Summary:
Mammalian cyclic nucleotide-gated (CNG) channels play an essential role in the signal transduction of the visual and olfactory sensory systems. Here we reveal the structural mechanism of ligand gating in human rod CNGA1 channel by determining its cryo-EM structures in both the apo closed and cGMP-bound open states. Distinct from most other members of voltage-gated tetrameric cation channels, CNGA1 forms a central channel gate in the middle of the membrane, occluding the central cavity. Structural analyses of ion binding profiles in the selectivity filters of the wild-type channel and the E365Q filter mutant allow us to unambiguously define the two Ca2+ binding sites inside the selectivity filter, providing structural insights into Ca2+ blockage and permeation in CNG channels. The structure of the E365Q mutant also reveals two alternative side-chain conformations at Q365, providing a plausible explanation for the voltage dependent gating of CNG channel acquired upon E365 mutation.
Keywords: cyclic nucleotide-gated, CNG channel, cGMP activation, Ca2+ blockage and permeation, signal transduction, visual and olfactory sensory system
eTOC Blurb:
Xue et al. present the high resolution structures of human rod CNGA1 in the presence and absence of cGMP ligand. These structures reveal how cGMP regulate the opening and closing of the channel. The study also provides structural insights into Ca2+ blockage and permeation in CNG channels.
Introduction:
Mammalian cyclic nucleotide-gated (CNG) channels are central to numerous signal transduction pathways, most classically in the visual and olfactory sensory systems (Bradley et al., 2005; Craven and Zagotta, 2006; Finn et al., 1996; Kaupp and Seifert, 2002; Matulef and Zagotta, 2003; Varnum and Dai, 2015; Yau and Baylor, 1989; Zagotta and Siegelbaum, 1996). Although classified as members of the voltage-gated ion channel superfamily, CNG channels are gated by ligands rather than voltage. Like voltage-gated K+ channels, they function as tetramers with each subunit comprised of six membrane-spanning segments and a C-terminal cytoplasmic cyclic nucleotide binding domain (CNBD) that modulates channel opening (Gordon and Zagotta, 1995b; Liu et al., 1996; Varnum and Zagotta, 1996). Native CNG channels form heterotetramers consisting of homologous alpha (CNGA) and beta (CNGB) subunits (Bonigk et al., 1993; Bradley et al., 1994; Chen et al., 1993; Dhallan et al., 1990; Gerstner et al., 2000; Kaupp et al., 1989; Korschen et al., 1995; Liman and Buck, 1994; Peng et al., 2004; Weitz et al., 2002; Zheng and Zagotta, 2004; Zhong et al., 2002). The α-subunits (CNGA1, A2 or A3), being the major components of CNG channels, can by themselves form functional homotetrameric channels and therefore have been commonly used to characterize various ion selectivity and ligand gating properties of CNG channels (Kaupp and Seifert, 2002; Richards and Gordon, 2000). Among the three most commonly studied CNG channels, rod CNGA1 and cone A3 in photoreceptors are activated by cGMP, whereas CNGA2 in olfactory sensory neurons can be activated by both cAMP and cGMP (Dhallan et al., 1990; Gordon and Zagotta, 1995a; Ildefonse et al., 1992; Nakamura and Gold, 1987; Varnum et al., 1995).
Like K+ channels, the last two membrane-spanning segments of CNG channels form the ion conduction pore that controls ion permeation. The CNG channel pore shares some sequence similarity with that of K+ channels, but its selectivity filter has a sequence of TIGE, lacking the conserved tyrosine and glycine residues of the TVGYGD signature sequence in the K+ channel selectivity filter (Heginbotham et al., 1992; Heginbotham et al., 1994; Zhou et al., 2001). Consequently, the selectivity filters of CNG channels are expected to adopt a different structure that likely contributes to their non-selective nature, allowing the conduction of most group I cations as well as Ca2+ (Baumann et al., 1994; Finn et al., 1997; Frings et al., 1995; Hackos and Korenbrot, 1999; Picones and Korenbrot, 1995). In permeating Ca2+, the ion binds to a site along the conduction pathway with high enough affinity to effectively block monovalent cations (Colamartino et al., 1991; Dzeja et al., 1999; Frings et al., 1995; Haynes and Yau, 1985; Haynes et al., 1986; Stern et al., 1987; Zimmerman and Baylor, 1992; Zufall et al., 1994). This apparent blockage of monovalent currents is of crucial physiological significance to visual transduction - it reduces the channel conductance, giving rise to low membrane potential noise and allows photoreceptor cells to detect light with high sensitivity (Yau and Baylor, 1989). Mutagenesis work pinpointed the role of the conserved acidic residue (usually a glutamate) in the selectivity filter of CNG channels that mediates this Ca2+ binding and subsequent block of monovalent currents (Eismann et al., 1994; Gavazzo et al., 2000; Park and MacKinnon, 1995; Root and MacKinnon, 1993; Seifert et al., 1999).
While CNG channels have been extensively studied for many years with a plethora of biochemical and physiological data, their structural information has been lagging behind as compared to other members of voltage-gated ion channel superfamily. In the absence of CNG channel structures, structures of other members of CNBD channels, such as the hyperpolarization-activated cyclic nucleotide-gated (HCN) channels and some prokaryotic cyclic nucleotide-gated K+ channels, were used as structural models for the CNG channels (Clayton et al., 2004; James et al., 2017; James and Zagotta, 2018; Lee and MacKinnon, 2017; Rheinberger et al., 2018; Zagotta et al., 2003). In addition, the bacterial non-selective NaK channel was used to recapitulate and model the selectivity of CNG channels (Alam et al., 2007; Derebe et al., 2011; Shi et al., 2006). Recently, the structures of the C. elegans CNG channel, TAX-4, in the presence and absence of cGMP ligand were determined, providing the first structural view of a CNG family channel (Li et al., 2017; Zheng et al., 2020). Here we determined the structures of human rod CNGA1 homotetramer in both the apo closed and cGMP-bound open states, revealing a unique position of the channel gate that is distinct from most canonical voltage-gated ion channels. Structural analysis of ion binding in CNGA1 also allows us to reveal the Ca2+ binding sites within the filter which, unexpectedly, is different from the mutagenesis-based prediction of external site formed by the conserved acidic residue.
Results:
1. Function analysis and structural determination of CNGA1
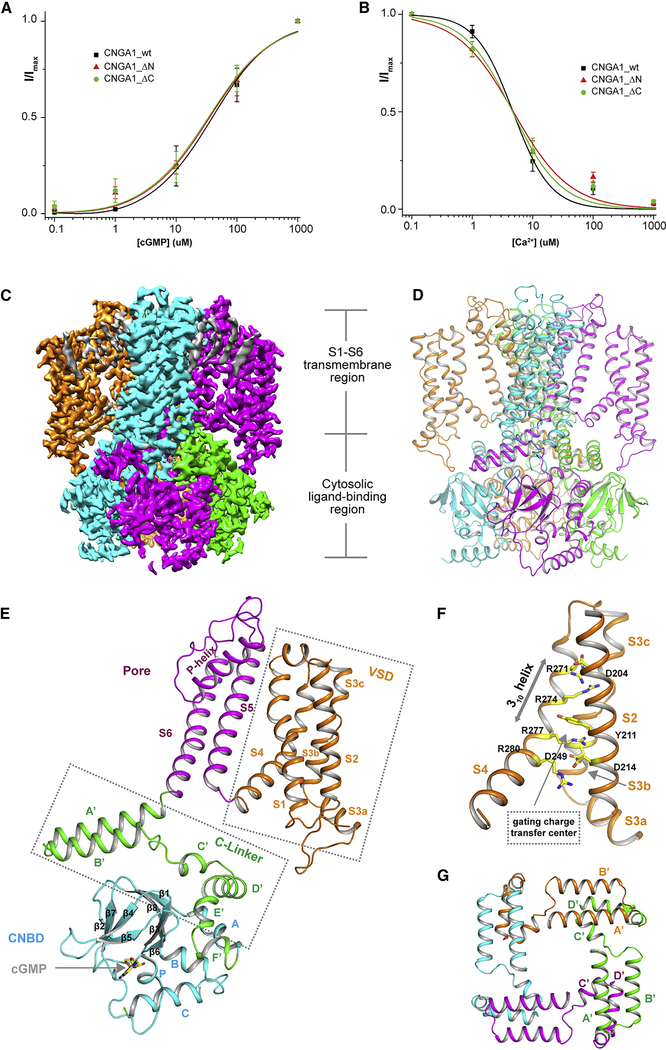
Human rod CNGA1 was expressed in HEK293F cells using the BacMam system and purified in digitonin detergent as a homo-tetramer (Methods). The N-terminal 144 residues of CNGA1 was removed in the final protein construct used for structural determination, as the deletion mutation significantly improved the protein expression and stability. Patch clamp recordings showed that the truncated channel exhibits similar cGMP activation and Ca2+ blockage properties as compared to the full-length channel, suggesting that the removal of the N-terminal region exerts no obvious effect on channel gating and conduction (Figure 1A and 1B, Figure S1). The single particle cryo-EM structures of CNGA1 homotetramer in the apo closed and cGMP-bound open states were determined to the resolutions of 2.64 and 2.91 Å, respectively (Figure S2 and S3, Table S1). The EM density maps of both structures are of high quality, allowing for accurate model building for the major part of the protein containing residues 156–605 (Figure 1C and 1D, Figure S4 and S5). In addition, electron density peaks from closely attached lipid molecules were clearly visible at the transmembrane region of the channel and were modeled as cholesterol or phospholipid molecules in the final structures. The C-terminal 85 residues of CNGA1 are disordered in our structures. Similar to what was observed in the N-terminal deletion construct, the deletion of the C-terminal disorder region has no significant effect on the gating and conduction of the channel (Figure 1A and 1B, Figure S1).
