Abstract
Neofusicoccum parvum, in the family Botryosphaeriaceae, was identified as the causal agent of bot gummosis of lemon (Citrus × limon) trees, in the two major lemon-producing regions in Italy. Gummy cankers on trunk and scaffold branches of mature trees were the most typical disease symptoms. Neofusicoccum parvum was the sole fungus constantly and consistently isolated from the canker bark of symptomatic lemon trees. It was identified on the basis of morphological characters and the phylogenetic analysis of three loci, i.e., the internal transcribed spacer of nuclear ribosomal DNA (ITS) as well as the translation elongation factor 1-alpha (TEF1) and β-tubulin (TUB2) genes. The pathogenicity of N. parvum was demonstrated by wound inoculating two lemon cultivars, ‘Femminello 2kr’ and ‘Monachello’, as well as citrange (C. sinensis × Poncirus trifoliata) ‘Carrizo’ rootstock. In artificial inoculations, the fungus was very aggressive on lemons and weakly virulent on citrange, consistently with symptoms observed in the field as a consequence of natural infections. This is the first report of N. parvum, both in a wide and in a strict taxonomic sense, as a pathogen of lemon in Italy.
Keywords: Botryosphaeriaceae, ITS, TEF1, TUB2, citrus, trunk and branch cankers, pathogenicity, Italy
1. Introduction
Trunk and branch cankers caused by species of Botryosphaeriaceae on citrus were traditionally referred to as Diplodia gummosis and Dothiorella gummosis [1,2]. More recently the comprehensive term bot gummosis was introduced to indicate trunk and branch cankers of citrus caused by fungi in the Botryosphaeriaceae family in California [3]. All common names of the disease refer to the most typical symptom, i.e., the gum exudate oozing from the bark cankers. Another typical symptom of the disease is a chocolate brown to dark brown discoloration of the wood which becomes evident after peeling the bark. Originally, the fungi causing Diplodia gummosis and Dothiorella gummosis were identified as Diplodia natalensis and Dothiorella gregaria (syn. Botryosphaeria ribis), respectively [1]. However, identifications of Botryosphaeriaceae species prior to the application of taxonomic criteria based on DNA sequencing and phylogenetic inference should be re-examined carefully as the taxonomy of this family of fungi has substantially changed during the last decade and is still evolving rapidly. As a matter of fact, very recently new species have been described and species that were previously separated on the basis of both multi-loci phylogenetic analysis and morphological characters have been reduced to synonymy on the basis of same genetic markers previously regarded as discriminatory [4,5,6,7]. After the introduction of modern molecular taxonomy, several Botryosphaeriaceae species were reported to be associated to bot gummosis in California, including Diplodia mutila, D. seriata, Dothiorella iberica (very recently indicated as a possible synonim of Do. sarmentorum), Spencermartinsia viticola (syn. Do. viticola), Lasiodiplodia parva, Neofusicoccum australe, N. luteum, N. mediterraneum, N. parvum and Neoscytalidium dimidiatum (syn. Ne. hyalinum, formerly Hendersonula toruloidea) [3,8,9]. Neofusicoccum parvum and Diaporthe foeniculina were reported to be responsible for shoot blight and branch cankers of citrus in Greece, but in pathogenicity tests N. parvum was significantly more virulent than D. foeniculina [10]. Dothiorella viticola was identified as the causal agent of bot gummosis of citrus in Tunisia [11]. Moreover, symptoms of bot gummosis were reproduced by artificially inoculating citrus trees with Neofusicoccum batangarum (very recently reduced to synonymy with N. ribis), the causal agent of a destructive disease of cactus pear in minor islands of Sicily [12,13]. In the island of Malta, about 80 marine miles south of Sicily, symptoms very similar to bot gummosis were reported on lemon (Citrus × limon) trees to be caused by species of Diaporthe [14].
During recent surveys in citrus growing areas in Sicily and Calabria, the two major citrus producer regions of Italy, symptoms of bot gummosis were observed to be common on mature, fruit-bearing lemon trees. This study was aimed at determining the etiology and the distribution of the disease in the major producing areas of lemon in Italy.
2. Materials and Methods
2.1. Fungal Collection and Isolation
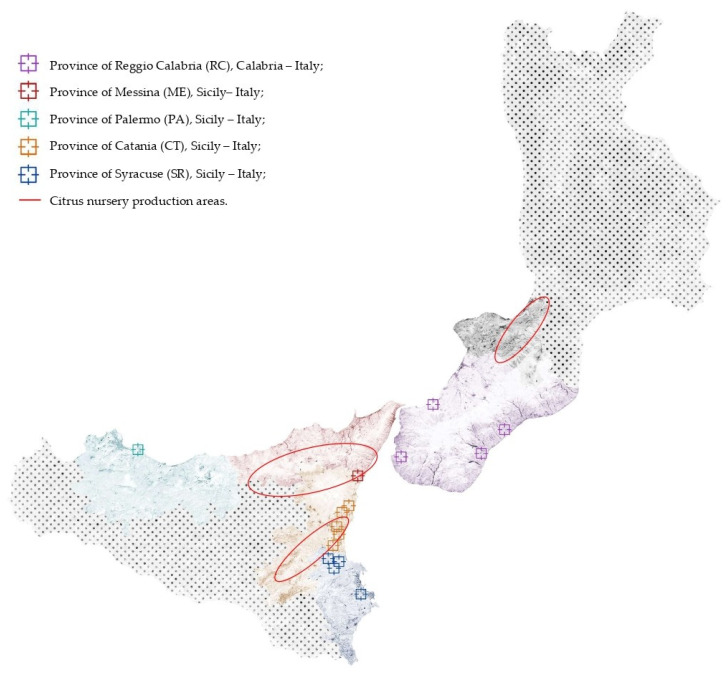
Five citrus-growing areas in southern Italy, four in Siciliy, including the province of Palermo and the three Sicilian districts in the provinces of Catania, Messina, and Syracuse to which the Protected Geographical Indication trademark was granted by EU, and one in Calabria (province of Reggio Calabria), were surveyed in 2017 and 2018 (Figure 1). The geographical map in Figure 1 also shows the close proximity between the surveyed areas and the major districts of Calabria and Sicily where citrus nursery plants production is concentrated.
Figure 1.
Map of southern Italy, including Sicily and Calabria, where sampling sites are indicated with colored squares (each color corresponds to a different province) while major citrus nursery production areas are indicated with circles.
Approximately 15 to 20 bark pieces from trunk and branch cankers were collected from 25 randomly selected symptomatic trees (approximately 5 to 60-yr-old). Samples were transported in a cooler to the laboratory of Molecular Plant Pathology at the Department of Agriculture, Food, and Environment (Di3A) of the University of Catania, Italy.
Bark pieces with the typical brown discoloration of the wood were rinsed with deionized water to remove organic debris, blotted dry with paper towels, then briefly flamed after dipping in 95% ethanol for 3 s. Small sections (approximately 2 to 3 mm) from the inner face of necrotic bark were cut with a sterile scalpel and placed in Petri dishes onto potato dextrose agar (PDA; Difco Laboratories, Detroit, MI, USA) amended with 100 µg/ml streptomycin (Sigma-Aldrich, Darmstadt, Germany). Cultures were incubated in the dark for 4 days at 25 °C.
Pure cultures of fungal isolates were obtained by monoconidial cultures. The list of isolates characterized in this study is reported in Table 1.
Table 1.
