Abstract
As one of the common abiotic stresses, chilling stress has negative effects on rice growth and development. Minimization of these adverse effects through various ways is vital for the productivity of rice. Nanoparticles (NPs) serve as one of the effective alleviation methods against abiotic stresses. In our research, zinc oxide (ZnO) NPs were utilized as foliar sprays on rice leaves to explore the mechanism underlying the effect of NPs against the negative impact of chilling stress on rice seedlings. We revealed that foliar application of ZnO NPs significantly alleviated chilling stress in hydroponically grown rice seedlings, including improved plant height, root length, and dry biomass. Besides, ZnO NPs also restored chlorophyll accumulation and significantly ameliorated chilling-induced oxidative stress with reduced levels of H2O2, MDA, proline, and increased activities of major antioxidative enzymes, superoxide dismutase (SOD), catalase (CAT), and peroxidase (POD). We further found that foliar application of ZnO NPs induced the chilling-induced gene expression of the antioxidative system (OsCu/ZnSOD1, OsCu/ZnSOD2, OsCu/ZnSOD3, OsPRX11, OsPRX65, OsPRX89, OsCATA, and OsCATB) and chilling response transcription factors (OsbZIP52, OsMYB4, OsMYB30, OsNAC5, OsWRKY76, and OsWRKY94) in leaves of chilling-treated seedlings. Taken together, our results suggest that foliar application of ZnO NPs could alleviate chilling stress in rice via the mediation of the antioxidative system and chilling response transcription factors.
Keywords: zinc oxide nanoparticles, chilling stress, rice, chilling response, antioxidative system
1. Introduction
Rice (Oryza sativa L.), a major crop in the world and the staple food for over half of the world’s population, is mostly a tropical crop, but its distribution also extends to temperate and subtropical regions [1,2]. However, rice usually encounters environmental stresses such as extreme temperatures, drought, flooding, and high salinity [3], which greatly influence the growth and development of rice. Temperature is one of the vital factors for the production of rice, as the ideal temperature for rice growth ranges from 25 to 30 °C [4]. As previously reported, the growth and germination of the rice plant is negatively influenced when the temperature of the environment declines below 15 °C, and chilling stress that occurs below 20 °C in the stages of grain-filling, booting, and flowering would result in an impossibility to grow rice on about 7 × 106 hectares of land in Southeast and South Asia [5,6,7]. Therefore, discovering the underlying mechanisms of the chilling response in rice will offer a theoretical basis for breeding chilling-tolerant varieties.
Chilling stress adversely affects the metabolic and physiological functions of rice, and thus reduces the yield [8,9]. The effects of chilling stress on rice growth and development involve different physiological pathways: affecting photosynthesis through the inhibition of chloroplast formation and chlorophyll biosynthesis in rice leaves [10]; inducing the accumulation of signal substances, e.g., malondialdehyde (MDA) and reactive oxygen species (ROS) [11,12]; accumulating compatible osmolytes, e.g., free proline [13] and impacting antioxidants, such as superoxide dismutase (SOD), catalase (CAT), and peroxidase (POD) [14,15].
Besides, there are numerous transcription factors involved in the chilling response and tolerance of rice, including bZIP (basic leucine zipper), MYB (v-myb avian myeloblastosis viral oncogene homolog), NAC (NAM, ATAF, and CUC), and WRKY (tryptophan-arginine-lysine-tyrosine) [16]. A recent study revealed that one bZIP transcription factor, OsbZIP52, suppresses chilling resistance by inhibiting chilling-stress-related genes (e.g., OsTPP1) [16]. Multiple studies reported that MYB transcription factors play a vital role in chilling tolerance. For instance, the R2R3-type OsMYB4 transcription factor regulates the rice chilling stress response independently via OsDREBs [17,18]. OsMYB30 inhibits the chilling tolerance by down-regulating the gene expression of β-AMYLASE (BMY) via interaction with JASMONATE ZIM-DOMAIN 9 (OsJAZ9) [19]. Moreover, a NAC transcription factor encoding gene, OsNAC5, enhances the tolerance of transgenic rice seedlings to chilling stress [20]. Furthermore, WRKY transcription factors, OsWRKY76 and OsWRKY94, confer chilling tolerance [21,22].
Minimization of chilling stress through various ways is vital for the productivity of rice. Nanoparticles (NPs) serve as an effective alleviation method against abiotic stresses. Advances in nanomaterials (NMs) can rise the production of crops in the present opposing environment. Present studies have shown that nanotechnology is able to enhance the development and growth of crops and alleviate the negative effects of abiotic stress [23]. Mohammadi et al. found that the application of TiO2 NPs not only alleviated the membrane damage under cold stress but also prevented oxidative stress in chickpeas [24]. TiO2 treatment also increased the crop tolerance to cold stress via maintaining the stability of carotenoid and chlorophyll accumulations, inducing the activity of superoxide dismutase, ascorbate peroxidase, and catalase [25], and enhancing the gene expression of chlorophyll- and rubisco-binding proteins [26].
Nonetheless, previous studies have shown that ZnO NPs could alleviate abiotic stress (e.g., toxicity of cadmium) in plants [27], while the function of ZnO NPs in mitigating chilling stress is largely unknown. For this reason, we sprayed ZnO NPs to observe the effect of ZnO NPs on alleviating chilling stress in this research. Finally, we found that foliar application of ZnO NPs mitigated the chilling stress in hydroponically grown rice seedlings with induced chlorophyll accumulation and reduced oxidative stress. Potential underlying mechanisms of ZnO NPs on alleviation of chilling stress are also discussed.