Figure 1. Function and overall structure of human CNGA1.
See also Figure S1–S6 and Table S1
(A) cGMP activation of CNGA1 and its truncation mutants measured at 100 mV. Curves are least square fits to the Hill equation with EC50=39.4±5.8 μM, n=0.90±0.11 for CNGA1_wt, EC50=35.4±7.3 μM, n=0.85±0.13 for CNGA1_DN and EC50=33.8±7.7 μM, n=0.84±0.14 for CNGA1_DC. Data points are mean ± SEM (n=5 independent experiments).
(B) Ca2+ inhibition of CNGA1 and its truncation mutants measured at −100 mV. Curves are least square fits to the Hill equation with Ki=4.71±0.96 μM, n=1.41±0.30 for CNGA1_wt, Ki=4.78±1.34 μM, n=0.93±0.18 for CNGA1_DN and Ki=4.63±0.86 μM, n=1.09±0.17 for CNGA1_DC. Data points are mean ± SEM (n=5 independent experiments).
(C) Side view of 3-D reconstruction of apo CNGA1 with each subunit individually colored. Lipid density is shown in grey.
(D) Side view of cartoon representation of the apo CNGA1 structure.
(E) Structure of a single subunit of cGMP-bound CNGA1 with each domain individually colored.
(F) Zoomed-in view of VSD with S1 helix removed for clarity.
(G) Formation of a gating ring by C-linker domain in a channel tetramer with each subunit individually colored.
CNGA1 consists of two structural components: the membrane-spanning S1-S6 region and the cytosolic ligand gating apparatus (Figure 1C–1E). Like other classical 6-TM tetrameric cation channels, the transmembrane S1-S6 of each subunit is separated into an S1-S4 voltage-sensor domain (VSD) and an S5-S6 pore domain. However, distinct from the majority of 6-TM voltage-gated channels, the tetramer assembly of CNGA1 is not domain-swapped – its VSD domain interacts directly with the pore domain from the same subunit instead of the neighboring subunit; and the VSD is connected to the pore domain with a short loop instead of an S4-S5 linker helix (Figure 1E). This non-domain-swapped channel assembly is also observed in other CNBD channels (James and Zagotta, 2018). Although CNGA1 is not voltage gated, its S1-S4 VSD structure is very similar to that of human HCN1 channel in depolarized state (Lee and MacKinnon, 2017) and also contains several key features of a canonical voltage sensor, including the presence of the conserved gating charge transfer center formed by Y211 and D214 from S2 and D249 from S3, multiple S4 arginine residues with R277 positioned in the charge transfer center, and the formation of a 310 helix in the arginine-containing part of S4 (Figure 1F and Figure S6). It is unclear what structure feature contributes to the loss of voltage sensitivity in the VSD of CNG channels.
The cytosolic soluble fragment of CNGA1 forms the gating apparatus in a channel tetramer responsible for cGMP activation of the channel. Following the same nomenclature and domain definition used for HCN channel (Zagotta et al., 2003), the soluble fragment of each subunit consists of the C-terminal CNBD that is specific for cGMP binding in CNGA1 and the C-linker domain that connects the CNBD to the channel pore (Figure 1E). The CNBD contains helices A, P, B, C and a β-roll (β1-β8) between helices A and B; cGMP binds inside the β-roll and is stabilized by the C-helix. The C-linker domain contains Helices A’-F’ and forms a gating ring architecture in a channel tetramer through the so-called elbow-on-shoulder inter-subunit interactions between helix A’-turn-helix B’(elbow) of one subunit and helix C’-turn-helix D’(should) of a neighboring subunit (Figure 1G) (Zagotta et al., 2003). The C-linker gating ring plays an important role in coupling ligand binding to pore opening.
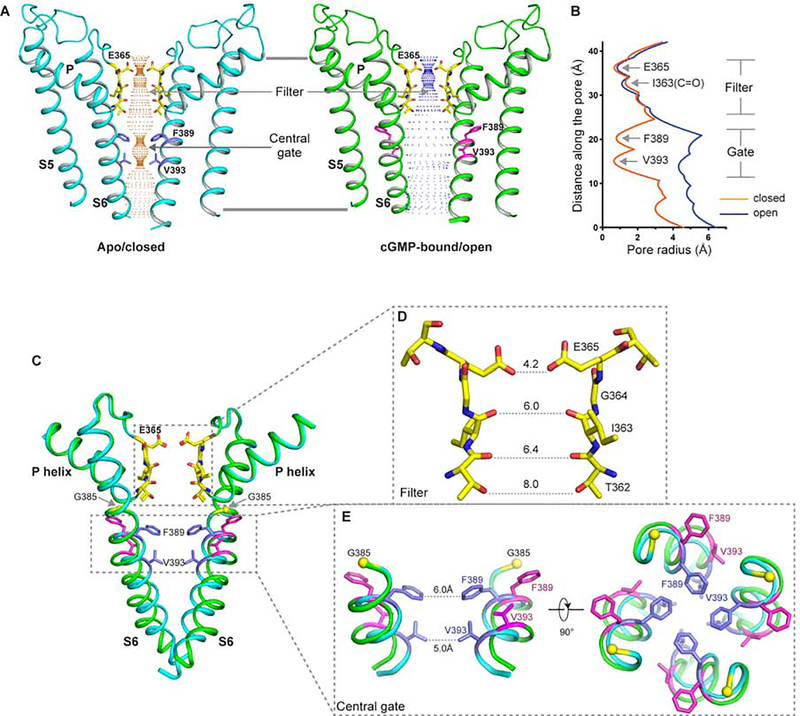
2. The ion conduction pore of CNGA1
The CNGA1 ion conduction pore, consisting of S5, S6 and pore (P) helix, adopts a closed conformation in the apo structure and an open conformation in the cGMP-bound structure (Figure 2A and 2B). In the apo structure, the four pore-lining S6 helices form a bundle-crossing in the middle of the membrane right below the selectivity filter with two layers of hydrophobic residues, F389s and V393s, forming the constriction points that prevent the passage of hydrated cations (Figure 2A and 2B). In the cGMP-bound state, the S6 helices undergo outward movement along with rotational motion hinged at Gly385 (Figure 2C and 2E, movie S1). Consequently, the two layers of constriction-forming residues dilate and rotate away from the central axis, resulting in a much wider opening at the central gate. This pore opening mechanic is triggered by the conformational change at the cytosolic gating apparatus upon cGMP binding and will be further described later.
Figure 2. Ion conduction pore of human CNGA1.
See also Figure S7
(A) The ion conduction pore of the apo closed (left) and ligand-bound open (right) CNGA1 with front and rear subunits removed for clarity. Central ion pathway is marked with dotted mesh. Key gating and filter residues are shown as sticks.
(B) Pore radius along the central axis in the open (in blue) and closed (in orange) states.
(C) Structural comparison of the pore region between the open (green) and closed (cyan) states. Only the pore helices, filters and S6 helices from two diagonal subunits are shown in the superposition. Yellow spheres mark the Cα atoms of G385s.
(D) Selectivity filter of CNGA1 with atom-to-atom distances in Å.
(E) Structural comparison of the central gate between the apo closed (cyan) and cGMP-bound open state (green) in side (left) and down (right) views.
In most canonical tetrameric cation channels such as K+ channels, the channel gate is formed by the inner-helix bundle-crossing near the cytosolic side of the membrane and the channel pore encloses a solvent-filled central cavity in the middle of the membrane (Zhou et al., 2001) (Figure S7A). In CNGA1, however, the shift of the closed gate to the middle of the membrane effectively occludes the central cavity space present in other channels and the ion conduction pathway becomes pinched shut right below the filter. The central gate formation appears to be a unique feature of the CNG family channels and was also observed in the TAX-4 structures (Zheng et al., 2020).
The selectivity filter of CNGA1remains unchanged between the open and the closed structures and therefore is unlikely to gate the channel. In a channel tetramer, the filter residues of 362TIGE365 line the ion pathway with the oxygen atoms from the sidechain hydroxyl groups of T362s, the backbone carbonyls of T362s and I363s, and the sidechain carboxylates of E365s (Figure 2D). At the external entrance to the selectivity filter, the side chains of highly conserved E365s, shown to be important for Ca2+ binding in CNG channels, form the narrowest point of the filter pathway with a cross distance of about 4.6 Å. While it is intuitive to suggest that E365s are well positioned to directly bind Ca2+ at the external entrance of CNGA1, ion binding analysis, however, shows no or little external Ca2+ binding as will be discussed in the next section. Beneath the E365 constriction point, the ion pathway of the filter has a wider dimension, indicating that the permeating cations likely pass through the filter in a partially hydrated form. Although the CNG channel filter shares some sequence similarity to the K+ channels, the overall filter dimension of human CNGA1 is much wider than that of a K+, but is narrower than that of TAX-4 (Figure S7B).
