SUMMARY
Soluble “SOSIP”-stabilized envelope (Env) trimers are promising HIV-vaccine immunogens. However, they induce high-titer responses against the glycan-free trimer base, which is occluded on native virions. To delineate the effect on base responses of priming with immunogens targeting the fusion peptide (FP) site of vulnerability, here, we quantify the prevalence of trimer-base antibody responses in 49 non-human primates immunized with various SOSIP-stabilized Env trimers and FP-carrier conjugates. Trimer-base responses account for ~90% of the overall trimer response in animals immunized with trimer only, ~70% in animals immunized with a cocktail of SOSIP trimer and FP conjugate, and ~30% in animals primed with FP conjugates before trimer immunization. Notably, neutralization breadth in FP-conjugate-primed animals correlates inversely with trimer-base responses. Our data provide methods to quantify the prevalence of trimer-base responses and reveal that FP-conjugate priming, either alone or as part of a cocktail, can reduce the trimer-base response and improve the neutralization outcome.
In brief
The exposed base region of soluble HIV-1 Env trimers elicits strong non-neutralizing antibody responses. Corrigan et al. quantify plasma anti-base responses in immunized NHPs and observe a reduction in anti-base responses with fusion-peptide priming. The percentage of anti-base responses correlates inversely with neutralization breadth, providing insights for improving vaccination strategies.
Graphical Abstract

INTRODUCTION
A major hurdle to an effective HIV vaccine is the inability of current vaccine immunogens to induce prevalent immune responses that target vulnerable conserved neutralization epitopes on the trimeric HIV-1 envelope (Env) glycoprotein, which is masked by dense glycosylation (Lee et al., 2016; Stewart-Jones et al., 2016; Wei et al., 2003). However, after years of infection, a substantial portion of chronically infected individuals develop broadly neutralizing antibodies (bNAbs) with various levels of breadth and potency targeting conserved Env epitopes, thus demonstrating the potential of the immune system to generate effective bNAbs (Hraber et al., 2014; Li et al., 2007; Simek et al., 2009; Walker et al., 2010). Substantial efforts have been made to develop immunogens that can induce cross-reactive neutralizing antibody responses similar to those found in some HIV-1-infected individuals. This has led to promising advances in vaccine development, including the use of engineered Env trimers, stabilized by a disulfide (SOS) linking gp120 and gp41 subunits and an Ile to Pro (IP) mutation to maintain a prefusion native-like conformation and to present critical bNAb epitopes (Sanders et al., 2013; Sanders and Moore, 2017). However, soluble SOSIP-stabilized trimers expose the Env base region, which lacks shielding glycans and is most likely occluded on HIV-1 virions by the viral membrane (Hu et al., 2015; Pancera et al., 2014). As a result, immunization with SOSIP-stabilized trimers leads to a dominant antibody response targeting the base region of the trimer with minimal neutralizing activity in mice and rabbits (Bianchi et al., 2018; Hu et al., 2015) and non-human primates (NHPs) (Cottrell et al., 2020; Martin et al., 2020; Nogal et al., 2020). Furthermore, simian-HIV (SHIV)-infected NHPs do not develop detectable base responses, confirming that anti-base responses are associated with soluble trimer immunization (Nogal et al., 2020). In addition, it is possible the dominant response to the trimer base may inhibit the development of less-dominant responses to neutralizing epitope regions of Env.
In addition to SOSIP-stabilized Env trimers, another promising class of HIV-1 immunogens involves the fusion peptide (FP) site of vulnerability. FP is a hydrophobic peptide at the N terminus of the gp41 subunit that is a critical part of the Env-entry machinery. FP embeds itself in the target cell membrane and initiates the fusion of viral and target cell membranes. Several bNAbs targeting the FP region, including VRC34.01, PGT151, and ASC202 (Falkowska et al., 2014; Kong et al., 2016; van Gils et al., 2016), have been isolated from chronically infected individuals. Immunization with FP-carrier conjugates and SOSIP-stabilized Env trimers in mice, guinea pigs, and NHPs elicits cross-clade, FP-directed neutralizing responses with isolated monoclonal antibodies (mAbs) neutralizing up to 59% of a cross-clade panel of 208 diverse strains (Cheng et al., 2019, 2020; Kong et al., 2019; Xu et al., 2018). Additionally, comparison of NHP immunization regimens has revealed FP-conjugate priming, alone or as part of a cocktail with an SOSIP-stabilized Env trimer, to elicit B cells reactive with both FP and Env trimer, and prevalence of such double-positive B cells is predictive of neutralizing activity about 1 year later (Cheng et al., 2020). Although these studies using FP conjugates and Env trimers are providing insight into how FP conjugates can enhance neutralizing activity, the effect on anti-base responses has not yet been investigated.
We hypothesized that priming with an FP-carrier conjugate, either alone or as part of a cocktail with a SOSIP-stabilized trimer, would focus the immune response to FP and decrease the prevalence of plasma antibody responses targeting the Env-trimer base. To elucidate the effect of the immunization regimen on the elicitation of trimer-base responses, we developed three different methods to quantify trimer-base responses. We used these to analyze plasma antibody responses from 49 NHPs immunized with various SOSIP-stabilized Env trimers and FP-carrier conjugates, including a self-assembling FP-conjugate nanoparticle, and we also analyzed correlations between elicited responses and vaccination outcomes.
RESULTS
Selection of NHPs immunized with FP conjugates and SOSIP-stabilized trimers
We selected for analysis 49 NHPs in eight groups with five immunization regimens (Figure 1). These NHPs could be divided into three immunization categories: trimer only, cocktail of trimer and FP, and FP-prime-trimer boost. The trimer-only category comprised four groups totaling 23 NHPs, which received trimer immunization at week 0 and week 4, with plasma samples taken at week 6 for analysis. Trimer-only immunogens included BG505 DS-SOSIP or CH505 DS-SOSIP deglycan variants with (1) three glycans (N230, N241, and N611) removed around the fusion peptide, (2) four glycans (N88, N230, N241, and N611) removed around the fusion peptide, or (3) three glycans (N197, N276, and N462) removed around the CD4-binding site (CD4bs) (the native CH505 trimer is missing the glycan at N362 near CD4bs) (Figure 1A and 1B) (Cheng et al., 2020). The CH505 DS-SOSIP immunogens were constructed as chimeras in which portions of the N and C termini of gp120 as well as the whole gp41 subunit were replaced by sequences from BG505. As a result, the base of the CH505 and BG505 immunogens were identical. Of note, NHPs in the trimer-only group were previously analyzed for FP-specific responses (Cheng et al., 2020; Kong et al., 2019; Xu et al., 2018), but anti-trimer base responses had not been analyzed.
Figure 1. Immunization regimens of 49 NHPs analyzed for plasma anti-trimer base responses; for all regimens, NHP plasma was analyzed 2 weeks after the second trimer immunization (indicated with a red arrow).
(A) Three groups of NHPs were immunized twice with CH505 DS-SOSIP immunogens as indicated. The CH505 DS-SOSIP immunogens were FP-degly3, with three glycans (N230, N241, and N611) removed around the fusion peptide (FP); FP-degly4, with four glycans (N88, N230, N241, and N611) removed around the FP; and CD4bs-degly4, with four glycans (N197, N276, N462, and N362) removed around the CD4-binding site.
