Abstract
Background:
Acute lymphoblastic leukemia (ALL) is a highly heterogeneous malignancy that accounts for nearly 75% of leukemias in children. While the exact mechanism of ALL is not fully understood, some genetic variants have been implicated as associated with ALL susceptibility. The association between some genetic variants in miRNA genes and ALL risk has been described previously. A previous study suggested that mir-612 rs12803915 G> A may be associated with pediatric ALL risk. High-resolution melting (HRM) analysis is a reliable method that can be applied for polymorphism detection.
Methods:
This retrospective study was performed on 100 B-ALL patients (52 males and 48 females; age 4.6 ± 3.2 years) and 105 age- and sex-matched healthy controls (48 males and 57 females; age 5.1 ± 3 years). We used HRM to identify mir-612 rs12803915 genotypes. Sanger sequencing was applied to validate the HRM results.
Results:
High resolution melting analysis was used to genotype the mir-612 rs12803915 polymorphism. We found no association between rs12803915 allele A and B-ALL risk in any inheritance models (p> 0.05).
Conclusion:
HRM is a suitable method to detect SNP rs12803915 in the mir-612 gene; however, we found no significant association between the rs12803915 polymorphism and ALL risk.
Key Words: Childhood ALL, Hsa-mir-612, High-Resolution Melting (HRM), MicroRNA, Polymorphism
Introduction
Acute lymphoblastic leukemia (ALL) is regarded as the most prevalent cancer in children under 15 years of age. Acute lymphoblastic leukemia is a leading cause of cancer mortality among children, especially in developed countries. However, ALL is less common in adults. The early disease onset and the high incidence of ALL in individuals with some congenital genetic syndromes indicates a remarkable genetic association in its origin. Several previous studies have identified inherited variants that are associated with pediatric ALL risk (1-6).
MicroRNAs (miRNAs) are sequences of 18-25 nucleotides that regulate the expression of approximately 30-50% of all human coding genes through binding to the 3’-untranslated regions of their target mRNAs. MiRNAs are involved in various cellular processes, including proliferation, differentiation, and apoptosis; hence, alterations in miRNAs can be associated with various cancers, including ALL (7, 8). Single nucleotide polymorphisms (SNPs) are the most commonly-known variants in the human genome that account for remarkable heterogeneity among different individuals (9). Genetic polymorphisms in miRNAs, known as MirSNPs, can be found in miRNA precursors, including Pri-miRNAs, Pre-miRNAs, as well as in mature-miRNAs. MirSNPs may influence miRNA biogenesis, processing, expression levels, and binding to target mRNAs (10).
Several studies suggested that miRNA polymorphisms may be associated with susceptibility to ALL. However, the results were controversial. In 2014, Hassani et al. reported that mir-146a rs2910164 was associated with pediatric ALL risk (11). However, another study found no relationship between mir-146a and ALL risk (12). Gutierrez et al. found that both mir-3117 rs12402181 and mir-3689d2 rs62571442 increased B-ALL risk (13). Also, two independent studies suggested that Pri-miR-34b/c rs4938723 polymorphism decreased ALL risk (14, 15). Similarly, Xue Y et al. showed that miR-100 rs543412 was a protective factor from childhood ALL (16).
Several reliable genetic screening methods have been used to investigate point mutations and polymorphisms; these include DNA sequencing, TaqMan PCR assay, polymerase chain reaction-restriction-fragment-length polymorphism (PCR-RFLP), amplification refractory mutation systems (ARMS), allele-specific PCR, and high-performance liquid chromatography (HPLC). Recently, high resolution melting (HRM) analysis was introduced as a reliable technique for mutational screening. High resolution melting is a real-time-based PCR method that uses a fluorescent dye to recognize melting curve differences to identify mutations in nucleic acid sequences. The benefits of HRM include reduced time due to the lack of requirement for post-PCR procedures, and lower cost than other methods (17-21).
The mir-612 gene resides on chromosome 11q13 and consists of 1 exon. According to the NCBI dbSNP, multiple polymorphisms are present within mir-612, including rs12803915. A previous study by Gutierrez et al. suggested that mir-612 rs12803915 significantly reduced B-ALL risk in a cohort of Spanish children. However, this study was the only report that examined the association between mir-612 rs12803915 and any type of cancer risk (22, 23).
