Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) caused a global pandemic of coronavirus disease in 2019 (COVID-19). Genome surveillance is a key method to track the spread of SARS-CoV-2 variants. Genetic diversity and evolution of SARS-CoV-2 were analyzed based on 260,673 whole-genome sequences, which were sampled from 62 countries between 24 December 2019 and 12 January 2021. We found that amino acid (AA) substitutions were observed in all SARS-CoV-2 proteins, and the top six proteins with the highest substitution rates were ORF10, nucleocapsid, ORF3a, spike glycoprotein, RNA-dependent RNA polymerase, and ORF8. Among 25,629 amino acid substitutions at 8484 polymorphic sites across the coding region of the SARS-CoV-2 genome, the D614G (93.88%) variant in spike and the P323L (93.74%) variant in RNA-dependent RNA polymerase were the dominant variants on six continents. As of January 2021, the genomic sequences of SARS-CoV-2 could be divided into at least 12 different clades. Distributions of SARS-CoV-2 clades were featured with temporal and geographical dynamics on six continents. Overall, this large-scale analysis provides a detailed mapping of SARS-CoV-2 variants in different geographic areas at different time points, highlighting the importance of evaluating highly prevalent variants in the development of SARS-CoV-2 antiviral drugs and vaccines.
Keywords: SARS-CoV-2, COVID-19, global pandemic, genetic diversity, genetic variant
1. Introduction
Since December 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused a global pandemic. As of 16 January 2021, more than 93 million cases and 2 million deaths have been reported, according to the daily update of the World Health Organization. As a virus of the Coronaviridae family, SARS-CoV-2 is a positive-sense, single-stranded RNA virus whose genome contains approximately 30,000 nucleotides, making it one of the largest genomes among RNA viruses. The coding region of the SARS-CoV-2 genome encodes for nonstructural proteins (NSPs) and structural proteins in different open reading frames (ORFs). The genetic makeup of polyproteins pp1a and pp1ab, encoded by ORFs 1a and 1b at the 5′-end, contains 16 nonstructural proteins (numbered from NSP1 to NSP16). The 3′-end of the genome (21 to 29 kb) encodes for at least 6 accessory proteins (ORF3a, ORF6, ORF7a, ORF7b, ORF8, ORF10) [1] and 4 structural proteins: spike glycoprotein (S), envelope protein (E), membrane glycoprotein (M), and nucleocapsid protein (N) (Figure 1A). Since viral proteins such as RNA-dependent RNA polymerase and spike glycoprotein are key drug targets [2], genetic variations in the genome may exert an important impact on the efficacy of current vaccines and antiviral treatments [3].
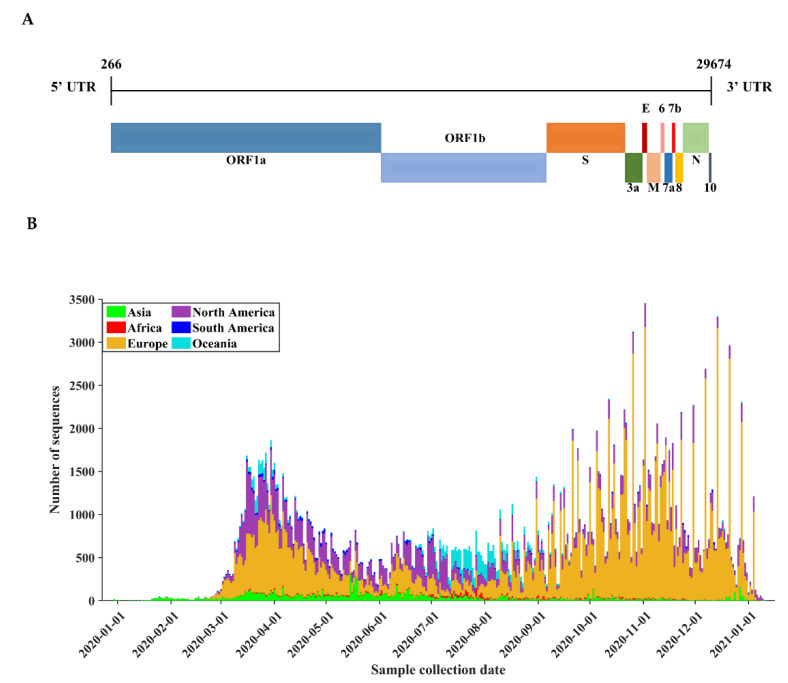
Figure 1.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genome organization and the distribution of the SARS-CoV-2 genome sequence used in this study. (A) Genomic architecture of SARS-CoV-2 based on the reference sequence (Wuhan-Hu-1, NCBI accession NC_045512). (B) The temporal and geographic distribution of all sequences. All temporal analyses were based on the date of sequence collection.
Global surveillance of SARS-CoV-2 genetic variants is important to control the coronavirus disease 2019 (COVID-19) pandemic. Previous analyses of SARS-CoV-2 genomes have characterized proximal origin [4,5,6], evolutionary rates [7], spatiotemporal distributions [8], transmission dynamics in many countries [9,10,11,12,13,14,15], phylogenetic networks of dominant strains [16,17,18,19], intragenomic diversity [20], epistatic interactions between viral genes [21], genetic variants in response to disease severity and immune responses [22,23,24,25], and the impact of spike variants on current vaccines and antibodies [26,27,28,29,30]. Despite its long genome structure, SARS-CoV-2 encodes a 3′-to-5′ exoribonuclease called NSP14 that provides the proofreading activity to correct errors made by the viral RNA-dependent RNA polymerase, thereby promoting a high fidelity of genome replication [31,32]. Nevertheless, some variants, such as D614G in spike, can escape proofreading and significantly shape the global dispersal of SARS-CoV-2 infections [33]. Moreover, circulating variants may exert a potential impact on the development of vaccines and antiviral drugs to control the COVID-19 pandemic [34]. Due to the dynamic dispersal of viral strains worldwide, it remains important to continue the surveillance of SARS-CoV-2 variants from a global perspective [35].
Here, we have conducted global surveillance of circulating variants in SARS-CoV-2 based on a large-scale dataset of whole-genome sequences from 62 countries worldwide. For the development of novel vaccines and antiviral drugs, this study contributes to a deeper understanding of diversity and global surveillance of key viral variants of SARS-CoV-2.
2. Materials and Methods
2.1. Data Collection
On 16 January 2021, we extracted SARS-CoV-2 complete genome sequences with high coverage (with <1% undefined bases and <0.05% unique amino acid (AA) substitutions and no insertion/deletion unless verified by submitter) from the GISAID database (https://www.gisaid.org/, the accessed date: 16 January 2021). We also downloaded the sequence metadata regarding the geographical locations and collection times. We acknowledge all contributors who have kindly deposited and shared their genome data on GISAID. A list of genome sequence acknowledgments is provided in Data S1. The complete genome of the Wuhan-Hu-1 isolate (NCBI accession NC_045512) was also collected as a reference strain.
Sequence quality control was conducted as follows: (i) one sequence per patient was sampled, while multiple sequences sampled from one patient were screened using the metadata regarding patient ID, authors, lab origin, and country origin; and (ii) sequences from nonhuman origins were removed.
2.2. Multiple Sequence Alignment
Nucleotide sequences were aligned to the reference sequence (Wuhan-Hu-1) using the option of “addfragments” in MAFFT version 7.471 [36], and output alignments were manually scrutinized by Seaview version 5.0 (http://doua.prabi.fr/software/seaview, the accessed date: 16 January 2021). The 5′ and 3′ untranslated regions were trimmed, and only sequences that contained full-length coding regions were retained. Inhouse MATLAB code (available at https://github.com/MiaoMiaorrk/sequence_analysis, the accessed date: 16 January 2021) was prepared to extract the nucleotide regions of 26 viral proteins, while the coding region of the viral polymerase NSP12 was concatenated by considering programmed −1 ribosomal frameshifting. The corresponding amino acid sequences were subsequently obtained for analyzing protein diversity and amino acid variants.
2.3. Protein Sequence Diversity
Polymorphic sites are amino acid or nucleotide positions that have at least one substitution compared with the reference Wuhan-Hu-1, ignoring gaps and ambiguities [30]. Given multiple sequence alignments, the amino acid substitution rate was defined as:
| (1) |
| (2) |
is the element at the position of the sequence, and is the element at the position of the reference Wuhan-Hu-1. is the length of the protein, and is the number of sequences except for the reference sequence. is the total number of invalid positions containing gaps or ambiguous letters. is the function that checks if two elements and are equal.