(B) One group of five NHPs was immunized twice with BG505 DS-SOSIP.
(C) Two groups of eight animals each were primed with FP8-KLH + BG505 DS-SOSIP or FP8-rTTHc nanoparticles + BG505 DS-SOSIP.
(D) One group of five animals was immunized with long intervals, primed with FP8-KLH at weeks 0, 4, and 20, followed by an FP7-KLH immunization at week 32, an FP6-KLH immunization at week 44, and two BG505 DS-SOSIP immunizations at weeks 56 and 64.
(E) One group of five animals was immunized with short intervals, primed with FP8-KLH at weeks 0, 4, and 8, followed by an FP7-KLH immunization at week 12, an FP6-KLH immunization at week 20, and two BG505 DS-SOSIP immunizations at weeks 24 and 28.
See also Figure S1.
The cocktail category comprised two groups each of eight NHPs, which received two immunizations, at week 0 and week 4, of a cocktail consisting of a carrier-conjugated FP and BG505 DS-SOSIP (Figure 1C). For one of the cocktail groups, the FP-carrier conjugate comprised the most prevalent sequence of eight N-terminal amino acids of FP (FP8v1) coupled to keyhole limpet hemocyanin (KLH), which we had shown previously to prime for broad FP responses (Xu et al., 2018). For the other cocktail group, we developed a nanoparticle version of the FP-carrier conjugate by first coupling the recombinant tetanus toxoid heavy chain (rTTHC) through SpyCatcher/SpyTag (Zakeri et al., 2012) to encapsulin (EN), a self-assembling nanoparticle, and then coupling FP8v1 to the rTTHC-EN nanoparticle through a bifunctional crosslinker Sulfo SIAB (Ou et al., 2020) (Figure S1).
The FP-prime trimer-boost category comprised two groups each of five NHPs, which received five FP v1-KLH priming immunizations and then were boosted twice with the BG505 DS-SOSIP trimer. One of the FP-primed groups was subjected to long immunization intervals and the other to short intervals (Figures 1D and 1E). As with other groups, plasma samples were collected 2 weeks after the second trimer immunization, corresponding to week 66 for the long interval group and to week 30 for the short interval group. Of note, NHPs in these two groups have been analyzed previously, although at different time points (Cheng et al., 2020; Kong et al., 2019; Xu et al., 2018), and anti-trimer base responses had not been investigated.
Although it would likely require more trimer immunizations to achieve breadth, we selected the 2 weeks after the second trimer immunization to analyze the plasma anti-base responses in all NHPs, to compare the effect of priming on the elicitation of anti-base responses because we wanted to focus on the early signature of the base response, which we believe might be affecting the development of the neutralization activity and breadth later on.
Anti-base Fabs block BG505 DS-SOSIP base recognition by base-binding mAbs and NHP plasma but do not affect bNAbs targeting major sites of Env vulnerability
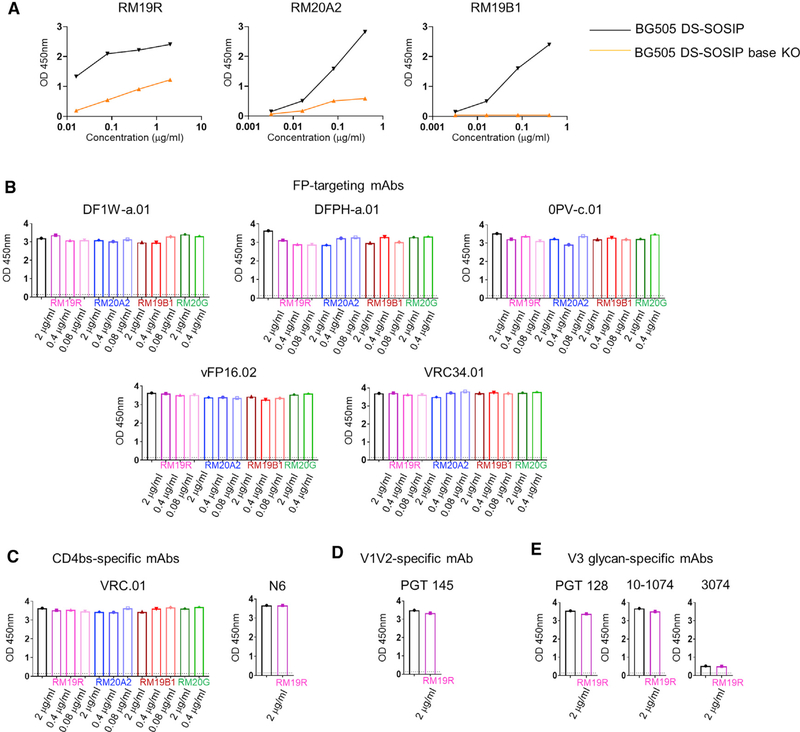
To assess plasma antibody responses to the base region of the Env trimers, we first set out to identify base-binding antibodies suitable for competition ELISA that would block binding to the base region but would not interfere with antibodies binding to other conserved regions. Previous studies have shown antigen-binding fragments (Fabs) of various NHP antibodies to bind to the base region of the trimer, including antibodies RM19R, RM20A2, RM19B1, and RM20G (Cottrell et al., 2020). We performed additional negative-stain-electron microscopy (ns-EM) and biolayer interferometry (BLI) analyses, which confirmed all four antibodies bound to the base region of the BG505 DS-SOSIP trimer (Figure S2A) and that RM19R and RM20A2 bound more strongly than did RM19B1 and RM20G (Figure S2B). Moreover, a BG505 DS-SOSIP mutant with two added base-region glycans at N502 and N660 had reduced ELISA binding to RM19R, RM20A2, and RM19B1, confirming these Fabs targeted the trimer-base region (Figure 2A). Next, we assessed whether these base-specific Fabs would interfere with the binding of bNAbs targeting other regions of the trimer. We analyzed 11 bNAbs, five of them targeting the FP region, three targeting the V3 region, two targeting the CD4-binding site (CD4bs), and one targeting the V1V2 region (Figures 2B–2E). No significant differences were observed by ELISA in the total BG505 DS-SOSIP responses for any of the 11 bNAbs when anti-base Fabs were added, indicating that these four base-binding Fabs do not interfere with binding of the assessed bNAbs to the BG505 DS-SOSIP trimer.
Figure 2. Anti-base Fabs showed base-specific ELISA binding and do not block mAbs targeting the FP, V3 glycan, CD4bs, or V1V2 region.
(A) RM19R, RM20A2, and RM19B1 IgG showed reduced binding to BG505 DS-SOSIP base KO, which has glycans added at N502 and N660 of the base region.
(B–E) Competition ELISA (OD450) for the binding of various epitope-specific mAbs to BG505 DS-SOSIP in the absence (black bars) or presence of base-binding Fabs: RM19R (pink), RM20A2 (blue), RM19B1 (red), or RM20G (green), at concentrations as indicated.
(B) FP-specific mAbs DF1W-a.01, DFPH-a.01, 0PV-c.01, vFP16.02, and VRC34.01.
(C) CD4-binding-site-specific mAbs VRC01 and N6.
