Abstract
Synaptic vesicle fusion is mediated by SNARE proteins - VAMP2 on the vesicle and Syntaxin-1/SNAP25 on the pre-synaptic membrane. Chaperones Munc18–1 and Munc13–1 co-operatively catalyze SNARE assembly via an intermediate ‘template’ complex containing Syntaxin-1 and VAMP2. How SNAP25 enters this reaction remains a mystery. Here we report that Munc13–1 recruits SNAP25 to initiate the ternary SNARE complex assembly by direct binding, as judged by bulk FRET spectroscopy and single-molecule optical tweezer studies. Detailed structure-function analyses show that the binding is mediated by the Munc13–1 MUN domain and is specific for the SNAP25 ‘linker’ region that connects the two SNARE motifs. Consequently, freely diffusing SNAP25 molecules on phospholipid bilayers are concentrated and bound in ~1:1 stoichiometry by the self-assembled Munc13–1 nano-clusters.
Neuronal communication involves the controlled release of neurotransmitters stored in synaptic vesicles (SV) into the neuronal synapse [1–4]. This process is tightly regulated to ensure that the message is timely and precise [3, 4]. SV fusion is catalyzed by the synaptic SNARE (soluble N-ethyl maleimide sensitive factor attachment protein) proteins - VAMP2 on the vesicle membrane (v-SNARE) and Syntaxin-1 and SNAP25 on the plasma membrane (t-SNAREs) [5, 6]. When the vesicle approaches the plasma membrane (PM), the helical SNARE motifs of the cognate v- and t-SNAREs constitutively assemble into a ternary complex that initially bridges and ultimately fuses the two membranes [5–7]. The SNARE complex nucleates into a four-helix bundle at its membrane distal end and progressively assembles (“zippers”) towards the membranes, exerting a potent force that ultimately drives the membranes together to fuse into one [6, 8, 9]. Though efficient, under in vitro conditions, the fusion process is artificially slow with its rate being limited by the intrinsic rate of nucleation of SNARE complex [10–12]. In vivo, nucleation is greatly accelerated by two co-operating, specialized molecular chaperones, Munc13 and Munc18 [3, 4, 13, 14]. How precisely these chaperones and their homologues achieve this, be it in the highly specialized context of neurotransmission or more broadly for vesicle fusion driving intracellular protein transport, is a major open question despite important insights that have been gleaned.
Structural studies indicate that Munc18–1 binds and stabilizes monomeric Syntaxin-1 in a ‘closed’ (auto-inhibited) conformation [15, 16]. In doing so, it prevents its assembly into SNARE complexes or engagement into off-pathway reactions [13, 15, 16]. This interaction is associated with Munc18–1 chaperone function and is critical for trafficking of Syntaxin-1 from endoplasmic reticulum (ER) to the PM [17–19]. Munc18–1 has also been shown to promote SNARE complex assembly and to accelerate the resultant vesicular fusion [13, 20]. This activator function is attributed to Munc18–1’s ability to bind and sequester partially-assembled SNARE complexes, termed SNAREpins [13]. Indeed, recent structural and biophysical studies suggest that Munc18–1 likely serves as a template for the formation of an early SNARE assembly intermediate (termed a ‘template complex’) wherein Syntaxin-1 and VAMP2 are properly oriented and aligned for nucleation into a four helix bundle [21–24]. Thus, the dual-binding modes of Munc18–1 to monomeric Syntaxin-1 and SNARE complexes allow it to both negatively-regulate and positively-assist SNARE complex assembly.
A key function of the other dedicated chaperone, Munc13–1, is to catalyze the overall process of nucleation by “opening” the closed Syntaxin-1/Munc18 complex to enable it to rapidly assemble with VAMP2 to form the template complex [14, 25–28]. This catalytic function has been mapped to the large central module within Munc13 called the MUN domain [26, 27], but the precise mechanism is not known. Independent of Munc18–1, Munc13–1 has been shown to promote the proper alignment of Syntaxin-1 and VAMP2 [10] thereby accelerating and stabilizing the Munc18/Syntaxin-1/VAMP2 template complex [22]. The mechanism involved was recently clarified with the findings that Munc13–1 independently binds both VAMP2 [29] and Syntaxin-1 [28, 30] at distinct sites of its MUN domain potentially enabling it to simultaneously interact with and orient these two SNAREs in molecular proximity to Munc18–1.
Munc13–1 is a large (~20 nm) multi-domain, multi-functional protein [24]. Independent of chaperoning SNAREpin assembly, Munc13–1 plays a key role in local tethering of SV at dedicated active zones of the pre-synaptic PM [3, 31, 32]. Local tethering of SV to the PM, occurs prior to nucleation of SNAREpins, and is required to form the pool of release ready vesicles [33]. It has been proposed that local tethering occurs when Munc13–1 itself forms molecular bridges linking the SV and PM through interactions involving the C1-C2B region and the C2C domain on opposite ends of the MUN domain [33–36]. Specifically, the C1 and C2B domains bind the PM through interactions with their physiological ligands diacylglycerol (DAG) and phosphatidylinositol 4,5-bisphosphate (PIP2) respectively, while the C-terminal C2C domain interacts with the SV membrane, at least in part via the acidic phospholipid, phosphatidylserine (PS) [33–35, 37, 38].
Overall, Munc18–1 and Munc13–1 co-operatively catalyze efficient SNAREpin nucleation via a template complex involving choreographed binding of Syntaxin-1 and VAMP-2. Of course, SNAREpins cannot nucleate without two of their four helices. Remarkably, especially for such a heavily investigated subject, how SNAP25 is chaperoned into nascent SNAREpins remains a mystery. It is known that VAMP2-containing SVs dock to PIP2-enriched regions on the PM, guided by Munc13–1 and SV-associated protein, Synaptotagmin [39, 40]. Syntaxin-1 is sequestered into these PIP2 microdomains, mediated by electrostatic interactions involving the Syntaxin-1 juxta-membrane region [41]. However, SNAP25, is highly dispersed on the PM surface and in fact, largely excluded from the PIP2 clusters under resting conditions [42, 43]. Exactly how SNAP25 is recruited to the site of vesicle docking to initiate SNARE complex assembly remains enigmatic.
Recently, a single-molecule force spectroscopy study revealed that the Munc13–1 MUN domain enhances the probability of SNAP25 binding to the Syntaxin-1/VAMP2/Munc18 template complex and subsequently complete assembly of the ternary SNARE complex [22]. This suggested that Munc13–1 may play a role in recruiting SNAP25 to initiate SNARE complex formation. Here we test and confirm this hypothesis. We find that the Munc13–1 MUN domain directly and specifically binds the SNAP25 linker region and this interaction forms the molecular basis of Munc13–1 function in stimulating SNARE complex assembly both on its own and in conjunction with Munc18–1.
