Abstract
Simple Summary
We here review liquid biopsy methods and their use in the diagnostics and treatment of patients with solid cancers. More specifically, circulating tumor DNA, circulating tumor cells, and their current and future clinical applications are considered. Important factors for further integration of liquid biopsy methods in clinical practice are discussed, with a special focus on a Nordic Healthcare system.
Abstract
Liquid biopsies have emerged as a potential new diagnostic tool, providing detailed information relevant for characterization and treatment of solid cancers. We here present an overview of current evidence supporting the clinical relevance of liquid biopsy assessments. We also discuss the implementation of liquid biopsies in clinical studies and their current and future clinical role, with a special reference to the Nordic healthcare systems. Our considerations are restricted to the most established liquid biopsy specimens: circulating tumor DNA (ctDNA) and circulating tumor cells (CTC). Both ctDNA and CTCs have been used for prognostic stratification, treatment choices, and treatment monitoring in solid cancers. Several recent publications also support the role of ctDNA in early cancer detection. ctDNA seems to provide more robust clinically relevant information in general, whereas CTCs have the potential to answer more basic questions related to cancer biology and metastasis. Epidermal growth factor receptor-directed treatment of non-small-cell lung cancer represents a clinical setting where ctDNA already has entered the clinic. The role of liquid biopsies in treatment decisions, standardization of methods, diagnostic performance and the need for further research, as well as cost and regulatory issues were identified as factors that influence further integration in the clinic. In conclusion, substantial evidence supports the clinical utility of liquid biopsies in cancer diagnostics, but further research is still required for a more general application in clinical practice.
Keywords: liquid biopsies, cancer, circulating tumor DNA, circulating tumor cells
1. Introduction
Examination of body fluids to diagnose disease has accompanied medicine from its very beginning. The timeline stretches from Hippocrates around 300 B.C., who suggested tasting the patient’s urine [1], to the plethora of diagnostic tests available in a medical biochemistry laboratory today [2]. The more recent advent of the term liquid biopsy relates to the development of new technologies that enable access to tumor cells and the tumor genome through samples of body fluids [3], analyses which have traditionally been performed using tissue biopsies.
A liquid biopsy is in principle a sample of any body fluid that may contain genetic material from a tumor; for instance, blood, urine, feces, saliva or cerebrospinal fluid [4]. The entities typically searched for are intact tumor cells, cell-free nucleic acids and tumor-derived extracellular vesicles. The applications reach beyond tumor genome characterization and include almost all aspects of cancer diagnostics; detection, characterization, prognostic stratification, therapy choices and response monitoring [5]. Here, we review blood-based liquid biopsy techniques and give examples of clinical applications and challenges in solid cancers, with a special focus on implementation in clinical studies and current and future roles in the health services. Our considerations will be restricted to the most established liquid biopsy specimens: circulating tumor DNA (ctDNA) and circulating tumor cells (CTC).
2. Methodological Approaches to Liquid Biopsies
2.1. Detection and Characterization of ctDNA
Tumor-derived DNA fragments present in the circulation of cancer patients have received much focus in recent clinical cancer research [6]. A major challenge in the detection of ctDNA is that it co-exists with wild-type cell-free DNA (cfDNA), predominantly shedded from hematopoietic cells. Whereas ctDNA can make up the majority of the total cfDNA in many late-stage cancers, it comprises only minute amounts of the total cfDNA in patients with early-stage cancers [7]. Thus, sensitive detection methods are necessary to distinguish ctDNA from background hematopoietic cell-derived cfDNA. Other challenges in ctDNA detection are the overall low concentration of cfDNA in plasma, its high degree of fragmentation, and the heterogeneity in cancer-specific mutations.
Tumor-associated aberrations such as point mutations [7,8,9], insertion/deletions [10,11], copy number variations (CNV) [8,9], translocations [10], and epigenetic changes such as methylations [11,12], have all been targeted in order to detect and characterize ctDNA (Table 1). The most commonly used method for ctDNA detection is to target point mutations in genes frequently mutated in cancer. This is usually performed using highly sensitive detection methods such as droplet digital PCR (ddPCR) and BEAMing, either by screening for hotspot mutations or by using a tumor tissue-informed approach [13,14,15]. For patients where the mutation profile is not known due to lack of tumor tissue, or where subclonal evolution or acquired resistance to treatment is investigated, simultaneous detection of multiple mutations by deep sequencing of large gene panels is a potent tool to detect and characterize ctDNA on a larger scale [16,17,18]. Early deep sequencing-based methods had low sensitivity due to PCR and sequencing errors and were not able to detect low fractions of ctDNA (<1%). The addition of unique molecular identifiers (UMI) to reduce these errors has largely defeated this issue. The sensitivity of deep sequencing is now comparable to ddPCR, which has long been considered the “gold standard” for ctDNA detection, albeit at a higher cost per sample and longer turnaround time [19,20]. A challenge with ctDNA sequencing is that the results can be confounded by somatic mutations in non-malignant hematopoietic cells, known as clonal hematopoiesis of indeterminate potential (CHIP), a phenomenon that increases with age [21,22]. It is therefore highly recommended to analyze matched cell-free and white blood cell DNA to remove CHIP variants, in addition to germline variants.
Table 1.
ctDNA markers and detection methods.
| Marker | Detection Chemistry | Advantages | Disadvantages |
|---|---|---|---|
| Point mutations | PCR/digital PCR | High sensitivity, quick, inexpensive | Few targets per analysis, only pre-identified mutations can be analyzed |
| Point mutations | Deep sequencing | High-throughput, detection of previously unknown mutations, tumor mutation burden (TMB) assessment | Moderate sensitivity, expensive, time-consuming |
| Copy number variations/trans-locations | PCR/digital PCR | High sensitivity, quick, inexpensive | Custom-designed assays needed, only pre-identified aberrations |
| Copy number variations/trans-locations | Low-pass whole-genome sequencing, targeted sequencing | Many and unknown targets | Low sensitivity, expensive, time-consuming |
| Hyper-methylation | Methylation-specific PCR | High sensitivity, low cost | Few targets per analysis, only pre-identified targets |
| Hyper-methylation | Bisulfite-sequencing | High-throughput, many targets | Low sensitivity for single markers, expensive |
For patients without detectable point mutations and cancers prone to CNVs, it might be useful to investigate CNVs as markers for ctDNA detection, although the sensitivity for CNV detection is low compared to point mutations and insertions/deletions [9,23]. Furthermore, recent reports have suggested that targeting differentially methylated regions (DMRs) can improve the detection of ctDNA [11,12]. In contrast to somatic mutations, which are often limited in number, epigenetic changes such as DMRs are frequent and also tissue- and cancer type-specific [24]. The sensitivity can be further improved by increasing the number of DMRs analyzed, enabling detection of ctDNA even at low variant allele frequencies [12].
2.2. Detection of CTCs
CTCs are tumor cells that have left the primary tumor or metastatic sites and entered the bloodstream by intravasation [25]. CTCs are extremely rare compared to other cells in the blood of patients with cancer (110,000 per liter) [26], so extensive enrichment is required to detect them. Current CTC-enrichment methods (Table 2) are based on either physical properties (size, density, deformability, and electrical charge) or biological features (cell-surface protein expression, invasive capacity, and viability) [27]. The CellSearch system, which is based on biological features, remains the gold standard for enrichment and detection of CTCs. Paramagnetic beads coupled with antibodies against the epithelial cell adhesion molecule (EpCAM) are used for CTC enrichment by this method. CTC detection in addition requires presence of epithelial cytokeratins and absence of the leukocyte marker CD45 in nucleated cells. Enumeration of CTCs using the CellSearch method has been approved for clinical use by the Food and Drug Administration (FDA) [28]. A limitation of the CellSearch method is that mesenchymal-type CTCs go undetected as the EpCAM marker becomes down-regulated following epithelial-to-mesenchymal transition (EMT), an important step of tumor cell dissemination [29]. To counteract this limitation, alternative antibody-based CTC capture methods have targeted mesenchymal and stem cell markers [30], but several challenges with this approach remain to be solved, e.g., when mesenchymal markers are expressed on circulating benign cell types [31].
Table 2.
Circulating tumor cell (CTC) enrichment methods.
| CTC Feature | Enrichment Method | Advantages | Disadvantages |
|---|---|---|---|
| Size | Size filtration | Simple, inexpensive, marker-independent | Low purity, loss of small CTCs |
| Size and deformability | Microfluidics | Easily automatable, marker-independent | Varying purity, loss of small CTCs, time-consuming |
| Density | Density gradient centrifugation | Simple, inexpensive, marker-independent | Low purity, potential loss of CTC clusters |
| Electrical charge | Dielectrophoresis | Marker-independent | Low CTC recovery, low purity, low throughput |
| Surface markers | Positive immunomagnetic selection | High recovery, high purity. | Marker dependent |
| Absence of surface markers | Immunomagnetic depletion | Marker-independent, high recovery, high purity | Loss of CTCs adhering to white blood cells |
CTC enrichment, however, can be independent of marker expression in the CTCs, either by negative depletion-based strategies or by utilizing differences in physical properties [32]. The exciting prospects of CTCs in cancer management, along with their rarity, has spurred technology development. In particular, advances in nanotechnology, microfluidics, and single-cell sequencing have found applications for CTC enrichment and analyses ex vivo and in vivo [33,34,35,36,37,38,39]. In spite of the plethora of different technological innovations, it remains a great challenge to capture the heterogeneity of CTCs and to preserve their viability and functional characteristics for experimental analyses [37].