(D) V1V2- specific mAb PGT145.
(E) V3 glycan-specific mAbs PGT128, 10–1074, and 3074.
See also Figure S2.
We then analyzed the ability of these Fabs to block trimer base responses from NHP plasma. We initially used plasma from one NHP, A13V009, which was immunized twice with CH505 DS-SOSIP, a trimer with three glycans around the FP removed (Cheng et al., 2020). We tested plasma at a starting dilution of 1:100 in a competition ELISA with each of the four base-binding Fabs at three concentrations, 2 μg/mL, 0.4 μg/mL, and 0.08 μg/mL (Figures 3A–3D) to assess dose dependency along with the ability to block responses targeting the Env-base region. RM19R and RM20A2 demonstrated the strongest base blocking, with 94% and 84% of trimer response blocked at 2 μg/mL for A13V009, respectively (Figure 3E). In contrast, RM19B1 and RM20G were unable to sufficiently block the base responses, with only 46% and 7.9% of trimer response blocked at 2 μg/mL for A13V009 (Figure 3E). Additionally, RM19R was the most dose dependent, with 94% of trimer response blocked at 2 μg/mL, 81% at 0.4 μg/mL, and 20% at 0.08 μg/mL for A13V009. In contrast, RM20A2 showed a difference of only 0.82% between 2 μg/mL and 0.4 μg/mL for A13V009 (Figure 3E). Based on these data, we decided to proceed with RM19R as the base-binding Fab for further experiments.
Figure 3. Anti-base Fabs block anti-BG505 DS-SOSIP responses in NHP immunized with trimer.
(A–D) ELISA for the binding of NHP A13V009 plasma to BG505 DS-SOSIP competing with base-binding Fabs at concentrations as indicated, starting at a dilution of 1:100: (A) RM19R, (B) RM20A2, (C) RM19B1, and (D) RM20G.
(E) Summary of the competition ELISA as a percentage of the BG505 DS-SOSIP plasma response blocked by base-binding Fabs.
(F) Raw OD values taken at 450 nm for the base-binding Fab RM19R at 2 μg/mL, 1 μg/mL, and 0.4 μg/mL in a competition ELISA with RM19R IgG.
To determine the concentration of RM19R Fab needed to most efficiently block the RM19R mAb response to BG505 DS-SOSIP base region, we tested RM19R as a full immunoglobulin G (IgG), serially diluted from a starting concentration of 2 μg/mL in a competition ELISA with RM19R Fab at three concentrations: 2 μg/mL, 1 μg/mL, and 0.4 μg/mL. At a concentration of 2 μg/mL, RM19R Fab was able to block the RM19R mAb trimer recognition by more than 95% at the lowest dilution; thus, a starting concentration of 2 μg/mL was chosen for the competition ELISA with NHP plasma (Figure 3F).
Priming with FP-carrier conjugates, either alone or in a cocktail with DS-SOSIP trimer, decreases plasma response to the trimer base
To evaluate Env base responses elicited by the 49 NHPs described above, a competition ELISA was performed using the RM19R Fab. Serially diluted plasma was assessed for binding to BG505 DS-SOSIP in the absence or presence of RM19R Fab. NHPs immunized with only CH505 DS-SOSIP or BG505 DS-SOSIP (trimer-only category) had significantly reduced responses to the BG505 DS-SOSIP trimer in the presence of the RM19R Fab (Figures 4A, 4B, and S3). NHPs primed with FP conjugates alone before the trimer immunizations (FP-prime-trimer -boost category) had the least reduction in BG505 DS-SOSIP responses by RM19R competition, and NHPs immunized with a cocktail of FP conjugate and Env trimer (cocktail category) elicited responses between those of the trimer-only and FP-prime-trimer-boost categories (Figures 4C, 4D, and S3).
Figure 4. Plasma binding to BG505 DS-SOSIP trimer was mostly blocked by RM19R Fab for NHPs primed with a SOSIP trimer but mostly maintained for NHPs primed with FP.
(A–D) Geometric mean ELISA responses (OD450) with 95% confidence interval for each group are plotted against plasma dilution factors in the presence (purple) or absence (gray) of RM19R Fab.
(A) 18 NHPs primed with CH505 DS-SOSIP trimers that had three or four glycans removed around the FP (FP-degly3 and FP-degly4), or four glycans removed around the CD4 binding site (CD4bs-degly4).
(B) 5 NHPs primed with BG505 DS-SOSIP trimers.
(C) 16 NHPs primed with a FP + BG505 DS-SOSIP trimer cocktail.
(D) 10 NHPs primed with FP8-KLH followed by sequential immunization with FP8–7-6-KLH and two BG505 DS-SOSIP trimers.
(E) RM19R mAb as a control.
See also Figure S3.
To quantify the total trimer responses and the percentage of trimer-base responses in these NHPs, we analyzed the raw ELISA data in two ways, assessing either the optical density at 450 nm (OD450) values for the wells representing a plasma dilution of 1:500 or the total area under the curve (AUC) for all seven dilutions. The value corresponding to the dilution of 1:500 was chosen because it was the dilution at which the RM19R Fab was able to block more than 95% of the BG505 DS-SOSIP response to its corresponding mAb (Figure 3F). Results from both methods were strongly correlated (r = 0.9844, p < 0.0001) (Figures S4 and S5). Ultimately, we chose to use the OD450 values at a 1:500 dilution for further data analysis because they were simpler to calculate than the AUC.
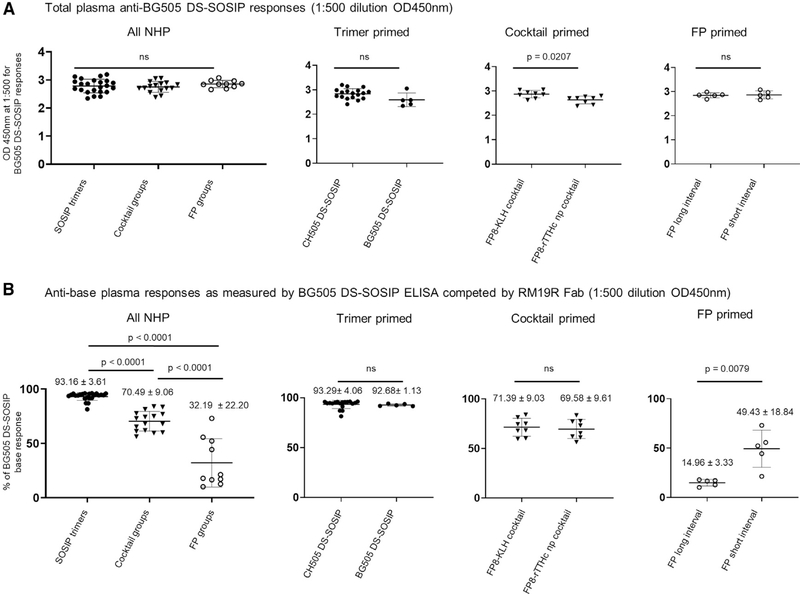
With the exception of the FP8-KLH + BG505 DS-SOSIP cocktail group having slightly higher total trimer responses than the FP8-rTTHc nanoparticle + BG505 DS-SOSIP cocktail group, as assessed by the OD450 value at 1:500 dilution, all NHPs elicited similar total trimer responses without RM19R competition (Figure 5A). To calculate the percentage of base responses, the difference in OD450 values at 1:500 dilution for the plasma responses with and without RM19R competition was divided by the value for the plasma responses without RM19R competition and multiplied by 100. This provided a numerical value by which to compare the base responses in NHPs from various immunization regimens. We observed no significant difference in base responses between the 18 NHPs that were immunized with a variant of CH505 DS-SOSIP and the five NHPs that were immunized with BG505 DS-SOSIP (Figure 5B, second panel from left). Therefore, the base response values of these 23 NHPs were combined for further analysis as the trimer-primed category, with the average percentage of base responses being 93% (Figure 5B, left panel). The trimer-primed category had significantly greater base responses than all other groups (p < 0.0001) (Figure 5B, left panel). The two cocktail-immunized groups, FP8-KLH + BG505 DS-SOSIP cocktail and FP-rTTHc nanoparticles + BG505 DS-SOSIP cocktail, had no significant difference in base responses (Figure 5B, second panel from right). The average base response of all cocktail immunized NHPs was 70%, which was significantly greater than that of the FP-prime-trimer-boost category (average base response of 32%) (Figures 5B, left panel). Interestingly, there was a significant difference between the two groups of the FP-prime-trimer-boost category. The five NHPs with long immunization intervals had base responses (average of 15%) significantly lower (p = 0.0079) than the five NHPs with short intervals (average of 49%) (Figures 5B, right panel).