MATERIALS AND METHODS
Materials
The cDNA constructs used in this study include full-length mouse His6-SNAP25b, residues 1–206 in a pET28 vector with a N-terminal Thrombin cut site; His12-Munc13L (Munc13–1 residues 529–1735 with residues 1408–1452 replaced by the sequence EF) in a modified pCMV-AN6 vector which includes a PreScission cut site following the His12 tag, GST-MUN (residues 859–1407, △1408–1452, EF) cloned into a pGEX vector with a N-terminal Thrombin cut site (a kind gift from Dr. Josep Rizo, UT Southwestern). We used a QuikChange mutagenesis kit (Agilent Technologies, Santa Clara, CA) to generate the N1128A/F1131A in the Munc13L background (Munc13LNF). SNAP25 linker domain mutants were synthesized (Genewiz Inc., Plainfield, NJ) with the SNAP25 residues 83–142 replaced with (GGGGS)10 repeats (SNAP25G4S) or SNAP23 residues 83–142 (referred to as SNAP25SNAP23) and cloned into the pET28 vector similar to the SNAP25WT construct. For fluorescent labeling experiments, we introduced a Halo tag at the c-terminus of the Munc13L and MUN constructs in the aforementioned vector background. We used a single-cysteine (S28C) version of SNAP25WT [44] and SNAP25G4S (synthesized from Genewiz Inc., Plainfield, NJ). Constructs used in the optical tweezer experiments (cytoplasmic domains of rat syntaxin 1A residues 1–265, R198C and VAMP2 residues 1–96, N29C) and the bulk FRET experiments (VAMP2 residues 1–96, S28C, SNAP25 residues 1–206, Q20C, Syntaxin 1 residues 1–258) have been described previously [22, 45]. Lipids, 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-dioleoylsn-glycero-3-phospho-L-serine) (DOPS), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-[amino(polyethylene glycol)-2000] (DOPE-PEG2000), phosphatidylinositol 4, 5-bisphosphate (PIP2), diacylglycerol (DAG) and 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2–1,3 benzoxadiazol-4-yl) (NBD-PE) were purchased from Avanti Polar Lipids (Alabaster, AL). The polystyrene beads (2.1 μm in diameter) with anti-digoxigenin coating used in optical tweezer experiments were purchased from Spherotech (Green Oaks, IL).
Protein production and purification
The MUN domain (with and without Halo tag) and all SNARE proteins were expressed and purified as described previously [22, 26, 46]. Briefly, the proteins were expressed in Escherichia coli strain BL21(DE3) (Novagen, Darmstadt, Germany) and cells were lysed with a cell disruptor (Avestin, Ottawa, Canada) in lysis buffer containing 50 mM HEPES, pH 7.4, 400 mM KCl, 4% TritonX-100, 10% glycerol, 0.5 mM Tris (2-carboxyethyl) phosphine hydrochloride (TCEP) and 1 mM phenylmethylsulfonyl fluoride. Samples were clarified using a 45 Ti rotor (Beckman Coulter, Brea, CA) at 140,000 × g for 30 minutes and incubated with Ni-NTA agarose (Thermofisher, Waltham, MA) or Glutathione agarose (Thermofisher, Waltham, MA) for 4–16 h at 4°C. The resin was subsequently washed in the HEPES buffer, without TritonX-100 and supplemented with 50 mM Imidazole for the Ni-NTA purification. After the washes (three column volumes) the protein was cleaved of the resin with Thrombin for 4–16 h at 4°C. The protein was purified using gel filtration (Superdex75 or 200 Hi-load column, GE Healthcare, Chicago, IL). The peak fractions were pooled and concentrated using filters of appropriate cut-offs (EMD Millipore, Burlington, MA). NOTE: Syntaxin-1 construct used in optical tweezer studies was biotinylated in situ using previously described biotin ligase protocol [45].
Munc13L (with or without Halo-tag) was expressed in ExpiHEK-293 cell cultures using Expifectamine as a transfection reagent (Thermofisher, Waltham, MA). Briefly, thawed cells were passaged three times prior to transfection and were grown for 72 hours before being spun down and rinsed in ice cold PBS. The pellet was re-suspended in the HEPES lysis buffer and lysed using a Dounce homogenizer. The sample was clarified using at 140,000 × g for 30 minutes at 4°C and the supernatant incubated overnight with Ni-NTA beads, in the presence of DNAse 1, RNAse A and Benzonase to remove nucleotide contamination. The protein was further washed in the lysis buffer (without TritonX-100) before being cleaved with PreScission Protease for 2 hours at room temperature. The eluted proteins were further purified via gel filtration (Superdex200, GE Healthcare Chicago, IL).
In all cases, the protein concentration was determined using a Bradford Assay (BioRad, Hercules, CA,) with BSA as a standard and protein purity was verified using SDS/PAGE analysis with Coomassie stain. All proteins were flash-frozen and stored at -80°C for long-term storage.
The SNAP25 and Munc13-Halo proteins were fluorescently labeled as described previously [44]. Typically, the proteins were incubated with 5X excess dye (AlexaFluor 488, 555 or 660 as noted) for 1h at room temperature (or overnight at 4°C) and the unbound dye was removed using spin columns (Thermofisher, Waltham, MA). The labeling efficiency was estimated using absorbance values (488 or 555 or 660 nm/280 nm) and in all cases, was verified to be >90%.
Microscale Thermophoresis Interaction Analysis
Halo-tagged Munc13–1 constructs (Munc13L, Munc13LNF and MUN) were labeled with AlexaFluor 660 and MST analysis was carried out with 50 nM of fluorescently labeled Munc13–1 constructs in a premium-coated glass capillary with 0.5 mg/ml BSA and 0.05% Tween-20 included in the HEPES buffer (50 mM HEPES, 140 mM KCl, 1 mM TCEP, 1 mM MgCl2) to prevent non-specific adsorption to the glass surface. The samples were measured in a Monolith NT.115 (Nanotemper Technologies, San Francisco, CA) and MST curves obtained with titration of SNAP25 (1 nM – 125 μM range) were fitted with a non-linear curve (sigmoidal) analysis to estimate the apparent dissociation constant (Kd).
Lipid Bilayer experiments
Liposomes were made using brain physiological composition (73% DOPC, 15% DOPS, 3% PIP2, 2% DAG, 5% DOPE-PEG 2000, 2% NBD-PE) using extrusion method with HEPES buffer (50 mM HEPES, 140 mM KCl, 1 mM TCEP, pH 7.4). Lipid bilayers were created by Mg2+ (5mM) induced bursting liposomes in ibidi glass-bottom chambers (ibidi GmbH, Germany). The bilayer was extensively washed with HEPES buffer and the fluidity of the lipid bilayer was verified using fluorescence recovery after photo-bleaching (FRAP) using the NBD fluorescence. FRAP measurements were analyzed on Wolfram Mathematica using a previously published algorithm [47]. Consistent with a fluid bilayer, the average diffusion coefficient of the lipid was calculated to be 1.6 ± 0.3 μm2/sec (Supplementary Figure 1). SNAP25 and Munc13L were added to the pre-washed supported bilayer and incubated for 30 minutes. For the clustering experiments (Figure 2), we used 160 nM SNAP25WT or SNAP25G4S (residue S28C) labeled with AlexaFluor555 with increasing concentrations (50 nm – 2μM) of unlabeled Munc13L. FRAP analysis showed that the membrane-bound AlexaFluor555-labeled SNAP25 was fairly mobile, with an average diffusion coefficient of 1.0 ± 0.2 μm2/sec (Supplementary Figure 1). For the co-localization experiments (Figure 3), we used 160 nM SNAP25WT (residue S28C) labeled with AlexaFluor555 with either 1 μM or 50 nM of Munc13L-Halo labeled with AlexaFluor488. Mander’s coefficients were calculated using ImageJ (Fiji) with the JACOP plugin [48]. The stoichiometry analysis were carried out under 50 nM Munc13L condition. Fluorescence profiles of individual ROIs corresponding to SNAP25 (550 nm) and Munc13–1 (647) from the same cluster (~70 in total) were plotted and number of bleaching steps were manually counted. For all experiments, the chambers were washed thoroughly (after 30 min incubation) to remove any non-specifically or weakly bound proteins and the samples were imaged on a TIRF microscope (Nikon) with a 63× oil objective.
