Abstract
An improved understanding of stem cell niches, organogenesis, and disease models has paved the way for developing a three-dimensional (3D) organoid culture system. Organoid cultures can be derived from primary tissues (single cells or tissue subunits), induced pluripotent stem cells (iPSCs), or embryonic stem cells (ESCs). As a significant technological breakthrough, 3D organoid models offer a promising approach for understanding the complexities of human diseases ranging from the mechanistic investigation of disease pathogenesis to therapy. Here, we discuss the recent applications, advantages, and limitations of organoids as in vitro models for studying metabolomics, drug development, infectious diseases, and the gut microbiome. We further discuss the use of organoids in cancer modeling using high throughput sequencing approaches.
1. INTRODUCTION
A significant development in scientific methods over time has led to the generation of three-dimensional (3D) organoid culture and 3D printed scaffolds that can recapitulate human biology and diseases more precisely. In 1946, Smith and Cochrae first used the term ‘organoid,’ which means ‘resembling an organ,’ to describe a case of cystic teratoma [1]. However, now the term ‘organoid’ has a more restricted definition- i.e., organoids are self-assembled in vitro 3D structures, primarily generated from primary tissues or stem cells such as adult stem cells, induced pluripotent stem cells (iPSCs), and embryonic stem cells (ESCs). In all situations, the generation of organoids depends on the self-assembly and differentiation of cells and the signaling indications from the extracellular matrix (ECM) and the conditioned media [1]. Once the 3D structures assemble, they can mimic the complex aspects of their organ counterparts, can be expanded long term, cryopreserved, and genetically modified.
Sato et al. developed the first 3D epithelial organoid in 2009 from a single leucine-rich repeat-containing G-protein coupled receptor (LGR5+) intestinal stem cell. The study showed that stem cells could be used to develop near-physiological, stable epithelial 3D structures when supplied with proper culture conditions (that mimic the in vivo stem cell niche) and embedded into a matrigel [2]. Later, organoids have been developed from various organs, such as the lung, liver, kidney, brain, pancreas, prostate, breast, taste buds, bladder, ovary, and stomach. Recently, Mameishvili et. al. has developed an organoid specifically from Aldh1b1 expressing mouse pancreatic cells and showed the Kras mutation, which is essential for pancreatic cancer progression [3]. They have also demonstrated the role of Aldh1b1 for tumor development in the mouse pancreatic cancer model [3]. Like organoids, spheroids are also multi-cellular 3D structures that are mostly used for studies related to cancer stem cells [4]. However, unlike the organoid culture system, spheroids are simple clusters of freely floating cell aggregates that are cultured under non-adherent, ultra-low attachment plates. Spheroids can be generated from cancer cell lines or tumor fragments, whereas organoids are specifically developed from tissue stem cells embedded within an ECM matrix. Another significant difference between the organoids and spheroids culture is the driving force’s nature for their development. Organoids are formed mostly due to internal developmental processes, whereas spheroids are primarily developed via cell-to-cell adhesion [5]. Furthermore, spheroids contain a high proportion of poorly differentiated cells (a feature that indicates cancer stem cell characteristics), making them an excellent model system for cancer stem cell studies [5]. Since organoids represent a higher order of complexity and recapitulate their parent organ more nearly than spheroids, researchers are more inclined towards organoid technology for disease modeling and optimizing drug discovery and regenerative medicine [6]. Organoids have become a regular practice in the everyday lab, owing to their wide use in many biological research applications and the relatively fast pace of technological developments in this field. However, certain critical information is necessary before starting any organoid culture. Essential considerations are to choose the appropriate organ or cells (tissue chunks, iPSCs, or ESCs) and the suitable culture protocol, extracellular matrix, growth factors, and morphogens. Acquisition of in-depth knowledge pertaining to these considerations, together with an advancement in the protocols for culturing organoids, is bound to greater applicability of these 3D models in diverse fields. Here, we discuss the recent development in organoid culture techniques and the emerging applications of organoids in translational research. However, in this review, we did not particularly focus on a specific organoid; instead, we highlighted the currently known advancements in organoid culture techniques and evaluated its application in different biological fields.
2. ORGANOID CULTURE SYSTEM AND ITS INGREDIENTS
The major foundation for developing an organoid culture depends on the ability of the homogenous cells to undergo self-organization and reconstitute the structure and function of the tissue of origin. The process of self-organization can be divided into a series of self-patterning or cell differentiation processes that are highly regulated through various morphogenic rearrangements. The traditional way of organoid development involves the digestion of tissue stem cells with collagenase, followed by culturing it in a 3D scaffold with growth factor conditioned media (Figure 1). Various morphogens, like Wnt-3a, R-spondin, epidermal growth factor (EGF), fibroblast growth factor (FGF), noggin, and gastrin are used for the culture of organoids (Table 1 and 2). However, a single specific media would not be helpful for culturing all different types of organoids from different tissue of origin. Therefore, several studies have used development biology research for deciding the media components according to the specific organ.
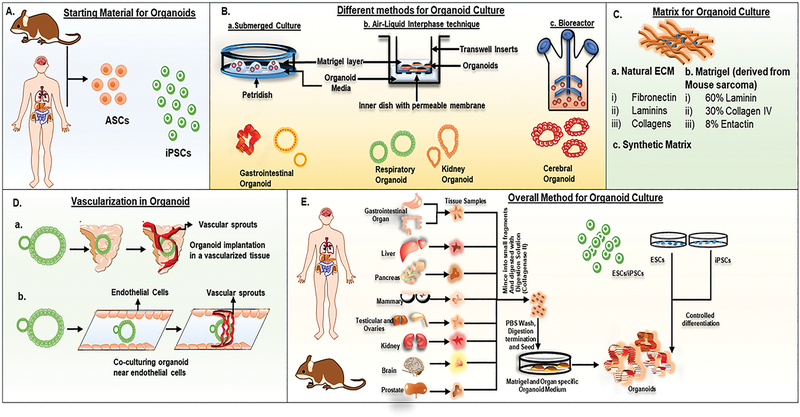
Figure 1: Overview of culture techniques and bioengineering advancements for an organoid generation:
A. Organoids can be generated from adult stem cells (ASCs) isolated from human/animal tissue samples or induced pluripotent stem cells (iPSCs). B. The culture method is important for the proper development of organoid. The essential methods are (a) submerged culture, (b) air-liquid interface, and (c) bioreactor technique. The submerged culture method is widely used for the generation of organoids of gastrointestinal origin. In the ALI method, organoids are grown on an inner dish, and the media is placed under the inner dish. This technique is widely used for the development of respiratory and kidney organoids. Further, this technique is also explored for the interaction study of organoids with the tumor-microenvironment. The disadvantage of submerged culture and the ALI method involves the limitation of nutrient availability once the organoid reaches a specific size. This limitation is overcome by bioreactors (3D suspension culture) that enhance the organoid’s availability of nutrients. This technique is mostly explored for cerebral organoids. C. Matrix is another important material that propelled the self-organization of cells towards a specific structure and led to organoid growth. Promising matrices that are well explored for organoid generation are: (a) natural extracellular matrix (ECM), (b) Matrigel; derived from mouse sarcoma, and (c) synthetic matrices. Bioengineering advancements have designed various synthetic matrices, which provide spatiotemporally controllable 3D matrices that can be exploited to give the desired architecture to the organoids. D. Vascularization of organoids can provide a near-physiological way to provide nutrients to the organoid cells. To common techniques explored for the vascular network in organoids are (a) implantation of organoids in a vascularized tissue and (b) co-culturing the organoids in between two layers of endothelial cells, which further lead to the generation of vascular sprouts in the organoids. E. Explains the overall approach for the generation of organoids from tissue sections or iPSCs/ESCs. In iPSCs/ESCs, the cells are first to undergo controlled differentiation to specific organ cell types and then followed by mixing with matrigel to form organ-specific organoids.
Table 1:
Summary of specific media compositions for different organoids
| Cerebral Organoids | Intestinal Organoids | Liver Organoids | Pancreas Organoids | Prostate Organoids | Lung Organoids | Mammary Organoids |
|---|---|---|---|---|---|---|
| Neurobasal-Medium NEAA Ratinoic Acid R Spondin Noggin |
DMEM/F12 FBS EGF R Spondin Noggin Wnt |
DMEM/F12 FBS EGF R Spondin FGF HGF Nicotinamide Insulin |
DMEM/F12 FGF EGF B27 R-Spondin FGF Nicotinamide Noggin Wnt A-8301 Gastrin |
DMEM/F12 FBS EGF R Spondin Noggin Dihydrotestosterone |
DMEM/F12 FBS B27 N2 GlutaMAX FGF Noggin SB431542 CHIR99021 |
DMEM/F12 FBS ITS-Seliniem X EGF Hydrocortisone T3 β Estrdiol Isoproterenol Hydrocholoride Ethanolamine O-phosphorylethanolamine |
| Ovary & Fallopian tube Organoids | Kidney Organoids | Bladder Organoids | Esophageal Organoids | |||
| DMEM/F12 FBS R-Spondin FGF EGF Nicotinamide Noggin Wnt |
DMEM High Glucose FBS NEAA GlutaMAX Hesparin APEL media FGF9 SB431542 CHIR99021 |
Hepatocyte Media EGF FBS GlutaMAX Primocin |
DMEM/F12 EGF Noggin R-Spondin Wnt SB202190 A83–01 Nicotinamide Gastrin |
|||
Table 2:
Summary of functions of growth factors and small molecule inhibitors applied in organoid cultures.
| GROWTH FACTORS | FUNCTIONS |
|---|---|
| EGF | • EGF stimulates the proliferation by binding to EGF receptors. • EGF induces hyperplastic changes and stimulate proliferation of cancer cells. |
| FGFs | • FGFs are a group of signaling protein play crucial role in normal development. • FGFs are mainly mitogens and also alternately known as pluripotent and promiscuous growth factors due to their multiple functions on multiple cell types. |
| Wnt | • Wnt plays a major role in embryonic development by its main functions on axis patterning, cell fate specification, cell proliferation and cell migration. |
| Noggin | • Noggin play a role in germ layer-specific derivation of specialized cells and acts as a regulator of bone morphogenetic proteins (BMPs) during development. |
| R-Spondin | • R-spondin proteins are secreted agonist of Wnt/β-catenin signaling pathway. |
| Gastrin | • Gastrin act as a growth factor in organoid culture and stimulates the proliferation of cells. |
| A83–01 | • Functions as a potent inhibitor of TGF-β I receptor superfamily and inhibits the differentiation. |
| Nicotinamide | • Nicotinamide is a type of vitamin B3 and act as part of the coenzyme nicotinamide adenine dinucleotide. |
| SB202190 | • Function as a p38 inhibitor |
| CHIR99021 | • A small molecule inhibitor of GSK3 signaling molecule, and thus promotes proliferation and growth by increasing the β-catenin and c-Myc protein stabilization. |
| Y27632 | • A Rho kinase inhibitor that inhibits the anoikis of dissociated stem cells. |
In addition to establishing appropriate growth-factor conditioned media, extracellular matrix (ECM) is another critical factor for organoid development. In the natural setting, the ECM is mainly composed of fiber-forming proteins, like collagen, elastin, laminin, fibronectin, and glycosaminoglycans, and release various growth factors, cytokines, and chemokines, thus regulates tissue development and homeostasis [7]. Therefore, while developing the 3D organoids model in vitro, it is necessary to mimic the ECM properties present in the tissue. Various matrices are available, from which the most commonly used matrix is ‘Matrigel’ (Figure 1C). The matrigel is a gelatinous protein mixture that resembles the ECM and is secreted by Engelbreth-Holm-Swarm mouse sarcoma cells [7]. It primarily contains a mixture of ECM components, including laminin, collagen type IV, entactin, heparin sulfate proteoglycans, and some growth factors such as TGFβ and FGF [7]. Another type of matrix that has been recently developed is ‘Synthetic Hydrogels,’ consisting of water-swollen networks made up of natural or synthetic polymers (Figure 1C) [8]. Compared to natural ECM gels, synthetic polymers are highly customizable and can be modified according to specific cultures by incorporating different proteins and other molecules. Recently, Gjoresvki et al. has also shown the potential of covalently cross-linked hydrophilic polymers (like PEG) hydrogels as an alternative to animal-derived 3D matrices, and they highlighted the effects of different ECM properties on organoid culture [9]. Despite this progress, synthetic hydrogels remain primitive compared to matrigel as they lack the critical property of cell-driven remodeling. In general, covalently linked hydrogels are inert and do not interact with cells unless they are not modified chemically. Integrin-binding peptides, such as Arg-Gly-Asp (RGD), has been shown to be grafted into the hydrogel networks to impart them with cell-adhesive property [10]. Since the synthetic hydrogels are constituted of the linear elastic network, they fail to recapitulate the physical characteristics of ECM, which lead to immature organoid growth and morphogenesis [10]. Furthermore, these covalently linked synthetic hydrogels’ elastic network are often undergone irreversible breakdown in the covalent bonds, either through easter-based hydrolysis or MMP-based proteolytic cleavage, thus limit the long-term culture of organoids [10].
Finally, the characteristics of the organoid depend on the starting cell type. Studies have shown the establishment of organoids from ASC, iPSCs, or ESCs (Figure 1A). Different cell types follow different developmental pathways; therefore, choosing an appropriate starting cell-population is essential for successfully generating an organoid culture. Organoids generated from ASCs, or adult tissue fragments, have been shown to highly recapitulate the homeostasis and regenerative capacity of the tissue of origin [11]. However, organoids derived from iPSCs hardly recapitulate the adult tissue stage and resembles the fetal tissue stage [12, 13]. This characteristic of iPSCs-derived organoids makes them an excellent system for studying developmental events and organogenesis. Interestingly, organoids of endoderm origin have been shown to be derived from both ASCs and iPSCs. On the other hand, organoids of ectoderm origin have mainly been generated from ASCs. Certain organoids such as cerebral organoids, neuroectodermal organoids, and mesodermal kidney organoids have only been shown to be derived from iPSCs [14]. When ASCs and iPSCs are highly used for the different organoids’ generation, only a few studies have shown the use of fetal progenitors to derive the organoids [15, 16]. Greggio et al. has demonstrated the fetal progenitor-derived-pancreatic organoids from mouse fetal (embryonic age 10.5) dorsal pancreata [17]. Different germ layers in the embryo lead to the development of organoids with varying types of cells. This fascinating cell complexity can be used in vitro to understand the interaction among different cell types during the developmental process. In contrast, iPSCs-derived organoids resemble mostly the early post-implantation embryonic development, and the ASCs-derived organoids are mostly composed of a specific subset of cell types. Since the ASCs derived organoids are believed to mimic the adult tissue of origin, they are extensively used for disease modeling, like cancer and neurodegenerative diseases [18]. In comparison, iPSCs-derived organoids are mostly exploited to understand diseases related to developmental defects, such as microcephaly [18].