To our knowledge, no study has examined the relationship between the mir-612 rs12803915 polymorphism and pediatric B-ALL risk in Iran. Therefore, we conducted the present case-control study to investigate any possible association between the mir-612 rs12803915 polymorphism and B-ALL susceptibility using HRM.
Materials and Methods
Patients
The current study included 100 B-ALL patients (52 males and 48 females; age 4.6 ± 3.2 years) and 105 age and sex-matched healthy controls (48 males and 57 females; age 5.1 ± 3 years). All subjects referred to Cancer Molecular Pathology Research Center of Ghaem Hospital, Mashhad, during 2017-2020. Patients’ medical records were used to gather clinical information, including age, sex, and complete blood count at presentation, cytogenetic abnormalities, and immunophenotyping data (Table 1). Also, the frequencies of patients’ initial symptoms were obtained. To diagnose ALL, peripheral blood or bone marrow smears were examined by a pathologist. Next, a monoclonal antibody panel, including Tdt, CD3, CD5, CD10, CD19, CD20, and cytoplasmic IgM, was used to confirm the initial diagnosis. The presence of recurrent ALL translocations, including t (12; 21), t (9; 22), t (4; 11), and t (1; 19) was evaluated by real-time PCR. Also, numerical and structural chromosomal abnormalities were analyzed by conventional karyotyping. It was approved by the ethics committee of IRAN University of Medical Sciences (ethical code: IR.IUMS.REC.1398.470) and performed under the principles of the Declaration of Helsinki. Written informed consent was obtained from all subjects.
Table 1.
Subjects’ demographic and clinical features.
| Patients | Controls | |
|---|---|---|
| No of Individuals | 100 | 105 |
| Mean age (years) ± SD | 4.6±3.2 | 5.1±3 |
| Sex | ||
| Male | 52 (52%) | 48 (45.7%) |
| Female | 48 (48%) | 57 (54.3%) |
| CBC parameters | ||
| WBC count (x1000/ul) | 40.148±38.476 | 7.935± 0.960 |
| Hemoglobin (gr/dl) | 7.533±1.681 | 13.241±1.253 |
| PLT Count (x1000/ul) | 96.495±62.206 | 226.588±39.656 |
| Cytogenetic alterations | ||
| Hyperdiploid | 23 (23%) | ------------ |
| Hypodiploid | 3 (3%) | ------------ |
| Pseudodiploid | 28 (28%) | ------------ |
| Normal karyotype | 29 (29%) | ------------ |
| ETV6-RUNX1 (n, %) | 14 (14%) | ------------ |
| BCR-ABL | 3 (3%) | ------------ |
| AF4-MLL | 3 (3%) | ------------ |
| E2A-PBX1 | 7 (7%) | ------------ |
| Immunophenotyping data | ||
| Pro-B-ALL | 21 (21%) | ------------ |
| Early-Pre-B | 15 (15%) | ------------ |
| Pre-B-ALL | 64 (64%) | ------------ |
DNA Extraction
Genomic DNA was extracted using a FavorPrepTM Genomic DNA Extraction Kit (Favorgen, Taiwan) according to the manufacturer’s instructions. The purity and concentration of the extracted DNA were determined with agarose gel electrophoresis and a NanoDrop spectrophotometer (Thermo, USA).
HRM analysis
Mir-612 rs12803915 was genotyped using HRM. A light Cycler® 96 Real-Time PCR system (Roche Diagnostics) was used for PCR amplification and HRM. Data was analyzed using Light Cycler 96 software version 1.1. Duplicate 15 μL reactions were prepared using 20 ng of DNA, 3 μL of 5x HOT FIREPol® EvaGreen® HRM master mix (CAT number: 08-31-00001), 9 μL of ddH2O, and 1 μL of each primer (10 pmol/µ). Forward and reverse primers were 5’CAGGGCTTCTGAGCTCCTTA3’ and 5’ TGAGAGTCCTGTCCTGGCTG3’, respectively. The gene amplification protocol was as follows: pre-incubation at 95 °C for 15 min by ramp rate 4.4 °C/sec, 2-step amplification program consisted of 40 cycles of denaturation at 95 °C for 10 sec by ramp rate of 4.4 °C/sec and annealing phase at 60 °C for 20 sec by ramp rate of 2.2 °C/sec. The melting program was: denaturation at 95 °C for 1 min, 40 °C for 1 min, and a subsequent melting that included a continuous fluorescent reading from 60 °C to 95 °C at the rate of 20 acquisitions per °C. To confirm the patients’ HRM results, Sanger sequencing was performed. Each HRM curve was compared with the melting curve of confirmed samples.