Figure 5. Immunization regimens with FP in the prime elicited substantial responses targeting regions outside the base of the trimer.
(A) Comparison of the total ELISA response to BG505 DS-SOSIP between all NHPs with trimer prime, cocktail prime, or FP prime, CH505 DS-SOSIP versus BG505 DS-SOSIP primed groups, FP8-KLH + BG505 DS-SOSIP cocktail versus FP8-rTTHc np + BG505 DS-SOSIP cocktail primed groups, and FP long interval versus FP short interval primed groups revealed no significant difference among various groups. The ELISA OD450 values for total BG505 DS-SOSIP binding at a dilution of 1:500 plasma were plotted for individual animals.
(B) Comparison of anti-base Fab competition ELISA response between all NHPs with trimer prime, cocktail prime, and FP only prime groups, CH505 DS-SOSIP versus BG505 DS-SOSIP primed groups, FP8-KLH + BG505 DS-SOSIP cocktail versus FP8-rTTHc np + BG505 DS-SOSIP cocktail primed groups, and FP long interval versus FP short interval primed groups for the percentage of plasma response targeting the base region of the BG505 DS SOSIP trimer.
The percentage of BG505 DS-SOSIP responses blocked was calculated by taking the difference between OD450 values without and with RM19R Fab at a plasma dilution of 1:500 divided by the OD450 values at a dilution of 1:500 without RM19R Fab blocking and multiplied by 100. Statistical analysis was performed with a two-tailed Mann-Whitney nonparametric test to assess p values for means ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; ns, not significant. Correlation analysis was performed using a two-tailed Pearson correlation coefficiency test.
See also Figures S3–S7.
Base responses inversely correlate with neutralization breadth for NHPs primed with FP-carrier conjugates
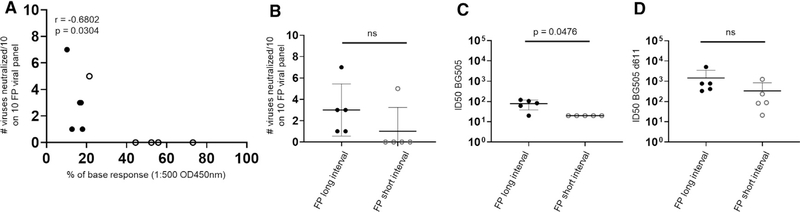
Because the two FP-primed groups elicited the lowest percentage of base responses and differed in their respective percentages, we analyzed those 10 NHPs further. Specifically, we analyzed neutralization by plasma from those 10 NHPs two weeks after the second trimer immunization against the BG505 strain and a mutant BG505 strain with glycan 611 removed (BG505Δ611), as well as a panel of 10 wild-type Env-pseudotyped viruses, each with sensitivity to FP-directed antibodies. The long-interval FP-primed group showed greater neutralization activity against BG505, a slightly greater neutralization titer against BG505Δ611, and better overall breadth (Figures 6B–6D) (although that was not statistically significant). Notably, neutralization breadth was found to correlate inversely with the percentage of base responses (r = −0.6802, p = 0.0304) (Figure 6A). These data suggest that FP priming leads to reduced base-binding responses, which appeared to be indicative of increased neutralization breadth.
Figure 6. Anti-base responses for the 10 FP-primed NHPs correlate inversely with the neutralization breadth and the long-interval group showed greater autologous neutralization titers than the short-interval group.
(A) Correlation between percentages of anti-base responses based on OD450 values at a dilution of 1:500 and neutralization breadth defined as the number of wild-type FP-sensitive HIV-1 viruses neutralized out of a 10-virus panel.
(B–D) Neutralization breadth (B) and ID50 titers on BG505 (C) and BG505 Δ611 (D) for the long-interval (black circles) and short-interval (hollow circles) FP-primed NHPs.
Statistical analysis was carried out with a two-tailed Mann-Whitney nonparametric test to assess p values for means ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; ns, not significant. Correlation analysis was performed using a two-tailed Pearson correlation coefficiency test.
See also Figure S6.
The plasma neutralizing activity against the BG505 strain for trimer primed and cocktail primed NHPs at week 6 was sporadic (Figure S6A). As only one animal showed weak autologous titer out of 23 trimer primed NHPs, we could not perform correlation analysis between the plasma base response and the 50% inhibitory dilution (ID50) titers on the BG505 strain for the trimer-primed NHPs. Of note, the CH505-primed 18 animals with either CD4bs deglycan or FP-site deglycan did not elicit any neutralization activity against CH505 (not shown) or BG505 wild-type viruses (Figure S6A). We did not see correlation between plasma base-binding responses and autologous neutralization ID50 titers against BG505 or BG505Δ611 in cocktail-primed NHPs (Figures S6B and S6C) or in FP-primed NHPs (Figures S6D and S6E).
Biolayer interferometry and MSD analysis confirm base-response results from ELISA competition assays
To confirm the base-response data generated by BG505 DS-SOSIP competition ELISA with the RM19R Fab, we performed a similar competition assay using biolayer interferometry (BLI) with biotinylated BG505 DS-SOSIP and RM19R Fab. Similar to the ELISA results, the RM19R Fab blocked RM19R mAb binding to BG505 DS-SOSIP completely and did not affect binding of CD4bs mAb VRC01 (Figure S7B, far right panel). We then analyzed all 49 NHP samples with BLI for binding to the trimer with and without RM19R Fab competition. Similar to the results from ELISA, all NHP groups, on average, showed similar total trimer responses (Figure S7A). In the presence of RM19R Fab, NHPs immunized with trimer only showed minimal binding to the trimer, NHPs immunized with a cocktail had slightly increased binding, and NHPs immunized with FP prime and trimer boost had the greatest responses to the trimer (Figure S7B). These results indicate that FP priming can reduce the base responses elicited by trimer immunization. Furthermore, on average, NHPs with long immunization intervals had the most binding to the trimer in the presence of the RM19R Fab, consistent with the ELISA results (Figure S7B, third panel from left).