Though both ASCs and iPSCs derived organoids have their applicability and advantages, they both suffer from certain disadvantages. One of the critical challenges in iPSCs derived organoids is their maturation and scalability [19]. The organoids from iPSCs can only be grown to micrometer to millimeter scale due to limited culture conditions (such as oxygen and nutrient supply to the organoid core), thus limiting the life span and functionality of the organoids. Further, iPSCs-derived organoids are highly dependent on self-organization and require lengthy and precise protocols to generate appropriate progenitor cells, resulting in cellular heterogeneity and organoid-to-organoid batch variation [19, 20]. On the other side, organoids generated from ASCs were illustrated to be genetically stable when cultured for more extended periods [21, 22]. However, cancer-derived organoids have been shown to grow poorly compared with organoids generated from normal tissues or early stages of tumors [23]. Further, scientists have demonstrated the variability in optimal culture conditions between normal and tumor organoid cultures [23]. Another inherent problem with the technology is the requirement of matrigel for culturing the organoids, limiting the use of organoids in regenerative medicine [24]. In summary, the culture techniques for organoid generation from iPSCs or ASCs are still budding and are highly driven by empirical considerations.
3. BIOENGINEERING APPROACHES OF NEW GENERATION OF ORGANOIDS FOR DISEASE MODELING
The recent development in organoid culture techniques has made the organoid represent more molecular and physical similarities to their tissue origin. However, the organoid-based system constitutes some key challenges. One of the significant limitations is the restrained growth of the organoids and the development of the necrotic core. As mentioned above, iPSC-derived organoids reach only a specific maturation and resemble the fetal tissues. The phenomenon can be because of the lack of in vivo environment factors necessary for organoid development. Different approaches have been initiated to recreate the tissue microenvironment, which can enhance tumor growth and development. In this section of the review, we are briefly discussing some of the bioengineering approaches for the better culture of the organoids (Figure 1B).
3.1. Bioreactors:
One of the major obstacles in organoid continuous growth is the proper diffusion of nutrients and oxygen to all the organoid cells. To overcome this, studies have exploited the bioreactor, whose constant spinning improves the aeration and nutrient uptake of the organoids in suspension culture [25–27] (Figure 1B (c)). Furthermore, the stirred-tank bioreactors have been shown to encourage more complex structures and increase the differentiation yield. A study using the brain organoid culture in the bioreactor system demonstrates that bioreactors can increase the organoid culture duration up to a year [25]. Other studies using the same system have yield iPSC-derived retinal organoids, kidney organoids, and brain region-specific organoids [26–28]).
3.2. Air-Liquid Interphase (ALI):
In an attempt to mimic the tumor microenvironment during the in vitro culture of organoids, scientists have developed the air-liquid interphase culture method (Figure 1B (b)). The technique involves the co-culturing of tumor epithelia with tumor fibroblast or with tumor-infiltrating lymphocytes [29]. In this method, the minced tumor tissue is resuspended in Type I collagen gel and layered on top of a pre-solidified collagen gel, i.e., prepared on an inner transwell insert [29]. The culture media is present in the outer dish and diffuses to the collagen, i.e., present in the inner container via the permeable membrane. ALI culture system is mostly recognized and used for lung organoids [30].
Along with the respiratory organoid system, the method has been exploited for other organs, including the pancreas, renal cell carcinoma, melanoma, gastrointestinal tissue, and female reproductive tract [31, 32]. The significant advantage of the ALI method is its ability to incorporate both the epithelial and mesenchymal/stromal components, thus accurately recapitulating the stem cell niche in the organoid culture system [32]. Researchers further utilized this method to examine the in vitro interaction of tumor and immune cells in the tumor microenvironment [29]. However, it was demonstrated that immune and fibroblast stromal cells, when co-cultured with organoids through the ALI method, undergo apoptosis after a period of 1 or 2-month.
3.3. Vascularization:
Another major challenge in organoid culture is the lack of functional blood vessels, the absence of which leads to the development of immature organoids, the necrotic inner core, and premature differentiation. Different strategies involving sacrificial molding and laser ablation were explored to generate vascularized organoids [33, 34]. Both of these techniques allow the building of channels within the culture scaffold, which can host the endothelial cells to form a microvascular network. Another attempt to generate vascularized organoids involves the co-culturing of organoids together with vascular endothelial cells (Figure 1D (b)). This protocol has been successfully applied to generate vascularization in different organ-specific organoids, such as the pancreas and liver [35]. Furthermore, the organ-specific vascular endothelial cells can be grown separately from the iPSC cells, and later the endothelial cells can be used to vascularize the organoid [35]. It was further shown that iPSC-derived brain organoids could be vascularized when implanted in the mouse brain [36]. When organoids were transplanted in different organs, such as the liver, intestine, and pancreas, they showed increased growth and maturation when exposed to in vivo factors [37–39] (Figure 1D (a)).
3.4. CRISPR/Cas9 genome-editing in Organoids:
Though in a very initial stage, scientists have started exploring the CRISPR/Cas9 technique to either knocking or knockout genes from organoids. Combining CRISPR/Cas9 with organoids can open new doors to understanding the role of specific genes in organ development and human diseases. CRISPR has been shown to be applied in mouse small intestinal organoids to cause frame-shift mutations in the two APC (known as a tumor suppressor and Wnt signaling negative regulator) alleles. The resulting organoids revealed growth in a Wnt-independent fashion. Further research has utilized CRISPR to introduce mutation in the KRAS, APC, p53, and SMAD4 genes in the colon cancer organoids [39, 40]. The technology has also been applied to iPSC-derived organoids. Freedman et al. had indicated that CRISPR could be used to model the disease phenotype. Their study generated a human iPSC-derived polycystic kidney disease (PKD) organoid and investigated the function by introducing truncating mutations in PKD1 or PKD2 genes [41]. Scientists have also started exploring genome-wide CRISPR screens in organoids. Ringel et al. have used the genome-scale CRISPR screening method in human intestinal organoids to investigate the TGF-β tumor suppressor pathway [42].
The CRISPR system can also be applied to introduce reporter sequences into the organoid genome. With the aid of CRISPR, fluorescent-labeled kidney organoids have been generated to uncover the role of nephron progenitor cells during nephrogenesis [43]. In the study, the researcher has developed the kidney organoids from the iPSCs cells that harbor Cre recombinase in the SIX2 locus and a loxP-flanked fluorescent cassette. Moreover, Jung et al. has used the same approach to visualize the different stages of intestinal differentiation in iPSC-derived intestinal organoids [44]. Though valuable, this technique has its limitation. Most importantly, the placing of the reporter gene, whose genomic location can hamper the gene expression. Although the technique has some limitations, it contains some major advantages. One of the significant advantages is to observe the differentiation of organoid cells in real-time. Further transplanting the reporter labeled organoid in the mouse can be tracked with fluorescence imaging [43, 44].
4. APPLICATION OF ORGANOIDS IN BIOLOGICAL SCIENCES
Following the establishment of organoids, the system has been exploited in different areas of biology and human diseases. For example, organoids developed from disease stem cells can be used to understand the genes involved and their molecular impact on disease progression study. Further, organoids can provide an excellent platform for investigating the viruses and pathogens that infect the human body. Here we discuss various areas of biological sciences where organoids have found their extensive application.
4.1. ORGANOIDS IN METABOLOMIC STUDY
Recent pivotal studies support the notion that metabolic dysfunctions have a major effect on different diseases. Further, metabolic reprogramming at present is considered a hallmark of tumorigenesis. Advancements in 3D culture approaches have enabled organoids to study various genomic, transcriptomic, and proteomic alterations both at normal and disease conditions. We discuss the less explored application of organoids and their usefulness for studying cellular metabolism (Figure 2A).
Figure 2. Application of organoids in different biology or biomedical sciences:
A. Organoids can be used to understand the metabolomics regulation or nutrient transport mechanism of cells under disease or normal conditions. B. Intestinal/gut organoids could be used to study the effect of gut-microbiome on cell physiology and growth. Different culture techniques are explored to study the gut-microbiome in organoids: (i) Microinjection involves injecting the gut-microbes or gut-microbiome-derived metabolites in the apical surface of the organoids. (ii) Mixing the organoids and gut-microbes with matrigel and culturing them together in the ECM matrix. (iii) Co-culturing the organoids with gut-microbes by putting the microbes in the organoid medium. C. Organoids represent useful tools for the study of infectious diseases through co-culturing the diseases related viruses with the organoids. D. Organoids provide a good platform to understand the role of cell niche or microenvironment in cell growth and differentiation. Techniques such as (i) embedding the organoids together with tumor microenvironment cells in the matrigel or (ii) adding the microenvironmental factors in the culture media or through (iii) Air-liquid interphase were explored to study the cell-niche along with organoid. E. (i) In drug screening and (ii) personalized medicine, patient-derived organoids can help to identify the specific/best drug for a specific disease or each patient.
Lu and colleagues demonstrated a culture protocol to establish 3D liver organoids for studying the hepatic glucose metabolism under normal and stress conditions [45]. Their work has shown that the hepatocyte organoids recapitulate the in vivo functions better than the hepatic monolayer or sandwich cultures [45]. The group has developed a hepatic bioreactor system, where the hepatic cells and collagen mixture was infused into hollow fibers to generate the liver organoids [45]. The developed liver organoids showed cuboidal-shaped hepatocytes and increased intracellular contact and bile-canaliculi-like structures. The same group also demonstrated that the entrapment of gel in the hollow fibers plays a critical role in the persistence of the cell functions [46]. The study suggests that the hollow fibers might mimic the blood vessels and provide a hepatocyte microenvironment for promoting hepatic cell assembly in collagen [46]. Another benchmark study by Zietek et al. has validated the use of intestinal organoids to assess nutrient transport, nutrient sensing, and hormone secretion [47]. Sodium-dependent glucose transporter SGLT1/SLC5A1, proton-coupled peptide transporter PEPT1/SLC15A1, and bile acid receptor TGR5 in the apical brush border side and facilitated glucose transporter GLUT2 in the basolateral side of the intestinal organoid. Altogether, Zietek et al. highlighted the robust applicability and usefulness of organoids in the field of metabolism.
Organoids have also been implicated in studying stem cell niche metabolism in the small intestine. For instance, researchers have developed mini-guts organoids with defined crypt domains and Wnt-conditioned organoids (resembling embryonic small intestinal organoids) to study mitochondrial oxidative role phosphorylation during differentiation and crypt formation [48]. Rodriguez-Colman et al. have used the standard media (containing epidermal growth factor, noggin, and R-spondin1; ENR) and Wnt3a-conditioned media (WENR) to generate the mini-gut organoids and Wnt-conditioned organoids, respectively [48]. Additionally, using the same organoid model, the study demonstrated that the glycolytic phenotype in Paneth cells and mitochondrial oxidative phosphorylation in Lgr5+ crypt base columnar cells is necessary for supporting both niche and stem cell function [48]. In another study, the use of human pluripotent stem cell-derived cardiac organoids (hCOs) was reported to explore the maturation of the mammalian hearts during postnatal life [49]. The hCOs model revealed that the metabolic switch from carbohydrate to fatty acids acts as a critical driver for cardiac maturation [49]. Organoids have also been exploited to examine the role of amino acids on stem cell functions and growth. Sean et al. investigated the critical function of L-alanyl-L-glutamine (a stable L-glutamine (Gln) dipeptide) in maintaining intestinal epithelial homeostasis [50]. Sean and co-authors have shown that Gln promotes intestinal stem cells’ expansion and mTOR signaling in jejunum-derived mouse organoids. Furthermore, Saito et al. illustrated the effect of deficiency of essential amino acids on enteroid stem cell differentiation to intestinal enteroendocrine cells, enterochromaffin cells, goblet cells, and Paneth cells [51]. Human liver organoids are composed of multiple hepatic cell types: epithelial hepatocyte cells, hepatic stellate cells, and kupffer cells have been established from human pluripotent stem cells. Using this model system, it was illustrated that free fatty acid exposure to the liver organoids leads to increased inflammation and triggers a physiological condition known as stenosis [52].
With the recent emphasis on learning the metabolomics of cancer stem cells using 3D organoid models, various matrigel-based extracellular flux analysis bioassays were developed [53] [54]. These assays help to determine the crypt metabolomic profile by measuring the oxygen consumption and extracellular acidification rates of the organoids and thus provide an excellent opportunity to define the role of metabolism during tumorigenesis. Furthermore, imaging techniques such as optical metabolic imaging (OMI) have been developed to provide an attractive non-destructive platform to quantify cellular metabolism in organoids. OMI utilizes the NAD (P)H and FAD autofluorescence intensity and a lifetime to detect the metabolic states of the cells. Using this method in breast, pancreatic, and head and neck tumor organoids, researchers have shown the metabolomic changes of organoids in response to cancer drugs [55–57]. To further explore the distribution of drugs and their metabolites, matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI-MSI) system was used in patient-derived colon tumor organoids [58]. The use of such mass spectrometry-based approaches to study metabolic profiles in 3D has led to the emergence of a new era of organoid research that encompasses its application to high throughput technologies, as discussed in the next sections.
