Abstract
Introduction
Researching carbapenem-resistant isolates enables the identification of carbapenemase-producing bacteria and prevents their spread.
Methods
P. aeruginosa isolates were recovered from Medicine Faculty of Recep Tayyip Erdoğan University and identified by conventional methods and the automated Vitek 2 Compact system. Antimicrobial susceptibility experiments were performed in accordance with CLSI criteria and the automated Vitek 2 Compact system. The PCR method was investigated for the presence of β-lactamase resistance genes. PFGE typing was performed to show clonal relation among samples.
Results
Seventy P. aeruginosa isolates were isolated from seventy patients. Of the patients, 67.1% had contact with the health service in the last 90 days and 75.7% of the patients had received antimicrobial therapy in the previous 90 days. Twenty-four isolates were carbapenem resistant, 2 isolates were multidrug-resistant except colistin, and none of the samples had colistin resistance. The gene encoding β-lactamase or metallo-β-lactamase was found in a total of 36 isolates. The blaVEB and blaPER genes were identified in 1 and 5 isolates alone or 17 and 13 isolates in combination with other resistance genes, respectively. The blaNDM was the most detected metallo-β-lactamase encoding gene (n=18), followed by blaKPC (n=12). blaIMP and blaVIM were detected in 5 and 1 isolates, respectively. Also, the association of blaVEB-blaPER and blaVEB-blaKPC-blaNDM was found to be very high. Much more resistance genes and co-occurrence were detected in hospital-acquired samples than community-acquired samples. No difference was found between the community and hospital-associated isolates according to PFGE results. Simultaneously from 6 patients, other microorganisms were also isolated and 5 of them died.
Conclusion
The average length of stay (days) was found to be significantly higher in HAI group than CAI group. The death of 5 patients with fewer or no resistance genes showed that the co-existence of other microorganisms in addition to resistance genes was important on death.
Keywords: Pseudomonas aeruginosa, antibiotic resistance genes, epidemiology, PFGE
Introduction
P. aeruginosa is an opportunistic gram-negative bacteria commonly found on many surfaces. Its rapid colonization on living and non-living areas leads to a wide range of diseases, including community and health-associated infections.1,2 P. aeruginosa is very difficult to control in the hospital environment due to its versatile and ubiquitous features.3 Infections caused by P. aeruginosa have been increased significantly in healthcare units, affecting specially patient with chronic diseases or with immunocompromised system.1,3
Infections caused by P. aeruginosa are difficult to treat because of its intrinsic resistance to many antipseudomonal agents and the ability to easily acquire antibiotic resistance.4 In hospital infections caused by P. aeruginosa isolates with multiple drug resistance, mortality, hospitalization, and treatment costs increase, especially in intensive care units. Mortality decreases significantly if early and appropriate antipseudomonal treatment is initiated.5,6 Carbapenems are powerful broad-spectrum-β-lactam antibiotics commonly used in the treatment of P. aeruginosa. However, increased carbapenem resistance among these organisms has been found worldwide, and carbapenem-resistant P. aeruginosa ranges from 10% to 50% in most countries. Few antibiotic options are available for treatment, and multidrug resistance is much more common in patients infected with carbapenem-resistant P. aeruginosa.7,8
In this study, it was aimed to determine the antimicrobial resistance profile of P. aeruginosa isolates isolated from culture samples sent from various polyclinics and services in a university hospital and thought to be an infectious agent of community (CAI) and hospital-associated (HAI), to screen the resistance genes and to investigate the clonal relationship between isolates.
Materials and Methods
Bacterial Isolates and Identification
Seventy clinical P. aeruginosa isolates were recovered from different units and samples (blood, urine, wound swab sample, sputum, tracheal aspirate, cerebrospinal fluid (CSF), bronchoalveolar lavage) in Medicine Faculty of Recep Tayyip Erdoğan University between April 2015 and October 2016. The clinical data of the patients were obtained retrospectively from file records and medical tables. One sample of each patient classified as a hospital (HAI, n=27) and community-associated (CAI, n=43) was evaluated. The isolates were identified by conventional methods and the automated Vitek 2 Compact (BioMerieux, France) system.
Antibiotic Susceptibility Testing
Antimicrobial susceptibility experiments were performed in accordance with CLSI (Clinical and Laboratory Standards Institute) criteria using both the Kirby–Bauer disc diffusion method and the automated Vitek 2 Compact (BioMerieux, France) system. In this context, amikacin (AK), gentamicin (GN), ciprofloxacin (CIP), ceftazidime (CAZ), piperacillin (PRL), piperacillin-tazobactam (TZP), cefoperazone (CEP), cefoperazone/sulbactam (CES) aztreonam (ATM), meropenem (MEM), cefepime (FEP), imipenem (IPM), levofloxacin (LEV) and colistin (CT) sensitivity was evaluated. P. aeruginosa ATCC 27853 was used as a quality control isolate.