To further confirm the data observed by both ELISA and BLI, we chose six NHP plasma samples for Meso Scale Discovery (MSD) assay with Avi-tagged BG505 DS-SOSIP in competition with a cocktail of the base-binding antibodies RM19R and RM20A2 in equal molar ratios. Specifically, we chose two NHP samples from each of the trimer-primed groups (NHP nos. A12V163 and A12V193), the cocktail-primed groups (NHP nos. 36337 and TT0), and the FP-primed groups (NHP nos. DF3R and DFPE) at the same time points as in the ELISA and BLI analyses (2 weeks after the final boost). We analyzed the difference between the total trimer responses and the trimer responses after competition with the base-binding antibodies by electrochemiluminescence (ECL) and obtained results consistent with those from ELISA and BLI described above. The trimer-primed NHPs showed a greater decrease in total trimer responses after RM19R + RM20A2 competition, in comparison to those primed with a cocktail of FP + BG505 DS-SOSIP or FP only, with the FP-primed group showing the smallest decrease in responses by anti-base antibody competition (Figures S7C–S7H). Furthermore, the percentage of base responses, calculated as described above but using AUC values, was found to correlate with the results above from ELISA (Figures S7J and S7K). These results confirm the base responses found by ELISA and validate the use of all three assays to assess responses targeting the exposed trimer base in NHP plasma.
DISCUSSION
An effective HIV-1 vaccine will likely comprise prime and boost immunogens to develop and enhance the elicitation of the bNAb responses with sufficient breadth and potency to confer protection. To date, soluble SOSIP-stabilized trimers derived from various HIV-1 clades and strains have been tested in preclinical vaccination studies. However, although the trimer base on the HIV-1 virion is not easily accessible, the base region on soluble trimers is exposed, free of glycan shielding and should, thus, be highly immunogenic. To understand the effect of base responses, we sought to quantify such responses relative to the overall anti-trimer responses in 49 NHPs immunized with varied regimens. Specifically, we developed methods to quantify the base responses with ELISA, MSD, and BLI using anti-base antibody competition. Our results confirm that immunization with SOSIP-stabilized trimers leads to dominant anti-base antibody responses.
We observed no significant difference in the prevalence of base responses among groups receiving wild-type or deglycosylated SOSIP-stabilized trimers, indicating that deglycosylation around the CD4bs or FP regions of the SOSIP-stabilized trimer does not significantly affect the dominant immunogenicity of the base region. We note that in prior studies, NHPs immunized with CD4bs deglycosylated trimers did show enhanced neutralizing activity against CD4bs deglycosylated viruses but not against wild-type viruses, indicating that the induced immune responses may be focused on glycan holes that do not exist on the wild-type Env trimers (Zhou et al., 2017). However, because we tested the base responses with the BG505 DS-SOSIP wild-type trimer, and not with the autologous deglycosylated trimer, our detection methods may not have assessed plasma-binding responses to CD4bs or FP region glycan holes that only exist on the deglycosylated trimers. Nonetheless, we did observe immunizations with a cocktail of FPand SOSIP trimer to reduce base immune responses when compared with immunizations with SOSIP trimers alone; and FP primes followed by trimer boosts further reduced the base response. These results confirmed our hypothesis, i.e., that priming with an FP-carrier conjugate, either alone or in a cocktail with a SOSIP-stabilized trimer, focuses the immune response to FP and decreases antibody responses targeting the Env-trimer base.
As a corollary, we observed reduced base responses to correlate with improved FP-directed neutralizing responses, in which the FP specificity of these antibody responses was demonstrated primarily by the ability of soluble FP to reduced neutralization (Cheng et al., 2020; Kong et al., 2019). It was unclear whether the results indicated a threshold rather than a correlation because the breadth disappeared when the base responses were greater than ~20%–30% (Figure 6A). Such a correlative or threshold response was also observed in prior results (Cheng et al., 2020), in which NHPs with frequencies of FP+/BG505+ B cells greater than 0.35% at pre-FP8–7-6 time points were more likely to have a greater neutralization breadth at the end of the study. Whether correlative or threshold, reduced off-target base responses and enhanced FP-specific responses are likely to contribute to better neutralizing activity. In contrast, priming with a trimer alone only weakly elicited such antibody responses. Moreover, we have isolated FP-specific cross-neutralizing antibodies from the short-interval FP-primed group (Kong et al., 2019) and from long-interval FP-primed and cocktail-primed NHPs (unpublished data), and the isolated monoclonals have neutralizing activity that matches the serum neutralization. Combining these prior B cell analyses with the anti-base response findings of the current study, the data suggest that FP priming skews the immune response away from the otherwise dominant response to the base and enhances on-target response in the vulnerable regions, with FP-specific cross-neutralizing activity elicited, as described in a schematic shown in Figure 7. Importantly, the prevalence of the base response inversely correlated with neutralization breadth on a 10-strain panel sensitive to FP-targeting antibodies, suggesting that reducing the immunodominant base response could increase the neutralization outcome. Moreover, because we noted that the reduction of base response and enhancement of neutralization activity and breadth appeared to be further improved by increasing the length of the interval between FP and trimer immunizations, we speculate that the increased interval may allow FP-specific antibodies to develop more fully and mature to increase the relative prevalence of this response versus that targeting the immunogenically dominant trimer base.
Figure 7. Schematic showing that priming with FP, a conserved subregion of the SOSIP trimer, provided a means to reduce the induction of trimer-base-specific antibodies in NHPs.
Priming with SOSIP trimers with base-region exposed elicited dominant trimer-base responses (90% of total response, antibodies in gray), and cocktail priming with trimer/FP reduced the anti-base responses (70% of total response), and elicited more antibodies targeting non-base region on the trimer (antibodies in green). Priming with FP elicited anti-FP specific antibodies, including those recognizing FP in trimer context (antibodies in red) and those that can only recognize FP in linear conformation (antibodies in black), and trimer boost after FP prime elicited least anti-base antibodies (30% of total responses) and most non-base region antibodies. Anti-FP specific antibodies are included in the anti-trimer non-base region antibodies. Elicitation of FP+trimer+ B cells in NHPs was characterized previously (Cheng et al., 2020).
Other soluble trimeric immunogens derived from type 1 fusion proteins, such as influenza virus hemagglutinin and respiratory syncytial virus fusion glycoprotein, also have exposed trimer-base regions. The anti-base quantification methods described here should be applicable to the quantification of base responses against such surface glycoproteins from these or similar pathogens. Notably, priming with FP alone or with FP + trimer cocktail elicited higher frequency of FP+trimer+ B cells at an early stage compared with trimer-alone priming, which correlated with the neutralizing activity about 1 year later (Cheng et al., 2020). These results indicate the neutralization outcome is likely determined by multiple factors, including the overall level of anti-base and anti-FP responses and by antibody responses targeting other regions of the Env, areas of investigation that need to be further explored.