3. Clinical Relevance of Liquid Biopsies in Solid Cancers
3.1. Early Detection of Disease
Early detection of cancer is pivotal for the treatment outcome of most cancers. However, the task is notoriously challenging [40]. Despite this, a few recent, seminal publications have provided evidence that liquid biopsies can become a tool for successful cancer screening [41,42]. The CancerSEEK assay, which is based on detection of both ctDNA and protein biomarkers in blood samples, was tested on 1005 symptomatic patients with eight different solid non-metastatic cancers and 812 healthy control persons [41]. The overall sensitivity at optimal biomarker thresholds was 62% and the specificity 99%. However, the sensitivity for cancer types diagnosed at a late stage (98% for ovarian cancer) was much higher than for cancer types predominantly diagnosed at earlier stages (33% for breast cancer). Caution has also been raised regarding the low false-positive rate reported (1%), as the controls in this study were healthy individuals, whereas a real screening setting would also include persons with inflammatory and other diseases, potentially increasing this rate. Recently, an early version of the CancerSEEK method combined with diagnostic PET-CT was successfully used to identify new cancer cases among 9911 previously cancer-free women [43]. Of the 143 with positive blood testing, 127 (95%) were subjected to PET-CT imaging and 64 (50%) obtained images indicating potential malignancies. Twenty-five of them were subsequently shown to have cancer through biopsy. A new version of the CancerSEEK assay is currently being tested in a multicenter, prospective, observational study aiming to include 1000 subjects with known or suspected cancer in addition to 2000 subjects without known cancer (ClinicalTrials.gov Identifier: NCT04213326) to validate the updated classification algorithm [44].
The Circulating Cell-free Genome Atlas (CCGA) consortium recently reported data from an even larger prospective trial, comprising 2484 patients with 50 different cancers and 4207 individuals without cancer [42]. Using targeted bisulfite sequencing to detect ctDNA, they obtained overall sensitivities ranging from 18% in all stage I cancers to 93% in stage IV cancers and a false-positive rate around 1%. For some cancers, such as pancreatic cancer, the sensitivities were considerably higher, yielding hope for a future clinical utility of this technology. Interestingly, a similar approach is currently being investigated in several large ongoing studies (e.g., STRIVE, PATHFINDER, SUMMIT) [45]. Other groups have also demonstrated promising cancer detection using methylation sequencing [12,46]. A research group demonstrated good distinction between 199 cancer patients and 62 healthy control persons in their validation cohort, reporting areas under receiver operating characteristic (ROC) curves from 0.91 to 0.98 [12]. More recently, the PanSeer ctDNA methylation blood test was reported to correctly classify 95% of 98 cancer patients and 96% of 207 samples from healthy control persons [46].
3.2. Prognostic Stratification
The evidence for the prognostic value of ctDNA in solid cancers is extensive and has been demonstrated for several different cancers, including breast [47,48], colorectal [49,50], lung [51,52,53], melanoma [14,54], and pancreatic cancer [13,55]. Several studies have indicated an inverse relation between ctDNA level and survival, with increasing levels of ctDNA being significantly associated with poor survival [7,47]. Further, early dynamic changes in ctDNA levels have also been shown to predict survival [52,53]. The evidence of a prognostic value of ctDNA is strongest for patients with inoperable disease [13,49,50], but there is also some evidence for resected patients [13,48]. A major challenge in using ctDNA to predict prognosis in a clinical setting is, however, the lack of standardized cutoff values for ctDNA positivity.
The prognostic relevance of CTCs has primarily been investigated using the CellSearch system, which has FDA approval for prognostication of metastatic breast, colon and prostate cancer. This application has been extended to additional solid cancer types, such as lung and urogenital cancers and malignant melanoma. Associations between ≥5 CTCs and inferior progression-free and overall survival have been demonstrated especially in the metastatic setting [5,56,57,58,59]. However, there is also evidence for prognostic relevance of CTCs in non-metastatic cancer, usually by applying a lower cutoff for CTC positivity (≥1 CTC per 7.5 mL blood) [60,61,62]. Prognostic relevance has also been shown for morphological features of CTCs, such as the association between CTC clusters in the circulation and adverse prognosis [63,64,65,66].
EMT and the reverse process mesenchymal-to-epithelial transition (MET) have been postulated as critical programs for efficient metastasis [29]. A link between CTCs with mesenchymal features and prognosis has been indicated in several studies [31,67,68,69]. However, the evidence for a prognostic value of mesenchymal-like CTCs is currently not as extensive as for the CellSearch system, and further research is required to clarify the role of CTC phenotypes [5,70].
3.3. Choosing Therapy Based on Liquid Biopsies
The ultimate utility of liquid biopsies in cancer is to guide clinical decision-making. Current evidence supports a predictive value of both CTCs and ctDNA with regard to various cancer treatments (recently thoroughly reviewed in [5,70]), but the data are most convincing for targeted therapies. The best established example is the role of ctDNA Epidermal Growth Factor Receptor (EGFR) mutations in the treatment of advanced non-small-cell lung cancer (NSCLC), as emphasized by the FDA approval of the cobas® EGFR Mutation Test v2 (Roche Diagnostics, Indianapolis, IN, USA) and the FoundationOne Liquid CDx tests (Foundation Medicine, Inc., Cambridge, MA, USA). Activating mutations in the EGFR gene, either in the primary tumor or in ctDNA, predicts treatment effect of EGFR tyrosine kinase inhibitors, whereas the EGFR T790M mutation commonly causes resistance [71,72,73,74]. Moreover, resistance-causing mutations (in the KRAS, ESR1, or PIK3CA genes) in relation to other targeted treatments have been detected in ctDNA from several cancer types, suggesting a future clinical utility of ctDNA in multiple treatment settings [75,76,77,78].
Molecular analysis of CTCs has proven to be predictive in relation to targeted treatment. For example, detection of nuclear-localized androgen receptor (AR) splice variant 7 protein (AR-V7) in CTCs (FDA-approved test) from patients with metastatic castration-resistant prostate cancer predicted a survival benefit if they were treated with taxanes rather than AR directed therapy [79,80]. Moreover, detection of AR-V7 or the neuroendocrine marker synaptophysin on CTCs both suggest resistance to androgen deprivation and AR inhibitor treatments and appears as useful to guide therapy of advanced prostate cancer [81,82]. Other promising cases include ER-expression on CTCs in ER-directed treatment of breast cancer and ALK rearrangements in CTCs from NSCLC patients treated with ALK inhibitors [83,84]. Moreover, methods to culture CTCs and establish CTC-derived explants has opened new possibilities for personalized therapy selection [85].
Furthermore, a survival benefit of radiotherapy in two independent cohorts of operable breast cancer patients was only demonstrated in CTC positive patients [86]. In addition, a survival benefit of using CTC counts to choose between anti-hormone treatment and chemotherapy in metastatic breast cancer has also been suggested [87], whereas copy number aberrations in CTCs from small-cell lung cancer patients were associated with chemo-refractiveness in another study [88].
Immune checkpoint inhibitors have received much attention due to convincing treatment effects for some cancer types [89]. Tumor mutational burden (TMB) in tissue biopsies is an established predictive factor for immunotherapy in some cancers and ctDNA mutations can be used as a surrogate [90,91]. PD-L1 expression on CTCs also seems to have potential for predicting treatment response to checkpoint inhibitors [92].
3.4. Disease Monitoring and Early Detection of Relapse
Serial analysis of ctDNA and CTCs has the potential to provide information on disease progression, therapeutic effect and resistance, and tumor evolution. Monitoring of tumor burden for early detection of disease progression and therapeutic effect is currently performed by analysis of conventional protein biomarkers in blood and radiological imaging, methods which suffer from low specificity and/or sensitivity [93]. However, recent studies have demonstrated that an increase in the ctDNA level may reveal disease relapse or progression at the same time or earlier than radiological imaging [48,94,95,96,97]. Moreover, longitudinal monitoring of ctDNA levels is shown to predict treatment efficacy in several cancer types [77,98,99]. Molecular characterization of ctDNA during follow-up may also reveal mutations causing therapy resistance, information that may help guiding treatment decisions in the future [7,47,77].
There are indications that longitudinal changes in CTC enumeration can predict treatment response [59,100], but overall it has been challenging to prove a strong clinical value of CTC monitoring, possibly related to their low numbers [101]. Thus, in the near future we expect ctDNA assessment to be more useful than CTCs for monitoring disease burden, treatment response and early detection of relapse.
4. Implementing Liquid Biopsies in Clinical Studies
Experimental evidence for the clinical utility of a liquid biopsy test, and not only its clinical relevance, is a prerequisite for its incorporation in clinical practice [102]. As described above, only a few tests currently have this level of evidence, encouraging the design of new interventional studies where the role of liquid biopsy tests for making treatment choices is evaluated. A large number of ongoing clinical trials now includes liquid biopsies in their trial design (Clinicaltrials.gov). However, only a few of the studies are interventional. Accordingly, the massive focus on prognostic value and monitoring potential in the liquid biopsy field should be shifted towards randomized clinical trials [103]. However, incorporation in such trials depends on promising observational data, emphasizing the cascade of evidence needed for a biomarker to make its way to the clinic [104].