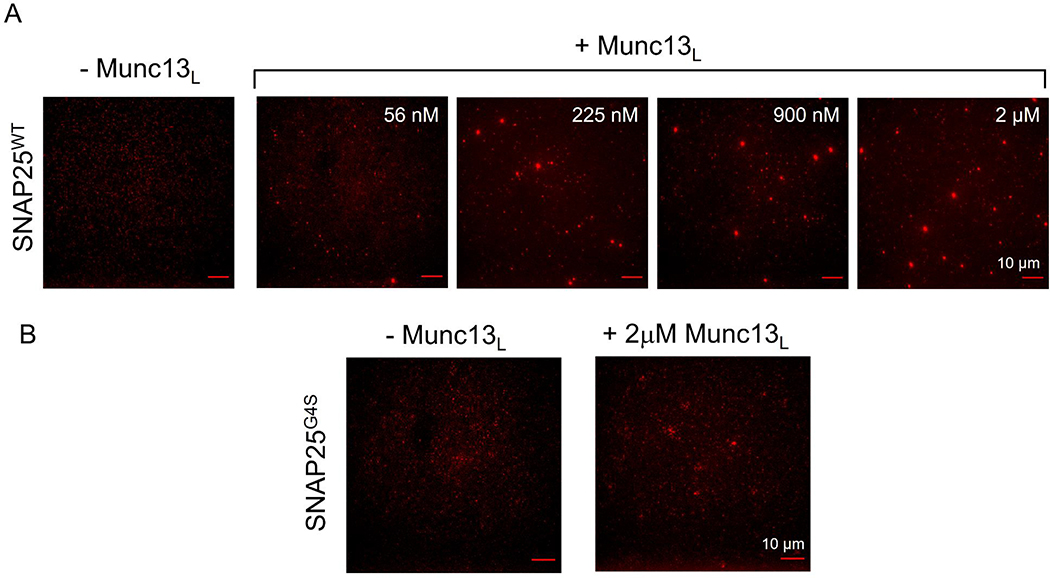
Figure 2. Munc13–1 binds and clusters SNAP25 on lipid bilayer surface.
(A) Distribution of AlexaFluor 488 labeled SNAP25WT (~150 nM) on supported lipid bilayer under physiological lipid and buffer composition in the absence or presence of Munc13L was visualized using TIRF microscopy. Munc13L induces clustering of SNAP25WT molecules and the number/size of the SNAP25 clusters directly correlates to concentration of Munc13L added. (B) Munc13L fails to cluster SNAP25 linker domain mutant (SNAP25G4S) even at the highest concentration (2 μM) tested. This suggests that MUN-SNAP25 linker domain interaction is feasible on lipid bilayers surfaces and results in clustering of otherwise diffusely distributed SNAP25 molecules. Representative fluorescent images are shown.
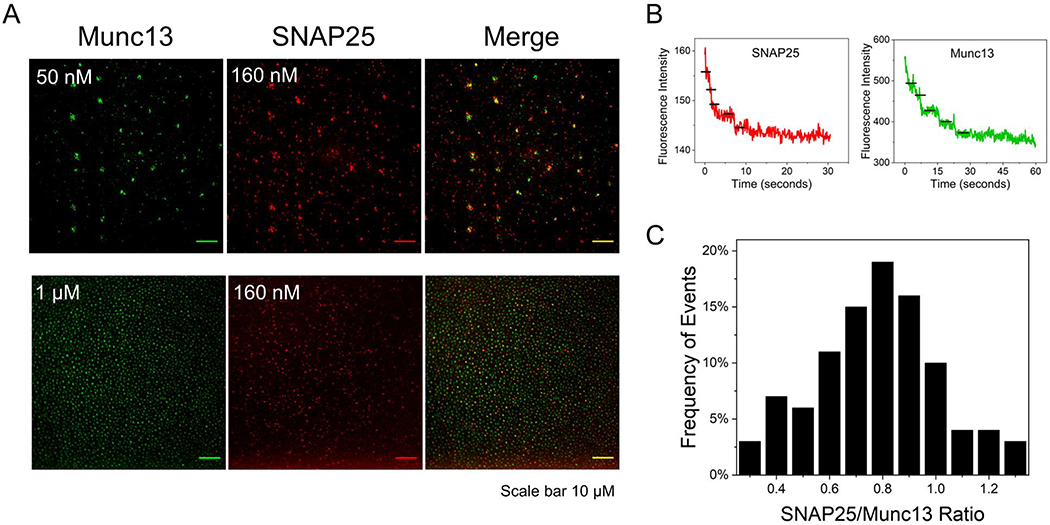
Figure 3. Munc13–1 co-clusters with SNAP25 at 1:1 stoichiometry.
(A) Representative fluorescence images showing that AlexaFluor 555 labeled SNAP25WT co-localizes with clusters of AlexaFluor 488 labeled Munc13L on lipid bilayer surface and the extent of co-localization depends on the concentration of Munc13L added. SNAP25 (160 nM) shows limited co-localization (Manders’ coefficient ~ 0.2) at 50 nM of Munc13L but has high overlap (Manders’ coefficient ~ 0.7) at 1μM of Munc13L. (B) Stoichiometry of SNAP25 and Munc13L in the co-clusters under low Munc13L concentration was estimated using step-bleaching analysis. Representative step-bleaching traces for SNAP25 (red) and Munc13L (green) are shown and the ratio of SNAP25:Munc13L quantified from ~70 co-clusters is shown as the histogram. Under these conditions, each cluster contains ~5–7 molecules at almost 1:1 ratio of Munc13L:SNAP25. This indicates that molecular interaction between the MUN domain and the SNAP25 linker region is likely responsible for the observed co-clustering of Munc13–1 and SNAP25.
FRET Experiment
We used a FRET-based assay to track N-terminal SNARE complex formation. For site-specific labeling with fluorophores, cysteines were introduced into cytoplasmic domains of SNAP25 (residues 20) and VAMP2 (residue 28) and was labeled with thiol-reactive fluorescent probes AlexaFluor488 maleimide and AlexaFluor555 maleimide respectively. 1.5 μM of labeled SNAP25 and unlabeled Syantaxin-1 were pre-mixed with or without 3 μM MUN domain and incubated at 37°C for 10 min. 500 nM of pre-warmed labeled VAMP2 was added to start the reaction and fluorescence at 580 nm was measured in plate reader (Molecular Devices) for 60 min with 15 sec intervals. In control experiments, we left out Syntaxin-1 from the reaction mixture.