Identity of the Neofusicoccum parvum isolates sourced from gummy cankers of lemon (Citrus × limon) trees in southern Italy characterized in this study, and corresponding GenBank accession numbers.
| Isolate | Species | Origin | Host, Variety | Accession Numbers | ||
|---|---|---|---|---|---|---|
| ITS | β-Tubulin | EF-1α | ||||
| BOT 1A | Neofusicoccum parvum | Catania, Siciliy-Italy | Citrus ×limon | MW727244 | MW789889 | MW789904 |
| BOT 1B | N. parvum | Catania, Siciliy-Italy | C. ×limon | MW727245 | MW789890 | MW789905 |
| BOT 1C | N. parvum | Catania, Siciliy-Italy | C. ×limon | MW727246 | MW789891 | MW789906 |
| BOT 1D | N. parvum | Catania, Siciliy-Italy | C. ×limon | MW727247 | MW789892 | MW789907 |
| BOT 1E | N. parvum | Catania, Siciliy-Italy | C. ×limon | MW727248 | MW789893 | MW789908 |
| BOT 2A | N. parvum | Syracuse, Siciliy-Italy | C. ×limon | MW727249 | MW789894 | MW789909 |
| BOT 2B | N. parvum | Syracuse, Siciliy-Italy | C. ×limon | MW727250 | MW789895 | MW789910 |
| BOT 2C | N. parvum | Syracuse, Siciliy-Italy | C. ×limon | MW727251 | MW789896 | MW789911 |
| BOT 2D | N. parvum | Syracuse, Siciliy-Italy | C. ×limon | MW727252 | MW789897 | MW789912 |
| BOT 2E | N. parvum | Syracuse, Siciliy-Italy | C. ×limon | MW727253 | MW789898 | MW789913 |
| BOT 4A | N. parvum | Messina, Siciliy-Italy | C. ×limon | MW788562 | MW789929 | MW789919 |
| BOT 4B | N. parvum | Messina, Siciliy-Italy | C. ×limon | MW788563 | MW789930 | MW789920 |
| BOT 4C | N. parvum | Messina, Siciliy-Italy | C. ×limon | MW788564 | MW789931 | MW789921 |
| BOT 4D | N. parvum | Messina, Siciliy-Italy | C. ×limon | MW788565 | MW789932 | MW789922 |
| BOT 4E | N. parvum | Messina, Siciliy-Italy | C. ×limon | MW788566 | MW789933 | MW789923 |
| BOT 5A | N. parvum | Palermo, Siciliy-Italy | C. ×limon | MW788567 | MW789934 | MW789924 |
| BOT 5B | N. parvum | Palermo, Siciliy-Italy | C. ×limon | MW788568 | MW789935 | MW789925 |
| BOT 5C | N. parvum | Palermo, Siciliy-Italy | C. ×limon | MW788569 | MW789936 | MW789926 |
| BOT 5D | N. parvum | Palermo, Siciliy-Italy | C. ×limon | MW788570 | MW789937 | MW789927 |
| BOT 5E | N. parvum | Palermo, Siciliy-Italy | C. ×limon | MW788571 | MW789938 | MW789928 |
| BOT 3A | N. parvum | Reggio Calabria, Calabria-Italy | C. ×limon | MW727254 | MW789899 | MW789914 |
| BOT 3B | N. parvum | Reggio Calabria, Calabria-Italy | C. ×limon | MW727255 | MW789900 | MW789915 |
| BOT 3C | N. parvum | Reggio Calabria, Calabria-Italy | C. ×limon | MW727256 | MW789901 | MW789916 |
| BOT 3D | N. parvum | Reggio Calabria, Calabria-Italy | C. ×limon | MW727257 | MW789902 | MW789917 |
| BOT 3E | N. parvum | Reggio Calabria, Calabria-Italy | C. ×limon | MW727258 | MW789903 | MW789918 |
2.2. Morphological Characteristics and Cardinal Temperatures for Growth of the Isolates
The isolates were induced to sporulate by plating them on PDA containing sterilized pine needles [4], and incubating at room temperature (ca. 20 to 25 °C) under diffused day light or near-UV light, until pycnidia developed. For microscopy, pycnidia and conidia were mounted in sterile distilled water or 100% lactic acid and observed microscopically at ×40 and ×100 magnifications, with an Axioskop (Zeiss) microscope. Images were captured with an AxioCam MRc5 camera (Zeiss), and measurements were made with the software AxioVision. For each isolate, 50 conidia were randomly selected, and their lengths, widths, and shape were recorded. For the dimension of pycnidia, 20 measurements were made. Colony characters and pigment production were noted after 4 to 6 d of growth on PDA or malt extract agar (MEA) at 25 °C, in the dark.
Colony colors (upper and lower surfaces) were rated and the conidial characteristics observed were compared with those that were reported in previous studies [15,16,17,18,19].
Radial growth rate and cardinal temperatures for radial growth were determined by growing the isolates on PDA in Petri dishes (9 cm diam.), and incubating them at 5, 10, 15, 20, 25, 30, and 35 °C, in the dark. Means of radial growth at the different temperatures were adjusted to a regression curve using Statgraphics Plus 5.1 software (Statgraphics Technologies Inc., The Plains, VA, USA), and the best polynomial model was chosen based on parameter significance (p < 0.05) and coefficient of determination (R2) to estimate the optimum growth temperature for each isolate. Four replicates of each isolate were evaluated and each experiment was repeated twice.
2.3. Amplification and Sequencing
Genomic DNA was isolated from 1-week-old cultures grown on PDA at 25 °C in the dark using the procedure of Schena and Cooke [20]. The internal transcribed spacer (ITS) region of the ribosomal DNA was amplified and sequenced with primers ITS1/ITS4 [21], part of the translation elongation factor 1 alpha gene (tef1) was sequenced and amplified with primers EF1-728F/EF1-986R [22], and the β-tubulin gene (tub2) was sequenced and amplified with Bt2a and Bt2b [23].
The PCR amplifications were performed on a GeneAmp PCR System 9700 (Applied Biosystems, Monza-Brianza, Italy). All PCRs were performed by using the Taq DNA polimerase recombinant (Invitrogen™, Carlsbad, 254 CA, USA) and carried out in a total volume of 25 μL containing the following: PCR Buffer (1X), dNTP mix (0.2 mM), MgCl2 (1.5 mM), forward and reverse primers (0.5 μM each), Taq DNA Polymerase (1 U), and 1 μL of genomic DNA.
The reaction protocol for ITS and β-tubulin included an initial preheat at 94 °C for 2 min; followed by 35 cycles of denaturation at 94 °C for 15 s, annealing at 58 °C for 15 s, and extension at 72 °C for 45 s; and final extension at 72 °C for 10 min. The translation EF α-1 included an initial denaturation at 95 °C for 8 min; followed by 35 cycles of 95, 58, and 72 °C for 15, 20, and 60 s, respectively; and a final extension at 72 °C for 10 min. The amplicons were detected in 1% agarose gel and purified products were sequenced by Macrogen Europe (Amsterdam, The Netherlands). Sequences were analyzed by using FinchTV v.1.4.0 [24] and MEGA7 [25] and the consensus sequences were deposited in Genbank.
2.4. Molecular Identification and Phylogenetic Analyses
Sequences of isolates were associated to a species by the application of BLAST algorithm on the NCBI nucleotide database. Species identity was confirmed by a multi-loci phylogenetical analysis including sequences from taxa that best matched the blast species identity [26,27,28]. The list of isolates used as reference is reported in Table 2. * Netto et al., unpublished.
Table 2.