Experiences from a Norwegian Regional Research Program on Liquid Biopsies
We recently finished a consortium-based five-year research program on liquid biopsies entitled “Personalized cancer therapy–biomarkers in clinical trials”, funded by the Western Norway Regional Health Authorities and involving research groups from three Norwegian hospitals. Several observational and interventional research projects were initiated and performed during the project period, providing considerable experience on the implementation of liquid biopsies in clinical studies [14,55,105,106,107,108,109]. The most obvious challenge was the plethora of available techniques for both ctDNA and CTC assessment, and the difficulty to standardize techniques across projects including different cancer types and treatment settings. Standardization was restricted by the continuous development of new technology in the field and a lack of consensus on what to prioritize when choosing technologies [110]. We would also like to emphasize the importance of standardization of pre-analytical conditions like sampling tubes, time from sampling to first centrifugation for ctDNA, storage conditions and times, and transport routines [102]. The requirement for rapid CTC enrichment after sample collection (2–96 h, depending on tube) makes the pre-analytical processing more complicated for CTCs than ctDNA; an argument to prefer ctDNA over CTCs in clinical studies [102,111,112]. During the development of the projects, we also learned that larger sample volumes (>10 mL blood) are preferable to increase analytical sensitivities, especially in non-metastatic settings [113].
To ensure successful integration of liquid biopsies in clinical studies, optimal standard operating procedures (SOP) need to be in place before the start of recruitment [110,114]. This will highlight the methodological needs, the choice of sample material, as well as staff needs, for implementing liquid biopsies. It is important to have personnel dedicated for sample handling and processing directly after blood draw, as the half-lives of ctDNA and CTCs are short, and the sample quality will deteriorate rapidly. To ensure efficient analysis of results, it is also important to allocate enough resources for bioinformatics.
5. Liquid Biopsies in Clinical Practice
5.1. Liquid Biopsies Currently Applied in Hospital Settings in the Nordic Countries
The implementation of new diagnostic tests in routine analysis in Nordic hospitals is often slower than in other regions because of the strict regulations and reimbursement system used in their public healthcare systems (further discussed in Section 5.2.5). The most established liquid biopsy test in clinical practice is testing for EGFR mutations in non-small-cell lung cancer (Table 3). As described above, several sensitizing mutations in this gene are strongly associated with response to EGFR tyrosine kinase inhibitors (reviewed in [115]). Traditionally, tumor tissue has been tested for EGFR-mutations, but it is not always possible to collect sufficient tumor samples without placing patients at risk of complications [116]. This is often even more challenging at the time of progression, when a rebiopsy ideally should be collected to individualize subsequent therapy [117]. Fortunately, several studies clearly demonstrate that EGFR mutations may be detected in cfDNA for a high proportion of patients with known EGFR mutations in their tumor. The response rates to EGFR TKIs are similar for these patients as for patients who are offered this treatment based on the detection of EGFR mutations in tissue samples [71,72,73,74,118]. Thus, it is well accepted that peripheral blood tests may replace tissue testing when tumor samples are unavailable or the risk associated with a biopsy is high. A similar approach is also accepted when testing for the most common resistance mutation among patients on EGFR TKIs, the T790M mutation [74].
Table 3.
Liquid biopsy applications currently in use or near use in Nordic clinical practice.
| Clinical Status | Liquid Biopsy Test | Method | Clinical Application | References |
|---|---|---|---|---|
| Currently in use | cfDNA EGFR gene mutation testing | Quantitative PCR | Predictive for EGFR-directed treatment (TKI inhibitors) of advanced lung cancer | [71,72,73,74,115,117,118] |
| Food and Drug Ad-ministration (FDA) approved |
FoundationOne Liquid CDx multigene panel (incl. EGFR, ALK, PIK3CA, BRCA genes) | Targeted sequencing | Predictive for targeted treatment of metastatic lung, prostate and breast cancer | [119,120,121] |
| FDA approved |
cfDNA KRAS and NRAS gene mutation testing | BEAMing, Digital PCR | Predictive for EGFR-directed treatment of metastatic colorectal cancer | [75] |
| FDA approved |
cfDNA TMB testing | Targeted sequencing | Predictive for treatment of several solid cancers with immune checkpoint inhibitors | [90,91] |
| FDA approved |
AR-V7 splice variant testing in CTCs | Immunofluorescence | Treatment choices for metastatic castration-resistant prostate cancer | [79,80] |
| FDA approved |
CellSearch CTC enumeration | Immuno- logical enrichment/ staining |
Prognostic stratification of breast, prostate and colorectal cancer | [58,60] |
5.2. Challenges for Further Integration of Liquid Biopsies in Clinical Practice
5.2.1. Evidence of Treatment Relevance Needed
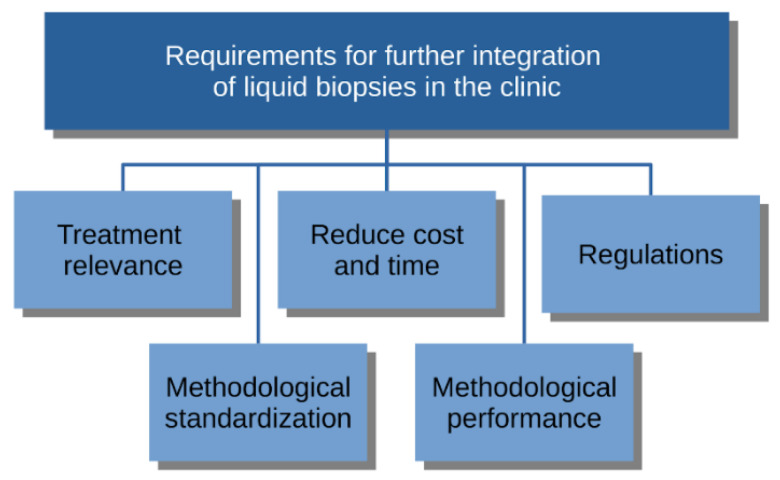
The multitude of prognostic cancer markers never reaching the clinic emphasizes the main criteria for successful integration in the clinic: relevance for clinical decision-making (Figure 1). We have already described promising research supporting the applicability of liquid biopsies in the choice of therapy and monitoring of treatment effect. Excellent evidence for such utility has been provided for NSCLC and ctDNA, explaining its current role in the clinic. However, for other cancers and treatment settings, there is still a need for more clinical trials [102]. CTC enumerations are not performed routinely in Nordic hospitals, despite the FDA approval, probably because of weak evidence for a predictive value.
Figure 1.
Overview of the most important identified requirements for further integration of liquid biopsies in the clinic.
The fact that mutations predicting therapy response or resistance can also be used to detect ctDNA emphasizes that targeted biological drugs might be the most likely candidates for development of accompanying liquid biopsy-diagnostics. We anticipate increasing clinical utilization of such ctDNA application, including detection of resistance mutations in relation to EGFR-directed therapy of colorectal cancer and ESR1 mutations in relation to anti-hormone therapy in breast cancer [75,76]. TMB assessment in relation to immune therapy is another promising candidate for useful application of liquid biopsies in clinical work [90,91]. Recently, several high-impact articles have suggested a role for ctDNA detection in early cancer detection with the possible benefit of treating at a non-metastatic disease stage [11,12,41,43]. Early detection screening programs have been questioned by the risk of overdiagnosis [122] and the establishment of early cancer detection described in these studies is very extensive. However, we believe that the approaches described in these articles can at least be beneficial for high-risk groups.
5.2.2. Methodological Standardization
The analytical performance of liquid biopsy tests must be assessed in prospective studies using validated protocols to decide clinical validity and utility. So far, most of our knowledge is based on retrospective analysis with poorly described and often quite disparate procedures. Standardization of the complete workflow is pivotal for implementation in routine diagnostics [114]. To address this, the European Liquid Biopsy Society [123] and BloodPAC [124] (USA) have been formed, joining experts from academic and clinical research with diagnostic companies and the pharmaceutical industry. These initiatives seek to establish standardized protocols, mutual validation of methods, data sharing and development of new technology.
The need for standardization includes both the pre-analytical steps, the analysis itself, interpretation of results and reporting. Proper reference materials are useful for both the pre-analytical and analytical validation (further discussed in [125]). The need for improvement, particularly regarding ctDNA, was shown in a recent proof-of-principle study [126]. Important questions are how to report ctDNA levels, and how to determine diagnostic thresholds. Recently, the National Cancer Institute’s Biorepositories and Biospecimen Research Branch has developed cfDNA-specific guidelines in an effort to standardize optimal procedures for sample collection and processing [127].
5.2.3. Cost and Time Requirements
For ctDNA, detection by deep sequencing is far more expensive than techniques based only on PCR, but it is superior for mutation profiling and early detection, as ‘a priori’ mutation knowledge is not required. A major contribution to the cost is the sequencing, which depends on the number of samples sequenced simultaneously, sequencing depth, sequencing reagents and instruments. In sparsely populated areas, such as the Nordic countries, few patient samples can delay turnaround time if sequencing costs are to be kept to a minimum. ctDNA detection by PCR-based methods is faster, cheaper and easier to perform on a small scale, although it is more difficult to generalize for several cancer types. For treatment monitoring, the level of a particular mutation using ddPCR/BEAMing is from our point of view the only affordable approach with sufficient sensitivity at the moment, even if it is limited to patient-specific mutations. However, this may change as sequencing costs are further reduced. Sample collection and processing for CTC analyses, as well as sample shipment in many hospital settings, is also associated with considerable cost and time requirements.
5.2.4. Liquid Biopsy Performance Compared to Routine Diagnostics
Massive evidence for EGFR mutation testing in ctDNA from NSCLC patients demonstrates good concordance with primary tumor assessment and clinical utility of ctDNA in relation to targeted treatment [71,72,73,74]. ctDNA is also more likely to reflect tumor heterogeneity and resistance mechanisms, particularly when multiple metastases are present [76,128]. Conversely, if targeted treatment is chosen based on a minor tumor clone found in ctDNA, a treatment effect on the bulk of the tumor (not harboring the clonal mutation) cannot be expected [102]. Based on published research and clinical experience with genetic diagnostic testing of aggressive acute myeloid leukemia, we foresee a scenario for solid cancers, where multiple mutations represent the oligoclonal repertoire of the tumor [129,130]. Treatment guided by response evaluation through ctDNA can then be tailored to remove the multiple clones by targeted therapy and chemotherapy.
