Abstract
Objective:
Gut microbiota respond to host physiological phenomena, yet little is known regarding shifts in the gut microbiome due to menopausal hormonal and metabolic changes in women. HIV infection impacts menopause and may also cause gut dysbiosis. We therefore sought to determine the association between menopausal status and gut microbiome composition in women with and without HIV.
Methods:
Gut microbiome composition was assessed in stool from 432 women (99 pre-menopausal HIV+, 71 pre-menopausal HIV−, 182 post-menopausal HIV+, 80 post-menopausal HIV−) via 16S rRNA gene sequencing. We examined cross-sectional associations of menopause with gut microbiota overall diversity and composition, and taxon and inferred metagenomic pathway abundance. Models were stratified by HIV serostatus and adjusted for age, HIV-related variables, and other potential confounders.
Results:
Menopause, i.e. post- vs. pre-menopausal status, was associated with overall microbial composition only in women with HIV (permutational MANOVA of Jensen Shannon Divergence: p=0.01). In women with HIV, menopause was associated with enrichment of gram-negative order Enterobacteriales, depletion of highly abundant taxa within Prevotella copri, and alterations in other low-abundance taxa. Additionally, menopause in women with HIV was associated with enrichment of metagenomic pathways related to Enterobacteriales, including degradation of amino acids and phenolic compounds, biosynthesis of enterobactin, and energy metabolism pathways. Menopause-related differences in some low-abundance taxa were also observed in women without HIV.
Conclusions:
A changing gut microbiome may be an overlooked phenomenon of reproductive aging in women with HIV. Longitudinal assessments across all reproductive stages are necessary to confirm these findings and identify health implications.
Keywords: menopause, gut microbiome, HIV
INTRODUCTION
Life expectancy for people living with HIV has increased dramatically due to improvements in antiretroviral therapy1. As a result, aging-related conditions are becoming a priority in HIV clinical care and research. For women with HIV, this includes consideration of the menopausal transition2. Menopause marks the end of the reproductive phase of a woman’s life, at which complete depletion of ovarian follicles leads to loss of ovarian estrogen production3. Estrogens play a protective role in the physiology of a number of chronic conditions, including dyslipidemia4, cardiovascular disease5, 6, and osteoporosis7, which are more prevalent in persons with HIV infection compared to those without HIV8, 9. There is some evidence that menopause symptomatology and menopause-related physiologic changes are heightened in women living with HIV, either due to HIV infection itself or HIV treatment2, 9, 10. The menopausal transition and its attendant disease risks are therefore important health considerations in women with HIV.
Little is known about how the menopausal transition may affect the gut microbiome – the community of microorganisms residing in the human gut that play important roles in metabolism and immunity, and are associated with risk for a range of human diseases11, 12. The menopausal transition could influence gut microbiome composition via loss of estrogen, as certain bacteria possess metabolic activity towards estrogen in the form of glucuronide deconjugation13. In short, bacteria become exposed to conjugated estrogens (and other hormones) via the enterohepatic circulation; in the gut, certain bacteria can deconjugate estrogens for intestinal reabsorption, while estrogens that remain conjugated are excreted14. It is also plausible that menopausal changes in mucosal immunity15 modulate the bacterial milieu in the gut.
A few small human studies have observed differences in the gut microbiome between pre- and post-menopausal women16–18. A study of 48 women in China (24 pre- and 24 post-menopause) reported lower gut microbiome diversity, higher relative abundance of phylum Bacteroidetes and genus Tolumonas, and lower relative abundance of phylum Firmicutes and genus Roseburia, in post-menopausal compared to pre-menopausal women17. A study of 37 women in Spain (17 pre- and 20 post-menopause) also observed higher Firmicutes relative abundance in post-menopausal compared to pre-menopausal women, as well as higher relative abundance of genera Lachnospira and Roseburia, and lower relative abundance of genera Parabacteroides, Prevotella, and Bilophila16. Another study of 89 women in Spain (44 pre- and 45 post-menopausal) observed that the gut microbiota of post-menopausal women was more similar to men than to pre-menopausal women, with steroid biosynthesis and degradation pathways enriched in pre-menopausal women and differential abundance of many taxa18. Yet, no studies have examined menopause-related microbiome differences in the context of HIV.
An understanding of gut microbiome changes in menopause will be important to determine any roles that gut microbes may play in post-menopausal disease risk, especially in women with HIV who may have gut dysbiosis19, disturbed mucosal immunity20, and are already at increased risk of cardiovascular disease21. Drawing participants from the Women’s Interagency HIV Study (WIHS), we aimed to examine differences between pre- and post-menopausal women with and without HIV in gut microbiome overall diversity and taxonomic and functional composition.
METHODS
Study cohort.
The WIHS is a prospective cohort of women with and at risk for HIV from 10 clinical sites across the United States, previously described in detail22, 23. From 1994-2019, WIHS participants attended semiannual study visits in which they underwent a comprehensive physical examination, provided biological specimens, and completed an interviewer-administered questionnaire assessing health behaviors, medical history, and medication use.
Gut microbiome study participants.
In 2016, WIHS participants from the Bronx, Brooklyn, and Chicago sites began home collection of stool samples for gut microbiome analysis using study provided kits with detailed instructions, described previously24, 25. Samples were mailed to the laboratory of Dr. Robert D. Burk by either the participant or WIHS research staff. We excluded samples with low microbiome sequencing depth as well as those collected while pregnant or taking post-menopausal hormone therapy (in the past 6 months), those with discordant self-report and STRAW+10 menopause data and/or missing menopause status or surgical history, bilateral oophorectomy, and partial or total hysterectomy (due to inability to assess menopausal status through menstrual patterns). From 572 cross-sectional samples, these exclusions resulted in a sample size of 432 samples for analysis, 281 of whom were from women living with HIV.
Menopause status.
Menopause was categorized using two survey sections from the WIHS bi-annual questionnaires: (1) self-report of menopause with the question “Have you been through menopause (the change of life)?”, defined to participants as not having menstruated for 12 or more months, and (2) self-report of STRAW+10 criteria26 with questions on menstrual bleeding patterns. Briefly, the Stages of Reproductive Aging Workshop +10 (STRAW+10) criteria, based on menstrual bleeding patterns, represent a consensus staging system agreed upon by experts in the field26. STRAW+10 defines three major stages of reproductive aging, with further subdivisions in each stage: the reproductive stage, characterized by normal to slightly irregular menstrual cycles; the menopausal transition stage, characterized by a ≥7 day difference in cycle length of consecutive menstrual cycles (early menopausal transition) or an interval of amenorrhea ≥60 days (late menopausal transition); and the post-menopause stage, determined retrospectively when 12 months have passed since a woman’s last menses. We defined women as pre-menopausal if they reported having a period in the last 12 months according to both self-report and STRAW+10 categorization, and as post-menopausal if they reported not having a period in the last 12 months according to self-report and STRAW+10 categorization. Women categorized as pre-menopausal who reported 12 months of amenorrhea at the immediately subsequent visit (i.e. 6 months later) and all later visits (according to both self-report and STRAW+10) were recategorized as post-menopausal. Similarly, women categorized as post-menopausal who reported a period at a later visit(s) (according to both self-report and STRAW+10) were re-categorized as pre-menopausal. In a further categorization of pre-menopausal women, we defined women as ‘reproductive stage’ or ‘menopausal transition’ based on the STRAW+10 categorization, but did not additionally break down into ‘early’ and ‘late’ transition due to small sample size.
Microbiome measurement.
Stool samples were collected at home by study participants, using tubes containing RNAlater, and gut microbiome was assessed by 16S rRNA gene sequencing; validity of our methods has been described previously24, 25. This assay was conducted blind to menopause status or any other participant characteristics. Briefly, DNA was extracted with the DNeasy PowerLyzer PowerSoil DNA Isolation Kit (QIAGEN, Valencia, CA), following the manufacturer’s instructions. PCR amplification was performed on the 16S rRNA gene V4 hypervariable region using the 515F and 806R primers, with a 12-bp unique Golay barcoding27. PCR reactions were performed with an initial denaturation of 95 °C for 5 min, followed by 15 cycles of 95 °C for 1 min, 55 °C for 1 min, and 68 °C for 1 min, followed by 15 cycles of 95 °C for 1 min, 60 °C for 1 min, and 68 °C for 1 min, and a final extension for 10 min at 68 °C on a GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA). The PCR products were purified using a QIAquick Gel Extraction Kit (QIAGEN, Valencia, CA) and quantified using a Qubit 2.0 Fluorometric High Sensitivity dsDNA Assay (Life Technologies, Carlsbad, CA). KAPA LTP Library Preparation Kit (KAPA Biosystems, Wilmington, MA) was used on the combined purified PCR products according to the manufacturer’s protocol and the size integrity of the amplicons with Illumina indices was validated with a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) at the Genomics Core at Albert Einstein College of Medicine. High-throughput amplicon sequencing was conducted on a MiSeq (Illumina, San Diego, CA. RRID:SCR_016379) using 2 × 300 paired-end fragments, to capture the entire 16S V4 region, at the Albert Einstein College of Medicine Sequencing Core.
Bioinformatics processing.
Illumina reads were preprocessed to remove bases that fell below PHRED quality score of 25 using prinseq28. Processed reads were then demultiplexed using sample specific barcode combinations using novobarcode29. Demultiplexed reads were combined into a QIIME2 file for further processing in QIIME230. Paired-end reads were joined, followed by quality filtering as described in Bokulich et al.31. Next the Deblur workflow was applied, which uses sequence error profiles to obtain putative error-free DNA sequences, referred to as amplicon sequence variants (ASVs)32. ASVs were assigned taxonomy using a naïve Bayes classifier pre-trained on the Greengenes33 13_8 99% OTUs, where the sequences have been trimmed to only include 250 bases from the 16S V4 region, bound by the 515F/806R primer pair. A phylogenetic tree was constructed via sequence alignment with MAFFT34, filtering the alignment, and applying FastTree35 to generate the tree. PICRUSt236 was used to infer the metagenomic functions of the microbial communities using the default reference database (IMG Nov. 2017 database). Number of observed ASVs and the Shannon Diversity Index were calculated in 100 iterations at 40 different rarefied sequencing depths (from 20 to 35,000 sequence reads per sample), and averaged for each subject at each depth, to generate rarefaction curves (Supplementary Figure 1). We excluded samples with sequencing depths <2000 sequence reads per sample after the Deblur workflow (n=11), based on visual observation in the rarefaction curves that this depth sufficiently represents the diversity of the samples in this study (Supplementary Figure 1). The sequencing depths after Deblur for the 432 women included in the analysis ranged from 2,507 to 60,932 (Q1=7,491, Q2=9,839, Q3=19,609). Quality control was assessed from duplicate samples run on the same PCR plate (intra-plate) or across plates (inter-plate), using coefficients of variation for the number of observed ASVs and the Shannon Diversity Index, and dissimilarity according to the Jensen Shannon Divergence (JSD). Mean coefficients of variation for the number of observed ASVs and the Shannon Diversity Index at 2,507 sequence reads per sample were 4.4% and 1.8% for intra-plate duplicates and 11.5% and 5.1% for inter-plate duplicates, respectively. Mean JSD dissimilarity among intra-plate and inter-plate duplicates was 0.10 and 0.12, while mean dissimilarity for non-duplicates was 0.47, consistent with high repeatability.
Statistical analysis.
General principles.
Data for analysis were pulled from the closest WIHS core visit to the time of stool sample receipt (median: 1.4 weeks). Demographic and clinical characteristics were compared between pre- and post-menopausal women using the Wilcoxon rank-sum test and Fisher’s exact test for continuous and categorical variables, respectively. Statistical models were adjusted for a priori variables – age (continuous), race (White, Black, other or multi-racial), ethnicity (Hispanic, non-Hispanic), HIV serostatus (positive, negative), hepatitis C virus antibody status (HCV status; positive, negative), hormonal contraceptive use (yes, no), HIV antiretroviral therapy (yes, no), viral load (undetectable, detectable) – as well as variables that were associated with menopause (p<0.10) independent of age, in the entire (unstratified) study population or in HIV+ or HIV− strata. Associations with menopause independent of age were assessed using likelihood ratio tests comparing a logistic model with age and the variable of interest to a model with age alone. Additional variables considered for inclusion in models were study site (Bronx, Brooklyn, Chicago), country of birth (U.S., Puerto Rico or other U.S. territories, other countries), employment status (employed, not employed), alcohol use (abstainer, 1-7 drinks/week, 8-12 drinks/week, or >12 drinks/week of alcoholic beverages), smoking status (never, former, current), injected or non-injected recreational drug use including marijuana (yes, no), body mass index (BMI in kg/m2; continuous), diabetes (ever any fasting glucose≥126 mg/dL, hemoglobin A1C≥6.5%, self-reported diabetes, or diabetes medication; yes, no), and any hypertension (systolic blood pressure ≥140 mmHg, diastolic blood pressure ≥90 mmHg, self-report, or use of anti-hypertensive medication; yes, no). Missing data for covariates were imputed based on the immediately prior study visit with available data. All analyses were conducted among the entire sample (unstratified), and stratified by HIV status. Analyses were conducted using R (version 3.5.1).
Within-subject (α-) and between-subject (β-) diversity.
α-diversity (within-subject diversity) was assessed by the average number of observed ASVs (richness) and the average Shannon Diversity Index over 100 iterations at the rarefied depth of 2507 sequences per sample; data were not rarefied for any other analyses. Multivariate linear regression was used to examine the difference in α-diversity by menopausal status, adjusting for covariates. β-diversity (between-subject diversity) was assessed using the Jensen Shannon Divergence (JSD), and permutational multivariate analysis of variance (PERMANOVA) was used to assess differences in overall microbiome composition by menopausal status, adjusting for covariates.
Taxa and functional pathways.
Taxa and functional pathways were analyzed in two stages: first using the Analysis of Composition of Microbiomes (ANCOM2) method37, followed by confirmatory multivariate linear regression, described below. ANCOM2 was used to detect taxa and functional pathways associated with menopause status, adjusting for covariates. We tested taxa at the different taxonomic levels (phylum, class, order, family, genus, species, ASV) separately, controlled the false discovery rate (FDR) at 10%, and excluded taxa or pathways from testing if they were present in <20% of the study population. A total of 10 phyla, 17 classes, 21 orders, 41 families, 84 genera, 131 species, 660 ASVs, and 964 functional pathways were tested. An ANCOM detection level ≥0.6 was considered significant – this level indicates that the ratios of the taxon or pathway to at least 60% of other taxa or pathways were detected to be significantly different (FDR q<0.10) between pre- and post-menopausal women. For the ANCOM-selected taxa and pathways, we constructed multivariate linear regression models, with centered log ratio (clr)-transformed taxon/pathway abundance as outcome, and menopausal status as the main predictor, adjusting for covariates.
Sensitivity analysis.
To further control for potential confounding that may result from differences in age and other variables between pre- and post-menopausal women, we next examined significant findings relating menopause with taxa and functional pathways using overlap weighted38 regression analyses with propensity score weights. Propensity scores for menopause status were derived from logistic regression, including all available covariates with standardized mean difference >10% and no missing data, so that all 432 women in our analysis would receive a propensity score; these variables included those defined above (age, race, ethnicity, HIV serostatus, HCV status, hormonal contraceptive use, study site, country of birth, employment status, alcohol use, smoking status, recreational drug use, BMI, diabetes, hypertension), as well as BMI category (BMI<25 kg/m2, 25 kg/m2 ≤BMI<30 kg/m2, BMI≥30 kg/m2), marijuana use (yes, no), use of hallucinogens or other drugs not including marijuana, crack, cocaine, heroin, illicit methadone, or methamphetamines (yes, no), health insurance status (yes, no), and diabetes medication (yes, no). Standardized mean differences of covariates, including age, between pre- and post-menopausal women were generally reduced after propensity score weighting, indicating that this method achieved balance between the groups (Supplementary Figure 2).
RESULTS
Participant characteristics.
The study population was comprised of 170 pre-menopausal women (99 living with HIV) and 262 post-menopausal women (182 living with HIV). Among the pre-menopausal women, 107 were in the reproductive stage and 63 were in the menopausal transition according to the STRAW+10 criteria. Post-menopausal women were older than pre-menopausal women, as expected, resulting in many age-related differences between pre- and post-menopausal women (e.g. higher prevalence of diabetes and hypertension in post-menopausal women) (Table 1). After adjusting for age, post-menopausal women were more likely to have been born in the U.S., less likely to be currently employed, and more likely to have formerly smoked compared with pre-menopausal women; on average, post-menopausal women had lower BMI than pre-menopausal women (Table 1). Menopausal differences in these characteristics were generally more apparent in women with HIV compared to women without HIV (Table 2).
Table 1.
Characteristics of the Women’s Interagency HIV Study participants by menopausal status.
| Pre-menopausal (N=170) | Post-menopausal (N=262) | P-valuea | Age-adjusted P-valueb | |
|---|---|---|---|---|
| Age, y (mean ± SD) | 44.6 ± 5.6 | 57.4 ± 5.9 | <0.0001 | |
| Site (%) | 0.77 | 0.09 | ||
| Bronx | 46.5 | 49.6 | ||
| Brooklyn | 27.6 | 27.1 | ||
| Chicago | 25.9 | 23.3 | ||
| Race (%) | 0.20 | 0.29 | ||
| Black/African-American | 58.8 | 63.7 | ||
| White | 4.7 | 7.3 | ||
| Other or multi-racial | 36.5 | 29.0 | ||
| Hispanic (%) | 32.4 | 23.3 | 0.05 | 0.30 |
| Country of birth (%) | 0.004 | 0.07 | ||
| U.S. | 73.5 | 84 | ||
| Puerto Rico or other U.S. territories | 3.5 | 5.3 | ||
| Other countries | 22.9 | 10.7 | ||
| Education (%) | 0.96 | 0.28 | ||
| Grade 6 or less | 1.2 | 2.3 | ||
| Grade 7-11 | 40.6 | 41.2 | ||
| Completed high school | 30.0 | 29.8 | ||
| Some college | 22.9 | 22.5 | ||
| Completed college or any graduate school | 4.7 | 4.2 | ||
| Missing | 0.6 | 0 | ||
| Employed (%) | 47.1 | 23.3 | <0.0001 | 0.007 |
| Alcohol use (%) | 0.01 | 0.72 | ||
| Abstainer | 45.3 | 61.1 | ||
| >0-7 drinks/wk | 44.7 | 32.8 | ||
| >7-12 drinks/wk | 4.7 | 3.1 | ||
| >12 drinks/wk | 5.3 | 3.1 | ||
| Cigarette smoking history (%) | 0.005 | 0.04 | ||
| Never smoker | 28.8 | 17.9 | ||
| Current smoker | 40.6 | 37.8 | ||
| Former smoker | 30.6 | 44.3 | ||
| Recreational drug use (%) | 30.6 | 17.2 | 0.001 | 0.86 |
| Hormonal contraceptive use (%) | 6.5 | 0.4 | 0.0002 | 0.46 |
| HIV-positive (%) | 58.2 | 69.5 | 0.02 | 0.73 |
| Detectable HIV viral loadc (% among HIV+) | 28.3 | 21.4 | 0.24 | 0.75 |
| HIV therapy (% among HIV+) | 90.9 | 96.2 | 0.10 | 0.43 |
| HCV antibody detectable (%) | 7.1 | 34.0 | <0.0001 | 0.19 |
| BMI, kg/m2 (mean ± SD) | 32.5 ± 8.1 | 30.4 ± 7.6 | 0.004 | 0.02 |
| Diabetes (%) | 17.6 | 34.7 | 0.0001 | 0.17 |
| Hypertension (%) | 36.5 | 65.3 | <0.0001 | 0.16 |
P-values from Wilcoxon rank-sum test for continuous variables and Fisher’s exact test for categorical variables.
P-values are from likelihood ratio tests comparing a logistic model with age and the variable of interest to a model with age alone.
Lower limit of detection = 20 copies/mL
Table 2.
Characteristics of pre- and post-menopausal women, stratified by HIV status.
| HIV+ | HIV− | |||||||
|---|---|---|---|---|---|---|---|---|
| Pre-menopausal (N=99) | Post-menopausal (N=182) | P-valuea | Age-adjusted P-valueb | Pre-menopausal (N=71) | Post-menopausal (N=80) | P-valuea | Age-adjusted P-valueb | |
| Age, y (mean ± SD) | 45.4 ± 5.6 | 57.1 ± 5.8 | <0.0001 | 43.6 ± 5.6 | 57.9 ± 6.2 | <0.0001 | ||
| Site (%) | 0.52 | 0.23 | 1 | 0.29 | ||||
| Bronx | 42.4 | 48.9 | 52.1 | 51.2 | ||||
| Brooklyn | 30.3 | 28.0 | 23.9 | 25.0 | ||||
| Chicago | 27.3 | 23.1 | 23.9 | 23.8 | ||||
| Race (%) | 0.08 | 0.65 | 0.32 | 0.07 | ||||
| Black/African-American | 58.6 | 63.2 | 59.2 | 65.0 | ||||
| White | 4.0 | 9.9 | 5.6 | 1.2 | ||||
| Other or multi-racial | 37.4 | 26.9 | 35.2 | 33.8 | ||||
| Hispanic (%) | 30.3 | 26.4 | 0.49 | 0.95 | 35.2 | 16.2 | 0.009 | 0.08 |
| Country of birth (%) | 0.05 | 0.04 | 0.03 | 0.43 | ||||
| U.S. | 71.7 | 82.4 | 76.1 | 87.5 | ||||
| Puerto Rico or other U.S. territories | 5.1 | 6.0 | 1.4 | 3.8 | ||||
| Other countries | 23.2 | 11.5 | 22.5 | 8.8 | ||||
| Education (%) | 0.78 | 0.39 | 0.16 | 0.70 | ||||
| Grade 6 or less | 2.0 | 3.3 | 0 | 0 | ||||
| Grade 7-11 | 41.4 | 38.5 | 39.4 | 47.5 | ||||
| Completed high school | 32.3 | 27.5 | 26.8 | 35.0 | ||||
| Some college | 20.2 | 25.3 | 26.8 | 16.2 | ||||
| Completed college or any graduate school | 4.0 | 5.5 | 5.6 | 1.2 | ||||
| Missing | 0 | 0 | 1.4 | 0 | ||||
| Employed (%) | 44.4 | 25.3 | 0.001 | 0.007 | 50.7 | 18.8 | <0.0001 | 0.44 |
| Alcohol use (%) | 0.10 | 0.52 | 0.03 | 0.85 | ||||
| Abstainer | 48.5 | 60.4 | 40.8 | 62.5 | ||||
| >0-7 drinks/wk | 41.4 | 35.2 | 49.3 | 27.5 | ||||
| >7-12 drinks/wk | 6.1 | 3.3 | 2.8 | 2.5 | ||||
| >12 drinks/wk | 4.0 | 1.1 | 7.0 | 7.5 | ||||
| Cigarette smoking history (%) | 0.005 | 0.04 | 0.28 | 0.80 | ||||
| Never smoker | 33.3 | 20.3 | 22.5 | 12.5 | ||||
| Current smoker | 39.4 | 34.1 | 42.3 | 46.2 | ||||
| Former smoker | 27.3 | 45.6 | 35.2 | 41.2 | ||||
| Recreational drug use (%) | 21.2 | 14.8 | 0.19 | 0.73 | 43.7 | 22.5 | 0.009 | 0.96 |
| Hormonal contraceptive use (%) | 10.1 | 0.5 | 0.0002 | 0.43 | 1.4 | 0 | 0.47 | 0.91 |
| Detectable HIV viral loadc (% among HIV+) | 28.3 | 21.4 | 0.24 | 0.75 | NA | NA | ||
| HIV therapy (% among HIV+) | 90.9 | 96.2 | 0.10 | 0.43 | NA | NA | ||
| HCV antibody detectable (%) | 8.1 | 34.1 | <0.0001 | 0.61 | 5.6 | 33.8 | 0.00001 | 0.10 |
| BMI, kg/m2 (mean ± SD) | 33.8 ± 8.5 | 29.9 ± 7.4 | <0.0001 | 0.001 | 30.8 ± 7.2 | 31.5 ± 8.1 | 0.54 | 0.43 |
| Diabetes (%) | 22.2 | 34.6 | 0.041 | 0.20 | 11.3 | 35.0 | 0.001 | 0.66 |
| Hypertension (%) | 38.4 | 61.5 | 0.0003 | 0.74 | 33.8 | 73.8 | <0.0001 | 0.05 |
P-values from Wilcoxon rank-sum test for continuous variables and Fisher’s exact test for categorical variables.
P-values are from likelihood ratio tests comparing a logistic model with age and the variable of interest to a model with age alone.
Lower limit of detection = 20 copies/mL
Gut microbiome diversity and composition by menopausal status.
Post- vs. pre-menopausal differences in overall gut microbiome diversity (α-diversity) and composition (β-diversity) were only observed in women with HIV (Supplementary Table 1), though significant effect modification by HIV serostatus was not observed (all p>0.05). The Shannon diversity index was higher in post-menopausal women with HIV compared to pre-menopausal women with HIV, adjusting for age, study site, race, ethnicity, country of birth, hormonal contraceptive use, BMI, employment status, HCV status, smoking status, hypertension, HIV status, HIV viral load, and HIV therapy (B [95% CI] = 0.31 [0.04 to 0.57], p = 0.02) (Supplementary Table 1; Supplementary Figure 3). The same pattern was observed for the number of observed ASVs (i.e. gut microbiome richness; p = 0.09). Post-menopausal women with HIV also differed in overall microbiome composition from pre-menopausal women with HIV, adjusting for covariates (R2 = 0.33%, p = 0.01) (Figure 1a; Supplementary Table 1). Consistent with this finding, principal coordinate analysis of the JSD in women with HIV revealed some separation of post-menopausal and pre-menopausal women (Figure 1b), while complete overlap was observed in women without HIV (Figure 1c). Gut microbiome overall diversity and composition did not differ significantly by menopausal status in the unstratified study population or in women without HIV. Additionally, pre-menopausal women in the menopausal transition did not differ from reproductive stage women in the α- or β-diversity measures (Supplementary Table 1); thus for increased power these were considered as the combined pre-menopausal group for downstream analyses. We also explored whether HIV-related gut dysbiosis was dependent on menopausal status (i.e. whether gut microbiome compositional differences between HIV+ and HIV− women were more apparent in pre- or post-menopause); however, HIV-related variation in the gut microbiome was similar in pre-menopausal and post-menopausal women (Supplementary Figure 4).
Figure 1. Differences in overall microbiome composition by menopausal status in the Women’s Interagency HIV Study.
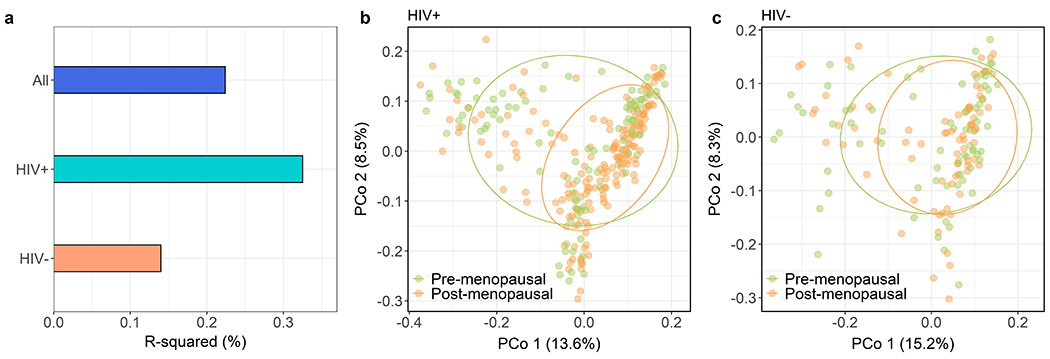
(a) Percent variation explained (R2) in the Jensen Shannon Divergence (JSD) by menopausal status (post- vs. pre-menopausal), according to PERMANOVA models. Models were adjusted for age, study site, race, ethnicity, country of birth, hormonal contraceptive use, BMI, employment status, HCV status, smoking status, hypertension, HIV status, HIV viral load, and HIV therapy; dummy variables for menopausal status by HIV status were used to obtain comparable R-squared values. P-values: all (p = 0.43), HIV+ (p = 0.01), HIV− (p = 0.49) (b) Principal coordinate analysis of the JSD in women with HIV, showing 75% data ellipses. (c) Principal coordinate analysis of the JSD in women without HIV, showing 75% data ellipses.
Differential abundance of microbial taxa by menopausal status.
Using the ANCOM2 method, adjusting for covariates, we identified several highly abundant taxa that differed between pre- and post-menopausal women with HIV. Order Enterobacteriales was enriched, while three ASVs from Prevotella copri were depleted, in post-menopausal women with HIV compared to pre-menopausal women with HIV (Figure 2a–b; Supplementary Tables 2–3; Supplementary Figure 5). Other minor taxa also differed in abundance by menopausal status in women with HIV, including ASVs from Ruminococcus torques, Bacteroides (unspecified species), and Parabacteroides distasonis (enriched in post-menopausal women with HIV) and species Bacteroides acidifaciens (depleted in post-menopausal women with HIV). We additionally observed menopause-related differences for minor taxa in the unstratified study population and in women without HIV (Figure 2a–b; Supplementary Tables 2–3), though these taxa largely did not overlap with the menopause-related taxa in women with HIV. For the majority of menopause-related taxa, results were consistent in models weighted with overlap propensity score weights (Supplementary Table 2). Further, effect modification by HIV status was observed for the effect of menopause status on certain taxa, including order Enterobacteriales (p-interaction = 0.001) and an ASV from Bacteroides (unspecified species; p-interaction = 0.001). Menopause-related taxa were generally not correlated with each other, with the exception of taxa from the same species or clade (Supplementary Figure 6), and the correlations were consistent in women with and without HIV (not shown).
Figure 2. Microbial taxa related to menopausal status in the Women’s Interagency HIV Study.
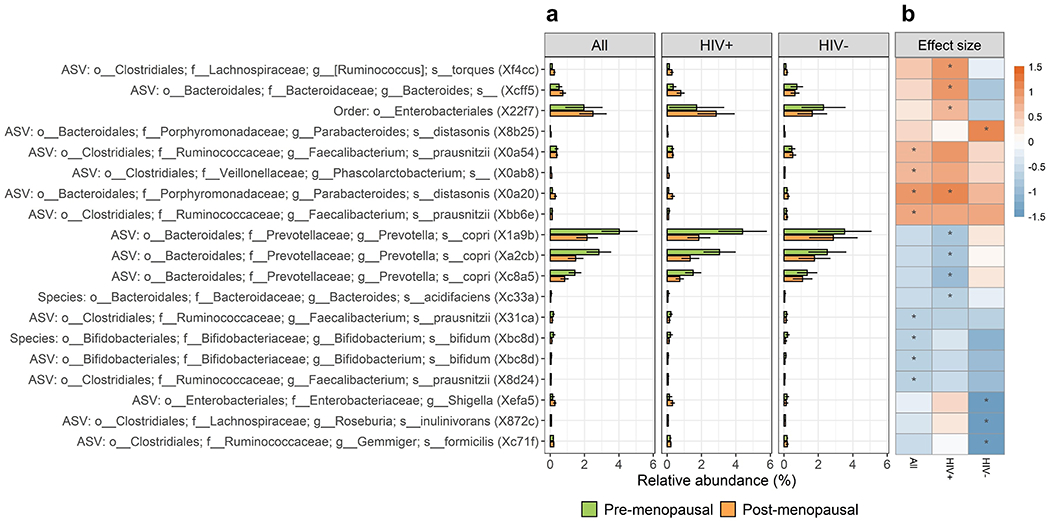
(a) Barplots of mean (95% CI) relative abundance for taxa associated with menopausal status (post- vs. pre-menopausal) in either the unstratified or HIV-stratified ANCOM2 analysis. Taxa names are shown as “Taxonomic level: order; family; genus; species (internal study ID)”. (b) Heatmap of linear regression estimates for the effect of menopausal status on taxon abundance. Taxon abundance was clr-transformed for linear regression, and models were adjusted for age, study site, race, ethnicity, country of birth, hormonal contraceptive use, BMI, employment status, HCV status, smoking status, hypertension (and HIV status in the unstratified analysis; HIV therapy and viral load in HIV+ models). Effect size (beta) represents the difference in clr-transformed abundance between post- and pre-menopausal women. Asterisk (*) indicates significant detection in ANCOM2 analysis.
Differential abundance of functional pathways by menopausal status.
Based on an a priori hypothesis, we first evaluated whether the β-glucuronidase pathway was related to menopausal status, but did not observe any significant associations (Supplementary Table 4). In ANCOM2 analysis, adjusting for covariates, we observed twenty functional pathways that differed in abundance between pre- and post-menopausal women with HIV (Supplementary Tables 4–5). In women with HIV, pathways related to degradation of amino acids (arginine and ornithine), aromatic/phenolic compounds, glycerol, alcohol, and sulfoquinovose, were enriched in post-menopausal compared to pre-menopausal women. Post-menopausal women with HIV also showed enrichment in biosynthesis of uridine diphosphate-sugars and enterobactin, as well as pathways related to energy metabolism (glycolysis, the tricarboxylic acid cycle, and glyoxylate bypass), and depletion of the ethylmalonyl-coenzyme A pathway. We additionally observed one menopause-related functional pathway (biotin biosynthesis) in the unstratified study population, and one (reductive acetyl coenzyme A pathway) in women without HIV (Supplementary Tables 4–5).
Associations of menopause-related pathways with menopause-related taxa.
Abundance of the order Enterobacteriales, enriched in post-menopausal women with HIV, was strongly correlated with the majority of menopause-related pathways in women with HIV (Figure 3), suggesting that alterations in this taxon may be responsible for the many enriched functional pathways in post-menopausal women with HIV. Indeed, menopausal status was significantly related to Enterobacteriales-specific abundance of many functional pathways in women with HIV (Supplementary Table 6).
Figure 3. Correlations between taxa and functional pathways associated with menopause in women with HIV, the Women’s Interagency HIV Study.
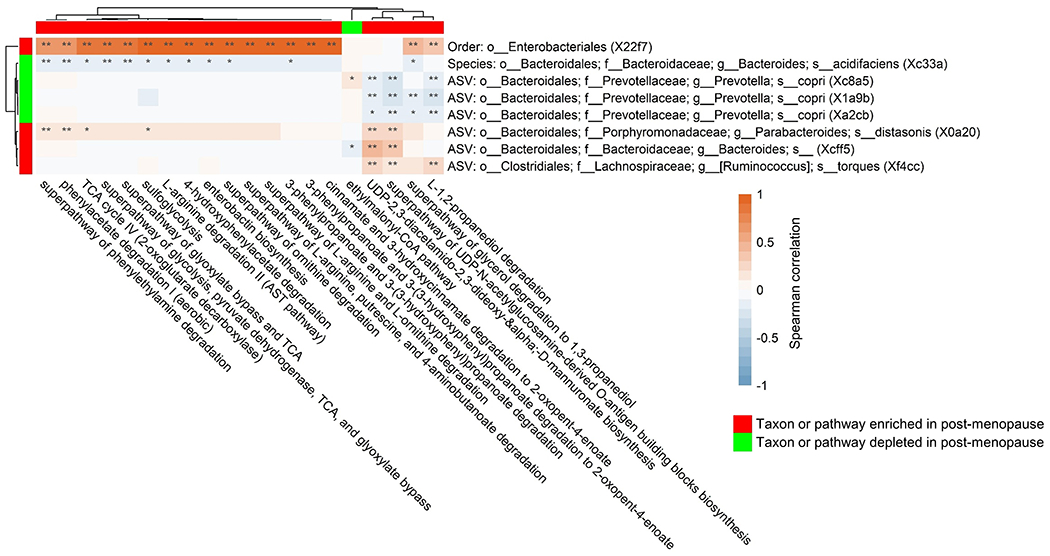
Heatmap shows Spearman’s correlations between taxa associated with menopausal status in women with HIV, and functional pathways associated with menopausal status in women with HIV. Correlations were calculated in women with HIV, on relative abundance of taxa and functional pathways.
DISCUSSION
In this cross-sectional analysis of women with or without HIV at pre- or post-menopausal stages of reproductive aging, we observed a significant difference in overall gut microbiome composition between pre- and post-menopausal women with HIV, but not in women without HIV. Menopause in women with HIV explained 0.33% of gut microbiome variation, an effect size similar to that of significant disease, medication, and dietary factors in large, extensively phenotyped studies of the human gut microbiome39, 40. The menopausal differences in women with HIV involved alterations in highly abundant gut bacterial taxa, including enrichment of the order Enterobacteriales and depletion of taxa from Prevotella copri, as well as alterations in a variety of degradative, biosynthetic, and energy-related bacterial functional pathways. A changing gut microbiome may be an overlooked and potentially clinically meaningful effect of the menopausal transition in women with HIV.
Few human studies have directly examined the association of menopausal status with the gut microbiota, reviewed in the Introduction. Our findings do not replicate these prior studies, but comparison with these studies may be limited due to the small sample sizes in prior studies, making their findings more subject to chance, and the differing study populations. Ours is the first study to examine associations of menopausal status with the gut microbiome in women with or at risk for HIV. To our knowledge, no studies have longitudinally examined within-woman changes in the gut microbiome pre- to post-menopause.
Here, we observed significant enrichment of order Enterobacteriales and depletion of three ASVs from Prevotella copri in post-menopausal women with HIV, as well as alterations in other minor taxa. These taxa are highly abundant in this study population, suggesting that the menopause-related compositional changes may be important, though the consequences require further investigation. Enterobacteriales is a gram-negative bacterial order that includes many pathogenic bacteria, and is implicated in antibiotic resistance41. Several studies have shown enrichment of Enterobacteriales in HIV infection, and association of Enterobacteriales with systemic immune activation and inflammation in people living with HIV42. Enterobacteriales may exert systemic effects via microbial translocation from the gut to the circulation43, which may contribute to the persistent immune activation observed in HIV44. The enrichment of Enterobacteriales in post-menopausal women with HIV correlated with enrichment of Enterobacteriales-related functional pathways, including pathways of amino acid and phenolic compound degradation, enterobactin biosynthesis, and energy metabolism. The genus Prevotella has been previously associated with HIV infection45, though this observation is believed to relate to male sexual behavior rather than HIV46 and is not observed in the WIHS cohort19. Prevotella copri comprises four distinct clades with different functional potential, that tend to be more prevalent in non-Westernized populations with high fiber diets47, and has been associated with insulin resistance48 and with inflammatory autoimmune diseases such as rheumatoid arthritis49 and ankylosing spondylitis50. We suspected that potential shifts in the gut microbiome during menopause would arise from bacterial metabolism of estrogen via glucuronide deconjugation13, but we did not observe an association of menopausal status with the bacterial β-glucuronidase pathway. However, metagenomic function was not measured directly in this study but rather it was inferred from taxonomy.
The observation of an association of menopause with the gut microbiome primarily in women with HIV, but not in uninfected women, could be due to the smaller sample of women without HIV. However, it may also be due to differences in sex hormones, the menopausal transition, or immunity in women with HIV. Pre-menopausal women with HIV may have lower estradiol and testosterone than uninfected women51, 52, which could modify their hormonal trajectories during the menopausal transition53, although this has not been studied in women with HIV. Women with HIV also may be more likely to experience ovarian dysfunction, in the form of more frequent amenorrhea and anovulation54, and have lower levels of anti-mullerian hormone55, a marker of ovarian reserve. Some research suggests that women with HIV have heightened menopause-related symptomatology (e.g. vasomotor symptoms), and menopause-related physiologic changes (e.g. body composition, bone mineral density)2, 9, 10. Lastly, people with HIV experience persistent immune activation and inflammation, which may be exacerbated in women after menopause: estrogen is known to have an enhancing effect on the immune system, which is lost during menopause, resulting in increased inflammation and an attenuated immune response in post-menopausal women56, 57. Further, gut permeability and microbial translocation may increase during the menopausal transition, leading to inflammation58. Inflammation in the gut can lead to overgrowth of opportunistic bacteria, such as Enterobacteriales59; this may explain our observation of a menopause-related increase in Enterobacteriales in women with HIV, who are already at risk of persistent inflammation60. Taken together, it is possible that HIV infection modulates the menopausal transition and its related microbial consequences. Yet it is important to note that the women without HIV in this cohort do not represent the general population, but rather a population with risk factors and behaviors similar to women with HIV. Therefore, the lack of a menopausal association in these women may not translate to other populations of women without HIV.
This study was strengthened by the large sample size, the comprehensive assessment of gut microbiome taxonomic composition via 16S rRNA gene sequencing, and the well-characterized WIHS cohort providing detailed information on menopause and other covariates. Our study also had several limitations. Self-report of menopause based on recall of menstrual cycles may be subject to measurement error61, however the longitudinal assessment of menopausal status every 6 months in WIHS reduces likelihood of misclassification. The observational, cross-sectional nature of the data did not allow us to draw causal conclusions or examine within-woman changes in the gut microbiome through the menopausal transition. Further, because menopausal status is so closely intermingled with age, adjustment for age in statistical models may not fully control for the effects of chronological aging. Similarly, the known close relationship of menopausal status with body composition and insulin sensitivity may drive microbiome associations. Our covariate adjustment strategy and sensitivity analysis with propensity score weighting suggests that our findings were independent of age and other potential confounders; however, as this is an observational study, potential for residual confounding remains. The stratified analysis in women without HIV suffered from less power, due to smaller sample size than the women with HIV. Lack of shotgun metagenomic data precluded analysis of true metagenomic functional modules with menopausal status, and lack of plasma hormone data precluded analysis of whether menopause-related taxa are also related to sex hormones that change during menopause (e.g. estradiol, follicle stimulating hormone). Finally, our findings in women with or at-risk for HIV in the U.S. may have limited generalizability to other populations.
CONCLUSIONS
In summary, we observed a significant difference in gut microbiome composition between pre-menopausal and post-menopausal women with HIV. Further research is warranted to determine potential mechanisms of these microbiome changes (e.g. hormone-related, immune-related), functional implications of these changes (e.g. with shotgun metagenomics or metatranscriptomics), and implications for health and disease. With the ongoing success of antiretroviral therapy and the lengthened life expectancy of women with HIV, gut microbiome changes during the menopausal transition may have a long-lasting impact on post-menopausal health and disease risk.
Supplementary Material
Acknowledgments
Sources of funding: This work was supported by a Women’s Interagency HIV Study (WIHS) substudy grant from the National Heart, Lung, and Blood Institute (R01HL140976). Data in this manuscript were collected by the WIHS, now the MACS/WIHS Combined Cohort Study (MWCCS). The contents of this publication are solely the responsibility of the authors and do not represent the official views of the National Institutes of Health (NIH). MWCCS (Principal Investigators): Bronx CRS (Kathryn Anastos and Anjali Sharma), U01-HL146204; Brooklyn CRS (Deborah Gustafson and Tracey Wilson), U01-HL146202; Data Analysis and Coordination Center (Gypsyamber D’Souza, Stephen Gange and Elizabeth Golub), U01-HL146193; Chicago-Cook County CRS (Mardge Cohen and Audrey French), U01-HL146245. The MWCCS is funded primarily by the National Heart, Lung, and Blood Institute (NHLBI), with additional co-funding from the Eunice Kennedy Shriver National Institute Of Child Health & Human Development (NICHD), National Institute On Aging (NIA), National Institute Of Dental & Craniofacial Research (NIDCR), National Institute Of Allergy And Infectious Diseases (NIAID), National Institute Of Neurological Disorders And Stroke (NINDS), National Institute Of Mental Health (NIMH), National Institute On Drug Abuse (NIDA), National Institute Of Nursing Research (NINR), National Cancer Institute (NCI), National Institute on Alcohol Abuse and Alcoholism (NIAAA), National Institute on Deafness and Other Communication Disorders (NIDCD), National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute on Minority Health and Health Disparities (NIMHD), and in coordination and alignment with the research priorities of the National Institutes of Health, Office of AIDS Research (OAR).
Financial Disclosures/Conflicts of Interest: NS, consultant with Ansh Labs and ASTELLAS/Ogeda, grant support from Menogenix, Inc. outside the submitted work; PCT, grant support from Merck outside the submitted work. AS, grant support from Gilead sciences outside the submitted work.
REFERENCES
- 1.Trickey A, May MT, Vehreschild J-J, et al. Survival of HIV-positive patients starting antiretroviral therapy between 1996 and 2013: a collaborative analysis of cohort studies. The Lancet HIV. 2017;4(8):e349–e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andany N, Kennedy VL, Aden M, Loutfy M. Perspectives on menopause and women with HIV. Int J Womens Health. 2016;8:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Santoro N The menopausal transition. The American journal of medicine. 2005;118 Suppl 12B:8–13. [DOI] [PubMed] [Google Scholar]
- 4.Palmisano BT, Zhu L, Eckel RH, Stafford JM. Sex differences in lipid and lipoprotein metabolism. Molecular Metabolism. 2018;15:45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iorga A, Cunningham CM, Moazeni S, Ruffenach G, Umar S, Eghbali M. The protective role of estrogen and estrogen receptors in cardiovascular disease and the controversial use of estrogen therapy. Biology of sex differences. 2017;8(1):33-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clegg D, Hevener AL, Moreau KL, et al. Sex Hormones and Cardiometabolic Health: Role of Estrogen and Estrogen Receptors. Endocrinology. 2017;158(5):1095–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manolagas SC, O’Brien CA, Almeida M. The role of estrogen and androgen receptors in bone health and disease. Nature Reviews Endocrinology. 2013;9(12):699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaplan RC, Hanna DB, Kizer JR. Recent Insights Into Cardiovascular Disease (CVD) Risk Among HIV-Infected Adults. Curr HIV/AIDS Rep. 2016;13(1):44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Looby SED. Menopause-associated metabolic manifestations and symptomatology in HIV infection: a brief review with research implications. J Assoc Nurses AIDS Care. 2012;23(3):195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tariq S, Delpech V, Anderson J. The impact of the menopause transition on the health and wellbeing of women living with HIV: A narrative review. Maturitas. 2016;88:76–83. [DOI] [PubMed] [Google Scholar]
- 11.Young VB. The role of the microbiome in human health and disease: an introduction for clinicians. BMJ. 2017;356:j831. [DOI] [PubMed] [Google Scholar]
- 12.Lynch SV, Pedersen O. The Human Intestinal Microbiome in Health and Disease. New England Journal of Medicine. 2016;375(24):2369–79. [DOI] [PubMed] [Google Scholar]
- 13.Baker JM, Al-Nakkash L, Herbst-Kralovetz MM. Estrogen-gut microbiome axis: Physiological and clinical implications. Maturitas. 2017;103:45–53. [DOI] [PubMed] [Google Scholar]
- 14.Kwa M, Plottel CS, Blaser MJ, Adams S. The Intestinal Microbiome and Estrogen Receptor-Positive Female Breast Cancer. Journal of the National Cancer Institute. 2016;108(8):djw029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grishina I, Fenton A, Sankaran-Walters S. Gender differences, aging and hormonal status in mucosal injury and repair. Aging and disease. 2014;5(2):160–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santos-Marcos JA, Rangel-Zuniga OA, Jimenez-Lucena R, et al. Influence of gender and menopausal status on gut microbiota. Maturitas. 2018;116:43–53. [DOI] [PubMed] [Google Scholar]
- 17.Zhao H, Chen J, Li X, Sun Q, Qin P, Wang Q. Compositional and functional features of the female premenopausal and postmenopausal gut microbiota. 2019;593(18):2655–64. [DOI] [PubMed] [Google Scholar]
- 18.Mayneris-Perxachs J, Arnoriaga-Rodríguez M, Luque-Córdoba D, et al. Gut microbiota steroid sexual dimorphism and its impact on gonadal steroids: influences of obesity and menopausal status. Microbiome. 2020;8(1):136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, Usyk M, Sollecito CC, et al. Altered Gut Microbiota and Host Metabolite Profiles in HIV-infected Women. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu H, Wang X, Veazey RS. Mucosal immunology of HIV infection. Immunol Rev. 2013;254(1):10–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanna DB, Ramaswamy C, Kaplan RC, et al. Sex- and Poverty-Specific Patterns in Cardiovascular Disease Mortality Associated With Human Immunodeficiency Virus, New York City, 2007–2017. Clinical Infectious Diseases. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bacon MC, von Wyl V, Alden C, et al. The Women’s Interagency HIV Study: an observational cohort brings clinical sciences to the bench. Clinical and diagnostic laboratory immunology. 2005;12(9):1013–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adimora AA, Ramirez C, Benning L, et al. Cohort Profile: The Women’s Interagency HIV Study (WIHS). International journal of epidemiology. 2018;47(2):393–4i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Zolnik CP, Qiu Y, et al. Comparison of Fecal Collection Methods for Microbiome and Metabolomics Studies. Frontiers in cellular and infection microbiology. 2018;8:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moon JY, Zolnik CP, Wang Z, et al. Gut microbiota and plasma metabolites associated with diabetes in women with, or at high risk for, HIV infection. EBioMedicine. 2018;37:392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harlow SD, Gass M, Hall JE, et al. Executive summary of the Stages of Reproductive Aging Workshop + 10: addressing the unfinished agenda of staging reproductive aging. Menopause (New York, NY). 2012;19(4):387–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caporaso JG, Lauber CL, Walters WA, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proceedings of the National Academy of Sciences of the United States of America. 2011;108 Suppl 1:4516–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics (Oxford, England). 2011;27(6):863–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hercus CJWhwnc. Novocraft short read alignment package. 2009.
- 30.Bolyen E, Rideout JR, Dillon MR, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology. 2019;37(8):852–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bokulich NA, Subramanian S, Faith JJ, et al. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nature methods. 2013;10(1):57–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amir A, McDonald D, Navas-Molina JA, et al. Deblur Rapidly Resolves Single-Nucleotide Community Sequence Patterns. mSystems. 2017;2(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and environmental microbiology. 2006;72(7):5069–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Molecular biology and evolution. 2009;26(7):1641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Douglas GM, Maffei VJ, Zaneveld J, et al. PICRUSt2: An improved and extensible approach for metagenome inference. 2019:672295. [Google Scholar]
- 37.Mandal S, Van Treuren W, White RA, Eggesbo M, Knight R, Peddada SD. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microbial ecology in health and disease. 2015;26:27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas LE, Li F, Pencina MJ. Overlap Weighting: A Propensity Score Method That Mimics Attributes of a Randomized Clinical Trial. Jama. 2020;323(23):2417–8. [DOI] [PubMed] [Google Scholar]
- 39.Falony G, Joossens M, Vieira-Silva S, et al. Population-level analysis of gut microbiome variation. 2016;352(6285):560–4. [DOI] [PubMed] [Google Scholar]
- 40.Zhernakova A, Kurilshikov A, Bonder MJ, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science (New York, NY). 2016;352(6285):565–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iredell J, Brown J, Tagg K. Antibiotic resistance in Enterobacteriaceae: mechanisms and clinical implications. 2016;352:h6420. [DOI] [PubMed] [Google Scholar]
- 42.Williams B, Landay A, Presti RM. Microbiome alterations in HIV infection a review. Cellular microbiology. 2016;18(5):645–51. [DOI] [PubMed] [Google Scholar]
- 43.Klase Z, Ortiz A, Deleage C, et al. Dysbiotic bacteria translocate in progressive SIV infection. Mucosal immunology. 2015;8(5):1009–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brenchley JM, Price DA, Schacker TW, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12(12):1365–71. [DOI] [PubMed] [Google Scholar]
- 45.Liu J, Williams B, Frank D, Dillon SM, Wilson CC, Landay AL. Inside Out: HIV, the Gut Microbiome, and the Mucosal Immune System. 2017;198(2):605–14. [DOI] [PubMed] [Google Scholar]
- 46.Armstrong AJS, Shaffer M, Nusbacher NM, et al. An exploration of Prevotella-rich microbiomes in HIV and men who have sex with men. Microbiome. 2018;6(1):198-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tett A, Huang KD, Asnicar F, et al. The Prevotella copri Complex Comprises Four Distinct Clades Underrepresented in Westernized Populations. Cell host & microbe. 2019;26(5):666–79.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pedersen H, Guðmundsdóttir V, Nielsen H, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535. [DOI] [PubMed] [Google Scholar]
- 49.Scher JU, Sczesnak A, Longman RS, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife. 2013;2:e01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wen C, Zheng Z, Shao T, et al. Quantitative metagenomics reveals unique gut microbiome biomarkers in ankylosing spondylitis. Genome biology. 2017;18(1):142-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karim R, Mack WJ, Kono N, et al. Gonadotropin and sex steroid levels in HIV-infected premenopausal women and their association with subclinical atherosclerosis in HIV-infected and -uninfected women in the women’s interagency HIV study (WIHS). The Journal of clinical endocrinology and metabolism. 2013;98(4):E610–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sinha-Hikim I, Arver S, Beall G, et al. The Use of a Sensitive Equilibrium Dialysis Method for the Measurement of Free Testosterone Levels in Healthy, Cycling Women and in Human Immunodeficiency Virus-Infected Women1. The Journal of Clinical Endocrinology & Metabolism. 1998;83(4):1312–8. [DOI] [PubMed] [Google Scholar]
- 53.Tepper PG, Randolph JF Jr., McConnell DS, et al. Trajectory Clustering of Estradiol and Follicle-Stimulating Hormone during the Menopausal Transition among Women in the Study of Women’s Health across the Nation (SWAN). The Journal of Clinical Endocrinology & Metabolism. 2012;97(8):2872–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yalamanchi S, Dobs A, Greenblatt RM. Gonadal Function and Reproductive Health in Women with Human Immunodeficiency Virus Infection. Endocrinology and Metabolism Clinics of North America. 2014;43(3):731–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wessman M, Korsholm AS, Bentzen JG, et al. Anti-müllerian hormone levels are reduced in women living with human immunodeficiency virus compared to control women: a case-control study from Copenhagen, Denmark. Journal of virus eradication. 2018;4(2):123–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ghosh M, Rodriguez-Garcia M, Wira CR. The immune system in menopause: pros and cons of hormone therapy. The Journal of steroid biochemistry and molecular biology. 2014;142:171–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gameiro CM, Romão F, Castelo-Branco C. Menopause and aging: changes in the immune system--a review. Maturitas. 2010;67(4):316–20. [DOI] [PubMed] [Google Scholar]
- 58.Shieh A, Epeldegui M, Karlamangla AS, Greendale GA. Gut permeability, inflammation, and bone density across the menopause transition. JCI insight. 2020;5(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zeng MY, Inohara N, Nuñez G. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal immunology. 2017;10(1):18–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Appay V, Sauce D. Immune activation and inflammation in HIV-1 infection: causes and consequences. The Journal of pathology. 2008;214(2):231–41. [DOI] [PubMed] [Google Scholar]
- 61.Taffe J, Dennerstein L. Retrospective self-report compared with menstrual diary data prospectively kept during the menopausal transition. Climacteric. 2000;3(3):183–91. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