2.4. Selective Pressure Analysis
To analyze the selective pressure on the protein-coding regions of SARS-CoV-2, the dN/dS ratio that measures the nonsynonymous (dN) to synonymous (dS) substitution rates at the coding region was calculated using KaKs_Calculator 2.0 with the method of modified YN (MYN) and the genetic code in Table 1 (standard code) [37]. The positive selection was defined by dN/dS > 1.
Table 1.
Basic characteristics of SARS-CoV-2 proteins.
| Protein | Gene | Protein Location | Protein Length | Polymorphic Sites | Substitution Rate (%) |
|---|---|---|---|---|---|
| NSP1 | ORF1a | 266–805 | 180 | 177 | 0.03 |
| NSP2 | ORF1a | 806–2719 | 638 | 608 | 0.07 |
| NSP3 | ORF1a | 2720–8554 | 1945 | 1752 | 0.03 |
| NSP4 | ORF1a | 8555–10,054 | 500 | 419 | 0.02 |
| 3C-like protease (NSP5) | ORF1a | 10,055–10,972 | 306 | 251 | 0.04 |
| NSP6 | ORF1a | 10,973–11,842 | 290 | 256 | 0.06 |
| NSP7 | ORF1a | 11,843–12,091 | 83 | 72 | 0.04 |
| NSP8 | ORF1a | 12,092–12,685 | 198 | 176 | 0.03 |
| NSP9 | ORF1a | 12,686–13,024 | 113 | 93 | 0.03 |
| NSP10 | ORF1a | 13,025–13,441 | 139 | 105 | 0.01 |
| NSP11 | ORF1a | 13,442–13,480 | 13 | 11 | 0.01 |
| RNA-dependent RNA polymerase (NSP12) | ORF1b | 13,442–13,468 13,468–16,236 | 932 | 715 | 0.13 |
| Helicase (NSP13) | ORF1b | 16,237–18,039 | 601 | 478 | 0.04 |
| 3′-to-5′ exonuclease (NSP14) | ORF1b | 18,040–19,620 | 527 | 444 | 0.03 |
| endoRNAse (NSP15) | ORF1b | 19,621–20,658 | 346 | 313 | 0.04 |
| 2′-O-ribose methyltransferase (NSP16) | ORF1b | 20,659–21,552 | 298 | 245 | 0.03 |
| Spike glycoprotein (S) | ORF2 | 21,563–25,384 | 1273 | 1096 | 0.14 |
| ORF3a | ORF3a | 25,393–26,220 | 275 | 273 | 0.20 |
| Envelope protein (E) | ORF4 | 26,245–26,472 | 75 | 72 | 0.02 |
| Membrane protein (M) | ORF5 | 26,523–27,191 | 222 | 168 | 0.02 |
| ORF6 | ORF6 | 27,202–27,387 | 61 | 60 | 0.03 |
| ORF7a | ORF7a | 27,394–27,759 | 121 | 121 | 0.04 |
| ORF7b | ORF7b | 27,756–27,887 | 43 | 43 | 0.07 |
| ORF8 | ORF8 | 27,894–28,259 | 121 | 121 | 0.12 |
| Nucleocapsid protein (N) | ORF9 | 28,274–29,533 | 419 | 378 | 0.31 |
| ORF10 | ORF10 | 29,558–29,674 | 38 | 37 | 0.72 |
2.5. Phylogenetic Analysis
At each sample collection date, from 24 December 2019 to 12 January 2021, three genomic sequences were randomly subsampled for each continent, while the sequences were all collected if the number of sequences was less than 3. This led to a subset of SARS-CoV-2 genome sequences (n = 5488). Multiple sequence alignments of this subset were subsequently imported to reconstruct a maximum-likelihood phylogenetic tree using NextStrain tools (https://nextstrain.org, the accessed date: 16 January 2021).
3. Results
3.1. Basic Characteristics of SARS-CoV-2 Genome Sequences
Our study analyzed a large-scale dataset of SARS-CoV-2 genome sequences (n = 260,673) that fulfilled the quality control criteria. During the pandemic of COVID-19, from 24 December 2019 to 12 January 2021, these full-length sequences were sampled from 62 countries located in Europe (n = 166,443), North America (n = 61,232), Asia (n = 13,709), Oceania (n = 13,166), South America (n = 3216), and Africa (n = 2907). The collection time of SARS-CoV-2 genome sequences is visualized in Figure 1B. Most sequences were collected during the first wave (March to May 2020) before the summer in the Northern Hemisphere and during the second wave (September 2020 to January 2021) in Europe and North America, where the COVID-19 pandemic remains severe.
3.2. Amino Acid Diversity of SARS-CoV-2 Proteins
The SARS-CoV-2 genome encodes 26 functional proteins that are potentially evolving during the global pandemic. Given 260,673 sequences, we measured the average amino acid diversity of SARS-CoV-2 proteins (Equation (1), Table 1). Among 26 viral proteins, the highest amino acid diversity was observed in ORF10 (0.72%), followed by nucleocapsid (0.31%), ORF3a (0.20%), spike (0.14%), NSP12 (0.13%), and ORF8 (0.12%), while the other proteins had low protein diversity (<0.1%). In contrast, two nonstructural proteins (NSP10 and NSP11) and two structural proteins (envelope and membrane proteins) had a low protein diversity (<0.02%).
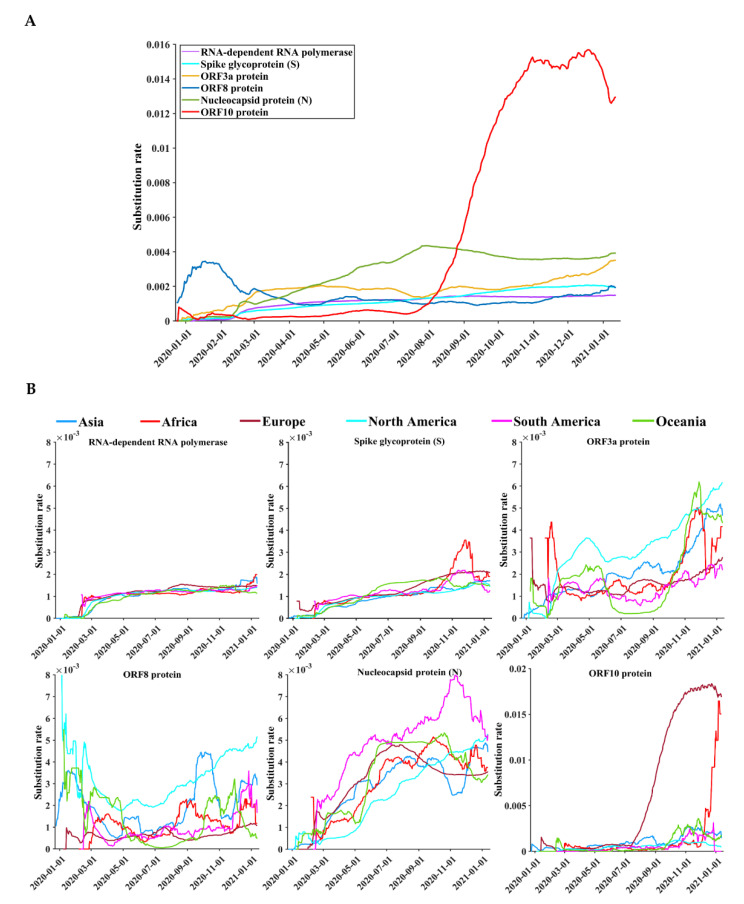
Due to the worldwide spread of SARS-CoV-2 infections, dynamic patterns of protein diversity were observed over time, especially in six viral proteins: ORF10, nucleocapsid, ORF3a, spike, NSP12, and ORF8 (Figure 2A). Unlike other proteins, whose protein diversities increase gradually over time, the protein diversity of ORF10 has increased sharply since July 2020. Figure 2B shows that the highest substitution rate of the ORF10 protein appeared in Europe, followed by Africa. Although the increasing patterns of protein diversity of the remaining four proteins, from December 2019 to January 2021, were roughly the same, the temporal patterns of spike and ORF10 diversity were different between Africa and other continents (Figure 2B). Protein diversity of spike and ORF10 reached their peaks in November and December 2020, respectively.
Figure 2.
