To the Editor:
Every year, ~8 million Americans require advanced care for nonhealing wounds that collectively are estimated to cost between $28-96 billion (Sen, 2019). Complications in healing disproportionally afflict the elderly who commonly suffer from comorbidities like vascular insufficiency and diabetes mellitus (Gosain and DiPietro, 2004, Gould et al., 2015) that disrupt wound closure. Histological staining of skin tissue sections with hematoxylin and eosin (H&E) can provide insight into cellular infiltration into the wound, infection, hyperproliferation at the edge of the wound, and/or fibrosis and serves as a critical technique in the research laboratory to understand wound pathophysiology and evaluate new wound care products (Eming et al., 2014, Gantwerker and Hom, 2012). However, the analysis of wound histology is time-intensive, reliant on subjective user input, and largely qualitative. The goal of this study was to develop an objective and automated method to quantitatively assess H&E stained wound sections to aid in wound healing research.
Recently, convolutional neural networks (CNNs) have been applied to many biomedical applications and demonstrated an ability to classify and segment large quantities of image data rapidly and accurately (Calderon-Delgado et al., 2018, Kose et al., 2020, Oskal et al., 2019, Rivenson et al., 2019, Ronneberger et al., 2015, Tang et al., 2019). CNNs typically utilize a deep-learning approach that allows them to learn features unique to different image regions and delineate them from other distinct regions of an image. This is accomplished using supervised learning in which a CNN learns image features from user traced image segmentation it considers as ground truth. This contrasts with unsupervised approaches that do not require labeled data, and instead find intrinsic patterns and features within the data set provided. While unsupervised approaches are immune to any potential training biases, it is difficult to control what patterns the network will choose to delineate. Supervised learning benefits from being able to teach a network a known number of relevant classes leading to its applications in biomedical image segmentation. Training a neural network can be a significant time investment, as it requires hundreds or more training images and significant processing power to learn to classify data accurately. Once trained, however, CNNs they produce repeatable, consistent results rapidly across datasets. In the last five years, networks employing U-Net architectures (Ronneberger et al., 2015) have proven capable of segmenting images on a pixel-by-pixel basis with accuracies greater than 90% in OCT images of skin (Calderon-Delgado et al., 2018) and uninjured H&E skin sections (Oskal et al., 2019). This pixel-wise accuracy makes U-Net CNNs ideal for collecting automated dimensional measurements and could provide quantitative metrics to evaluate pathological delays in healing.
In this study, we trained a CNN capable of segmenting morphologically distinct and clinically relevant regions of wound tissue for the automated calculation of wound depth, wound width, epidermal/dermal thicknesses, and re-epithelialization percentage. To accomplish this, a U-Net segmentation network was trained and evaluated using images of H&E stained murine skin tissue containing full-thickness, excisional wounds from animals between 4-24 months of age, with and without streptozotocin-induced diabetes (Jones et al., 2018). Animal studies were conducted in accordance with University of Arkansas IACUC protocols #16001 and #17063. Full details on the methods can be found in the Supplementary Material.
The U-Net architecture was comprised of 4 symmetric encoding and decoding layers created using the deep learning toolbox in MATLAB 2019a (Figure S1a). To train the network, 395 unique 512x512 pixel images from 25 H&E stained murine tissue sections were collected at days 3 (n=8), 5 (n=8), and 10 (n=9) post-wounding. Custom written MATLAB code was used to manually segment 7 regions including the epidermis, dermis/hypodermis, granulation tissue, scab, hair follicles, skeletal muscle, and background. The 395 images were each augmented by reflection to improve network accuracy and robustness (Perez and Wang, 2017), thus increasing the size of the image set to 790. 70% of the images were randomly assigned to a training set, 20% to a validation set, and 10% to a testing set (Figure S1b). Training was performed with an initial learning rate of 10−3 using an Adam optimizer and a cross-entropy loss function. Training continued for 100 epochs and was terminated when validation loss stopped decreasing to prevent overfitting.
Once trained, an independent test set of images was segmented by the network and its output masks were compared on a pixel-by-pixel basis directly to the corresponding user segmented image masks (Figure 1). The granulation tissue (GT), epidermis (E), dermis (D), muscle (M), and background (BG) all were classified with accuracies ≥ 90%, while the scab (S) and hair follicle (HF) classes were slightly lower with some misclassification along their boundaries with surrounding tissue regions (Figure 2b). Overall, the network had a classification accuracy of 92.5% when compared to the user-defined images in the test set, performing similarly to published segmentation networks for other applications (Calderon-Delgado et al., 2018, Oskal et al., 2019, Roy et al., 2017, Tang et al., 2019).
Figure 1.