Another important clinical setting in which liquid biopsies have the potential to replace or reduce routine diagnostics is in the monitoring of tumor development during treatment [6]. Repeated blood sampling is feasible and many of the liquid biopsy-based analyses are both more sensitive and less resource-demanding than radiological imaging [97,131,132]. Nevertheless, this approach is insufficient for a subgroup of patients in most published studies, either because some patients lack liquid biopsy markers or because the increase in marker levels detected by liquid biopsy is delayed compared to radiological evidence of recurrence. By using personalized ctDNA and CTC markers, more sensitive assays, and larger volumes of blood, the detection rates can hopefully be optimized [96,97]. However, it is difficult to imagine a total replacement of radiological imaging in the surveillance of patients with solid cancers. We believe a combination of surveillance methods might be more likely, preferably with more frequent ctDNA assessments than radiological examinations.
5.2.5. Regulations
In Norway, the healthcare system is publically funded and new expensive methods need to be approved by a national health technology assessment before implementation in routine diagnostics. The main topic considered in these processes is the clinical utility in regard to sample type, cancer type, risk of complications, clinical application, and related treatment options. In the evaluation of liquid biopsy methods for predictive purposes both the stakeholders’ perspectives and the availability of targeted drugs can affect the interpretation of evidence and the conclusions [133]. With the multitude of newly suggested liquid biopsy-based approaches, a national health technology assessment is undoubtedly needed, but the guidelines to act upon need to be carefully defined.
6. Conclusions and Future Perspectives
Liquid biopsies are currently mostly research tools in clinical trials and in the development of novel diagnostics. The first disease that may take advantage of ctDNA analysis is NSCLC, where it already guides targeted treatment. Recent evidence suggests further application of liquid biopsies in several solid cancers, but validation is required for regulatory approval and establishment of clinical use, not the least to justify the high cost of advanced sample processing and accompanying bioinformatic analysis. Randomized clinical trials using liquid biopsies for treatment decisions are therefore required for further integration of the methods into the clinic. ctDNA seems to have the best potential for becoming a widespread marker in routine diagnostics, whereas CTCs are challenging due to their difficult detection and sampling issues, but may answer more basic questions connected to cancer biology and metastasis.
The required expenses represent a limitation to provide liquid biopsy as a robust tool globally. Clearly, technological progress in microfluidics, computing and electronics will reduce the price level of the diagnostic technology. We need an open discussion about open licenses to secure the benefit of knowledge in all regions. The expected revolution in diagnostics should encourage a focus on this topic for international organizations like the World Health Organization in their effort against cancer.
The full potential for liquid biopsies seems to be in concert with other diagnostic modalities [41]. Imaging diagnostics in cancer is also rapidly developing, and provides anatomical and various functional information about the tumor. Liquid biopsy and functional imaging may be the diagnostics tool combination needed to accelerate precision medicine in cancer.
Author Contributions
Conceptualization, O.N., R.B.F., B.G., B.T.G., and R.H.; data collection and manuscript preparation, all authors. All authors have read and agreed to the published version of the manuscript.
Funding
This work was part of a strategic program called “Personalized medicine—biomarkers and clinical studies”, which was supported by the Western Norway Health Authorities, grant 912009.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data sharing not applicable. No new data were created or analyzed in this study.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Berger D. A brief history of medical diagnosis and the birth of the clinical laboratory. Part 1--Ancient times through the 19th century. MLO Med. Lab. Obs. 1999;31:28–30. [PubMed] [Google Scholar]
- 2.Lindberg M., Garmo G., Hardang I., Monsen A.-L.B. Nasjonal Brukerhåndbok i Medisinsk Biokjemi. Norsk Forbund for Medisinsk Biokjemi; Oslo, Norway: 2018. [Google Scholar]
- 3.NCI Dictionary of Cancer Terms—National Cancer Institute. [(accessed on 29 December 2020)]; Available online: https://www.cancer.gov/publications/dictionaries/cancer-terms/
- 4.Siravegna G., Marsoni S., Siena S., Bardelli A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017;14:531–548. doi: 10.1038/nrclinonc.2017.14. [DOI] [PubMed] [Google Scholar]
- 5.Pantel K., Alix-Panabières C. Liquid biopsy and minimal residual disease—Latest advances and implications for cure. Nat. Rev. Clin. Oncol. 2019;16:409–424. doi: 10.1038/s41571-019-0187-3. [DOI] [PubMed] [Google Scholar]
- 6.Nordgård O., Tjensvoll K., Gilje B., Søreide K. Circulating tumour cells and DNA as liquid biopsies in gastrointestinal cancer. Br. J. Surg. 2018;105:e110–e120. doi: 10.1002/bjs.10782. [DOI] [PubMed] [Google Scholar]
- 7.Bettegowda C., Sausen M., Leary R.J., Kinde I., Wang Y., Agrawal N., Bartlett B.R., Wang H., Luber B., Alani R.M., et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci. Transl. Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heitzer E., Ulz P., Belic J., Gutschi S., Quehenberger F., Fischereder K., Benezeder T., Auer M., Pischler C., Mannweiler S., et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med. 2013;5:30. doi: 10.1186/gm434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rothwell D.G., Ayub M., Cook N., Thistlethwaite F., Carter L., Dean E., Smith N., Villa S., Dransfield J., Clipson A., et al. Utility of ctDNA to support patient selection for early phase clinical trials: The TARGET study. Nat. Med. 2019;25:738–743. doi: 10.1038/s41591-019-0380-z. [DOI] [PubMed] [Google Scholar]
- 10.Gawroński A.R., Lin Y.-Y., McConeghy B., LeBihan S., Asghari H., Koçkan C., Orabi B., Adra N., Pili R., Collins C.C., et al. Structural variation and fusion detection using targeted sequencing data from circulating cell free DNA. Nucleic Acids Res. 2019;47:e38. doi: 10.1093/nar/gkz067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu L., Toung J., Jassowicz A., Vijayaraghavan R., Kang H., Zhang R., Kruglyak K., Huang H., Hinoue T., Shen H., et al. Targeted methylation sequencing of plasma cell-free DNA for cancer detection and classification. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2018;29:1445–1453. doi: 10.1093/annonc/mdy119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen S.Y., Singhania R., Fehringer G., Chakravarthy A., Roehrl M.H.A., Chadwick D., Zuzarte P.C., Borgida A., Wang T.T., Li T., et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nat. Cell Biol. 2018;563:579–583. doi: 10.1038/s41586-018-0703-0. [DOI] [PubMed] [Google Scholar]
- 13.Pietrasz D., Pécuchet N., Garlan F., Didelot A., Dubreuil O., Doat S., Imbert-Bismut F., Karoui M., Vaillant J.-C., Taly V., et al. Plasma Circulating Tumor DNA in Pancreatic Cancer Patients Is a Prognostic Marker. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017;23:116–123. doi: 10.1158/1078-0432.CCR-16-0806. [DOI] [PubMed] [Google Scholar]
- 14.Forthun R.B., Hovland R., Schuster C., Puntervoll H., Brodal H.P., Namløs H.M., Aasheim L.B., Meza-Zepeda L.A., Gjertsen B.T., Knappskog S., et al. ctDNA detected by ddPCR reveals changes in tumour load in metastatic malignant melanoma treated with bevacizumab. Sci. Rep. 2019;9:1–15. doi: 10.1038/s41598-019-53917-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tabernero J., Lenz H.-J., Siena S., Sobrero A., Falcone A., Ychou M., Humblet Y., Bouché O., Mineur L., Barone C., et al. Analysis of circulating DNA and protein biomarkers to predict the clinical activity of regorafenib and assess prognosis in patients with metastatic colorectal cancer: A retrospective, exploratory analysis of the CORRECT trial. Lancet Oncol. 2015;16:937–948. doi: 10.1016/S1470-2045(15)00138-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chabon J.J., Simmons A.D., Lovejoy A.F., Esfahani M.S., Newman A.M., Haringsma H.J., Kurtz D.M., Stehr H., Scherer F., Karlovich C.A., et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat. Commun. 2016;7:11815. doi: 10.1038/ncomms11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parikh A.R., Leshchiner I., Elagina L., Goyal L., Levovitz C., Siravegna G., Livitz D., Rhrissorrakrai K., Martin E.E., Van Seventer E.E., et al. Liquid versus Tissue Biopsy for Detecting Acquired Resistance and Tumor Heterogeneity in Gastrointestinal Cancers. Nat. Med. 2019;25:1415–1421. doi: 10.1038/s41591-019-0561-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Namløs H.M., Boye K., Mishkin S.J., Barøy T., Lorenz S., Bjerkehagen B., Stratford E.W., Munthe E., Kudlow B.A., Myklebost O., et al. Noninvasive Detection of ctDNA Reveals Intratumor Heterogeneity and Is Associated with Tumor Burden in Gastrointestinal Stromal Tumor. Mol. Cancer Ther. 2018;17:2473–2480. doi: 10.1158/1535-7163.MCT-18-0174. [DOI] [PubMed] [Google Scholar]