Temporal distributions of the substitution rates of the six SARS-CoV-2 proteins. The proteins were the top six proteins with the highest substitution rates. (A) The global substitution rate curves of SARS-CoV-2 proteins based on a time sliding window. (B) The substitution rate curves of SARS-CoV-2 proteins based on a time sliding window on different continents. The vertical axis represents the moving-window substitution rate, calculated by dividing the total number of polymorphic sites of a protein contained in the sequences, 15 days before and after a specific date, by the total number of all positions of the protein in the period.
3.3. Positive Selection Is Rare in SARS-CoV-2 Protein-Coding Genes
To analyze selective pressure on SARS-CoV-2 protein-coding regions, we estimated the dN/dS ratios of nonsynonymous and synonymous substitution rates (Figure S1). The median dN/dS ratio for all protein-coding genes was 0.31 (IQR: 0.24 to 0.65). All viral protein-coding genes had a median dN/dS ratio less than 1, and the highest level of median dN/dS ratio was observed in ORF9 (0.65, IQR: 0.49 to 0.65), followed by ORF6 (0.56) and ORF7b (0.50) (Table S1).
3.4. Circulating Variants of SARS-CoV-2
Given the large-scale dataset of 260,673 sequences, we analyzed the characteristics of nucleotide and the amino acid variants of SARS-CoV-2. The nucleotide composition of the coding region of the reference sequence Wuhan-Hu-1 was A (29.94%), T (32.08%), C (18.37%), and G (19.61%). In the coding region of the SARS-CoV-2 genome, 22,964 of 29,409 nucleotide positions (78.08%) were polymorphic, with at least one single-nucleotide polymorphism (SNP) compared with the reference Wuhan-Hu-1. Most nucleotide substitutions (94.40%) were SNPs, while indels (5.60%) were rare. Moreover, the most common nucleotide substitutions were C to T (46.27%), G to T (13.47%), A to G (11.26%), T to C (6.66%), and others (22.34%).
We next analyzed the amino acid variants of SARS-CoV-2. There were 8484 (86.95%) polymorphic sites calculated as the number of sites having one or more substitutions compared to the reference sequence (Wuhan-Hu-1, NCBI accession NC_045512) across the 9757 amino acid positions in 26 concatenated proteins. In total, 25,629 substitutions were identified at 8484 amino acid positions. Notably, 587 of 1273 fully conserved positions were mostly located in the enzymatic proteins of NSP12 (17.05%), NSP3 (15.16%), and spike (13.90%). The frequency of variants on different continents was calculated by dividing the number of sequences carrying a given variant on a specific continent by the total number of sequences on that continent; 115 polymorphic sites occurred with a frequency greater than 0.5%. Highly polymorphic sites that occurred with a frequency greater than 1% were mostly observed in nucleocapsid (3.10%), ORF10 (2.63%), ORF8 (2.48%), ORF3a (2.18%), spike (0.71%), and NSP12 (0.64%).
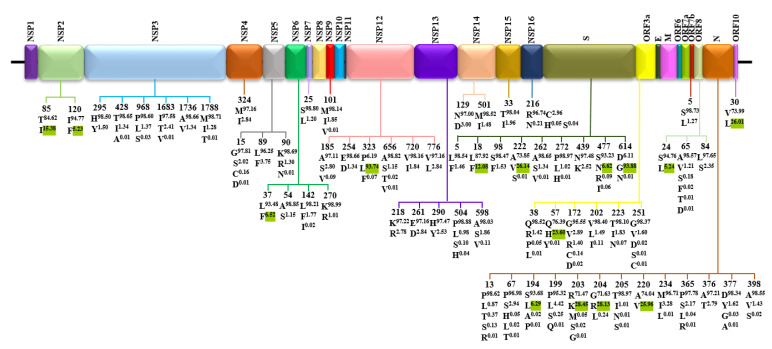
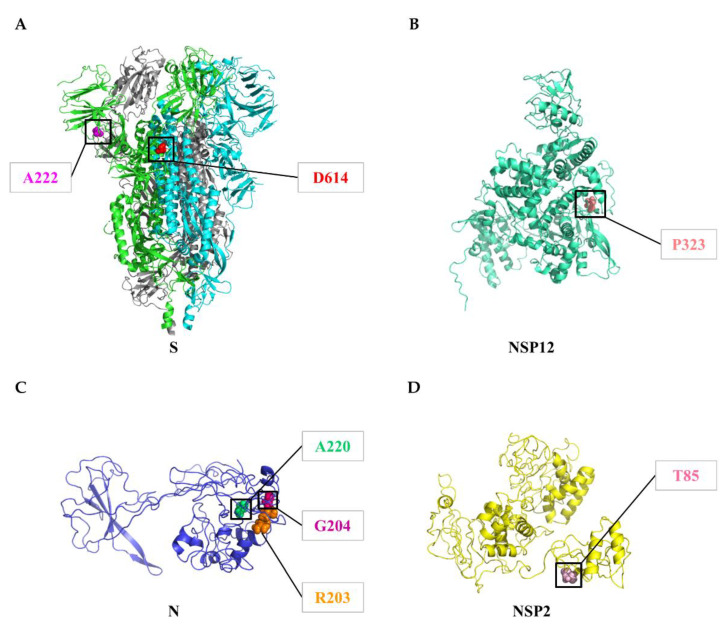
In addition, the major variants of each SARS-CoV-2 protein are mapped in Figure 3. For each position, the Wuhan-Hu-1 index is shown at the top, followed by the variants with a frequency >1%. Fifteen variants with the prevalence >5% were identified, including (i) four variants in nucleocapsid: S194L (6.29%), R203K (28.45%), G204R (28.13%), and A220V (25.96%); (ii) four variants in spike: L18F (12.08%), A222V (26.14%), S477N (6.62%), and D614G (93.88%); (iii) two variants in NSP2: T85I (15.38%) and I120F (5.23%); (iv) five variants in each of the 5 proteins: L37F (6.52%) in NSP6, P323L (93.74%) in NSP12, Q57H (23.60%) in ORF3a, S24L (5.24%) in ORF8, and V30L (26.01%) in ORF10.
Figure 3.
Distribution of variants at positions of SARS-CoV-2 proteins. For each site, the reference index is shown at the top, followed by variants with a frequency >1%. Variants highlighted with green superscripts were those with frequencies >5%.
3.5. Geographical and Temporal Trends of the Frequent Variants
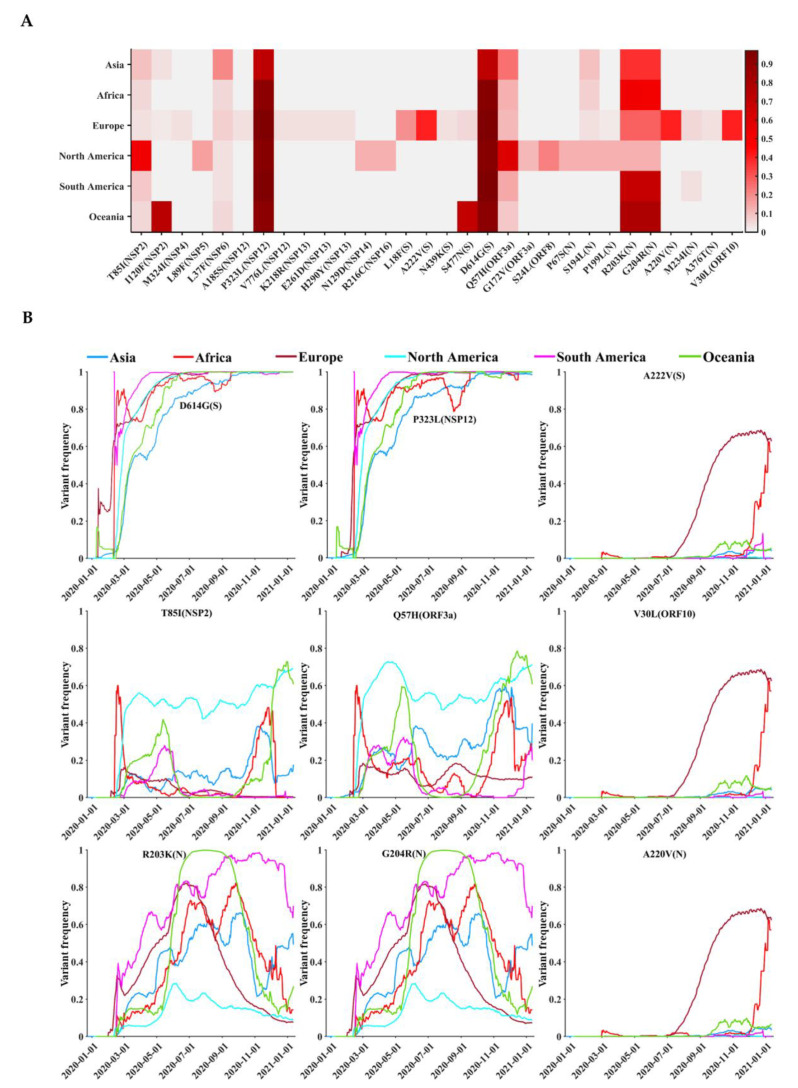
The geographic distribution and temporal progression of frequent variants (n = 30), with a high frequency greater than 2.5%, are shown in Figure 4. Figure 4A shows that these variants were found mainly in nucleocapsid (P67S, S194L, P199L, R203K, G204R, A220V, M234I, and A376T), spike (L18F, A222V, N439K, S477N, and D614G), NSP12 (A185S, P323L, and V776L), NSP13 (K218R, E261D, and H290Y), NSP2 (T85I and I120F), and ORF3a (Q57H and G172V), respectively. In addition, there was one variant in each of the seven proteins NSP4 (M324I), NSP5 (L89F), NSP6 (L37F), NSP14 (N129D), NSP16 (R216C), ORF8 (S24L), and ORF10 (V30L). Among all frequent variants, D614G in spike and P323L in NSP12, with a high frequency, were common on the six continents (Figure 4A). D614G (97.26%) and P323L (97.29%) (Table 2) in South America exhibited the highest variant prevalence. R203K and G204R in nucleocapsid showed different prevalence on the six continents. Seven common variants were mainly prevalent on a single continent, including the NSP2 variant T85I in North America, the NSP2 variant I120F in Oceania, the spike variant A222V in Europe, the spike variant S477N in Oceania, the ORF3a variant Q57H in North America, the nucleocapsid variant A220V in Europe, and the ORF10 variant V30L in Europe. The prevalence of the other 19 variants was generally similar across six continents.