Optical Tweezer experiments
Single molecule force spectroscopy experiments were carried out as described previously [22, 23, 45, 49]. Briefly, ternary SNARE complexes were assembled by mixing the purified SNARE proteins and incubating at 4°C overnight. Assembled SNARE complexes were purified by binding to Ni-NTA-agarose and cross-linked with DTDP (2,2'-dithiodipyridine disulfide) treated DNA handles (50:1 molar ratio) in PBS buffer (100 mM phosphate buffer, 500 mM NaCl, pH 8.5). Dual-trap optical tweezers and basic protocols for single-molecule experiments have been described in detail elsewhere [45, 49]. To follow SNARE assembly/disassembly pathways, samples of the 2 DNA handles, one cross-linked with the SNARE complex and the other bound by a streptavidin molecule, were separately bound to anti-digoxigenin antibody coated polystyrene beads. The beads were injected into a microfluidic channel and trapped. The 2 beads are then brought close to allow a single SNARE complex to be tethered between them. All manipulation experiments were carried out in PBS buffer supplemented with the oxygen scavenging system. All single molecules were pulled and relaxed by increasing and decreasing, respectively, the trap separation at a speed of 10 nm/s or held at constant mean forces by keeping the trap separation constant.
RESULTS
Munc13–1 MUN domain directly binds SNAP25 in solution
To test if there is direct molecular interaction between Munc13–1 and SNAP25, we employed Microscale Thermophoresis (MST), a highly sensitive technique that allows one to quantify molecular interactions with limited amounts of protein under varying buffer conditions [50, 51]. Informed by earlier reports [26, 29, 30, 33, 34], we focused on the conserved C-terminal portion of Munc13–1 consisting of the contiguous C1-C2B-MUN-C2C domains (Munc13–1 residues 529–1735; referred to as Munc13L), except for residues 1408–1452 within a non-conserved loop in the MUN domain, which was deleted and replaced with EF residues to minimize dimerization/oligomerization [27, 28]. To facilitate the MST analysis, we introduced a C-terminal Halo tag to the Munc13L fragment to enable covalent attachment of a fluorescent tag. Titration of full-length wild-type SNAP25 (residues 1–206, SNAP25WT) into AlexaFluor 660 labeled Munc13L yielded a classical single-site dose-response curve with a dissociation constant (Kd) of 26 ± 2 μM (Figure 1A). This indicated that that Munc13–1 directly binds SNAP25, albeit with a modest affinity under soluble conditions. This binding affinity is comparable to that reported (25–100 μM) for specific binding of the MUN domain to Syntaxin-1, VAMP2, and the fully assembled SNARE complex when these proteins are also free in solution [10, 28–30]. Therefore, the novel binding we observe between Munc13–1 and SNAP25 is similarly likely to be physiologically relevant at the high local concentrations that exist in active zones.
Figure 1. Dissecting Munc13–1 interaction with SNAP25 using Microscale Thermophoresis (MST) analysis.
(A) Titration of full-length SNAP25 into AlexaFluor 660 labeled Munc13L (C1-C2B-MUN-C2C domains) produced a concentration-dependent change in the MST signal, yielding an apparent dissociation constant (Kd) = 26 ± 2 μM implying that Munc13L directly binds to SNAP25. Munc13–1 domain likely binds to the SNAP25 linker region as replacing the SNAP25 linker domain that connects that SNARE helical motifs with a GGGGS sequence (SNAP25G4S) completely abrogates Munc13-SNAP25 interaction (red curve). This interaction is highly specific as swapping the SNAP25 linker region with closely associated SNAP23 linker (SNAP25SNAP23) also disrupts the binding (blue curve).(B) MUN domain (orange curve) alone is sufficient to bind SNAP25 albeit with slightly reduced affinity (Kd = 48 ± 3 μM) and SNAP25 binding site is unique and different from the Syntaxin-1 binding site, as the Syntaxin non-binding Munc13–1 mutant (Munc13LNFAA) is capable of binding SNAP25 (purple curve). Average and standard deviations from a minimum of three independent experiments are shown.
We then sought to identify the regions within Munc13–1 and SNAP25 responsible for their binding. The central MUN domain of Munc13–1 contains the distinct binding sites for both VAMP2 and Syntaxin-1, each of which is required for its chaperone function [29, 30]. So, we expressed and purified the isolated MUN domain (residues 859–1531, △1408–1452, EF), and labeled it with AlexaFluor 660 at a C-terminal Halo tag. MST analysis showed that the MUN domain alone can bind SNAP25WT with an apparent Kd of 48 ± 3 μM (Figure 1B). This indicated that the MUN domain serves as the primary binding site for SNAP25 and that the flanking C1-C2B and C2C portions might weakly augment the primary MUN-SNAP25 binding site, be it directly or indirectly.
Binding SNAP25 involves the linker region between its two SNARE motifs
Since the isolated MUN domain promotes the nucleation of SNARE complexes [27], we reasoned that the Munc13–1 binding site on SNAP25 is likely outside of the SNARE helices, as these would need to be free to assemble with the SNARE motifs of VAMP2 and Syntaxin-1. This logic would make the 60 residue ‘linker’ region that concatenates the SNARE helical motifs the best candidate for productive binding by Munc13–1. Therefore, we designed and tested a SNAP25 mutant (SNAP25G4S) in which the linker region was replaced with an inert, flexible, non-specific sequence (GGGGS repeats) yet maintaining the overall length. Even though SNAP25G4S successfully replaced SNAP25WT in a bulk vesicle fusion assay (Supplementary Figure 2) and has been shown to assemble in SNARE complex [52], it failed to bind to Munc13L even at the highest SNAP25G4S concentration (100 μM) tested (Figure 1A). This suggests that the MUN domain likely binds to the SNAP25 linker region.
SNAP25 binding to Munc13–1 is specific and unique
To assess the specificity of binding, we replaced the SNAP25 linker region with that of its close homologue, SNAP23, producing a chimeric protein termed SNAP25SNAP23. SNAP23 is structurally and functionally similar to SNAP25 and is expressed in all tissues [53–55], where it associates with Syntaxin-1 and VAMP2 and functions as part of the ubiquitous cellular fusion machinery [53]. Notably, the linker region of SNAP23 (compared with SNAP25) is markedly less conserved (~45% identical) than either SNARE motif (~70%). Indeed, SNAP25SNAP23 failed to bind to Munc13L (Figure 1A), confirming that Munc13–1 binding to SNAP-25 is isoform specific and that specificity resides in the linker region.
Munc13–1 has been shown to bind and activate Syntaxin-1 via a hydrophobic pocket near the middle of the MUN domain [30]. To assess if SNAP25 binding overlaps with the Syntaxin site, we tested a previously described mutation in Munc13–1 (N1128A/F1131A; Munc13LNF) that disrupts binding to Syntaxin-1 [30]. These mutations had no effect and Munc13LNF bound SNAP25 with a Kd of 29 ± 10 μM (Figure 1B, purple). This indicates that the SNAP25 binding site within the MUN domain is distinct from the Syntaxin-1 binding pocket.
Munc13–1 binds SNAP25 on lipid membrane surface at 1:1 stoichiometry
SNAP25 and Munc13 are both naturally concentrated within active zones of the pre-synaptic plasma membrane, where they are restricted to a common surface by distinct mechanisms. SNAP25 is anchored by the direct insertion of its multiple palmitoylated cysteines in the linker region [52, 56, 57]. As mentioned, Munc13–1 is anchored by binding to lipid headgroups by C1-C2B domains [34–36]. We thus investigated the SNAP25-Munc13 interaction on a lipid bilayer surface. We anticipated that restriction of these two proteins to a two-dimensional surface would increase the association rate, resulting overall in markedly greater stability of the Munc13-SNAP25 complexes thereby enabling them to be imaged and quantified on the supported bilayers using total internal reflection fluorescence (TIRF) microscopy.