4.2. ORGANOIDS FOR GUT MICROBIOME STUDY
Studies have shown the potential of organoids for understanding the dynamic interaction between diet, microbiota, and the host intestinal epithelium [59, 60] (Figure 2B). The gut microbiota, which consists of trillions of microorganisms in the gastrointestinal tract, plays significant functions in determining human health. These microorganisms have been shown to regulate the lifespan, bioavailability, chemical structure, and biological activities of the ingested compounds via diet. Besides, microbial metabolites have been shown to facilitate multiple intestinal signaling pathways, thus making this metabolic-microbiota and host interaction a potential target for developing therapeutic drugs [61]. To date, the effect of microbiota on host metabolism has been studied mostly in human 2D cell lines. Since 2D monolayer cultures minimally recapitulate the cellular complexity and organization, 3D organoid models were used to enumerate the role of microbial metabolism on host disease pathogenesis. Lukovac and colleagues have examined the influence of microbially produced short-chain fatty acids on intestinal function by using mouse gut organoids [62]. The bacterial-conditioned media for culturing the organoids were used to prove the concept. The different gut-microbiota members stimulate different responses in the organoids by modulating the genes involved in various metabolic pathways [62]. The microbially generated short-chain fatty acids play a crucial function in maintaining gut health [63]. Interestingly, intestinal organoids have shown their potential to study the beneficial effect of gut microbiota-derived short-chain fatty acids on mitigating graft-versus-host-diseases [64]. Additionally, Schilderink et al. determined the impact of short-chain fatty acid, butyrate, inhuman, and mouse intestinal organoids [65]. The activation of ALDH1A1 and ALDH1A3 expression through butyric acid plays a vital role in converting epithelial retinol to retinoic acid, thus maintaining gut homeostasis [65]. Furthermore, the intestinal organoid system has addressed the role of enterochromaffin (EC) cells as chemosensors that detect stimuli from ingested chemicals, gut microorganisms, and endogenous regulatory pathways [66]. Similarly, Sarah and co-authors have investigated the function of microbially-derived acetate, propionate, and butyrate on intestinal stem cells using duodenum organoids [67].
Organoids have also been implicated in studying the influence of diet on tumorigenesis. A study of flavon-3-ols (extracted from grape) on human and mouse-derived colon tumor organoids showed the inhibitory effect of flavon-3-ols on colorectal cancer [68]. Flavon-3-ols available to the gut-microbiota and are metabolized to low-molecular-weight phenolic products [69–71]. Another very recent study has highlighted the gut microbiome’s role in determining a cancer mutation [60]. The study found contrasting effects (both tumor-initiating and tumor-suppressive) of mutant p53 in different segments of the gut and mouse-derived tumor organoids. However, he showed that supplementing the organoids with gut microbiota-gallic acid can suppress the tumor-suppressive effect of p53 and lead to the tumor phenotype in organoids.
Even though 3D organoid models provide an excellent platform to ascertain the role of gut-microbiome and host interaction, this technology has several challenges. First, the healthy and disease-derived organoids’ responses can be different and should be taken into account. Further, the microbes interact with the apical epithelial surfaces, i.e., the lumen which faces the interior of the enteroids. Thus, for evaluating the gut microbially-derived metabolites in intestinal organoids, the polarity of the cells should be considered. To achieve this, organoids are disrupted into single or clumps of cells followed by embedding into the extracellular matrix and directly culturing with the media containing the desired microorganism. Since dissociating the organoids expose the apical side of the cells; therefore, it provides a suitable interaction between the microorganism and the intestinal organoid cells [72]. Another technique is microinjection, which employed the introduction of microbes into the organoid lumen (Figure 2B (i)). However, the collection of cell debris and mucus within the enclosed organoid lumen can make this technique confounding and labor-intensive. A new technique has been introduced to overcome this problem, which reverses the apical polarity of the organoids by manipulating the extracellular matrix composition. In this current method, the organoids were first developed in matrigel equivalent basement membrane extract [2] and then transferred to suspension culture growth media (in low attachment plate) without dissociating the organoid into single cells [73]. Using this technique, the authors showed that the organoids in the suspension culture lack a lumen, and thus the boundaries of epithelial cells become indiscernible.
Studies have suggested that most of the microbial communities in the gut environment are comprised of anaerobic microbes, which are highly sensitive to oxygen. Hence, the high throughput microinjection technique was used to study the interaction of anaerobic microbes with the intestinal organoids. In a recent study, the authors have developed a semi-automated high-throughput microinjection device that transports the microbes or cargo directly into the lumen of the organoids [74]. Since the organoid lumen is satisfactorily hypoxic, the authors showed the survival of an oxygen-sensitive anaerobe, B adolescents, in the organoid lumen over four days.
Altogether, these studies corroborate the concept that intestinal organoids recapitulate the gut microenvironment and, therefore, can serve as a suitable model to examine the microbe-gut-host interaction.
4.3. ORGANOIDS FOR INFECTIOUS DISEASES
For many decades, single-cell culture and animal models have provided the stage for understanding the host-pathogen interaction under disease conditions. These models also advance the overall development of drugs and vaccines for treating infectious diseases. However, the absence of complex cellular context and the inherent differences of the immune system between humans and animals limit the use of single-cell culture and animals. Advancements in 3D organoid cultures have boosted drug development and helped understand infectious diseases and host-cell responses (Figure 2C). One of the uses of organoids in infectious diseases involved the study of Zika virus infection (ZIKV) using the brain organoid. In 2016, the ZIKV was infected into the human pluripotent stem cell-derived cerebral organoids to comprehend its association with neurological complications in newborns. Using brain organoid, ZIKV was found to induce neural cell death in the early stages of brain development [75]. Further, Gabriel et al. showed that ZIKV induces premature differentiation of infected neural progenitor cells and generates mitotic defects [76]. Dang et al. had used human embryonic stem cell-derived organoid to recapitulate the early stage of fetal brain development and demonstrated a mechanistic connection between disrupted neurogenesis and ZIKV mediated TLR3 activation [77].
Organoids, such as intestinal organoids, have shown their robustness for the assessment of noroviruses and rotaviruses infections [78, 79]. A study using intestinal organoids infected with noroviruses showed bile as a critical factor for strain-dependent noroviruses replication [78]. Human-derived small intestine and lung organoids were exploited to define the Cryptosporidium infection mechanism. Cryptosporidium is a protozoan parasite and is the leading cause of diarrhea and child mortality worldwide. To understand the pathophysiology of Cryptosporidium, Heo et al. have analyzed the Cryptosporidium transcriptome during its infection and demonstrated that Cryptosporidium could replicate in the organoids and completes its entire life cycle [80].
Organoids are also highly used for understanding the infections related to the respiratory tract. Generally, the first contact of pathogenic microbes and the respiratory tract is through epithelial cells, and epithelial cells can be easily grown as organoids. During the COVID-19 pandemic, lung and gut organoids are highly used for understanding the SARS-CoV-2 infection and biology. Studying SARS-CoV-2 with intestinal organoids showed that the angiotensin-converting enzyme 2 (ACE2) receptor (through which the virus enters the cell) is highly expressed in the brush border of intestinal organoids. Furthermore, it was found that the SARS-CoV-2 virus can infect both high and low-expressing ACE2 cells. One of the early studies by Lamers et al. has demonstrated that airway and gut organoids can be used as a model to study the SARS-CoV2 infection in the lung and gut [81]. The study revealed that SARS-CoV2 mainly targets and infects the ciliated cells but not the goblet cells. Additionally, a recent study has utilized the human iPSC derived brain organoids to understand the direct involvement of SARS-CoV2 in the central nervous system [82]. The authors detected increased viral antigen (SARS-CoV2 nucleocapsid) protein and viral particles in the cryosectioned brain organoids. The localization of SARS-CoV2 to TUJ1- and NESTIN-positive cells of the brain organoids were also shown, thus suggesting the direct infection of cortical neurons and NPCs SARS-CoV2. Furthermore, another group of researchers has developed region-specific brain organoids to investigate and understand the ability of SARS-CoV2 to infect and impair the function of brain cells [83]. Though the adverse outcome of COVID-19 is respiratory failure, often, many of the patients develop additional complications involving gastrointestinal, neurological, and metabolic systems. A retrospective study on 7,337 COVID-19 patients has shown a strong correlation between the poor outcomes in COVID-19 patients and type 2 diabetes [84]. Liuliu and the group have generated liver and pancreas organoids to investigate the viral tropism of SARS-CoV2 [85]. This organoid-based study provided evidence that pancreatic alpha and beta cells and liver organoids are permissive to SARS-CoV2 infection. Zhao et al. has shown the infection of liver ductal organoids by SARS-CoV2 virus and demonstrated increased cholangiocyte damage upon SARS-CoV2 infection [86]. Organoids were also used robustly to study the efficacy of drugs that are developed against SARS-CoV2. A recent study has highlighted the effect of clinical-grade soluble human-recombinant ACE2 in blocking the growth of SARS-CoV2 infection in human blood vessels organoids and kidney organoids [87].
Besides this, an ACE2 (+) expression was studied in multiple organoids using single-cell RNA sequencing analysis. The analysis revealed that ACE2 is highly expressed in intestinal, lung, and retinal organoids, and limited-expression was found in brain and prostate organoids [88]. Further, SARS-CoV2 causes multiple organ damage, with acute kidney injury (AKI) as the second leading organ damage in COVID-19 patients. Xia and the group have established long-term cultures of normal human kidney proximal tubule epithelial cells (KPTECs) by co-culturing the KPTECs organoid with irradiated J2 fibroblast feeder cells to investigate the infection of SARS-CoV2 in kidney cells [89]. Another study in human long organoids has tested the therapeutic potential of 25-hydrocholesterol (25HC) against SARS-CoV2 infection [90]. In COVID-19 patients, SARS-CoV2 infection stimulates the cholesterol 25-hydroxlase gene, which then mediates cholesterol conversion to 25HC. 25HC was found to inhibit the viral membrane fusion and thus block its entry into human lung organoids.
Organoids have also been used to understand the human reproductive tract infections, such as herpes simplex virus type 2 (HSV-2), chlamydia, gonorrhea, syphilis, and human immunodeficiency virus (HIV) infection [91]. HSV-2 infection in females predominantly occurs in the lower female reproductive tract (FRT). The mucosal lining of the vagina, which is made of stratified squamous epithelial cells, interacts with the FRT pathogenic organisms. A significant challenge to study infectious diseases is the recreation of a microenvironment of native tissue. The air-liquid interface (ALI) organoid cultures of vaginal epithelial cells were developed to overcome this problem and determine the mechanism of HSV-2 infection [92]. The study focused on examining the effect of progesterone on the cells of the vaginal epithelial organoid when they were exposed to HSV-2 infection. When compared to the traditional liquid-liquid interphase culture of vaginal epithelial cells, ALI closely mimics the in vivo condition of the FRT compared to liquid-liquid interphase, therefore provide a better model to study the host-pathogen interaction in FRT. Recently, Kessler and the group have utilized the human fallopian tube organoids to analyze the genital Chlamydia trachomatis (Ctr) infection and its effect on epithelial homeostasis [93]. Furthermore, the author elucidated an increase in LIF signaling and increased stemness in the organoids when infected with Ctr. Moreover, DNA methylation pattern changes of the genomic DNA from the infected organoids showed that Ctr enhances the DNA hypermethylation. These studies highlight progress in the organoid culture establishments for investigating the essential aspects of host-pathogen interaction during infection. Moreover, the list of pathogens that can be grown and organoids to study their infection mechanism is still growing.
4.4. ORGANOIDS FOR TUMOR MICROENVIRONMENT
The in vitro cultures of neoplastic cells as an organoid typically consist of epithelial cells and matrigel that mimic the tumor microenvironment. However, a significant part of the solid tumor comprises the tumor microenvironment, involving tumor-infiltrating immune cells, vasculature, and tumor-stroma. Several studies have demonstrated the mechanism for tumor microenvironment involvement in tumor growth. However, there is a dearth of in vitro models that reiterate tumor cells and tumor microenvironment interaction. Advent in organoid culture opens new approaches to understanding tumor microenvironment in cancer (Figure 2D). Using ALI methodology, it has been demonstrated that culturing patient-derived organoids with tumor-infiltrating lymphocytes can successfully recapitulate the PD1/PD-L1-dependent immune checkpoint. Co-culturing cancer-associated fibroblast (CAFs) with tumor cells as a single stroma-attached organoid is another method to understand the involvement of tumor microenvironment in cancer. The method involves the aggregation of dissociated tumor cells and CAF and the matrigel and put as a single dome in a cell-culture well plate [94, 95]. Seino et al. used the single stroma-attached organoid to reveal physical attachment or the juxtracrine interaction of CAFs with pancreatic ductal adenocarcinoma cells (PDAC) is necessary for CAFs to support the growth of PDAC organoids under Wnt3A deprived condition. In addition, co-culturing of PDAC organoids with pancreatic stellate cells discloses the presence of two distinct CAF subtypes, characterized by either IL-6 expressing inflammatory CAFs and a-smooth muscle actin (a-SMA) expressing myofibroblastic CAF [95]. IL-1 and transforming growth factor (TGF)β are identified as the tumor-secreted ligands, which promote the distinct inflammatory and myofibroblastic subtype [95]. In addition, DeNardo and the group utilized mammary submerged organoid cultures to investigate the involvement of the adaptive immune system (CD4+ T cells) in macrophage-induced mammary invasive behavior [96]. Treatment of non-invasive mammary organoids with tumor-associated macrophages (TAMs) (isolated from mammary carcinomas) and TH2-type cytokines (IL-4 or IL-13) resulted in organoid disruption and formation of invasive structures. Further, to demonstrate the direct involvement of CD4+ T cells in regulating TAMs induced mammary tumor cell invasion, the aforementioned culture was tri-cultured with CD4+ T cells. Another study of PDAC organoids and CAFs and peripheral blood lymphocytes co-culturing has shown the lymphocyte infiltration and movement of the myofibroblast-like CAFs, towards the tumor cells [97].
Tumor-induced fibroblast support tumor growth in breast cancer; however, much less is known about the role of normal fibroblast in tumor progression. Organoid co-cultures were initiated with estrogen receptor-positive (ER+) breast cancer cells with normal breast fibroblasts (NAF) and tumor-associated fibroblast (TAF) [98]. Co-culturing the breast tumor organoids with NAFs revealed the tumor-induced secretion of pro-proliferative factor IL1β from NAFs. Furthermore, the co-culturing of organoids with TAF led to the secretion of CCL7 from TAFs, thus enhancing the release of PDGF-BB from the tumor cells, which then induced the secretion of IL1β from NAFs [98]. Overall, it described the paracrine signaling involvement between ER+ breast cancer cells, NAFs, and TAFs that boosted the ER+ breast tumor cell proliferation. Co-culturing of human endothelial cells with mouse breast organoids has shown the development of a functional capillary vessel network that could unite to mice circulatory system [99].
Furthermore, current organoids protocols are depending on animal-derived matrigel or collagens for the matrix. However, both collagen and matrigel are composed of complex components, such as collagen IV, entactin, laminin, and various growth factors. The presence of these undefined growth factors can influence the growth and cellular activity of the organoids. Therefore, researchers have identified and established new generation matrices with known components to increase the potential application of organoids.