Statistical analysis
Statistics were analyzed using SPSS software version 26. The Mann-Whitney U test and the chi-square test (χ2) were used to analyze continuous and categorical data, respectively. The association between the polymorphism and ALL risk was analyzed by logistic regression test and odds ratios (ORs) with 95% confidence intervals (CIs). Any deviation from Hardy-Weinberg equilibrium was analyzed by the χ2 test. P values less than 0.05 were considered to be statistically significant. Also, the sample size was determined according to the mentioned reference (24).
Results
The most common findings at presentation were fever (69.5%), lethargy and malaise (61%), hepatomegaly or splenomegaly (40%), bone and joint pain (29%), and lymphadenopathy (25%). Also, skin complications, including petechia, purpura, and ecchymosis were documented for 18% of the patients. Data regarding hematological parameters, cytogenetic abnormalities, and immunophenotyping stage are shown in Table 1.
High-resolution melting was applied to genotype the mir-612 rs12803915 polymorphism. The melting curve data displayed temperature-shift differences between the mutated and normal samples (Figs. 1 and 2). Each genotype was accurately detected by the difference plot curve (Fig. 3). Also, Sanger sequencing data was presented in Figure.4.
Fig. 1.
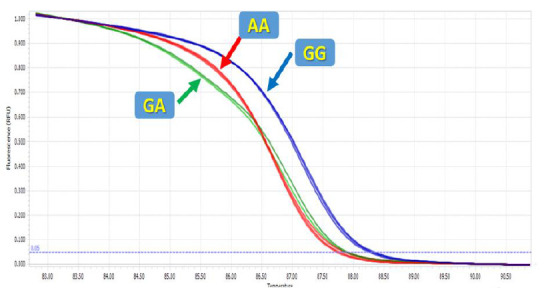
High resolution melting curves for rs12803915.
Fig. 2.
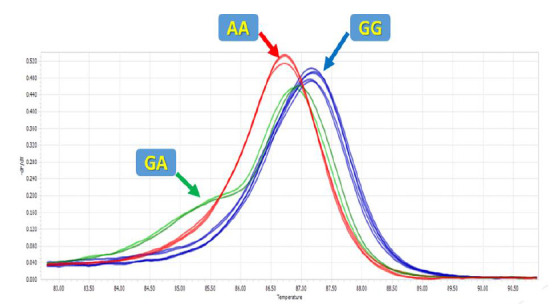
High resolution melting peaks for rs12803915.
Fig. 3.
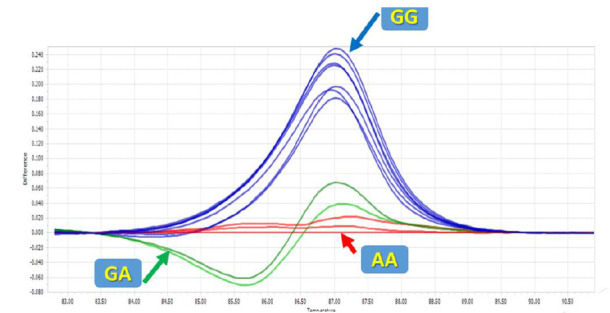
HRM difference plots for rs12803915.
Fig. 4.