2. Results
2.1. Details of ZnO NPs
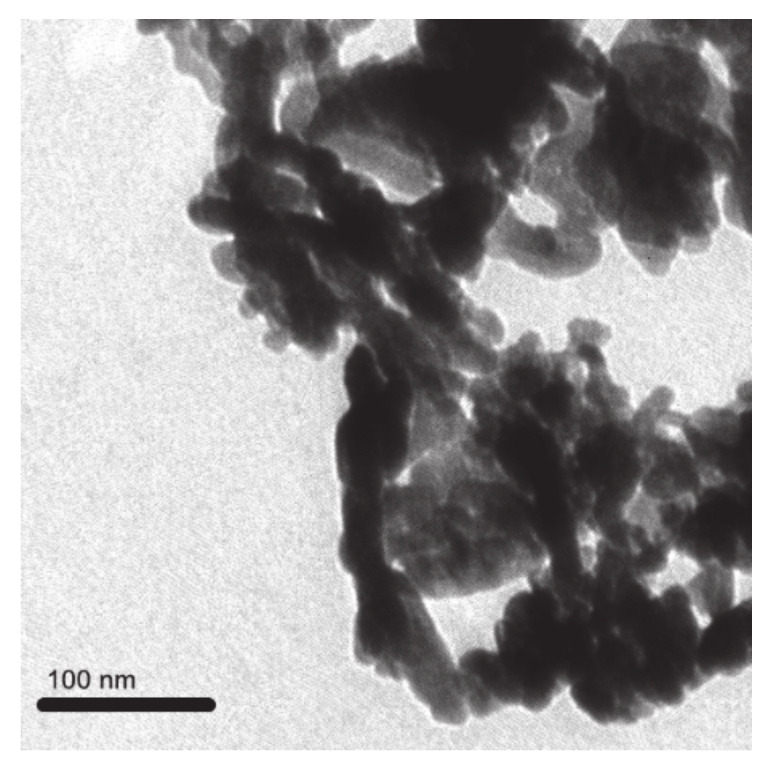
The NPs of ZnO were purchased from Chaowei Nano Technology Co., Ltd. (Shanghai, China). The particle morphology and size distribution of ZnO NPs were observed by transmission electron microscopy (TEM; Tecnai F20 S-TWIN, FEI Co., Hillsboro, OR, USA). The complete details of the ZnO NPs are shown in Table S1. The TEM images (Figure 1) revealed that the ZnO NPs were spherical with regular diameters of 30 nm. The size distribution of the ZnO NPs in media (50 mg/L TX-10) with a concentration of 100 mg/L was determined with a Malvern Zetasizer (Nano ZS, Malvern, UK). To minimize agglomeration, we sonicated the different concentrations of ZnO NPs suspensions for 1 h before use.
Figure 1.
Characterization of the zinc oxide nanoparticles (ZnO NPs). Transmission electron microscopy (TEM) images of the ZnO NPs. Scale bars, 100 nm.
2.2. ZnO NPs Significantly Mitigated Chilling Stress in Rice
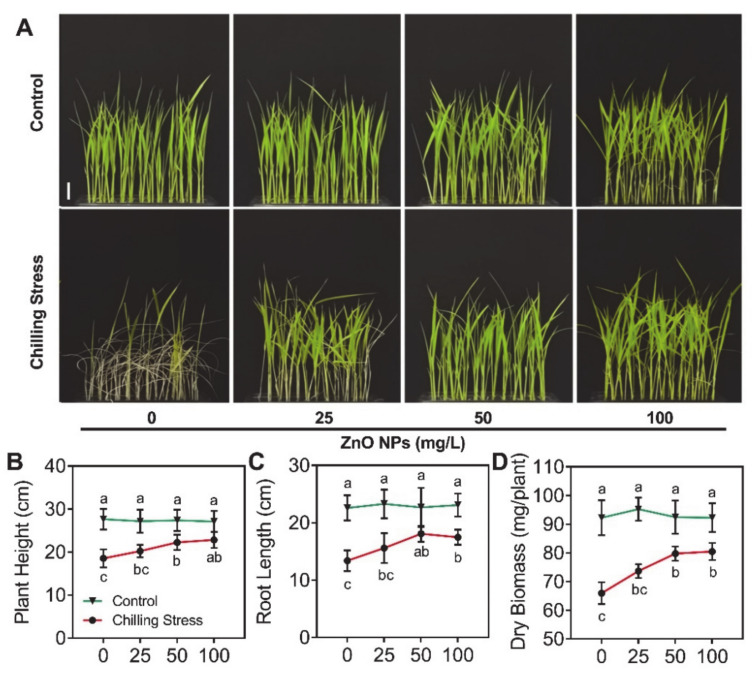
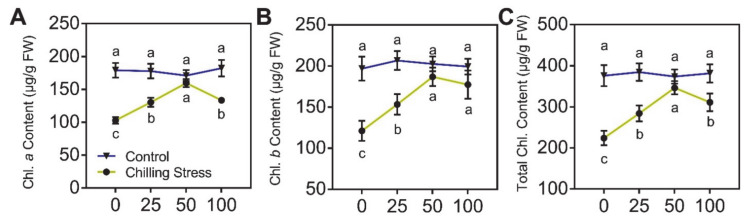
To evaluate the effect of ZnO NPs application on chilling stress, 20-day-old rice plants were exposed to the chilling condition (16 h light/8 h dark, at 10 °C). The low temperature (10 °C) caused toxicity in rice seedlings (Figure 2A). After 5 days of chilling stress treatment and 10 days of recovery, the plant height, root length, and dry biomass significantly decreased by 32.9%, 40.7%, and 28.5%, respectively compared with those of untreated control (Figure 2B–D). Rice seedlings also exhibited visible chlorosis in young leaves, and the content of total chlorophyll dramatically decreased by 40.4% after chilling stress treatment (Figure 3), compared to untreated control.
Figure 2.
ZnO NPs alleviate chilling stress in rice. Phenotype (A), plant height (B), root length (C), and dry biomass (D) of rice seedlings subjected to 0, 25, 50, and 100 mg/L of ZnO NPs under chilling stress were measured, respectively. The tests were repeated six times. The data given are the averages of six replicates, with the standard deviation (SD) shown by the error bars. Different letters above or below error bars show the differences at p < 0.05. Scale bar = 5 cm (A).
Figure 3.
Effects of ZnO NPs on the content of chlorophyll (Chl.) in rice seedlings under chilling stress. Chl. a (A), Chl. b (B), and total Chl. (C) in leaves of rice seedlings subjected to 0, 25, 50, and 100 mg/L of ZnO NPs under chilling stress were measured, respectively. The tests were repeated three times. The data given are the averages of three replicates, with the standard deviation (SD) shown by the error bars. Different letters above or below error bars show the differences at p < 0.05. FW, fresh weight.
