.K., Yuan M. Isolation, identification and drug sensitivity of the pathogens of the skin ulceration disease in Apostichopus japonicus. J. Northwest A F Univ. (Nat. Sci. Ed.) 2007;35:87–90. [Google Scholar]
- 52.Nerva L., Forgia M., Ciuffo M., Chitarra W., Chiapello M., Vallino M., Varese G., Turina M. The mycovirome of a fungal collection from the sea cucumber Holothuria polii. Virus Res. 2019;273:197737. doi: 10.1016/j.virusres.2019.197737. [DOI] [PubMed] [Google Scholar]
- 53.MacTavish T., Stenton-Dozey J., Vopel K., Savage C. Deposit-Feeding Sea Cucumbers Enhance Mineralization and Nutrient Cycling in Organically-Enriched Coastal Sediments. PLoS ONE. 2012;7:e50031. doi: 10.1371/journal.pone.0050031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li L., Li Q. Effects of stocking density, temperature, and salinity on larval survival and growth of the red race of the sea cucumber Apostichopus japonicus (Selenka) Aquac. Int. 2009;18:447–460. doi: 10.1007/s10499-009-9256-4. [DOI] [Google Scholar]
- 55.Liu X.F., Xue C.H., Wang Y.M., Li H.Y. Comparative analysis of nutritive composition in body wall and internal organs of sea cucumber (Apostichopus japonicus)at Rushan. J. Fish. China. 2011;35:587–593. [Google Scholar]
- 56.Yang H., Hamel J.F., Mercier A. The Sea Cucumber Apostichopus japonicus: History, Biology and Aquaculture. Academic Press; Cambridge, MA, USA: 2015. [Google Scholar]
- 57.Purcell S.W., Lovatelli A., Vasconcellos M., Ye Y. Managing Sea Cucumber Fisheries with An Ecosystem Approach. FAO; Rome, Italy: p. 157. FAO Fisheries and Aquaculture Technical Paper 520. [Google Scholar]
- 58.Gu M., Ma H., Mai K., Zhang W., Ai Q., Wang X., Bai N. Immune response of sea cucumber Apostichopus japonicus coelomocytes to several immunostimulants in vitro. Aquaculture. 2010;306:49–56. doi: 10.1016/j.aquaculture.2010.05.024. [DOI] [Google Scholar]
- 59.Liu H., Zheng F., Sun X., Hong X., Dong S., Wang B., Tang X., Wang Y. Identification of the pathogens associated with skin ulceration and peristome tumescence in cultured sea cucumbers Apostichopus japonicus (Selenka) J. Invertebr. Pathol. 2010;105:236–242. doi: 10.1016/j.jip.2010.05.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.