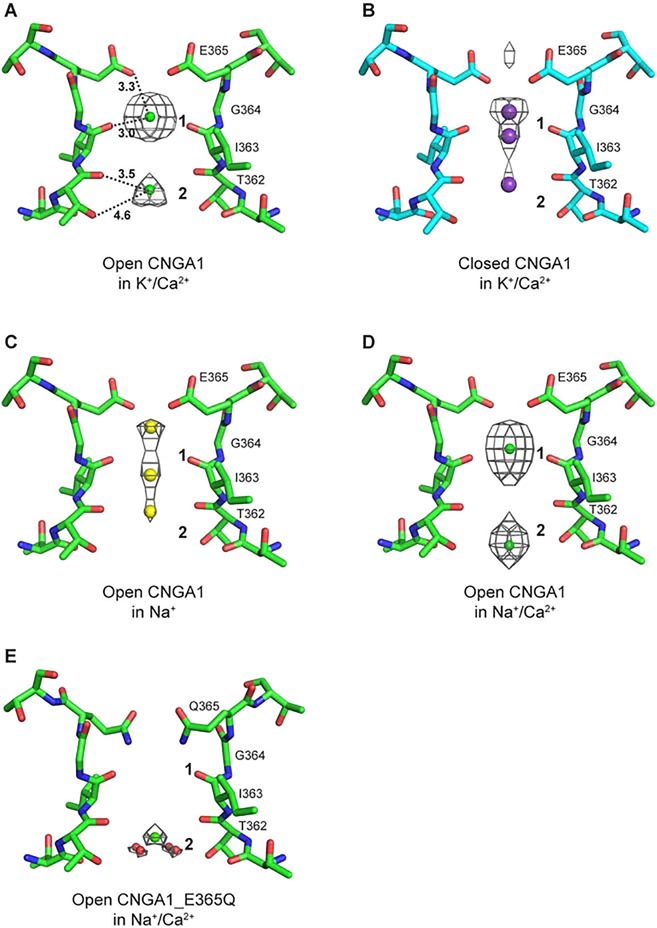
3. Ion binding profile in CNGA1 selectivity filter
The high quality density maps at the pore region of CNGA1 revealed clear electron density in the filter likely from the bound ions, allowing us to analyze the ion binding profile in the channel. It is worth noting that the protein samples used to obtain the open and closed structures were both purified in 150 mM KCl and supplemented with 1 mM Ca2+ before plunge freezing vitrification. The only difference between the two is the presence or absence of cGMP (Methods). Intriguingly, despite almost identical ionic conditions, the ion binding profile in the filter is quite different between the two structures. In the open structure, we observed two discrete density peaks in the filter, a particularly strong one (assigned as site 1) surrounded by the carbonyl oxygen atoms from I363s and a relatively weaker peak (assigned as site 2) surrounded by the carbonyls and hydroxyls of T362s (Figure 3A). In the closed structure, on the other hand, the electron density in the filter is more diffusive with three weak peaks: two adjacent ones just above and below site 1 and the third one at the upper end of site 2 (Figure 3B). Although the conserved filter acidic residue was predicted to form the main Ca2+ site at the external entrance of CNG channels based on mutagenesis, we observed no density at E365 site in the open structure and only weak density in the closed structure despite the presence of 1 mM Ca2+, indicating no or weak Ca2+ binding, if any, at the external entrance of CNGA1.
Figure 3. Ion binding profile in the selectivity filter of CNGA1.
Shown are electron density maps of filter ions in the structures of the cGMP-bound open and apo closed CNGA1 in various salt conditions. All maps are scaled and contoured at 7 σ.
(A) Open CNGA1 in 150 mM KCl and 1 mM CaCl2.
(B) Closed CNGA1 in 150 mM KCl and 1 mM CaCl2.
(C) Open CNGA1 in 150 mM NaCl.
(D) Open CNGA1 in 150 mM NaCl and 1 mM CaCl2.
(E) Open CNGA1 E365Q mutant in 150 mM NaCl and 1 mM CaCl2.
The presence of both K+ and Ca2+ and their equivalent number of electrons make it difficult to delineate the contribution of the two ions to those filter electron density peaks in the open and closed pores. To unambiguously define how ions, particularly Ca2+, bind in the CNG channel filter, we set to determine the CNGA1 structures with different salt conditions. We reasoned that under physiological conditions the main permeating ions for CNG channels are Na+ and Ca2+, and the role of Ca2+ as a permeating blocker will only be physiologically relevant when the channel is in an open, conductive state. We therefore focused the ion binding analysis on the open channel and determined the structures of cGMP-bound CNGA1 samples that were purified in 150 mM NaCl and supplemented with or without 1 mM Ca2+ before vitrification (Methods and Table S1). The structure obtained in the Na+ only condition shows diffusive and weak electron density distribution in the filter attributable to Na+ binding (Figure 3C). In the structure obtained with the presence of 1 mM Ca2+, we observed two discrete electron density peaks at sites 1 and 2 that have much higher intensity than those in the Na+ only condition, indicating the replacement of Na+ by Ca2+ in the filter (Figure 3D). The electron density at site 1 is stronger than that at site 2, indicating a higher affinity Ca2+ binding at site 1. The ion density in the filter of open CNGA1 under Na+/Ca2+ condition is very similar to that under K+/Ca2+ condition (Figure 3A vs 3D), suggesting that the two filter density peaks observed in the initial open channel structure are also contributed by the bound Ca2+. Interestingly, the density distribution within the filter of the closed CNGA1 structure obtained under K+/Ca2+ condition is strikingly similar to that of the open channel in Na+ only condition (Figure 3B vs 3C), suggesting that the filter density in the closed CNGA1 is mainly from K+, and Ca2+ failed to replace those filter K+ ions when the pore is closed. This is an unexpected observation which would imply an open state-dependent Ca2+ binding in CNGA1. We suspect that a late-stage Ca2+ introduction to the protein sample together with the special location of the closed channel gate contributes to the lack of Ca2+ binding in the closed CNGA1 in our study as will be further speculated in the Discussion.
Several conclusions can be drawn based on the ion binding profile in the filter of CNGA1. Firstly, the highly conserved E365 does not form the previously suggested external Ca2+ site. Secondly, primary Ca2+ binding occurs at site 1 and is coordinated by the carbonyl oxygen atoms of Ile363s with an ion-ligand distance of about 3.0 Å. Thirdly, the carbonyl and hydroxyl groups of T362s enclosed a secondary Ca2+ site (site 2) with a minimum distance of about 3.5 Å between Ca2+ and a nearest oxygen atom, suggesting that the site 2 Ca2+ has to be hydrated. Fourthly, the density of monovalent ions (both K+ and Na+) in the filter is weak and diffusive, an indication of lower affinity non-specific binding; on the contrary, the density peaks from Ca2+ are strong and discrete, indicating higher affinity specific Ca2+ binding, particularly at the primary site.
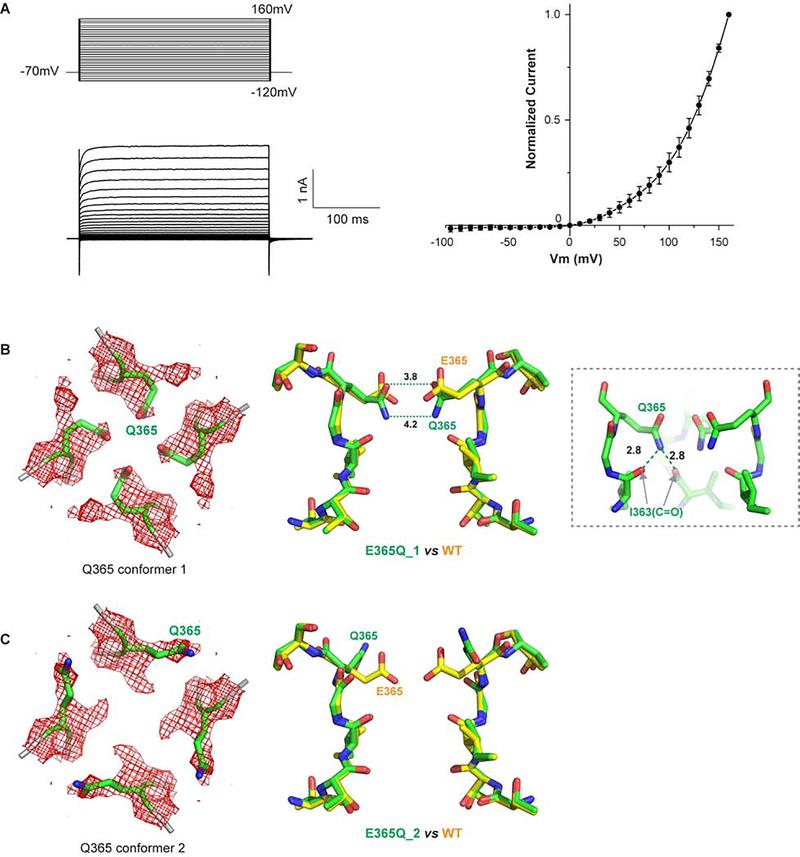
4. Loss of Ca2+ binding and gain of voltage-dependent gating in E365Q mutant
The conserved filter acidic residue has been shown to be essential for Ca2+ binding and blockage in CNG channels from previous mutagenesis and electrophysiology studies (Eismann et al., 1994; Gavazzo et al., 2000; Park and MacKinnon, 1995; Root and MacKinnon, 1993; Seifert et al., 1999). However, in our CNGA1 structures, the filter acidic residue E365 does not form the expected external Ca2+ binding site. To test if E365 is important for the observed Ca2+ binding inside the CNGA1 filter, we also determined the structure of the cGMP-bound E365Q mutant in the presence of 150 mM Na+ and 1 mM Ca2+. The mutation completely abolishes site 1 Ca2+ binding and we only observe weak Ca2+ density at site 2 (Figure 3E), indicating that the negatively charged E363 is indeed important for Ca2+ binding at the primary site. The high resolution map of the mutant structure (~2.73 Å) also reveals some satellite density associated with the site 2 Ca2+ likely from the water in the hydration shell. In mutagenesis studies, CNG channels have been shown to retain Ca2+ binding and blockage when the filter glutamate was replaced by aspartate but not any other amino acids, implying that a negatively charged side chain is necessary to stabilize the Ca2+ likely through electrostatic interaction. Considering that the distance of about 3.3 Å between E365 carboxylate oxygen and site 1 Ca2+is too far for optimal Ca2+ chelation (~ 2.4 Å) (Figure 3A), it is reasonable to suggest that the electrostatic interaction between E365 and Ca2+ is mediated by water.