Although we were able to show that FP-carrier priming reduced base responses, the extent to which those base responses directly hinder neutralization activity and breadth is not entirely clear. Additionally, the longevity and severity of the base responses found in vaccine-test animals primed with either trimer or FP + SOSIP trimer cocktails warrants further investigation. In general, the presence of highly dominant, base-specific responses early in the immunization regimen would be expected to inhibit the development of responses against other regions of the trimer, such as those targeting epitopes recognized by neutralizing antibody lineages. Indeed, the inverse correlation between base responses and neutralization breadth in FP primed NHPs does suggest that reducing plasma-base responses may be helpful to achieving more-optimal vaccine outcomes. The FP-primed group with long immunization intervals elicited the least base response, but still showed an average of 15% of the total trimer responses to target the base region. It will be interesting to see whether further reductions in the anti-base response will further improve vaccination outcomes. Trimer immunogens, such as BG505 DS-SOSIP base knockout (KO) with glycans covering the base as shown in Figure 2A or BG505 MD39-based trimers with glycan-masking of the base region (Kulp et al., 2017) or trimer immunogens presented on nanoparticles (Antanasijevic et al., 2020), on virus-like particles (VLPs) (Tong et al., 2012) or coupled to alum (Moyer et al., 2020) may reduce base responses, although further evaluation is needed to assess other aspects of these immunogens. In general, our results indicate that non-neutralizing base responses should be assessed whenever trimers are used as immunogens and that this assessment may be critical to efforts geared toward developing an effective HIV-1 vaccine.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by John R. Mascola (john.mascola@nih.gov ).
Materials availability
All new reagents are available by MTA for non-commercial research.
Data and code availability
The published article includes all data generated or analyzed during this study. This study did not generate new code.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
NHP studies
Local, state, federal and institute policies were implemented and maintained while caring for the animals involved in this study in an American Association for Accreditation of Laboratory Animal Care-accredited facility (Bioqual Inc, MD). The Animal Care and Use Committee of the Vaccine Research Center, NIAID, NIH approved each of the studies involved, which included those listed under protocols VRC #16–667.1 for the CH505 DS-SOSIP FP deglycosylated groups, #16–666.1 for the CH505 DS-SOSIP CD4bs deglycosylated groups, #16–667.2 for the FP primed groups, #16–450.5 for the BG505 DS-SOSIP groups, and #16–808.2 for the FP + SOSIP cocktail groups. Healthy (B-virus, SIV, SRV, and STLV negative) Indian rhesus macaques without previous exposure to HIV or SHIV and without prior involvement in procedures or drug experimentation of both sexes, aged 2–14 years, and with body weights ranging from 41–109 kg were evenly distributed to different groups in each of the studies based on body weights. 100 μg of specified, filter-sterilized immunogen mixed in a total volume of 1ml of PBS and 200 μL of Adjuplex (Sigma-Aldrich Inc, MO or Adjuplex equivalent formulated based on US Patent 6,676,958 B2) used for each immunization regardless of immunogen (in case of cocktail, the two immunogens were mixed at 50 μg each). Immunizations at 500 μL each were given via a needle syringe to the caudle thigh of the two hind legs. Two weeks after each immunization, whole blood was collected, and plasma and peripheral blood mononuclear cells (PBMCs) were isolated using Ficoll density gradient centrifugation.
Cell lines
Expi293F cells (cat# A14257) and FreeStyle 293-F cells (cat# R79007) were purchased from ThermoFisher Scientific Inc. Cells were maintained in FreeStyle 293 Expression Medium. The cell line was used directly from the commercial sources and cultured following manufacturer suggestions as described in Method Details below.
METHOD DETAILS
Base-binding antibody and Fab production
Sequences for base-binding antibodies RM19R, RM19B1, RM20A2, RM20G were previously reported (Cottrell et al., 2020). The variable regions of each antibody were synthesized (GenScript) with human light chain constant regions (κ or λ as indicated by original sequence) or human IgG CH1 with a C-terminal AviTag/HRV 3C protease/6xHis tag and subcloned into pVRC8400 for expression of Fabs. The RM19R heavy variable region was also synthesized with a human IgG1 heavy constant backbone for expression of mAb. Heavy and light chain pairs were co-transfected (1:1 mass ratio) in Expi293F cells (Thermo Fisher) using ExpiFectamine 293 according to the manufacturer’s protocol (Thermo Fisher). Six days post-transfection, culture supernatants were harvested. Kappa Fabs were loaded onto a CaptureSelect KappaXL Affinity column (Thermo Fisher), lambda Fabs were loaded onto a LambdaFabSelect column (Cytiva), and mAb was loaded onto a rProtein A column (Cytiva). All resins were washed with PBS and antibodies were eluted with a low pH buffer appropriate for the resin and buffer exchanged into PBS. Proteins were then polished by loading onto a Superdex 200 16/600 SEC column (Cytiva) to remove any dimers or higher order aggregates.
Base-binding BG505 DS-SOSIP competition ELISA for NHP samples
The base-binding BG505 DS-SOSIP competition ELISA was modified from a previously reported method of an anti-trimer lectin-captured ELISA (Cheng et al., 2020). Ninety-six well Coastar half plates (Costar High Binding Half-Area; Corning, Kennebunk, ME) were coated (50 μl/well) with 2 μg/ml snowdrop lectin in PBS overnight at 4°C. The following morning, they were washed five times with PBS-T (0.05% tween/PBS) and then blocked with 5% skim milk/PBS blocking buffer (50 μl/well) for one hour at room temperature. The plates were then washed fix times with PBS-T. For the trimer capture, the plates were coated with BG505 DS-SOSIP (50 μl/well) at 2 μg/ml in 10% FBS PBS for two hours at room temperature and were subsequently washed five times with PBS-T. The plates were then incubated with either 2 μg/ml RM19R Fab/blocking buffer (50 μl/well) or blocking buffer alone (50 μl/well) for one hour at room temperature. Serially diluted epitope-specific mAbs starting at 2 μg/ml, or serially diluted NHP sera starting at a 1:100 dilution was then additionally added to the wells for one hour at room temperature (without mixing). The plates were then washed five times with PBS-T. HRP-conjugated goat anti-monkey IgG secondary antibodies were added to the plate at 1:5000 dilution for one hour at room temperature. The plates were then washed five times with PBS-T. The plates were then developed with tetramethylbenzidine (TMB) (50 μl/well) substrate for ten minutes at room temperature in the dark. The reaction was stopped by adding sulfuric acid (50 μl/well) and the optical density (OD) of each well was read at 450nm.
Base-binding BG505 DS-SOSIP competition ELISA for epitope-specific antibodies
The FP specific mAbs include NHP derived DF1W-a.01, DFPH-a.01 and OPV.c-1, mouse derived vFP16.02, and VRC34.01 which was isolated from a chronically HIV-1 infected individual (Kong et al., 2016, 2019; Xu et al., 2018). The V3 specific mAbs include PGT128, 10–1074 and 3074, and the CD4-binding site specific bNAbs include VRC01 and N6, all of which were isolated from chronically HIV-1 infected individuals (Doores et al., 2015; Hioe et al., 2010; Huang et al., 2016; Jiang et al., 2010; Walker et al., 2011; Wu et al., 2010). The V1V2 specific bNAb is PGT145, which was also isolated from a chronically infected individual (Lee et al., 2017). Each of these antibodies were tested in a BG505 DS-SOSIP ELISA competition at 2 μg/ml (same method as for NHP samples) with each of the four base binding Fabs at three concentrations, 2 μg/ml, 0.4 μg/ml and 0.08 μg/ml, with the exception of RM20G which was only tested at concentrations of 2 μg/ml and 0.4 μg/ml. Additionally, N6 and PGT145 were only tested against RM19R at 2 μg/ml.
