Abstract
Viroids are single-stranded circular noncoding RNAs that infect plants. The noncoding nature indicates that viroids must harness their RNA genomes to redirect host machinery for infection. Therefore, the viroid model provides invaluable opportunities for delineating fundamental principles of RNA structure-function relationships and for dissecting the composition and mechanism of RNA-related cellular machinery. There are two viroid families, Pospiviroidae and Avsunviroidae. Members of both families replicate via the RNA-based rolling-circle mechanism with some variations. Viroid replication is generally divided into three steps: transcription, cleavage, and ligation. Decades of studies have uncovered numerous viroid RNA structures with a regulatory role in replication and multiple enzymes critical for the three replication steps. This review discusses these findings and highlights the latest discoveries. Future studies will continue to elucidate regulatory factors and mechanism of host machinery exploited by viroids and provide new insights into host-viroid interactions in the context of pathogenesis.
Keywords: viroid, DNA-dependent RNA polymerase, RNA-templated transcription, TFIIIA-7ZF, RPL5, RNA promoter, DNA ligase I, chloroplastic tRNA ligase
Introduction
Viroids are single-stranded circular noncoding RNAs that replicate and systemically traffic in plants [1,2]. To date, there are more than 30 viroids grouped into two families, Pospiviroidae and Avsunviroidae [3]. Members of the two families are categorized by the distinct features in overall genome structure, replication sites and processes, and whether they possess ribozymes [1,2,4]. By and large, members of Pospiviroidae have a rod-shaped RNA genome, replicate via the asymmetric rolling-circle model in the nucleus, and do not have intrinsic ribozyme activity. PSTVd is the type species of Pospiviroidae. In contrast, members of Avsunviroidae adopt a highly branched structure at one end of their RNA genome, replicate via the symmetric rolling-circle model in chloroplasts, and possess hammerhead ribozymes. Avocado sunblotch viroid (ASBVd) is the type species of Avsunviroidae.
Viroids, as noncoding RNAs, must co-opt host factors to complete infection. Therefore, the viroid replication system offers excellent opportunities to unravel the function and mechanism of RNA-related host machinery. For instance, viroids of Pospiviroidae are substrates of DNA ligase I, uncovering an unexpected function of this deeply conserved enzyme [5]. In addition, all viroids redirect host DNA-dependent RNA polymerases (DdRPs) for RNA-templated replication [1,2,4]. It was later found that the human Hepatitis delta virus exploits this RNA-dependent RNA polymerase activity of DdRPs for replication [6]. Moreover, this RNA-templated activity regulates gene expression in bacteria [7] and mammalian cells [8] as a widely used mechanism in gene regulation.
Rolling-circle replication of viroids
After entering a cell through opportunistic mechanical wounds or with the aid of insect vectors, viroids traffic to specific organelles for replication. The replication process can largely be divided into three steps: transcription, cleavage, and ligation (Figure 1) [4]. Members of Pospiviroidae replicate in the nucleus starting from the circular (+)-RNA genome, via oligomeric (−)-RNA intermediates, to the generation of oligomeric (+)-RNAs. The oligomeric (+)-RNAs are cleaved into unit-length (+)-strands and ligated to yield progeny. Members of Avsunviroidae replicate in chloroplasts using circular (+)-RNA templates to generate oligomeric (−)-RNAs, which are cleaved and ligated to unit-length circular (−)-RNAs. Circular (−)-RNAs then serve as templates for producing oligomeric (+)-RNAs that are cleaved and subsequently ligated to generate unit-length progeny circular molecules.