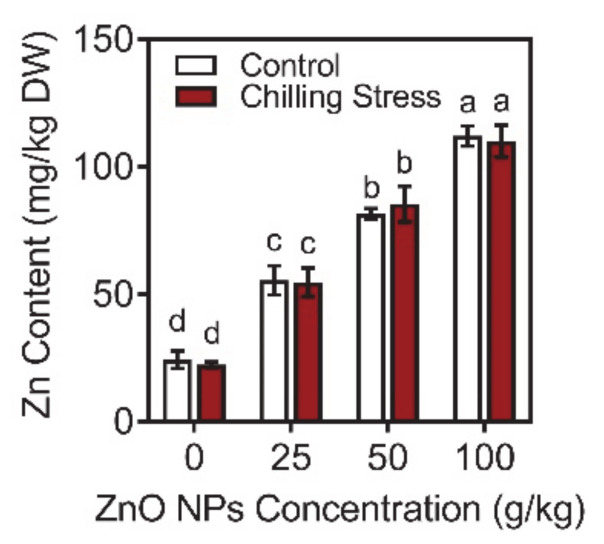
In contrast, the inhibition of growth of rice seedlings under chilling stress was alleviated after foliar application of ZnO NPs with different concentrations. The content of the Zn element in rice leaves was analyzed (Figure 4) after removal of ZnO NPs, which were physically adsorbed on the surfaces of leaves using a KI/I2 solution. After the application of 25, 50, and 100 mg/L ZnO NPs, the Zn content in leaves dramatically increased, namely 42.5-, 20.3-, and 45.0-fold, compared to the application of 0 mg/L ZnO NPs under chilling stress (Figure 4). Similar results were displayed without chilling stress (Figure 4). The supplementation with ZnO NPs displayed a good performance in the alleviation of chilling stress in rice seedlings, which was reflected in a significantly higher value of plant height, root length, and dry biomass of rice plants. Moreover, by spraying with different concentrations of ZnO NPs, the symptoms of chlorosis in rice leaves were significantly relieved and the accumulation of total chlorophyll was partially (25 mg/L) or totally (50 and 100 mg/L) restored (Figure 3).
Figure 4.
Zn accumulation in rice leaves. Zn accumulation of rice seedlings subjected to 0, 25, 50, and 100 mg/L of ZnO NPs under chilling stress were measured, respectively. The tests were repeated three times. The data given are the averages of three replicates, with the SD shown by the error bars. Different letters above error bars show the differences at p < 0.05. DW, dry weight.
2.3. Application of ZnO NPs Alleviated Chilling-Induced Oxidative Stress
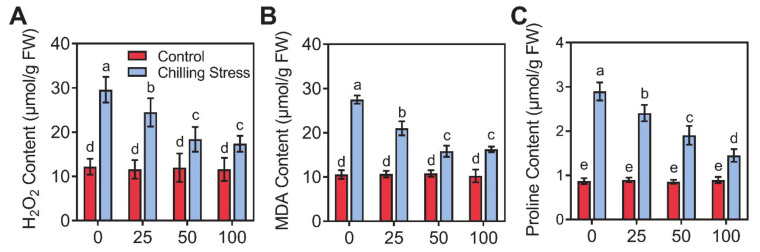
To further understand how ZnO NPs alleviated the chilling stress, we measured the contents of H2O2, MDA, and proline, and the activities of antioxidant enzymes (SOD, CAT, and POD) in rice plants (Figure 5 and Figure 6). On one hand, we found that chilling stress caused severe oxidative stress, as indicated by the increased ROS in non-radical form H2O2 (Figure 5A), which was severely toxic and caused serious damage to the plant cells. Chilling stress treatment strongly increased the levels of H2O2, MDA, and proline in rice leaves by 2.42-, 2.60-, and 3.33-fold, respectively, compared to control (Figure 5). The supplementation of 100 mg/L ZnO NPs significantly reduced H2O2, MDA, and proline contents in leaves by 41.2%, 40.9%, and 50.0%, respectively, compared to single chilling stress (Figure 5). On the other hand, we found that chilling stress greatly reduced the antioxidant enzymes (SOD, CAT, and POD) activity in leaves by 45.8%, 41.6%, and 47.7%, respectively, compared to control (Figure 6). The application of 100 mg/L ZnO NPs greatly increased the activities of SOD, CAT, and POD in chilling-treated rice seedlings compared to those under only chilling stress (Figure 6).
Figure 5.
Effects of ZnO NPs on oxidative damage in rice leaves under chilling stress. Hydrogen peroxide (H2O2, A), malondialdehyde (MDA, B), and proline (C) of rice seedlings subjected to 0, 25, 50, and 100 mg/L of ZnO NPs under chilling stress were measured, respectively. The tests were repeated three times. The data given are the averages of three replicates, with the SD shown by the error bars. Different letters above error bars show the differences at p < 0.05. FW, fresh weight.
Figure 6.
Effects of ZnO NPs on antioxidant enzyme activity in rice leaves under chilling stress. Superoxide dismutase (SOD, A), catalase (CAT, B), and peroxidase (POD, C) of rice seedlings subjected to 0, 25, 50, and 100 mg/L of ZnO NPs under chilling stress were measured, respectively. The tests were repeated three times. The data given are the averages of three replicates, with the SD shown by the error bars. Different letters above error bars show the differences at p < 0.05. FW, fresh weight.
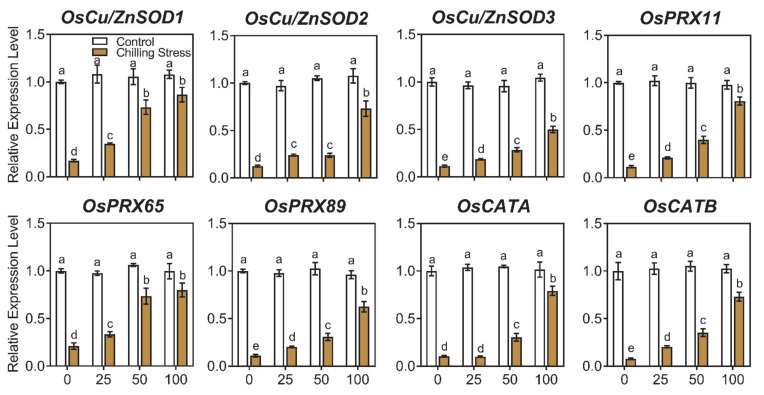
To investigate the molecular basis of how ZnO NPs reduces oxidative stress in rice, the gene expression of antioxidant enzymes containing OsCu/ZnSOD1, OsCu/ZnSOD2, OsCu/ZnSOD3, OsPRX11, OsPRX65, OsPRX89, OsCATA, and OsCATB was measured. Chilling stress significantly decreased the gene expression of antioxidant enzymes (SOD, CAT, and POD) compared to control (Figure 7 and Figure S1), while the application of different concentrations of ZnO NPs greatly increased the gene expression of SOD, CAT, and POD, in chilling-treated rice plants compared to only chilling stress (Figure 7 and Figure S1). These results indicated that ZnO NPs application could relieve oxidative stress in rice seedlings exposed to chilling stress by reducing ROS and enhancing the expression of antioxidant enzymes.