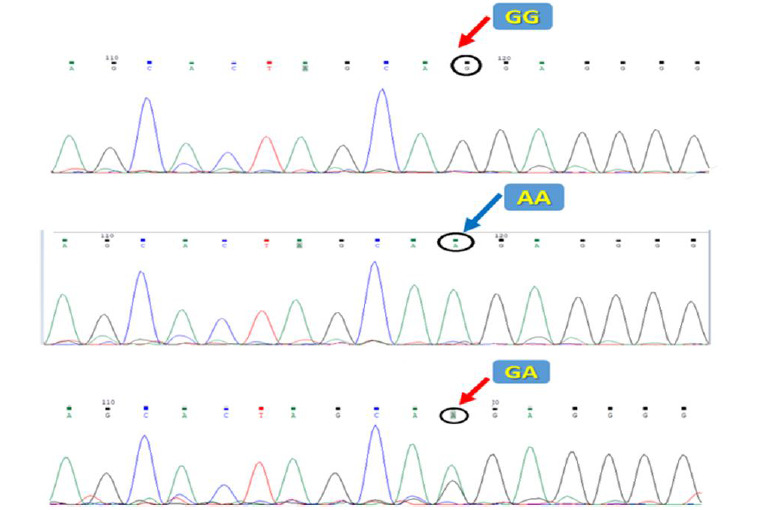
Sanger sequencing for rs12803915.
The genotypes and allelic frequencies of the mir-612 rs12803915 polymorphism in both B-ALL patients and healthy controls are shown in Table 2. The allele frequency for mir-612 rs12803915 allele A was 15%. We found no association between allele A and B-ALL risk in any inheritance models (p> 0.05).
Table 2.
Genotyping and allele frequency of mir-612 rs12803915.
| Gene polymorphism | patients N (%) | controls N (%) | OR (95% CI) | p value |
|---|---|---|---|---|
| Recessive | ||||
| GA+GG | 98 (98%) | 102 (97.1%) | ||
| AA | 2 (2%) | 3 (2.9%) | 0.694 (0.113-4.242) | 0.692 |
| Dominant | ||||
| GG | 72 (72%) | 75 (71.4%) | ||
| GA+AA | 28 (28%) | 30 (28.6%) | 0.972 (0.529-1.786) | 0.928 |
| Codominant | ||||
| GG | 72 (72%) | 75 (71.4%) | ||
| GA | 26 (26%) | 27 (25.7%) | 1.003 (0.535-1.880) | 0.992 |
| AA | 2 (2%) | 3 (2.9%) | 0.694 (0.113-4.278) | 0.694 |
| Allele | ||||
| G | 170 (85%) | 177 (84.3%) | ||
| A | 30 (15%) | 33 (15.7%) | 0.947 (0.553-1.620) | 0.841 |
Moreover, the mir-612 rs12803915 polymorphism genotypes in both cases and controls were in Hardy-Weinberg equilibrium (p= 0.84 and 0.76, respectively).
Discussion
In the current study, we showed that mir-612 rs12803915 could be genotyped by HRM analysis. However, Gutierrez et al. used a TaqMan assay to characterize this polymorphism (23). Of the various genotyping methods, Sanger sequencing is regarded as the most reliable for detecting polymorphisms; however, this method is both time-consuming and relatively expensive. The TaqMan assay requires probes that are more expensive than DNA-binding dyes. PCR-RFLP requires enzymes and multiple post-PCR steps, including restriction enzyme digestion and electrophoresis. HRM allows genotyping in a relatively short time at low cost. However, DNA samples used for HRM should be normalized in concentration using the same dilution buffer to prevent false results (17, 25-30). In the present study, we developed a protocol for the first time to genotype rs12803915 using HRM.
Several previous studies suggested HRM as a fast, convenient, and reliable assay with high sensitivity and specificity. For example, Zhang et al. found HRM sensitivity and specificity to be> 99% when compared with Sanger sequencing. Another study showed that for PCR products of 300 bp or less, all genotypes were correctly identified by HRM; however, when the PCR products were greater than 400 bp, the sensitivity and specificity decreased to 96.1 and 99.4%, respectively. In our protocol, the PCR product was 75 bp to avoid false-negative results (31, 32).
We also examined and found no association between rs12803915 in pre-mir-612 at the chromosome 11q13 locus and pediatric B-ALL risk.
Previous genome-wide association studies suggested the 11q13 locus as associated with risk of several cancers, including prostate, colon, and breast. This locus is a gene-poor region that contains multiple miRNA genes, including mir-612 (22). Dysregulation of mir-612 expression was shown in various cancers, including melanoma, bladder and colon cancers. For instance, Sheng L et al. reported that mir-612 expression was significantly less in colorectal cancer tissue than in normal tissue. They also found that increased mir-612 expression significantly reduced the proliferation and invasiveness of a colorectal cell line. Also, Zhu Y et al., and Liu M et al., showed similar tumor suppressor effects for mir-612 in melanoma and bladder cancers, respectively (33-35).