GenBank accession numbers of sequences of Neofusicoccum spp. isolates of different geographical and host origins used as reference in phylogenetic analyses.
| Species | Isolate | Origin | Host | Source | GenBank Accession Number | ||
|---|---|---|---|---|---|---|---|
| ITS | tef1 | β-Tubulin | |||||
| N. algeriense | CAA 322 | Portugal | Malus domestica | [29] | KX505906 | KX505894 | KX505916 |
| N. algeriense | CBS 137504 | Algeria | Vitis vinifera | [29] | KJ657702 | KX505893 | KX505915 |
| N. batangarum | CBS 124922 | Cameroon | Terminalia catappa | [29] | FJ900606 | FJ900652 | FJ900633 |
| N. batangarum | CBS 143023 | Italy, Favignana | Opuntia ficus-indica | [13] | MF414730 | MF414768 | MF414749 |
| N. batangarum | CBS 143025 | Italy, Linosa | Opuntia ficus-indica | [13] | MF414747 | MF414785 | MF414766 |
| N. batangarum | CBS 127348 | USA, Florida | Schinus terebinthifolius | [28] | HM357636 | KX464674 | KX464952 |
| N. batangarum | CMM4553 | Brasil | Anacardium sp. | Unpublished | KT728917 | KT728921 | KT728913 |
| N. batangarum | CBS 124923 | Cameroon | Terminalia catappa | [29] | FJ900608 | FJ900654 | FJ900635 |
| N. batangarum | CBS 124924 | Cameroon | Terminalia catappa | [29] | FJ900607 | FJ900653 | FJ900634 |
| N. batangarum | CBS 143026 | Italy, Lampedusa | Opuntia ficus-indica | [13] | MF414748 | MF414786 | MF414767 |
| N. batangarum | CBS 143024 | Italy, Ustica | Opuntia ficus-indica | [13] | MF414738 | MF414776 | MF414757 |
| N. brasiliense | CMM1285 | Brazil | Mangifera indica | [29] | JX513628 | JX513608 | KC794030 |
| N. cordaticola | CBS 123634 | South Africa | Syzygium cordatum | [29] | EU821898 | EU821868 | EU821838 |
| N. kwambonambiense | CBS 123639 | South Africa | Syzygium cordatum | [29] | EU821900 | EU821870 | EU821840 |
| N. macroclavatum | CBS 118223 | Australia | Eucalyptus globulus | [29] | DQ093196 | DQ093217 | DQ093206 |
| N. parvum | CBS 111994 | Australia | Telopea sp. | [28] | AF452519 | KX464702 | KX464982 |
| N. parvum | CAA 192 | Portugal | Ferula communis | [29] | KX505905 | KX505892 | KX505913 |
| N. parvum | CBS 112930 | South Africa | Vitis vinifera | [28] | AY343467 | AY343359 | KX464983 |
| N. parvum | CBS 121486 | Spain | Vitis vinifera cv. Parellada | [28] | EU650672 | KX464707 | KX464992 |
| N. parvum | CBS 114472 | USA | Leucadendron sp. | [28] | AF452523 | FJ150710 | KX464987 |
| N. parvum | CBS 111524 | USA, Hawaii | Protea cynaroides | [28] | AF452524 | FJ150709 | KX465009 |
| N. parvum | CBS 124492 | Zambia | Syzygium guineense | [28] | FJ655000 | KX464684 | KX464962 |
| N. parvum | CBS 110301(ex-type) | Portugal | Vitis vinifera | [29] | AY259098 | AY573221 | EU673095 |
| N. parvum | CMW9081 (ex-type) | New Zealand | Populus nigra | [29] | AY236943 | AY236888 | AY236917 |
| N. ribis | CBS 115475 | USA | Ribes sp. | [29] | AY236935 | AY236877 | AY236906 |
| N. umdonicola | CBS 123646 | South Africa | Syzygium cordatum | [29] | EU821905 | EU821875 | EU821845 |
For all isolates, ITS, tef1, and tub2 sequences were trimmed to a common length and concatenated using the Sequence Matrix software [30]. Concatenated sequences were aligned with MUSCLE [31] as implemented in MEGA7 [25], and edited manually for checking indels and single nucleotide polymorphisms. Phylogenetic analyses were performed in Mega with the maximum likelihood method using the Tamura–Nei model and 1000 bootstrap replications [32].
2.5. Pathogenicity Tests
The pathogenicity of an isolate of Neofusicoccum parvum (BOT 1D), sourced from lemon in Sicily, was assessed on twigs and stem of 2-year-old trees of three citrus varieties, Citrus × limon var. Monachello, Citrus × limon var. Femminello and citrange ‘Carrizo’ (Citrus sinensis Osbeck × Poncirus trifoliata Raf.), grown in a greenhouse maintained at 20 to 26 °C.
Two plants per variety were inoculated in three points, on two twigs and on stem per tree. A hole in each twig was made with a 3 mm cork-borer. For the inoculation, a 3 mm diam. mycelium plug from 5-d-old PDA culture of the test isolate (BOT 1D) was placed on the freshly wounded surface; the wound was covered with the excised bark disk and sealed with Parafilm®. The stem of each tree was inoculated 10 cm above the soil level (a single hole per stem) using the method above reported. Two trees per each citrus variety were inoculated with sterile agar (Controls).
The length and breadth of each resulting lesion were recorded 30 days after inoculation (d.a.i.), and the outer surface areas of the bark cankers on twigs and stems were calculated as ellipses.
Re-isolation was made from lesions and the identity of resulting fungal colonies was confirmed by both morphological and molecular analysis (ITS, tef1, and tub2 genes were sequenced) as described above.
3. Results
3.1. Symptoms
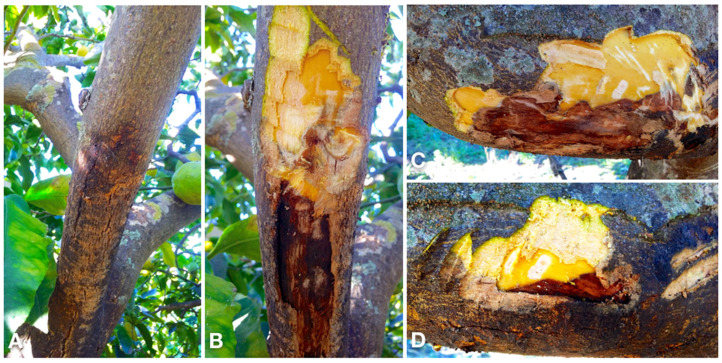
Bot gummosis was detected on mature, fruit-bearing lemon trees both in commercial orchards and in trees planted singularly or in small groups in home gardens where trees were grown for both ornamental purposes and domestic consumption of fruits. Among the 25 sampled trees, one, in the province of Syracuse, was about 5-yr-old; five trees, of which three in two distinct sites of the province of Catania, one in the province of Messina and one in the province of Palermo were over 40-yr-old. The rest of the trees were between approximately 15 and 30 years old. The disease was common, but usually in commercial orchards symptomatic trees were scattered and the proportion of symptomatic trees in each orchard did not exceed 5%. Exceptionally, in an orchard in the municipality of Lentini (Syracuse) 30 out of 400, 20-yr-old trees were symptomatic. Typically, symptoms, i.e. cankers with an abundant gummous exudate, were observed on the main trunk and scaffold branches (Figure 2A–E) of trees. Another typical symptom was a chocolate to dark brown discoloration of the wood and the cambial face of the bark, which was visible after removing the bark (Figure 2C and Figure 3B–D). Cankers usually originated at the level of main branch scaffolding and progressively expanded upward to the secondary branches and downward to the grafting line between the scion and the rootstock (Figure 2A,C,D). However, in most cases they were restricted to the lemon scion and only exceptionally and in any case to a limited extent they expanded on sour orange (Citrus × aurantium) or citrange (Citrus sinensis × Poncirus trifoliata) rootstock. In a few cases, as a consequence of chronic infections the cankers girdled the trunk. Old cankers either ceased to produce gum or produced it only on the expanding edge; they showed longitudinal bark splittings on secondary branches and irregular cracking and scaling of the bark on trunk and scaffold branches (Figure 2E and Figure 3A,D). Leaf yellowing, defoliation, and dieback of single branches occurred commonly. Severely affected trees showed decline symptoms, including leaf chlorosis, severe crown thinning, and branch and twig blight and dieback (Figure 4).
Figure 2.
Symptoms of bot gummosis on lemon trees: (A,B) cankers with abundant gummous exudate on scaffold branches of ‘Femminello Siracusano 2Kr’ lemon (Citrus × limon) tree; (C) typical tan to brown discoloration of the wood beneath the bark of a canker; the discolored wood is visible after removing the bark; (D) gummous canker on a branch of lemon tree; (E) old canker with bark scaling at the level of main branch scaffolding, expanding upward to the secondary branches and downward to the grafting line between the scion and the rootstock.