Figure 4.
Distribution of frequent variants across the SARS-CoV-2 proteins in different geographic areas. (A) Frequencies of the top 30 variants, with the highest variant frequencies in different continents. (B) Temporal and geographic dynamics of frequent SARS-CoV-2 variants. The vertical axis represents the moving-window variant frequency calculated by dividing the number of sequences containing a specific variant, 15 days before and after a specific date, by the total number of sequences in the period (31 days).
Table 2.
The overall prevalence of SARS-CoV-2 variants in a large-scale dataset of 260,673 sequences.
| Genetic Variant | Variant Frequency (%) | ||||||
|---|---|---|---|---|---|---|---|
| Asia | Africa | Europe | North America | South America | Oceania | Total | |
| T85I(NSP2) | 10.23 | 5.72 | 3.26 | 52.63 | 8.84 | 5.96 | 15.38 |
| I120F(NSP2) | 3.43 | 0.00 | 2.12 | 0.02 | 0.00 | 72.98 | 5.23 |
| M324I(NSP4) | 0.25 | 0.41 | 4.38 | 0.08 | 0.09 | 0.11 | 2.84 |
| L89F(NSP5) | 0.23 | 0.03 | 0.08 | 15.73 | 0.06 | 0.33 | 3.75 |
| L37F(NSP6) | 19.93 | 4.93 | 6.84 | 3.28 | 3.09 | 4.78 | 6.52 |
| A185S(NSP12) | 0.15 | 0.34 | 4.35 | 0.04 | 0.00 | 0.12 | 2.81 |
| P323L(NSP12) | 71.54 | 91.83 | 96.04 | 92.87 | 97.29 | 91.22 | 93.74 |
| V776L(NSP12) | 0.18 | 0.34 | 4.33 | 0.24 | 0.00 | 0.12 | 2.84 |
| K218R(NSP13) | 0.15 | 0.34 | 4.32 | 0.02 | 0.00 | 0.11 | 2.78 |
| E261D(NSP13) | 0.20 | 0.58 | 4.39 | 0.08 | 0.03 | 0.11 | 2.84 |
| H290Y(NSP13) | 0.55 | 0.17 | 3.80 | 0.25 | 0.03 | 0.22 | 2.53 |
| N129D(NSP14) | 0.17 | 0.00 | 0.05 | 12.61 | 0.03 | 0.32 | 3.00 |
| R216C(NSP16) | 0.20 | 0.00 | 0.07 | 12.40 | 0.03 | 0.33 | 2.96 |
| L18F(S) | 0.32 | 0.28 | 18.78 | 0.30 | 0.34 | 0.12 | 12.08 |
| A222V(S) | 0.50 | 0.79 | 40.78 | 0.17 | 0.34 | 0.53 | 26.14 |
| N439K(S) | 0.45 | 0.03 | 3.90 | 0.01 | 0.06 | 0.06 | 2.52 |
| S477N(S) | 0.19 | 0.66 | 4.82 | 0.14 | 0.12 | 71.22 | 6.62 |
| D614G(S) | 71.91 | 93.91 | 96.11 | 93.10 | 97.26 | 91.26 | 93.88 |
| Q57H(ORF3a) | 24.97 | 12.34 | 11.86 | 59.86 | 14.94 | 8.08 | 23.59 |
| G172V(ORF3a) | 0.20 | 0.07 | 0.07 | 12.07 | 0.06 | 0.31 | 2.89 |
| S24L(ORF8) | 0.23 | 0.17 | 0.15 | 21.78 | 0.03 | 0.90 | 5.24 |
| P67S(N) | 0.17 | 0.03 | 0.11 | 12.21 | 0.06 | 0.33 | 2.94 |
| S194L(N) | 9.64 | 6.54 | 4.27 | 12.50 | 0.19 | 1.19 | 6.29 |
| P199L(N) | 0.29 | 0.21 | 2.29 | 12.49 | 0.06 | 0.40 | 4.42 |
| R203K(N) | 36.96 | 52.15 | 28.16 | 13.35 | 69.12 | 78.12 | 28.45 |
| G204R(N) | 36.73 | 51.62 | 27.71 | 13.31 | 69.07 | 78.08 | 28.13 |
| A220V(N) | 0.45 | 1.10 | 40.53 | 0.13 | 0.09 | 0.35 | 25.96 |
| M234I(N) | 0.50 | 0.55 | 4.77 | 0.61 | 3.64 | 0.30 | 3.28 |
| A376T(N) | 0.15 | 0.35 | 4.33 | 0.02 | 0.00 | 0.11 | 2.78 |
| V30L(ORF10) | 0.58 | 0.69 | 40.64 | 0.08 | 0.13 | 0.35 | 26.01 |
We next explored the temporal and geographic dynamics of the variants with a frequency >15%, including: D614G (93.88%) and A222V (26.14%) in spike; A220V (25.96%), R203K (28.45%), and G204R (28.13%) in nucleocapsid; T85I (15.38%) in NSP2; Q57H (23.60%) in ORF3a; P323L (93.74%) in NSP12; V30L (26.01%) in ORF10 (Table 2). Figure 4B visualizes the temporal and geographical dynamics of 9 common variants on six continents during the COVID-19 pandemic, from December 2019 to January 2021. (i) Since the emergence of the spike variant D614G (Figure 5A) in January 2020 and the NSP12 variant P323L (Figure 5B) in January 2020, they have become the dominant variants across all continents, with an increasing prevalence over time (Figure 4B). (ii) During the pandemic, from July to December 2020, the prevalence of A222V in spike, A220V in nucleocapsid (Figure 5C), and V30L in ORF10 increased continuously in Europe and Africa, while their emergence in other continents was less common. (iii) Two neighboring variants in nucleocapsid, R203K and G204R (Figure 5C), showed roughly similar frequencies over time in the same geographic areas. (iv) Temporal trends of the NSP2 variant T85I (Figure 5D) and the ORF3a variant Q57H were distinct on different continents. From March to December 2020, the prevalence of T85I and Q57H was higher in North America than on other continents.
Figure 5.
Variants in the SARS-CoV-2 proteins. (A) Site 222 (magenta) and Site 614 (red) of spike. Almost all sequences showed a variant (D614G). Site 614 is located at the interface between two subunits. (B) Site 323 (deep salmon) of the NSP12 protein. Many sequences showed a variant (P323L). (C) Site 203 (orange), Site 204 (hot pink), and Site 220 (lime green) of the nucleocapsid protein. The frequencies of variants R203K, G204R, and A220V were high. (D) Site 85 (light pink) of the NSP2 protein. Many sequences showed a variant (T85I). The structures of spike, NSP12, the nucleocapsid protein, and NSP2 were collected from https://zhanglab.ccmb.med.umich.edu/COVID-19/. The protein structural figures were generated by the software PyMOL (http://www.pymol.org/, the accessed date: 16 January 2021).