In a previous study of bovine CNGA1, the neutralization mutation of the filter glutamate (E363Q) has also been shown to render the channel voltage dependent with outwardly rectifying macroscopic current in the presence of saturating cGMP (Martinez-Francois et al., 2009). It was suggested that this voltage gating is caused by voltage dependent conformational change in the selectivity filter of the mutant. Our recordings of the equivalent E365Q mutant of human CNGA1 confirm the voltage-dependent activation of the mutant in the presence of 1 mM cGMP (Figure 4A). Interestingly, the structure of the E365Q mutant reveals two alternative conformations at residue Q365 (Figure 4B and 4C), providing a plausible explanation for the voltage dependent gating in the filter of the mutant. In one conformation, the Q365 side chain moves downward and also closer to the central axis with its amine group positioned within hydrogen-bonding distance (~ 2.8 Å) to the carbonyl oxygen of Ile363 from its own subunit as well as the neighboring one. Consequently, the amide and the carbonyl groups of Q365 side chain form two narrow constriction points with similar cross distances of about 4.2 Å and 3.8 Å, respectively, narrower than the single constrict formed by E365 in the wild type channel (Figure 4B). With the formation of a narrower external entrance and the loss of negative charge to stabilize the permeating cations, the four Q365 side chains likely plug the ion conduction pathway in this conformation. In the other conformation, the Q365 side chain flip away from the center, creating a much wider opening at the external entrance (Figure 4C). It is worth noting that the EM density map of Q365 represents an averaged picture of the two conformations and it is possible that both conformations co-exist in a channel tetramer. Having said that, the block of mutant channel may not require all four Q365 side chains to be in the plugged conformation and two diagonal Q365s are probably sufficient to plug the channel. Other than the two alternative conformations at Q365, the mutant channel has virtually identical structure to the cGMP-bound wild type CNGA1. We suspect that the transition between the two conformations contributes to the voltage dependent gating in the mutant and a depolarizing membrane potential may favor the latter conductive conformation in Q365. This voltage dependent conformational change could be driven simply by the outward movement of permeating cations at depolarizing membrane potential, which would push the Q356 side chain to flip outwardly from its plugged conformation to the conductive conformation and thereby unblock the channel.
Figure 4. Voltage dependent gating of CNGA1 E365Q mutant.
(A) Sample traces and I-V curves of voltage activation of CNGA1 E365Q mutant. Whole cell currents were recorded with 1 mM cGMP in the pipette (cytosolic). I-V curves were obtained from the peak currents at various activation potentials. Data points are mean ± SEM (n=5 independent experiments).
(B) Down view (left) of Q365 in conformation 1 with EM density map contoured at 5.0 σ and side view (right) of the mutant filter (green) in comparison with the wide-type filter (yellow). Inset: H-bonds between Q365 side chain and carbonyls of two neighboring I363s.
(C) Down view (left) of Q365 in conformation 2 and side view (right) of the mutant filter in comparison with the wide-type filter (right). The mutant filter is conductive in this conformation.
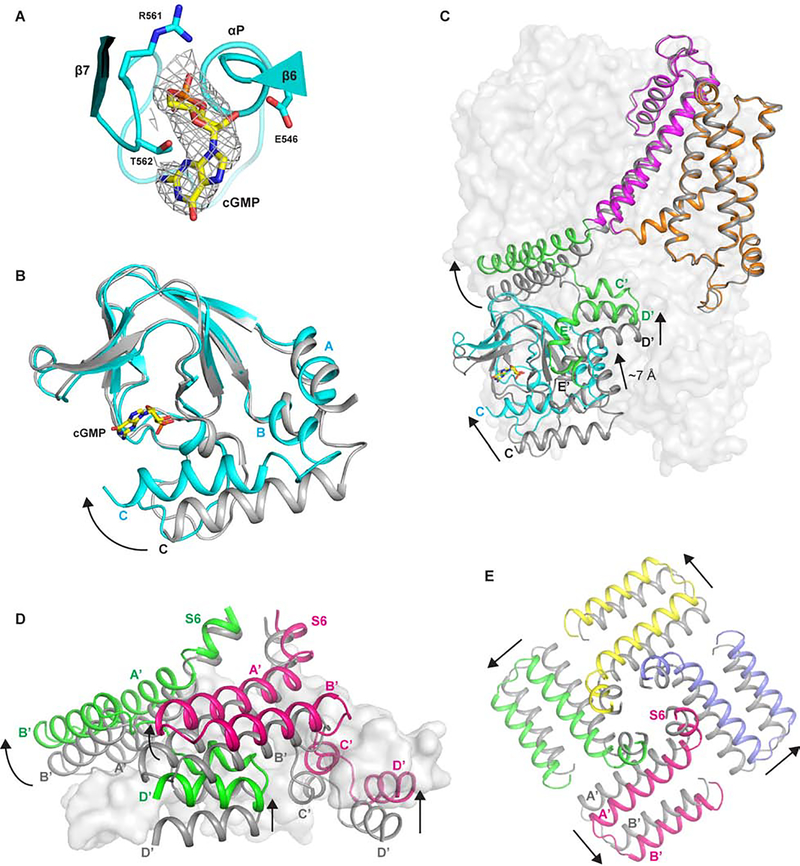
5. cGMP binding and conformational change at the cytosolic gating apparatus
The bound cGMP can be unambiguously identified from the EM density maps of the CNGA1 structures obtained in the presence of the ligand (Figure 5A). Same as other CNBD channels, cGMP is situated in the pocket formed by the β-roll and capped by the C-helix in CNGA1 (Figure 5B). To explain how ligand-induced conformational change at CNBD propagates to the distal ion conduction pore and opens the channel, we did two sets of structural comparison between the open and closed states, one locally at CNBD and the other globally with the whole channel. The local structural comparison at CNBD shows that the major movement triggered by GMP binding is the swing of C-helix closer to the ligand binding pocket (Figure 5B). Due to the tight intra- and inter-subunit interactions between CNBD and the C-linker domain, this local movement at CNBD propagates to the other part of the protein through the C-linker helices, resulting in a much larger upward movement of the entire cytosolic ligand-gating apparatus as illustrated in a global structural comparison (Figure 5C). The sequence of conformational changes from cGMP binding to pore opening can be described as following: the swing of C-helix triggered by GMP binding leads to an upward translation of about 7 Å at helices C’-F’ of the C-linker domain (Figure 5C); through the elbow-on-shoulder inter-subunit packing between helix C’-turn-helix D’ and helix A’-turn-helix B’, the movement of helices C’-F’ is translated to a swaying motion at helix A’-turn- helix B’ in both upward and outward directions (Figure 5D and 5E); the movement of helix A’ is directly coupled to S6 through a tight covalent connection, resulting in the rotation and dilation of the pore lining S6 hinged at Gly385, which in turn opens the channel (Figure 2C, movie S2). While the ligand binding triggers quite significant conformational changes at the cytosolic gating apparatus, it barely introduces any movement at the transmembrane part of the channel other than the S6 segment after the glycine gating hinge (Figure 5C).
Figure 5. cGMP activation of CNGA1.
(A) Zoomed-in view of cGMP binding site in CNBD. Density for cGMP is contoured at 5 σ. cGMP and key ligand interacting residues are shown in stick.
(B) Local structural comparison at CNBD between the apo closed (grey) and cGMP-bound open CNGA1 (cyan). Arrow indicates the movement of C-helix.
(C) Global structural comparison between the apo closed (grey) and cGMP-bound open CNGA1 (domains are colored individually). Only the front subunits are shown in cartoon representation. Arrows indicate the ligand-induced movements from closed to open state at various parts of cytosolic region.
(D) Side view of conformational changes at C-linker domain from closed (grey) to open (green and red) state. Only helices A’-D’ from two front subunits are shown in cartoon representation for clarity.