Figure 3.
(A) Old canker on a secondary branch of lemon tree with longitudinal bark splitting at the base of the branch and the gum exudate in the advancing upward front, indicating the canker is still active; (B) the same as (A) after peeling the bark to show the dark brown discoloration of the wood beneath the bark; (C) typical chocolate brown discoloration of the wood underneath the bark; (D) a detail showing the necrotic bark of the canker exuding gum and the typical chocolate brown wood discoloration beneath the bark.
Figure 4.
In the foreground, a lemon tree approximately 20-yr-old with yellowing of the canopy as a consequence of bot gummosis on scaffold branches.
3.2. Fungus Isolation and Morphological Identification
A fungus with pale gray and rapidly growing colonies on potato-dextrose-agar (PDA) was constantly (from all samples and in all sampling sites) and consistently (around 100% of isolations) recovered from the inner bark with the typical brown discoloration. Based on the morphotype, i.e., colony morphology and conidial characteristics, all isolates were assigned to Neofusicoccum, a genus in the Botryosphaeriaceae family (Figure 5A,C). A set of 25 isolates, each from a distinct tree, was characterized further.
Figure 5.
(A) Colony morphology of an isolate of Neofusicoccum parvum recovered from lemon on potato-dextrose-agar after 5 d incubation at 25 °C; (B) Pycnidia developed on MEA containing sterilized pine needles; (C) Unicellular, ellipsoidal, thin-walled hyaline conidia of Neofusicoccum parvum mounted in sterile distilled water.
Colonies on MEA of all 17 isolates developed an abundant grayish aerial mycelium, which produced black pycnidia after two weeks of incubation at 25 °C (Figure 5B). On PDA mycelium was white and became smoky gray to gray-olivaceous after 5 d (Figure 4A). The mycelium was fast-growing and covered the 9 cm diam. Petri dishes after 5 d incubation at 25 °C in the dark. Optimum temperature for radial colony growth was between 25 and 30 °C for all the isolates tested (Table 3). Little growth was observed at 10 or 35 °C. Stromatic conidiomata were produced in pine needle cultures within 14 d. The conidiomata were globose and non-papillate to pyriform with a short, acute papilla, entire locule lined with conidiogenous cells, and measured 150–250 μm in diameter. Conidia were ellipsoidal with apex round and base flat, unicellular, and hyaline and measured 16.9–17.3 × 5.4–5.6 μm, with a mean length to width ratio = 3.2 (Figure 5C). Old conidia becoming 1–2-septate hyaline, or light brown with the middle cell darker than the terminal cells.
Table 3.
Mean radial growth rates of colonies of representative Neofusicoccum parvum isolates of different geographical origin on PDA at three different temperatures, as determined after 3 d of incubation.
| Isolates of N. parvum | 15 °C (mm d-1) ± S.D. a | 25 °C (mm d-1) ± S.D. a | 30 °C (mm d-1) ± S.D. a |
|---|---|---|---|
| BOT 1A | 3.73 ± 0.07 | 7.90 ± 1.08 | 6.71 ± 0.27 |
| BOT 1D | 3.11 ± 0.91 | 7.73 ± 0.28 | 7.38 ± 0.47 |
| BOT 2A | 3.41 ± 0.05 | 7.55 ± 0.32 | 7.06 ± 0.28 |
| BOT 2D | 3.53 ± 0.62 | 6.68 ± 0.90 | 5.58 ± 0.28 |
| BOT 3A | 3.45 ± 0.39 | 7.06 ± 0.28 | 6.23 ± 0.20 |
| BOT 3D | 3.30 ± 0.17 | 6.83 ± 0.32 | 6.63 ± 0.46 |
| BOT 4A | 3.42 ± 0.06 | 6.58 ± 0.62 | 7.38 ± 0.47 |
| BOT 5D | 3.28± 0.17 | 7.22 ± 0.42 | 5.37 ± 0.38 |
a Mean of four replicate Petri dishes.
3.3. Molecular Identification
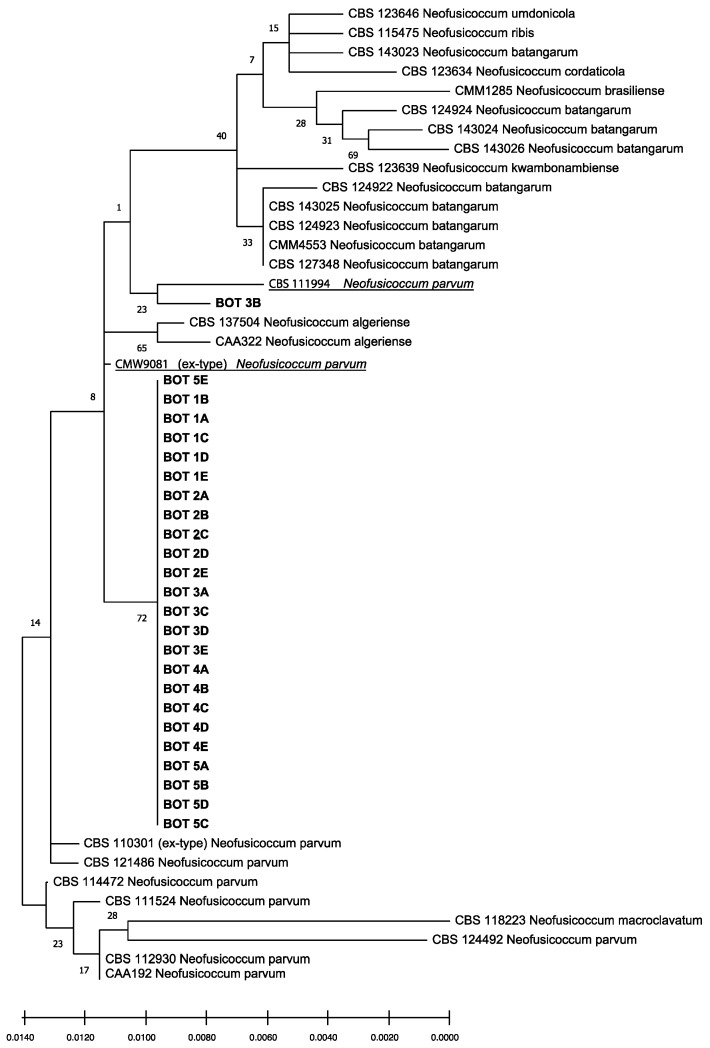
The isolates obtained had identical ITS, tef1, and tub2 sequences. Preliminary BLAST analyses of these three gene regions yielded several identical sequences belonging to Neofusicoccum spp. but deposited with different taxa names. Consequently, this analysis enabled the identification at the genus level, but did not provide reliable information on the species. The phylogenetic analysis of the combined data set of sequences from ITS, tef1, and tub2 sequences (Figure 6) produced trees with a high concordance with those reported by Lopes et al. [27,29], Yang et al. [28], and Zhang et al. [7]. According to this analysis, isolates obtained from gummy cankers of Citrus × limon were identified as Neofusicoccum parvum sensu stricto (s.s.), since they clearly clustered with the ex-type (CMW9081 from Populus nigra [29]) and other reference isolates of this species (CBS 111994 from Telopea sp. [28]) and were differentiated from other Neofusicoccum isolates in the Neofusicoccum species complex [7].
Figure 6.
Phylogenetic tree of isolates of Neofusicoccum parvum sourced from lemon trees in this study (in bold), and reference isolates of Neofusicoccum parvum (underlined) and other closely related Neofusicoccum species and isolates (Table 1 and Table 2). The tree was built using concatenated sequences of ITS-5.8S-ITS2 region, tef1-α and β-tubulin genes. Numbers on nodes indicate the posterior probabilities from the maximum likelihood method.