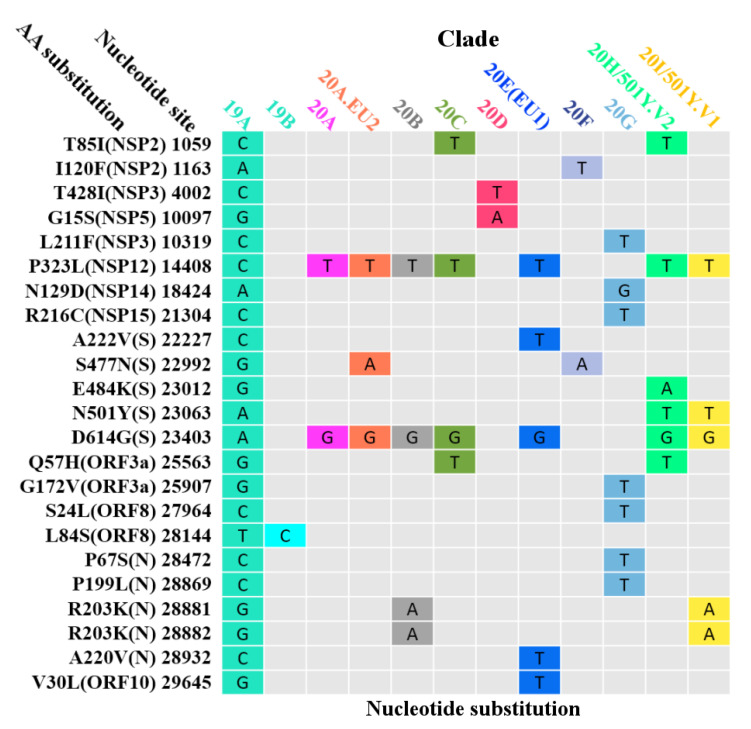
3.6. Global Surveillance of SARS-CoV-2 Clades
The phylogenetic tree (Figure S2), performed based on the tree topology and variant clusters of 5488 complete SARS-CoV-2 nucleotide sequences, was divided into at least 12 different clades (19A, 19B, 20A, 20A.EU2, 20B, 20C, 20D, 20E (EU1), 20F, 20G, 20H/501Y.V2, and 20I/501Y.V1), as of January 2021. SARS-CoV-2 clades are classified based on nucleotide substitutions in the genome (Figure 6). For example, the 20A clade is featured with the variants D614G (spike) and P323L (NSP12), while the 20A.EU2 clade is defined by three variants of P323L (NSP12), S477N (spike), and D614G (spike). Furthermore, T85I (NSP2) and Q57H (ORF3a) were circulating with a high frequency in North America (Figure 4B). These two variants, plus P323L (NSP12) and D614G (spike), define the 20C clade. Moreover, the 20E (EU1) clade, with P323L (NSP12), A222V (spike), D614G (spike), A220V (nucleocapsid), and V30L (ORF10), gradually gained their predominance in Europe over time.
Figure 6.
The substitution matrix at the nucleic acid level. Nucleotide substitutions showed in this figure are the main substitutions characterizing the SARS-CoV-2 clades.
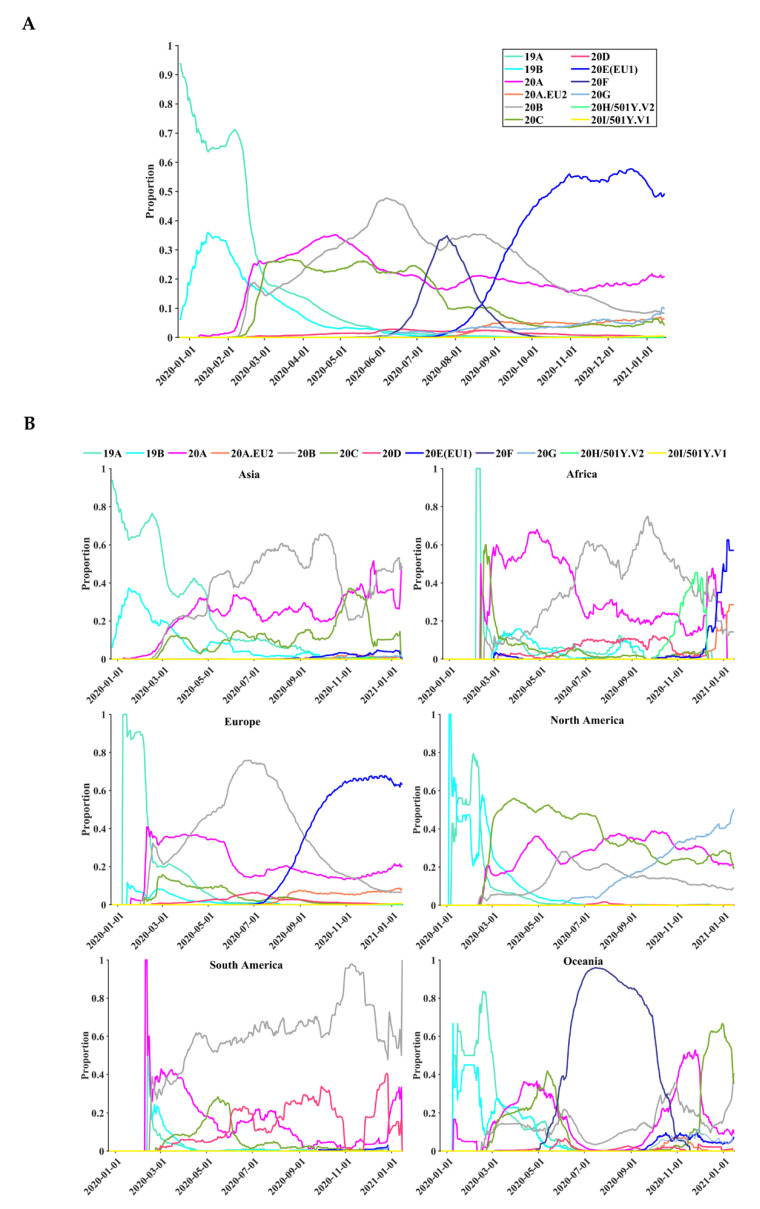
Temporal and geographical dynamics of 12 SARS-CoV-2 clades were observed on six continents based on 260,673 full-length genome sequences (Figure 7). Among the 12 different clades, the most prevalent clade was 20E (EU1) (n = 68,173, 26.15%), followed by 20B (n = 60,278, 23.12%), 20A (n = 56,801, 21.79%), 20C (n = 32,608, 12.51%), and others (n = 42,813, 16.43%) (Table S2). The 19A and 19B clades dominated the global infections from December 2019 to March 2020 (Figure 7A). Their dominant role in the global pandemic was subsequently replaced by the 20A clade. Since mid-September 2020, Clade 20E (EU1) became the dominant group worldwide. Furthermore, different continents exhibited different temporal dynamics (Figure 7B). For instance, the dominant clade in January 2021 was 20B in Asia, 20E (EU1) in Africa, 20E (EU1) in Europe, 20G in North America, 20B in South America, and 20C in Oceania. Clades 20H/501Y.V2 and 20I/501Y.V1, also known as B.1.351 and B.1.1.7, the two newly emerged clades, were mainly circulating in Africa and Europe, respectively.
Figure 7.
Temporal distributions of the 12 clades based on 260,673 complete SARS-CoV-2 nucleotide sequences. (A) The global distribution of the 12 clades over time. (B) The distribution of the 12 clades over time on six continents. The vertical axis represents the moving-window proportion, calculated by dividing the number of sequences belonging to a specific clade, 15 days before and after a specific date, by the total number of sequences in the period (31 days).
4. Discussion
This study presents global surveillance of SARS-CoV-2 genetic diversity and circulating variants based on a large-scale dataset of 260,673 sequences sampled from December 2019 to January 2021. Based on this first one-year survey of genetic diversity, our study describes the temporal and geographical trends of circulating variants on different continents. Despite the proofreading activity of viral exoribonuclease, an increasing prevalence of key genetic variants, such as D614G in viral spike and P323L in NSP12, was observed in many countries and continents. Early surveillance of potentially evolving viral strains with higher transmission fitness and greater infectivity plays an important role in controlling the ongoing COVID-19 pandemic.