Figure 7.
Effects of ZnO NPs on gene expression of antioxidant enzymes in rice leaves under chilling stress. Gene expression of rice seedlings subjected to 0, 25, 50, and 100 mg/L of ZnO NPs under chilling stress was measured, respectively. Gene expression was first normalized to the UBQ5 gene in rice as an internal control and executed relative to each gene’s expression of control (assigned a value of 1). The data given are the averages of three replicates, with the SD shown by the error bars. Different letters above error bars show the differences at p < 0.05.
2.4. ZnO NPs Reduced Gene Expression of Chilling Response Transcription Factors under Chilling Stress in Rice
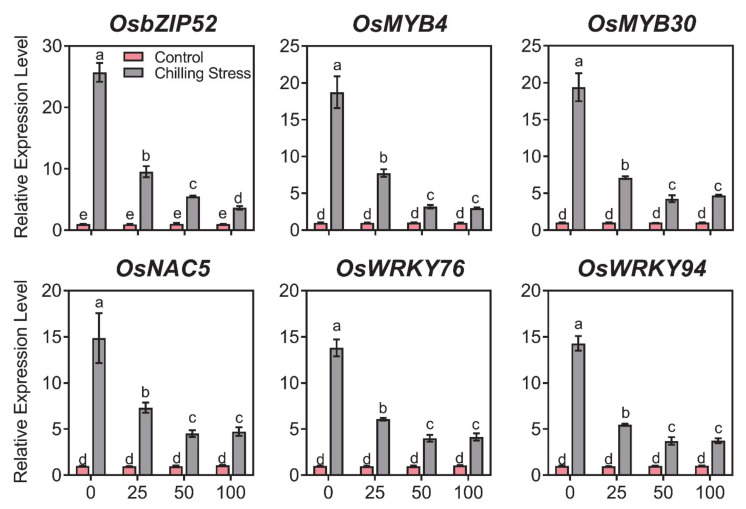
To investigate the molecular basis of how ZnO NPs alleviates chilling stress in rice, the genes of chilling response transcription factors including OsbZIP52, OsMYB4, OsMYB30, OsNAC5, OsWRKY76, and OsWRKY94 were chosen for gene expression analysis in rice leaves (Figure 8 and Figure S2). Exposure to chilling stress significantly induced the expression of OsbZIP52, OsMYB4, OsMYB30, OsNAC5, OsWRKY76, and OsWRKY94 in rice leaves. Application of ZnO NPs restored the expression of all above-mentioned genes to control level after chilling stress in rice leaves (Figure 8 and Figure S2). These results demonstrated that the reduced gene expression of the chilling response may be an important mechanism for the ZnO NPs-produced alleviation.
Figure 8.
Effects of ZnO NPs on gene expression of chilling response transcription factors in rice leaves under chilling stress. Gene expression of rice seedlings subjected to 0, 25, 50, and 100 mg/L of ZnO NPs under chilling stress was measured, respectively. Gene expression was first normalized to the UBQ5 gene in rice as an internal control and executed relative to each gene’s expression of control (assigned a value of 1). The data given are the averages of three replicates, with the SD shown by the error bars. Different letters above error bars show the differences at p < 0.05.
3. Discussion
Applications of ZnO NPs to resistance to stresses (drought stress, salinity stress, cadmium and lead toxicity) have been reported in several plants. The supplementation of ZnO NPs increased the seed germination percentage and rate, and decreased the seed fresh and dry weight of soybean (Glycine max L.) under drought stress [28]. Besides, under salinity stress, feeding with ZnO NPs also greatly improve plant growth and development in sunflower (Helianthus annuus L.) [29] and wheat (Triticum aestivum L.) [30]. Also, ZnO NPs alleviated cadmium (Cd) toxicity in wheat by enhancing wheat growth and Zn concentrations, while reducing the Cd concentration in plants [27]. ZnO NPs alleviated the bioavailability of cadmium and lead and changed the uptake of iron in hydroponically grown lettuce (Lactuca sativa L.) [31]. However, no related research has reported the effects of ZnO NPs on rice seedlings under chilling stress. In this study, we found that foliar application of ZnO NPs efficiently mitigated chilling stress in rice seedlings. In particular, ZnO NPs application significantly increased the resistance to chilling stress in rice by restoring chlorophyll biosynthesis, reducing the level of ROS, enhancing the activity of antioxidant enzymes, and regulating the gene expression of chilling stress response transcription factors.
Firstly, to confirm that the ZnO NPs were absorbed by rice plants, we measured the content of Zn element in rice leaves with different treatments (Figure 4). Consistent with previous reports on plants [32,33], our results revealed that the accumulation of ZnO NPs greatly increased under both control and chilling stress conditions in rice seedlings with different supplements of ZnO NPs (Figure 4), while there was no difference in the content of Zn element between control and chilling stress treatment under different supplements of ZnO NPs (Figure 4). These results demonstrated that with the increase in the amount of ZnO NPs sprayed, the Zn concentration in the organs of rice leaves increased but was not affected by chilling stress.
Chilling stress adversely affects the metabolic and physiological functions of rice, and thus reduces the yield [8,9,34,35]. Chilling stress impacts rice growth and development by affecting photosynthesis, with the inhibition of chloroplast formation and chlorophyll biosynthesis in rice leaves [10,36,37]. Similar to previous studies, rice seedlings also exhibited visible chlorosis in young leaves, and the content of total chlorophyll (chlorophyll a and b) was significantly reduced after the chilling stress treatment (Figure 3) compared to untreated control. Furthermore, by spraying with ZnO NPs, the symptoms of chlorosis in rice leaves were significantly relieved, and the accumulation of total chlorophyll was restored (Figure 3).