- 19.Newman A.M., Lovejoy A.F., Klass D.M., Kurtz D.M., Chabon J.J., Scherer F., Stehr H., Liu C.L., Bratman S.V., Say C., et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 2016;34:547–555. doi: 10.1038/nbt.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phallen J., Sausen M., Adleff V., Leal A., Hruban C., White J., Anagnostou V., Fiksel J., Cristiano S., Papp E., et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med. 2017;9:eaan2415. doi: 10.1126/scitranslmed.aan2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu Y., Ulrich B.C., Supplee J., Kuang Y., Lizotte P.H., Feeney N.B., Guibert N.M., Awad M.M., Wong K.-K., Jänne P.A., et al. False-Positive Plasma Genotyping Due to Clonal Hematopoiesis. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018;24:4437–4443. doi: 10.1158/1078-0432.CCR-18-0143. [DOI] [PubMed] [Google Scholar]
- 22.Razavi P., Li B.T., Brown D.N., Jung B., Hubbell E., Shen R., Abida W., Juluru K., De Bruijn I., Hou C., et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat. Med. 2019;25:1928–1937. doi: 10.1038/s41591-019-0652-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heitzer E., Ulz P., Geigl J.B. Circulating Tumor DNA as a Liquid Biopsy for Cancer. Clin. Chem. 2015;61:112–123. doi: 10.1373/clinchem.2014.222679. [DOI] [PubMed] [Google Scholar]
- 24.Hoadley K.A., Yau C., Wolf D.M., Cherniack A.D., Tamborero D., Ng S., Leiserson M.D., Niu B., McLellan M.D., Uzunangelov V., et al. Multiplatform Analysis of 12 Cancer Types Reveals Molecular Classification within and across Tissues of Origin. Cell. 2014;158:929–944. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pantel K., Speicher M.R. The biology of circulating tumor cells. Oncogene. 2016;35:1216–1224. doi: 10.1038/onc.2015.192. [DOI] [PubMed] [Google Scholar]
- 26.Stoecklein N.H., Fischer J.C., Niederacher D., Terstappen L.W.M.M. Challenges for CTC-based liquid biopsies: Low CTC frequency and diagnostic leukapheresis as a potential solution. Expert Rev. Mol. Diagn. 2015;16:147–164. doi: 10.1586/14737159.2016.1123095. [DOI] [PubMed] [Google Scholar]
- 27.Alix-Panabières C., Pantel K. Technologies for detection of circulating tumor cells: Facts and vision. Lab Chip. 2013;14:57–62. doi: 10.1039/C3LC50644D. [DOI] [PubMed] [Google Scholar]
- 28.Millner L.M., Linder M.W., Valdes R. Circulating Tumor Cells: A Review of Present Methods and the Need to Identify Heterogeneous Phenotypes. Ann. Clin. Lab. Sci. 2013;43:295–304. [PMC free article] [PubMed] [Google Scholar]
- 29.De Craene B., Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 30.Ferreira M.M., Ramani V.C., Jeffrey S.S. Circulating tumor cell technologies. Mol. Oncol. 2016;10:374–394. doi: 10.1016/j.molonc.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Werner S., Stenzl A., Pantel K., Todenhöfer T. Expression of Epithelial Mesenchymal Transition and Cancer Stem Cell Markers in Circulating Tumor Cells. Adv. Exp. Med. Biol. 2017;994:205–228. doi: 10.1007/978-3-319-55947-6_11. [DOI] [PubMed] [Google Scholar]
- 32.Alix-Panabières C., Pantel K. Challenges in circulating tumour cell research. Nat. Rev. Cancer. 2014;14:623–631. doi: 10.1038/nrc3820. [DOI] [PubMed] [Google Scholar]
- 33.Siemer S., Wünsch D., Khamis A., Lu Q., Scherberich A., Filippi M., Krafft M.P., Hagemann J., Weiss C., Ding G.-B., et al. Nano Meets Micro-Translational Nanotechnology in Medicine: Nano-Based Applications for Early Tumor Detection and Therapy. Nanomaterials. 2020;10:383. doi: 10.3390/nano10020383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dan Z., Daxiang C., Zou D., Cui D. Advances in isolation and detection of circulating tumor cells based on microfluidics. Cancer Biol. Med. 2018;15:335–353. doi: 10.20892/j.issn.2095-3941.2018.0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu X., Jiang Z., Wang J., Ren Y., Wu A. Microfluidic applications on circulating tumor cell isolation and biomimicking of cancer metastasis. Electrophoresis. 2020;41:933–951. doi: 10.1002/elps.201900402. [DOI] [PubMed] [Google Scholar]
- 36.Bin Lim S., Lim C.T., Lim W.-T. Single-Cell Analysis of Circulating Tumor Cells: Why Heterogeneity Matters. Cancers. 2019;11:1595. doi: 10.3390/cancers11101595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gribko A., Künzel J., Wünsch D., Lu Q., Nagel S.M., Knauer S.K., Stauber R.H., Ding G.-B. Is small smarter? Nanomaterial-based detection and elimination of circulating tumor cells: Current knowledge and perspectives. Int. J. Nanomed. 2019;14:4187–4209. doi: 10.2147/IJN.S198319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou J., Kulasinghe A., Bogseth A., O’Byrne K., Punyadeera C., Papautsky I. Isolation of circulating tumor cells in non-small-cell-lung-cancer patients using a multi-flow microfluidic channel. Microsyst. Nanoeng. 2019;5:1–12. doi: 10.1038/s41378-019-0045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kulasinghe A., Kapeleris J., Kimberley R., Mattarollo S.R., Thompson E.W., Thiery J.-P., Kenny L., O’Byrne K., Punyadeera C. The prognostic significance of circulating tumor cells in head and neck and non-small-cell lung cancer. Cancer Med. 2018;7:5910–5919. doi: 10.1002/cam4.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alix-Panabières C., Pantel K. Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as Liquid Biopsy. Cancer Discov. 2016;6:479–491. doi: 10.1158/2159-8290.CD-15-1483. [DOI] [PubMed] [Google Scholar]
- 41.Cohen J.D., Li L., Wang Y., Thoburn C., Afsari B., Danilova L., Douville C., Javed A.A., Wong F., Mattox A., et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926–930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu M.C., Oxnard G.R., Klein E.A., Swanton C., Seiden M.V., CCGA Consortium Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 2020;31:745–759. doi: 10.1016/j.annonc.2020.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lennon A.M., Buchanan A.H., Kinde I., Warren A., Honushefsky A., Cohain A.T., Ledbetter D.H., Sanfilippo F., Sheridan K., Rosica D., et al. Feasibility of blood testing combined with PET-CT to screen for cancer and guide intervention. Science. 2020;369:eabb9601. doi: 10.1126/science.abb9601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Detecting Cancers Earlier Through Elective Plasma-Based CancerSEEK Testing. [(accessed on 29 December 2020)]; Available online: https://clinicaltrials.gov/ct2/show/NCT04213326.
- 45.GRAIL Is Conducting One of the Largest Clinical Study Programs Ever. [(accessed on 29 December 2020)]; Available online: https://grail.com/clinical-studies/
- 46.Chen X., Gole J., Gore A., He Q., Lu M., Min J., Yuan Z., Yang X., Jiang Y., Zhang T., et al. Non-invasive early detection of cancer four years before conventional diagnosis using a blood test. Nat. Commun. 2020;11:1–10. doi: 10.1038/s41467-020-17316-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dawson S.-J., Tsui D.W., Murtaza M., Biggs H., Rueda O.M., Chin S.-F., Dunning M.J., Gale D., Forshew T., Mahler-Araujo B., et al. Analysis of Circulating Tumor DNA to Monitor Metastatic Breast Cancer. N. Engl. J. Med. 2013;368:1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 48.Garcia-Murillas I., Schiavon G., Weigelt B., Ng C., Hrebien S., Cutts R.J., Cheang M., Osin P., Nerurkar A., Kozarewa I., et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci. Transl. Med. 2015;7:302ra133. doi: 10.1126/scitranslmed.aab0021. [DOI] [PubMed] [Google Scholar]
- 49.Garlan F., Laurent-Puig P., Sefrioui D., Siauve N., Didelot A., Sarafan-Vasseur N., Michel P., Perkins G., Mulot C., Blons H., et al. Early Evaluation of Circulating Tumor DNA as Marker of Therapeutic Efficacy in Metastatic Colorectal Cancer Patients (PLACOL Study) Clin. Cancer Res. 2017;23:5416–5425. doi: 10.1158/1078-0432.CCR-16-3155. [DOI] [PubMed] [Google Scholar]