We created a supported lipid bilayer with functionally-relevant lipid composition (73% PC, 15% PS, 3% PIP2, 2% DAG) by bursting pre-formed vesicles onto a glass coverslip. We had anticipated that it would be necessary to use palmitoylated SNAP25 in this experiment, but instead observed that even non-palmitoylated SNAP25, added at low (150 nM) concentrations in solution, bound to the bilayer. This is likely due to the amphipathic nature of two SNARE motifs of SNAP25 [7] as it is well-established that amphipathic peptides associate strongly to phospholipid surfaces as their hydrophobic side chains could insert into the lipid bilayers [58, 59]. On its own, freely-diffusing AlexaFluor 555-labeled SNAP25 molecules thus bound were monomeric and uniformly distributed on the lipid bilayer surface (Figure 2A, Supplementary Figure 1). When Munc13L was present, SNAP25 was no longer monodisperse, but rather located in numerous distinct clusters of variable intensity (Figure 2A) and the clustering efficiency directly correlated to the concentration of Munc13L (Figure 2A, Supplementary Figure 3).
Considering that Munc13–1 has been shown to self-associate into clusters on lipid bilayers under both in vitro and physiological conditions [60, 61], we reasoned that SNAP25 puncta are a consequence of its association with Munc13–1 clusters. To confirm, we co-localized AlexaFluor555-labeled SNAP25 with AlexaFluor488-labeled Munc13L using two color TIRF microscopy (Figure 3A). As expected, Munc13L on its own formed clusters on lipid bilayer surface (Figure 3A) and SNAP25 increasingly co-localized with these clusters as the concentration of Munc13L was increased. When Munc13L was added at 50 nM bulk concentration the Manders’ coefficient (SNAP25:Munc13L) was 0.2 ± 0.07, which increased to 0.6 ± 0.05 at 1 μM Munc13L, consistent with a concentration-dependent binding process. As a control, we employed membrane-anchored (palmitoylated) SNAP25 and obtained similar results (Supplementary Figure 4).
To establish the specificity of this binding, we employed the linker mutant, SNAP25G4S. As expected, no clustering of SNAP25G4S was observed, even at the highest concentration (2 μM) of Munc13L (Figure 2B). This suggests that similar to the soluble conditions, SNAP25 binding to Munc13L nanoclusters on membranes involves the SNAP25 linker region. Finally, to establish the stoichiometry of binding of SNAP25 to the Munc13L nanoclusters, we performed two color photo-bleaching experiments (Fig. 3B). Reliable identification of single steps by this method can only be made when relatively small (<10) numbers of molecules are present. Therefore, we performed these experiments at a low Munc13L concentration (50 nM) designed to limits the copy number of Munc13L in the nanoclusters. Among these co-clusters, independent of their size, we observed a broad distribution of the apparent mole ratio of SNAP25 to Munc13L centering on ~0.8 mole/mole (Figure 3B). Considering the high and similar efficiency of labeling of both SNAP25 and Munc13L, the ratio of fluorescence likely reflects the actual mole ratio. This suggests that Munc13–1 binds SNAP25 at approximately 1:1 stoichiometry.
Linker-specific binding by MUN is necessary for SNAP25 to enter the SNARE complex
We then examined the potential functional relevance of the MUN-SNAP25 linker domain interaction to Munc13–1’s established and independent role [10, 22, 34] in accelerating nucleation of SNAREpins. We employed a fluorescence resonance energy transfer (FRET) assay with fluorescent probes (AlexaFluor 488 & AlexaFluor 555) introduced at the N-termini of SNAP25 (residue Q20) and VAMP2 (residue S28) respectively to directly track the effect of the MUN domain on the initiation of SNARE complex assembly.
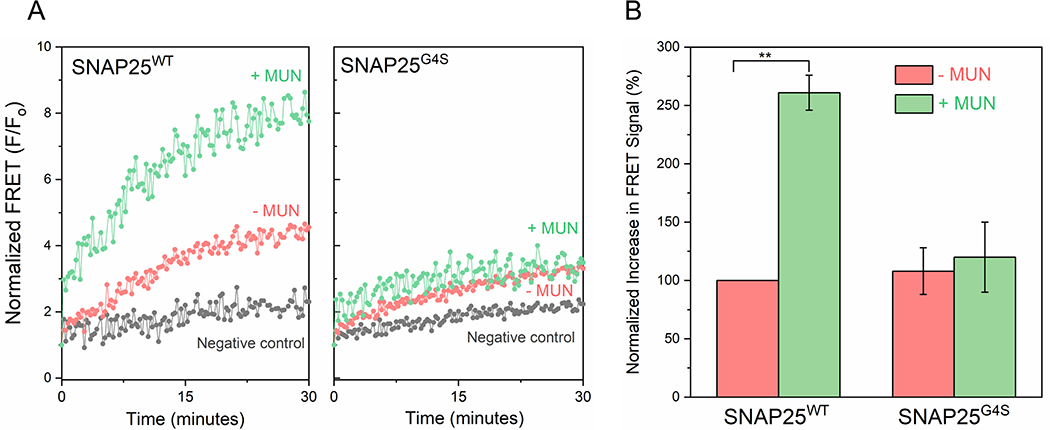
In the absence of the MUN domain, SNAP25WT spontaneously assembled into a complex with VAMP2 and unlabeled Syntaxin-1 over the course of an hour as evident from the gradual increase of the acceptor fluorescence intensity (Figure 4A). We did not detect FRET in control experiments when Syntaxin-1 was omitted confirming that the observed FRET signal corresponds to the formation of a ternary SNARE complex (Figure 4A). In line with the bulk-fusion assays (Supplementary Figure 2) and previous reports [52, 57], mutation of the linker region had very little effect on the spontaneous rate of SNAP25 assembly into SNARE complexes (Figure 4A), with comparable increase in acceptor (FRET) signal with SNAP25G4S mutant (Figure 4A). The MUN domain strongly stimulated the rate of association of SNAP25WT with the cognate SNAREs (~3-fold enhancement of FRET signal after 60 min) but had no effect on the rate of assembly of SNAP25G4S mutant (Figures 4A and 4B). This data forcefully suggests that binding of SNAP25 (via its linker) is required for Munc13–1 to chaperone SNARE complex assembly.
Figure 4. Munc13 interaction with SNAP25 is essential for its chaperone function.
FRET between AlexaFluor 488 labeled SNAP25 and AlexaFluor 555 labeled soluble VAMP2 introduced in the N-terminus (SNAP25 residue 20 & VAMP2 residue 28) shows that in the presence of cytoplasmic Syntaxin-1, the MUN domain activates the initial engagement of the SNARE complex. Negative control wherein Syntaxin-1 was left out confirms the observed FRET signal corresponds to formation of the ternary SNARE complex. The stimulatory effect of MUN domain was not observed with SNAP25 linker mutant (SNAP25G4S) indicating that the MUN-SNAP25 linker domain interaction is crucial for Munc13–1 ability to promote SNARE complex formation. Representative fluorescence traces for first 30 min of the reaction (left) and end-point quantification at 60 min (right) from two independent trials are shown. Statistical significance was determined using student t-test (** p<0.005).