Meso scale discovery assay
We assessed the antigenicity assays for the immunogens with the Meso Scale Discovery (MSD) platform. Standard MSD 384 well streptavidin coated SECTOR®Imager 6000 plates were blocked with 35 μL of 5% (W/V) MSD Blocker A and incubated for 1 hr at room temperature (RT) on a Heidolph Titramax 100 vibrational plate shaker at 650 rpm. The plates were washed thrice with 0.05%Tween PBS (wash buffer) and were coated with biotinylated BG505 DS-SOSIP FPV1 10ln QQ protein at an optimized concentration of 1 μg/mL for 1 hour. 1% MSD Blocker A was used as the diluent in the assay. Plates were washed as described above and wells were incubated with 10 μL per well of either assay diluent or assay diluent containing 2 μg/mL trimer base-directed mAb cocktail (equal parts RM19R and RM20A2 with mouse Fc). Duplicate wells of serial 4-fold dilutions covering the range of 100– 409,600 of the test samples were prepared in dilution plates. After 1 hour of incubation, the plates were again washed with the wash buffer and the serial dilution samples plates were added to MSD plates such that each sample is tested with n of 4 on wells treated with assay diluent and n of 4 on wells treated with blocking antibodies. After an hour of incubation with the samples, the plates were washed again, and SULFO-TAG conjugated goat anti-mouse secondary detection antibody were used for detection at an optimized concentration of 1 μg/mL on 2 sample replicates pretreated with assay diluent and two sample replicates pretreated with blocking antibodies. SULFO-TAG conjugated goat anti-Hu/NHP secondary detection antibody was used for detection at an optimized concentration of 1 μg/mL on 2 sample replicates pretreated with assay diluent and two sample replicates pretreated with blocking antibodies. After an additional hour of incubation, the unbound secondary detection antibody was washed off the plates and the plates were read using 1X MSD Read Buffer on the MSD Sector Imager 6000.
For sample wells treated with assay diluent and detected with anti-Hu/NHP detect, the binding response is evaluated as the “unblocked” readout of trimer-binding antibodies present in that NHP sample. For sample wells treated with blocking antibodies and detected with anti-NHP detect the binding response is evaluated as the “blocked” readout of non-base-directed trimer binding antibodies present in that NHP sample. For sample wells treated with assay diluent and detected with anti-mouse detect that readout is a negative control. For wells treated with blocking antibodies and detected with anti-mouse detect the binding response is a positive control for stability of the mouse base blocking antibodies during the assay.
Streptavidin 384 well plates, Blocker A, Read Buffer and SULFO-TAG anti-species were purchased from Meso-Scale Discovery. Base-Directed mouse mAbs RM19R and RM20A2 were produced and supplied by Christopher Gonelli of the Vaccine Research Center.
Base-binding BG505 DS-SOSIP competition biolayer interferometry assay
The base-binding octet assay was performed with biolayer interferometry using a FortéBio Octet HTX instrument. Biotinylated BG505 DS-SOSIP trimer (5 μg/ml in PBS + 1% BSA) was loaded onto streptavidin (SA) biosensors (18–5019, FortéBio) for 300 s. The biosensors were then equilibrated in buffer (PBS + 1% BSA) for 60 s before probing RM19R Fab (5 μg/ml) to block the base of the trimer or buffer to leave the trimer unblocked for 300 s. The biosensors were then equilibrated again in buffer for 60 s before probing either RM19R or VRC01 mAb (25 μg/ml), NHP sera at a 1:50 dilution or buffer for a control for 300 s. Finally, the biosensors were equilibrated again in buffer for 300 s to observe any dissociation. All of the traces were aligned to the baseline following the loading of the biotinylated trimer. Data were exported from Octet analysis software and analyzed on GraphPad Prism. To note, two of the trimer primed NHP were omitted from analysis due to octet biosensor issues (NHP# 05D214 and A11V069).
Neutralization assays
Neutralization assays using a single round of infection Env-pseudovirus were performed as previous described using TZM-Bl target cells and heat inactivated NHP sera (Cheng et al., 2020; Kong et al., 2019). Briefly, 293T cells cotransfected with an Env expression plasmid and a sPG3ΔEnv backbone were used to generate the Env-pseudovirus stocks used in the assay. The sera were assessed at various dilutions, using an 8-point 4-fold dilution method which began at a dilution factor of 1:20. For one hour, the heat-inactivated sera and virus stocks were mixed and incubated at 37°C, at which point TZM-bl cells were added to the mixture for additional incubation at the same temperature. The following day, cDMEM was used as a means of feeding the cells. The following and final day, the cells were lysed to assess luciferase activity (RLU) after fitting the data to a 5-parameter hill slope equation by nonlinear regression. Finally, the 50% inhibitory dilutions (ID50) were determined and were used to assess breadth across the 10 FP-sensitive HIV-1 strains tested and to test for any correlative relationship with the percentage of base response, as determined by ELISA.
FP-KLH immunogens
All FP-KLH immunogens were prepared as previously described (Kong et al., 2019; Xu et al., 2018). Briefly, FP7-KLH and FP6-KLH were synthesized (GenScript) with the removal of the N-terminal amino acid (AVGIGAVF and AVGIGAV respectively) from FP8-KLH’s sequence (AVGIGAVF). KLH was used as a carrier protein (Thermo-Scientific) after activation and ligation to the FP peptides’ Cys thiol group. Antigenicity tests were performed using FP-specific antibodies previously described (Cheng et al., 2020).
FP-rTTHc nanoparticle immunogens
We used the SpyTag/SpyCatcher system to create the rTTHC-nanoparticles, because of solubility/yield issues with the direct rTTHC-coupling. The gene of encapsulin with spyTag (EN-spyT) was codon optimized and cloned into a pET11a vector (Novagen). The plasmid was transformed into Shuffle T7(New England Biolab) cells. The cells were grown to an OD of 0.6 before induction with 0.2mM IPTG overnight at 18°C. The harvested bacterial cells were lysed by sonication and supernatant was heated at 56°C for 15min. Supernatant was clarified by centrifuge and saturated (NH4)2SO4 solution was added to final 20% saturation. The precipitations were harvested and stored at −80°C before the carrier protein rTTHC coupled to the nanoparticle. The protein EN-spyT were re-suspended in PBS and endotoxin were removed using established procedure (Aida and Pabst, 1990).
rTTHC with spyCatcher (rTTHC-spyC) were produced in transiently transfected Expi 293 cells. Proteins were purified from the supernatant using Ni-NTA followed by size exclusion column on Superdex 200 Increase 10/300 GL in PBS.