(E) Down view of the conformational change at the gating ring-forming helix A’-turn-helix B’. All subunits are colored grey in the closed state and individually colored in the open state.
Discussion
The structures of human CNGA1 homotetramer in the open and closed states as well as in different salt conditions reveal several distinct features of CNG channels. In most voltage-gated tetrameric cation channels, the transmembrane part of the channel assembly is domain-swapped and the channel pore contains a solvent-filled central cavity with a cytosolic gate near the membrane surface. The CNG channel assembly, on the other hand, is not domain-swapped and its pore has a central gate in the middle of the membrane which effectively eliminates the central cavity in the closed state. Contrary to previous suggestions that CNG channel selectivity filter may function as a gate based on accessibility studies (Contreras and Holmgren, 2006; Contreras et al., 2008), the CNGA1 selectivity filter remains identical in both the open and closed structures, and therefore unlikely to form the gate. Our ion binding analysis reveals two Ca2+ sites inside the CNG filter, but shows no obvious Ca2+ binding at the predicted site at the external entrance formed by the conserved filter acidic residue (E365 in HsCNGA1).
In our ion binding analysis, it is interesting to note that Ca2+ can readily replace the monovalent ions in the filter when the channel is open, but no obvious Ca2+ binding is observed when the channel is closed. We speculate that two combined factors contribute to this observation of open state-dependent Ca2+ binding in our study. One is the experimental procedure used for preparing Ca2+-containing samples, in which Ca2+ was added to the channel proteins right before (~ 0.5–1h) they were vitrified using plunge freezing. The other factor is likely related to the location of the central gate and the resulting loss of cavity space in a closed CNG channel. It is expected that the vacancy of pre-occupied monovalent ions is prerequisite to the binding of Ca2+ inside the filter. This ion exchange process can be efficiently achieved in the filter of an open channel as Ca2+ entrance from one side can expel the bound monovalent ions from the opposite side. In a closed CNG pore, however, there is no central cavity space to accommodate the displaced monovalent ions and the ion replacement can be significantly hindered because the external entrance of the filter provides the only passage for both the exit and entrance of filter ions, and Ca2+ can only enter after pre-occupied monovalent ions are vacated. This hindered ion exchange process, combined with the channel’s relatively short exposure to Ca2+, may significantly reduce the Ca2+ occupancy in the closed CNG channel filter.
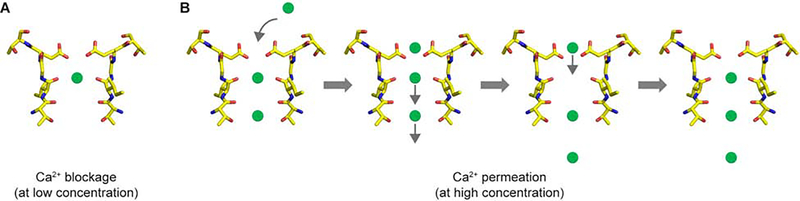
The importance of the conserved filter acidic residue for Ca2+ binding has been extensively studied by mutagenesis in various CNG channels. In human CNGA1, this acidic residue, E365, does not directly form a predicted Ca2+ site at the external entrance, but is indeed important for site 1 Ca2+ binding within the filter. The lack of external Ca2+ binding in CNGA1 is intriguing as the four E365s in a channel tetramer are well positioned to coordinate Ca2+ similarly to those acidic residues in other Ca2+ selective channels such as CaV (Zhao et al., 2019) and TRPV6 (Saotome et al., 2016). While the dominant Ca2+ occupation at site 1 may prevent the external Ca2+ binding due to the close proximity, we suspect that the four filter glutamates still form a lower affinity external Ca2+ site that can transiently attract Ca2+ at higher concentration. In the closed CNGA1 structure obtained in the presence of Ca2+, we indeed observed a weak density peak at the external entrance that could potentially come from low occupancy Ca2+ binding (Figure 3B). Similar density was observed in the Tax-4 structure and was also attributed to Ca2+ binding (Zheng et al., 2020). Having a lower affinity transient Ca2+ site at the external entrance may be necessary to form a multi-site, single file pore for selective Ca2+ permeation in CNG channels, which is conceptually similar to the classical ion conduction model for voltage-gated Ca2+ channel (Almers, 1984; Hess, 1984). At lower concentrations, Ca2+ primarily binds to site 1 and blocks the Na+ permeation in the CNG channel (Figure 6A). At higher external Ca2+ concentration, a 2nd Ca2+ ion may start to occupy site 2 and destabilize the Ca2+ at site 1 by electrostatic repulsion. The transient Ca2+ binding at external entrance promotes Ca2+ permeation by providing the necessary repulsive force to knock off site 1 Ca2+, which in turn moves to site 2 while pushing the site 2 Ca2+ into the cytosol. Site 1 will then be refilled by the external Ca2+ and the filter is ready for the next cycle of Ca2+ permeation (Figure 6B).
Figure 6. Working model for Ca2+ blockage (A) and permeation (B) in CNG channels.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact:
Further information and requests for resources and reagents should be directed to Youxing Jiang (youxing.jiang@utsouthwestern.edu)
Materials availability:
All unique reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and code availability:
The cryo-EM density maps of the human CNGA1 and its mutant have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-23306 for the apo state in K+/Ca2+, EMD-23307 for the cGMP-bound state in K+/Ca2+, EMD-23309 for the cGMP-bound state in Na+, EMD-23308 for the cGMP-bound state in Na+/Ca2+, and EMD-23310 for the cGMP-bound E365Q mutant in Na+/Ca2+. Atomic coordinates have been deposited in the Protein Data Bank under accession numbers 7LFT for the apo structure in K+/Ca2+, 7LFW for the cGMP-bound structure in K+/Ca2+, 7LFY for the cGMP-bound structure in Na+, 7LFX for the cGMP-bound structure in Na+/Ca2+, and 7LG1 for the cGMP-bound E365Q structure in Na+/Ca2+.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Lines
HEK293F cells (Thermo Fisher Scientific) were maintained in FreeStyle 293 Expression Medium (Gibco) at 37°C, 8% CO2, 130 rpm in a Reach-In CO2 Incubator (Thermo Fisher Scientific) throughout. HEK293 cells (ATCC) used for electrophysiology were grown in DMEM media (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS, Thermo Fisher Scientific) and kept in a 37°C, 5 % CO2 incubator.
METHOD DETAILS
Protein expression and purification
Truncated Homo sapiens CNGA1 145–690aa (HsCNGA1, NCBI accession: NP_000078.2) containing an N-terminal Flag tag was cloned into a pEZT-BM vector (Morales-Perez et al., 2016) and expressed in HEK293F cells using the BacMam system (Thermo Fisher Scientific). Bacmids were synthesized using E. coli DH10bac cells (Thermo Fisher Scientific) and baculoviruses were produced in Sf9 cells using Cellfectin II reagent (Thermo Fisher Scientific). For protein expression, cultured HEK293F cells were infected with the baculoviruses at a ratio of 1:40 (virus:HEK293F, v/v) and supplemented with 10mM sodium butyrate to boost protein expression level. Cells were cultured in suspension at 37°C for 48 hr and then harvested by centrifugation at 4,000 g for 15 min. All purification procedures were carried out at 4°C. The cell pellet was resuspended in buffer A (25 mM HEPES pH 7.4, 150 mM KCl or NaCl) supplemented with 50 μM cGMP and a protease inhibitor cocktail (2 μg/ml DNase, 0.5 μg/ml pepstatin, 2 μg/ml leupeptin, and 1 μg/ml aprotinin and 0.1 mM PMSF). After homogenization by sonication, HsCNGA1 was extracted with 1% (w/v) Lauryl Maltose Neopentyl Glycol (LMNG, Anatrace) by gentle agitation for 1.5 hr. After extraction, the supernatant was collected by centrifugation at 40,000 g for 30 min and incubated with anti-Flag M2 affinity resin (Genescript) by gentle agitation for 1 hr. The resin was then collected on a disposable gravity column (Bio-Rad) and washed with 20 column volume of Buffer A supplemented with 0.06% (w/v) Digitonin and 50 uM cGMP. HsCNGA1 was eluted in Buffer A with 0.06% (w/v) Digitonin, 50 uM cGMP and 0.1 mg/ml Flag peptide. The protein eluate was concentrated and further purified by size-exclusion chromatography on a Superose 6 10/300 GL column (GE Healthcare) in Buffer A with 0.06% (w/v) Digitonin. Note that cGMP ligand was removed in the final purification step of size-exclusion chromatography. The peak fractions were collected and concentrated to 6 mg/ml for cryo-EM analysis. To prepare the protein samples in various conditions for cryo-EM analysis, such as with or without cGMP and Ca2+, 1 mM cGMP and/or 1 mM CaCl2 were added to the protein samples (purified in 150 mM KCl or NaCl) about 1 hour before they were applied to the carbon grids and plunge frozen. The HsCNGA1_E365Q mutant construct was generated using QuikChange (Agilent). The same procedure was used to express and purify HsCNGA1_E365Q for cryo-EM analysis.