Figure 1. Rolling-circle replication mechanism.
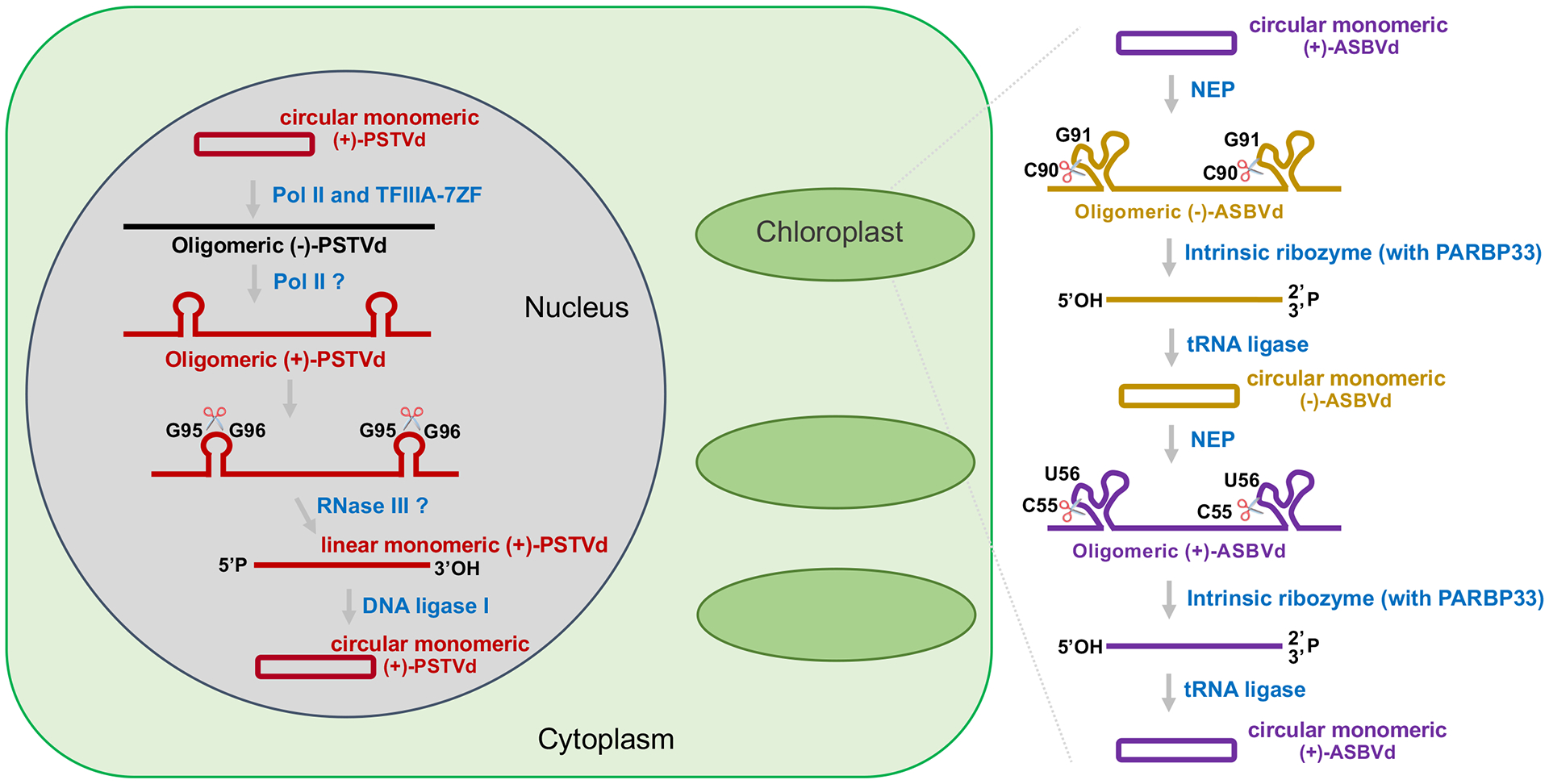
PSTVd and ASBVd are selected to represent members of Pospiviroidae and Avsunviroidae, respectively. Red and black lines refer to (+)- and (−)-strands of PSTVd, respectively. Purple and yellow lines refer to (+)- and (−)-strands of ASBVd, respectively. Pol II, DNA-dependent RNA polymerase II. NEP, nuclear-encoded plastid RNA polymerase. P, phosphate terminus. OH, hydroxyl terminus.
The presence of circular viroids [9–11], longer-than-unit-length (−)-RNAs [12–19] and (+)-RNAs [12,15,17,18,20,21], as well as duplexes composed of one unit-length (+)-RNA and one longer-than-unit-length (−)-RNA [12,14,22,23] led to the rolling-circle replication model [12,14,17,23]. The finding that circular (−)-PSTVd does not exist established the asymmetric rolling-circle mechanism for members of Pospiviroidae [24]. The aforementioned double-stranded duplexes may serve as an efficient trigger of plant RNA silencing activity for small RNA (vd-sRNA) generation [25]. Interestingly, a novel homology-independent bioinformatics approach exploits vd-sRNAs to assemble viroid genomes [26]. Notably, some vd-sRNAs, derived from viroids of both families, are mapped to junction regions of oligomers, implying the existence of oligomeric duplexes during replication [26].
DNA-dependent RNA polymerases for RNA-templated transcription
Early studies reported that multiple polymerases could transcribe the PSTVd RNA genome in vitro [27–29]. However, mounting evidence supports DNA-dependent RNA Polymerase II (Pol II) as the authentic transcription enzyme for viroids of Pospiviroidae. First, purified tomato Pol II complex could transcribe the (+)-PSTVd RNA templates in vitro [27,30,31]. Second, a low concentration of α-amanitin, known to specifically impair Pol II activity, inhibits PSTVd transcription in nuclear extracts [32] and the transcription from (+)-PSTVd to (−)-PSTVd by partially purified Pol II [27]. In addition, the low concentration of α-amanitin inhibits the replication of PSTVd [33,34], cucumber pale fruit viroid [33], hop stunt viroid (HSVd) [35], and citrus exocortis viroid (CEVd) [36–39] in cells. Third, RNA-immunoprecipitation demonstrated that the largest subunit of Pol II interacts with both (−)- and (+)-strands of CEVd [40] and PSTVd [30] in vivo. Furthermore, Pol II preferentially interacts with circular (+)-PSTVd in plants [30]. Whether Pol II recognizes oligomeric (−)-strands as templates remains unclear due to the discrepancy in the literature [34,41]. Notably, transcription is a continuous process in vivo as the products from circular (+)-strands readily serve as templates for producing oligomeric (+)-RNAs. Therefore, uncoupling the two steps is required to confirm the authentic enzyme using oligomeric (−)-RNAs as templates. For all members of Avsunviroidae, their transcription is probably catalyzed by the single-subunit nuclear-encoded plastid RNA polymerase (NEP) [4], based on the observation that ASBVd in vivo replication is sensitive to NEP inhibitor targetoxin [42].