Detection of β-Lactamase Encoding Resistance Genes
The presence of β-lactamase coding genes was investigated by the PCR method (Table 1). To obtain template DNA, a single colony bacterial isolates were inoculated into 3 mL of Luria–Bertani (LB) broth medium (1% tryptone, 0.5% yeast extract, 0.5% NaCl, pH 7.4) at 37 °C for 16 hours at 200 rpm. One and half mL of the grown culture was transferred to eppendorf tubes and centrifuged at 10,000 rpm for 5 minutes in microcentrifuge. The supernatant part was poured and 1 mL of sterile distilled water was added to the pellet and vortexed (twice). One mL of sterile distilled water was added to the pellet and the bacterial suspension was boiled at 95 °C for 10 minutes. After boiling, the bacteria suspension was centrifuged at 13,000 rpm for 5 minutes. Three µL of the supernatant part was used as template DNA in PCR. Standard PCR mixtures were prepared as follows: 1.5 units Taq DNA polymerase I (Solis Biodyne), 5 μL DNA, 10 μL 5X DNA polymerase buffer (Solis Biodyne), 3 μL of 25 mM MgCl2, 2.5 μL of 4 mM each dNTP and 1 μL each primer stock (10 pmol/μL) and sterile deionized water was added to complete 50 μL of final volume. PCR reactions occurred with the following amplification conditions: 3 min 94 °C initial denaturation, followed by 34 cycles of denaturation at 94 °C 45 sec, annealing at 55 °C 1 min, extension at 70 °C 3 min, and final extension step 5 min at 70 °C. Amplification products were run in 1.5–2% agarose gel and visualized with UV light. The oligonucleotides used in PCR and their melting temperature (Tm) are given in Table 1.
Table 1.
Primers Used in the Amplification of Selected Genes
| Primer | 5′-3′ Sequence | Amplicon Size (bp) | Tm (°C) | Reference |
|---|---|---|---|---|
| VEB | F- ATTTCCCGATGCAAAGCGT | 542 | 55 | [9] |
| R- TTATTCCGGAAGTCCCTGT | ||||
| PER | F- ATGAATGTCATCACAAAAT | 927 | 50 | [10] |
| R- TCAATCCGGACTCACT | ||||
| IMP | F:CATGGTTTGGTGGTTCTTGT | 488 | 56 | [11] |
| R:ATAATTTGGCGGACTTTGGC | ||||
| VIM | F:ATTGGTCTATTTGACCGCGTC | 780 | 58 | [11] |
| R:TGCTACTCAACGACTGAGCG | ||||
| NDM | F:TGGAATTGCCCAATATTATGC | 813 | 54 | [12] |
| R:TCAGCGCAGCTTGTCGGCCATGC | ||||
| KPC | F: CGTTCTTGTCTCTCATGGCC | 796 | 52 | [13] |
| R: CCTCGCTGTGCTTGTCATCC | ||||
| OXA-48 | F:CAGTCAAGTTCAACCCAACCG | 438 | 55 | [14] |
| R:GCGTGGTTAAGGATGAACAC | ||||
| OXA- 51 | F: TAATGCTTTGATCGGCCTTG | 353 | 52 | [15] |
| R:TGGATTGCACTTCATCTTGG | ||||
| OXA–23 | F:GATCGGATTGGAGAACCAGA | 501 | 52 | [15] |
| R:ATTTCTGACCGCATTTCCAT |
Abbreviations: Tm, melting temperature; bp, base pairs.
Molecular Typing of the P. aeruginosa Isolates
PFGE typing was performed on 70 isolates to show clonal relation among samples. Isolation and deproteinization of the genomic DNA were done following the protocol of Durmaz et al with minor modifications.16 Briefly, P. aeruginosa colonies on Mueller Hinton agar were suspended in 1 mL HST (100mm Tris-HCl, 100mm EDTA, pH 8.0) and the optical density was adjusted to 0.8 (590 nm). The cells were embedded into low (2%) melting agarose. Molds were prepared with the help of agarose and kept in the refrigerator at 4 ° C for 10 minutes. The prepared isolate molds were added HLS-1 (50 mm Tris-HCl, 50 mm EDTA, pH 8.0, 2.5 mg/mL Lysozyme, 1.5 mg/mL Proteinase K) and incubated at 37 °C for 1 hour in a water bath. At the end of the period, HLS-1 was removed. HLS-2 (0.5 M EDTA, 1% sarcosyl, 400 µg/mL Proteinase K) was placed on molds and incubated in a water bath at 55 °C for 2 hours. After the lysis step, the molds were washed 3 times each with dH2O and 1XTE (Tris-EDTA). After digestion of the cells and washing of the plugs, genomic DNA in the agarose plugs was restricted by 10 U of SpeI (Thermo) for 3 hours at 37 °C in a incubator. DNA fragments were separated on 1% pulse-field certified agarose (BIO-RAD) gels run in 0.5X Tris-borate-EDTA buffer (44.5 mM/LTris, 44.5 mM/L boric acid, 1 mM/L EDTA [pH8.0]) by using a CHEF-DR II system (Bio-Rad Laborato-ries). The electrophoresis conditions were 14°C at 6V/cm2 for 18 hours. The initial and final switch times were 5 seconds and 20 seconds, respectively. The gel was stained with ethidium bromide (5mg/mL) for 30 mins and photographed under UV light. The DNA band profiles were analyzed with Gel Compar software (version 3.0; Applied Maths, Sint-Martens-Latem, Belgium) dendrogram and Dice. According to the interpretative criteria of Tenover et al, the clinical isolates were aligned according to PFGE profile, where the profiles sharing 100% similarity is grouped in clusters.17
Statistical Analysis
SPSS 21.0 (NY IBM Corp., 2012) program was used. SPSS 21.0 (NY IBM Corp., 2012) program was used. Categorical variables were expressed in terms of frequency (n) and percentage (%).The χ2 test and Fisher’s exact test were used to compare categorical data. P <0.05 significance level was accepted.