HEK293 (CRL-1573) cell lines were purchased from and authenticated by the American Type Culture Collection (ATCC, Manassas, VA). HEK293F cells (RRID: CVCL_D603) were purchased from and authenticated by Thermo Fisher Scientific. The cell lines were tested negative for mycoplasma contamination.
Cryo-EM sample preparation and data acquisition
Purified HsCNGA1samples in various conditions at 6mg/ml were applied to a glow-discharged Quantifoil R1.2/1.3 300-mesh gold holey carbon grid (Quantifoil, Micro Tools GmbH, Germany), blotted for 4.0 s under 100% humidity at 4°C and plunged into liquid ethane using a Mark IV Vitrobot (FEI).
For the data of HsCNGA1 samples in NaCl, movies were acquired on a Titan Krios microscope (FEI) operated at 300 kV with a K3 Summit direct electron detector (Gatan), using a slit width of 20 eV on a GIF-Quantum energy filter. Images were recorded with Serial EM in super-resolution counting mode with a super resolution pixel size of 0.4165 Å. The defocus range was set from −0.9 to −2.2 μm. Each movie was dose-fractionated to 40 frames under a dose rate of 21 e−/pixel/s, with a total exposure time of 2 s, resulting in a total dose of about 60.5 e−/Å2.
For the data of HsCNGA1 samples in KCl collected at Janelia EM facility, movies were acquired similarly to the samples in NaCl with the following changes. Data were collected using CDS (Correlated Double Sampling) mode of the K3 camera with a super resolution pixel size of 0.422 Å. Each movie was dose-fractionated to 60 frames with a dose rate of 1e−/Å2/frame for a total dose of 60e−/Å2. The total exposure time was between 5 to 6 seconds.
Image processing
Movie frames were motion corrected and binned two times and dose-weighted using MotionCor2 (Zheng et al., 2017). The CTF parameters of the micrographs were estimated using the GCTF program (Zhang, 2016). The rest of the image processing steps were carried out using RELION 3.1 (Nakane et al., 2020; Scheres, 2012; Zivanov et al., 2018). All resolution was reported according to the gold-standard Fourier shell correlation (FSC) using the 0.143 criterion (Henderson et al., 2012). Local resolution was estimated using Relion.
Aligned micrographs were manually inspected to remove those with ice contamination and bad defocus. Particles were selected using Gautomatch (K. Zhang, MRC LMB, http://www.mrc-lmb.cam.ac.uk/kzhang/) and extracted using a binning factor of 3. 2D classification was performed in Relion 3.1. Selected particles after 2D classification were subjected to one around of 3D classification. The C. elegans TAX-4 map (EMD-6656) (Li et al., 2017) low-pass filtered to 60 Å was used as the initial model. Classes that showed clear features of the CNG channel were combined and subjected to 3D auto-refinement and another round of 3D classification without performing particle alignment using a soft mask around the protein portion of the density. The best resolving class was then re-extracted with the original pixel size and further refined. Beam tilt, anisotropic magnification, and per-particle CTF estimations and Bayesian polishing were performed in Relion 3.1 to improve the resolution of the final reconstruction.
Model building, refinement and validation
All EM maps of HsCNGA1 in the open and closed states show high quality density for de novo model building in Coot (Emsley et al., 2010), facilitated by previous cryo-EM structure of CeTAX-4 (PDB:6wej; PDB:6wek) (Zheng et al., 2020). The model was manually adjusted in Coot and refined against the map by using the real space refinement module with secondary structure and non-crystallographic symmetry restraints in the Phenix package (Adams et al., 2010).
The final structural model of each subunit contains residues 156–605. The N-terminal residues of 145–155 and the C-terminal residues of 606–690 were disordered in the structure. The binding of cGMP ligand appears to make the CNBD region more dynamic, resulting in a less well-resolved EM density map at CNBD in the ligand-bound structure as compared to that in the apo structure. Therefore, CNBD from the apo HsCNGA1 structure as well as CNBD from ligand-bound TAX-4 structure was used to facilitate the model building for the CNBD part of the cGMP-bound HsCNGA1 structure. The regions that did not fit to the density map were adjusted manually in Coot. The statistics of the geometries of the models were generated using MolProbity (Chen et al., 2010). All the figures were prepared in PyMol (Schrödinger, LLC.), UCSF Chimera (Pettersen et al., 2004) and UCSF ChimeraX (Goddard et al., 2018). Pore radii were calculated using the HOLE program (Smart et al., 1996).
Electrophysiology
2 μg of pEZT-BM vector containing HsCNGA1 or its mutant was transfected into HEK293 cells using Lipofectamine 2000 (Life Technology). To facilitate cell selection for patch clamp,0.2 μg of pNGFP-EU vector containing GFP was co-transfected into HEK293 cells (Kawate and Gouaux, 2006). 48 hours after transfection, cells were dissociated by trypsin treatment and kept in complete serum-containing medium and re-plate on 35 mm tissue culture dishes in tissue culture incubator until recording. Channel currents were recorded using both whole-cell and inside-out configurations.
Patch pipettes were pulled from borosilicate glass (Harvard Apparatus) and heat-polished to a resistance of 3–5 MΩ. After the patch pipette attached to the cell membrane, a giga-seal (>10GΩ) was formed by gentle suction. The whole-cell configuration was formed by short zap or suction to rupture the patch. The inside-out configuration was formed by pulling the pipette away from the cell, and the pipette tip was exposed to the air for a short period in some cases. Data were acquired using an AxoPatch 200B amplifier (Molecular Devices) with a low-pass analogue filter set to 1 kHz. The current signal was sampled at a rate of 20 kHz using a Digidata 1322A digitizer (Molecular Devices) and further analyzed with pClamp11 software (Molecular Devices).
The cGMP activation of HsCNGA1 was recorded in excised patches (inside-out patches). the pipette solution contained 140 mM NaCl, 4 mM KCl, 1 mM EGTA, and 10 mM HEPES, pH 7.4, whereas the bath solution contained 140 mM KCl, 5 mM EGTA, and 10 mM HEPES, pH 7.4. Various concentrations of cGMP were added to the bath solution. The extracellular Ca2+ blockage of HsCNGA1 was recorded using patch clamp in whole-cell configuration. The pipette solution contained 140 mM KCl, 5 mM EGTA, 10 mM HEPES, pH 7.4, and 1 mM cGMP. The bath solution contained 140 mM NaCl, 4 mM KCl, 1 mM EGTA, and 10 mM HEPES, pH 7.4. Free calcium concentrations in the bath solution were controlled by mixing 1 mM EGTA with appropriate amounts of CaCl2 calculated using MAXCHELATOR (http://maxchelator.stanford.edu). To generate the current and voltage relationship, the membrane potential was held at 0 mV, followed by voltage pulses ramp from −100 to +100 mV over 800 ms duration. Voltage-dependence of HsCNGA1_E365Q was studied using patch clamp in whole-cell configuration with 1mM cGMP in the pipette solution. The currents were elicited by stepping the voltage from the holding potential (−70 mV) to various command potentials (−120 mV to 160 mV) for 1 s and then stepped back to −70 mV.
Supplementary Material
Movie S1. Conformational change at central gate between apo closed and cGMP-bound open states, Related to Figure 2
Movie S2. Overall conformational changes of CNGA1 upon cGMP binding, Related to Figure 2 and Figure 5
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| Top10 | Thermo Fisher Scientific | Cat# 18258012 |
| DH5α | Thermo Fisher Scientific | Cat# C404003 |
| DH10Bac | Thermo Fisher Scientific | Cat# 10361012 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Sodium Butyrate | Sigma-Aldrich | Cat# 303410 |
| Lauryl Maltose Neopentyl Glycol | Anatrace | Cat# NG310 |
| Digitonin | Acros Organics | Cat# 11024-24-1 |
| Anti-DYKDDDDK G1 Affinity Resin | GenScript | Cat# L00432 |
| Flag peptide | Sigma-Aldrich | Cat# F3290 |
| Antibiotic Antimycotic Solution | Sigma-Aldrich | Cat# A5955 |
| Sf-900™ II SFM medium | Thermo Fisher Scientific | Cat# 10902088 |
| FreeStyle™ 293 Expression Medium | Thermo Fisher Scientific | Cat# 12338018 |
| Critical Commercial Assays | ||
| Superose 6 Increase 10/300 GL | GE healthcare | Cat# 29091596 |
| Deposited Data | ||
| Coordinates of Apo CNGA1 in K+/Ca2+ | This paper | PDB: 7LFT |
| Coordinates of cGMP-bound CNGA1 in K+/Ca2+ | This paper | PDB: 7LFW |
| Coordinates of cGMP-bound CNGA1 in Na+ | This paper | PDB: 7LFY |
| Coordinates of cGMP-bound CNGA1 in Na+/Ca2+ | This paper | PDB: 7LFX |
| Coordinates of cGMP-bound CNGA1_E365Q in Na+/Ca2+ | This paper | PDB: 7LG1 |
| Cryo-EM map of Apo CNGA1 in K+/Ca2+ | This paper | EMDB: 23306 |
| Cryo-EM map of cGMP-bound CNGA1 in K+/Ca2+ | This paper | EMDB: 23307 |
| Cryo-EM map of cGMP-bound CNGA1 in Na+ | This paper | EMDB: 23309 |
| Cryo-EM map of cGMP-bound CNGA1 in Na+/Ca2+ | This paper | EMDB: 23308 |
| Cryo-EM map of cGMP-bound CNGA1_E365Q in Na+/Ca2+ | This paper | EMDB: 23310 |
| Experimental Models: Cell Lines | ||
| Sf9 cells | Thermo Fisher Scientific | Cat# 11496015 |
| HEK293F cells | Thermo Fisher Scientific | Cat# R79007 |
| HEK293 cells | ATCC | CRL-1573 |
| Oligonucleotides | ||
| CNGA1_F_primer: gatataGCTAGCaaagataagaaagaagaggagaag | This paper | N/A |
| CNGA1_R_primer: gatataGCGGCCGCtcatgtagagtcgatgggcccactttc | This paper | N/A |
| Recombinant DNA | ||
| pEZT-BM vector | Morales-Perez et al., 2016 | Plasmid # 74099 (Addgene) |
| pEZT-BM-Flag-CNGA1 and mutations | This paper | N/A |
| pNGFP-EU | Kawate and Gouaux, 2006 | N/A |
| Software and Algorithms | ||
| MotionCor2 | Zheng et al., 2017 | http://msg.ucsf.edu/em/software/motioncor2.html |
| GCTF | Zhang, 2016 | https://www.mrc-lmb.cam.ac.uk/kzhang/Gctf/ |
| RELION | Scheres, 2012 | http://www2.mrc-lmb.cam.ac.uk/relion |
| Chimera | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera/ |
| Coot | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot |
| MolProbity | Chen et al., 2010 | http://molprobity.biochem.duke.edu/ |
| PyMol | ||
| PHENIX | Adams et al., 2010 | https://www.phenix-online.org |
| HOLE | Smart et al., 1996 | http://www.holeprogram.org/ |
| Others | ||
| Quantifoil R1.2/1.3 Au 300 mesh | Electron Microscopy Sciences | Cat# Q3100AR1.3 |
Highlights:
Structures of human CNGA1 channel in ligand-bound open and apo closed states.