- 50.Grasselli J., Elez E., Caratù G., Matito J., Santos C., Macarulla T., Vidal J., Garcia M., Viéitez J., Paéz D., et al. Concordance of blood- and tumor-based detection of RAS mutations to guide anti-EGFR therapy in metastatic colorectal cancer. Ann. Oncol. 2017;28:1294–1301. doi: 10.1093/annonc/mdx112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwaederlé M.C., Patel S.P., Husain H., Ikeda M., Lanman R.B., Banks K.C., Talasaz A., Bazhenova L., Kurzrock R. Utility of Genomic Assessment of Blood-Derived Circulating Tumor DNA (ctDNA) in Patients with Advanced Lung Adenocarcinoma. Clin. Cancer Res. 2017;23:5101–5111. doi: 10.1158/1078-0432.CCR-16-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raja R., Kuziora M., Brohawn P.Z., Higgs B.W., Gupta A., Dennis P.A., Ranade K. Early Reduction in ctDNA Predicts Survival in Patients with Lung and Bladder Cancer Treated with Durvalumab. Clin. Cancer Res. 2018;24:6212–6222. doi: 10.1158/1078-0432.CCR-18-0386. [DOI] [PubMed] [Google Scholar]
- 53.Phallen J., Leal A., Woodward B.D., Forde P.M., Naidoo J., Marrone K.A., Brahmer J.R., Fiksel J., Medina J.E., Cristiano S., et al. Early Noninvasive Detection of Response to Targeted Therapy in Non–Small Cell Lung Cancer. Cancer Res. 2019;79:1204–1213. doi: 10.1158/0008-5472.CAN-18-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee R., Gremel G., Marshall A., Myers K., Fisher N., Dunn J., Dhomen N., Corrie P., Middleton M., Lorigan P., et al. Circulating tumor DNA predicts survival in patients with resected high-risk stage II/III melanoma. Ann. Oncol. 2018;29:490–496. doi: 10.1093/annonc/mdx717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tjensvoll K., Lapin M., Buhl T., Oltedal S., Berry K.S.-O., Gilje B., Søreide J.A., Javle M., Nordgård O., Smaaland R. Clinical relevance of circulating KRAS mutated DNA in plasma from patients with advanced pancreatic cancer. Mol. Oncol. 2015;10:635–643. doi: 10.1016/j.molonc.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dianat-Moghadam H., Azizi M., Eslami-S Z., Cortés-Hernández L.E., Heidarifard M., Nouri M., Alix-Panabières C. The Role of Circulating Tumor Cells in the Metastatic Cascade: Biology, Technical Challenges, and Clinical Relevance. Cancers. 2020;12:867. doi: 10.3390/cancers12040867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cristofanilli M., Pierga J.-Y., Reuben J., Rademaker A., Davis A.A., Peeters D.J., Fehm T., Nolé F., Gisbert-Criado R., Mavroudis D., et al. The clinical use of circulating tumor cells (CTCs) enumeration for staging of metastatic breast cancer (MBC): International expert consensus paper. Crit. Rev. Oncol. Hematol. 2019;134:39–45. doi: 10.1016/j.critrevonc.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 58.Bidard F.-C., Peeters D.J., Fehm T., Nolé F., Gisbert-Criado R., Mavroudis D., Grisanti S., Generali D., Garcia-Saenz J.A., Stebbing J., et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: A pooled analysis of individual patient data. Lancet Oncol. 2014;15:406–414. doi: 10.1016/S1470-2045(14)70069-5. [DOI] [PubMed] [Google Scholar]
- 59.Cohen S.J., Punt C.J.A., Iannotti N., Saidman B.H., Sabbath K.D., Gabrail N.Y., Picus J., Morse M., Mitchell E., Miller M.C., et al. Relationship of Circulating Tumor Cells to Tumor Response, Progression-Free Survival, and Overall Survival in Patients with Metastatic Colorectal Cancer. J. Clin. Oncol. 2008;26:3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 60.Janni W.J., Rack B., Terstappen L.W.M.M., Pierga J.-Y., Taran F.-A., Fehm T., Hall C., De Groot M.R., Bidard F.-C., Friedl T.W., et al. Pooled Analysis of the Prognostic Relevance of Circulating Tumor Cells in Primary Breast Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016;22:2583–2593. doi: 10.1158/1078-0432.CCR-15-1603. [DOI] [PubMed] [Google Scholar]
- 61.Trapp E., Janni W., Schindlbeck C., Jückstock J., Andergassen U., De Gregorio A., Alunni-Fabbroni M., Tzschaschel M., Polasik A., Koch J.G., et al. Presence of Circulating Tumor Cells in High-Risk Early Breast Cancer During Follow-Up and Prognosis. J. Natl. Cancer Inst. 2019;111:380–387. doi: 10.1093/jnci/djy152. [DOI] [PubMed] [Google Scholar]
- 62.Uenosono Y., Arigami T., Kozono T., Yanagita S., Hagihara T., Haraguchi N., Matsushita D., Hirata M., Arima H., Funasako Y., et al. Clinical significance of circulating tumor cells in peripheral blood from patients with gastric cancer. Cancer. 2013;119:3984–3991. doi: 10.1002/cncr.28309. [DOI] [PubMed] [Google Scholar]
- 63.Balakrishnan A., Koppaka D., Anand A., Deb B., Grenci G., Viasnoff V., Thompson E.W., Gowda H., Bhat R., Rangarajan A., et al. Circulating Tumor Cell cluster phenotype allows monitoring response to treatment and predicts survival. Sci. Rep. 2019;9:7933. doi: 10.1038/s41598-019-44404-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sawabata N., Susaki Y., Nakamura T., Kawaguchi T., Yasukawa M., Taniguchi S. Cluster circulating tumor cells in surgical cases of lung cancer. Gen. Thorac. Cardiovasc. Surg. 2020;68:975–983. doi: 10.1007/s11748-020-01308-3. [DOI] [PubMed] [Google Scholar]
- 65.Szczerba B.M., Castro-Giner F., Vetter M., Krol I., Gkountela S., Landin J., Scheidmann M.C., Donato C., Scherrer R., Singer J., et al. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nat. Cell Biol. 2019;566:553–557. doi: 10.1038/s41586-019-0915-y. [DOI] [PubMed] [Google Scholar]
- 66.Gkountela S., Castro-Giner F., Szczerba B.M., Vetter M., Landin J., Scherrer R., Krol I., Scheidmann M.C., Beisel C., Stirnimann C.U., et al. Circulating Tumor Cell Clustering Shapes DNA Methylation to Enable Metastasis Seeding. Cell. 2019;176:98–112.e14. doi: 10.1016/j.cell.2018.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Satelli A., Brownlee Z., Mitra A., Meng Q.H., Li S., Mitra A. Circulating Tumor Cell Enumeration with a Combination of Epithelial Cell Adhesion Molecule- and Cell-Surface Vimentin–Based Methods for Monitoring Breast Cancer Therapeutic Response. Clin. Chem. 2015;61:259–266. doi: 10.1373/clinchem.2014.228122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yokobori T., Iinuma H., Shimamura T., Imoto S., Sugimachi K., Ishii H., Iwatsuki M., Ota D., Ohkuma M., Iwaya T., et al. Plastin3 Is a Novel Marker for Circulating Tumor Cells Undergoing the Epithelial–Mesenchymal Transition and Is Associated with Colorectal Cancer Prognosis. Cancer Res. 2013;73:2059–2069. doi: 10.1158/0008-5472.CAN-12-0326. [DOI] [PubMed] [Google Scholar]
- 69.Hou J., Guo C., Lyu G. Clinical significance of epithelial–mesenchymal transition typing of circulating tumour cells in colorectal cancer. Colorectal Dis. 2020;22:581–587. doi: 10.1111/codi.14927. [DOI] [PubMed] [Google Scholar]
- 70.Kilgour E., Rothwell D.G., Brady G., Dive C. Liquid Biopsy-Based Biomarkers of Treatment Response and Resistance. Cancer Cell. 2020;37:485–495. doi: 10.1016/j.ccell.2020.03.012. [DOI] [PubMed] [Google Scholar]
- 71.Douillard J.-Y., Ostoros G., Cobo M., Ciuleanu T., Cole R., McWalter G., Walker J., Dearden S., Webster A., Milenkova T., et al. Gefitinib Treatment in EGFR Mutated Caucasian NSCLC: Circulating-Free Tumor DNA as a Surrogate for Determination of EGFR Status. J. Thorac. Oncol. Off. Publ. Int. Assoc. Study Lung Cancer. 2014;9:1345–1353. doi: 10.1097/JTO.0000000000000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mayo-De-Las-Casas C., Jordana-Ariza N., Garzón-Ibañez M., Balada-Bel A., Bertrán-Alamillo J., Viteri-Ramírez S., Reguart N., Muñoz-Quintana M.A., Lianes-Barragan P., Camps C., et al. Large scale, prospective screening of EGFR mutations in the blood of advanced NSCLC patients to guide treatment decisions. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2017;28:2248–2255. doi: 10.1093/annonc/mdx288. [DOI] [PubMed] [Google Scholar]
- 73.Wu Y.-L., Lee V., Liam C.-K., Lu S., Park K., Srimuninnimit V., Wang J., Zhou C., Appius A., Button P., et al. Clinical utility of a blood-based EGFR mutation test in patients receiving first-line erlotinib therapy in the ENSURE, FASTACT-2, and ASPIRATION studies. Lung Cancer. 2018;126:1–8. doi: 10.1016/j.lungcan.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 74.Jenkins S., Yang J.C.-H., Ramalingam S.S., Yu K., Patel S., Weston S., Hodge R., Cantarini M., Jänne P.A., Mitsudomi T., et al. Plasma ctDNA Analysis for Detection of the EGFR T790M Mutation in Patients with Advanced Non–Small Cell Lung Cancer. J. Thorac. Oncol. Off. Publ. Int. Assoc. Study Lung Cancer. 2017;12:1061–1070. doi: 10.1016/j.jtho.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 75.Siravegna G., Mussolin B., Buscarino M., Corti G., Cassingena A., Crisafulli G., Ponzetti A., Cremolini C., Amatu A., Lauricella C., et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat. Med. 2015;21:795–801. doi: 10.1038/nm.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schiavon G., Hrebien S., Garcia-Murillas I., Cutts R.J., Pearson A., Tarazona N., Fenwick K., Kozarewa I., Lopez-Knowles E., Ribas R., et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci. Transl. Med. 2015;7:313ra182. doi: 10.1126/scitranslmed.aac7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.O’Leary B., Hrebien S., Morden J.P., Beaney M., Fribbens C., Huang X., Liu Y., Bartlett C.H., Koehler M., Cristofanilli M., et al. Early circulating tumor DNA dynamics and clonal selection with palbociclib and fulvestrant for breast cancer. Nat. Commun. 2018;9:1–10. doi: 10.1038/s41467-018-03215-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.André F., Ciruelos E., Rubovszky G., Campone M., Loibl S., Rugo H.S., Iwata H., Conte P., Mayer I.A., Kaufman B., et al. Alpelisib for PIK3CA-Mutated, Hormone Receptor–Positive Advanced Breast Cancer. N. Engl. J. Med. 2019;380:1929–1940. doi: 10.1056/NEJMoa1813904. [DOI] [PubMed] [Google Scholar]