4.5. ORGANOIDS FOR THERAPEUTICS AND DRUG DEVELOPMENT STUDIES
According to precision medicine, every patient should be treated differentially based on their genetic and transcriptomic landscape. However, failure to understand individual patients’ landscapes leads to treatment failures or resistance towards the drug. The majority of the disease modeling and drug screening studies involved using the 2D monolayer cultures as their model system; however, 2D cell lines exhibit multiple culture-induced mutations or contaminations with other cell lines. This has fueled organoid’s increased application in early drug discovery programs and toxicity screens [49]. Furthermore, patient-derived organoids can be generated rapidly even with a limited amount of harvested tissue, and it highly maintains the heterogeneity and genetic features of their original tissues.
Organoids developed from the patient’s tissue samples can also be used for personalized treatment regimens (Figure 2E). Growing the matched healthy and diseased organoids from patient tissue samples allows the clinical screening of various drug combinations that will efficiently and specifically target the diseased tissue [51]. In cystic fibrosis, the drug effectiveness depends on the mutational type of the CFTR gene. However, characterizing the mutation and the specific drug treatment for the cystic fibrosis patient is time-consuming and costly. Dekkers et al. demonstrated that rectal organoids could be useful in predicting cystic fibrotic patients who can potentially respond to the drug [100]. Furthermore, the Bear group has utilized CFTR patient-derived respiratory organoids for examining a fluorescent-based assay, which measures the patient-specific drug responses based on the CFTR gene mutations [101].
Organoids biobank has been established for various types of tumors: colon, pancreas, prostate, and liver cancer. Using the two biobanks derived from colon cancer was exploited to screen 83 clinically used drugs [102]. Vlachogiannis and the group have developed live organoid biobanks from patients with metastatic gastrointestinal cancer and compared the drug responses of the organoids with the patient response in the clinical trials [103]. Further research on patient-derived pancreatic cancer organoids has investigated the sensitivity of chemotherapeutic drugs based on gene expression signatures [104]. Moreover, the same study has reported new driver mutations in the KRAS wild-type pancreatic tumors that involve ERBB2S310F, MAP2K1Q58-E62del, and PIK3CAE110del. The authors suggested alternative targeted treatment regimes, such as afatinib (for ERBBS310F mutations) and everolimus (for PIK3CAE110del mutations) for chemo-refractory pancreatic organoids. Screening of chemotherapeutic agents; temozolomide (TMZ) and bis-choloethylnitrosourea (BCNU), and ionizing radiation on glioma stem cells (GSC) and cerebral organoid glioma (COG) showed high resistance activity of GSC towards drug and radiation-induced damage [105]. However, when GSC was co-cultured with 2D cerebral cells exhibited more genotoxic stress on drug and radiation treatment [105]. Overall, the study revealed that COG recapitulates the in vivo disease condition and treatment responses more closely than 2D cultures. Phan and the group have established a mini-rings geometry assay system to perform high-throughput drug screening of 3D tumor organoids [105]. The method involves the establishment of organoids in a ring shape around the rim of a 96-well plate. The assay provides the advantage of changing the media or addition of drugs to be easily performed by pipetting directly in the center of the well, thus preventing the disruption of the gel. Using this technology, Phan et al. has tested 240 kinase inhibitors (FDA approved or in clinical development) in patient-derived organoids and suggested specific drugs to individual patients. A study using colon organoids has shown the effectiveness of afatinib and other EGFR inhibitors in advanced-stage colon cancer with APC mutation [106]. Foundation Hubrecht Organoid Technology (HUB; Utrecht, The Netherlands) had conducted a multi-centered cohort study on organoids derived from the biopsy of the metastatic colon, breast, and non-small cell lung cancers. The study demonstrated a positive correlation between the drug responses on the tumor organoids with clinical responses of patients [107]. EGFR inhibitors were also explored in pancreatic cancer by Kaushik et al. [108]. In the study, authors have utilized pancreatic tumor organoids to demonstrate the selective inhibition of pancreatic tumor stem cells by afatinib (a pan-EGFR inhibitor) and gemcitabine combination treatment. Further, it was shown that afatinib and gemcitabine synergistically decrease metastasis in pancreatic cancer.
Another benchmark study in drug discovery using the organoid system was presented by Greka et al. research. Greka’s group has identified misfolded mutant mucin 1 (MUC1-fs) protein trapped in the tubules of Mucin 1 kidney disease patient’s organoids [109]. With this knowledge, the same group has identified a potential drug, BRD4780 (a small molecule), to treat Mucin 1 kidney disease. As a mechanism, it was shown that MUC1-fs is trapped in TMED9 cargo receptor-containing vesicles, and treatment with BRD4780 leads to the release of MUC1-fs by binding to TMED9.
The major limitation in drug screening is the differences in experimental protocol and the approaches for data analysis. The development of a more standardized screening and data analysis method can increase the robustness of drug-screening data. To achieve this, Driehuis et al. have provided an in-depth protocol for establishing head and neck squamous cell carcinoma (HNSCC) organoids and a subsequent semi-automated therapy screen method [110]. By improving the traditional organoids protocol, Arlotta’s group has generated long-growing organoids that resemble the rich diversity of cell types of the human cerebral cortex [111]. With this improved protocol, the Arlotta group suggests that brain development’s dynamic process can be modeled in organoids and can provide a valuable model to investigate developmental abnormalities associated with human neurological diseases. This type of work put the foundation to understand the genetic alterations related to neurological diseases and the development of drugs to target these mutations.
Organoids are just not limited to cancer, other notable diseases such as polycystic kidney disease, microcephaly, psychiatric disorders, and aforementioned infectious diseases can also be studied through this model system.
4.6. ORGANOIDS IN HIGH THROUGHPUT SEQUENCING APPLICATIONS
Organoids serve as a 3D platform to model difficult to study processes temporally and spatially. Examples include the onset of neuropsychiatric disorders in the human fetal brain, early epigenomic changes leading to cancer initiation, among other developmental pathologies. The organoid model transcriptome and epigenome study help identify functional elements that may facilitate disease onset [112]. Furthermore, organoids function as a translational bridge between in vitro and in vivo models, enabling rapid transition of in vitro discoveries into the clinics. This is particularly relevant for drug screening in case of aggressive malignancies wherein administering a drug suited to the patient’s genetic profile is critical for better prognosis and disease management. Organoids, therefore, can play a central role in personalized medicine. Given organoids can essentially function as miniature organs in a dish, evaluating the transcriptome and epigenome of these in vitro models through high-throughput platforms such as analyses at the single-cell level would enable us to dissect development and pathophysiology comprehensively. Recent advances in the methodology to isolate and culture tissues as three-dimensional (3D) organoids have led to profound improvements in the success rate of deriving organoids from normal and diseased human tissues. However, it remains unclear if organoids can precisely recapitulate the cell state-specific transcriptomic and epigenomic landscapes of the tissues intend to model.
With recent advances in high throughput and omics profiling technologies, it is now possible to characterize organoids at the transcriptomic, proteomic, and epigenomic levels (Figure 3). Such high throughput applications, both bulk and single-cell sequencing technologies. While bulk sequencing applications have provided many insights through comparison of normal and diseased states, single-cell applications hold tremendous potential to be of use in areas that require understanding cellular heterogeneity. In addition, intercepting the proteomic milieu of physiological and disease states using mass spectrometry is gaining momentum in recent years [113]. In the section below, we discuss some of the recent applications of single-cell RNA-seq and other epigenomic technologies in organoids. We discuss the challenges and future applications of the intersection of the 3D culture and high-throughput single-cell technologies in various tissues.
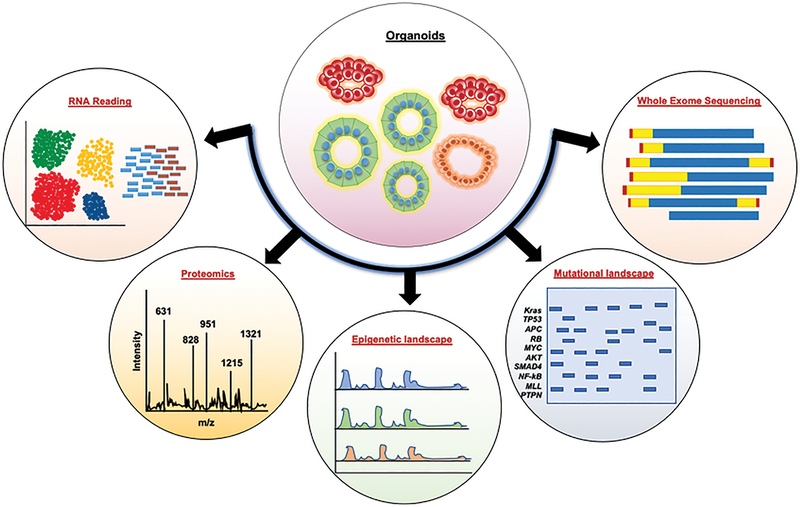
Figure 3. High-throughput sequencing approaches in organoids:
High-throughput sequencing approaches have been extensively explored in organoids to study different molecular mechanisms during cancer or other diseases. (i) RNA-sequencing and single-cell RNA sequencing in tumor organoids are used to profile the total transcriptomic status of the cells during tumor development or metastasis. (ii) Proteomics analysis on organoid through mass-spec illustrate the changes in various proteins during disease vs. normal. (iii) ChIP-Seq or ATAC seq analysis in the organoids demonstrate different epigenetic profile. (iv) Patient-derived organoids are extensively used to study the mutational landscape or load during tumor development. Various mutations such as Kras, TP53, APC, etc., and their oncogenic functions have been profiled in tumor organoids. (v) Whole-exome sequencing in organoids is used to understand the changes or mutations in the protein-coding regions of the genes.
Gut:
Most applications of the single-cell RNA-seq have been restricted to mouse small intestine organoids to date, and these studies have provided the most insight into the cellular composition of adult stem cell-derived organoids [114–117]. Grun et al. sequenced the transcriptome of hundreds of randomly selected cells from mouse intestinal organoids to identify rare cell types from the intestinal epithelium [116]. An algorithm called RaceID for rare cell type identification in complex populations of single cells. Using this algorithm, Reg4 was identified as a unique marker of a rare population of hormone-producing enteroendocrine cells. Not only did RaceID confirmed the existence of known enteroendocrine lineages, but it also discovered novel subtypes, which were subsequently validated in vivo. A rare population of Lgr5-positive secretory cells within the ex vivo-isolated Lgr5-positive stem cells and their direct progeny were identified. In another study, the mouse intestine atlas was generated by profiling 53,193 individual epithelial cells from the mouse small intestine and organoids [117]. Through single-cell RNA-seq, the authors could classify as cell type or state as enterocyte, goblet, Paneth, enteroendocrine, or tuft cells. A new enteroendocrine cell taxonomy was generated by comparing genes in each cell cluster with the canonical classification markers. Interestingly, heterogeneity in the tuft cell population was split into two distinct subtypes Tuft-1 and Tuft-2. Tuft1 and Tuft2 cell populations were enriched in unique signatures related to neuronal development and immune-related genes. Furthermore, the authors compared cell composition and gene expression landscapes in organoids after exposure to different microbe populations, providing insights into how cell types and gene expression profiles in the intestine evolve with microbial infections. In another seminal study, Biton et al. applied single-cell sequencing analysis and organoid culture to understand the contribution of adaptive immunity in intestinal stem cell maintenance [118]. A nonconventional antigen-presenting role of Lgr5+ intestinal stem cells and the influence of Th cells on epithelial cell renewal was deciphered. They found that Lgr5+ intestinal stem cells interacted with activating naive Th cells via major histocompatibility complex class II (MHCII) antigen presentation. Treg and interleukin-10 (IL-10) were found to promote the renewal of the intestinal stem cell compartment, while Th1, Th2, and Th17 cells and derived cytokines (i.e., IL-13 or IL-17) depleted intestinal stem cells and induced organoid differentiation. In addition, luminal signals (bacteria or parasite) influenced the intestinal stem cell-Th axis and shaped epithelial cell composition. This study is a classic example of how epithelial and immune cell crosstalk can act as a regulated response to maintain intestinal stem cell self-renewal or promote differentiation. The aforementioned studies were conducted using murine organoids. Extended to humans, single-cell RNA-seq will allow controlled experimentation of human intestinal cell types to diverse dietary, microbial, or pharmaceutical manipulations. However, improved organoid culture methods are needed to enable the long-term culture of progenitors and differentiated cells isolated from the human gut within a 3D structure.
Liver:
Generating liver in vitro from pluripotency via recapitulating the growth pathways has been challenging due to the dynamic developmental, structural, and cellular heterogeneity of the organ. iPSCs in 2D monocultures can be differentiated to hepatic endoderm and then toward ‘hepatocyte-like’ cells [119, 120]. However, these ‘hepatocyte-like’ cells do not function as mature, metabolically competent hepatocytes. Cells with only modest similarity to human hepatocytes have been generated using certain widely used protocols, which may even be off-target cells with similarity to the intestinal epithelium [121]. Incorporating additional signals such as the mesoderm-derived paracrine signals that promote hepatocyte maturation in liver organoids and a 3D microenvironment that mediates cell-to-cell surface contact has shown great promise in generating hepatic organoids [37, 122]. It has been shown that 2D monocultures of primary adult human hepatocytes can only be maintained short-term due to dedifferentiation and cell death [123]. However, with the advancement in protocols for isolation of hepatic stem cells (HSCs) from adult tissues and culture HSCs in 3D matrix environments, the differentiated hepatic cells can be maintained as genetically stable organoids [22]. Furthermore, primary liver tumor organoids have successfully been established using tumor resections from liver cancer patients [124].
Broutier et al. showed that the primary patient-derived liver cancer organoids maintain the expression profile of the corresponding tissue-of-origin and tumor subtype using genome-wide transcriptomic (RNA-seq) analysis [124]. Each tumor organoid was subjected to whole-exome sequencing (WES) analysis following short (<2 months, early passage) or extended (>4 months, late passage) periods in culture to ascertain whether the different organoids retain the parent tumor’s mutational landscape. The organoids retained >90% of the patient’s tissue variants for early cultures and >80% after long-term culture to compare the global variant profile. Further, the single nucleotide variants (SNVs), indels, and the distribution of base substitutions in the original tissue were retained in culture. Additionally, novel genes with prognostic value in liver cancer cohorts were identified through comparative gene expression profiling of tumor organoids versus healthy liver organoids. These findings demonstrate the potential of high-throughput analyses conducted using organoids to identify novel genes of importance in human liver cancer. Single-cell RNA-seq analyses on the adult liver could, in principle, map the transcriptome states of the hepatic progenitors, hepatic stem cells, and mature hepatocytes in vivo, which can be compared to the transcriptome of the adult stem-cell-derived organoids maintained in vitro. Such comparisons would enable assessment of the accuracy and precision of organoid culture ex vivo and would reveal the dynamic changes in the transcriptome of these organoids with time. In any case, current protocols lack cellular diversity (e.g., kupffer cells, stellate cells, bile ducts, portal endothelium, etc.) and the structural organization of the human liver. In the future, modifications to the existing protocols to support the in vitro growth of diverse hepatic cell populations would lead to liver organoid technology benchmarking.