Linker-dependent binding by MUN recruits SNAP25 to the template complex
To independently test this conclusion in the fuller context of Munc13–1’s co-operation with Munc18–1 in chaperoning SNARE complex assembly, we employed an established single molecule assay involving optical tweezers (Figure 5A) [22, 23, 45, 49]. Here, Syntaxin-1 and VAMP2 were cross-linked at their N-termini (between Syntaxin R198C and VAMP2 N29C) via a disulfide bond and pulled from their C-termini via two DNA handles attached to two optically trapped polystyrene beads [22, 23]. We first pulled a single SNARE complex in the presence of Munc18–1 and SNAP25WT (0.5 μM & 0.6 μM respectively added into the solution). The resultant force-extension curve (FEC) indicated that a single SNARE complex unfolds in a characteristic stepwise manner among different states when pulled (Figure 5B, FEC #1 & #2 grey curves with different regions indicated by their associated state numbers). We observed reversible C-terminal domain (CTD) transitions at about 15 pN (between state 1 and state 2, regions marked by gray ovals), followed by an irreversible unfolding of the N-terminal domain (NTD) at ~16–17 pN (from state 2 to state 3, extension jumps marked by gray arrows), and finally irreversible unfolding of the Syntaxin-SNAP25 (t-SNARE) complex (marked by green arrows) with the attendant dissociation of SNAP-25 (state 5).
Figure 5: Munc13–1 recruits SNAP25 to Munc18–1/Syntaxin-1/VAMP2 template complex.
(A) Experimental setup for optical tweezers. A single SNARE complex was pulled from the C termini of Syntaxin 1A (red) and VAMP2 (blue) via two DNA handles attached to two optically trapped polystyrene beads. The N-termini of Syntaxin 1A and VAMP2 were cross-linked via a disulfide bond. Munc18–1 (gray) and the MUN domain of Munc13–1 (yellow) were added in the solution. (B) Representative force extension curves (FECs) obtained for SNARE complexes assembled with SNAP25WT or SNAP25G4S in the presence of Munc18–1 and MUN domain as indicated. The Syntaxin–VAMPs conjugate was pulled (grey FECs) or relaxed (black FECs) by changing the separation between two optical traps at a speed of 10 nm/s or held at constant mean force around 5 pN (red FECs). The molecular states associated with different FEC regions are indicated by the corresponding state numbers (see Supplementary Figure 5 for their molecular diagrams) (C) Probability of full SNARE complex assembly from the Munc18–1/Syntaxin-1/VAMP2 template complex observed within 100s at 5 pN constant mean force. MUN domain ability to stimulate SNARE complex assembly is significantly diminished with the SNAP25 linker domain mutant (SNAP25G4S) as compared to SNAP25WT (p = 0.065). The N value refers to the total number of trials or pulling/relaxation rounds and average and standard error of the means are shown the error bar indicates the standard deviation (σ) of the corresponding probability (p) calculated as σ = √(p(1−p)/N).
Relaxing the force applied to the SNARE proteins then revealed two reversible transitions along the relaxation curves (Figure 5B FEC #1 black curve). The first transition from 14 to 6 pN (marked by blue rectangle) was caused by the folding of partially closed Syntaxin-1 (state 6) and the lower force transition at ~3–6 pN resulted from the formation of Munc18–1/Syntaxin-1/VAMP2 template complex (state 7).
To detect the ternary SNARE complex assembly, we held the template complex at a constant mean force around 5 pN for 100 seconds to await SNAP25WT binding (Figure 5B FEC#2 red curve). Indeed, in some cases, SNAP25WT from the solution was able to bind to the template complex resulting in reassembly of the full SNARE complex (state 1). This reflects the intrinsic rate of productive association of free SNAP25 with the template complex.
To test whether the rate of productive association of SNAP25 to the template complex is accelerated by Munc13–1, we included the MUN domain (1 μM) in addition to Munc18–1 and SNAP25WT in solution (Figure 5B FEC #3 & #4). Indeed, the MUN domain stabilized the template complex and promoted the rate of re-assembly of the SNARE complex from the template complex. Statistically, the MUN domain nearly doubled the probability of SNARE assembly from the template complex (Figure 5C). All of these observations are consistent with our previous reports [22, 23].
To test whether binding of SNAP25 to Munc13–1 via the SNAP25 linker is required for Munc13–1 to chaperone SNAP25 in this context, we repeated the above experiment with linker domain mutant, SNAP25G4S. With Munc18–1 only, the probability of SNARE complex formation with the SNAP25G4S was similar to that observed with SNAP25WT (Figure 5C). This further confirmed the alteration of the liker domain does not affect the inherent ability to SNAP25 helices to bind its cognate SNARE partners. Addition of the MUN domain along with Munc18–1 (Figure 5B, FEC #5, #6, #7) stabilized the template complex [22] but did not appreciably increase the probability of the SNAP25G4S to complete SNARE assembly as compared to SNAP25WT (p = 0.065) (Figure 5C). This strongly indicates that MUN-SNAP25 binding (via the linker domain) contributes to the enhanced assembly of the four-helix bundle from the Munc18–1/Syntaxin-1/VAMP2 template complex.
DISCUSSION
Altogether, we have identified a novel, functionally relevant interaction between the Munc13–1 MUN domain and the SNAP25 linker region. Our findings are supported by several previous studies [52, 57, 62, 63]. For example, it has been demonstrated that yeast exocyst protein Sec6p, which is strikingly similar to the c-terminal half of Munc13–1 MUN domain, binds Sec9 a SNAP25 homologue at the linker region. Indeed, disrupting the Sec6p-Sec9 interaction results in significant defects in exocytosis and overall cell growth [62, 63]. Furthermore, the SNAP25 linker domain has been shown to play a crucial role in regulating vesicle priming and the Ca2+-regulated exocytosis [52, 57]. Strikingly, functional analysis in chromaffin cells revealed that SNAP25 linker-domain mutant (similar to the SNAP25G4S used in this study) fails to rescue secretion and significantly reduces the size of primed vesicle pools even though it has the innate capacity to form productive SNARE complexes in vitro [52]. While not conclusive, this is consistent with impairments in Munc13–1 chaperoned SNARE complex assembly under these conditions, and in line with our FRET and optical-tweezer experiments (Figures 4 & 5).
This newly uncovered feature of Munc13–1 solves an enduring mystery – how is SNAP25 chaperoned into the SNARE complex – while being compatible with the several established roles of this remarkably multi-functional protein. We now see that Munc13–1 can potentially bind all three synaptic SNAREs simultaneously to sterically position them along with Munc18–1 to maximize the rate of their productive assembly into SNAREpins [10, 14]. It is well-established that the inherently cytoplasmic Munc13–1 translocates to DAG & PIP2-rich domains on the PM in response to neuronal activity and elevated intracellular Ca2+ concentration [33, 37]. On the PM, Munc13–1 initially functions as a tethering factor for local capture of SVs [3, 31, 33, 61]. At this stage, Munc13–1 is thought to adopt an ‘erect’ topology with the C1/C2B region anchored on the PM and extended (~ 15 nm) MUN domain reaching out to tether the SV via the C2C domain at its apex [24, 33, 64]. However, this erect topology seems incompatible with Munc13–1 role in catalyzing the nucleation of SNAREpins because in this extended configuration, Munc13–1 would prevent the membrane from coming closer and any nucleated SNAREpin would be inherently unstable when the involved bilayers are separated by more than ~ 10 nm [9, 46].