Transcription on viroid RNA templates starts at defined positions. Pol II initiates transcription at C1 or U359 position using circular (+)-PSTVd templates [32], whereas the initiation site from oligomeric (−)-PSTVd to oligomeric (+)-PSTVd awaits to be mapped. Moreover, the initiation sites of other viroids of Pospiviroidae remain to be elucidated. As the type species of Avsunviroidae, the transcription initiation sites of ASBVd were mapped to U121 and U119 in (+)- and (−)-strands, respectively, both residing in the right terminal loop [43]. However, the in vivo initiation sites in another chloroplastic viroid, peach latent mosaic viroid (PLMVd), were mapped to C51 and A286 for the (+)- and (−)-strand templates, respectively [44,45]. The PLMVd initiation sites in both strands locate at similar double-stranded motifs containing the conserved GUC triplet proximal to the cleavage sites [44,45]. Taken together, chloroplastic viroids adopt their specific strategies for transcription initiation during infection. Viroid transcription initiation at defined positions can be explained by the de novo transcription mode, akin to DNA-dependent transcription. In line with this assumption, a recent report provides empirical evidence supporting the de novo transcription of Pol II on PSTVd circular (+)-RNA templates [31].
Regulatory mechanism underlying RNA-templated transcription
Recent studies unraveled the first host transcription factor dedicated to viroid RNA-templated transcription. Transcription factor IIIA (TFIIIA) was shown to directly bind PSTVd in a gel shift assay [46]. Following this work, a recent study showed that a splicing variant of TFIIIA, TFIIIA-7ZF, is the critical transcription factor for RNA-templated transcription catalyzed by Pol II [30]. TFIIIA-7ZF interacts with Pol II and both (+)- and (−)-PSTVd RNAs, modulates PSTVd replication in plants and directly enhances Pol II processivity when transcribing PSTVd RNA templates [30]. It is noteworthy that TFIIIA-7ZF also binds with HSVd [47] and has been suggested to play a role in the replication of apple fruit crinkle viroid [48]. Thus, TFIIIA-7ZF-based replication catalyzed by Pol II is possibly conserved for members of Pospiviroidae.
Purified Pol II alone, without general transcription factors, cannot initiate DNA promoter-dependent transcription [31,49–51]. When mixing with TFIIIA-7ZF and circular PSTVd RNA templates, however, purified Pol II can initiate de novo transcription generating longer-than-unit-length products. This observation suggests that TFIIIA-7ZF acts as a general transcription factor in RNA-templated transcription [31]. Moreover, a conserved general transcription factor TFIIS, which is critical for DNA-dependent transcription, is dispensable for PSTVd RNA-templated transcription in cells. This observation implies that distinct Pol II machinery is formed for RNA-templated transcription [31]. It will be critical to dissect the functional components of Pol II and NEP machinery on viroid templates to achieve a comprehensive understanding of RNA-templated transcription.
Viroid RNA-templated transcription is error-prone. Chloroplastic viroids display the highest mutation rate among all biological entities [52,53], whereas nuclear-replicating viroids possess a mutation rate similar to some RNA viruses [53]. Mutations in accumulated (−)-PSTVd intermediates are nearly absent from the regions corresponding to the central conserved region (CCR) in (+)-PSTVd [54], reflecting the biased Pol II fidelity in transcribing distinct regions [54] or the selection pressure from RNA 3D structure-based constraint [55].
Cleavage and ligation steps in viroid biogenesis
Members of Pospiviroidae do not possess intrinsic ribozymes, so they must rely on a host ribonuclease for cleavage. Using CEVd as a model, dimeric (+)-CEVd is cleaved between G96 and G97 in the CCR in vivo, specifically the upper strand of CCR [56]. The equivalent cleavage site on oligomeric (+)-PSTVd is mapped between G95 and G96 in vitro [57]. Thus, the cleavage site is probably conserved in members of Pospiviroidae. The cleavage products of CEVd possess a 5’-phosphomonoester and 3’-hydroxyl termini, hinting at the involvement of a host RNase III-type enzyme [4,58] yet to be identified. Interestingly, the ligation of PSTVd and several relatives are catalyzed by DNA ligase I [5]. To date, viroids of Pospiviroidae are the only known RNA substrates of DNA ligase I, raising the question of whether this enzyme accepts any endogenous RNA substrates and, if so, the biological significance.
ASBVd and all members of Avsunviroidae possess the intrinsic hammerhead ribozyme to cleave their oligomeric intermediates to unit-length linear products [2,4]. The cleavages occur between C55-U56 in (+)-ASBVd and between C90-G91 in (−)-ASBVd [59]. Despite possessing ribozyme activities, ASBVd interacts with a host factor, PARBP33, to enhance cleavage efficiency [60]. The cleavage products possess the 5’-hydroxyl and 2’,3’-cyclic phosphodiester termini structures [59]. It is generally accepted that the chloroplastic tRNA ligase is responsible for the ligation of chloroplastic viroids [61]. Whether viroid replication impairs or reduces the endogenous function of this tRNA ligase as one of the causes of symptoms deserves further investigation.
Critical RNA motifs that interact with host factors in replication
In the past 40 years, many RNA structures in various viroids have been found to be critical for one or multiple steps in infection. Some of those RNA structures are known to interact with host factors. Recently, the TFIIIA-7ZF binding site has been established as an RNA promoter [31]. This promoter resides at the lower strand of the left terminal region spanning loops 3–5 [30], which is proximal to the transcription initiation site [32], overlapping with the Pol II binding region [62], and critical for replication [63]. This promoter also overlaps with a GC box that was regarded as a potential promoter based on the mapping of the transcription initiation site and mutational analysis [32]. However, the left terminal region of PSTVd may fold into two distinct conformations [64–66]. Since it is unclear which confirmation is involved in the transcription initiation, the structural basis of this RNA promoter remains to be determined [47].
Genome-wide functional analysis has identified two regions in PSTVd genome critical for replication [63]. One region, spanning loops 1–4, has been identified as the binding sites for TFIIIA-7ZF [30] and Pol II [62]. The other region in the CCR, spanning loops 13–15, has been shown to be critical for RPL5 (ribosomal protein L5) binding [67]. Notably, RPL5 is a splicing regulator suppressing the generation of TFIIIA-7ZF [68,69]. PSTVd interaction with RPL5 impairs the splicing regulation activity of RPL5 and is critical to modulate the expression of TFIIIA-7ZF, thereby influencing PSTVd transcription (Figure 2) [67]. It awaits to be clarified whether related viroids of Pospiviroidae employ the same RPL5/TFIIIA-7ZF regulatory cascade in regulating replication.
Figure 2. PSTVd modulating TFIIIA-7ZF expression through interaction with RPL5.
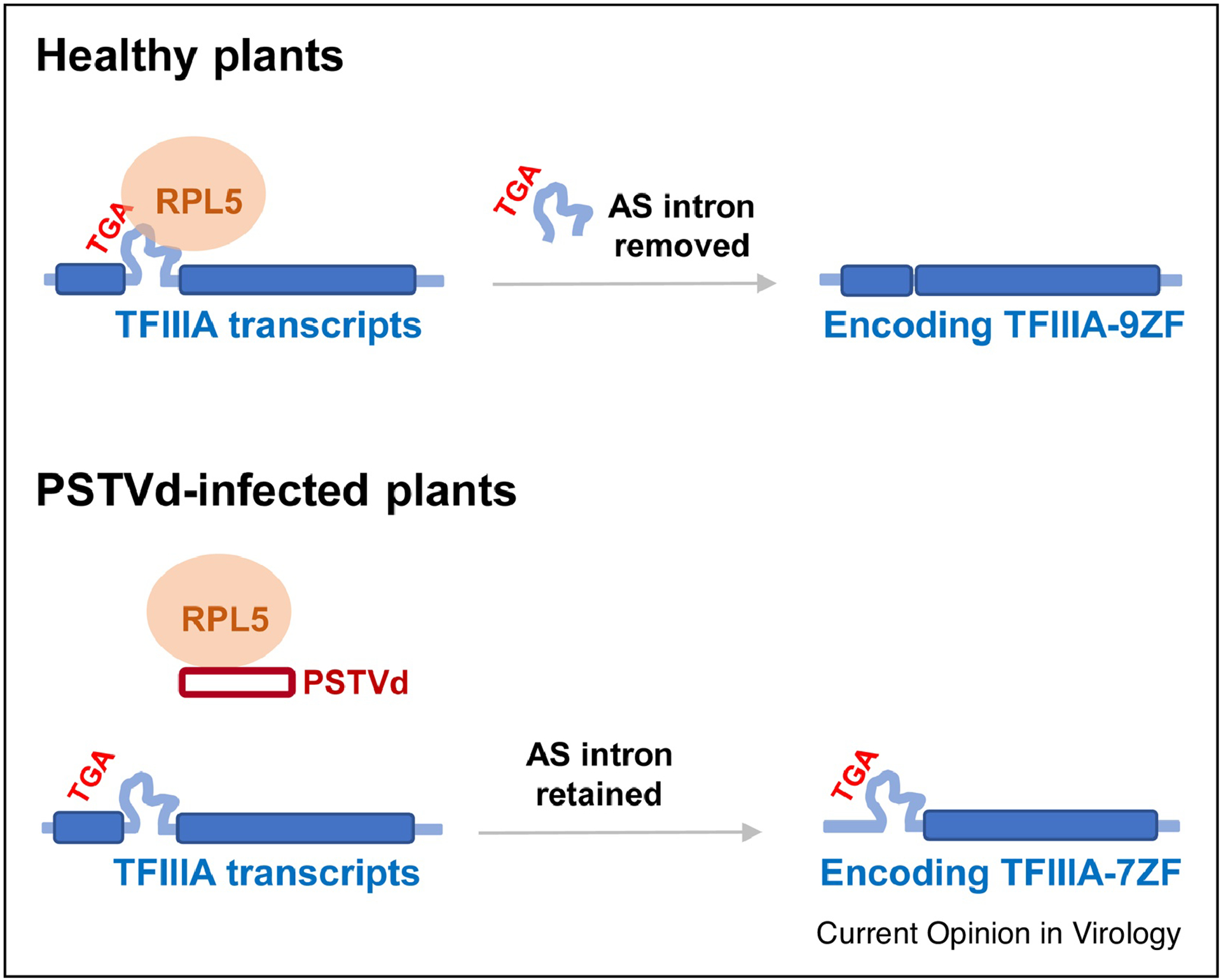
PSTVd directly binds RPL5 and reduces the intron removal of TFIIIA transcripts, resulting in optimized expression of TFIIIA-7ZF. AS, alternative splicing.
The CCR is also critical for processing (i.e. cleavage and ligation) [4]. Particularly, hairpin I, which is conserved in all five genera in Pospiviroidae, is critical for cleavage [56]. Hairpin I is formed by the rearrangement of the upper strand of the CCR and consists of a tetraloop, an internal symmetric loop of 1–3 nt in each strand, and occasionally a 1 nt symmetric or asymmetric loop flanked by short stems [4]. Two adjacent hairpin I structures in oligomeric intermediates form a kissing loop via their palindromic tetraloops, providing a double-stranded structure for cleavage [56]. The cleavage results in two protruding nucleotides in each strand, which is the characteristic feature of RNase III activity [56]. The cleaved products then form the loop E structure for DNA ligase I-mediated ligation [56]. Since loop E is not conserved in Pospiviroidae, it remains to be clarified whether DNA ligase I relies on loop E or a more general structure in the CCR.
Summary and perspectives
Several seminal discoveries regarding viroid replication were reported in the last decade. Discoveries of ligases for members of both families and the terminal structures of cleavage products provide new insights into the ligation steps. The dedicated transcription factor, TFIIIA-7ZF, has been uncovered to be critical for Pol II-catalyzed transcription using PSTVd RNA templates. PSTVd can modulate its replication through interaction with RPL5, leading to an optimized expression of TFIIIA-7ZF.
Due to technical constraints, genetic approaches were rarely applied in viroid research, which may be essential to corroborate the function of host factors harnessed by viroids. The progress in understanding the host machinery for viroids replication also presents new opportunities to elucidate the significance of such interactions in the context of viroid pathogenesis. It is well demonstrated that the viroid model can help dissect various RNA-related host machinery. From this prospect, future studies on RNA promoters and auxiliary factors dedicated to RNA templates may shed a novel light on RNA structure-function relationships.
Acknowledgements
This review is dedicated to the memory of late Professor Biao Ding (Ohio State University). The author expresses gratitude to Bin Liu, Shachinthaka D. Dissanayaka Mudiyanselage, Junfei Ma, Bansi Patel, and Laxmi Kharel at Mississippi State University for critical reading of the manuscript. The author apologizes to colleagues whose work was not cited in this review due to the page limit. Work from the author’s laboratory was supported by research grants from the National Science Foundation (IOS-1564366 and MCB-1906060) and the US National Institutes of Health (R15GM135893). Funders had no role in the study design, in the collection, analysis and interpretation of the data, in the writing of the report or in the decision to submit the article for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The author declares no conflict of interest.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as
• of special interest
•• of outstanding interest
References
- 1.Ding B: The biology of viroid-host interactions. Annu Rev Phytopathol 2009, 47:105–131. [DOI] [PubMed] [Google Scholar]
- 2.Flores R, Gago-Zachert S, Serra P, Sanjuan R, Elena SF: Viroids: survivors from the RNA world? Annu Rev Microbiol 2014, 68:395–414. [DOI] [PubMed] [Google Scholar]
- 3.Giguère T, Perreault JP: Classification of the Pospiviroidae based on their structural hallmarks. PLoS One 2017, 12:e0182536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flores R, Gas ME, Molina-Serrano D, Nohales MÁ, Carbonell A, Gago S, De la Peña M, Daròs JA: Viroid Replication: Rolling-Circles, Enzymes and Ribozymes. Viruses 2009, 1:317–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5**.Nohales MÁ, Flores R, Daròs JA: Viroid RNA redirects host DNA ligase 1 to act as an RNA ligase. Proc Natl Acad Sci U S A 2012, 109:13805–13810. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identifies the ligase for viroids of Pospiviroidae.
- 6.Taylor JM: Hepatitis D Virus Replication. Cold Spring Harb Perspect Med 2015, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wassarman KM, Saecker RM: Synthesis-mediated release of a small RNA inhibitor of RNA polymerase. Science 2006, 314:1601–1603. [DOI] [PubMed] [Google Scholar]
- 8.Wagner SD, Yakovchuk P, Gilman B, Ponicsan SL, Drullinger LF, Kugel JF, Goodrich JA: RNA polymerase II acts as an RNA-dependent RNA polymerase to extend and destabilize a non-coding RNA. EMBO J 2013, 32:781–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Owens RA, Erbe E, Hadidi A, Steere RL, Diener TO: Separation and infectivity of circular and linear forms of potato spindle tuber viroid. Proc Natl Acad Sci U S A 1977, 74:3859–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McClements WL, Kaesberg P: Size and secondary structure of potato spindle tuber viroid. Virology 1977, 76:477–484. [DOI] [PubMed] [Google Scholar]
- 11.Sänger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK: Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A 1976, 73:3852–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Branch AD, Robertson HD: A replication cycle for viroids and other small infectious RNA’s. Science 1984, 223:450–455. [DOI] [PubMed] [Google Scholar]
- 13.Rohde W, Sänger HL: Detection of complementary RNA intermediates of viroid replication by Northern blot hybridization. Biosci Rep 1981, 1:327–336. [DOI] [PubMed] [Google Scholar]
- 14.Owens RA, Diener TO: RNA intermediates in potato spindle tuber viroid replication. Proc Natl Acad Sci U S A 1982, 79:113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bruening G, Gould AR, Murphy PJ, Symons RH: Oligomers of avocado sunblotch viroid are found in infected avocado leaves. FEBS Lett 1982, 148:71–78. [Google Scholar]
- 16.Branch AD, Robertson HD, Dickson E: Longer-than-unit-length viroid minus strands are present in RNA from infected plants. Proc Natl Acad Sci U S A 1981, 78:6381–6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hutchins CJ, Keese P, Visvader JE, Rathjen PD, McInnes JL, Symons RH: Comparison of multimeric plus and minus forms of viroids and virusoids. Plant Mol Biol 1985, 4:293–304. [DOI] [PubMed] [Google Scholar]
- 18.Spiesmacher E, Mühlbach HP, Schnólzer M, Haas B, Sänger HL: Oligomeric forms of potato spindle tuber viroid (PSTV) and of its complementary RNA are present in nuclei isolated from viroid-infected potato cells. Biosci Rep 1983, 3:767–774. [DOI] [PubMed] [Google Scholar]
- 19.Mühlbach HP, Faustmann O, Sänger HL: Contitions for optimal growth of a PSTV-infected potato cell suspension and detection of viroid-complementary longer-than-unit-length RNA in these cells. Plant Mol Biol 1983, 2:239–247. [DOI] [PubMed] [Google Scholar]
- 20.Haseloff J, Mohamed NA, Symons RH: Viroid RNAs of cadang-cadang disease of coconuts. Nature 1982, 299:316–321. [Google Scholar]
- 21.Mohamed NA, Haseloff J, Imperial JS, Symons RH: Characterization of the Different Electrophoretic Forms of the Cadang-Cadang Viroid. J Gen Virol 1982, 63:181–188. [Google Scholar]
- 22.Hadidi A, Hashimoto J, Diener TO: Potato spindle tuber viroid-specific double-stranded RNA in extracts from infected tomato leaves. Ann l’Institut Pasteur / Virol 1982, 133:15–31. [Google Scholar]
- 23.Ishikawa M, Meshi T, Ohno T, Okada Y, Sano T, Ueda I, Shikata E: A revised replication cycle for viroids: The role of longer than unit length RNA in viroid replication. Mol Gen Genet MGG 1984, 196:421–428. [DOI] [PubMed] [Google Scholar]
- 24.Branch AD, Benenfeld BJ, Robertson HD: Evidence for a single rolling circle in the replication of potato spindle tuber viroid. Proc Natl Acad Sci U S A 1988, 85:9128–9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding SW, Voinnet O: Antiviral immunity directed by small RNAs. Cell 2007, 130:413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26*.Wu Q, Wang Y, Cao M, Pantaleo V, Burgyan J, Li WX, Ding SW: Homology-independent discovery of replicating pathogenic circular RNAs by deep sequencing and a new computational algorithm. Proc Natl Acad Sci U S A 2012, 109:3938–3943. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study suggests the existence of oligomeric dsRNA intermediates during viroid replication.
- 27.Rackwitz HR, Rohde W, Sänger HL: DNA-dependent RNA polymerase II of plant origin transcribes viroid RNA into full-length copies. Nature 1981, 291:297–301. [DOI] [PubMed] [Google Scholar]
- 28.Boege F, Rohde W, Sänger HL: In vitro transcription of viroid RNA into full-length copies by RNA-dependent RNA polymerase from healthy tomato leaf tissue. Biosci Rep 1982, 2:185–194. [DOI] [PubMed] [Google Scholar]
- 29.Rohde W, Rackwitz H-R, Boege F, Sänger HL: Viroid RNA is accepted as a template for in vitro transcription by DNA-dependent DNA polymerase I and RNA polymerase from Escherichia coli. Biosci Rep 1982, 2:929–939. [DOI] [PubMed] [Google Scholar]
- 30**.Wang Y, Qu J, Ji S, Wallace AJ, Wu J, Li Y, Gopalan V, Ding B: A land plant-specific transcription factor directly enhances transcription of a pathogenic noncoding RNA template by DNA-dependent RNA polymerase II. Plant Cell 2016, 28:1094–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identifies TFIIIA-7ZF as the first eukaryotic transcription factor for (PSTVd) RNA-templated transcription.
- 31*.Dissanayaka Mudiyanselage SD, Wang Y: Evidence supporting that RNA polymerase II catalyzes de novo transcription using potato spindle tuber viroid circular RNA templates. Viruses 2020, 12:371. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study supports the de novo transcription activity of Pol II when using viroid RNA templates and defines an RNA promoter in circular (+)-PSTVd.
- 32.Kolonko N, Bannach O, Aschermann K, Hu KH, Moors M, Schmitz M, Steger G, Riesner D: Transcription of potato spindle tuber viroid by RNA polymerase II starts in the left terminal loop. Virology 2006, 347:392–404. [DOI] [PubMed] [Google Scholar]
- 33.Mühlbach HP, Sänger HL: Viroid replication is inhibited by alpha-amanitin. Nature 1979, 278:185–188. [DOI] [PubMed] [Google Scholar]
- 34.Schindler IM, Mühlbach HP: Involvement of nuclear DNA-dependent RNA polymerases in potato spindle tuber viroid replication: a reevaluation. Plant Sci 1992, 84:221–229. [Google Scholar]
- 35.Yoshikawa N, Takahashi T: Inhibition of hop stunt viroid replication by α-amanitin. J Plant Dis Prot 1986, 93:62–71. [Google Scholar]
- 36.Flores R: Synthesis of RNAs specific to Citrus exocortis viroid by a fraction rich in nuclei from infected Gynura aurantiaca: Examination of the nature of the products and solubilization of the polymerase-template complex. J Gen Virol 1989, 70:2695–2706. [Google Scholar]
- 37.Flores R, Semancik JS: Properties of a cell-free system for synthesis of citrus exocortis viroid. Proc Natl Acad Sci U S A 1982, 79:6285–6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rivera-Bustamante RF, Semancik JS: Properties of a viroid-replicating complex solubilized from nuclei. J Gen Virol 1989, 70:2707–2716. [Google Scholar]
- 39.Semancik JS, Harper KL: Optimal conditions for cell-free synthesis of citrus exocortis viroid and the question of specificity of RNA polymerase activity. Proc Natl Acad Sci U S A 1984, 81:4429–4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warrilow D, Symons RH: Citrus exocortis viroid RNA is associated with the largest subunit of RNA polymerase II in tomato in vivo. Arch Virol 1999, 144:2367–2375. [DOI] [PubMed] [Google Scholar]
- 41.Spiesmacher E, Mühlbach HP, Tabler M, Sänger HL: Synthesis of (+) and (−) RNA molecules of potato spindle tuber viroid (PSTV) in isolated nuclei and its impairment by transcription inhibitors. Biosci Rep 1985, 5:251–265. [DOI] [PubMed] [Google Scholar]
- 42.Navarro JA, Vera A, Flores R: A chloroplastic RNA polymerase resistant to tagetitoxin is involved in replication of avocado sunblotch viroid. Virology 2000, 268:218–225. [DOI] [PubMed] [Google Scholar]
- 43.Navarro JA; Flores R: Characterization of the initiation sites of both polarity strands of a viroid RNA reveals a motif conserved in sequence and structure. EMBO J 2000, 19:2662–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Delgado S, Martínez de Alba AE, Hernández C, Flores R: A short double-stranded RNA motif of Peach latent mosaic viroid contains the initiation and the self-cleavage sites of both polarity strands. J Virol 2005, 79:12934–12943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Motard J, Bolduc F, Thompson D, Perreault JP: The peach latent mosaic viroid replication initiation site is located at a universal position that appears to be defined by a conserved sequence. Virology 2008, 373:362–375. [DOI] [PubMed] [Google Scholar]
- 46.Eiras M, Nohales MA, Kitajima EW, Flores R, Daròs JA: Ribosomal protein L5 and transcription factor IIIA from Arabidopsis thaliana bind in vitro specifically Potato spindle tuber viroid RNA. Arch Virol 2011, 156:529–533. [DOI] [PubMed] [Google Scholar]
- 47.Dissanayaka Mudiyanselage SD, Qu J, Tian N, Jiang J, Wang Y: Potato spindle tuber viroid RNA-templated transcription: factors and regulation. Viruses 2018, 10:503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48*.Matousek J, Steinbachova L, Drabkova LZ, Kocabek T, Potesil D, Mishra AK, Honys D, Steger G: Elimination of viroids from tobacco pollen involves a decrease in propagation rate and an increase of the degradation processes. Int J Mol Sci 2020, 21:3029. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study supports that the RPL5/TFIIIA-7ZF regulatory cascade may be involved in the replication of apple fruit crinkle viroid, which is in the same family as PSTVd but belonging to a distinct genus.
- 49.Thomas MC, Chiang CM: The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol 2006, 41:105–178. [DOI] [PubMed] [Google Scholar]
- 50.Hantsche M, Cramer P: Conserved RNA polymerase II initiation complex structure. Curr Opin Struct Biol 2017, 47:17–22. [DOI] [PubMed] [Google Scholar]
- 51.Vannini A, Cramer P: Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol Cell 2012, 45:439–446. [DOI] [PubMed] [Google Scholar]
- 52**.Gago S, Elena SF, Flores R, Sanjuán R: Extremely high mutation rate of a hammerhead viroid. Science 2009, 323:1308. [DOI] [PubMed] [Google Scholar]; This report illustrats that a chloroplastic viroid possesses the highest mutation rate among all biological entities for the first time.
- 53*.López-Carrasco A, Ballesteros C, Sentandreu V, Delgado S, Gago-Zachert S, Flores R, Sanjuán R: Different rates of spontaneous mutation of chloroplastic and nuclear viroids as determined by high-fidelity ultra-deep sequencing. PLOS Pathog 2017, 13:e1006547. [DOI] [PMC free article] [PubMed] [Google Scholar]; This report reports that a nuclear-replicating viroid possesses a mutation rate similar to some RNA viruses.
- 54*.Wu J, Bisaro D: Biased Pol II fidelity contributes to conservation of functional domains in the Potato spindle tuber viroid genome. PLoS Pathog In press. [DOI] [PMC free article] [PubMed] [Google Scholar]; This report suggests a different error rate of Pol II in transcribing certain functional domain in potato spindle tuber viroid RNA genome.
- 55*.Wang Y, Zirbel CL, Leontis NB, Ding B: RNA 3-dimensional structural motifs as a critical constraint of viroid RNA evolution. PLoS Pathog 2018, 14:e1006801. [DOI] [PMC free article] [PubMed] [Google Scholar]; This report summarizes a novel mechanism that RNA 3D motifs effectively constrain viroid evolution and selectively retain replication products with certain mutations in the quasispecies.
- 56.Gas M-E, Hernández C, Flores R, Daròs JA: Processing of nuclear viroids in vivo: An interplay between RNA conformations. PLoS Pathog 2007, 3:e182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baumstark T, Schrüder AR., Riesner D: Viroid processing: switch from cleavage to ligation is driven by a change from a tetraloop to a loop E conformation. EMBO J 1997, 16:599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58**.Gas ME, Molina-Serrano D, Hernández C, Flores R, Daròs JA: Monomeric linear RNA of citrus exocortis viroid resulting from processing in vivo has 5’-phosphomonoester and 3’-hydroxyl termini: implications for the RNase and RNA ligase involved in replication. J Virol 2008, 82:10321–10325. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identifies a conserved structure critical for the cleavage of viroids of Pospiviroidae, suggests the involvement of an RNase III-type enzyme, and mapped the termini structures of the cleavage products.
- 59.Hutchins CJ, Rathjen PD, Forster AC, Symons RH: Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res 1986, 14:3627–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Daròs JA, Flores R: A chloroplast protein binds a viroid RNA in vivo and facilitates its hammerhead-mediated self-cleavage. EMBO J 2002, 21:749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61**.Nohales MA, Molina-Serrano D, Flores R, Daròs JA: Involvement of the chloroplastic isoform of tRNA ligase in the replication of viroids belonging to the family Avsunviroidae. J Virol 2012, 86:8269–8276. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identifies the ligase for viroids of Avsunviroidae.
- 62.Bojic T, Beeharry Y, Zhang DJ, Pelchat M: Tomato RNA polymerase II interacts with the rod-like conformation of the left terminal domain of the potato spindle tuber viroid positive RNA genome. J Gen Virol 2012, 93:1591–1600. [DOI] [PubMed] [Google Scholar]
- 63*.Zhong X, Archual AJ, Amin AA, Ding B: A genomic map of viroid RNA motifs critical for replication and systemic trafficking. Plant Cell 2008, 20:35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study reveals the function of local RNA motifs in PSTVd replication and trafficking.
- 64.Gast FU, Kempe D, Spieker RL, Sänger HL: Secondary structure probing of potato spindle tuber viroid (PSTVd) and sequence comparison with other small pathogenic RNA replicons provides evidence for central non-canonical base-pairs, large A-rich loops, and a terminal branch. J Mol Biol 1996, 262:652–670. [DOI] [PubMed] [Google Scholar]
- 65.Riesner D, Henco K, Rokohl U, Klotz G, Kleinschmidt AK, Domdey H, Jank P, Gross HJ, Sänger HL: Structure and structure formation of viroids. J Mol Biol 1979, 133:85–115. [DOI] [PubMed] [Google Scholar]
- 66.Dingley AJ, Steger G, Esters B, Riesner D, Grzesiek S: Structural characterization of the 69 nucleotide potato spindle tuber viroid left-terminal domain by NMR and thermodynamic analysis. J Mol Biol 2003, 334:751–767. [DOI] [PubMed] [Google Scholar]
- 67**.Jiang J, Smith HN, Ren D, Dissanayaka SM, Dawe AL, Wang L, Wang Y: Potato spindle tuber viroid modulates its replication through a direct interaction with a splicing regulator. J Virol 2018, 92:e01004–18. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study reveals that PSTVd can actively modulate its replication by optimizing the expression of TFIIIA-7ZF, which is achieved by the direct interaction between PSTVd and the splicing regulator, RPL5.
- 68.Hammond MC, Wachter A, Breaker RR: A plant 5S ribosomal RNA mimic regulates alternative splicing of transcription factor IIIA pre-mRNAs. Nat Struct Mol Biol 2009, 16:541–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Layat E, Cotterell S, Vaillant I, Yukawa Y, Tutois S, Tourmente S: Transcript levels, alternative splicing and proteolytic cleavage of TFIIIA control 5S rRNA accumulation during Arabidopsis thaliana development. Plant J 2012, 71:35–44. [DOI] [PubMed] [Google Scholar]