3.4. Pathogenicity Tests
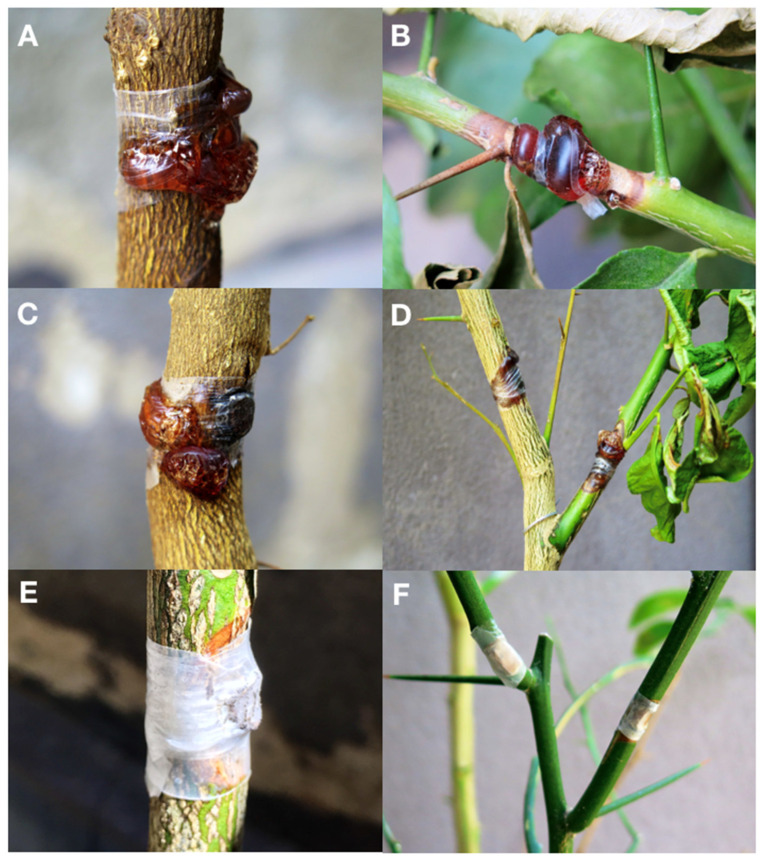
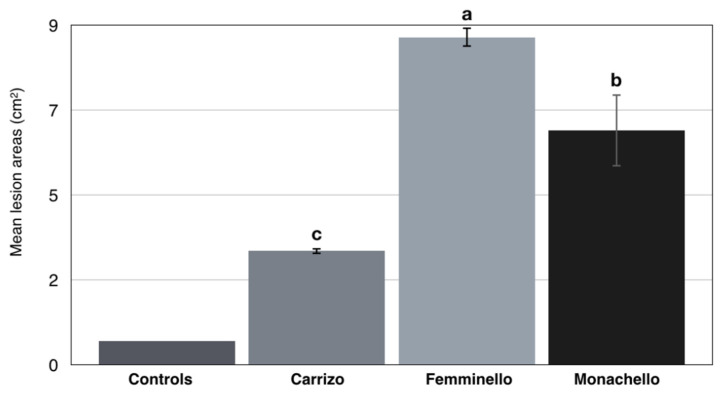
In pathogenicity tests, N. parvum (isolate BOT 1D) induced gummy canker on twigs and stem of all three citrus varieties tested. Symptoms appeared at 14 d.a.i.; they were more severe on lemon cultivars ‘Femminello 2kr’(lesion areas ranging from 8.4 to 8.9 cm2 30 d.a.i.), and ‘Monachello’ (lesion areas ranging from 6.5 to 6.8 cm2 30 d.a.i.) and included abundant gummosis (Figure 7A–D), while gummosis was much less abundant on Citrange ‘Carrizo’ (lesion areas ranging from 3.1 to 3.3 cm2 30 d.a.i.) (Figure 7E,F). Differences in mean lesion size between the three citrus varieties tested were significant, according to Tukey‘s HSD (Honestly Significant Difference) test (p < 0.05) (Figure 8). No symptoms were observed on the controls.
Figure 7.
Gum exudate induced by wound-inoculation of Neofusicoccum parvum on stems and twigs of lemon ‘Femminello Siracusano 2kr’ (A,B) and ‘Monachello’ (C,D) 14 d a.i.; necrotic lesions without gummous flux on inoculated Citrange ‘Carrizo’ (E,F).
Figure 8.
Mean lesion areas (cm2) on the stem of citrange (Citrus sinensis × Poncirus trifoliata) ‘Carrizo’ and lemon (Citrus × limon) cultivars ‘Femminello 2kr’ and ‘Monachello’, wound-inoculated with isolate BOT1D of Neofusicoccum parvum, 30 d.a.i. Controls (‘Femminello 2kr’ lemon, ‘Monachello’ lemon and ‘Carrizo’ citrange) stem inoculated with a sterile agar plug, showed no symptoms. Values sharing different letters are statistically different according to Tukey’s honestly significant difference, for p = 0.05.
The N. parvum test isolate was re-isolated from inoculated plants and identified by sequencing and multi-loci phylogenetic analysis, while no fungal pathogens were isolated from control plants, thus fulfilling Koch’s postulates.
4. Discussion
Bot gummosis, traditionally regarded as a minor disease in citrus orchards [33], was found to be common and widespread in lemon groves of southern Italy. Although it is not as destructive as the malsecco disease caused by the mitosporic fungus Plenodomus tracheiphilus (syn. Phoma tracheiphila) [34], bot gummosis can be regarded as a major constraint for the lemon industry in this production area as it causes premature aging of trees and reduces their productivity. In this respect, the impact of bot gummosis in commercial lemon orchards is as severe as wood rot disease caused by species of Basidiomycetes and unlike wood rot, bot gummosis may affect trees under the age of 25 years [35]. The present study provided evidence that Neofusicoccum parvum in the family Botryosphaeriaceae is responsible for bot gummosis of lemon trees in Sicily and Calabria, the two major lemon producing Italian regions. This fungus was the only species of this family associated to the typical disease symptoms in all surveyed areas and the two most popular Italian lemon cultivars, ‘Femminello 2kr’ and ‘Monachello’, were shown to be very susceptible to the infections by this pathogen in pathogenicity assays. Consistently with symptoms observed on naturally infected trees in the field, pathogenicity tests revealed that N. parvum was very aggressive on ‘Femminello 2kr’ and ‘Monachello’, which reacted to the infection with the abundant production of gum exudate, while it was weakly virulent on ‘Carrizo’ citrange, commonly used as a citrus rootstock. These marked differences in susceptibility to bot gummosis among different citrus genotypes are in agreement with results of other Authors [10]. In pathogenicity trials carried out in Greece the citrumelo ‘Swingle’ (Poncirus trifoliata ×C. paradisi), commonly used as a rootstock, was proved to be the least susceptible to N. parvum and D. foeniculina infections among nine different citrus species [10]. Neofusicoccum parvum is a generalist, cosmopolitan pathogen, occurring in various environments with a temperate, Mediterranean, or subtropical climate. The host range of this species encompasses at least 90 botanical entities, especially woody angiosperms, including conifers and many horticultural plants [4,36,37,38,39,40,41,42,43,44]. In association with other fungal pathogens, including other Botryosphaeriaceae species, N. parvum is involved in the trunk disease complex of grapevine, also known as Botryosphaeria dieback, and is regarded as one of the most aggressive causal agent of this disease, which is responsible for severe economic losses in vineyards worldwide [45,46]. It was also reported as a pathogen of citrus from Australia, California, and Europe [8,47,48]. However, this is the first report of N. parvum as a pathogen of citrus in Italy.