With respect to the mir-612 rs12803915 polymorphism, Kim HK et al. cloned both allelic forms of rs12803915 in pre-mir-612 into expression vectors and found that the rs12803915 SNP affected mature mir-612 expression in a cell-type-specific manner. The SNP increased mature mir-612 expression in prostate cancer cell lines, decreased it in colon cancer cell lines, and had no effect in breast cancer cell lines (22).
Gutierrez et al. examined the association between the mir-612 rs12803915 polymorphism and pediatric ALL risk in Spain. To our knowledge, this was the only study that investigated an association between this polymorphism with ALL risk. They showed that allele A was significantly protective against pediatric ALL (p= 0.007, OR: 0.61); however, after applying the restrictive Bonferroni correction, a significant p value was not obtained (23).
In our study, no significant relationship was found between the mir-612 rs12803915 polymorphism and ALL risk. The discrepancies between the studies may be related to racial and environmental differences between the populations. One limitation of the present study is the relatively small sample size, which may limit the statistical power of our analysis. To clarify the effect of this polymorphism on cancer risk, especially B-ALL, studies with different ethnicities and larger sample sizes are needed.
In summary, we successfully used HRM to detect rs12803915 in the mir-612 gene. However, the SNP was not associated with pediatric B-ALL risk in our population.
Acknowledgements
This paper was funded by Iran University of Medical Sciences (grant number: #14298). Also, we thank and appreciate Mashhad University of Medical Science (MUMS) and staff of Cancer Molecular Pathology department of GHAEM Hospital for their valuable assistance.
All of the authors declare that there are no conflicts of interests.
References
- 1.Esparza SD, Sakamoto KM. Topics in pediatric leukemia–acute lymphoblastic leukemia. MedGenMed. 2005;7(1):23. [PMC free article] [PubMed] [Google Scholar]
- 2.Silva-Junior AL, Alves FS, Kerr MW, Xabregas LA, Gama FM, Rodrigues MG, et al. Acute lymphoid and myeloid leukemia in a Brazilian Amazon population: Epidemiology and predictors of comorbidity and deaths. . PloS one. 2019;14(8):e0221518. doi: 10.1371/journal.pone.0221518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiemels JL, Walsh KM, de Smith AJ, Metayer C, Gonseth S, Hansen HM, et al. GWAS in childhood acute lymphoblastic leukemia reveals novel genetic associations at chromosomes 17q12 and 8q24.21. . Nat Commun. 2018;9(1):286. doi: 10.1038/s41467-017-02596-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo LM, Xi JS, Ma Y, Shao L, Nie CL, Wang GJ. ARID5B gene rs10821936 polymorphism is associated with childhood acute lymphoblastic leukemia: a meta-analysis based on 39,116 subjects. . Tumour Biol. 2014;35(1):709–13. doi: 10.1007/s13277-013-1097-0. [DOI] [PubMed] [Google Scholar]
- 5.Woo JS, Alberti MO, Tirado CA. Childhood B-acute lymphoblastic leukemia: a genetic update. Experimental hematology & oncology. 2014;3:16. doi: 10.1186/2162-3619-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esmaili MA, Kazemi A, Zaker F, Faranoush M, Rezvany MR. Effects of Reduced Mir-24 Expression on Plasma Methotrexate Levels, Therapy-Related Toxicities, and Patient Outcomes in Pediatric Acute Lymphoblastic Leukemia. . Reports of Biochemistry & Molecular Biology. . 2020;8(4):358. [PMC free article] [PubMed] [Google Scholar]
- 7.Mishra PJ, Banerjee D, Bertino JR. MiRSNPs or MiR-polymorphisms, new players in microRNA mediated regulation of the cell: Introducing microRNA pharmacogenomics. . Cell cycle. 2008;7(7):853–8. doi: 10.4161/cc.7.7.5666. [DOI] [PubMed] [Google Scholar]