- 79.Antonarakis E.S., Lu C., Wang H., Luber B., Nakazawa M., Roeser J.C., Chen Y., Mohammad T.A., Chen Y., Fedor H.L., et al. AR-V7 and Resistance to Enzalutamide and Abiraterone in Prostate Cancer. N. Engl. J. Med. 2014;371:1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Scher H.I., Graf R.P., Schreiber N.A., Jayaram A., Winquist E., McLaughlin B., Lu D., Fleisher M., Orr S., Lowes L., et al. Assessment of the Validity of Nuclear-Localized Androgen Receptor Splice Variant 7 in Circulating Tumor Cells as a Predictive Biomarker for Castration-Resistant Prostate Cancer. JAMA Oncol. 2018;4:1179–1186. doi: 10.1001/jamaoncol.2018.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Armstrong A.J., Halabi S., Luo J., Nanus D.M., Giannakakou P., Szmulewitz R.Z., Danila D.C., Healy P., Anand M., Rothwell C.J., et al. Prospective Multicenter Validation of Androgen Receptor Splice Variant 7 and Hormone Therapy Resistance in High-Risk Castration-Resistant Prostate Cancer: The PROPHECY Study. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2019;37:1120–1129. doi: 10.1200/JCO.18.01731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pal S.K., He M., Chen L., Yang L., Pillai R., Twardowski P., Hsu J., Kortylewski M., Jones J.O. Synaptophysin expression on circulating tumor cells in patients with castration resistant prostate cancer undergoing treatment with abiraterone acetate or enzalutamide. Urol. Oncol. Semin. Orig. Investig. 2018;36:162.e1–162.e6. doi: 10.1016/j.urolonc.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Paoletti C., Schiavon G., Dolce E.M., Darga E.P., Carr T.H., Geradts J., Hoch M., Klinowska T., Lindemann J., Marshall G., et al. Circulating Biomarkers and Resistance to Endocrine Therapy in Metastatic Breast Cancers: Correlative Results from AZD9496 Oral SERD Phase I Trial. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018;24:5860–5872. doi: 10.1158/1078-0432.CCR-18-1569. [DOI] [PubMed] [Google Scholar]
- 84.Pailler E., Adam J., Barthélémy A., Oulhen M., Auger N., Valent A., Borget I., Planchard D., Taylor M., André F., et al. Detection of Circulating Tumor Cells Harboring a Unique ALK Rearrangement in ALK-Positive Non–Small-Cell Lung Cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2013;31:2273–2281. doi: 10.1200/JCO.2012.44.5932. [DOI] [PubMed] [Google Scholar]
- 85.Hodgkinson C.L., Morrow C.J., Li Y., Metcalf R.L., Rothwell D.G., Trapani F., Polanski R., Burt D.J., Simpson K.L., Morris K., et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat. Med. 2014;20:897–903. doi: 10.1038/nm.3600. [DOI] [PubMed] [Google Scholar]
- 86.Goodman C.R., Seagle B.-L.L., Friedl T.W.P., Rack B., Lato K., Fink V., Cristofanilli M., Donnelly E.D., Janni W., Shahabi S., et al. Association of Circulating Tumor Cell Status With Benefit of Radiotherapy and Survival in Early-Stage Breast Cancer. JAMA Oncol. 2018;4:e180163. doi: 10.1001/jamaoncol.2018.0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bidard F.-C., Jacot W., Dureau S., Bachelot T., Pierga J.-Y., Bourgeois H., Goncalves A., Ladoire S., Naman H., Dalenc F., et al. Efficacy of Circulating Tumor Cell Count-Driven vs Clinician-Driven First-line Therapy Choice in Hormone Receptor-Positive, ERBB2-Negative Metastatic Breast Cancer: The STIC CTC Randomized Clinical Trial. JAMA Oncol. 2021;7:34–41. doi: 10.1001/jamaoncol.2020.5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Carter L., Rothwell D.G., Mesquita B., Smowton C., Leong H.S., Fernandez-Gutierrez F., Li Y., Burt D.J., Antonello J., Morrow C.J., et al. Molecular analysis of circulating tumor cells identifies distinct copy-number profiles in patients with chemosensitive and chemorefractory small-cell lung cancer. Nat. Med. 2017;23:114–119. doi: 10.1038/nm.4239. [DOI] [PubMed] [Google Scholar]
- 89.Postow M.A., Callahan M.K., Wolchok J.D. Immune Checkpoint Blockade in Cancer Therapy. J. Clin. Oncol. 2015;33:1974–1982. doi: 10.1200/JCO.2014.59.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gandara D.R., Paul S.M., Kowanetz M., Schleifman E., Zou W., Li Y., Rittmeyer A., Fehrenbacher L., Otto G., Malboeuf C., et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat. Med. 2018;24:1441–1448. doi: 10.1038/s41591-018-0134-3. [DOI] [PubMed] [Google Scholar]
- 91.Wang Z., Duan J., Cai S., Han M., Dong H., Zhao J., Zhu B., Wang S., Zhuo M., Sun J., et al. Assessment of Blood Tumor Mutational Burden as a Potential Biomarker for Immunotherapy in Patients with Non–Small Cell Lung Cancer With Use of a Next-Generation Sequencing Cancer Gene Panel. JAMA Oncol. 2019;5:696–702. doi: 10.1001/jamaoncol.2018.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Janning M., Kobus F., Babayan A., Wikman H., Velthaus J.-L., Bergmann S., Schatz S., Falk M., Berger L.-A., Böttcher L.-M., et al. Determination of PD-L1 Expression in Circulating Tumor Cells of NSCLC Patients and Correlation with Response to PD-1/PD-L1 Inhibitors. Cancers. 2019;11:835. doi: 10.3390/cancers11060835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Diaz L.A., Bardelli A. Liquid Biopsies: Genotyping Circulating Tumor DNA. J. Clin. Oncol. 2014;32:579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Newman A.M., Bratman S.V., To J., Wynne J.F., Eclov N.C.W., Modlin L.A., Liu C.L., Neal J.W., Wakelee H.A., Merritt R.E., et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014;20:548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Diehl F., Schmidt K., Choti M.A., Romans K.E., Goodman S.N., Li M., Thornton K., Agrawal N., Sokoll L.J., Szabo S.A., et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schøler L.V., Reinert T., Ørntoft M.-B.W., Kassentoft C.G., Árnadóttir S.S., Vang S., Nordentoft I., Knudsen M., Lamy P., Andreasen D., et al. Clinical Implications of Monitoring Circulating Tumor DNA in Patients with Colorectal Cancer. Clin. Cancer Res. 2017;23:5437–5445. doi: 10.1158/1078-0432.CCR-17-0510. [DOI] [PubMed] [Google Scholar]
- 97.Garcia-Murillas I., Chopra N., Comino-Méndez I., Beaney M., Tovey H., Cutts R.J., Swift C., Kriplani D., Afentakis M., Hrebien S., et al. Assessment of Molecular Relapse Detection in Early-Stage Breast Cancer. JAMA Oncol. 2019;5:1473–1478. doi: 10.1001/jamaoncol.2019.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Goldberg S.B., Narayan A., Kole A.J., Decker R.H., Teysir J., Carriero N.J., Lee A., Nemati R., Nath S.K., Mane S.M., et al. Early Assessment of Lung Cancer Immunotherapy Response via Circulating Tumor DNA. Clin. Cancer Res. 2018;24:1872–1880. doi: 10.1158/1078-0432.CCR-17-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dagogo-Jack I., Brannon A.R., Ferris L.A., Campbell C.D., Lin J.J., Schultz K.R., Ackil J., Stevens S., Dardaei L., Yoda S., et al. Tracking the Evolution of Resistance to ALK Tyrosine Kinase Inhibitors Through Longitudinal Analysis of Circulating Tumor DNA. JCO Precis. Oncol. 2018;2018:1–14. doi: 10.1200/PO.17.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li Y., Gong J., Zhang Q., Lu Z., Gao J., Li Y., Cao Y., Shen L. Dynamic monitoring of circulating tumour cells to evaluate therapeutic efficacy in advanced gastric cancer. Br. J. Cancer. 2016;114:138–145. doi: 10.1038/bjc.2015.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schochter F., Friedl T.W.P., DeGregorio A., Krause S., Huober J., Rack B., Janni W. Are Circulating Tumor Cells (CTCs) Ready for Clinical Use in Breast Cancer? An Overview of Completed and Ongoing Trials Using CTCs for Clinical Treatment Decisions. Cells. 2019;8:1412. doi: 10.3390/cells8111412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Merker J.D., Oxnard G.R., Compton C., Diehn M., Hurley P., Lazar A.J., Lindeman N., Lockwood C.M., Rai A.J., Schilsky R.L., et al. Circulating Tumor DNA Analysis in Patients with Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2018;36:1631–1641. doi: 10.1200/JCO.2017.76.8671. [DOI] [PubMed] [Google Scholar]