In a previous study, it was shown that N. parvum is more aggressive than other Botryosphaeriaceae species associated to bot gummosis of citrus [3]. This may explain its prevalence on lemon, which is more susceptible to bot gummosis than other citrus species [49]. Like other species of Neofusicoccum, N. parvum produces phytotoxins, which probably act as virulence factors but none of them is host-specific [50,51,52]. Different Neofusicoccum species cause similar effects on plants, perhaps due to the production of these secondary metabolites. Neofusicoccum parvum, e.g., when inoculated on cactus pear cladodes was able to induce on this host the same symptoms as N. batagarum (13) and conversely in artificial inoculations N. batangarum induced on citrus plants the same symptoms as N. parvum [13]. Genes encoding for virulence factors that allow N. parvum to colonize the wood of the trunk and the transcriptional dynamics of grapevine genes during the stem colonization by this fungal pathogen have been investigated [53,54]. In general, the Botryosphaeriaceae, like other opportunistic albeit aggressive plant pathogens such as Colletotrichum spp., behave as entophytes, saprobes or latent pathogens shifting to an aggressive pathogenic life style when the host-plant is stressed [55,56,57,58,59]. It has been suggested that the widespread occurrence of N. parvum and related species is due to their polyphagy and cross-infection potential as well as to their behavior as endophytes or latent pathogens favoring a global spread through the movement of asymptomatic plants and plant propagation material [40,56,60,61]. According with this hypothesis, it can be speculated that the widespread occurrence of N. parvum in lemon growing areas of southern Italy may also depend on their close proximity with districts where the nursery production of citrus plants is concentrated. As a consequence, it can be assumed that most lemon trees in these areas have a common origin and N. parvum has been spread as a latent pathogen with nursery plants. This study provided evidence that restricted outbreaks of bot gummosis may occur in lemon groves of southern Italy. Local emergence probably due to environmental stresses is typical of diseases caused by Botryosphaeriaceae [13,42,62]. Extreme temperatures and water deficit are suspected to be the most frequent stressors favoring bot gummosis of citrus. These environmental factors condition not only the disease onset and development, but also the severity of symptom expression. A better understanding of the factors favoring the emergence of the bot gummosis in lemon orchards would be useful to adopt appropriate management strategies to prevent the disease.
5. Conclusions
The constant association between Neofusicoccum parvum and symptoms of bot gummosis on lemon trees and its aggressiveness towards two of the most popular Italian lemon cultivars (one traditional and one selected quite recently) clearly indicate the fungus is responsible for this disease in major lemon-growing areas of southern Italy. The occurrence of bot gummosis in a relatively vast geographical area where its presence has been documented since the past century, albeit with different common names, suggests this record of N. parvum on lemon is not occasional even if it is the first in Italy. Nursery plants are suspected to be the main vehicle for the wide distribution of this fungus whose life style as latent pathogen might have favored its spread through asymptomatic plants. It is noteworthy this study provided evidence of local outbreaks of bot gummosis triggered by environmental factors. The incidence and frequency of these outbreaks could be expected to increase in the future as a consequence of both climate change and the introduction of more susceptible lemon cultivars. With regard to the latter aspect, the marked difference in susceptibility shown by the two lemon cultivars in pathogenicity trials would suggest to extend the test to other lemon genotypes.
Acknowledgments
We are grateful to V. Lo Giudice for collaboration and helpful suggestions during field surveys and A. Davies for the English revision of the text.
Author Contributions
Conceptualization, S.O.C., F.A., M.R., and A.P.; methodology, A.P., S.O.C., M.R., and F.A.; software, F.A., M.R., and R.P.; validation, S.O.C. and A.P.; formal analysis, F.A., M.R., and R.P.; investigation, F.A., M.R., and R.P.; resources, S.O.C. and A.P.; data curation, F.A. and M.R.; writing—original draft preparation, F.A. and M.R.; writing—review and editing, S.O.C. and A.P.; visualization, S.O.C.; supervision, S.O.C. and A.P.; project administration, S.O.C. and A.P.; funding acquisition, S.O.C. and A.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the University of Catania, Italy “Investigation of phytopathological problems of the main Sicilian productive contexts and eco-sustainable defense strategies (MEDIT-ECO)” PiaCeRi - PIAno di inCEntivi per la Ricerca di Ateneo 2020-22 linea 2” “5A722192155 ”, by the project “Smart and innovative packaging, postharvest rot management and shipping of organic citrus fruit (BiOrangePack)” Partnership for Research and Innovation in the Mediterranean Area (PRIMA)-H2020 (E69C20000130001); “F.A. has been granted a Ph.D. fellowship “Scienze Agrarie, Alimentari, Forestali e Ambientali—XXXIII cycle”, University of Palermo; M.R. has been granted a fellowship by CREA “OFA” (Rende, Italy), this study is part of his activity as PhD, in “Agricultural, Food, and Forestry Science”, University Mediterranea of Reggio Calabria, XXXV cycle”.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Klotz L.J. Fungal, Bacterial, and Nonparasitic Diseases and Injuries Originating in the Seedbed, Nursery, and Orchard. In: Reuther W., Calavan E.C., Carman G.E., editors. The Citrus Industry. Volume 4. University of California-Division of Agricultural Sciences; Riverside, CA, USA: 1978. pp. 1–66. [Google Scholar]
- 2.Scaramuzzi G. Le Malattie degli Agumi. Edizioni Agricole; Bologna, Italy: 1965. pp. 12–17. [Google Scholar]
- 3.Adesemoye A.O., Mayorquin J.S., Wang D.H., Twizeyimana M., Lynch S.C., Eskalen A. Identification of Species of Botryosphaeriaceae Causing Bot gummosis in Citrus in California. Plant Dis. 2014;98:55–61. doi: 10.1094/PDIS-05-13-0492-RE. [DOI] [PubMed] [Google Scholar]
- 4.Phillips A.J.L., Alves A., Abdollahzadeh J., Slippers B., Wingfield M.J., Groenewald J.Z., Crous P.W. The Botryosphaeriaceae: Genera and Species Known from Culture. Stud. Mycol. 2013;76:51–167. doi: 10.3114/sim0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dissanayake A.J., Camporesi E., Hyde K.D., Phillips A.J.L., Fu C.Y., Yan J.Y., Li X. Dothiorella Species Associated with Woody Hosts in Italy. Mycosphere. 2016;7:51–63. doi: 10.5943/mycosphere/7/1/6. [DOI] [Google Scholar]
- 6.Phillips A.J.L., Hyde K.D., Alves A., Liu J.K. Families in Botryosphaeriales: A Phylogenetic, Morphological and Evolutionary Perspective. Fungal Divers. 2019;94:1–22. doi: 10.1007/s13225-018-0416-6. [DOI] [Google Scholar]