Results and Discussion
Epidemiological Results
One sample from each patient was accepted into the study and seventy P. aeruginosa isolates were obtained from seventy patients (Table 2).
Table 2.
Distribution of Cases According to Service and Intensive Care Units
| Total (n=70) | HAI (n=27) | CAI (n=43) | |
|---|---|---|---|
| Services, n (%) | |||
| IMICU | 14 (20.0) | 11 (40.7) | 3 (7.0) |
| SICU | 12 (17.2) | 8 (29.6) | 4 (9.3) |
| ARICU | 4 (5.7) | 3 (11.1) | 1 (2.3) |
| CVSICU | 3 (4.3) | 1 (3.7) | 2 (4.7) |
| CICU | 2 (2.8) | 1 (3.7) | 1 (2.3) |
| Other services | 35 (50.0) | 3 (11.1) | 32 (74.4) |
Abbreviations: IMUCU, Internal Medicine Intensive Care Unit; SICU, Surgery Intensive Care Unit; ARICU, Anesthesia and Reanimation Intensive Care Unit; CVSICU, Cardiovascular Surgery Intensive Care Unit; CICU, Coronary Intensive Care Unit.
One sample from each patient was accepted into the study and seventy P. aeruginosa isolates were obtained from seventy patients (Table 2). The average age of 70 cases was 66 with minimum 17 and maximum 92 years old. The average age of HAI and CAI was 38.6% (n=27) and 61.4% (n=43) respectively. The two groups were similar in terms of age (z=−1.829, p=0.067) and gender distribution (χ2 (1)= 0.509, p=0.476). The average length of stay (days) was found to be significantly higher in the group with HAI than in the group with CAI (22 vs 18 days). Thirty-three isolates (47.1%) were isolated from respiratory secretions and most of them were respiratory infections (n=16 community-acquired pneumonia (CAP) and n=13 ventilator-associated pneumonia (VAP)). The ratio of the patients contacted with the health service in the last 90 days was 67.1% and 75.7% of the patients had received antimicrobial therapy in the previous 90 days. The most common comorbidity was cardiovascular diseases (38.6%). Respiratory pathologies constituted 27.1% (n=19) of the patients in the distribution of the first (preliminary) diagnosis at the time of admission to the hospital, other diagnoses were urinary system infections (18.6%, n=13), neurological pathologies (12.9%, n=9) Gastro Intestinal System (GIS) pathologies (11.4%, n=8), Cardiovascular System (CVS) pathologies (8.6% n=6), trauma (8.6% n=6) skin-soft tissue pathologies (7.1% n=5) and oncological pathologies (5.7% n=4) (Table 3). When the two groups were evaluated in terms of the distribution of hospitalization indications, it was found that the most complaint was respiratory failure (n=14) and the presence of mental change was significant in the HAI group (χ2 (1) =6.756, Fisher’s exact test, p=0 0.019). The history of hospital contact and use of antibiotics in the last 3 months was high in all patients (81.5% and 72.1% in HAI and CAI, respectively). While 33.3% (n=9) of patients with HAI had an operative history in the last one month, this rate was 14% (n=6) in cases with TAI (χ2 (1) =3.700, p=0.054). The detailed descriptions of demographic and pathological characteristics of HAI and CAI could be found in Table 3.
Table 3.
Comparison of Demographic and Pathological Characteristics of HAI (n=27) and CAI (n=43) Groups
| Total | HAI | CAI | Statistics | ||
|---|---|---|---|---|---|
| n=70 | n=27 | n=43 | z, χ2 | p value | |
| Age (year) | 66 (17–92) | 73 (18–92) | 64 (17–89) | −1.829 | 0.067 |
| Over 65 years old | 16 (81±6.9) | 20 (75±6.6) | (%95 confidal) | 0.0013 | |
| Duration of stay (days) | 19.5 (1–72) | 22 (14–72) | 18 (1–39) | −2.921 | 0.003 |
| Gender | |||||
| Women | 27 (38.6) | 9 (33.3) | 18 (41.9) | 0.509 | 0.476 |
| Male | 43 (61.4) | 18 (66.7) | 25 (58.1) | ||
| Surgery history (last 1 month) | 15 (21.4) | 9 (33.3) | 6 (14.0) | 3.700 | 0.054 |
| Hospital history (last 3 months) | 47 (67.1) | 16 (59.3) | 31 (72.1) | 1.238 | 0.266 |
| Antibiotic utilization (last 3 months) | 53 (75.7) | 22 (81.5) | 31 (72.1) | 0.795 | 0.373 |
| First (Pre) diagnosis, n (%) | – | – | |||
| Respiratory pathologies | 19 (27.1) | 10 (37.0) | 9 (20.9) | ||
| Urinary pathologies | 13 (18.6) | 0 | 13 (30.2) | ||
| Neurological pathologies | 9 (12.9) | 5 (18.5) | 4 (9.3) | ||
| GIS pathologies | 8 (11.4) | 5 (18.5) | 3 (7.0) | ||
| CVS pathologies | 6 (8.6) | 2 (7.4) | 4 (9.3) | ||
| Trauma | 6 (8.6) | 3 (11.1) | 3 (7.0) | ||
| Soft-tissue pathologies | 5 (7.1) | 0 | 5 (11.6) | ||
| Oncological pathologies | 4 (5.7) | 2 (7.4) | 2 (4.7) | ||
| Hospitalization indication, n (%) | |||||
| Respiratory Failure | 14 (20.0) | 6 (22.2) | 8 (18.6) | 0.136 | 0.713 |
| Mental changes | 4 (5.7) | 4 (14.8) | 0 | 6.756* | 0.019 |
| Hemodynamic instability | 4 (5.7) | 1 (3.7) | 3 (7.0) | 0.330* | 0.498 |
| Post CPR | 7 (10.0) | 4 (14.8) | 3 (7.0) | 1.132* | 0.253 |
Note: *Fisher’s exact test.