Reveal the structural mechanism of cGMP-dependent gating in CNGA1.
Define the Ca2+ binding sites within the selectivity filter of CNGA1.
Acknowledgments:
Single particle cryo-EM data were collected at the University of Texas Southwestern Medical Center Cryo-EM Facility that is funded by the CPRIT Core Facility Support Award RP170644 and the Howard Hughes Medical Institute Janelia Cryo-EM Facility. We thank Xiaowei Zhao and Shixin Yang at the Janelia Cryo-EM Facility for help in microscope operation and data collection. This work was supported in part by the Howard Hughes Medical Institute (to Y.J.) and by grants from the National Institute of Health (GM079179 to Y.J.), and the Welch Foundation (Grant I-1578 to Y.J.).
Footnotes
Declaration of interests: The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam A, Shi N, and Jiang Y (2007). Structural insight into Ca2+ specificity in tetrameric cation channels. Proc Natl Acad Sci U S A 104, 15334–15339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almers W, McCleskey EW (1984). Non-selective conductance in calcium channels of frog muscle: calcium selectivity in a single-file pore. J Physiol 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann A, Frings S, Godde M, Seifert R, and Kaupp UB (1994). Primary structure and functional expression of a Drosophila cyclic nucleotide-gated channel present in eyes and antennae. Embo J 13, 5040–5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonigk W, Altenhofen W, Muller F, Dose A, Illing M, Molday RS, and Kaupp UB (1993). Rod and cone photoreceptor cells express distinct genes for cGMP-gated channels. Neuron 10, 865–877. [DOI] [PubMed] [Google Scholar]
- Bradley J, Li J, Davidson N, Lester HA, and Zinn K (1994). Heteromeric olfactory cyclic nucleotide-gated channels: a subunit that confers increased sensitivity to cAMP. Proc Natl Acad Sci U S A 91, 8890–8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley J, Reisert J, and Frings S (2005). Regulation of cyclic nucleotide-gated channels. Curr Opin Neurobiol 15, 343–349. [DOI] [PubMed] [Google Scholar]
- Chen TY, Peng YW, Dhallan RS, Ahamed B, Reed RR, and Yau KW (1993). A new subunit of the cyclic nucleotide-gated cation channel in retinal rods. Nature 362, 764–767. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton GM, Silverman WR, Heginbotham L, and Morais-Cabral JH (2004). Structural basis of ligand activation in a cyclic nucleotide regulated potassium channel. Cell 119, 615–627. [DOI] [PubMed] [Google Scholar]
- Colamartino G, Menini A, and Torre V (1991). Blockage and permeation of divalent cations through the cyclic GMP-activated channel from tiger salamander retinal rods. J Physiol 440, 189–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras JE, and Holmgren M (2006). Access of quaternary ammonium blockers to the internal pore of cyclic nucleotide-gated channels: implications for the location of the gate. J Gen Physiol 127, 481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras JE, Srikumar D, and Holmgren M (2008). Gating at the selectivity filter in cyclic nucleotide-gated channels. Proc Natl Acad Sci U S A 105, 3310–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven KB, and Zagotta WN (2006). CNG and HCN channels: two peas, one pod. Annu Rev Physiol 68, 375–401. [DOI] [PubMed] [Google Scholar]
- Derebe MG, Zeng W, Li Y, Alam A, and Jiang Y (2011). Structural studies of ion permeation and Ca2+ blockage of a bacterial channel mimicking the cyclic nucleotide-gated channel pore. Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhallan RS, Yau KW, Schrader KA, and Reed RR (1990). Primary structure and functional expression of a cyclic nucleotide-activated channel from olfactory neurons. Nature 347, 184–187. [DOI] [PubMed] [Google Scholar]
- Dzeja C, Hagen V, Kaupp UB, and Frings S (1999). Ca2+ permeation in cyclic nucleotide-gated channels. Embo J 18, 131–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eismann E, Muller F, Heinemann SH, and Kaupp UB (1994). A single negative charge within the pore region of a cGMP-gated channel controls rectification, Ca2+ blockage, and ionic selectivity. Proc Natl Acad Sci U S A 91, 1109–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn JT, Grunwald ME, and Yau KW (1996). Cyclic nucleotide-gated ion channels: an extended family with diverse functions. Annu Rev Physiol 58, 395–426. [DOI] [PubMed] [Google Scholar]
- Finn JT, Solessio EC, and Yau KW (1997). A cGMP-gated cation channel in depolarizing photoreceptors of the lizard parietal eye. Nature 385, 815–819. [DOI] [PubMed] [Google Scholar]
- Frings S, Seifert R, Godde M, and Kaupp UB (1995). Profoundly different calcium permeation and blockage determine the specific function of distinct cyclic nucleotide-gated channels. Neuron 15, 169–179. [DOI] [PubMed] [Google Scholar]
- Gavazzo P, Picco C, Eismann E, Kaupp UB, and Menini A (2000). A point mutation in the pore region alters gating, Ca(2+) blockage, and permeation of olfactory cyclic nucleotide-gated channels. J Gen Physiol 116, 311–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstner A, Zong X, Hofmann F, and Biel M (2000). Molecular cloning and functional characterization of a new modulatory cyclic nucleotide-gated channel subunit from mouse retina. J Neurosci 20, 1324–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, and Ferrin TE (2018). UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci 27, 14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SE, and Zagotta WN (1995a). Localization of regions affecting an allosteric transition in cyclic nucleotide-activated channels. Neuron 14, 857–864. [DOI] [PubMed] [Google Scholar]
- Gordon SE, and Zagotta WN (1995b). Subunit interactions in coordination of Ni2+ in cyclic nucleotide-gated channels. Proc Natl Acad Sci U S A 92, 10222–10226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackos DH, and Korenbrot JI (1999). Divalent cation selectivity is a function of gating in native and recombinant cyclic nucleotide-gated ion channels from retinal photoreceptors. J Gen Physiol 113, 799–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes L, and Yau KW (1985). Cyclic GMP-sensitive conductance in outer segment membrane of catfish cones. Nature 317, 61–64. [DOI] [PubMed] [Google Scholar]
- Haynes LW, Kay AR, and Yau KW (1986). Single cyclic GMP-activated channel activity in excised patches of rod outer segment membrane. Nature 321, 66–70. [DOI] [PubMed] [Google Scholar]
- Heginbotham L, Abramson T, and MacKinnon R (1992). A functional connection between the pores of distantly related ion channels as revealed by mutant K+ channels. Science 258, 1152–1155. [DOI] [PubMed] [Google Scholar]
- Heginbotham L, Lu Z, Abramson T, and MacKinnon R (1994). Mutations in the K+ channel signature sequence. Biophys J 66, 1061–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R, Sali A, Baker ML, Carragher B, Devkota B, Downing KH, Egelman EH, Feng Z, Frank J, Grigorieff N, et al. (2012). Outcome of the first electron microscopy validation task force meeting. Structure 20, 205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess P, Tsien RW (1984). Mechanism of ion permeation through calcium channels. Nature 301, 453–456. [DOI] [PubMed] [Google Scholar]
- Ildefonse M, Crouzy S, and Bennett N (1992). Gating of retinal rod cation channel by different nucleotides: comparative study of unitary currents. J Membr Biol 130, 91–104. [DOI] [PubMed] [Google Scholar]
- James ZM, Borst AJ, Haitin Y, Frenz B, DiMaio F, Zagotta WN, and Veesler D (2017). CryoEM structure of a prokaryotic cyclic nucleotide-gated ion channel. Proc Natl Acad Sci U S A 114, 4430–4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James ZM, and Zagotta WN (2018). Structural insights into the mechanisms of CNBD channel function. J Gen Physiol 150, 225–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaupp UB, Niidome T, Tanabe T, Terada S, Bonigk W, Stuhmer W, Cook NJ, Kangawa K, Matsuo H, Hirose T, et al. (1989). Primary structure and functional expression from complementary DNA of the rod photoreceptor cyclic GMP-gated channel. Nature 342, 762–766. [DOI] [PubMed] [Google Scholar]