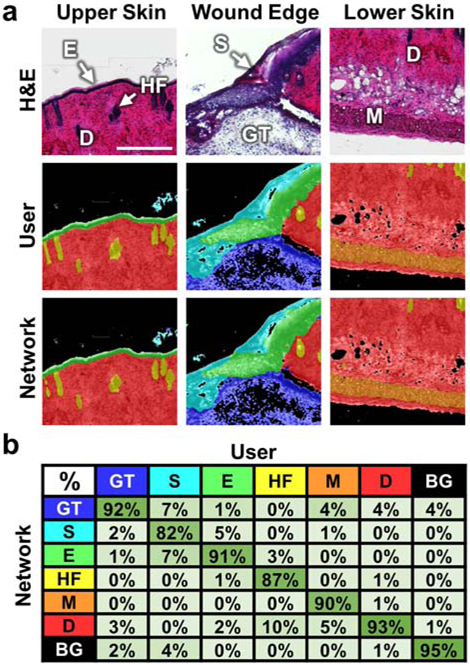
Validation of the CNN. (a) Three representative test set images from distinct areas of skin wound tissue are shown with brightfield H&E images in the top row, the user segmented ground truth masks in the middle row, and the corresponding network segmented masks in the bottom row. (b) A confusion matrix summarizes the pixel classification accuracy of the network for each wound region based on comparisons to the user traced masks. Color-coded classes included granulation tissue (blue), scab (teal), epidermis (green), hair follicles (yellow), muscle (orange), dermis (red), and background (black). Scale bar is 250 μm.
Figure 2.
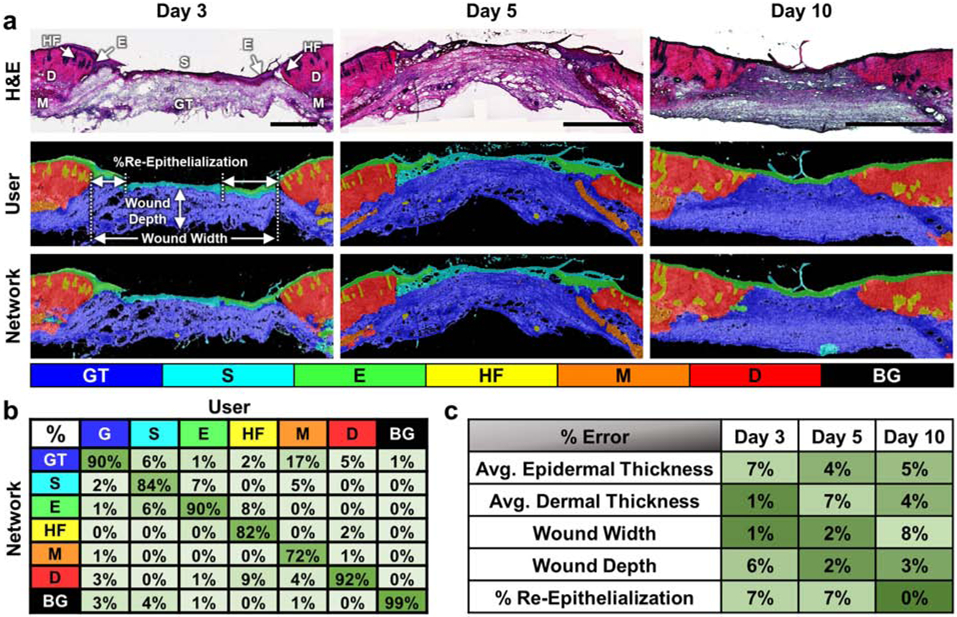
Automated network segmentation and quantification of whole wound sections. (a) Representative H&E stained sections of skin wound tissue from days 3, 5 and 10 post-wounding (top row) with segmentation results from manual user tracing (middle row) and the CNN (bottom row) demonstrate the network’s ability to accurately segment full-thickness wounds. (b) The network demonstrated good accuracy across different wound regions and had an overall accuracy of 94.06%. (c) Automated measurements using the wound segmentation results revealed only small errors between the network and user-defined ground truth results. All scale bars are 500 μm.
To assess the ability of the network to segment and quantify whole wound sections, an additional test set of 6 whole sections from days 3, 5, and 10 post wounding were manually traced and then segmented by the trained network (Figure 2a). Accuracy of the whole section classification was 94.06% (Figure 2b), which was similar to the original test set evaluation (Figure 1). The segmentation accuracies of individual slides all fell in a range between 92.32-96.22%. Based on the segmented regions, measurements of wound depth, wound width, epidermal/dermal thicknesses, and % re-epithelialization were automatically quantified from the whole tissue sections. Minimum separation distances based on the pixel-wise locations of epidermis and dermis classes were used to define wound width and the percentage of re-epithelialization, while wound depth was assessed using the depth of the granulation at the midpoint (Figure 2a). The average thicknesses of the epithelium -including the migrating epithelial tongue- and dermis/hypodermis were calculated using Euclidean distance transform measurements. Percent error was calculated for each metric based on the results from the network segmentation relative to the measurements of the user traced sections (Figure 2c). Overall error in these measurements was 4.3±2.7%, with no time-point demonstrating substantially different (>2 S.D.) levels of error. Additionally, thickness measurements along the length of the wound sections were strongly correlated between the user and network defined masks (R=0.91±0.04 for the epidermis and R=0.98±0.02 for the dermis) (Figure S2).