The oxidative bursts produced by chilling stress can induce the accumulation of signal substances, e.g., malondialdehyde (MDA) and reactive oxygen species (ROS) [11,12]; accumulate compatible osmolytes, e.g., free proline [13] and impact antioxidants, such as superoxide dismutase (SOD), catalase (CAT), and peroxidase (POD) [14,15]. However, the responses change in different higher plants and display dose-, tissue-, development-, and genotype-dependency [38]. The activity of CAT in rice markedly decreased, and the contents of H2O2 and MDA greatly increased, by chilling stress treatment compared to control [15]. The activity of SOD and CAT in some varieties of rice seedlings displayed small changes due to chilling stress, and other varieties of rice showed greatly reduced levels; however, the contents of H2O2 and MDA increased under chilling stress in most tested varieties of rice [39]. In our research, the activity of all of the antioxidant enzymes measured reduced after chilling stress (Figure 6), which was similar to the reports of Sohag et al. [15] and Wang et al. [39]. The gene expression of antioxidant enzymes containing OsCu/ZnSOD1, OsCu/ZnSOD2, OsCu/ZnSOD3, OsPRX11, OsPRX65, OsPRX89, OsCATA, and OsCATB was also induced with the application of ZnO NPs under chilling stress (Figure 7). Reduced SOD, CAT, and POD activity and gene expression might lead to an oxidative burst after chilling stress in rice seedlings, as revealed by the ROS level, such as H2O2 in chilling-treated rice, and finally caused induced lipid peroxidation, e.g., MDA (Figure 5, Figure 6 and Figure 7). Foliar application of ZnO NPs alleviated chilling-produced oxidative stress by enhancing antioxidant enzyme activity and reducing ROS content in rice seedlings. The oxidative stress was greatly mitigated by ZnO NPs, demonstrating that improving the antioxidant defense capacity may be an important mechanism for the ZnO NPs-produced alleviation.
Different transcription factors involved in the chilling tolerance and chilling response in rice, namely OsbZIP52 [16], OsMYB4 [17,18], OsMYB30 [19], OsNAC5 [20], OsWRKY76 [21], and OsWRKY94 [22], were reported to be greatly induced under chilling stress in related studies. In this study, all the above chilling response genes tested increased under chilling stress (Figure 8), which was similar to the former reports [16,17,18,19,20,21,22]. ZnO NPs reduced the expression of these chilling-stress-induced response genes, thereby alleviating various toxicities caused by chilling stress. Foliar application of ZnO NPs decreased the gene expression of the chilling response in rice seedlings, which demonstrated that the reduced gene expression of the chilling response may be an important mechanism for the ZnO NPs-produced alleviation.
In conclusion, we demonstrated that foliar application of ZnO NPs could efficiently mitigate the toxicity of chilling stress in rice. In particular, ZnO NPs significantly reduced the toxicity of chilling stress in rice leaves by regulating the gene expression of chilling stress response transcription factors, indicating ZnO NPs played a vital role in regulating the chilling response. ZnO NPs also had influential and multiple effects on plant growth and chlorophyll biosynthesis, finally enhancing the antioxidant potential and the abilities of ROS scavenging under chilling stress. Therefore, our findings not only illustrate the effect and mechanism of ZnO NPs in suppressing the toxicity of chilling stress in rice, but also provide a practicable solution for reducing chilling stress effects on crops.
4. Materials and Methods
4.1. Plant Materials and Treatments
The seeds of rice (Oryza sativa ssp Japonica cultivar Nipponbare) were sterilized with 20% (v/v) NaClO and then soaked in ultrapure water at 30 °C for 2 days. A total of 48 rice seeds were then transplanted into 96-well plastic hydroponic boxes including 1/2 Murashige-Skoog (MS) standard liquid medium, and planted in a growth chamber with 75% relative humidity (normal condition: 16 h light at 30 °C/8 h dark at 25 °C). For the foliage ZnO NPs spraying, 10-day-old rice plants grown in diverse boxes including 1/2 MS standard liquid medium under the normal condition were sprayed 50 mL ZnO NPs solution (25, 50, and 100 mg/L ZnO NPs with 50 mg/L TX-10) daily for 10 days. The 50 mg/L TX-10 replicates were utilized as controls. For the chilling stress treatment, 20-day-old rice plants were placed under the chilling condition (16 h light/8 h dark, at 10 °C) for 5 days. Then, the chilling-treated rice seedlings were moved to the normal condition for 1 week. The 32-day-old rice seedlings with or without ZnO NPs treatment were sampled for phenotypical, physiological, and molecular detections. Six replicates (the replicates here correspond to one plant) were tested for the measurements of plant height, root length, and dry biomass, and three replicates (the replicates here correspond to three to six plants depending on the weight) were detected for other molecular, physiological, and biochemical measurements.
4.2. Measurement of Zinc (Zn) Content
The concentration of Zn in rice leaves was measured according to a previous method [40]. Briefly, Zn absorbed on the surface of seedlings was eliminated by immersing the root in 20 mM Na2-EDTA (disodium ethylenediamine tetra-acetic acid) for 60 min, and then rinsing five times with distilled water. The sample of rice was dried at 105 °C for 120 min and at 60 °C for 48 h. The weight of whole seedlings was measured as dry biomass after reaching an unchanging weight. Dried plant samples (about 0.1 g) were mixed with 6 mL of HNO3 and kept in the reaction with a Multiwave (MARS 6, CEM Co., Matthews, NC, USA) at 140 °C for about 2 h. Finally, the content of Zn was measured with the atomic absorption spectrometer AA-7000 (Shimadzu, Tokyo, Japan).
4.3. Determination of Chlorophyll (Chl.)
The content of Chl. in rice leaves was quantified as previously reported [41]. About 100 mg of fresh rice leaves were mixed and ground with 10 mL 80% (v/v) acetone. After being kept in darkness for 2 h, the mixture was centrifuged at 12,000× g for 10 min. Finally, the absorbance at 663 and 645 nm was measured, respectively.
4.4. Hydrogen Peroxide (H2O2) Detection
The H2O2 content was measured using a hydrogen peroxide assay kit (Solarbio, Beijing, China) [42]. The content of H2O2 in rice leaves was detected by measuring the absorbance of titanium-peroxide complex formation at 415 nm.