Pancreas:
Applications of the organoid culture to the pancreas include the study of in vitro differentiation of pancreatic progenitors to insulin-producing islets and the pathogenesis of pancreatic cancer [15]. Several studies indicate that epigenomic reprogramming is an emerging mechanism of pancreatic ductal adenocarcinoma (PDAC) progression. Inactivating mutations of chromatin modifiers are frequent genetic events in pancreatic tumors [125]. Further, an association between the disruptions of large heterochromatin domains with the metastatic transition in pancreatic cancer has been demonstrated [126]. Given the importance of evaluating the epigenomic landscape changes during pancreatic cancer progression, organoids may serve as a robust and representative miniature in vitro models of studying pancreatic cancer metastasis. Roe et al. used organoids generated from primary pancreatic tumors (T) and matched metastatic (M) tissues derived from the autochthonous PDAC mouse model (Kras+/LSL-G12D; Trp53+/LSL-R172H; Pdx1-Cre) [127]. The authors found regions with consistent GAIN and LOSS of H3K27 acetylation in all M organoids compared to matched T organoids, irrespective of the anatomic site of the metastatic lesion. These GAIN regions with enhanced H3K27 acetylation, indicative of enhancer elements, were enriched at the corresponding locations in human PDAC cell lines and patient-derived PDAC organoids. Ontology analysis of genes located near GAIN regions revealed a significant enrichment of developmental functions and implicated FOXA1 in GAIN enhancer activation. Through high throughput approaches such as H3K27ac ChIP-seq and ATAC-Seq, the authors showed that FOXA1 mediated enhancer reprogramming drives PDAC aggressiveness, and manipulation of FOXA1 alters enhancer activity in PDAC cells and turn affects metastasis. This study underscores that applying high throughput approaches in organoids can lead to significant strides in uncovering disease pathogenesis mechanisms. In another study, patient-derived PDAC organoids were injected into the pancreatic duct of immunocompromised mice using a modified retrograde pancreatic intraductal injection method [128]. The authors referred to this in vivo model as the intraductal grafted organoid (IGO) model. Tuveson and colleagues had previously generated another in vivo model of PDAC that comprised injecting the patient-derived PDAC organoids directly into the interstitium of immunocompromised mice (referred to as an orthotopically grafted organoid model or OGO) [129]. Since PDAC progression is believed to occur in the pancreatic ductal system, the IGO model is better at recapitulating the progression from preinvasive cells within the duct system to extra ductal frank invasive tumors surrounding desmoplasia. To determine the influence of the different microenvironments on the transcriptome of the tumors, the authors performed RNA sequencing (RNA-seq) using pairs of tumors derived from the OGO and IGO organoids. The OGO xenografts were shown to exhibit the previously described “squamous PDAC” [130] and “basal-like” [131] gene signatures. In contrast, the IGO xenografts were enriched in “progenitor PDAC” [130] and “classical” [131] signatures using high throughput transcriptional and molecular analyses. Moreover, xenografts derived from the organoid models were classified as Fast- and Slow-progressor phenotypes correlated with the molecular analyses and subtype classifications of human PDAC. This study is a classic example of how organoids can be used as personalized models of PDAC and can provide insights into the heterogeneity of the disease.
Brain:
In vitro neural differentiation from induced pluripotent stem cells (iPSCs) and human embryonic stem cells (hESCs) recapitulates early brain development stages starting with the initial commitment to neuroepithelium followed by the sequential creation of neural progenitor cells and subsequent birth of early and late neurons. Complex structures in the developing brain, such as the progenitor zones along the apical-basal axis and a diverse variety of functional neurons, can be generated in organoids [132, 133]. Because human brain development is a highly dynamic process driven by a spatially and temporally controlled transcriptional program that functions synchrony with an epigenomic program, cerebral organoids serve as an ideal model for examining in vitro neural differentiation. Luo et al. used whole-genome methylome profiling and transcriptome sequencing to examine the epigenomic and transcriptomic changes during in vitro differentiation using cerebral organoids and the human fetal brain [134]. Transcriptomic signatures in organoids faithfully modeled gene expression profiles in the early-to-mid human fetal brain. The authors also found that regions with Non-CG Methylation overlapped with super-enhancers and were actively expressed during early fetal stages but became suppressed after the transition to postnatal development. Further, the cerebral organoids showed enrichment for common transcription factor binding motifs and promoter DNA methylation signatures as that of the human fetal brain. Overall, the remodeling of the methylome observed during cerebral organoid differentiation in this study underscored that the epigenomic state achieved in the cerebral organoids closely resembled the human fetal brain [134].
5. CONCLUSION
The accessibility and the property to imitate in vivo tissue biology have made the organoid a great platform to study different fields of disease modeling. This model has shown its potential for research, including therapeutic responses and drug discovery. Although there is immense development in the technology for 3D organoid culture systems, the researchers are still exploring and standardizing techniques for their robust use in translational research. A major limitation in the field involves that the researcher has no control over the function and behavior of the cells once they assemble into organoids. This drawback limits the use of organoids to full potential. However, collaborating with advanced bioengineering techniques allow the culture of organoids in a more defined environment. Bioengineered biomaterials and microtechnology can mimic the embryo environment by providing controlled geometric and/or mechanical inputs to the organoids. Moreover, utilizing the engineering technology makes it possible to deliver morphogens/growth factors in a controlled manner. These approaches will impart more systemic information in the field and lead to the defined growth of organoids. Finally, scientists are also exploring the bioreactor, ALI culture method, and vasculogenic technologies to address the microenvironment and overcome the nutrient availability problem. Given the rapid advance in organoid technology, we trust that organoids will provide better opportunities to study and be used in human health.
6. FUTURE PROSPECTS
Despite the extensive application of organoids in the various biology fields, there are still several diseases where the organoid culture technology has not been explored yet or is still in infancy. Most of the neurodevelopmental or neuropsychiatric conditions, such as schizophrenia, Parkinson or autism, are mostly studied using animal model systems [135]. The disorders like autism spectrum disorders or Parkinson’s constitute clinical heterogeneity (including epilepsy, sleep disruptions, and motor disturbances), making it difficult to investigate using an organoid culture system [136–138]. However, researchers have started developing protocols to generate more mature and complex brain organoids that can be actively used for neuropsychiatric studies [139]. Furthermore, organoids can be used to understand diseases, such as developmental brain injury and disorders (DBD) related to the early developmental stages of the brain. A lot of the diseases have been shown to be associated with stem cells. However, scientists are still confounded by how the anomalies are happening within the stem cells or how they know which type of specialized cell it has to make. Therefore, the organoids system can be used to answer the involvement of stem cells in a disease like emphysema, a type of lung disease, where lung stem cells fail to repair the damage [140]. Further, scientists have suggested the idea of using organoids to screen drugs that can enable the formation of specialized cell types for genetic diseases, like cystic fibrosis, where ciliated cells that eradicate mucus from the lung are not working appropriately [141]. Organoids from cystic fibrosis patient tissues can be generated, followed by developing a drug that might make the ciliated cells work better in the organoid culture system. Since scientists have developed the techniques to co-culture the organoids with different immune cells, organoid culture technology can also be explored to understand the mechanisms and screen drugs for autoimmune diseases. Moreover, the application of organoid technology can be exploited for immunotherapy studies on tumor organoids, where T-lymphocyte from a healthy donor can be activated and expanded with patient-derived organoids. Organoids generated from pluripotent stem cells mimic the early embryonic organs, therefore, contain both epithelium and mesenchymal cells [20]. Recent studies on organoids have been used to address the fibrosis mechanism in the lung and intestine [142]. Using the same protocol, organoids can also be used to study fibrosis in other organs, such as the liver and kidney. Though in a very initial stage, scientists have already started thinking of applying organoid technology in the above-mentioned diseases.
Acknowledgments
Funding
The authors in this article were supported primarily by the following grants from the National Institutes of Health P01 CA217798, R01 CA210637, R01 CA183459, R01 CA195586, R01 CA201444, R01 CA228524, F99 CA234962, U01 CA200466, and U01 CA210240, and the Nebraska Department of Health and Human Services LB595.
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCE
- [1].Kretzschmar K, Clevers H, Organoids: Modeling Development and the Stem Cell Niche in a Dish, Developmental cell 38(6) (2016) 590–600. [DOI] [PubMed] [Google Scholar]
- [2].Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H, Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche, Nature 459(7244) (2009) 262–5. [DOI] [PubMed] [Google Scholar]
- [3].Mameishvili E, Serafimidis I, Iwaszkiewicz S, Lesche M, Reinhardt S, Bölicke N, Büttner M, Stellas D, Papadimitropoulou A, Szabolcs M, Anastassiadis K, Dahl A, Theis F, Efstratiadis A, Gavalas A, Aldh1b1 expression defines progenitor cells in the adult pancreas and is required for Kras-induced pancreatic cancer, Proc Natl Acad Sci U S A 116(41) (2019) 20679–20688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Nath S, Devi GR, Three-dimensional culture systems in cancer research: Focus on tumor spheroid model, Pharmacol Ther 163 (2016) 94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Nunes AS, Barros AS, Costa EC, Moreira AF, Correia IJ, 3D tumor spheroids as in vitro models to mimic in vivo human solid tumors resistance to therapeutic drugs, Biotechnol Bioeng 116(1) (2019) 206–226. [DOI] [PubMed] [Google Scholar]
- [6].Fang Y, Eglen RM, Three-Dimensional Cell Cultures in Drug Discovery and Development, SLAS Discov 22(5) (2017) 456–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bonnans C, Chou J, Werb Z, Remodelling the extracellular matrix in development and disease, Nat Rev Mol Cell Biol 15(12) (2014) 786–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cruz-Acuña R, Quirós M, Farkas AE, Dedhia PH, Huang S, Siuda D, García-Hernández V, Miller AJ, Spence JR, Nusrat A, García AJ, Synthetic hydrogels for human intestinal organoid generation and colonic wound repair, Nat Cell Biol 19(11) (2017) 1326–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gjorevski N, Sachs N, Manfrin A, Giger S, Bragina ME, Ordóñez-Morán P, Clevers H, Lutolf MP, Designer matrices for intestinal stem cell and organoid culture, Nature 539(7630) (2016) 560–564. [DOI] [PubMed] [Google Scholar]
- [10].Blondel D, Lutolf MP, Bioinspired Hydrogels for 3D Organoid Culture, Chimia (Aarau) 73(1) (2019) 81–85. [DOI] [PubMed] [Google Scholar]
- [11].Kaushik G, Ponnusamy MP, Batra SK, Concise Review: Current Status of Three-Dimensional Organoids as Preclinical Models, Stem Cells 36(9) (2018) 1329–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhou T, Tan L, Cederquist GY, Fan Y, Hartley BJ, Mukherjee S, Tomishima M, Brennand KJ, Zhang Q, Schwartz RE, Evans T, Studer L, Chen S, High-Content Screening in hPSC-Neural Progenitors Identifies Drug Candidates that Inhibit Zika Virus Infection in Fetal-like Organoids and Adult Brain, Cell Stem Cell 21(2) (2017) 274–283.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Bräuninger M, Lewitus E, Sykes A, Hevers W, Lancaster M, Knoblich JA, Lachmann R, Pääbo S, Huttner WB, Treutlein B, Human cerebral organoids recapitulate gene expression programs of fetal neocortex development, Proc Natl Acad Sci U S A 112(51) (2015) 15672–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kim J, Koo BK, Knoblich JA, Human organoids: model systems for human biology and medicine, Nat Rev Mol Cell Biol 21(10) (2020) 571–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Balak JRA, Juksar J, Carlotti F, Lo Nigro A, de Koning EJP, Organoids from the Human Fetal and Adult Pancreas, Curr Diab Rep 19(12) (2019) 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mustata RC, Vasile G, Fernandez-Vallone V, Strollo S, Lefort A, Libert F, Monteyne D, Pérez-Morga D, Vassart G, Garcia MI, Identification of Lgr5-independent spheroid-generating progenitors of the mouse fetal intestinal epithelium, Cell Rep 5(2) (2013) 421–32. [DOI] [PubMed] [Google Scholar]