Among 26 viral proteins, the highest protein sequence diversity was observed in spike, nucleocapsid, ORF3a, and ORF10 (Table 1). As a critical target of many vaccines and neutralizing antibodies, the spike protein interacts with cellular receptors, such as angiotensin-converting enzyme 2 (ACE2), for viral entry [38,39]. Although whether the spike variants may weaken the performance of vaccines is still under evaluation [40,41], antibody therapeutics and vaccine development still need to consider the impact of spike variants on the antigenicity of the virus [42]. As an indispensable protein, nucleocapsid is the major target of primers and probes in many real-time reverse transcription polymerase chain reaction (RT-PCR) tests, while nucleotide substitutions in the primer binding region may affect the sensitivity of some diagnostic assays [43]. Although single nucleotide substitutions may not be an issue for solid assays that target multiple loci (n > 2) in the genome [44], close monitoring of SARS-CoV-2 nucleotide substitutions that are located within the primer and probe regions of RT-PCR assays remains important. ORF3a plays a role in virulence, ion channel formation, viral release, and apoptosis [45], while ORF10 is an accessory protein that is dispensable for viral replication in humans [46]. Our study presents that the highest substitution rate of the ORF10 protein appeared in Europe, followed by Africa (Figure 2B). Previous studies have reported the import of SARS-CoV-2 from Europe to Africa [47,48], which explains some similar patterns between Europe and Africa. To support surveillance, a paucity of SARS-CoV-2 sequence data from Africa requires further sequencing efforts on the African continent [47]. Additionally, other viral proteins are also known to interact with many host proteins that participate in multiple biological processes such as protein trafficking, translation, and transcription [49]. Genetic variants that promote viral fitness and escape may exert a potential impact on current diagnostic tests, vaccines, antiviral strategies, and immune responses [23,40,50]. Close monitoring of key viral variants is critical for the development and optimization of vaccines and antiviral therapies, according to previous experience from the management of viral infections such as influenza virus, respiratory syncytial virus, and human immunodeficiency virus [51,52,53,54,55,56,57].
Highly prevalent variants have been observed in SARS-CoV-2. A high prevalence of important variants, such as D614G (93.88%) in spike and P323L (93.74%) in NSP12, was confirmed by our large-scale study. Compared with wildtype D614, the D614G variant changes the dynamic structures of the viral spike to bind with human angiotensin-converting enzyme 2 [58] and increases transmission fitness with enhanced viral loads in the upper respiratory tract of SARS-CoV-2 cases [34]. The D614G variant is located at a region interfacing with the neighboring subunit, and it potentially alters the trimer stability of the viral spike to enhance membrane fusion and host entry [59]. In addition, D614G increases the replication fitness of SARS-CoV-2 in cell cultures and enhances viral transmission in hamsters [60]. Furthermore, D614G in spike is coevolving with P323L in NSP12, with strong allelic associations [16]. The coexistence of D614G (spike), P323L (NSP12), and C241U (5′-UTR) may contribute to increased transmission fitness [61]. The P323L variant might alter the secondary and tertiary structures of NSP12 to interact with NSP8, thereby affecting viral replication [62,63]. Additionally, other variants like S25L in NSP7, S194L, R203K, G204R, and T205I in nucleocapsid, T85I and I120F in NSP2, S477N in spike, and Q57H in ORF3a have also been confirmed by previous studies [62,64]. Although many SARS-CoV-2 variants have been observed, their associations with viral loads and infectivity need more investigation. Vaccines may need to be updated periodically to avoid the potential impact of newly arising SARS-CoV-2 variants on the clinical efficacy of the vaccines [65].
Key SARS-CoV-2 variants in certain clades may play an important role in viral transmission and adaptability. Monitoring the dynamics of SARS-CoV-2 clades is crucial because different viral clades, such as B.1.1.7, may be associated with viral loads and disease severity [66]. Due to the evolving nature of SARS-CoV-2, the emergence of some variants in B.1.1.7, B.1.351, and P.1 clades may affect viral transmissibility and antigenic profiles [65,67]. Based on large-scale full-length genome sequences, our findings showed distinct geographical and temporal patterns of SARS-CoV-2 clades on different continents (Figure 7). We observed the dynamic patterns of SARS-CoV-2 clades over time; for instance, Clade 20E (EU1) has become the dominant group worldwide since mid-September 2020. The 20H/501Y.V2 and 20I/501Y.V1 clades were observed in Africa in October 2020 and in Europe in November 2020, respectively. SARS-CoV-2 variants in different clades might exert an impact on some vaccines and antibodies [27,65,68]. Spike variants such as D614G and N501Y, contained in the 20H/501Y.V2 and 20I/501Y.V1 clades, may lead to changes in viral antigenicity that are harmful to monoclonal antibody therapies and vaccine protection [67], thereby mediating potential escape from vaccine response [41]. Since a cure against COVID-19 is still lacking, the existence of current and novel circulating clades that are globally overdispersed highlights the importance of rapid interventions to reduce viral transmissions [10].
There are limitations in our study. First, our analyses focused on genetic diversity over the past one-year period, while global surveillance of new clades or lineages is still needed during the ongoing COVID-19 pandemic. Second, patient information on disease severity, immune responses, and transmission history was largely lacking in our retrieved dataset. Future studies are needed to reveal potential associations of genetic variants with disease progression and transmission fitness. Third, a limited number of full-length genome sequences from Africa and Asia were deposited in the public databases, which limited our analyses to fully characterize the genetic diversity of SARS-CoV-2 in Africa and Asia. Future analyses should integrate more sequences from different countries and continents.
5. Conclusions
Despite its global spread, SARS-CoV-2 harbors only a small proportion of highly prevalent variants that determine the classification of viral clades or lineages. Based on a large-scale dataset of 260,673 genomic sequences, our study presents a comprehensive mapping of highly prevalent SARS-CoV-2 variants as well as the dynamic changes of clades during the one-year pandemic of COVID-19. Genomic surveillance is important for monitoring the genetic variants that may be resistant to current antiviral drugs and vaccines [35,69]. To effectively control the COVID-19 pandemic, further studies are still needed to evaluate the impact of highly prevalent variants on antiviral drugs and vaccines before their wide application.
Acknowledgments
We are very grateful to the numerous researchers who have generated and shared their genome data with the world through the Global Initiative on Sharing All Influenza Data (GISAID). A list of all submitted research is provided in Data S1.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/biomedicines9040412/s1. A list of SARS-CoV-2 genome sequence acknowledgments is provided in Data S1. Table S1: Selective pressure analysis, Table S2: Distribution of clades, Figure S1: dN/dS ratios, Figure S2: Phylogenetic tree are provided as supplementary materials.
Author Contributions
Conceptualization, M.M., E.D.C., and G.L.; data curation and analysis, M.M.; funding acquisition, G.L.; methodology, M.M., and E.D.C.; visualization, M.M.; writing— original draft, review and editing, M.M., E.D.C., and G.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Nature Science Foundation of China (grant number 31871324, 81730064, 31571368), the Hunan Youth Elite Project (grant number 2018RS3006), and the National Science and Technology Major Project (grant number 2018ZX10715004). The funding bodies had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Data Availability Statement
SARS-CoV-2 whole-genome sequences were obtained from the Global Initiative on Sharing All Influenza Data (GISAID) (https://www.gisaid.org/, the accessed date: 16 January 2021).