- 8.Luan C, Yang Z, Chen B. The functional role of microRNA in acute lymphoblastic leukemia: relevance for diagnosis, differential diagnosis, prognosis, and therapy. . Onco Targets Ther. 2015;8:2903–2914. doi: 10.2147/OTT.S92470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10(6):389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dzikiewicz-Krawczyk A. MicroRNA polymorphisms as markers of risk, prognosis and treatment response in hematological malignancies. Crit Rev Oncol Hematol. 2015;93(1):1–17. doi: 10.1016/j.critrevonc.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 11.Hasani SS, Hashemi M, Eskandari-Nasab E, Naderi M, Omrani M, Sheybani-Nasab M. A functional polymorphism in the miR-146a gene is associated with the risk of childhood acute lymphoblastic leukemia: a preliminary report. . Tumor Biol. . 2014;35(1):219–25. doi: 10.1007/s13277-013-1027-1. [DOI] [PubMed] [Google Scholar]
- 12.Jemimah Devanandan H, Venkatesan V, Scott JX, Magatha LS, Durairaj Paul SF, Koshy T. MicroRNA 146a polymorphisms and expression in Indian children with acute lymphoblastic leukemia. Lab Med. . 2019;50(3):249–253. doi: 10.1093/labmed/lmy074. [DOI] [PubMed] [Google Scholar]
- 13.Gutierrez-Camino A, Martin-Guerrero I, Dolzan V, Jazbec J, Carbone-Baneres A, Garcia de Andoin N, et al. Involvement of SNPs in miR-3117 and miR-3689d2 in childhood acute lymphoblastic leukemia risk. Oncotarget. 2018;9(33):22907–22914. doi: 10.18632/oncotarget.25144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tong N, Chu H, Wang M, Xue Y, Du M, Lu L, et al. Pri-miR-34b/c rs4938723 polymorphism contributes to acute lymphoblastic leukemia susceptibility in Chinese children. Leuk Lymphoma. . 2016;57(6):1436–41. doi: 10.3109/10428194.2015.1092528. [DOI] [PubMed] [Google Scholar]
- 15.Hashemi M, Bahari G, Naderi M, Sadeghi-Bojd S, Taheri M. Pri-miR-34b/c rs4938723 polymorphism is associated with the risk of childhood acute lymphoblastic leukemia. Cancer Genet. 2016;209(11):493–496. doi: 10.1016/j.cancergen.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 16.Xue Y, Yang X, Hu S, Kang M, Chen J, Fang Y. A genetic variant in miR-100 is a protective factor of childhood acute lymphoblastic leukemia. . Cancer Med. . 2019;8(5):2553–2560. doi: 10.1002/cam4.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin CY, Li MJ, Chang JG, Liu SC, Weng T, Wu KH, et al. High-resolution melting analyses for genetic variants in ARID5B and IKZF1 with childhood acute lymphoblastic leukemia susceptibility loci in Taiwan. Blood Cells Mol Dis. 2014;52(2-3):140–5. doi: 10.1016/j.bcmd.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Vali Z, Raz A, Bokharaei H, Nabavi M, Bemanian MH, Yazdi MS, et al. Development of a high-resolution melting analysis method based on SYBR Green-I for rs7216389 locus genotyping in asthmatic child patients. . Avicenna J Med Biotechnol. 2014;6(2):72–80. [PMC free article] [PubMed] [Google Scholar]
- 19.Qi JH, Wang J, Chen J, Shen F, Huang JT, Sen S, et al. High-resolution melting analysis reveals genetic polymorphisms in microRNAs confer hepatocellular carcinoma risk in Chinese patients. . BMC cancer. . 2014;14(1):643. doi: 10.1186/1471-2407-14-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ponchel F, Toomes C, Bransfield K, Leong FT, Douglas SH, Field SL, et al. Real-time PCR based on SYBR-Green I fluorescence: an alternative to the TaqMan assay for a relative quantification of gene rearrangements, gene amplifications and micro gene deletions. BMC Biotechnol. 