- [17].Greggio C, De Franceschi F, Figueiredo-Larsen M, Grapin-Botton A, In vitro pancreas organogenesis from dispersed mouse embryonic progenitors, J Vis Exp (89) (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Clevers H, Modeling Development and Disease with Organoids, Cell 165(7) (2016) 1586–1597. [DOI] [PubMed] [Google Scholar]
- [19].Lee CT, Bendriem RM, Wu WW, Shen RF, 3D brain Organoids derived from pluripotent stem cells: promising experimental models for brain development and neurodegenerative disorders, J Biomed Sci 24(1) (2017) 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].McCauley HA, Wells JM, Pluripotent stem cell-derived organoids: using principles of developmental biology to grow human tissues in a dish, Development 144(6) (2017) 958–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Behjati S, Huch M, van Boxtel R, Karthaus W, Wedge DC, Tamuri AU, Martincorena I, Petljak M, Alexandrov LB, Gundem G, Tarpey PS, Roerink S, Blokker J, Maddison M, Mudie L, Robinson B, Nik-Zainal S, Campbell P, Goldman N, van de Wetering M, Cuppen E, Clevers H, Stratton MR, Genome sequencing of normal cells reveals developmental lineages and mutational processes, Nature 513(7518) (2014) 422–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Huch M, Gehart H, van Boxtel R, Hamer K, Blokzijl F, Verstegen MM, Ellis E, van Wenum M, Fuchs SA, de Ligt J, van de Wetering M, Sasaki N, Boers SJ, Kemperman H, de Jonge J, Ijzermans JN, Nieuwenhuis EE, Hoekstra R, Strom S, Vries RR, van der Laan LJ, Cuppen E, Clevers H, Long-term culture of genome-stable bipotent stem cells from adult human liver, Cell 160(1–2) (2015) 299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fujii M, Shimokawa M, Date S, Takano A, Matano M, Nanki K, Ohta Y, Toshimitsu K, Nakazato Y, Kawasaki K, Uraoka T, Watanabe T, Kanai T, Sato T, A Colorectal Tumor Organoid Library Demonstrates Progressive Loss of Niche Factor Requirements during Tumorigenesis, Cell Stem Cell 18(6) (2016) 827–838. [DOI] [PubMed] [Google Scholar]
- [24].Drost J, Clevers H, Translational applications of adult stem cell-derived organoids, Development 144(6) (2017) 968–975. [DOI] [PubMed] [Google Scholar]
- [25].Qian X, Jacob F, Song MM, Nguyen HN, Song H, Ming GL, Generation of human brain region-specific organoids using a miniaturized spinning bioreactor, Nat Protoc 13(3) (2018) 565–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Przepiorski A, Sander V, Tran T, Hollywood JA, Sorrenson B, Shih JH, Wolvetang EJ, McMahon AP, Holm TM, Davidson AJ, A Simple Bioreactor-Based Method to Generate Kidney Organoids from Pluripotent Stem Cells, Stem Cell Reports 11(2) (2018) 470–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ovando-Roche P, West EL, Branch MJ, Sampson RD, Fernando M, Munro P, Georgiadis A, Rizzi M, Kloc M, Naeem A, Ribeiro J, Smith AJ, Gonzalez-Cordero A, Ali RR, Use of bioreactors for culturing human retinal organoids improves photoreceptor yields, Stem Cell Res Ther 9(1) (2018) 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hamad S, Derichsweiler D, Papadopoulos S, Nguemo F, Saric T, Sachinidis A, Brockmeier K, Hescheler J, Boukens BJ, Pfannkuche K, Generation of human induced pluripotent stem cell-derived cardiomyocytes in 2D monolayer and scalable 3D suspension bioreactor cultures with reduced batch-to-batch variations, Theranostics 9(24) (2019) 7222–7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Neal JT, Li X, Zhu J, Giangarra V, Grzeskowiak CL, Ju J, Liu IH, Chiou SH, Salahudeen AA, Smith AR, Deutsch BC, Liao L, Zemek AJ, Zhao F, Karlsson K, Schultz LM, Metzner TJ, Nadauld LD, Tseng YY, Alkhairy S, Oh C, Keskula P, Mendoza-Villanueva D, De La Vega FM, Kunz PL, Liao JC, Leppert JT, Sunwoo JB, Sabatti C, Boehm JS, Hahn WC, Zheng GXY, Davis MM, Kuo CJ, Organoid Modeling of the Tumor Immune Microenvironment, Cell 175(7) (2018) 1972–1988.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Choi J, Iich E, Lee JH, Organogenesis of adult lung in a dish: Differentiation, disease and therapy, Dev Biol 420(2) (2016) 278–286. [DOI] [PubMed] [Google Scholar]
- [31].Sekiya S, Kikuchi T, Shimizu T, Perfusion culture maintained with an air-liquid interface to stimulate epithelial cell organization in renal organoids in vitro, BMC Biomed Eng 1 (2019) 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Li X, Ootani A, Kuo C, An Air-Liquid Interface Culture System for 3D Organoid Culture of Diverse Primary Gastrointestinal Tissues, Methods Mol Biol 1422 (2016) 33–40. [DOI] [PubMed] [Google Scholar]
- [33].Hsieh YK, Chen SC, Huang WL, Hsu KP, Gorday KAV, Wang T, Wang J, Direct Micromachining of Microfluidic Channels on Biodegradable Materials Using Laser Ablation, Polymers (Basel) 9(7) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].He J, Mao M, Liu Y, Shao J, Jin Z, Li D, Fabrication of nature-inspired microfluidic network for perfusable tissue constructs, Adv Healthc Mater 2(8) (2013) 1108–13. [DOI] [PubMed] [Google Scholar]
- [35].Zhang YS, Arneri A, Bersini S, Shin SR, Zhu K, Goli-Malekabadi Z, Aleman J, Colosi C, Busignani F, Dell’Erba V, Bishop C, Shupe T, Demarchi D, Moretti M, Rasponi M, Dokmeci MR, Atala A, Khademhosseini A, Bioprinting 3D microfibrous scaffolds for engineering endothelialized myocardium and heart-on-a-chip, Biomaterials 110 (2016) 45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Mansour AA, Gonçalves JT, Bloyd CW, Li H, Fernandes S, Quang D, Johnston S, Parylak SL, Jin X, Gage FH, An in vivo model of functional and vascularized human brain organoids, Nat Biotechnol 36(5) (2018) 432–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, Zhang RR, Ueno Y, Zheng YW, Koike N, Aoyama S, Adachi Y, Taniguchi H, Vascularized and functional human liver from an iPSC-derived organ bud transplant, Nature 499(7459) (2013) 481–4. [DOI] [PubMed] [Google Scholar]
- [38].Phelps EA, Enemchukwu NO, Fiore VF, Sy JC, Murthy N, Sulchek TA, Barker TH, García AJ, Maleimide cross-linked bioactive PEG hydrogel exhibits improved reaction kinetics and cross-linking for cell encapsulation and in situ delivery, Adv Mater 24(1) (2012) 64–70, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Dye BR, Dedhia PH, Miller AJ, Nagy MS, White ES, Shea LD, Spence JR, A bioengineered niche promotes in vivo engraftment and maturation of pluripotent stem cell derived human lung organoids, Elife 5 (2016) September 28;5:e19732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, Watanabe T, Kanai T, Sato T, Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids, Nat Med 21(3) (2015) 256–62. [DOI] [PubMed] [Google Scholar]
- [41].Freedman BS, Brooks CR, Lam AQ, Fu H, Morizane R, Agrawal V, Saad AF, Li MK, Hughes MR, Werff RV, Peters DT, Lu J, Baccei A, Siedlecki AM, Valerius MT, Musunuru K, McNagny KM, Steinman TI, Zhou J, Lerou PH, Bonventre JV, Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids, Nat Commun 6 (2015) 8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ringel T, Frey N, Ringnalda F, Janjuha S, Cherkaoui S, Butz S, Srivatsa S, Pirkl M, Russo G, Villiger L, Rogler G, Clevers H, Beerenwinkel N, Zamboni N, Baubec T, Schwank G, Genome-Scale CRISPR Screening in Human Intestinal Organoids Identifies Drivers of TGF-β Resistance, Cell Stem Cell 26(3) (2020) 431–440.e8. [DOI] [PubMed] [Google Scholar]
- [43].Howden SE, Vanslambrouck JM, Wilson SB, Tan KS, Little MH, Reporter-based fate mapping in human kidney organoids confirms nephron lineage relationships and reveals synchronous nephron formation, EMBO Rep 20(4) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Jung KB, Lee H, Son YS, Lee JH, Cho HS, Lee MO, Oh JH, Lee J, Kim S, Jung CR, Kim J, Son MY, In vitro and in vivo imaging and tracking of intestinal organoids from human induced pluripotent stem cells, Faseb j 32(1) (2018) 111–122. [DOI] [PubMed] [Google Scholar]
- [45].Lu Y, Zhang G, Shen C, Uygun K, Yarmush ML, Meng Q, A novel 3D liver organoid system for elucidation of hepatic glucose metabolism, Biotechnol Bioeng 109(2) (2012) 595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Shen C, Zhang G, Meng Q, Enhancement of the predicted drug hepatotoxicity in gel entrapped hepatocytes within polysulfone-g-poly (ethylene glycol) modified hollow fiber, Toxicol Appl Pharmacol 249(2) (2010) 140–7. [DOI] [PubMed] [Google Scholar]
- [47].Zietek T, Rath E, Haller D, Daniel H, Intestinal organoids for assessing nutrient transport, sensing and incretin secretion, Scientific Reports 5(1) (2015) 16831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Rodriguez-Colman MJ, Schewe M, Meerlo M, Stigter E, Gerrits J, Pras-Raves M, Sacchetti A, Hornsveld M, Oost KC, Snippert HJ, Verhoeven-Duif N, Fodde R, Burgering BM, Interplay between metabolic identities in the intestinal crypt supports stem cell function, Nature 543(7645) (2017) 424–427. [DOI] [PubMed] [Google Scholar]
- [49].Mills RJ, Titmarsh DM, Koenig X, Parker BL, Ryall JG, Quaife-Ryan GA, Voges HK, Hodson MP, Ferguson C, Drowley L, Plowright AT, Needham EJ, Wang QD, Gregorevic P, Xin M, Thomas WG, Parton RG, Nielsen LK, Launikonis BS, James DE, Elliott DA, Porrello ER, Hudson JE, Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest, Proc Natl Acad Sci U S A 114(40) (2017) E8372–e8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Moore SR, Guedes MM, Costa TB, Vallance J, Maier EA, Betz KJ, Aihara E, Mahe MM, Lima AA, Oriá RB, Shroyer NF, Glutamine and alanyl-glutamine promote crypt expansion and mTOR signaling in murine enteroids, Am J Physiol Gastrointest Liver Physiol 308(10) (2015) G831–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Saito Y, Iwatsuki K, Hanyu H, Maruyama N, Aihara E, Tadaishi M, Shimizu M, Kobayashi-Hattori K, Effect of essential amino acids on enteroids: Methionine deprivation suppresses proliferation and affects differentiation in enteroid stem cells, Biochem Biophys Res Commun 488(1) (2017) 171–176. [DOI] [PubMed] [Google Scholar]
- [52].Ouchi R, Togo S, Kimura M, Shinozawa T, Koido M, Koike H, Thompson W, Karns RA, Mayhew CN, McGrath PS, McCauley HA, Zhang RR, Lewis K, Hakozaki S, Ferguson A, Saiki N, Yoneyama Y, Takeuchi I, Mabuchi Y, Akazawa C, Yoshikawa HY, Wells JM, Takebe T, Modeling Steatohepatitis in Humans with Pluripotent Stem Cell-Derived Organoids, Cell Metab 30(2) (2019) 374–384.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Fan YY, Davidson LA, Callaway ES, Wright GA, Safe S, Chapkin RS, A bioassay to measure energy metabolism in mouse colonic crypts, organoids, and sorted stem cells, Am J Physiol Gastrointest Liver Physiol 309(1) (2015) G1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Bas T, Augenlicht LH, Real time analysis of metabolic profile in ex vivo mouse intestinal crypt organoid cultures, J Vis Exp (93) (2014) e52026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Walsh AJ, Cook RS, Sanders ME, Aurisicchio L, Ciliberto G, Arteaga CL, Skala MC, Quantitative optical imaging of primary tumor organoid metabolism predicts drug response in breast cancer, Cancer Res 74(18) (2014) 5184–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Walsh AJ, Castellanos JA, Nagathihalli NS, Merchant NB, Skala MC, Optical Imaging of Drug-Induced Metabolism Changes in Murine and Human Pancreatic Cancer Organoids Reveals Heterogeneous Drug Response, Pancreas 45(6) (2016) 863–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Shah AT, Heaster TM, Skala MC, Metabolic Imaging of Head and Neck Cancer Organoids, PLoS One 12(1) (2017) e0170415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Liu X, Flinders C, Mumenthaler SM, Hummon AB, MALDI Mass Spectrometry Imaging for Evaluation of Therapeutics in Colorectal Tumor Organoids, J Am Soc Mass Spectrom 29(3) (2018) 516–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Nigro G, Hanson M, Fevre C, Lecuit M, Sansonetti PJ, Intestinal Organoids as a Novel Tool to Study Microbes-Epithelium Interactions, Methods Mol Biol 1576 (2019) 183–194. [DOI] [PubMed] [Google Scholar]
- [60].Kadosh E, Snir-Alkalay I, Venkatachalam A, May S, Lasry A, Elyada E, Zinger A, Shaham M, Vaalani G, Mernberger M, Stiewe T, Pikarsky E, Oren M, Ben-Neriah Y, The gut microbiome switches mutant p53 from tumour-suppressive to oncogenic, Nature 586(7827) (2020) 133–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Waldecker M, Kautenburger T, Daumann H, Busch C, Schrenk D, Inhibition of histone-deacetylase activity by short-chain fatty acids and some polyphenol metabolites formed in the colon, J Nutr Biochem 19(9) (2008) 587–93. [DOI] [PubMed] [Google Scholar]