4.5. Measurement of Malondialdehyde (MDA) Content
The accumulation of MDA was determined following a previously described protocol [43]. Approximately 100 mg of rice leaves were homogenized and mixed with 10 mL 10% (v/v) of trichloroacetic acid, and the mixture was then centrifuged at top speed (12,000× g) for 20 min. The supernatant was mixed with an equal volume of thiobarbituric acid and then kept at 95 °C for 30 min. The mixture was centrifuged at 12,000× g for 30 min after being quickly cooled on ice, and the supernatant absorbance at 450, 532, and 600 nm was measured, respectively.
4.6. Measurement of Free Proline (Pro)
The content of free proline was measured as previously reported [44]. About 1 g of rice leaves were ground with an aqueous solution of 3% (v/v) sulfosalicylic acid. After centrifugation at room temperature at 12,000× g for 15 min, 2 mL of supernatant was added to the same volumes of glacial acetic acid and acidic ninhydrin. This mixture was heated at 95 °C for 40 min, cooled on ice, and then followed by the addition of 4 mL of toluene to extract the colored reaction product separated from the aqueous phase. The absorbance of the toluene phase at 520 nm was determined, and the standard curve of proline was utilized to assay the accumulation of free proline.
4.7. Antioxidant Enzyme Activities Determination
The activity of the antioxidant enzymes containing superoxide dismutase (SOD), catalase (CAT), and peroxidase (POD) was measured using respective commercial kits (Solarbio, Beijing, China) as previously published [45,46,47]. The standardization of antioxidant enzymes activity was described: one unit of SOD activity was expressed as the suppression of photoreduction of 4-nitro blue tetrazolium chloride (NBT) by half under light; one unit of CAT activity was detected according to the decomposition of 1 nmol H2O2 per min; one unit of POD activity was assayed by determining an absorbance (470 nm) increase of 0.01 per min.
4.8. Gene Expression Assay
Relative gene expression was determined as previously published [48,49]. The RNAprep Pure Plant Kit (Tiangen, Beijing, China) and HiScript III 1st Strand cDNA Synthesis Kit (Vazyme Biotech Co., Ltd., Nanjing, China) were used to extract and reverse-transcribe the total RNA from rice leaves. AceQ qPCR SYBR Green Master Mix (Vazyme, Nanjing, China) was used to apply the quantitative real-time PCR (qRT-PCR). The qRT-PCR primers are listed in Table S2. The ACTIN gene in rice was used as an internal control (Table S2), and the level of gene expression was measured following the method of 2−ΔΔCt [50].
4.9. Statistical Analyses
The data in the figures and tables is presented as means with standard deviation (SD) because there were six or three replications. The mean differences were assessed using ANOVA. The statistical significance of differences between treatments was assessed using Bonferroni post-hoc tests.
Supplementary Materials
The following are available online. Figure S1: Expression profiles of antioxidant enzymes genes in rice leaves, Figure S2: Expression profiles of chilling response transcription factors genes in rice leaves, Table S1: Characterization details of ZnO NPs, Table S2: Primers used in the present study.
Author Contributions
Conceptualization, Y.S. and R.L.; methodology, M.J. and R.L.; software, H.Z.; validation, H.Z.; formal analysis, Y.S. and R.L.; investigation, Y.S., M.J., and R.L.; resources, R.L.; data curation, Y.S. and R.L.; writing—original draft preparation, Y.S.; writing—review and editing, M.J. and R.L.; visualization, M.J. and R.L.; supervision, R.L.; project administration, R.L.; funding acquisition, R.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Natural Foundation of Anhui Province (grant number: 1908085QC103), the Open Project Program of State Key Laboratory of Rice Biology (grant number: 20200104), Funding of Introduction & Stability for Talents supported by Anhui Agricultural University (grant number: yj2018-39), Postdoctoral Science Foundation of Zhejiang Province (grant number: ZJ2020157), and China Postdoctoral Science Foundation (grant number: 2020M680078).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in the article or Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds are not available from the authors.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sasaki T., Burr B. International rice genome sequencing project: The effort to completely sequence the rice genome. Curr. Opin. Plant Biol. 2000;3:138–141. doi: 10.1016/S1369-5266(99)00047-3. [DOI] [PubMed] [Google Scholar]
- 2.Kovach M.J., Sweeney M.T., McCouch S.R. New insights into the history of rice domestication. Trends Genet. 2007;23:578–587. doi: 10.1016/j.tig.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 3.Cramer G.R., Urano K., Delrot S., Pezzotti M., Shinozaki K. Effects of abiotic stress on plants: A systems biology perspective. BMC Plant Biol. 2011;11:163. doi: 10.1186/1471-2229-11-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Q., Chen Q., Wang S., Hong Y., Wang Z. Rice and cold stress: Methods for its evaluation and summary of cold tolerance-related quantitative trait loci. Rice. 2014;7:24. doi: 10.1186/s12284-014-0024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sales E., Viruel J., Domingo C., Marques L. Genome wide association analysis of cold tolerance at germination in temperate japonica rice (Oryza sativa L.) varieties. PLoS ONE. 2017;12:e0183416. doi: 10.1371/journal.pone.0183416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu C.T., Wang W., Mao B.G., Chu C.C. Cold stress tolerance in rice: Physiological changes, molecular mechanism, and future prospects. Hereditas (Beijing) 2018;40:171–185. doi: 10.16288/j.yczz.18-007. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 7.Sthapit B.R., Witcombe J.R. Inheritance of tolerance to chilling stress in rice during germination and plumule greening. Crop Sci. 1998;38:660–665. doi: 10.2135/cropsci1998.0011183X003800030007x. [DOI] [Google Scholar]
- 8.Ma Q., Dai X., Xu Y., Guo J., Liu Y., Chen N., Xiao J., Zhang D., Xu Z., Zhang X., et al. Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol. 2009;150:244–256. doi: 10.1104/pp.108.133454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arshad M.S., Farooq M., Asch F., Krishna J.S.V., Prasad P.V.V., Siddique K.H.M. Thermal stress impacts reproductive development and grain yield in rice. Plant Physiol. Biochem. 2017;115:57–72. doi: 10.1016/j.plaphy.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 10.Yang J.C., Li M., Xie X.Z., Han G., Sui N., Wang B.S. Deficiency of phytochrome B alleviates chilling-induced photoinhibition in rice. Am. J. Bot. 2013;100:1860–1870. doi: 10.3732/ajb.1200574. [DOI] [PubMed] [Google Scholar]
- 11.Sato Y., Masuta Y., Saito K., Murayama S., Ozawa K. Enhanced chilling tolerance at the booting stage in rice by transgenic overexpression of the ascorbate peroxidase gene, OsAPXa. Plant Cell Rep. 2011;30:399–406. doi: 10.1007/s00299-010-0985-7. [DOI] [PubMed] [Google Scholar]
- 12.Xie G., Kato H., Imai R. Biochemical identification of the OsMKK6-OsMPK3 signalling pathway for chilling stress tolerance in rice. Biochem. J. 2012;443:95–102. doi: 10.1042/BJ20111792. [DOI] [PubMed] [Google Scholar]
- 13.Huang J., Sun S., Xu D., Lan H., Sun H., Wang Z., Bao Y., Wang J., Tang H., Zhang H. A TFIIIA-type zinc finger protein confers multiple abiotic stress tolerances in transgenic rice (Oryza sativa L.) Plant Mol. Biol. 2012;80:337–350. doi: 10.1007/s11103-012-9955-5. [DOI] [PubMed] [Google Scholar]
- 14.Lu X., Zhou Y., Fan F., Peng J.H., Zhang J. Coordination of light, circadian clock with temperature: The potential mechanisms regulating chilling tolerance in rice. J. Integr. Plant Biol. 2019;62:737–760. doi: 10.1111/jipb.12852. [DOI] [PubMed] [Google Scholar]
- 15.Sohag A.A.M., Tahjib-Ul-Arif M., Afrin S., Khan M.K., Hannan M.A., Skalicky M., Mortuza M.G., Brestic M., Hossain M.A., Murata Y. Insights into nitric oxide-mediated water balance, antioxidant defence and mineral homeostasis in rice (Oryza sativa L.) under chilling stress. Nitric Oxide. 2020;100:7–16. doi: 10.1016/j.niox.2020.04.001. [DOI] [PubMed] [Google Scholar]
- 16.Liu C., Wu Y., Wang X. bZIP transcription factor OsbZIP52/RISBZ5: A potential negative regulator of cold and drought stress response in rice. Planta. 2012;235:1157–1169. doi: 10.1007/s00425-011-1564-z. [DOI] [PubMed] [Google Scholar]
- 17.Park M.R., Yun K.Y., Mohanty B., Herath V., Xu F., Wijaya E., Bajic V.B., Yun S.J., De Los Reyes B.G. Supraoptimal expression of the cold-regulated OsMyb4 transcription factor in transgenic rice changes the complexity of transcriptional network with major effects on stress tolerance and panicle development. Plant Cell Environ. 2010;33:2209–2230. doi: 10.1111/j.1365-3040.2010.02221.x. [DOI] [PubMed] [Google Scholar]
- 18.Tominaga-Wada R., Nukumizu Y. Expression analysis of an R3-Type MYB transcription factor CPC-LIKE MYB4 (TRICHOMELESS2) and CPL4-Related transcripts in Arabidopsis. Int. J. Mol. Sci. 2012;13:3478–3491. doi: 10.3390/ijms13033478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lv Y., Yang M., Hu D., Yang Z., Ma S., Li X., Xiong L. The OsMYB30 transcription factor suppresses cold tolerance by interacting with a JAZ protein and suppressing betaamylase expression. Plant Physiol. 2017;173:1475–1491. doi: 10.1104/pp.16.01725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song S.Y., Chen Y., Chen J., Dai X.Y., Zhang W.H. Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta. 2011;234:331–345. doi: 10.1007/s00425-011-1403-2. [DOI] [PubMed] [Google Scholar]
- 21.Yokotani N., Sato Y., Tanabe S., Chujo T., Shimizu T., Okada K., Yamane H., Shimono M., Sugano S., Takatsuji H., et al. WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J. Exp. Bot. 2013;64:5085–5097. doi: 10.1093/jxb/ert298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L., Zhao Y., Xu S., Zhang Z., Xu Y., Zhang J., Chong K. OsMADS57 together with OsTB1 coordinates transcription of its target OsWRKY94 and D14 to switch its organogenesis to defense for cold adaptation in rice. New Phytol. 2018;218:219–231. doi: 10.1111/nph.14977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shang Y., Hasan M.K., Ahammed G.J., Li M., Yin H., Zhou J. Applications of nanotechnology in plant growth and crop protection: A review. Molecules. 2019;24:2558. doi: 10.3390/molecules24142558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohammadi R., Amiri N.M., Mantri L. Effect of TiO2 nanoparticles on oxidative damage and antioxidant defense systems in chickpea seedlings during cold stress. Russ. J. Plant Physiol. 2013;61:768–775. doi: 10.1134/S1021443714050124. [DOI] [Google Scholar]
- 25.Mohammadi H., Esmailpour M., Gheranpaye A. Effects of TiO2 nanoparticles and water-deficit stress on morpho-physiological characteristics of dragonhead (Dracocephalum moldavica L.) plants. Environ. Toxicol. Chem. 2014;33:2429–2437. doi: 10.14720/aas.2016.107.2.11. [DOI] [Google Scholar]
- 26.Hasanpour H., Maali-Amiri R., Zeinali H. Effect of TiO2 nanoparticles on metabolic limitations to photosynthesis under cold in chickpea. Russ. J. Plant Physiol. 2015;62:779–787. [Google Scholar]
- 27.Hussain A., Ali S., Rizwan M., Rehman M.Z.U., Javed M.R., Imran M., Chatha S.A.S., Nazir R. Zinc oxide nanoparticles alter the wheat physiological response and reduce the cadmium uptake by plants. Environ. Pollut. 2018;242:1518–1526. doi: 10.1016/j.envpol.2018.08.036. [DOI] [PubMed] [Google Scholar]
- 28.Sedghi M., Hadi M., Toluie S.G. Effect of nano zinc oxide on the germination parameters of soybean seeds under drought stress. Ann. West Univ. Timis. 2013;16:73–78. [Google Scholar]
- 29.Torabian S., Zahedi M., Khoshgoftar A.H. Effects of foliar spray of two kinds of zinc oxide on the growth and ion concentration of sunflower cultivars under salt stress. J. Plant Nutr. 2016;39:172–180. doi: 10.1080/01904167.2015.1009107. [DOI] [Google Scholar]
- 30.Fathi A., Zahedi M., Torabian S., Khoshgoftar A. Response of wheat genotypes to foliar spray of ZnO and Fe2O3 nanoparticles under salt stress. J. Plant Nutr. 2017;40:1376–1385. doi: 10.1080/01904167.2016.1262418. [DOI] [Google Scholar]
- 31.Sharifan H., Ma X., Moore J.M., Habib M.R., Evans C. Zinc oxide nanoparticles alleviated the bioavailability of cadmium and lead and changed the uptake of iron in hydroponically grown lettuce (Lactuca sativa L. var Longifolia) ACS Sustain. Chem. Eng. 2019;7:16401–16409. doi: 10.1021/acssuschemeng.9b03531. [DOI] [Google Scholar]
- 32.Chen J., Liu X., Wang C., Yin S., Li X., Hu W., Simon M., Shen Z., Xiao Q., Chu C. Nitric oxide ameliorates zinc oxide nanoparticles-induced phytotoxicity in rice seedlings. J. Hazard. Mater. 2015;297:173–182. doi: 10.1016/j.jhazmat.2015.04.077. [DOI] [PubMed] [Google Scholar]
- 33.Yang Z., Chen J., Dou R., Gao X., Mao C., Wang L. Assessment of the phytotoxicity of metal oxide nanoparticles on two crop plants, maize (Zea mays L.) and rice (Oryza sativa L.) Int. J. Environ. Res. Public Health. 2015;12:15100–15109. doi: 10.3390/ijerph121214963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Badea C., Basu S. The effect of low temperature on metabolism of membrane lipids in plants and associated gene expression. Plant Omics. 2009;2:78–84. [Google Scholar]
- 35.Lv Y., Hussain M.A., Luo D., Tang N. Current understanding of genetic and molecular basis of cold tolerance in rice. Mol. Breed. 2019;39:159. doi: 10.1007/s11032-019-1073-5. [DOI] [Google Scholar]
- 36.Jeong S.W., Sun M.C., Dong S.L., Sang N.A., Park Y.I. Differential susceptibility of photosynthesis to light-chilling stress in rice (Oryza Sativa L.) depends on the capacity for photochemical dissipation of light. Mol. Cells. 2002;13:419–428. [PubMed] [Google Scholar]
- 37.Lu X., Song S., Xiao Y., Fan F., Peng J. Circadian clock-coordinated response to chilling stress in rice. Environ. Exp. Bot. 2021;185:104398. doi: 10.1016/j.envexpbot.2021.104398. [DOI] [Google Scholar]
- 38.Ryter S.W., Kim H.P., Hoetzel A., Park J.W., Nakahira K., Wang X., Choi A.M. Mechanisms of cell death in oxidative stress. Antioxid. Redox Sign. 2007;9:49–89. doi: 10.1089/ars.2007.9.49. [DOI] [PubMed] [Google Scholar]
- 39.Wang G.J., Miao W., Wang J.Y., Ma D.R., Li J.Q., Chen W.F. Effects of exogenous abscisic acid on antioxidant system in weedy and cultivated rice with different chilling sensitivity under chilling stress. J. Agron. Crop Sci. 2013;199:200–208. doi: 10.1111/jac.12004. [DOI] [Google Scholar]
- 40.Ding Y., Gong S., Wang Y., Wang F., Bao H., Sun J., Cai C., Yi K., Chen Z., Zhu C. MicroRNA166 modulates cadmium tolerance and accumulation in rice. Plant Physiol. 2018;177:1691–1703. doi: 10.1104/pp.18.00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan J., Shi M., Huang J.Z., Xu J., Wang Z.D., Guo D.P. Regulation of photosynthetic performance and antioxidant capacity by 60Co γ irradiation in Zizania latifolia plants. J. Environ. Radioactiv. 2014;129:33–42. doi: 10.1016/j.jenvrad.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 42.Jiang M., Jiang J., Li S., Li M., Tan Y.Y., Song S.Y., Shu Q.Y., Huang J.Z. Glutamate alleviates cadmium toxicity in rice via suppressing cadmium uptake and translocation. J. Hazard. Mater. 2020;384:121319. doi: 10.1016/j.jhazmat.2019.121319. [DOI] [PubMed] [Google Scholar]
- 43.Tang L., Cai H., Ji W., Luo X., Wang Z., Wu J., Wang X., Cui L., Wang Y., Zhu Y., et al. Overexpression of GsZFP1 enhances salt and drought tolerance in transgenic alfalfa (Medicago sativa L.) Plant Physiol. Bioch. 2013;71:22–30. doi: 10.1016/j.plaphy.2013.06.024. [DOI] [PubMed] [Google Scholar]
- 44.He J., Duan Y., Hua D., Fan G., Wang L., Liu Y., Chen Z., Han L., Qu L.J., Gong Z. DEXH Box RNA Helicase-mediated mitochondrial reactive oxygen species production in Arabidopsis mediates crosstalk between abscisic acid and auxin signaling. Plant Cell. 2012;24:1815–1833. doi: 10.1105/tpc.112.098707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aebi H. Catalase in vitro. Method Enzymol. 1984;105:121–126. doi: 10.1016/s0076-6879(84)05016-3. [DOI] [PubMed] [Google Scholar]
- 46.Hossain M.A., Hasanuzzaman M., Fujita M. Up-regulation of antioxidant and glyoxalase systems by exogenous glycinebetaine and proline in mung bean confer tolerance to cadmium stress. Physiol. Mol. Biol. Plants. 2010;16:259–272. doi: 10.1007/s12298-010-0028-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duan J., Zhang M., Zhang H., Xiong H., Liu P., Ali J., Li J., Li Z. OsMIOX, a myo-inositol oxygenase gene, improves drought tolerance through scavenging of reactive oxygen species in rice (Oryza sativa L.) Plant Sci. 2012;196:143–151. doi: 10.1016/j.plantsci.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 48.Jiang M., Wu X.J., Song Y., Shen H.Z., Cui H.R. Effects of OsMSH6 mutations on microsatellite stability and homeologous recombination in rice. Front. Plant Sci. 2020;11:220. doi: 10.3389/fpls.2020.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang M., Dai S., Wang B.Q., Xie Z.M., Li J.L., Wang L.Y., Li S., Tan Y.Y., Tian B., Shu Q.Y., et al. Gold nanoparticles synthesized using melatonin suppress cadmium uptake and alleviate its toxicity in rice. Environ. Sci-Nano. 2021 doi: 10.1039/D0EN01172J. [DOI] [Google Scholar]
- 50.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta c (t)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available in the article or Supplementary Materials.