2003;3:18. doi: 10.1186/1472-6750-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niya MH, Basi A, Koochak A, Tameshkel FS, Rakhshani N, Zamani F, et al. Sensitive high-resolution melting analysis for screening of KRAS and BRAF mutations in Iranian human metastatic colorectal cancers. . Asian Pac J Cancer Prev. 2016;17(12):5147–5152. doi: 10.22034/APJCP.2016.17.12.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim HK, Prokunina-Olsson L, Chanock SJ. Common genetic variants in miR-1206 (8q24. 2) and miR-612 (11q13. 3) affect biogenesis of mature miRNA forms. PloS one. 2012;7(10):e47454. doi: 10.1371/journal.pone.0047454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gutierrez-Camino A, Lopez-Lopez E, Martin-Guerrero I, Piñan MA, Garcia-Miguel P, Sanchez-Toledo J, et al. Noncoding RNA–related polymorphisms in pediatric acute lymphoblastic leukemia susceptibility. . Pediatric research. . 2014 ;75(6):767–773. doi: 10.1038/pr.2014.43. [DOI] [PubMed] [Google Scholar]
- 24.Pourhoseingholi MA, Vahedi M, Rahimzadeh M. Sample size calculation in medical studies. Gastroenterol Hepatol Bed Bench. . 2013;6(1):14–17. [PMC free article] [PubMed] [Google Scholar]
- 25.Er TK, Chang JG. High-resolution melting: applications in genetic disorders. . Clin Chim Acta. 2012;414:97–201. doi: 10.1016/j.cca.2012.09.012. [DOI] [PubMed] [Google Scholar]
- 26.Vossen RH, Aten E, Roos A, den Dunnen JT. High-Resolution Melting Analysis (HRMA)—More than just sequence variant screening. . Hum Mutat. . 2009;30(6):860–6. doi: 10.1002/humu.21019. [DOI] [PubMed] [Google Scholar]
- 27.Wittwer CT. High-resolution DNA melting analysis: advancements and limitations. . Hum Mutat. 2009;30(6):857–9. doi: 10.1002/humu.20951. [DOI] [PubMed] [Google Scholar]
- 28.Reed GH, Kent JO, Wittwer CT. High-resolution DNA melting analysis for simple and efficient molecular diagnostics. 2007;8(6) doi: 10.2217/14622416.8.6.597. [DOI] [PubMed] [Google Scholar]
- 29.Boyd EM, Bench AJ, van‘t Veer MB, Wright P, Bloxham DM, Follows GA, et al. High resolution melting analysis for detection of BRAF exon 15 mutations in hairy cell leukaemia and other lymphoid malignancies. Br J Haematol. 2011;155(5):609–12. doi: 10.1111/j.1365-2141.2011.08868.x. [DOI] [PubMed] [Google Scholar]
- 30.Montgomery JL, Sanford LN, Wittwer CT. High-resolution DNA melting analysis in clinical research and diagnostics. Expert review of molecular diagnostics. . 2010;10(2):219–40. doi: 10.1586/erm.09.84. [DOI] [PubMed] [Google Scholar]
- 31.Zhang L, Cui G, Li Z, Wang H, Ding H, Wang DW. Comparison of high-resolution melting analysis, TaqMan Allelic discrimination assay, and Sanger sequencing for Clopidogrel efficacy genotyping in routine molecular diagnostics. J Mol Diagn. 2013;15(5):600–6. doi: 10.1016/j.jmoldx.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 32.Reed GH, Wittwer CT. Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin Chem. . 2004;50(10):1748–54. doi: 10.1373/clinchem.2003.029751. [DOI] [PubMed] [Google Scholar]
- 33.Zhu Y, Zhang HL, Wang QY, Chen MJ, Liu LB. Overexpression of microRNA-612 restrains the growth, invasion, and tumorigenesis of melanoma cells by targeting Espin. . Mol Cells. 2018;41(2):119–126. doi: 10.14348/molcells.2018.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu M, Chen Y, Huang B, Mao S, Cai K, Wang L, et al. Tumor-suppressing effects of microRNA-612 in bladder cancer cells by targeting malic enzyme 1 expression. . International journal of oncology. 2018;52(6):1923–1933. doi: 10.3892/ijo.2018.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheng L, He P, Yang X, Zhou M, Feng Q. miR-612 negatively regulates colorectal cancer growth and metastasis by targeting AKT2. Cell death & disease. . 2015;6(7):e1808. doi: 10.1038/cddis.2015.184. [DOI] [PMC free article] [PubMed] [Google Scholar]