Munc13–1 is therefore predicted to change its orientation, ultimately to a ‘flat’ topology, in which the inter-membrane separation is reduced to ~5 nm (as observed in release-ready vesicles in the active zone) where SNAREpins can be meta-stably clamped at the approximately half-zippered state [65–67]. This reorientation is likely driven by rearrangement of the C1-C2B domains on the PM [34, 37], and may hypothetically occur via transitional states at intermediate separations that sterically favor the concerted reaction in which three SNAREs from two membranes and two molecular chaperones can favorably align. The precise molecular details of Munc13–1’s switch from a local tether to a SNARE chaperone remains to be determined. In this context, we cannot adjudicate whether Munc13–1 can sterically bind SNAP25 in the erect conformation or requires a conformational switch because the location of the binding site within the MUN domain is not yet known. Previous work has shown that that Munc18 function does not require the linker region of SNAP-25 [20], which suggests that Munc18 and Munc13 play complementary roles in chaperoning SNARE assembly.
DAG/PIP2 binding has been shown to induce recruitment and clustering of Munc13–1 on the PM surface [37, 60, 61] and nanoclusters containing ~5–10 Munc13–1 molecules have been shown to be intimately associated with SV in the readily-releasable pool by super-resolution microscopy [60, 68]. Recently, we have shown that possibly similar nanoclusters form by self-assembly of Munc13–1 protomers [61]. Local multi-valency likely augments Munc13’s chaperone functions as it does vesicle capture [33, 61]. It is easy to imagine that Munc13–1 clusters could help concentrate SNAP25 molecules at the vesicle docking site, similar to the co-cluster we observe (Figure 3) thus increasing the probability of productive SNARE complex formation. While the precise molecular details of Munc13–1 collective and overlapping interaction with the individual SNARE proteins remains to be determined, we address a long-standing question concerning the recruitment of SNAP25 to the vesicle docking site and in turn, highlight the pivotal role of Munc13–1 in orchestrating SNARE complex assembly. It is worth noting that Munc13 family of proteins are not universal for vesicular exocytosis, rather other Munc13-like factors (e.g. CAPS) have been implicated in tethering/chaperoning process. Thus, it would be interesting to also examine if the ability to bind SNAP25 (and other SNARE proteins) are conserved across different exocytosis pathways.
Supplementary Material
ACKNOWLEDGEMENT
This work was supported by National Institute of Health (NIH) grant DK027044 to JER/SSK and GM131714 to YZ.
REFERENCES
- 1.Jahn R and Fasshauer D, Molecular machines governing exocytosis of synaptic vesicles. Nature, 2012. 490(7419): p. 201–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rizo J and Xu J, The Synaptic Vesicle Release Machinery. Annu Rev Biophys, 2015. 44: p. 339–67. [DOI] [PubMed] [Google Scholar]
- 3.Sudhof TC, Neurotransmitter release: the last millisecond in the life of a synaptic vesicle. Neuron, 2013. 80(3): p. 675–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sudhof TC and Rothman JE, Membrane fusion: grappling with SNARE and SM proteins. Science, 2009. 323(5913): p. 474–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sollner T, et al. , A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell, 1993. 75(3): p. 409–18. [DOI] [PubMed] [Google Scholar]
- 6.Weber T, et al. , SNAREpins: minimal machinery for membrane fusion. Cell, 1998. 92(6): p. 759–72. [DOI] [PubMed] [Google Scholar]
- 7.Sutton RB, et al. , Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature, 1998. 395(6700): p. 347–53. [DOI] [PubMed] [Google Scholar]
- 8.Gao Y, et al. , Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science, 2012. 337(6100): p. 1340–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li F, et al. , Energetics and dynamics of SNAREpin folding across lipid bilayers. Nat Struct Mol Biol, 2007. 14(10): p. 890–6. [DOI] [PubMed] [Google Scholar]
- 10.Lai Y, et al. , Molecular Mechanisms of Synaptic Vesicle Priming by Munc13 and Munc18. Neuron, 2017. 95(3): p. 591–607 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weninger K, et al. , Single-molecule studies of SNARE complex assembly reveal parallel and antiparallel configurations. Proc Natl Acad Sci U S A, 2003. 100(25): p. 14800–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pobbati AV, Stein A, and Fasshauer D, N- to C-terminal SNARE complex assembly promotes rapid membrane fusion. Science, 2006. 313(5787): p. 673–6. [DOI] [PubMed] [Google Scholar]
- 13.Shen J, et al. , Selective activation of cognate SNAREpins by Sec1/Munc18 proteins. Cell, 2007. 128(1): p. 183–95. [DOI] [PubMed] [Google Scholar]
- 14.Ma C, et al. , Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science, 2013. 339(6118): p. 421–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dulubova I, et al. , Munc18–1 binds directly to the neuronal SNARE complex. Proc Natl Acad Sci U S A, 2007. 104(8): p. 2697–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Misura KM, Scheller RH, and Weis WI, Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature, 2000. 404(6776): p. 355–62. [DOI] [PubMed] [Google Scholar]
- 17.Shi L, et al. , Dual roles of Munc18–1 rely on distinct binding modes of the central cavity with Stx1A and SNARE complex. Mol Biol Cell, 2011. 22(21): p. 4150–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rowe J, et al. , Syntaxin 1A is delivered to the apical and basolateral domains of epithelial cells: the role of munc-18 proteins. J Cell Sci, 2001. 114(Pt 18): p. 3323–32. [DOI] [PubMed] [Google Scholar]
- 19.Arunachalam L, et al. , Munc18–1 is critical for plasma membrane localization of syntaxin1 but not of SNAP-25 in PC12 cells. Mol Biol Cell, 2008. 19(2): p. 722–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen J, et al. , SNARE bundle and syntaxin N-peptide constitute a minimal complement for Munc18–1 activation of membrane fusion. J Cell Biol, 2010. 190(1): p. 55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baker RW, et al. , A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly. Science, 2015. 349(6252): p. 1111–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shu T, et al. , Munc13–1 MUN domain and Munc18–1 cooperatively chaperone SNARE assembly through a tetrameric complex. Proc Natl Acad Sci U S A, 2020. 117(2): p. 1036–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiao J, et al. , Munc18–1 catalyzes neuronal SNARE assembly by templating SNARE association. Elife, 2018. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sitarska E, et al. , Autoinhibition of Munc18–1 modulates synaptobrevin binding and helps to enable Munc13-dependent regulation of membrane fusion. Elife, 2017. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Betz A, et al. , Direct interaction of the ratunc-13 homologue Munc13–1 with the N terminus of syntaxin. J Biol Chem, 1997. 272(4): p. 2520–6. [DOI] [PubMed] [Google Scholar]
- 26.Yang X, et al. , Syntaxin opening by the MUN domain underlies the function of Munc13 in synaptic-vesicle priming. Nat Struct Mol Biol, 2015. 22(7): p. 547–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basu J, et al. , A minimal domain responsible for Munc13 activity. Nat Struct Mol Biol, 2005. 12(11): p. 1017–8. [DOI] [PubMed] [Google Scholar]