- [62].Lukovac S, Belzer C, Pellis L, Keijser BJ, de Vos WM, Montijn RC, Roeselers G, Differential modulation by Akkermansia muciniphila and Faecalibacterium prausnitzii of host peripheral lipid metabolism and histone acetylation in mouse gut organoids, mBio 5(4) (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Russell WR, Hoyles L, Flint HJ, Dumas ME, Colonic bacterial metabolites and human health, Curr Opin Microbiol 16(3) (2013) 246–54. [DOI] [PubMed] [Google Scholar]
- [64].Mathewson ND, Jenq R, Mathew AV, Koenigsknecht M, Hanash A, Toubai T, Oravecz-Wilson K, Wu S-R, Sun Y, Rossi C, Fujiwara H, Byun J, Shono Y, Lindemans C, Calafiore M, Schmidt TM, Honda K, Young VB, Pennathur S, van den Brink M, Reddy P, Gut microbiome–derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease, Nature Immunology 17(5) (2016) 505–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Schilderink R, Verseijden C, Seppen J, Muncan V, van den Brink GR, Lambers TT, van Tol EA, de Jonge WJ, The SCFA butyrate stimulates the epithelial production of retinoic acid via inhibition of epithelial HDAC, Am J Physiol Gastrointest Liver Physiol 310(11) (2016) G1138–46. [DOI] [PubMed] [Google Scholar]
- [66].Bellono NW, Bayrer JR, Leitch DB, Castro J, Zhang C, O’Donnell TA, Brierley SM, Ingraham HA, Julius D, Enterochromaffin Cells Are Gut Chemosensors that Couple to Sensory Neural Pathways, Cell 170(1) (2017) 185–198.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Pearce SC, Weber GJ, van Sambeek DM, Soares JW, Racicot K, Breault DT, Intestinal enteroids recapitulate the effects of short-chain fatty acids on the intestinal epithelium, PLoS One 15(4) (2020) e0230231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Toden S, Ravindranathan P, Gu J, Cardenas J, Yuchang M, Goel A, Oligomeric proanthocyanidins (OPCs) target cancer stem-like cells and suppress tumor organoid formation in colorectal cancer, Sci Rep 8(1) (2018) 3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Ozdal T, Sela DA, Xiao J, Boyacioglu D, Chen F, Capanoglu E, The Reciprocal Interactions between Polyphenols and Gut Microbiota and Effects on Bioaccessibility, Nutrients 8(2) (2016) 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Zhang LS, Davies SS, Microbial metabolism of dietary components to bioactive metabolites: opportunities for new therapeutic interventions, Genome Med 8(1) (2016) 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Trošt K, Ulaszewska MM, Stanstrup J, Albanese D, De Filippo C, Tuohy KM, Natella F, Scaccini C, Mattivi F, Host: Microbiome co-metabolic processing of dietary polyphenols - An acute, single blinded, cross-over study with different doses of apple polyphenols in healthy subjects, Food Res Int 112 (2018) 108–128. [DOI] [PubMed] [Google Scholar]
- [72].Forbester JL, Goulding D, Vallier L, Hannan N, Hale C, Pickard D, Mukhopadhyay S, Dougan G, Interaction of Salmonella enterica Serovar Typhimurium with Intestinal Organoids Derived from Human Induced Pluripotent Stem Cells, Infect Immun 83(7) (2015) 2926–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Co JY, Margalef-Català M, Li X, Mah AT, Kuo CJ, Monack DM, Amieva MR, Controlling Epithelial Polarity: A Human Enteroid Model for Host-Pathogen Interactions, Cell Rep 26(9) (2019) 2509–2520.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Williamson IA, Arnold JW, Samsa LA, Gaynor L, DiSalvo M, Cocchiaro JL, Carroll I, Azcarate-Peril MA, Rawls JF, Allbritton NL, Magness ST, A High-Throughput Organoid Microinjection Platform to Study Gastrointestinal Microbiota and Luminal Physiology, Cell Mol Gastroenterol Hepatol 6(3) (2018) 301–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Garcez PP, Loiola EC, Madeiro da Costa R, Higa LM, Trindade P, Delvecchio R, Nascimento JM, Brindeiro R, Tanuri A, Rehen SK, Zika virus impairs growth in human neurospheres and brain organoids, Science 352(6287) (2016) 816–8. [DOI] [PubMed] [Google Scholar]
- [76].Gabriel E, Ramani A, Karow U, Gottardo M, Natarajan K, Gooi LM, Goranci-Buzhala G, Krut O, Peters F, Nikolic M, Kuivanen S, Korhonen E, Smura T, Vapalahti O, Papantonis A, Schmidt-Chanasit J, Riparbelli M, Callaini G, Krönke M, Utermöhlen O, Gopalakrishnan J, Recent Zika Virus Isolates Induce Premature Differentiation of Neural Progenitors in Human Brain Organoids, Cell Stem Cell 20(3) (2017) 397–406.e5. [DOI] [PubMed] [Google Scholar]
- [77].Dang J, Tiwari SK, Lichinchi G, Qin Y, Patil VS, Eroshkin AM, Rana TM, Zika Virus Depletes Neural Progenitors in Human Cerebral Organoids through Activation of the Innate Immune Receptor TLR3, Cell Stem Cell 19(2) (2016) 258–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, Neill FH, Blutt SE, Zeng XL, Qu L, Kou B, Opekun AR, Burrin D, Graham DY, Ramani S, Atmar RL, Estes MK, Replication of human noroviruses in stem cell-derived human enteroids, Science 353(6306) (2016) 1387–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Finkbeiner SR, Zeng XL, Utama B, Atmar RL, Shroyer NF, Estes MK, Stem cell-derived human intestinal organoids as an infection model for rotaviruses, mBio 3(4) (2012) e00159–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Heo I, Dutta D, Schaefer DA, Iakobachvili N, Artegiani B, Sachs N, Boonekamp KE, Bowden G, Hendrickx APA, Willems RJL, Peters PJ, Riggs MW, O’Connor R, Clevers H, Modelling Cryptosporidium infection in human small intestinal and lung organoids, Nat Microbiol 3(7) (2018) 814–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Lamers MM, Beumer J, van der Vaart J, Knoops K, Puschhof J, Breugem TI, Ravelli RBG, Paul van Schayck J, Mykytyn AZ, Duimel HQ, van Donselaar E, Riesebosch S, Kuijpers HJH, Schipper D, van de Wetering WJ, de Graaf M, Koopmans M, Cuppen E, Peters PJ, Haagmans BL, Clevers H, SARS-CoV-2 productively infects human gut enterocytes, Science 369(6499) (2020) 50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Zhang BZ, Chu H, Han S, Shuai H, Deng J, Hu YF, Gong HR, Lee AC, Zou Z, Yau T, Wu W, Hung IF, Chan JF, Yuen KY, Huang JD, SARS-CoV-2 infects human neural progenitor cells and brain organoids, Cell Res (2020) 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Jacob F, Pather SR, Huang WK, Wong SZH, Zhou H, Zhang F, Cubitt B, Chen CZ, Xu M, Pradhan M, Zhang DY, Zheng W, Bang AG, Song H, de ATJC, Ming GL, Human Pluripotent Stem Cell-Derived Neural Cells and Brain Organoids Reveal SARS-CoV-2 Neurotropism, bioRxiv (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Zhu L, She ZG, Cheng X, Qin JJ, Zhang XJ, Cai J, Lei F, Wang H, Xie J, Wang W, Li H, Zhang P, Song X, Chen X, Xiang M, Zhang C, Bai L, Xiang D, Chen MM, Liu Y, Yan Y, Liu M, Mao W, Zou J, Liu L, Chen G, Luo P, Xiao B, Zhang C, Zhang Z, Lu Z, Wang J, Lu H, Xia X, Wang D, Liao X, Peng G, Ye P, Yang J, Yuan Y, Huang X, Guo J, Zhang BH, Li H, Association of Blood Glucose Control and Outcomes in Patients with COVID-19 and Pre-existing Type 2 Diabetes, Cell Metab 31(6) (2020) 1068–1077.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Yang L, Han Y, Nilsson-Payant BE, Gupta V, Wang P, Duan X, Tang X, Zhu J, Zhao Z, Jaffré F, Zhang T, Kim TW, Harschnitz O, Redmond D, Houghton S, Liu C, Naji A, Ciceri G, Guttikonda S, Bram Y, Nguyen DT, Cioffi M, Chandar V, Hoagland DA, Huang Y, Xiang J, Wang H, Lyden D, Borczuk A, Chen HJ, Studer L, Pan FC, Ho DD, tenOever BR, Evans T, Schwartz RE, Chen S, A Human Pluripotent Stem Cell-based Platform to Study SARS-CoV-2 Tropism and Model Virus Infection in Human Cells and Organoids, Cell Stem Cell 27(1) (2020) 125–136.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Zhao B, Ni C, Gao R, Wang Y, Yang L, Wei J, Lv T, Liang J, Zhang Q, Xu W, Xie Y, Wang X, Yuan Z, Liang J, Zhang R, Lin X, Recapitulation of SARS-CoV-2 infection and cholangiocyte damage with human liver ductal organoids, Protein Cell 11(10) (2020) 771–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Monteil V, Kwon H, Prado P, Hagelkrüys A, Wimmer RA, Stahl M, Leopoldi A, Garreta E, Hurtado Del Pozo C, Prosper F, Romero JP, Wirnsberger G, Zhang H, Slutsky AS, Conder R, Montserrat N, Mirazimi A, Penninger JM, Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2, Cell 181(4) (2020) 905–913.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Mahalingam R, Dharmalingam P, Santhanam A, Kotla S, Davuluri G, Karmouty-Quintana H, Ashrith G, Thandavarayan RA, Single-cell RNA sequencing analysis of SARS-CoV-2 entry receptors in human organoids, J Cell Physiol. 2021. April; 236(4):2950–2958.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Xia S, Wu M, Chen S, Zhang T, Ye L, Liu J, Li H, Long Term Culture of Human Kidney Proximal Tubule Epithelial Cells Maintains Lineage Functions and Serves as an Ex vivo Model for Coronavirus Associated Kidney Injury, Virol Sin 35(3) (2020) 311–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Wang S, Li W, Hui H, Tiwari SK, Zhang Q, Croker BA, Rawlings S, Smith D, Carlin AF, Rana TM, Cholesterol 25-Hydroxylase inhibits SARS-CoV-2 and coronaviruses by depleting membrane cholesterol, Embo j (2020) e2020106057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Heidari-Khoei H, Esfandiari F, Hajari MA, Ghorbaninejad Z, Piryaei A, Baharvand H, Organoid technology in female reproductive biomedicine, Reprod Biol Endocrinol 18(1) (2020) 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Lee Y, Dizzell SE, Leung V, Nazli A, Zahoor MA, Fichorova RN, Kaushic C, Effects of Female Sex Hormones on Susceptibility to HSV-2 in Vaginal Cells Grown in Air-Liquid Interface, Viruses 8(9) (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Kessler M, Hoffmann K, Fritsche K, Brinkmann V, Mollenkopf HJ, Thieck O, Teixeira da Costa AR, Braicu EI, Sehouli J, Mangler M, Berger H, Meyer TF, Chronic Chlamydia infection in human organoids increases stemness and promotes age-dependent CpG methylation, Nat Commun 10(1) (2019) 1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Seino T, Kawasaki S, Shimokawa M, Tamagawa H, Toshimitsu K, Fujii M, Ohta Y, Matano M, Nanki K, Kawasaki K, Takahashi S, Sugimoto S, Iwasaki E, Takagi J, Itoi T, Kitago M, Kitagawa Y, Kanai T, Sato T, Human Pancreatic Tumor Organoids Reveal Loss of Stem Cell Niche Factor Dependence during Disease Progression, Cell Stem Cell 22(3) (2018) 454–467.e6. [DOI] [PubMed] [Google Scholar]
- [95].Biffi G, Oni TE, Spielman B, Hao Y, Elyada E, Park Y, Preall J, Tuveson DA, IL1-Induced JAK/STAT Signaling Is Antagonized by TGFβ to Shape CAF Heterogeneity in Pancreatic Ductal Adenocarcinoma, Cancer Discov 9(2) (2019) 282–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].DeNardo DG, Barreto JB, Andreu P, Vasquez L, Tawfik D, Kolhatkar N, Coussens LM, CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages, Cancer Cell 16(2) (2009) 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Tsai S, McOlash L, Palen K, Johnson B, Duris C, Yang Q, Dwinell MB, Hunt B, Evans DB, Gershan J, James MA, Development of primary human pancreatic cancer organoids, matched stromal and immune cells and 3D tumor microenvironment models, BMC Cancer 18(1) (2018) 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Chatterjee S, Bhat V, Berdnikov A, Liu J, Zhang G, Buchel E, Safneck J, Marshall AJ, Murphy LC, Postovit LM, Raouf A, Paracrine Crosstalk between Fibroblasts and ER(+) Breast Cancer Cells Creates an IL1β-Enriched Niche that Promotes Tumor Growth, iScience 19 (2019) 388–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Fernández-Periáñez R, Molina-Privado I, Rojo F, Guijarro-Muñoz I, Alonso-Camino V, Zazo S, Compte M, Alvarez-Cienfuegos A, Cuesta AM, Sánchez-Martín D, Alvarez-Méndez AM, Sanz L, Alvarez-Vallina L, Basement membrane-rich organoids with functional human blood vessels are permissive niches for human breast cancer metastasis, PLoS One 8(8) (2013) e72957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Dekkers JF, Berkers G, Kruisselbrink E, Vonk A, de Jonge HR, Janssens HM, Bronsveld I, van de Graaf EA, Nieuwenhuis EE, Houwen RH, Vleggaar FP, Escher JC, de Rijke YB, Majoor CJ, Heijerman HG, de Winter-de Groot KM, Clevers H, van der Ent CK, Beekman JM, Characterizing responses to CFTR-modulating drugs using rectal organoids derived from subjects with cystic fibrosis, Sci Transl Med 8(344) (2016) 344ra84. [DOI] [PubMed] [Google Scholar]
- [101].Ahmadi S, Bozoky Z, Di Paola M, Xia S, Li C, Wong AP, Wellhauser L, Molinski SV, Ip W, Ouyang H, Avolio J, Forman-Kay JD, Ratjen F, Hirota JA, Rommens J, Rossant J, Gonska T, Moraes TJ, Bear CE, Phenotypic profiling of CFTR modulators in patient-derived respiratory epithelia, NPJ Genom Med 2 (2017) 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Molecular biology of Homo sapiens, Cold Spring Harb Symp Quant Biol 51 Pt 1 (1986) 1–702. [PubMed] [Google Scholar]
- [103].Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernández-Mateos J, Khan K, Lampis A, Eason K, Huntingford I, Burke R, Rata M, Koh DM, Tunariu N, Collins D, Hulkki-Wilson S, Ragulan C, Spiteri I, Moorcraft SY, Chau I, Rao S, Watkins D, Fotiadis N, Bali M, Darvish-Damavandi M, Lote H, Eltahir Z, Smyth EC, Begum R, Clarke PA, Hahne JC, Dowsett M, de Bono J, Workman P, Sadanandam A, Fassan M, Sansom OJ, Eccles S, Starling N, Braconi C, Sottoriva A, Robinson SP, Cunningham D, Valeri N, Patient-derived organoids model treatment response of metastatic gastrointestinal cancers, Science 359(6378) (2018) 920–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Tiriac H, Belleau P, Engle DD, Plenker D, Deschênes A, Somerville TDD, Froeling FEM, Burkhart RA, Denroche RE, Jang GH, Miyabayashi K, Young CM, Patel H, Ma M, LaComb JF, Palmaira RLD, Javed AA, Huynh JC, Johnson M, Arora K, Robine N, Shah M, Sanghvi R, Goetz AB, Lowder CY, Martello L, Driehuis E, LeComte N, Askan G, Iacobuzio-Donahue CA, Clevers H, Wood LD, Hruban RH, Thompson E, Aguirre AJ, Wolpin BM, Sasson A, Kim J, Wu M, Bucobo JC, Allen P, Sejpal DV, Nealon W, Sullivan JD, Winter JM, Gimotty PA, Grem JL, DiMaio DJ, Buscaglia JM, Grandgenett PM, Brody JR, Hollingsworth MA, O’Kane GM, Notta F, Kim E, Crawford JM, Devoe C, Ocean A, Wolfgang CL, Yu KH, Li E, Vakoc CR, Hubert B, Fischer SE, Wilson JM, Moffitt R, Knox J, Krasnitz A, Gallinger S, Tuveson DA, Organoid Profiling Identifies Common Responders to Chemotherapy in Pancreatic Cancer, Cancer Discov 8(9) (2018) 1112–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Linkous A, Balamatsias D, Snuderl M, Edwards L, Miyaguchi K, Milner T, Reich B, Cohen-Gould L, Storaska A, Nakayama Y, Schenkein E, Singhania R, Cirigliano S, Magdeldin T, Lin Y, Nanjangud G, Chadalavada K, Pisapia D, Liston C, Fine HA, Modeling Patient-Derived Glioblastoma with Cerebral Organoids, Cell Rep 26(12) (2019) 3203–3211.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Pauli C, Hopkins BD, Prandi D, Shaw R, Fedrizzi T, Sboner A, Sailer V, Augello M, Puca L, Rosati R, McNary TJ, Churakova Y, Cheung C, Triscott J, Pisapia D, Rao R, Mosquera JM, Robinson B, Faltas BM, Emerling BE, Gadi VK, Bernard B, Elemento O, Beltran H, Demichelis F, Kemp CJ, Grandori C, Cantley LC, Rubin MA, Personalized In Vitro and In Vivo Cancer Models to Guide Precision Medicine, Cancer Discov 7(5) (2017) 462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Weeber F, Ooft SN, Dijkstra KK, Voest EE, Tumor Organoids as a Pre-clinical Cancer Model for Drug Discovery, Cell Chem Biol 24(9) (2017) 1092–1100. [DOI] [PubMed] [Google Scholar]
- [108].Kaushik G, Seshacharyulu P, Rauth S, Nallasamy P, Rachagani S, Nimmakayala RK, Vengoji R, Mallya K, Chirravuri-Venkata R, Singh AB, Foster JM, Ly QP, Smith LM, Lele SM, Malafa MP, Jain M, Ponnusamy MP, Batra SK, Selective inhibition of stemness through EGFR/FOXA2/SOX9 axis reduces pancreatic cancer metastasis, Oncogene (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Subramanian A, Sidhom EH, Emani M, Vernon K, Sahakian N, Zhou Y, Kost-Alimova M, Slyper M, Waldman J, Dionne D, Nguyen LT, Weins A, Marshall JL, Rosenblatt-Rosen O, Regev A, Greka A, Single cell census of human kidney organoids shows reproducibility and diminished off-target cells after transplantation, Nat Commun 10(1) (2019) 5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Driehuis E, Kretzschmar K, Clevers H, Establishment of patient-derived cancer organoids for drug-screening applications, Nat Protoc 15(10) (2020) 3380–3409. [DOI] [PubMed] [Google Scholar]
- [111].Velasco S, Kedaigle AJ, Simmons SK, Nash A, Rocha M, Quadrato G, Paulsen B, Nguyen L, Adiconis X, Regev A, Levin JZ, Arlotta P, Individual brain organoids reproducibly form cell diversity of the human cerebral cortex, Nature 570(7762) (2019) 523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Brazovskaja A, Treutlein B, Camp JG, High-throughput single-cell transcriptomics on organoids, Curr Opin Biotechnol 55 (2019) 167–171. [DOI] [PubMed] [Google Scholar]
- [113].Gonneaud A, Asselin C, Boudreau F, Boisvert FM, Phenotypic Analysis of Organoids by Proteomics, Proteomics 17(20) (2017). [DOI] [PubMed] [Google Scholar]
- [114].Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, Clevers H, Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium, Gastroenterology 141(5) (2011) 1762–72. [DOI] [PubMed] [Google Scholar]
- [115].Basak O, Beumer J, Wiebrands K, Seno H, van Oudenaarden A, Clevers H, Induced Quiescence of Lgr5+ Stem Cells in Intestinal Organoids Enables Differentiation of Hormone-Producing Enteroendocrine Cells, Cell Stem Cell 20(2) (2017) 177–190.e4. [DOI] [PubMed] [Google Scholar]
- [116].Grün D, Lyubimova A, Kester L, Wiebrands K, Basak O, Sasaki N, Clevers H, van Oudenaarden A, Single-cell messenger RNA sequencing reveals rare intestinal cell types, Nature 525(7568) (2015) 251–5. [DOI] [PubMed] [Google Scholar]
- [117].Haber AL, Biton M, Rogel N, Herbst RH, Shekhar K, Smillie C, Burgin G, Delorey TM, Howitt MR, Katz Y, Tirosh I, Beyaz S, Dionne D, Zhang M, Raychowdhury R, Garrett WS, Rozenblatt-Rosen O, Shi HN, Yilmaz O, Xavier RJ, Regev A, A single-cell survey of the small intestinal epithelium, Nature 551(7680) (2017) 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Biton M, Haber AL, Rogel N, Burgin G, Beyaz S, Schnell A, Ashenberg O, Su CW, Smillie C, Shekhar K, Chen Z, Wu C, Ordovas-Montanes J, Alvarez D, Herbst RH, Zhang M, Tirosh I, Dionne D, Nguyen LT, Xifaras ME, Shalek AK, von Andrian UH, Graham DB, Rozenblatt-Rosen O, Shi HN, Kuchroo V, Yilmaz OH, Regev A, Xavier RJ, T Helper Cell Cytokines Modulate Intestinal Stem Cell Renewal and Differentiation, Cell 175(5) (2018) 1307–1320.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Mallanna SK, Duncan SA, Differentiation of hepatocytes from pluripotent stem cells, Curr Protoc Stem Cell Biol 26 (2013) 1g.4.1–1g.4.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Asumda FZ, Hatzistergos KE, Dykxhoorn DM, Jakubski S, Edwards J, Thomas E, Schiff ER, Differentiation of hepatocyte-like cells from human pluripotent stem cells using small molecules, Differentiation 101 (2018) 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Cahan P, Li H, Morris SA, Lummertz da Rocha E, Daley GQ, Collins JJ, CellNet: network biology applied to stem cell engineering, Cell 158(4) (2014) 903–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Asai A, Aihara E, Watson C, Mourya R, Mizuochi T, Shivakumar P, Phelan K, Mayhew C, Helmrath M, Takebe T, Wells J, Bezerra JA, Paracrine signals regulate human liver organoid maturation from induced pluripotent stem cells, Development 144(6) (2017) 1056–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Kegel V, Deharde D, Pfeiffer E, Zeilinger K, Seehofer D, Damm G, Protocol for Isolation of Primary Human Hepatocytes and Corresponding Major Populations of Non-parenchymal Liver Cells, J Vis Exp (109) (2016) e53069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Broutier L, Mastrogiovanni G, Verstegen MM, Francies HE, Gavarró LM, Bradshaw CR, Allen GE, Arnes-Benito R, Sidorova O, Gaspersz MP, Georgakopoulos N, Koo BK, Dietmann S, Davies SE, Praseedom RK, Lieshout R, JNM IJ, Wigmore SJ, Saeb-Parsy K, Garnett MJ, van der Laan LJ, Huch M, Human primary liver cancer-derived organoid cultures for disease modeling and drug screening, Nat Med 23(12) (2017) 1424–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, Johns AL, Miller D, Nones K, Quek K, Quinn MC, Robertson AJ, Fadlullah MZ, Bruxner TJ, Christ AN, Harliwong I, Idrisoglu S, Manning S, Nourse C, Nourbakhsh E, Wani S, Wilson PJ, Markham E, Cloonan N, Anderson MJ, Fink JL, Holmes O, Kazakoff SH, Leonard C, Newell F, Poudel B, Song S, Taylor D, Waddell N, Wood S, Xu Q, Wu J, Pinese M, Cowley MJ, Lee HC, Jones MD, Nagrial AM, Humphris J, Chantrill LA, Chin V, Steinmann AM, Mawson A, Humphrey ES, Colvin EK, Chou A, Scarlett CJ, Pinho AV, Giry-Laterriere M, Rooman I, Samra JS, Kench JG, Pettitt JA, Merrett ND, Toon C, Epari K, Nguyen NQ, Barbour A, Zeps N, Jamieson NB, Graham JS, Niclou SP, Bjerkvig R, Grützmann R, Aust D, Hruban RH, Maitra A, Iacobuzio-Donahue CA, Wolfgang CL, Morgan RA, Lawlor RT, Corbo V, Bassi C, Falconi M, Zamboni G, Tortora G, Tempero MA, Gill AJ, Eshleman JR, Pilarsky C, Scarpa A, Musgrove EA, Pearson JV, Biankin AV, Grimmond SM, Whole genomes redefine the mutational landscape of pancreatic cancer, Nature 518(7540) (2015) 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].McDonald OG, Li X, Saunders T, Tryggvadottir R, Mentch SJ, Warmoes MO, Word AE, Carrer A, Salz TH, Natsume S, Stauffer KM, Makohon-Moore A, Zhong Y, Wu H, Wellen KE, Locasale JW, Iacobuzio-Donahue CA, Feinberg AP, Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis, Nat Genet 49(3) (2017) 367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Roe JS, Hwang CI, Somerville TDD, Milazzo JP, Lee EJ, Da Silva B, Maiorino L, Tiriac H, Young CM, Miyabayashi K, Filippini D, Creighton B, Burkhart RA, Buscaglia JM, Kim EJ, Grem JL, Lazenby AJ, Grunkemeyer JA, Hollingsworth MA, Grandgenett PM, Egeblad M, Park Y, Tuveson DA, Vakoc CR, Enhancer Reprogramming Promotes Pancreatic Cancer Metastasis, Cell 170(5) (2017) 875–888.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Miyabayashi K, Baker LA, Deschênes A, Traub B, Caligiuri G, Plenker D, Alagesan B, Belleau P, Li S, Kendall J, Jang GH, Kawaguchi RK, Somerville TDD, Tiriac H, Hwang CI, Burkhart RA, Roberts NJ, Wood LD, Hruban RH, Gillis J, Krasnitz A, Vakoc CR, Wigler M, Notta F, Gallinger S, Park Y, Tuveson DA, Intraductal Transplantation Models of Human Pancreatic Ductal Adenocarcinoma Reveal Progressive Transition of Molecular Subtypes, Cancer Discov 10(10) (2020) 1566–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Boj SF, Hwang CI, Baker LA, Chio II, Engle DD, Corbo V, Jager M, Ponz-Sarvise M, Tiriac H, Spector MS, Gracanin A, Oni T, Yu KH, van Boxtel R, Huch M, Rivera KD, Wilson JP, Feigin ME, Öhlund D, Handly-Santana A, Ardito-Abraham CM, Ludwig M, Elyada E, Alagesan B, Biffi G, Yordanov GN, Delcuze B, Creighton B, Wright K, Park Y, Morsink FH, Molenaar IQ, Borel Rinkes IH, Cuppen E, Hao Y, Jin Y, Nijman IJ, Iacobuzio-Donahue C, Leach SD, Pappin DJ, Hammell M, Klimstra DS, Basturk O, Hruban RH, Offerhaus GJ, Vries RG, Clevers H, Tuveson DA, Organoid models of human and mouse ductal pancreatic cancer, Cell 160(1–2) (2015) 324–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Somerville TDD, Xu Y, Miyabayashi K, Tiriac H, Cleary CR, Maia-Silva D, Milazzo JP, Tuveson DA, Vakoc CR, TP63-Mediated Enhancer Reprogramming Drives the Squamous Subtype of Pancreatic Ductal Adenocarcinoma, Cell Rep 25(7) (2018) 1741–1755.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SG, Hoadley KA, Rashid NU, Williams LA, Eaton SC, Chung AH, Smyla JK, Anderson JM, Kim HJ, Bentrem DJ, Talamonti MS, Iacobuzio-Donahue CA, Hollingsworth MA, Yeh JJ, Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma, Nat Genet 47(10) (2015) 1168–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, Yao B, Hamersky GR, Jacob F, Zhong C, Yoon KJ, Jeang W, Lin L, Li Y, Thakor J, Berg DA, Zhang C, Kang E, Chickering M, Nauen D, Ho CY, Wen Z, Christian KM, Shi PY, Maher BJ, Wu H, Jin P, Tang H, Song H, Ming GL, Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure, Cell 165(5) (2016) 1238–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA, Cerebral organoids model human brain development and microcephaly, Nature 501(7467) (2013) 373–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Luo C, Lancaster MA, Castanon R, Nery JR, Knoblich JA, Ecker JR, Cerebral Organoids Recapitulate Epigenomic Signatures of the Human Fetal Brain, Cell Rep 17(12) (2016) 3369–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Zhao X, Bhattacharyya A, Human Models Are Needed for Studying Human Neurodevelopmental Disorders, Am J Hum Genet 103(6) (2018) 829–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Greenland JC, Williams-Gray CH, Barker RA, The clinical heterogeneity of Parkinson’s disease and its therapeutic implications, Eur J Neurosci 49(3) (2019) 328–338. [DOI] [PubMed] [Google Scholar]
- [137].Erro R, Vitale C, Amboni M, Picillo M, Moccia M, Longo K, Santangelo G, De Rosa A, Allocca R, Giordano F, Orefice G, De Michele G, Santoro L, Pellecchia MT, Barone P, The heterogeneity of early Parkinson’s disease: a cluster analysis on newly diagnosed untreated patients, PLoS One 8(8) (2013) e70244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Jeste SS, Geschwind DH, Disentangling the heterogeneity of autism spectrum disorder through genetic findings, Nat Rev Neurol 10(2) (2014) 74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Yakoub AM, Cerebral organoids exhibit mature neurons and astrocytes and recapitulate electrophysiological activity of the human brain, Neural Regen Res 14(5) (2019) 757–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Balkissoon R, Stem Cell Therapy for COPD: Where are we?, Chronic Obstr Pulm Dis 5(2) (2018) 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].de Poel E, Lefferts JW, Beekman JM, Intestinal organoids for Cystic Fibrosis research, J Cyst Fibros 19 Suppl 1 (2020) S60–s64. [DOI] [PubMed] [Google Scholar]
- [142].Strikoudis A, Cieślak A, Loffredo L, Chen YW, Patel N, Saqi A, Lederer DJ, Snoeck HW, Modeling of Fibrotic Lung Disease Using 3D Organoids Derived from Human Pluripotent Stem Cells, Cell Rep 27(12) (2019) 3709–3723.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]