- 7.Zhang W., Groenewald J.Z., Lombard L., Schumacher R.K., Phillips A.J.L., Crous P.W. Evaluating Species in Botryosphaeriales. Pers. Mol. Phylogeny Evol. Fungi. 2021;46:63–115. doi: 10.3767/persoonia.2021.46.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adesemoye A.O., Eskalen A. First Report of Spencermartinsia viticola, Neofusicoccum australe, and N. parvum Causing Branch Canker of Citrus in California. Plant Dis. 2011;95:770. doi: 10.1094/PDIS-02-11-0092. [DOI] [PubMed] [Google Scholar]
- 9.Mayorquin J.S., Wang D.H., Twizeyimana M., Eskalen A. Identification, Distribution, and Pathogenicity of Diatrypaceae and Botryosphaeriaceae Associated with Citrus Branch Canker in the Southern California Desert. Plant Dis. 2016;100:2402–2413. doi: 10.1094/PDIS-03-16-0362-RE. [DOI] [PubMed] [Google Scholar]
- 10.Vakalounakis D.J., Ntougias S., Kavroulakis N., Protopapadakis E. Neofusicoccum parvum and Diaporthe foeniculina Associated with Twig and Shoot Blight and Branch Canker of Citrus in Greece. J. Phytopathol. 2019;167:527–537. doi: 10.1111/jph.12843. [DOI] [Google Scholar]
- 11.Hamrouni N., Nouri M.T., Trouillas F.P., Said A., Sadfi-Zouaoui N., Hajlaoui M.R. Dothiorella gummosis Caused by Dothiorella viticola, First Record from Citrus in Tunisia. New Dis. Rep. 2018;38:10. doi: 10.5197/j.2044-0588.2018.038.010. [DOI] [Google Scholar]
- 12.Schena L., Burruano S., Giambra S., Surico G., Pane A., Evoli M., Magnano Di San Lio G., Cacciola S.O. First Report of Neofusicoccum batangarum as Causal Agent of Scabby Cankers of Cactus Pear (Opuntia ficus-indica) in Minor Islands of Sicily. Plant Dis. 2018;102:445. doi: 10.1094/PDIS-07-17-1039-PDN. [DOI] [Google Scholar]
- 13.Aloi F., Giambra S., Schena L., Surico G., Pane A., Gusella G., Stracquadanio C., Burruano S., Cacciola S.O. New Insights into Scabby Canker of Opuntia ficus-indica, Caused by Neofusicoccum batangarum. Phytopathol. Mediterr. 2020;59:269–284. [Google Scholar]
- 14.Guarnaccia V., Crous P.W. Emerging Citrus Diseases in Europe Caused by Species of Diaporthe. IMA Fungus. 2017;8:317–334. doi: 10.5598/imafungus.2017.08.02.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phillips A.J.L. Botryosphaeria Species Associated with Diseases of Grapevines in Portugal. Phytopathol. Mediterr. 2002;41:3–18. [Google Scholar]
- 16.Slippers B., Fourie G., Crous P.W., Coutinho T.A., Wingfield B.D., Wingfield M.J. Multiple Gene Sequences Delimit Botryosphaeria australis sp. nov. from B. lutea. Mycologia. 2004;96:1030–1041. doi: 10.1080/15572536.2005.11832903. [DOI] [PubMed] [Google Scholar]
- 17.Crous P.W., Slippers B., Wingfield M.J., Rheeder J., Marasas W.F.O., Phillips A.J.L., Alves A., Burgess T., Barber P., Groenewald J.Z. Phylogenetic Lineages in the Botryosphaeriaceae. Stud. Mycol. 2006;55:235–253. doi: 10.3114/sim.55.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wright A.F., Harmon P.F. Identification of Species in the Botryosphaeriaceae Family Causing Stem Blight on Southern Highbush Blueberry in Florida. Plant Dis. 2010;94:966–971. doi: 10.1094/PDIS-94-8-0966. [DOI] [PubMed] [Google Scholar]
- 19.McDonald V., Eskalen A. Botryosphaeriaceae Species Associated with Avocado Branch Cankers in California. Plant Dis. 2011;95:1465–1473. doi: 10.1094/PDIS-02-11-0136. [DOI] [PubMed] [Google Scholar]
- 20.Schena L., Cooke D.E.L. Assessing the Potential of Regions of the Nuclear and Mitochondrial Genome to Develop a “Molecular Tool Box” for the Detection and Characterization of Phytophthora Species. J. Microbiol. Methods. 2006;67:70–85. doi: 10.1016/j.mimet.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 21.White T.J., Bruns T., Lee S., Taylor J.W. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J., editors. PCR Protocols: A Guide to Methods and Applications. Volume 18. Academic Press, Inc.; San Diego, CA, USA: 1990. pp. 315–322. [Google Scholar]
- 22.Carbone I., Kohn L.M. A Method for Designing Primer Sets for Speciation Studies in Filamentous Ascomycetes. Mycologia. 1999;91:553–556. doi: 10.1080/00275514.1999.12061051. [DOI] [Google Scholar]
- 23.Glass N.L., Donaldson G.C. Development of Primer Sets Designed for Use with the PCR to Amplify Conserved Genes from Filamentous Ascomycetes. Appl. Environ. Microbiol. 1995;61:1323–1330. doi: 10.1128/AEM.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.FinchTV v.1.4.0. [(accessed on 16 February 2021)]; Available online: https://digitalworldbiology.com/FinchTV.
- 25.Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slippers B., Boissin E., Phillips A.J.L., Groenewald J.Z., Lombard L., Wingfield M.J., Postma A., Burgess T., Crous P.W. Phylogenetic Lineages in the Botryosphaeriales: A Systematic and Evolutionary Framework. Stud. Mycol. 2013;76:31–49. doi: 10.3114/sim0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopes A., Phillips A.J.L., Alves A. Mating Type Genes in the Genus Neofusicoccum: Mating Strategies and Usefulness in Species Delimitation. Fungal Biol. 2017;121:394–404. doi: 10.1016/j.funbio.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 28.Yang T., Groenewald J.Z., Cheewangkoon R., Jami F., Abdollahzadeh J., Lombard L., Crous P.W. Families, Genera, and Species of Botryosphaeriales. Fungal Biol. 2017;121:322–346. doi: 10.1016/j.funbio.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 29.Lopes A., Barradas C., Phillips A.J.L., Alves A. Diversity and Phylogeny of Neofusicoccum Species Occurring in Forest and Urban Environments in Portugal. Mycosphere. 2016;7:906–920. doi: 10.5943/mycosphere/si/1b/10. [DOI] [Google Scholar]
- 30.Vaidya N.H., Hadjicostis C.N., Dominguez-Garcia A.D. Distributed Algorithms for Consensus and Coordination in the Presence of Packet-Dropping Communication Links–Part II: Coefficients of Ergodicity Analysis Approach. arXiv. 20111109.6392 [Google Scholar]
- 31.Edgar R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Timmer L.W., Garnsey S.M., Graham J.H. Compendium of Citrus Diseases. 2nd ed. American Phytopathological Society Press; Saint Paul, MN, USA: 2000. p. 92. [Google Scholar]
- 34.Migheli Q., Cacciola S.O., Balmas V., Pane A., Ezra D., Magnano di San Lio G. Mal Secco Disease Caused by Phoma tracheiphila: A Potential Threat to Lemon Production Worldwide. Plant Dis. 2009;93:852–867. doi: 10.1094/PDIS-93-9-0852. [DOI] [PubMed] [Google Scholar]
- 35.Roccotelli A., Schena L., Sanzani S.M., Cacciola S.O., Mosca S., Faedda R., Ippolito A., Magnano di San Lio G. Characterization of Basidiomycetes Associated with Wood Rot of Citrus in Southern Italy. Phytopathology. 2014;104:851–858. doi: 10.1094/PHYTO-10-13-0272-R. [DOI] [PubMed] [Google Scholar]
- 36.Golzar H., Burgess T.I. Neofusicoccum parvum, a Causal Agent Associated with Cankers and Decline of Norfolk Island Pine in Australia. Australas. Plant Pathol. 2011;40:484. doi: 10.1007/s13313-011-0068-4. [DOI] [Google Scholar]
- 37.Iturritxa E., Slippers B., Mesanza N., Wingfield M.J. First Report of Neofusicoccum parvum Causing Canker and Die-back of Eucalyptus in Spain. Australas. Plant Dis. Notes. 2011;6:57. doi: 10.1007/s13314-011-0019-5. [DOI] [Google Scholar]