- 28.Ma C, et al. , Munc13 mediates the transition from the closed syntaxin-Munc18 complex to the SNARE complex. Nat Struct Mol Biol, 2011. 18(5): p. 542–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang S, et al. , Munc18 and Munc13 serve as a functional template to orchestrate neuronal SNARE complex assembly. Nat Commun, 2019. 10(1): p. 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang S, et al. , Conformational change of syntaxin linker region induced by Munc13s initiates SNARE complex formation in synaptic exocytosis. EMBO J, 2017. 36(6): p. 816–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rizo J and Sudhof TC, The membrane fusion enigma: SNAREs, Sec1/Munc18 proteins, and their accomplices--guilty as charged? Annu Rev Cell Dev Biol, 2012. 28: p. 279–308. [DOI] [PubMed] [Google Scholar]
- 32.Camacho M, et al. , Heterodimerization of Munc13 C2A domain with RIM regulates synaptic vesicle docking and priming. Nat Commun, 2017. 8: p. 15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quade B, et al. , Membrane bridging by Munc13–1 is crucial for neurotransmitter release. Elife, 2019. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu X, et al. , Functional synergy between the Munc13 C-terminal C1 and C2 domains. Elife, 2016. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shin OH, et al. , Munc13 C2B domain is an activity-dependent Ca2+ regulator of synaptic exocytosis. Nat Struct Mol Biol, 2010. 17(3): p. 280–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Basu J, et al. , Munc13–1 C1 domaina ctivation lowers the energy barrier for synaptic vesicle fusion. J Neurosci, 2007. 27(5): p. 1200–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michelassi F, et al. , A C1-C2 Module in Munc13 Inhibits Calcium-Dependent Neurotransmitter Release. Neuron, 2017. 95(3): p. 577–590 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kabachinski G, et al. , CAPS and Munc13 utilize distinct PIP2-linked mechanisms to promote vesicle exocytosis. Mol Biol Cell, 2014. 25(4): p. 508–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park Y, et al. , Synaptotagmin-1 binds to PIP(2)-containing membrane but not to SNAREs at physiological ionic strength. Nat Struct Mol Biol, 2015. 22(10): p. 815–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.vandenBogaart G, et al. , Phosphatidylinositol 4,5-bisphosphate increases Ca2+ affinity of synaptotagmin-1 by 40-fold. J Biol Chem, 2012. 287(20): p. 16447–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Honigmann A, et al. , Phosphatidylinositol 4,5-bisphosphate clusters act as molecular beacons for vesicle recruitment. Nat Struct Mol Biol, 2013. 20(6): p. 679–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chamberlain LH, Burgoyne RD, and Gould GW, SNARE proteins are highly enriched in lipid rafts in PC12 cells: implications for the spatial control of exocytosis. Proc Natl Acad Sci U S A, 2001. 98(10): p. 5619–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salaun C, Gould GW, and Chamberlain LH, The SNARE proteins SNAP-25 and SNAP-23 display different affinities for lipid rafts in PC12 cells. Regulation by distinct cysteine-rich domains. J Biol Chem, 2005. 280(2): p. 1236–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krishnakumar SS, et al. , Conformational dynamics of calcium-triggered activation of fusion by synaptotagmin. Biophys J, 2013. 105(11): p. 2507–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiao J, et al. , Single-Molecule Protein Folding Experiments Using High-Precision Optical Tweezers. Methods Mol Biol, 2017. 1486: p. 357–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang YJ, et al. , Snapshot of sequential SNARE assembling states between membranes shows that N-terminal transient assembly initializes fusion. Proc Natl Acad Sci U S A, 2016. 113(13): p. 3533–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pincet F, et al. , FRAP to Characterize Molecular Diffusion and Interaction in Various Membrane Environments. PLoS One, 2016. 11(7): p. e0158457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manders EMM, Verbeek FJ, and Aten JA, Measurement of Colocalization of Objects in Dual-Color Confocal Images. Journal of Microscopy-Oxford, 1993. 169: p. 375–382. [DOI] [PubMed] [Google Scholar]
- 49.Rebane AA, Ma L, and Zhang Y, Structure-Based Derivation of Protein Folding Intermediates and Energies from Optical Tweezers. Biophys J, 2016. 110(2): p. 441–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duhr S and Braun D, Why molecules move along a temperature gradient. Proc Natl Acad Sci U S A, 2006. 103(52): p. 19678–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wienken CJ, et al. , Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun, 2010. 1: p. 100. [DOI] [PubMed] [Google Scholar]
- 52.Shaaban A, et al. , The SNAP-25 linker supports fusion intermediates by local lipid interactions. Elife, 2019. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kunii M, et al. , Opposing roles for SNAP23 in secretion in exocrine and endocrine pancreatic cells. J Cell Biol, 2016. 215(1): p. 121–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ravichandran V, Chawla A, and Roche PA, Identification of a novel syntaxin- and synaptobrevin/VAMP-binding protein, SNAP-23, expressed in non-neuronal tissues. J Biol Chem, 1996. 271(23): p. 13300–3. [DOI] [PubMed] [Google Scholar]
- 55.Wang G, et al. , Syndet is a novel SNAP-25 related protein expressed in many tissues. J Cell Sci, 1997. 110 (Pt 4): p. 505–13. [DOI] [PubMed] [Google Scholar]
- 56.Greaves J, et al. , The hydrophobic cysteine-rich domain of SNAP25 couples with downstream residues to mediate membrane interactions and recognition by DHHC palmitoyl transferases. Mol Biol Cell, 2009. 20(6): p. 1845–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nagy G, et al. , The SNAP-25 linker as an adaptation toward fast exocytosis. Mol Biol Cell, 2008. 19(9): p. 3769–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wagle S, et al. , Interaction of SNARE Mimetic Peptides with Lipid bilayers: Effects of Secondary Structure, Bilayer Composition and Lipid Anchoring. Sci Rep, 2019. 9(1): p. 7708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gimenez-Andres M, Copic A, and Antonny B, The Many Faces of Amphipathic Helices. Biomolecules, 2018. 8(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sakamoto H, et al. , Synaptic weight set by Munc13–1 supramolecular assemblies. Nat Neurosci, 2018. 21(1): p. 41–49. [DOI] [PubMed] [Google Scholar]
- 61.Li F, et al. , Vesicle capture by discrete self-assembled clusters of membrane-bound Munc13. Biorxiv, 2020. [Google Scholar]
- 62.Dubuke ML, et al. , The Exocyst Subunit Sec6 Interacts with Assembled Exocytic SNARE Complexes. J Biol Chem, 2015. 290(47): p. 28245–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sivaram MV, et al. , Dimerization of the exocyst protein Sec6p and its interaction with the t-SNARE Sec9p. Biochemistry, 2005. 44(16): p. 6302–11. [DOI] [PubMed] [Google Scholar]
- 64.Gipson P, et al. , Morphologies of synaptic protein membrane fusion interfaces. Proc Natl Acad Sci U S A, 2017. 114(34): p. 9110–9115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li X, et al. , Symmetrical organization of proteins under docked synaptic vesicles. FEBS Lett, 2019. 593(2): p. 144–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hua SY and Charlton MP, Activity-dependent changes in partial VAMP complexes during neurotransmitter release. Nat Neurosci, 1999. 2(12): p. 1078–83. [DOI] [PubMed] [Google Scholar]
- 67.Prashad RC and Charlton MP, SNARE zippering and synaptic strength. PLoS One, 2014. 9(4): p. e95130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reddy-Alla S, et al. , Stable Positioning of Unc13 Restricts Synaptic Vesicle Fusion to Defined Release Sites to Promote Synchronous Neurotransmission. Neuron, 2017. 95(6): p. 1350–1364 e12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.