In summary, this work demonstrates that a CNN can be developed to accurately segment full H&E stained wound sections on a pixel-wise basis in less than 30 seconds using a desktop computer (Figure 1). These segmentation masks can be used to automatically measure wound geometry with minimal error (Figure 2). Automatic delineation of relevant wound regions also provides a foundation to further quantify other image features, and our network could be paired with additional neural networks or automated image processing techniques to quantify region-specific microvessel or cellular densities in the future. Furthermore, the network generated here for rapid segmentation and evaluation of H&E sections can be retrained via transfer learning (Shin et al., 2016) to develop future CNNs capable of quantifying wound features using substantially different staining protocols, imaging parameters, or sources of contrast.
Supplementary Material
Acknowledgements:
We would like to acknowledge Gianna Busch, Caila Hanes, and Ayman Yosef for their help with tissue sectioning and imaging of H&E stained tissue samples. This research was funded by NIH grant numbers R00EB017723 and R01AG056560, as well as the Arkansas Biosciences Institute.
Abbreviations Used:
- H&E
Hematoxylin and eosin
- CNN
Convolutional neural network
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest:
The authors state no conflict of interest.
Data Availability:
Code, example images, and trained network data related to this article can be found at [https://github.com/kylepquinn/JID_WoundSegmentation_2020], hosted at Github. Imaging datasets related to this article can be provided by the authors upon request.
References:
- Calderon-Delgado M, Tiju J-W, Lin M-Y, Huang S-L. High Resolution Human Skin Image Segmentation by means of Fully Convolutional Neural Networks. International Conference on Numerical Simulation of Optoelectronic Devices (NUSOD). Hong Kong 2018. p. 31–2. [Google Scholar]
- Eming SA, Martin P, Tomic-Canic M. Wound repair and regeneration: mechanisms, signaling, and translation. Sci Transl Med 2014;6(265):265sr6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantwerker EA, Hom DB. Skin: histology and physiology of wound healing. Clin Plast Surg 2012;39(1):85–97. [DOI] [PubMed] [Google Scholar]
- Gosain A, DiPietro LA. Aging and wound healing. World J Surg 2004;28(3):321–6. [DOI] [PubMed] [Google Scholar]
- Gould L, Abadir P, Brem H, Carter M, Conner-Kerr T, Davidson J, et al. Chronic wound repair and healing in older adults: current status and future research. J Am Geriatr Soc 2015;63(3):427–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Ramser HE, Woessner AE, Quinn KP. In vivo multiphoton microscopy detects longitudinal metabolic changes associated with delayed skin wound healing. Commun Biol 2018;1:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kose K, Bozkurt A, Alessi-Fox C, Brooks DH, Dy JG, Rajadhyaksha M, et al. Utilizing Machine Learning for Image Quality Assessment for Reflectance Confocal Microscopy. J Invest Dermatol 2020;140(6):1214–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskal K, Risdal M, Janssen E, Undersrud E, Gulsrud T. A U-Net based approach to epidermal tissue segmentation in whole slide histopathological images. SN Applied Sciences 2019;1:672. [Google Scholar]
- Perez L, Wang J. The effectiveness of data augmentation in image classification using deep learning. arXiv preprint arXiv:171204621 2017.
- Rivenson Y, Wang H, Wei Z, de Haan K, Zhang Y, Wu Y, et al. Virtual histological staining of unlabelled tissue-autofluorescence images via deep learning. Nat Biomed Eng 2019;3(6):466–77. [DOI] [PubMed] [Google Scholar]
- Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Cham: Springer International Publishing; 2015. p. 234–41. [Google Scholar]
- Roy AG, Conjeti S, Karri SP, Sheet D, Katouzain A, Wachinger C, et al. ReLayNet: retinal layer and fluid segmentation of mascular optical coherence tomography using fully convolutional networks. Biomedical Optics Express 2017;8(8):3627–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen C. Human Wounds and Its Burden: An Updated Compendium of Estimates. Advances in Wound Care 2019;8(2):39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H-C, Roth H, Gao M, Lu L, Xu Z, Nogues I, et al. Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning. IEEE Transactions on Medical Imaging 2016;35:1285–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Yang F, Yuan S, Zhan Ca. A Multi-Stage Framework With Context Information Fusion Structure For Skin Lesion Segmentation. 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019). Venice, Italy2019. p. 1407–10. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Code, example images, and trained network data related to this article can be found at [https://github.com/kylepquinn/JID_WoundSegmentation_2020], hosted at Github. Imaging datasets related to this article can be provided by the authors upon request.