Considering the invasive procedures and applications during the P. aeruginosa isolation period, the presence of decubitus with hemodialysis, endotracheal tube application, nasogastric tube application, peripheric artery catheter application, urinary catheter and mechanical ventilator applications was found to be significantly higher in the HAI group compared to the CAI group (p <0.01). Bronchoscopy was performed on HAI patients but it was not performed on CAI patients. It was found to be significantly higher as a risk factor in patients with HAI (p = 0.038). The rate of performing cystostomy was similar in both groups. The detailed descriptions of diagnosis, clinical features and risk factors of HAI and CAI groups could be found in Table 4.
Table 4.
Diagnosis, Clinical Features and Risk Factors of HAI (n =27) and CAI (n =43) Groups
| Total | HAI | CAI | Statistics | ||
|---|---|---|---|---|---|
| n=70 | n=27 | n=43 | z, χ2 | p value | |
| Final diagnosis on the PA isolation date (%) | |||||
| VAP (ventilator-associated pneumonia) | 13 (18.6) | 13 (48.2) | 0 | - | - |
| KSI (blood stream infection) | 6 (8.6) | 6 (22.2) | 0 | ||
| HP (hospital pneumonia) | 6 (8.6) | 6 (22.2) | 0 | ||
| HUTI (hospital urinary tract infection) | 1 (1.4) | 1 (3.7) | 0 | ||
| UTI (urinary tract infection) | 16 (22.9) | 0 | 16 (37.2) | ||
| CAP (community-associated pneumatic) | 16 (22.9) | 0 | 16 (37.2) | ||
| STI (soft tissue infection) | 6 (8.5) | 0 | 6 (14.0) | ||
| IAI (intra-abdominal infection) | 3 (4.2) | 0 | 3 (6.9) | ||
| CBSI (community-bloodstream infection) | 2 (2.9) | 0 | 2 (4.7) | ||
| Meningitis | 1 (1.4) | 1 (3.7) | 0 | ||
| Comorbid patients, n (%) | |||||
| CVS diseases | 27 (38.6) | 12 (44.4) | 15 (34.9) | 0.640 | 0.424 |
| KOAH | 12 (17.1) | 5 (18.5) | 7 (16.3) | 0.059* | 0.526 |
| Neurological diseases | 9 (12.9) | 5 (18.5) | 4 (9.3) | 1.257* | 0.292 |
| Diabetes mellitus | 9 (12.9) | 3 (11.1) | 6 (14.0) | 0.120* | 0.517 |
| Malignant diseases | 7 (10.0) | 3 (11.1) | 4 (9.3) | 0.060* | 0.554 |
| Renal diseases | 6 (8.6) | 1 (3.7) | 5 (11.6) | 1.329* | 0.394 |
| Skin-soft tissue pathologies | 5 (7.1) | 2 (7.4) | 3 (7.0) | 0.005* | 0.645 |
| Invasive procedures and applications during the PA isolation period, n (%) | |||||
| Hemodialysis | 24 (34.3) | 19 (70.4) | 5 (11.6) | 25.402 | 0.000 |
| Endotracheal application | 27 (38.6) | 23 (85.2) | 4 (9.3) | 40.307 | 0.000 |
| Enteral nutrition | 49 (70.0) | 16 (59.3) | 33 (76.7) | 2.415 | 0.120 |
| Peripheral venous catheter | 48 (68.6) | 18 (66.7) | 30 (69.8) | 0.074 | 0.786 |
| Nasogastric | 26 (37.1) | 16 (59.3) | 10 (23.3) | 9.209 | 0.002 |
| Peripheral artery catheter | 24 (34.3) | 19 (70.4) | 5 (11.6) | 25.402 | 0.000 |
| Decubitus | 22 (31.4) | 17 (63.0) | 5 (11.6) | 20.281 | 0.000 |
| Central venous catheter | 21 (30.0) | 11 (40.7) | 10 (23.3) | 2.415 | 0.120 |
| Urinary catheter | 26 (37.1) | 16 (59.3) | 10 (23.3) | 9.209 | 0.002 |
| Mechanical ventilator | 21 (30.0) | 17 (63.0) | 4 (9.3) | 22.742 | 0.000 |
| Total parenteral nutrition | 14 (20.0) | 3 (11.1) | 11 (25.6) | 2.171 | 0.141 |
| Transfusion | 14 (20.0) | 6 (22.2) | 8 (18.6) | 0.136 | 0.713 |
| Cystostomy | 13 (18.6) | 6 (22.2) | 7 (16.3) | 0.387 | 0.534 |
| Bronchoscopy | 7 (10.0) | 7 (16.3) | 0 | 4.884* | 0.038 |
| Steroid | 7 (10.0) | 3 (11.1) | 4 (9.3) | 0.060* | 0.554 |
| Tracheotomy | 3 (4.3) | 1 (3.7) | 2 (4.7) | 0.036* | 0.671 |
| Chest tube | 3 (4.3) | 0 | 3 (7.0) | 1.968* | 0.279 |
Note: *Fisher’s exact test.