- Kaupp UB, and Seifert R (2002). Cyclic nucleotide-gated ion channels. Physiol Rev 82, 769–824. [DOI] [PubMed] [Google Scholar]
- Kawate T, and Gouaux E (2006). Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14, 673–681. [DOI] [PubMed] [Google Scholar]
- Korschen HG, Illing M, Seifert R, Sesti F, Williams A, Gotzes S, Colville C, Muller F, Dose A, Godde M, et al. (1995). A 240 kDa protein represents the complete beta subunit of the cyclic nucleotide-gated channel from rod photoreceptor. Neuron 15, 627–636. [DOI] [PubMed] [Google Scholar]
- Lee CH, and MacKinnon R (2017). Structures of the Human HCN1 Hyperpolarization-Activated Channel. Cell 168, 111–120 e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Zhou X, Wang S, Michailidis I, Gong Y, Su D, Li H, Li X, and Yang J (2017). Structure of a eukaryotic cyclic-nucleotide-gated channel. Nature 542, 60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liman ER, and Buck LB (1994). A second subunit of the olfactory cyclic nucleotide-gated channel confers high sensitivity to cAMP. Neuron 13, 611–621. [DOI] [PubMed] [Google Scholar]
- Liu DT, Tibbs GR, and Siegelbaum SA (1996). Subunit stoichiometry of cyclic nucleotide-gated channels and effects of subunit order on channel function. Neuron 16, 983–990. [DOI] [PubMed] [Google Scholar]
- Martinez-Francois JR, Xu Y, and Lu Z (2009). Mutations reveal voltage gating of CNGA1 channels in saturating cGMP. J Gen Physiol 134, 151–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matulef K, and Zagotta WN (2003). Cyclic nucleotide-gated ion channels. Annu Rev Cell Dev Biol 19, 23–44. [DOI] [PubMed] [Google Scholar]
- Morales-Perez CL, Noviello CM, and Hibbs RE (2016). Manipulation of Subunit Stoichiometry in Heteromeric Membrane Proteins. Structure 24, 797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, and Gold GH (1987). A cyclic nucleotide-gated conductance in olfactory receptor cilia. Nature 325, 442–444. [DOI] [PubMed] [Google Scholar]
- Nakane T, Kotecha A, Sente A, McMullan G, Masiulis S, Brown P, Grigoras IT, Malinauskaite L, Malinauskas T, Miehling J, et al. (2020). Single-particle cryo-EM at atomic resolution. Nature 587, 152–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CS, and MacKinnon R (1995). Divalent cation selectivity in a cyclic nucleotide-gated ion channel. Biochemistry 34, 13328–13333. [DOI] [PubMed] [Google Scholar]
- Peng C, Rich ED, and Varnum MD (2004). Subunit configuration of heteromeric cone cyclic nucleotide-gated channels. Neuron 42, 401–410. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Picones A, and Korenbrot JI (1995). Permeability and interaction of Ca2+ with cGMP-gated ion channels differ in retinal rod and cone photoreceptors. Biophys J 69, 120–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheinberger J, Gao X, Schmidpeter PA, and Nimigean CM (2018). Ligand discrimination and gating in cyclic nucleotide-gated ion channels from apo and partial agonist-bound cryo-EM structures. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards MJ, and Gordon SE (2000). Cooperativity and cooperation in cyclic nucleotide-gated ion channels. Biochemistry 39, 14003–14011. [DOI] [PubMed] [Google Scholar]
- Root MJ, and MacKinnon R (1993). Identification of an external divalent cation-binding site in the pore of a cGMP-activated channel. Neuron 11, 459–466. [DOI] [PubMed] [Google Scholar]
- Saotome K, Singh AK, Yelshanskaya MV, and Sobolevsky AI (2016). Crystal structure of the epithelial calcium channel TRPV6. Nature 534, 506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH (2012). RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert R, Eismann E, Ludwig J, Baumann A, and Kaupp UB (1999). Molecular determinants of a Ca2+-binding site in the pore of cyclic nucleotide-gated channels: S5/S6 segments control affinity of intrapore glutamates. Embo J 18, 119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi N, Ye S, Alam A, Chen L, and Jiang Y (2006). Atomic structure of a Na+- and K+-conducting channel. Nature 440, 570–574. [DOI] [PubMed] [Google Scholar]
- Smart OS, Neduvelil JG, Wang X, Wallace BA, and Sansom MS (1996). HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph 14, 354–360, 376. [DOI] [PubMed] [Google Scholar]
- Stern JH, Knutsson H, and MacLeish PR (1987). Divalent cations directly affect the conductance of excised patches of rod photoreceptor membrane. Science 236, 1674–1678. [DOI] [PubMed] [Google Scholar]
- Varnum MD, Black KD, and Zagotta WN (1995). Molecular mechanism for ligand discrimination of cyclic nucleotide-gated channels. Neuron 15, 619–625. [DOI] [PubMed] [Google Scholar]
- Varnum MD, and Dai G (2015). Cyclic Nucleotide–Gated Channels. In Handbook of Ion Channels, Zheng JT, Matthew C, ed. (CRC Press; ), pp. 381–402. [Google Scholar]
- Varnum MD, and Zagotta WN (1996). Subunit interactions in the activation of cyclic nucleotide-gated ion channels. Biophys J 70, 2667–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitz D, Ficek N, Kremmer E, Bauer PJ, and Kaupp UB (2002). Subunit stoichiometry of the CNG channel of rod photoreceptors. Neuron 36, 881–889. [DOI] [PubMed] [Google Scholar]
- Yau KW, and Baylor DA (1989). Cyclic GMP-activated conductance of retinal photoreceptor cells. Annu Rev Neurosci 12, 289–327. [DOI] [PubMed] [Google Scholar]
- Zagotta WN, Olivier NB, Black KD, Young EC, Olson R, and Gouaux E (2003). Structural basis for modulation and agonist specificity of HCN pacemaker channels. Nature 425, 200–205. [DOI] [PubMed] [Google Scholar]
- Zagotta WN, and Siegelbaum SA (1996). Structure and function of cyclic nucleotide-gated channels. Annu Rev Neurosci 19, 235–263. [DOI] [PubMed] [Google Scholar]
- Zhang K (2016). Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Huang G, Wu Q, Wu K, Li R, Lei J, Pan X, and Yan N (2019). Cryo-EM structures of apo and antagonist-bound human Cav3.1. Nature 576, 492–497. [DOI] [PubMed] [Google Scholar]
- Zheng J, and Zagotta WN (2004). Stoichiometry and assembly of olfactory cyclic nucleotide-gated channels. Neuron 42, 411–421. [DOI] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Fu Z, Su D, Zhang Y, Li M, Pan Y, Li H, Li S, Grassucci RA, Ren Z, et al. (2020). Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel. Nat Struct Mol Biol 27, 625–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, Molday LL, Molday RS, and Yau KW (2002). The heteromeric cyclic nucleotide-gated channel adopts a 3A:1B stoichiometry. Nature 420, 193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Morais-Cabral JH, Kaufman A, and MacKinnon R (2001). Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature 414, 43–48. [DOI] [PubMed] [Google Scholar]
- Zimmerman AL, and Baylor DA (1992). Cation interactions within the cyclic GMP-activated channel of retinal rods from the tiger salamander. J Physiol 449, 759–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, and Scheres SH (2018). New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufall F, Firestein S, and Shepherd GM (1994). Cyclic nucleotide-gated ion channels and sensory transduction in olfactory receptor neurons. Annu Rev Biophys Biomol Struct 23, 577–607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Conformational change at central gate between apo closed and cGMP-bound open states, Related to Figure 2
Movie S2. Overall conformational changes of CNGA1 upon cGMP binding, Related to Figure 2 and Figure 5
Data Availability Statement
The cryo-EM density maps of the human CNGA1 and its mutant have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-23306 for the apo state in K+/Ca2+, EMD-23307 for the cGMP-bound state in K+/Ca2+, EMD-23309 for the cGMP-bound state in Na+, EMD-23308 for the cGMP-bound state in Na+/Ca2+, and EMD-23310 for the cGMP-bound E365Q mutant in Na+/Ca2+. Atomic coordinates have been deposited in the Protein Data Bank under accession numbers 7LFT for the apo structure in K+/Ca2+, 7LFW for the cGMP-bound structure in K+/Ca2+, 7LFY for the cGMP-bound structure in Na+, 7LFX for the cGMP-bound structure in Na+/Ca2+, and 7LG1 for the cGMP-bound E365Q structure in Na+/Ca2+.