- 38.Spagnolo A., Marchi G., Peduto F., Phillips A.J.L., Surico G. Detection of Botryosphaeriaceae Species within Grapevine Woody Tissues by Nested PCR, with Particular Emphasis on the Neofusicoccum parvum/N. ribis complex. Eur. J. Plant Pathol. 2011;129:485–500. doi: 10.1007/s10658-010-9715-9. [DOI] [Google Scholar]
- 39.Úrbez-Torres J.R., Gubler W.D. Susceptibility of Grapevine Pruning Wounds to Infection by Lasiodiplodia theobromae and Neofusicoccum parvum. Plant Pathol. 2011;60:261–270. doi: 10.1111/j.1365-3059.2010.02381.x. [DOI] [Google Scholar]
- 40.Molina-Gayosso E., Silva-Rojas H.V., García-Morales S., Avila-Quezada G. First Report of Black Spots on Avocado Fruit Caused by Neofusicoccum parvum in Mexico. Plant Dis. 2012;96:287. doi: 10.1094/PDIS-08-11-0699. [DOI] [PubMed] [Google Scholar]
- 41.Sakalidis M.L., Slippers B., Wingfield B.D., Hardy G.E.S.J., Burgess T.I. The Challenge of Understanding the Origin, Pathways and Extent of Fungal Invasions: Global Populations of the Neofusicoccum parvum-N. ribis Species Complex. Divers. Distrib. 2013;19:873–883. doi: 10.1111/ddi.12030. [DOI] [Google Scholar]
- 42.Moral J., Morgan D., Trapero A., Michailides T.J. Ecology and Epidemiology of Diseases of Nut Crops and Olives Caused by Botryosphaeriaceae Fungi in California and Spain. Plant Dis. 2019;103:1809–1827. doi: 10.1094/PDIS-03-19-0622-FE. [DOI] [PubMed] [Google Scholar]
- 43.Elvira-Recuenco M., Cacciola S.O., Sanz-Ros A.V., Garbelotto M., Aguayo J., Solla A., Mullet M., Drenkhan T., Oskay F., Aday Kaya A.G., et al. Potential Interactions between Invasive Fusarium circinatum and Other Pine Pathogens in Europe. Forests. 2020;11:7. doi: 10.3390/f11010007. [DOI] [Google Scholar]
- 44.Haenzi M., Cochard B., Chablais R., Crovadore J., Lefort F. Neofusicoccum parvum, a New Agent of Sequoia Canker and Dieback Identified in Geneva, Switzerland. Forests. 2021;12:434. doi: 10.3390/f12040434. [DOI] [Google Scholar]
- 45.Úrbez-Torres J.R., Gubler W.D. Pathogenicity of Botryosphaeriaceae Species Isolated from Grapevine Cankers in California. Plant Dis. 2009;93:584–592. doi: 10.1094/PDIS-93-6-0584. [DOI] [PubMed] [Google Scholar]
- 46.Mondello V., Songy A., Battiston E., Pinto C., Coppin C., Trotel-Aziz P., Clément C., Mugnai L., Fontaine F. Grapevine Trunk Diseases: A Review of Fifteen Years of Trials for Their Control with Chemicals and Biocontrol Agents. Plant Dis. 2018;102:1189–1217. doi: 10.1094/PDIS-08-17-1181-FE. [DOI] [PubMed] [Google Scholar]
- 47.Cunnington J.H., Priest M.J., Powney R.A., Cother N.J. Diversity of Botryosphaeria Species on Horticultural Plants in Victoria and New South Wales. Austral. Plant Pathol. 2007:157–159. doi: 10.1071/AP07002. [DOI] [Google Scholar]
- 48.Bezerra J.D.P., Crous P.W., Aiello D., Gullino M.L., Polizzi G., Guarnaccia V. Genetic Diversity and Pathogenicity of Botryosphaeriaceae Species Associated with Symptomatic Citrus Plants in Europe. Plants. 2021;10:492. doi: 10.3390/plants10030492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salerno M., Cutuli G. Guida Illustrate di Patologia degli Agrumi. Edagrigole–Edizioni Agricole della Calderini; Bologna, Italy: 1994. pp. 30–35. [Google Scholar]
- 50.Abou-Mansour E., Débieux J.L., Ramírez-Suero M., Bénard-Gellon M., Magnin-Robert M., Spagnolo A., Chong J., Farine S., Bertsch C., L’Haridon F., et al. Phytotoxic Metabolites from Neofusicoccum parvum, a Pathogen of Botryosphaeria dieback of Grapevine. Phytochemistry. 2015;115:2017–2215. doi: 10.1016/j.phytochem.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 51.Masi M., Aloi F., Nocera P., Cacciola S.O., Surico G., Evidente A. Phytotoxic Metabolites Isolated from Neufusicoccum batangarum, the Causal Agent of the Scabby Canker of Cactus Pear (Opuntia ficus-indica L.) Toxins. 2020;12:126. doi: 10.3390/toxins12020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salvatore M.M., Alves A., Andolfi A. Secondary Metabolites Produced by Neofusicoccum Species Associated with Plants: A Review. Agriculture. 2021;11:149. doi: 10.3390/agriculture11020149. [DOI] [Google Scholar]
- 53.Blanco-Ulate B., Rolshausen P., Cantu D. Draft Genome Sequence of Neofusicoccum parvum Isolate UCR-NP2, a Fungal Vascular Pathogen Associated with Grapevine Cankers. Genome Announc. 2013;1:e00339-13. doi: 10.1128/genomeA.00339-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Massonnet M., Figueroa-Balderas R., Galarneau E.R., Miki S., Lawrence D.P., Sun Q., Wallis C.M., Baumgartner K., Cantu D. Neofusicoccum parvum Colonization of the Grapevine Woody Stem Triggers Asynchronous Host Responses at the Site of Infection and in the Leaves. Front. Plant. Sci. 2017;8:1117. doi: 10.3389/fpls.2017.01117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Slippers B., Wingfield M.J. Botryosphaeriaceae as Endophytes and Latent Pathogens of Woody Plants: Diversity, Ecology and Impact. Fungal Biol. Rev. 2007;21:90–106. doi: 10.1016/j.fbr.2007.06.002. [DOI] [Google Scholar]
- 56.Pavlic-Zupanc D., Wingfield M.J., Boissin E., Slippers B. The Distribution of Genetic Diversity in the Neofusicoccum parvum/N. ribis Complex Suggests Structure Correlated with Level of Disturbance. Fungal Ecol. 2015;13:93–102. doi: 10.1016/j.funeco.2014.09.002. [DOI] [Google Scholar]
- 57.Cacciola S.O., Gilardi G., Faedda R., Schena L., Pane A., Garibaldi A., Gullino M.L. Characterization of Colletotrichum ocimi Population Associated with Black Spot of Sweet Basil (Ocimum basilicum) in Northern Italy. Plants. 2020;9:654. doi: 10.3390/plants9050654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haridas S., Albert R., Binder M., Bloem J., LaButti K., Salamov A., Andreopoulos B., Baker S.E., Barry K., Bills G. 101 Dothideomycetes Genomes: A Test Case for Predicting Lifestyles and Emergence of Pathogens. Stud. Mycol. 2020;96:141–153. doi: 10.1016/j.simyco.2020.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Riolo M., Aloi F., Pane A., Cara M., Cacciola S.O. Twig and Shoot Dieback of Citrus, a New Disease Caused by Colletotrichum Species. Cells. 2021;10:449. doi: 10.3390/cells10020449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pancher M., Ceol M., Corneo P.E., Longa C.M.O., Yousaf S., Pertot I., Campisano A. Fungal Endophytic Communities in Grapevines (Vitis vinifera L.) Respond to Crop Management. Appl. Environ. Microbiol. 2012;78:4308–4317. doi: 10.1128/AEM.07655-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pérez C.A., Wingfield M.J., Slippers B., Altier N.A., Blanchette R.A. Endophytic and Canker-associated Botryosphaeriaceae Occurring on Non-native Eucalyptus and Native Myrtaceae Trees in Uruguay. Fungal Divers. 2010;41:53. doi: 10.1007/s13225-009-0014-8. [DOI] [Google Scholar]
- 62.Marsberg A., Kemler M., Jami F., Nagel J.H., Postma-Smidt A., Naidoo S., Wingfield M.J., Crous P.W., Spatafora J.W., Hesse C.N., et al. Botryosphaeria dothidea: A Latent Pathogen of Global Importance to Woody Plant Health. Mol. Plant. Pathol. 2017;18:477–488. doi: 10.1111/mpp.12495. [DOI] [PMC free article] [PubMed] [Google Scholar]