Antibiotic Susceptibility
Thirteen antibiotics were used to determine the resistance phenotype of 70 P. aeruginosa in this study (Table 5). While the highest resistance was observed against piperacillin (n = 36), colistin resistance was not observed in any isolate. Twenty-four (34.3%) isolates were carbapenem resistant.
Table 5.
Resistance Rates of P. aeruginosa Isolates
| Antibiotics | Resistant | Sensitive | Medium Sensitive |
|---|---|---|---|
| Ceftazidim | 30 (42.9) | 34 (48.6) | 6 (8.6) |
| Cefepim | 34 (48.6) | 29 (41,4) | 7 (10.0) |
| Cefoperazone | 12 (17.1) | 58 (82.9) | 0 |
| Cefoperazone-sulbactam | 10 (14.3) | 60 (85.7) | 0 |
| Piperacillin | 36 (51.4) | 30 (42.9) | 4 (5.7) |
| Piperacillin-tazobactam | 34 (48.6) | 29 (41.4) | 7 (10.0) |
| Amikacin | 10 (14.4) | 54 (77.0) | 6 (8.6) |
| Gentamicin | 12 (17.1) | 54 (77.0) | 4 (5.7) |
| Ciprofloxacin | 18 (25.7) | 47 (67.2) | 5 (7.1) |
| Levofloxacin | 28 (40.0) | 38 (54.3) | 4 (5.7) |
| Colistin | 0 | 70 (100) | 0 |
| Imipenem | 13 (18.6) | 56 (80.0) | 1 (1.4) |
| Meropenem | 18 (25.7) | 49 (70.0) | 3 (4.3) |
| Carbapenem resistance | 24 (34.3) | 46 (65.7) | 0 |
A meta-analysis compared the period between 2007 and 2016, evaluated the pooled resistance prevalence of P. aeruginosa to piperacillin-tazobactam, ceftazidime, cefepime, meropenem, imipenem, ciprofloxacin, gentamicin, amikacin, tobramycin, and colistin were 33.9%, 38.6%, 35.6%, 30.1%, 28.0%, 30.7%, 28.2%, 17.8%, 15.7%, and 2.2%, respectively.18 They found the resistance rates of piperacillin, piperacillin–tazobactam, imipenem, meropenem, amikacin and colistin significantly increased between 2012 and 2016; however, gentamicin, tobramycin and ciprofloxacin resistance rates significantly decreased. Similarly, our study showed that resistance to gentamicin and ciprofloxacin decreased, whereas resistance to piperacillin-tazobactam, piperacilline, meropenem, imipenem, and amikacin increased. The same meta–analysis18 showed that antibiotic resistance rates vary according to geographical areas and colistin resistance for the black sea region is between 0% and 4%. In this context, it is possible to say that there is no change in colistin resistance for the black sea region.
Detection of MBLs
Carbapenems are used as the last resort for the treatment of infections caused by pathogens. However, P. aeruginosa clinical isolates are capable of producing enzymes, such as MBL, that inactivate carbapenems. When the resistance genes of the isolates were scanned, the gene encoding β-lactamase or metallo-β-lactamase was found in a total of 36 isolates. The blaVEB gene was identified in only 1 isolate alone, but in combination with other resistance genes in a total of 17 isolates (Table 6). While the blaPER gene was detected in 5 samples alone, it was found in 13 samples in combination with other genes. Neither blaVEB nor blaPER was detected in 9 samples. Among the genes encoding metallo-β-lactamase, the most blaNDM positive was detected (n=18), followed by 12 positive samples of blaKPC. blaIMP and blaVIM were detected in 5 and 1 samples, respectively. In 2 isolates, blaVEB-blaPER-blaKPC-blaNDM-blaOXA-48 combination was found. Also, the association of blaVEB-blaPER and blaVEB-blaKPC-blaNDM was found to be very high. Much more resistance genes and associations were detected in hospital-acquired samples than community-acquired samples, both proportionally and in terms of co-occurrence (Table 6).
Table 6.
β-Lactamase Genes Identified in P. aeruginosa Isolates
| Genes | Isolate Numbers | Positive Number (%) |
|---|---|---|
| blaVEB | 3, 6, 16, 23*, 26, 28, 30, 33, 37, 38*, 46, 48, 50*, 53*, 56, 60, 61*, 65* | 18 (25.71) |
| blaPER | 3, 8, 4, 6, 7*, 13, 20*, 28, 29, 36*, 37, 55*, 56, 61*, 64*, 65*, 66*, 67* | 18 (25.71) |
| blaKPC | 3, 4, 6, 16, 28, 36*, 37, 38*, 43, 50*, 56, 66* | 12 (17.14) |
| blaNDM | 6, 7*, 9, 10, 11, 26, 27*, 28, 29, 30, 36*, 37, 38*, 43, 46, 53*, 56, 68 | 18 (25.71) |
| blaIMP | 7*, 23*, 30, 33, 36* | 5 (7.14) |
| blaVIM | 26 | 1 (1.42) |
| blaOXA-23 | 60 | 1 (1.42) |
| blaOXA-48 | 6, 7*, 9, 20*, 26, 28, 37, 38*, 46, 59, 60, 67* | 12 (17.14) |
Note: *Indicates community-associated samples.
Clonal Relationship
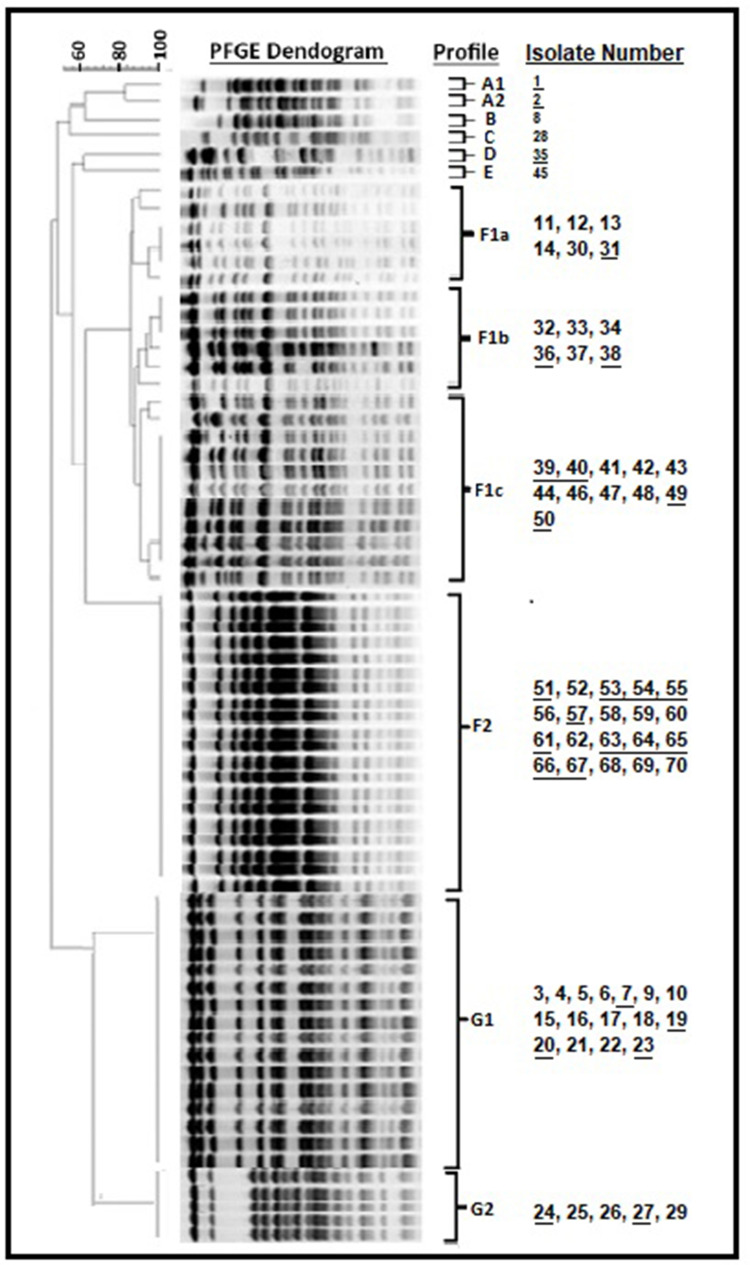
Clonal association was investigated with a total of 6 gel images and 7 major PFGE profiles were identified (Figure 1). When looked clonally, 43 isolates were found to be related under major F group. The F1 subgroup is divided into 3 patterns (F1a, F1b, F1c). When the relationship with the F1 subgroup is examined, it is also closely related to this group according to the 85% Tennover criteria. Group A, 2 (A1-A2); Group F, 2 (F1-F2); Group G is divided into 2 (G1-G2) subgroups. In particular, six isolates (65, 66, 67, 68, 69, 70 and 6 isolates) were included in the group of 20 isolates classified as F2 in the major F group. The 6 isolates mentioned above are 100% associated with 14 isolates in the F2 major group (51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64). Most community-associated isolates were collected in the F2 clade, while most hospital-associated isolates were collected in the G1 clade. However, no difference was found between the community and hospital-associated isolates according to PFGE results.
Figure 1.
PFGE analysis of P. aeruginosa isolates generated seven major clusters (profile 1 (A1, A2, B, C, D, E), F1a, F1b, F1c, F2, G1 and G2). Community-associated samples were shown as underlined.
Simultaneously, other microorganisms were also isolated from patients from which these 6 P. aeruginosa isolates were isolated. Six of the 70 patients were also co-infected with other microorganisms, namely with: Acinetobacter spp.,/Klebsiella pneumoniae (patients 65 and 68), Acinetobacter spp., (patients 66 and 69); Candida spp., (patient 67); Klebsiella spp., (patient 70). In opposition to patient nº70, the remained patient has deceased. While samples numbered 65, 66, and 67 of these patients were community-associated, samples 68, 69 and 70 were hospital-associated. Among these six samples, blaPER/blaVEB, blaKPC, blaOXA-51 and blaNDM resistance genes were detected in 65, 66, 67 and 68, respectively, while no resistance gene was detected in sample 69 and 70. Other information about the patients from whom 6 isolates are isolated as in Table 7. Compared to sample 28 and 37, which carried 5 β-lactamase coding genes, the death of these 5 patients with fewer or no resistance genes showed that the coexistence of other factors - especially other microorganisms in addition to resistance genes, was important.
Table 7.
Epidemic Characteristics of 6 Patients in Which Other Microorganisms Were Isolated in Addition to P. aeruginosa
| Sample No | 65 | 66 | 67 | 68 | 69 | 70 |
|---|---|---|---|---|---|---|
| Sample | CSF | TAC | Blood | Blood | TAC | Urine |
| Age | 42 | 82 | 84 | 88 | 87 | 47 |
| Gender | Man | Man | Man | Women | Man | Man |
| Holding dates | 30.07.2016 | 15.05.2016 | 08.04.2016 | 06.09.2016 | 11.07.2016 | 22.06.2016 |
| 22.12.2016 | 16.08.2016 | 24.04.2016 | 06.11.2016 | 01.11.2016 | 24.08.2016 | |
| Holding time (day) | 145 | 93 | 16 | 61 | 113 | 62 |
| Service | ARICU - IMICU PCU |
IMICU | IMICU | IMICU | IMICU | IDS |
| Comorbidity or additional disease | IVTA – TSH | CRI | PTC | PTC | KOAH | NBS |
| Positive simultaneous breeding: another example | Blood -TAC | Blood | TAC | Blood - Urine | TAC | Urine |
| Positive simultaneous growth: other microorganism. |
Acinetobacter spp. Klebsiella pneumoniae |
Acinetobacter spp. | Candida spp. |
Acinetobacter spp. Klebsiella spp. |
Acinetobacter spp. | Klebsiella spp. |
| History of using antibiotics in the last 3 months | NO | YES | YES | YES | YES | YES |
| The fate of the patient | EX | EX | EX | EX | EX | Following |
Since P. aeruginosa and Acinetobacter species producing extended-spectrum β-lactamase (ESBL) are resistant to most β-lactam antibiotics, monitoring and control of its spread is important when such β-lactamase occurs. Since a limited number of antibiotics such as antipseudomonal penicillins and cephalosporins, aminoglycosides, fluoroquinolones and carbapenems are effective on P. aeruginosa, it is important to monitor and control the spread of genes that cause resistance to these drugs.19 It is essential to define the main risk groups as well as the genetic data of the isolates.2 According to the European Center for Disease Prevention and Control (ECDC) in 2016, P. aeruginosa resistance in most European countries exceeded 10% of all antimicrobials studied. Moreover, the prevalence of MDR- P. aeruginosa is increasing globally, mainly due to nosocomial outbreaks and transferable resistance mechanisms, a phenomenon associated with the spread of high-risk clones, especially horizontally acquired (by multiple loci sequence types ST235, ST111, ST175). Despite its low antibiotic consumption comparable to other Northern European countries20 resistance rates of P. aeruginosa in Estonia, especially to carbapenems, are much higher than in other low-end use countries, and trends are becoming alarming. In 2012, 12.5% of the isolates reported to the ECDC were carbapenem resistant, but this had risen to over 20% by 2016.21,22 Although the main trigger associated with carbapenem resistant in P. aeruginosa is the production of plasmid-mediated β-lactamases/carbapenemases, mutational resistance mechanisms in chromosomal genes– for example, altered expression of outer membrane porins or efflux system and increased chromosomal cephalosporinase (AmpC) activity may all be affected.
Conclusion
In Turkey, the period of 2011–2016, the overall carbapenem resistance has varied between 42.9% - 30.26-%. In the University hospitals where our study was conducted, this ratio was similarly 36.55. The average length of stay (days) was significantly higher in the group with HAI than in the group with CAI. Compared to sample 28 and 37, which carried 5 β-lactamase coding genes, the death of these 5 patients with fewer or no resistance genes showed that the coexistence of other factors - especially other microorganisms in addition to resistance genes, was important.
Acknowledgment
This work was supported by Recep Tayyip Erdogan University Research Fund Grants (BAP).
Ethical Approval
This study was reviewed and approved by the Scientific and Ethical Committee of Rize Recep Tayyip Erdogan University Clinical Research Ethics Committee (Rize, Turkey) (40465587-32/2018-32). The data of patients’ clinical variables were collected from their medical records and did not contain name, address, or other personal information. The patients’ written informed consent was obtained from individual or guardian participants. This study was also in line with the guidelines outlined in the Declaration of Helsinki.
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
- 1.Pachori P, Gothalwal R, Gandhi P. Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit; a critical review. Genes Dis. 2019;6(2):109–119. doi: 10.1016/j.gendis.2019.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Telling K, Laht M, Brauer A, et al. Multidrug resistant Pseudomonas aeruginosa in Estonian hospitals. BMC Infect Dis. 2018;18(1):513. doi: 10.1186/s12879-018-3421-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chairat S, Ben Yahia H, Rojo-Bezares B, Sáenz Y, Torres C, Ben Slama K. High prevalence of imipenem-resistant and metallo-β-lactamase-producing Pseudomonas aeruginosa in the Burns Hospital in Tunisia: detection of a novel class 1 integron. J Chemother. 2019;31(3):120–126. doi: 10.1080/1120009X.2019.1582168 [DOI] [PubMed] [Google Scholar]
- 4.Hu Y, Cao J, Yang Q, et al. Risk factors for carbapenem-resistant Pseudomonas aeruginosa, Zhejiang Province, China. Emerg Infect Dis. 2019;25(10):1861–1867. doi: 10.3201/eid2510.181699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonnet V, Dupont H, Glorion S, et al. Influence of bacterial resistance on mortality in intensive care units: a registry study from 2000 to 2013 (IICU Study). J Hosp Infect. 2019;102(3):317–324. doi: 10.1016/j.jhin.2019.01.011 [DOI] [PubMed] [Google Scholar]
- 6.Tabak YP, Merchant S, Ye G, et al. Incremental clinical and economic burden of suspected respiratory infections due to multi-drug-resistant Pseudomonas aeruginosa in the United States. J Hosp Infect. 2019;103(2):134–141. doi: 10.1016/j.jhin.2019.06.005 [DOI] [PubMed] [Google Scholar]
- 7.Feng W, Sun F, Wang Q, et al. Epidemiology and resistance characteristics of Pseudomonas aeruginosa isolates from the respiratory department of a hospital in China. J Glob Antimicrob Resist. 2017;8:142–147. doi: 10.1016/j.jgar.2016.11.012 [DOI] [PubMed] [Google Scholar]
- 8.Karampatakis T, Tsergouli K, Politi L, et al. Molecular epidemiology of endemic carbapenem-resistant gram-negative bacteria in an intensive care unit. Microb Drug Resist. 2019;25(5):712–716. doi: 10.1089/mdr.2018.0266 [DOI] [PubMed] [Google Scholar]
- 9.Moubareck C, Bre´mont S, Conroy MC, Courvalin P, Lambert T. GES-11, a novel integron-associated GES variant in Acinetobacter baumannii. Antimicrob Agents Chemother. 2009;53(8):3579–3581. doi: 10.1128/AAC.00072-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Celenza G, Pellegrini C, Caccamo M, Segatore B, Amicosante G, Perilli M. Spread of blaCTX-M-type and blaPER-2 β-lactamase genes in clinical isolates from Bolivian hospitals. J Antimicrob Chemother. 2006;57(5):975–978. doi: 10.1093/jac/dkl055 [DOI] [PubMed] [Google Scholar]
- 11.Jeon BC, Jeong SH, Bae IK, Kwon SB, Lee K, Young D. Investigation of a nosocomial outbreak of imipenem-resistant Acinetobacter baumannii producing the OXA-23 β-lactamase in Korea. J Clin Microbiol. 2005;43(5):2241–2245. doi: 10.1128/JCM.43.5.2241-2245.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cicek AC, Saral A, Iraz M, et al. OXA and GES-type β-lactamases predominate in extensively drug-resistant Acinetobacter baumannii isolates from a Turkish University Hospital. Clin Microbiol Infect. 2014;20(5):410–415. doi: 10.1111/1469-0691.12338 [DOI] [PubMed] [Google Scholar]
- 13.Poirel N, He´ritier C, Tolun V, Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004;48(1):15–22. doi: 10.1128/aac.48.1.15-22.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119–123. doi: 10.1016/j.diagmicrobio.2010.12.002 [DOI] [PubMed] [Google Scholar]
- 15.Woodford N, Ellington MJ, Coelho JM, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006;27(4):351–353. doi: 10.1016/j.ijantimicag.2006.01.004 [DOI] [PubMed] [Google Scholar]
- 16.Durmaz R, Otlu B, Koksal F, et al. The optimization of a rapid pulsed-field gel electrophoresis protocol for the typing of Acinetobacter baumannii, Escherichia coli and Klebsiella spp. Jpn J Infect Dis. 2009;62(5):372–377. [PubMed] [Google Scholar]
- 17.Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial isolate typing. J Clin Microbiol. 1995;33(9):2233–2239. doi: 10.1128/JCM.33.9.2233-2239.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Acara A, Karaahmetoğlu G, Akalın H, Altay AF. Pooled prevalence and trends of antimicrobial resistance in Pseudomonas aeruginosa clinical isolates over the past 10 years in Turkey: a meta-analysis. J Glob Antimicrob Resist. 2019;18:64–70. doi: 10.1016/j.jgar.2019.01.032 [DOI] [PubMed] [Google Scholar]
- 19.Atilla A, Eroğlu C, Esen S, Sünbül M, Leblebicioğlu H. Investigation of the frequency of PER-1 type β-lactamase and antimicrobial resistance rates in nosocomial isolates of Pseudomonas aeruginosa. Mikrobiyol Bul. 2012;46(1):1–8. [PubMed] [Google Scholar]
- 20.URL-1, Antimicrobial consumption database (ESAC-Net). Available from: http://ecdc.europa.eu/en/antimicrobial-consumption/surveillance-and-disease-data/database. Accessed April9, 2021.
- 21.URL-2, Antimicrobial resistance surveillance in Europe 2016. Available from: http://ecdc.europa.eu/en/publications-data/antimicrobial-resistance-surveillance-europe-2016. Accessed April9, 2021.
- 22.URL-3, Antimicrobial resistance surveillance in Europe 2012. Available from: http://ecdc.europa.eu/en/publications-data/antimicrobial-resistance-surveillance-europe-2012. Accessed April9, 2021.