To prepare nanoparticle carrier(rTTHC-spy-EN), EN-spyT and rTTHC-spyC were mixed at 1:1.2 molar ratio and incubated at room temperature for approximately 2 hours. rTTHC-spy-EN particles were purified by size exclusion column on Superdex200 Increase 10/300 GL in PBS buffer. The FP conjugates were prepared with the bifunctional cross-linker Sulfo-SIAB using previously described procedures (Ou et al., 2020).
HIV-1 envelope trimer immunogens
All HIV-1 envelope trimer immunogens were prepared as previously described using transiently transfected 293F cells (Pancera et al., 2014; Sanders et al., 2013). bnAbs 2G12 or VRC01 were used in affinity chromatography to purify the trimers, along with gel filtration (Superdex200 16/60HL column) and a 447–52D affinity column functioning as a negative selection to remove V3-exposed trimers. Antigenicity tests were performed on the trimers using a Meso Scale Discovery (MSD) platform as previously described (Kwon et al., 2015). The trimer immunogens used in the FP long interval group at week 56 and 64 were produced as previously described in stable CHO cell lines, with purification by non-affinity chromatography (Cheng et al., 2020). Both methods of trimer productions produced extremely similar antigenicity results.
BG505 DS-SOSIP and base-binder binding kinetics
Base-binder Fabs (RM19R, RM20A2, RM19B1, RM20G) or VRC34.01 Fab were loaded onto buffer-equilibrated Anti-Human Fab-CH1 2nd Generation (FAB2G) biosensors (FortéBio) for 300 s at 0.15–25 μg/mL (concentration optimized for each Fab) in PBS + 0.1% BSA + 0.02% Tween-20 (same buffer used for subsequent steps). The biosensors were equilibrated in buffer for 60 s before measuring association with serial dilutions of BG505 DS-SOSIP (1:2 dilutions starting as a high as 200 nM) for 300 s (120 s for VRC34.01). Dissociation of the BG505 DS-SOSIP was then measured by returning the biosensors to buffer for 300 s (120 s for VRC34.01, 600 s for RM19R, RM20A2). Data Analysis v9.0 (FortéBio) was used to align the data measurements and for fitting of association and dissociation curves using a 1:1 binding model. The affinity (KD) was determined from the association (kon) and dissociation (koff) constants.
Electron microscopy and single particle analysis
To generate Env-Fab complexes for negative stain electron microscopy, 20μg BG505 DS-SOSIP was incubated overnight at 4°C with a 4-fold molar excess of base binder Fab (RM19R, RM20A2, RM19B1, or RM20G). Complexes were separated from the excess Fab by size exclusion chromatography using a Superose 6 Increase 10/300 GL column (Cytiva) using PBS as the elution buffer. Samples were diluted as appropriate, adsorbed to freshly glow-discharged carbon-coated copper grids, washed with several drops of buffer containing 10 mM HEPES, pH 7.0, and 150 mM NaCl, and negatively stained with 0.7% uranyl formate. Between 100 and 150 micrographs were recorded for each complex at a nominal magnification of 57,000 using a ThermoFisher Talos F200C G2 electron microscope operated at 200 kV and equipped with a Ceta 16M CCD camera. The pixel size was 2.53 Å. The EPU software was used for data collection. For single particle analysis, particles were picked automatically using in-house developed software (Y.T., unpublished data) and examined manually in EMAN2.1 (Tang et al., 2007). These particles were subjected to reference-free 2D classification using Relion 1.4 (Scheres, 2012). The resulting classes were inspected visually. Classes lacking sharp features or representing noise or sub-stoichiometric complexes were excluded. Initial models were generated in EMAN2.1 using Relion’s 2D class averages, low-pass filtered to 60 Å, and used as the reference volumes for three-dimensional reconstruction and refinement using reference projections in SPIDER (Shaikh et al., 2008) with imposed C3 symmetry. The refined maps were low-pass filtered and used, along with the corresponding particle stacks, as input for the 3D refinement procedure of FREALIGN (Grigorieff, 2007) with separation into 3D classes (Lyumkis et al., 2013). UCSF Chimera (Pettersen et al., 2004) was used for visualization of the maps and maps were segmented using Segger (Pintilie et al., 2010).
QUANTIFICATION AND STATISTICAL ANALYSES
Statistical analysis comparing percentage of total BG505 DS-SOSIP response and base response was performed by a two-tailed non-parametric Mann-Whitney tests comparing the mean and standard deviation. *p < 0.05, **p < 0.01, ***p < 0.0001. For ELISA, the percentage of base BG505 DS-SOSIP response was calculated by taking the difference between the raw OD values at 1:500 dilution of the wells receiving blocking buffer only and RM19R Fab, divided by the raw OD value of the well with RM19R Fab. Two-tailed Pearson Correlation Coefficiency Tests for analyzing mean and standard deviation were used to assess correlations between percentage of BG505 DS-SOSIP response blocked with RM19R and neutralization activity and breadth.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| VRC01 | Wu et al., 2010 | RRID: AB_2491019 |
| RM19R | Cottrell et al., 2020 | N/A |
| RM20A2 | Cottrell et al., 2020 | N/A |
| RM19B1 | Cottrell et al., 2020 | N/A |
| RM20G | Cottrell et al., 2020 | N/A |
| DF1W-a.01 | Kong et al., 2019 | N/A |
| DFPH-a.01 | Kong et al., 2019 | N/A |
| 0PV-c.01 | Kong et al., 2019 | N/A |
| vFP16.02 | Xu et al., 2018 | N/A |
| VRC34.01 | Kong et al., 2016 | RRID: AB_2819225 |
| N6 | Huang et al., 2016 | N/A |
| PGT145 | Lee et al., 2017 | RRID: AB_2491054 |
| PGT128 | Walker et al., 2011 | RRID: AB_2491047 |
| 10-1074 | Jiang et al., 2010 | N/A |
| 3074 | Hioe et al., 2010 | N/A |
| Bacterial and virus strains | ||
| BG505 | N/A | |
| BG505Δ611 | Kong et al., 2016 | N/A |
| 10-strain panel for neutralization assessments | Xu et al., 2018 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Superdex200 10/300GL Column | GE Healthcare Life Sciences | Cat# 28990944 |
| MabSelect SuRe Protein A Resin | GE Healthcare Life Sciences | Cat# 17543802 |
| KLH | ThermoFisher Scientific Inc. | Cat#77600 |
| MBS (m-maleimidobenzoyl-N-hydoxysuccinimide ester) | ThermoFisher Scientific Inc. | Cat#22311 |
| Cysteine-added FP8 peptide: AVGIGAVFC | Xu et al., 2018 | N/A |
| Cysteine-added FP7 peptide: AVGIGAVC | Xu et al., 2018 | N/A |
| Cysteine-added FP6 peptide: AVGIGAC | Xu et al., 2018 | N/A |
| BG505 DS-SOSIP | Kwon et al., 2015 | N/A |
| BG505 DS-SOSIP-Avi | Kong et al., 2016 | N/A |
| CH505 DS-SOSIP FP degly3 | Cheng et al., 2020 | N/A |
| CH505 DS-SOSIP FP degly4 | Cheng et al., 2020 | N/A |
| CH505 DS-SOSIP CD4bs degly | Cheng et al., 2020 | N/A |
| FP8v1-KLH | