Abstract
The large volume of data used in cancer diagnosis presents a unique opportunity for deep learning algorithms, which improve in predictive performance with increasing data. When applying deep learning to cancer diagnosis, the goal is often to learn how to classify an input sample (such as images or biomarkers) into predefined categories (such as benign or cancerous). In this article, we examine examples of how deep learning algorithms have been implemented to make predictions related to cancer diagnosis using clinical, radiological, and pathological image data. We present a systematic approach for evaluating the development and application of clinical deep learning algorithms. Based on these examples and the current state of deep learning in medicine, we discuss the future possibilities in this space and outline a roadmap for implementations of deep learning in cancer diagnosis.
Introduction:
Diagnosing cancer often relies on physicians using visual pattern recognition to identify concerning lesions clinically, radiologically, or pathologically. For example, during a clinical exam, a dermatologist notes the irregular border and abnormal colors of a lesion suspicious for melanoma. Similarly, a radiologist reviewing a mammogram notes atypia concerning for breast cancer. In both examples, as with most cancer diagnoses, biopsies are ultimately taken to aid in the diagnosis. A pathologist then reviews the tissue under a microscope to help make a diagnosis; both clinical and pathological information is considered. The aforementioned workflows depend on a human physician who has undergone years of training on large volumes of data to recognize the features that are concerning for malignancy.
The large volume of data used in cancer diagnosis presents a unique opportunity for machine learning algorithms, which improve in predictive performance with increasing experience and data.1 Machine learning algorithms can be supervised or unsupervised.2 Supervised algorithms rely on the use of labeled data (for example, labeled photos of normal and abnormal mammograms) to make predictions, while unsupervised algorithms find hidden patterns and relationships without labels.2
When applying machine learning to cancer diagnosis, the goal is often to learn how to classify an input sample (such as a clinical, radiologic, or pathology image) into predefined categories (such as benign or cancerous). These categories could be a specific diagnosis (e.g. melanoma) or a diagnostic category (e.g. malignant versus benign). In a supervised learning framework, these algorithms rely on a training set of labeled data to learn the representative features of each category or diagnosis.
For image classification tasks, deep learning, a subset of machine learning, has been particularly successful.2 Deep learning relies on multi-layer neural networks with many hidden layers, composed of connected artificial neurons that perform mathematical operations on input data.3 In particular, image tasks often rely on convolutional neural networks (CNNs), a type of neural network that is particularly adept at classifying images.4 Image classification has worked exceedingly well because CNNs, which mimic natural visual processing in the brain, are able to interpret dense information such as the relationship of nearby pixels and objects.4
In a medical setting, a deep learning algorithm may be trained to take in a photo of a skin lesion and be able to predict whether or not the lesion is indicative of skin cancer (Figure 1). During training, labeled image data is inputted to the neural network and undergoes filtering (i.e. convolutions) and subsampling (i.e. pooling) steps in order for the network to learn the image features. Just as the network learns the simple image features, it adjusts weights in later layers of the neural network in order to optimize the relationship between imaging features and classification of the input images.3
Figure 1:
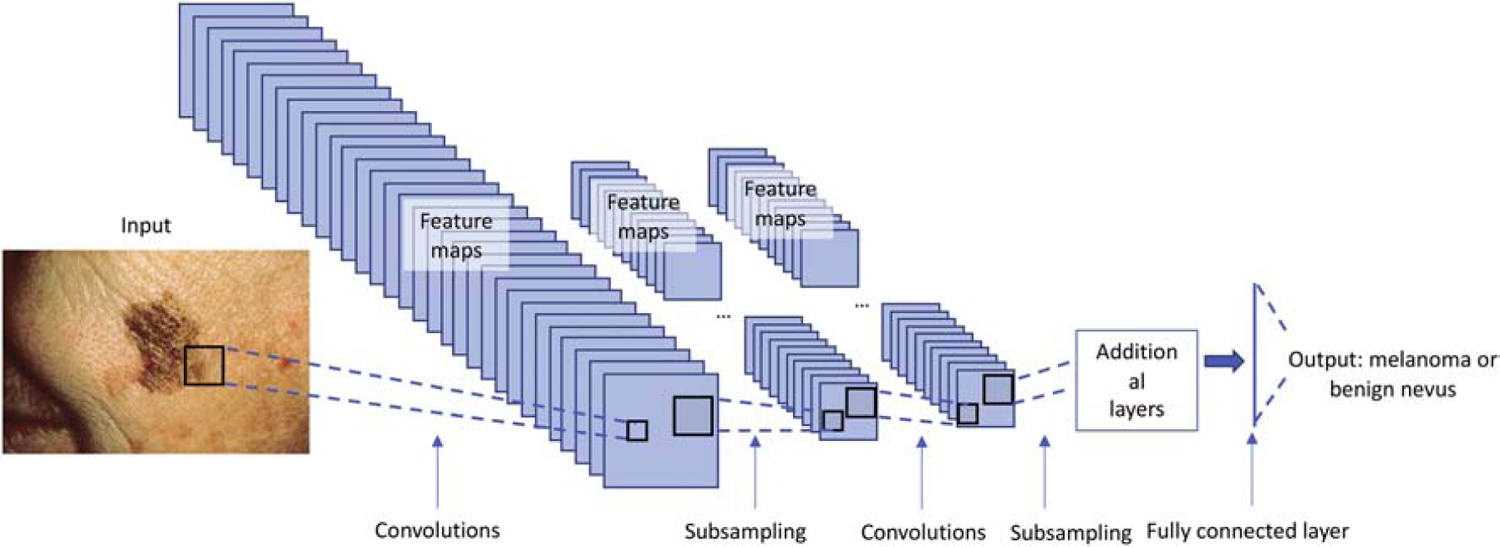
During training, labeled image data (in this case skin lesions) is inputted to the neural network. Images undergo feature extraction (i.e. convolutions) and subsampling (i.e. pooling) steps in order for the network to learn the image features. As the network learns the image features, it adjusts weights to these features in order to optimize the correct classification of the input images. In this example, the output is whether an image represents a benign nevus or a melanoma.
In Figure 2, we show an overview of how deep learning models are implemented for image classification tasks. In the first step, a model architecture is selected based on the task. For image classification tasks, deep CNNs pre-trained on ImageNet, a database of 1.28 million images representing over 1000 categories, are a common choice.3–5 The benefit of this pretraining is that the model already has representations of the basic lines and shapes needed for any image recognition task. This model is then trained using labeled training and validation data to optimize of the weights in the neural network which define the difference between the prediction and the known true label.3 By minimizing that distance or “loss”, the model learns the salient features and how to correctly classify the input images. A separate validation set is used during training to understand the performance of the model and ensure that the network does not overfit to the data.3 Once a model is trained, it is tested on an independent test set so that its final performance can be evaluated. For example, to check that a clinical deep learning algorithm is generalizable, the independent test set data may come from a different hospital system. In the future, these models can eventually be tested prospectively in a clinical environment in order to demonstrate their clinical utility in a real-world environment.
Figure 2:
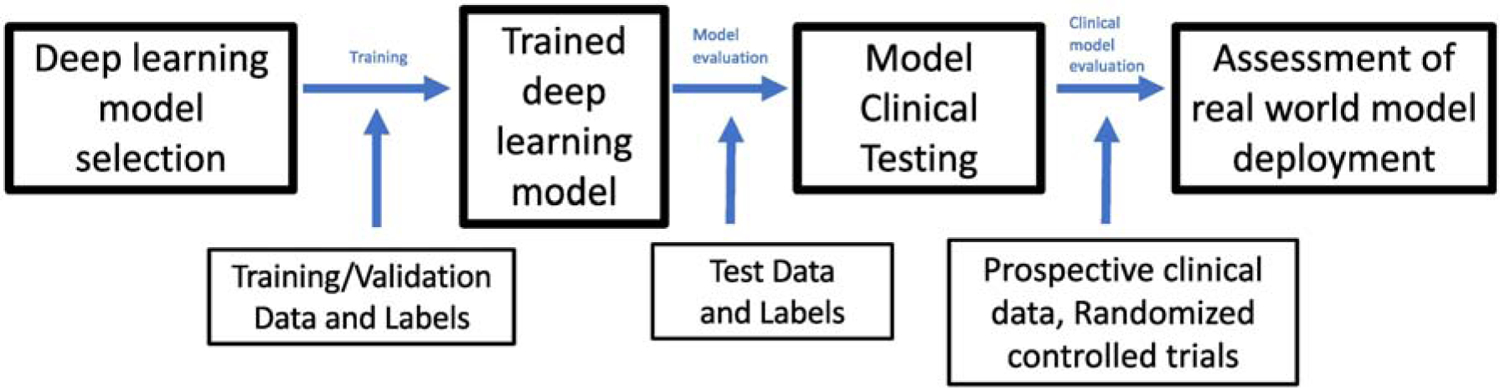
An overview of how deep learning models are implemented for image classification tasks.
In this article, we examine examples of how deep learning algorithms have been implemented to make predictions related to cancer diagnosis using clinical, radiological, and pathological image data. We approach this evaluation through four questions (Figure 3) that aid in dissecting the appropriate development and application of clinical deep learning algorithms. Based on these examples and the current state of deep learning in cancer diagnosis, we discuss the future possibilities in this space and outline a roadmap for future implementations of deep learning in cancer diagnosis.
Figure 3:
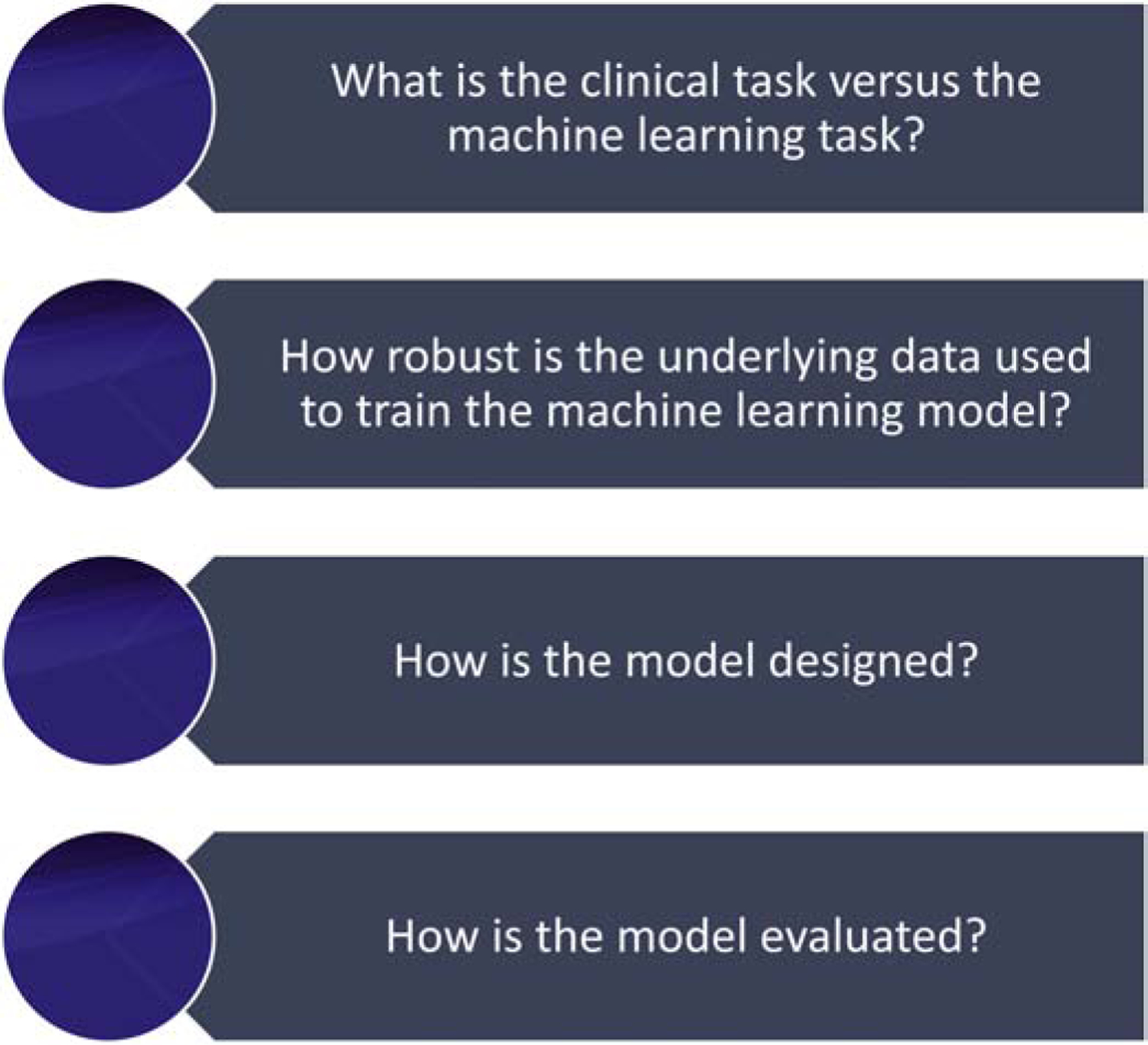
Four questions that aid in assessing the appropriate development and application of clinical deep learning algorithms
What is the clinical task versus the machine learning task?
The application of deep learning in medicine aims to ultimately automate a task currently carried out by humans either with greater speed and/or accuracy. To understand how well an algorithm is performing, its performance must be benchmarked against human decisions or another ‘ground truth’ set of diagnoses or classifications. In the clinical applications discussed here, deep learning is used in a supervised manner: the model is created using these labels to train the model. When evaluating deep learning papers, it is important to ask if the classification task is representative of the actual clinical workflow and whether it is a fair comparison.
For example, Esteva et al. created a deep learning model that was able to distinguish between benign versus malignant skin lesions.6 Their team used a deep CNN trained on 129,450 clinical images representing 2,032 different skin diseases to these images into 757 diseases classes.6 When testing their CNN, they looked at its ability to classify images into a three classes: benign lesions, malignant lesions, and non-neoplastic lesions.6 They also looked at the ability of the CNN to classify images into nine classes based on similar treatment modalities.6 Finally, they tested their CNN on biopsy-proven images to see if the algorithm could distinguish between malignant keratinocyte lesions and seborrheic keratoses and melanoma versus benign nevi.6 Esteva et al compared the performance of their CNN on these three test classification tasks to that of board-certified dermatologists performing the same task and concluded that the CNN performed at the level of the a board-certified dermatologist.6
While their results are promising, the tasks presented were not entirely representative of the clinical workflow in dermatology. A dermatologist performing a full body skin exam has to first identify lesions of interest prior to deciding whether or not such a lesion may represent malignancy. During the skin exam, tactile feedback provides additional information. Dermatologists also take into account a patient’s medical history, the history of the lesion, and other risk factors. In comparison, the CNN was presented with already identified lesions to make predictions. Identifying melanoma versus benign nevus or malignant keratinocyte lesions versus seborrheic keratoses only captures part of a clinician’s task. Melanoma can be phenotypically diverse: benign nevi, seborrheic keratoses, pigmented basal cells, hematomas, and vascular growths are just some of the lesions that may mimic melanoma.7 In Esteva et al’s paper, the three class (benign lesions, malignant lesions, and non-neoplastic lesions) and nine class (based on treatment modalities) classification tasks approximate some of the triage decisions that dermatologists make, but still do not encapsulate the full clinical workflow.6 More recently, Liu et. al developed a machine learning algorithm that generates top three differential diagnoses using patient images and medical history from across 26 different skin disease diagnoses, including melanoma, basal cell carcinoma, and squamous cell carcinoma.8 The incorporation of additional medical history, the expansion of diagnoses considered, and the generation of differential diagnoses brings the Liu et al algorithm closer to the clinical realm.8
In another example, Coudray et al trained a CNN to distinguish between adenocarcinoma of the lung, squamous cell carcinoma of the lung, and normal lung tissue using whole slide images obtained from the Cancer Genome Atlas.9 A total of 1,634 whole slide images representing 1,176 tumor tissues and 459 normal tissues were split into training, validation, and test sets to train and evaluate the model. Notably, because of the size of the images, each slide was split into tiles, resulting in large sample sizes for training/validation and testing. When testing on whole slide images, the CNN achieved an area under the curve (AUC) of 0.97.9 This algorithm was then tested on an independent cohort of whole slide images of frozen and formalin-fixed paraffin-embedded tissue and lung biopsies, which led to a drop in performance, with an AUC above 0.83 in all cases.9 In this case, the clinical workflow is similar to the deep learning task – pathologists make a diagnosis based off visualizing a slide, similarly the algorithm makes its classification based on slide visualization.9 Notably, the algorithm was trained on whole slide images from The Cancer Genome Atlas, and the authors demonstrated that there is a drop in AUC when the algorithm is tested on images derived from tissue processed in different ways at their academic center.9 The CNN was trained using only hematoxylin and eosin (H&E) stained slides, while in real clinical practice, additional immunohistochemical stains are often used to aid in diagnosis.9 However, there is the possibility that machine learning tools may be able to make diagnoses without the need for special stains; further validation will be needed to evaluate this possibility.
For both Esteva et al and Coudray et al’s algorithms, there are some mismatches between the framing of the machine learning task and the clinical context and auxiliary information frequently used by dermatologists and pathologists. It is important to understand the impact of this mismatch in evaluating the suitability of the trained machine learning algorithm and in designing deployment pilot studies.
How robust is the underlying data used to train the machine learning model?
When assessing the model performance, the data underlying the model is important to consider. Important factors include data adjudication, patient inclusion and exclusion criteria, and patient representation.
Deep learning algorithms rely on training/validation and testing data that have been properly labelled. Data adjudication is important as “junk in” can lead to “junk out” for algorithms. For each machine learning task, a gold standard should be set for proper data labeling. For example, with skin cancer, the gold standard in clinical care is pathological confirmation rather than simply clinical agreement. The two aforementioned deep learning papers in the skin cancer field had only a subset of their data pathologically confirmed, the rest were “clinically” diagnosed.6, 8 Labeling the training/validation and testing data appropriately is not always straightforward as the clinical task may differ from the endpoint of diagnosis. For example, radiologists aren’t trained based on the eventual biopsy results of patients who undergo imaging, but rather classify images based on standards of risk and confidence for which multiple readers might disagree. Even within pathology, there is debate about how pathological confirmation should be viewed.10 Even with pathology-based gold standards there are limitations, and pathologists have had interobserver disagreement documented across multiple cancer types, including melanoma.10 Adamson et al suggested a solution to this problem: re-considering the labeling of pathology images in cancer discrimination tasks into three bins based on a panel of experts – total agreement of cancer diagnosis, total agreement of cancer absence, and disagreement on diagnosis.11
For a model to be applicable to a broad patient population, the training data need to be representative of this population. To this end, detailed descriptions of the inclusion and exclusion criteria used to select patients or patient samples is necessary.12 Moreover, a detailed description of patient demographics is likewise important. Biases in the input dataset can be propagated in the training of the model.13 For example, skin cancer is far less prevalent in darker skin types; however, it still does occur. Basal cell carcinoma, the most common cutaneous malignancy, occurs at a yearly incidence 212 to 250 per 100,000 in Caucasians, 1–2 per 100,000 in African Americans, 5.8–6.4 per 100,000 in Chinese individuals, and 50–171 in Hispanic individuals.14 However, most of the studies using deep learning to predict skin cancer from image data have largely left out darker skin types from their training/validation and test data.11 For example, Liu et al’s predictive algorithm that classifies across 26 common skin conditions had only one individual of the darkest skin type and less than 3% of the second darkest skin type in the test data set.15 Not including a subset of the population in algorithm creation can perpetuate bias and health disparities.11
Another issue to consider when assessing the training/validation and testing data is whether the classes are balanced.16, 17 Class imbalance means a significant skew towards a particular category; as a result, algorithms trained on highly imbalanced data can end up over-classifying to the majority group.16 Class imbalance in the test set can also be an issue – for example, if a test contains 98% benign lesions and 2% malignant lesions, a simple classifier that simply labels all test samples as benign will have a 98% accuracy, though its performance will be poor in actual practice.16 When dealing with uncommon diseases, studies are at risk of having unbalanced datasets because the number of control samples available can be up to 1000 times higher than the number of cancer cases.17 There are methodologies, such as data augmentation, to deal with cases of class imbalance, and any algorithm that trains/validates or tests on imbalanced data should explicitly outline how this imbalance is addressed.17
How is the model designed?
The choice of the deep learning architecture can greatly impact the model’s performance. Most image classification tasks use CNNs, which do particularly well on this type of task due to their ability to learn image features. There are several standard architectures available, and they differ in the network depth and connection patterns that impose different priors over images. Popular architectures includes ResNet and Inception, which capture lessons learned from computer science and represent the state of the art in image classification with deep learning algorithms. The architecture selected for the algorithm should be clearly described within the paper.
When the architecture used is not clearly described, it can be difficult to understand the underlying algorithm or to make attempts to replicate it. For example, McKinney et al aimed to predict biopsy-proven breast cancer based on mammogram images using a deep learning model.18 However, the model is described as an ensemble of three different models, whose details are not fully described nor the contribution in performance for each model in the ensemble.18 A major concern in applying machine learning to medicine is the presence of over-engineering when a simpler or smaller model could do just as well. In general, if novel architectures are used within a paper, there should be a comparison to standard benchmarks to help justify the use of the novel architecture.
How is the model evaluated?
Understanding how a model is evaluated is important for understanding how well the model may generalize in the real world. A model that does well on the training data but does poorly on the test data may suffer from overfitting. Metrics for evaluating machine learning algorithms include sensitivity, specificity, and positive predictive value.2 Receiver operating characteristics (ROC) curve plots the sensitivity (the true positive rate) on the x-axis versus 1-specificity (the false positive rate) on the y-axis; a random classifier is represented by a diagonal line with a slope of 1 and an intercept at 0.2 The area under the curve (AUC) of the ROC curve demonstrates the classifier’s capability of distinguishing between classes.2
The evaluation metrics should be computed based on the model’s performance on an independent test set.2 Often this independent test set is retrospective in nature; however, in order to truly validate the generalizability of a deep learning algorithm on clinical tasks, prospective application and evaluation is required given differences between real world data and retrospective data. The gold standard for showing the efficacy of a medical intervention is the randomized controlled trial (RCT), and guidelines for RCTs using AI have been developed.12 To date, there have been a few RCTs using AI algorithms.19 Two recent trials were completed using a deep learning algorithm to aid in endoscopic tasks.20, 21 The adenoma detection rate (ADR) during colonoscopies is used as a quality indicator of colonoscopies; though it is a surrogate clinical metric for the clinical outcome that matters – reducing the number of colon cancers.21 In Wang et al’s trial, 1046 patients were enrolled in a double-blind sham-controlled trial using colonoscopy with computer-aided detection to evaluate ADR.21 In the computer-aided detection arm of the trial, endoscopists had a statistically significant higher ADR of 34%, compared to 28% in the sham-controlled arm.21
A second study evaluated the additional value in AI aided esophagogastroduodenoscopy (EGD) in assessing upper gastrointestinal diseases, including gastric cancer.20 During EGD, endoscopists must comprehensively visualize the stomach in order to avoid missing subtle gastric cancers; an area that is missed is a blind spot.20 Chen et al tested the ability of an AI assistive tool to help reduce the blind spot rate.20 Their prospective, single-blind, randomized trial was done at a single center with three parallel groups (unsedated ultrathin transoral endoscopy, unsedated conventional EGD, and sedated conventional EGD), each of which had an AI assistance arm and an unassisted arm.20 In all three groups, the blind spot rate of the AI assisted arm of the study was significantly lower than the blind spot rate of the group without AI assistance.20
While these results are promising, a higher level question needs to be investigated for AI assistance particularly used in cancer screening tasks. Like any tool used in cancer screening, AI should not increase the number of benign lesions that are misclassified as cancer, as this leads to unnecessary procedures nor should it miss clinically obvious lesions.22 Rather, a clinically useful AI algorithm would identify additional curable cancers that would have been otherwise missed while not overcalling early cancer lesions that would never progress or be life threatening.22 To understand whether AI is leading to clinically meaningful outcomes in cancer screening, additional longitudinal data is necessary.
Additionally, the recent randomized controlled trials demonstrate another important reality – in most cases, real clinical evaluation will require physicians working with an AI tool, rather than an AI tool replacing a physician.19 Clinical medicine always has a certain level of uncertainty, and a clinical case may not neatly fall into the categories predicted by the AI algorithm. Given the complexities of clinical care and the limitations of training data, creating an AI tool that completely replaces the clinical workflow of a physician is not currently feasible.
Next steps in the applications of AI for cancer diagnostics
A rigorously evaluated AI tool in the hands of a physician has the potential to improve workflows, improve accuracy, and cut costs. As reviewed above, there are several factors to consider when evaluating AI algorithms applied to tasks in cancer diagnostics. Even though the AI algorithms reviewed above do not perfectly approximate the clinical task, there is still ample opportunity for these algorithms to work hand in hand with healthcare workers. For example, skin lesions are not always seen by dermatologists initially, who are skin specialists, but are often first evaluated by primary care physicians or nurse practitioners and family doctors. Both Esteva et al and Liu et al’s dermatology algorithms can distinguish between benign lesions, malignant lesions, and non-neoplastic lesions with similar performance to board certified dermatologists; such an algorithm may serve as a useful triage tool for non-specialists making decisions about dermatology referrals.6, 15
AI presents the opportunity to not only build tools that will improve physician workflows, but also accomplish tasks that were previously not possible. AI algorithms are able to pick up patterns that are not discernable by humans, and this creates the potential for endless creative applications. For example, human pathologist cannot look at a histopathology slide without any special stains or testing and discern what genetic mutations might be present. However, Coudray et al demonstrated the ability of AI to predict multiple clinically relevant gene mutations in EGFR, STK11, FAT1, SETBP1, KRAS, and TP53 from H&E histopathology slides of lung cancer samples.9 This type of task could be replicated from other cancer types and provide feedback to pathologists about what additional testing might be indicated for a tissue sample.
Another problem in cancer diagnosis and treatment is identifying the primary tumor’s origin, particularly when the disease is metastatic and not well-differentiated.23 In fact, an estimated 3% of metastatic cancer cases have no obvious primary based on human assessment of the cancer.23 Jiao et al developed a deep learning algorithm that used somatic passenger mutations from whole genome sequencing of 24 tumor types to be able to predict the primary tumor of origin.23 They found this algorithm had an accuracy of 88% and 83% on independent primary and metastatic tumor samples.23 These early findings suggest that machine learning could be used with molecular data to learn new insights about tumors.
Artificial intelligence has the potential to revolutionize cancer diagnosis in several different domains. By thinking critically about how the machine learning algorithm aligns with how clinical workflow, how it is designed and tested, and what data is used, physicians can understand the potential of machine learning and play a key role in their development, evaluation, and deployment.
Acknowledgement:
Sam Finlayson for his feedback on Figure 2. JZ is supported by NSF CCF 1763191, NIH R21 MD012867-01, NIH P30AG059307, and grants from the Silicon Valley Foundation and the Chan-Zuckerberg Initiative. RD is supported by the NIH T32 5T32AR007422-38. DO is supported by American College of Cardiology / Merck Fellowship. BH is supported by NSF DGE-114747.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
☐ The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med. January 2019;25(1):44–56. 10.1038/s41591-018-0300-7 [DOI] [PubMed] [Google Scholar]
- 2.Liu Y, Chen PC, Krause J, Peng L. How to Read Articles That Use Machine Learning: Users’ Guides to the Medical Literature. JAMA. November 2019;322(18):1806–1816. 10.1001/jama.2019.16489 [DOI] [PubMed] [Google Scholar]
- 3.Zou J, Huss M, Abid A, Mohammadi P, Torkamani A, Telenti A. A primer on deep learning in genomics. Nat Genet. January 2019;51(1):12–18. 10.1038/s41588-018-0295-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rawat W, Wang Z. Deep Convolutional Neural Networks for Image Classification: A Comprehensive Review. Neural Comput. September 2017;29(9):2352–2449. 10.1162/NECO_a_00990 [DOI] [PubMed] [Google Scholar]
- 5.Russakovsky O, Deng J, Su H, et al. ImageNet Large Scale Visual Recognition Challenge. IJCV 2015.
- 6.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. February 2017;542(7639):115–118. 10.1038/nature21056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rongioletti F, Smoller BR. Unusual histological variants of cutaneous malignant melanoma with some clinical and possible prognostic correlations. J Cutan Pathol. October 2005;32(9):589–603. 10.1111/j.0303-6987.2005.00418.x [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, Jain A, Eng C, et al. A deep learning system for differential diagnosis of skin diseases. Nat Med. June 2020;26(6):900–908. 10.1038/s41591-020-0842-3 [DOI] [PubMed] [Google Scholar]
- 9.Coudray N, Ocampo PS, Sakellaropoulos T, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med. October 2018;24(10):1559–1567. 10.1038/s41591-018-0177-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adamson AS, Welch HG. Machine Learning and the Cancer-Diagnosis Problem - No Gold Standard. N Engl J Med. December 2019;381(24):2285–2287. 10.1056/NEJMp1907407 [DOI] [PubMed] [Google Scholar]
- 11.Adamson AS, Smith A. Machine Learning and Health Care Disparities in Dermatology. JAMA Dermatol. November 2018;154(11):1247–1248. 10.1001/jamadermatol.2018.2348 [DOI] [PubMed] [Google Scholar]
- 12.Liu X, Cruz Rivera S, Moher D, Calvert MJ, Denniston AK, Group S-AaC-AW. Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: the CONSORT-AI extension. Nat Med. September 2020;26(9):1364–1374. 10.1038/s41591-020-1034-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zou J, Schiebinger L. AI can be sexist and racist - it’s time to make it fair. Nature. July 2018;559(7714):324–326. 10.1038/d41586-018-05707-8 [DOI] [PubMed] [Google Scholar]
- 14.Gloster HM, Neal K. Skin cancer in skin of color. J Am Acad Dermatol. November 2006;55(5):741–60; 10.1016/j.jaad.2005.08.063 [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Jain A, Eng C, et al. A deep learning system for differential diagnosis of skin diseases. arXiv 2019. [DOI] [PubMed]
- 16.Johnson J, Khoshgoftaar T. Survey on deep learning with class imbalance. Journal of Big Data 2019.
- 17.Buda M, Maki A, Mazurowski MA. A systematic study of the class imbalance problem in convolutional neural networks. Neural Networks 2018. p. 249–259. [DOI] [PubMed]
- 18.McKinney SM, Sieniek M, Godbole V, et al. International evaluation of an AI system for breast cancer screening. Nature. January 2020;577(7788):89–94. 10.1038/s41586-019-1799-6 [DOI] [PubMed] [Google Scholar]
- 19.Picardo S, Ragunath K. Artificial intelligence in endoscopy: the guardian angel is around the corner. Gastrointest Endosc. February 2020;91(2):340–341. 10.1016/j.gie.2019.10.026 [DOI] [PubMed] [Google Scholar]
- 20.Chen D, Wu L, Li Y, et al. Comparing blind spots of unsedated ultrafine, sedated, and unsedated conventional gastroscopy with and without artificial intelligence: a prospective, single-blind, 3-parallel-group, randomized, single-center trial. Gastrointest Endosc. February 2020;91(2):332–339. 10.1016/j.gie.2019.09.016 [DOI] [PubMed] [Google Scholar]
- 21.Wang P, Liu X, Berzin TM, et al. Effect of a deep-learning computer-aided detection system on adenoma detection during colonoscopy (CADe-DB trial): a double-blind randomised study. Lancet Gastroenterol Hepatol. January 2020; 10.1016/S2468-1253(19)30411-X [DOI] [PubMed]
- 22.Prasad V. Twitter. Accessed February 22, 2020, https://twitter.com/VPrasadMDMPH/status/1212840987363442689?s=20
- 23.Jiao W, Atwal G, Polak P, et al. A deep learning system accurately classifies primary and metastatic cancers using passenger mutation patterns. Nat Commun. February 2020;11(1):728. 10.1038/s41467-019-13825-8 [DOI] [PMC free article] [PubMed] [Google Scholar]