Conflicts of Interest
None of Erik De Clercq’s drugs [70,71] were designed or approved for coronavirus treatment. The authors declare no conflict of interests.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Shang J., Han N., Chen Z., Peng Y., Li L., Zhou H., Ji C., Meng J., Jiang T., Wu A. Compositional diversity and evolutionary pattern of coronavirus accessory proteins. Briefings Bioinform. 2021;22:1267–1278. doi: 10.1093/bib/bbaa262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 3.Koyama T., Weeraratne D., Snowdon J.L., Parida L. Emergence of Drift Variants That May Affect COVID-19 Vaccine Development and Antibody Treatment. Pathogens. 2020;9:324. doi: 10.3390/pathogens9050324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lam T.T., Jia N., Zhang Y.W., Shum M.H., Jiang J.F., Zhu H.C., Tong Y.G., Shi Y.X., Ni X.B., Liao Y.S., et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020;583:282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 7.Duchene S., Featherstone L., Haritopoulou-Sinanidou M., Rambaut A., Lemey P., Baele G. Temporal signal and the phylodynamic threshold of SARS-CoV-2. Virus Evol. 2020;6:veaa061. doi: 10.1093/ve/veaa061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jia J.S., Lu X., Yuan Y., Xu G., Jia J., Christakis N.A. Population flow drives spatio-temporal distribution of COVID-19 in China. Nature. 2020;582:389–394. doi: 10.1038/s41586-020-2284-y. [DOI] [PubMed] [Google Scholar]
- 9.Popa A., Genger J.-W., Nicholson M.D., Penz T., Schmid D., Aberle S.W., Agerer B., Lercher A., Endler L., Colaço H., et al. Genomic epidemiology of superspreading events in Austria reveals mutational dynamics and transmission properties of SARS-CoV-2. Sci. Transl. Med. 2020;12:eabe2555. doi: 10.1126/scitranslmed.abe2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.du Plessis L., McCrone J.T., Zarebski A.E., Hill V., Ruis C., Gutierrez B., Raghwani J., Ashworth J., Colquhoun R., Connor T.R., et al. Establishment and lineage dynamics of the SARS-CoV-2 epidemic in the UK. Science. 2021;371:708–712. doi: 10.1126/science.abf2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Candido D.S., Claro I.M., De Jesus J.G., Souza W.M., Moreira F.R.R., Dellicour S., Mellan T.A., Du Plessis L., Pereira R.H.M., Sales F.C.S., et al. Evolution and epidemic spread of SARS-CoV-2 in Brazil. Science. 2020;369:1255–1260. doi: 10.1126/science.abd2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng X., Gu W., Federman S., Du Plessis L., Pybus O.G., Faria N.R., Wang C., Yu G., Bushnell B., Pan C.-Y., et al. Genomic surveillance reveals multiple introductions of SARS-CoV-2 into Northern California. Science. 2020;369:582–587. doi: 10.1126/science.abb9263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du P., Ding N., Li J., Zhang F., Wang Q., Chen Z., Song C., Han K., Xie W., Liu J., et al. Genomic surveillance of COVID-19 cases in Beijing. Nat. Commun. 2020;11:5503. doi: 10.1038/s41467-020-19345-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mercatelli D., Giorgi F.M. Geographic and Genomic Distribution of SARS-CoV-2 Mutations. Front. Microbiol. 2020;11:1800. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elizondo V., Harkins G.W., Mabvakure B., Smidt S., Zappile P., Marier C., Maurano M.T., Perez V., Mazza N., Beloso C., et al. SARS-CoV-2 genomic characterization and clinical manifestation of the COVID-19 outbreak in Uruguay. Emerg. Microbes Infect. 2021;10:51–65. doi: 10.1080/22221751.2020.1863747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang H.-C., Chen C.-H., Wang J.-H., Liao H.-C., Yang C.-T., Chen C.-W., Lin Y.-C., Kao C.-H., Lu M.-Y.J., Liao J.C. Analysis of genomic distributions of SARS-CoV-2 reveals a dominant strain type with strong allelic associations. Proc. Natl. Acad. Sci. USA. 2020;117:30679–30686. doi: 10.1073/pnas.2007840117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forster P., Forster L., Renfrew C., Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. USA. 2020;117:9241–9243. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rambaut A., Holmes E.C., O’Toole Á., Hill V., McCrone J.T., Ruis C., Du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomez-Carballa A., Bello X., Pardo-Seco J., Martinon-Torres F., Salas A. Mapping genome variation of SARS-CoV-2 worldwide highlights the impact of COVID-19 super-spreaders. Genome Res. 2020;30:1434–1448. doi: 10.1101/gr.266221.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Digard P., Lee H.M., Sharp C., Grey F., Gaunt E. Intra-genome variability in the dinucleotide composition of SARS-CoV-2. Virus Evol. 2020;6:veaa057. doi: 10.1093/ve/veaa057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng H.-L., Dichio V., Horta E.R., Thorell K., Aurell E. Global analysis of more than 50,000 SARS-CoV-2 genomes reveals epistasis between eight viral genes. Proc. Natl. Acad. Sci. USA. 2020;117:31519–31526. doi: 10.1073/pnas.2012331117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young B.E., Fong S.-W., Chan Y.-H., Mak T.-M., Ang L.W., E Anderson D., Lee C.Y.-P., Amrun S.N., Lee B., Goh Y.S., et al. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: An observational cohort study. Lancet. 2020;396:603–611. doi: 10.1016/S0140-6736(20)31757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ovsyannikova I.G., Haralambieva I.H., Crooke S.N., Poland G.A., Kennedy R.B. The role of host genetics in the immune response to SARS-CoV-2 and COVID-19 susceptibility and severity. Immunol. Rev. 2020;296:205–219. doi: 10.1111/imr.12897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang F., Gan R., Zhen Z., Hu X., Li X., Zhou F., Liu Y., Chen C., Xie S., Zhang B., et al. Adaptive immune responses to SARS-CoV-2 infection in severe versus mild individuals. Signal Transduct. Target. Ther. 2020;5:156. doi: 10.1038/s41392-020-00263-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J.C., Muecksch F., Rutkowska M., Hoffmann H.-H., Michailidis E., et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife. 2020;9:e61312. doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell. 2020;182:1284–1294.e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeng L., Li D., Tong W., Shi T., Ning B. Biochemical features and mutations of key proteins in SARS-CoV-2 and their impacts on RNA therapeutics. Biochem. Pharmacol. 2021:114424. doi: 10.1016/j.bcp.2021.114424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kemp S.A., Collier D.A., Datir R.P., Ferreira I.A.T.M., Gayed S., Jahun A., Hosmillo M., Rees-Spear C., Mlcochova P., Lumb I.U., et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021;592:277–282. doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dearlove B., Lewitus E., Bai H., Li Y., Reeves D.B., Joyce M.G., Scott P.T., Amare M.F., Vasan S., Michael N.L., et al. A SARS-CoV-2 vaccine candidate would likely match all currently circulating variants. Proc. Natl. Acad. Sci. USA. 2020;117:23652–23662. doi: 10.1073/pnas.2008281117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogando N.S., Zevenhoven-Dobbe J.C., Van Der Meer Y., Bredenbeek P.J., Posthuma C.C., Snijder E.J. The Enzymatic Activity of the nsp14 Exoribonuclease Is Critical for Replication of MERS-CoV and SARS-CoV-2. J. Virol. 2020;94:e01246-20. doi: 10.1128/JVI.01246-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robson F., Khan K.S., Le T.K., Paris C., Demirbag S., Barfuss P., Rocchi P., Ng W.-L. Coronavirus RNA Proofreading: Molecular Basis and Therapeutic Targeting. Mol. Cell. 2020;80:1136–1138. doi: 10.1016/j.molcel.2020.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Volz E., Hill V., McCrone J.T., Price A., Jorgensen D., O’Toole Á., Southgate J., Johnson R., Jackson B., Nascimento F.F., et al. Evaluating the Effects of SARS-CoV-2 Spike Mutation D614G on Transmissibility and Pathogenicity. Cell. 2021;184:64–75.e11. doi: 10.1016/j.cell.2020.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2020;592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore J.P., Offit P.A. SARS-CoV-2 Vaccines and the Growing Threat of Viral Variants. JAMA. 2021;325:821–822. doi: 10.1001/jama.2021.1114. [DOI] [PubMed] [Google Scholar]