- 103.Coakley M., Garcia-Murillas I., Turner N.C. Molecular Residual Disease and Adjuvant Trial Design in Solid Tumors. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019;25:6026–6034. doi: 10.1158/1078-0432.CCR-19-0152. [DOI] [PubMed] [Google Scholar]
- 104.Goossens N., Nakagawa S., Sun X., Hoshida Y. Cancer biomarker discovery and validation. Transl. Cancer Res. 2015;4:256–269. doi: 10.3978/j.issn.2218-676X.2015.06.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Thomsen L.C.V., Honoré A., Reisæter L.A., Almås B., Førde K., Kristoffersen E.K., Melve G.K., Biermann M., Helle S.I., Azeem W., et al. A prospective phase I trial of dendritic cell-based cryoimmunotherapy in metastatic castration-resistant prostate cancer. J. Clin. Oncol. 2020;38:3029. doi: 10.1200/JCO.2020.38.15_suppl.3029. [DOI] [Google Scholar]
- 106.Lapin M., Tjensvoll K., Oltedal S., Buhl T., Gilje B., Smaaland R., Nordgård O. MINDEC-An Enhanced Negative Depletion Strategy for Circulating Tumour Cell Enrichment. Sci. Rep. 2016;6:28929. doi: 10.1038/srep28929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lapin M., Tjensvoll K., Oltedal S., Javle M., Smaaland R., Gilje B., Nordgård O. Single-cell mRNA profiling reveals transcriptional heterogeneity among pancreatic circulating tumour cells. BMC Cancer. 2017;17:1–10. doi: 10.1186/s12885-017-3385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lapin M., Oltedal S., Tjensvoll K., Buhl T., Smaaland R., Garresori H., Javle M., Glenjen N.I., Abelseth B.K., Gilje B., et al. Fragment size and level of cell-free DNA provide prognostic information in patients with advanced pancreatic cancer. J. Transl. Med. 2018;16:1–10. doi: 10.1186/s12967-018-1677-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tjensvoll K., Nordgård O., Skjæveland M., Oltedal S., Janssen E.A.M., Gilje B. Detection of disseminated tumor cells in bone marrow predict late recurrences in operable breast cancer patients. BMC Cancer. 2019;19:1–11. doi: 10.1186/s12885-019-6268-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lampignano R., Neumann M.H.D., Weber S., Kloten V., Herdean A., Voss T., Groelz D., Babayan A., Tibbesma M., Schlumpberger M., et al. Multicenter Evaluation of Circulating Cell-Free DNA Extraction and Downstream Analyses for the Development of Standardized (Pre)analytical Work Flows. Clin. Chem. 2019;66:149–160. doi: 10.1373/clinchem.2019.306837. [DOI] [PubMed] [Google Scholar]
- 111.Ignatiadis M., Dawson S.-J. Circulating tumor cells and circulating tumor DNA for precision medicine: Dream or reality? Ann. Oncol. 2014;25:2304–2313. doi: 10.1093/annonc/mdu480. [DOI] [PubMed] [Google Scholar]
- 112.Haber D.A., Velculescu V.E. Blood-Based Analyses of Cancer: Circulating Tumor Cells and Circulating Tumor DNA. Cancer Discov. 2014;4:650–661. doi: 10.1158/2159-8290.CD-13-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Riva F., Dronov O.I., Khomenko D.I., Huguet F., Louvet C., Mariani P., Stern M.-H., Lantz O., Proudhon C., Pierga J.-Y., et al. Clinical applications of circulating tumor DNA and circulating tumor cells in pancreatic cancer. Mol. Oncol. 2016;10:481–493. doi: 10.1016/j.molonc.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Connors D., Allen J., Alvarez J., Boyle J., Cristofanilli M., Hiller C., Keating S., Kelloff G., Leiman L., McCormack R., et al. International liquid biopsy standardization alliance white paper. Crit. Rev. Oncol. Hematol. 2020;156:103112. doi: 10.1016/j.critrevonc.2020.103112. [DOI] [PubMed] [Google Scholar]
- 115.Herbst R.S., Morgensztern D., Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446–454. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 116.Heerink W.J., De Bock G.H., De Jonge G.J., Groen H.J.M., Vliegenthart R., Oudkerk M. Complication rates of CT-guided transthoracic lung biopsy: Meta-analysis. Eur. Radiol. 2017;27:138–148. doi: 10.1007/s00330-016-4357-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mok T.S., Wu Y.-L., Ahn M.-J., Garassino M.C., Kim H.R., Ramalingam S.S., Shepherd F.A., He Y., Akamatsu H., Theelen W.S., et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N. Engl. J. Med. 2016;376:629–640. doi: 10.1056/NEJMoa1612674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Luo J., Shen L., Zheng D. Diagnostic value of circulating free DNA for the detection of EGFR mutation status in NSCLC: A systematic review and meta-analysis. Sci. Rep. 2015;4:6269. doi: 10.1038/srep06269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.FDA Approves Liquid Biopsy NGS Companion Diagnostic Test for Multiple Cancers and Biomarkers. [(accessed on 29 December 2020)]; Available online: https://www.fda.gov/drugs/fda-approves-liquid-biopsy-ngs-companion-diagnostic-test-multiple-cancers-and-biomarkers.
- 120.de Bono J., Mateo J., Fizazi K., Saad F., Shore N., Sandhu S., Chi K.N., Sartor O., Agarwal N., Olmos D., et al. Olaparib for Metastatic Castration-Resistant Prostate Cancer. N. Engl. J. Med. 2020;382:2091–2102. doi: 10.1056/NEJMoa1911440. [DOI] [PubMed] [Google Scholar]
- 121.Necchi A., Grivas P., Bratslavsky G., Shapiro O., Jacob J., Sokol E., Vergilio J.-A., Killian K., Lin D.I., Ramkissoon S.H., et al. Comprehensive Genomic Profiling (CGP) in Post-Systemic Treatment (Post) Metastatic Sites (MET) and Pretreatment (Pre) Primary Tumors (PT) of Metastatic Prostate Cancer (MPC) J. Clin. Oncol. 2020;38:175. doi: 10.1200/JCO.2020.38.6_suppl.175. [DOI] [Google Scholar]
- 122.Srivastava S., Koay E.J., Borowsky A.D., De Marzo A.M., Ghosh S., Wagner P.D., Kramer B.S. Cancer overdiagnosis: A biological challenge and clinical dilemma. Nat. Rev. Cancer. 2019;19:349–358. doi: 10.1038/s41568-019-0142-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.UKE—European Liquid Biopsy Society (ELBS) [(accessed on 29 December 2020)]; Available online: https://www.uke.de/english/departments-institutes/institutes/tumor-biology/european-liquid-biopsy-society-elbs/index.html.
- 124.BloodPAC. [(accessed on 29 December 2020)]; Available online: https://www.bloodpac.org.
- 125.Geeurickx E., Hendrix A. Targets, pitfalls and reference materials for liquid biopsy tests in cancer diagnostics. Mol. Asp. Med. 2020;72:100828. doi: 10.1016/j.mam.2019.10.005. [DOI] [PubMed] [Google Scholar]
- 126.Peng R., Zhang R., Horan M.P., Zhou L., Chai S.Y., Pillay N., Tay K.H., Badrick T., Li J. From Somatic Variants Toward Precision Oncology: An Investigation of Reporting Practice for Next-Generation Sequencing-Based Circulating Tumor DNA Analysis. Oncologist. 2019;25:218–228. doi: 10.1634/theoncologist.2019-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Greytak S.R., Engel K.B., Parpart-Li S., Murtaza M., Bronkhorst A.J., Pertile M.D., Moore H.M. Harmonizing Cell-Free DNA Collection and Processing Practices through Evidence-Based Guidance. Clin Cancer Res. 2020;26:3104–3109. doi: 10.1158/1078-0432.CCR-19-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yang M., Topaloğlu U., Petty W.J., Pagni M., Foley K.L., Grant S.C., Robinson M., Bitting R.L., Thomas A., Alistar A.T., et al. Circulating mutational portrait of cancer: Manifestation of aggressive clonal events in both early and late stages. J. Hematol. Oncol. 2017;10:100. doi: 10.1186/s13045-017-0468-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pemovska T., Kontro M., Yadav B., Edgren H., Eldfors S., Szwajda A., Almusa H., Bespalov M.M., Ellonen P., Elonen E., et al. Individualized Systems Medicine Strategy to Tailor Treatments for Patients with Chemorefractory Acute Myeloid Leukemia. Cancer Discov. 2013;3:1416–1429. doi: 10.1158/2159-8290.CD-13-0350. [DOI] [PubMed] [Google Scholar]
- 130.Nakamura S., Yokoyama K., Shimizu E., Yusa N., Kondoh K., Ogawa M., Takei T., Kobayashi A., Ito M., Isobe M., et al. Prognostic impact of circulating tumor DNA status post–allogeneic hematopoietic stem cell transplantation in AML and MDS. Blood. 2019;133:2682–2695. doi: 10.1182/blood-2018-10-880690. [DOI] [PubMed] [Google Scholar]
- 131.Groot V.P., Mosier S., Javed A.A., Teinor J.A., Gemenetzis G., Ding D., Haley L.M., Yu J., Burkhart R.A., Hasanain A., et al. Circulating Tumor DNA as a Clinical Test in Resected Pancreatic Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019;25:4973–4984. doi: 10.1158/1078-0432.CCR-19-0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tie J., Wang Y., Tomasetti C., Li L., Springer S., Kinde I., Silliman N., Tacey M., Wong H.-L., Christie M., et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci. Transl. Med. 2016;8:346ra92. doi: 10.1126/scitranslmed.aaf6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lesko L.J., Zineh I., Huang S.-M. What Is Clinical Utility and Why Should We Care? Clin. Pharmacol. Ther. 2010;88:729–733. doi: 10.1038/clpt.2010.229. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable. No new data were created or analyzed in this study.