- 36.Katoh K., Standley D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang D., Zhang Y., Zhang Z., Zhu J., Yu J. KaKs_Calculator 2.0: A Toolkit Incorporating Gamma-Series Methods and Sliding Window Strategies. Genom. Proteom. Bioinform. 2010;8:77–80. doi: 10.1016/S1672-0229(10)60008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang J., Wang W., Chen Z., Lu S., Yang F., Bi Z., Bao L., Mo F., Li X., Huang Y., et al. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature. 2020;586:572–577. doi: 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- 39.Lee W.S., Wheatley A.K., Kent S.J., DeKosky B.J. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat. Microbiol. 2020;5:1185–1191. doi: 10.1038/s41564-020-00789-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vaidyanathan G. Vaccine makers in Asia rush to test jabs against fast-spreading COVID variant. Nature. 2021 doi: 10.1038/d41586-021-00041-y. [DOI] [PubMed] [Google Scholar]
- 41.Garcia-Beltran W.F., Lam E.C., Denis K.S., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021 doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu J., Peng P., Wang K., Fang L., Luo F.-Y., Jin A.-S., Liu B.-Z., Tang N., Huang A.-L. Emerging SARS-CoV-2 variants reduce neutralization sensitivity to convalescent sera and monoclonal antibodies. Cell. Mol. Immunol. 2021;18:1061–1063. doi: 10.1038/s41423-021-00648-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corman V.M., Drosten C. Authors’ response: SARS-CoV-2 detection by real-time RT-PCR. Eurosurveillance. 2020;25:2001035. doi: 10.2807/1560-7917.ES.2020.25.21.2001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Artesi M., Bontems S., Göbbels P., Franckh M., Maes P., Boreux R., Meex C., Melin P., Hayette M.-P., Bours V., et al. A Recurrent Mutation at Position 26340 of SARS-CoV-2 Is Associated with Failure of the E Gene Quantitative Reverse Transcription-PCR Utilized in a Commercial Dual-Target Diagnostic Assay. J. Clin. Microbiol. 2020;58:e01598-20. doi: 10.1128/JCM.01598-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Issa E., Merhi G., Panossian B., Salloum T., Tokajian S. SARS-CoV-2 and ORF3a: Nonsynonymous Mutations, Functional Domains, and Viral Pathogenesis. mSystems. 2020;5:e00266-20. doi: 10.1128/mSystems.00266-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pancer K., Milewska A., Owczarek K., Dabrowska A., Kowalski M., Łabaj P.P., Branicki W., Sanak M., Pyrc K. The SARS-CoV-2 ORF10 is not essential in vitro or in vivo in humans. PLoS Pathog. 2020;16:e1008959. doi: 10.1371/journal.ppat.1008959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Motayo B.O., Oluwasemowo O.O., Olusola B.A., Akinduti P.A., Arege O.T., Obafemi Y.D., Faneye A.O., Isibor P.O., Aworunse O.S., Oranusi S.U. Evolution and genetic diversity of SARS-CoV-2 in Africa using whole genome sequences. Int. J. Infect. Dis. 2021;103:282–287. doi: 10.1016/j.ijid.2020.11.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tegally H., Wilkinson E., Lessells R.J., Giandhari J., Pillay S., Msomi N., Mlisana K., Bhiman J.N., von Gottberg A., Walaza S., et al. Sixteen novel lineages of SARS-CoV-2 in South Africa. Nat. Med. 2021;27:440–446. doi: 10.1038/s41591-021-01255-3. [DOI] [PubMed] [Google Scholar]
- 49.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baric R.S. Emergence of a Highly Fit SARS-CoV-2 Variant. N. Engl. J. Med. 2020;383:2684–2686. doi: 10.1056/NEJMcibr2032888. [DOI] [PubMed] [Google Scholar]
- 51.Zhang M., Li G., Shang J., Pan C., Zhang M., Yin Z., Xie Q., Peng Y., Mao Q., Xiao X., et al. Rapidly decreased HBV RNA predicts responses of pegylated interferons in HBeAg-positive patients: A longitudinal cohort study. Hepatol. Int. 2020;14:212–224. doi: 10.1007/s12072-020-10015-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li G., Liu Y., Jing X., Wang Y., Miao M., Tao L., Zhou Z., Xie Y., Huang Y., Lei J., et al. Mortality risk of COVID-19 in elderly males with comorbidities: A multi-country study. Aging. 2020;13:27–60. doi: 10.18632/aging.202456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miao M., Jing X., De Clercq E., Li G. Danoprevir for the Treatment of Hepatitis C Virus Infection: Design, Development, and Place in Therapy. Drug Des. Dev. Ther. 2020;14:2759–2774. doi: 10.2147/DDDT.S254754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang C., Wang Y., Hu M., Wen L., Wen C., Wang Y., Zhu W., Tai S., Jiang Z., Xiao K., et al. Antibody seroconversion in asymptomatic and symptomatic patients infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin. Transl. Immunology. 2020;9:e1182. doi: 10.1002/cti2.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miao M., De Clercq E., Li G. Clinical significance of chemokine receptor antagonists. Expert Opin. Drug Metab. Toxicol. 2020;16:11–30. doi: 10.1080/17425255.2020.1711884. [DOI] [PubMed] [Google Scholar]
- 56.Zhou Z., Zhang M., Wang Y., Zheng F., Huang Y., Huang K., Yu Q., Cai C., Chen D., Tian Y., et al. Clinical characteristics of older and younger patients infected with SARS-CoV-2. Aging. 2020;12:11296–11305. doi: 10.18632/aging.103535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saunders K.O., Wiehe K., Tian M., Acharya P., Bradley T., Alam S.M., Go E.P., Scearce R., Sutherland L., Henderson R., et al. Targeted selection of HIV-specific antibody mutations by engineering B cell maturation. Science. 2019;366:eaay7199. doi: 10.1126/science.aay7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yurkovetskiy L., Wa Wang Y., De Clercq E., Li G. Current and emerging non-nucleoside reverse transcriptase inhibitors (NNRTIs) for HIV-1 treatment. Expert Opin. Drug Metab. Toxicol. 2019;15:813–829. doi: 10.1080/17425255.2019.1673367. [DOI] [PubMed] [Google Scholar]
- 59.Jiang X., Zhang Z., Wang C., Ren H., Gao L., Peng H., Niu Z., Ren H., Huang H., Sun Q. Bimodular effects of D614G mutation on the spike glycoprotein of SARS-CoV-2 enhance protein processing, membrane fusion, and viral infectivity. Signal Transduct. Target. Ther. 2020;5:268. doi: 10.1038/s41392-020-00392-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hou Y.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., Leist S.R., Schäfer A., Nakajima N., Takahashi K., et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370:1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kannan S.R., Spratt A.N., Quinn T.P., Heng X., Lorson C.L., Sönnerborg A., Byrareddy S.N., Singh K. Infectivity of SARS-CoV-2: There Is Something More than D614G? J. Neuroimmune Pharmacol. 2020;15:574–577. doi: 10.1007/s11481-020-09954-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Garvin M.R., Prates E.T., Pavicic M., Jones P., Amos B.K., Geiger A., Shah M.B., Streich J., Gazolla J.G.F.M., Kainer D., et al. Potentially adaptive SARS-CoV-2 mutations discovered with novel spatiotemporal and explainable AI models. Genome Biol. 2020;21:304. doi: 10.1186/s13059-020-02191-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chand G.B., Banerjee A., Azad G.K. Identification of novel mutations in RNA-dependent RNA polymerases of SARS-CoV-2 and their implications on its protein structure. PeerJ. 2020;8:e9492. doi: 10.7717/peerj.9492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu Y., Kang L., Shen Z., Li X., Wu W., Ma W., Fang C., Yang F., Jiang X., Gong S., et al. Dynamics of severe acute respiratory syndrome coronavirus 2 genome variants in the feces during convalescence. J. Genet. Genom. 2020;47:610–617. doi: 10.1016/j.jgg.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021 doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Davies N.G., Jarvis C.I., Edmunds W.J., Jewell N.P., Diaz-Ordaz K., Keogh R.H. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature. 2021 doi: 10.1038/s41586-021-03426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. Nature. 2021 doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 68.Thomson E.C., Rosen L.E., Shepherd J.G., Spreafico R., Filipe A.D.S., Wojcechowskyj J.A., Davis C., Piccoli L., Pascall D.J., Dillen J., et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021;184:1171–1187.e20. doi: 10.1016/j.cell.2021.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lauring A.S., Hodcroft E.B. Genetic Variants of SARS-CoV-2—What Do They Mean? JAMA. 2021;325:529–531. doi: 10.1001/jama.2020.27124. [DOI] [PubMed] [Google Scholar]
- 70.Li G., Xu M., Yue T., Gu W., Tan L. Life-long passion for antiviral research and drug development: 80th birthday of Prof. Dr. Erik De Clercq. Biochem. Pharmacol. 2021;185:114485. doi: 10.1016/j.bcp.2021.114485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li G., Yue T., Zhang P., Gu W., Gao L.J., Tan L. Drug Discovery of Nucleos(t)ide Antiviral Agents: Dedicated to Prof. Dr. Erik De Clercq on Occasion of His 80th Birthday. Molecules. 2021;26:923. doi: 10.3390/molecules26040923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
SARS-CoV-2 whole-genome sequences were obtained from the Global Initiative on Sharing All Influenza Data (GISAID) (https://www.gisaid.org/, the accessed date: 16 January 2021).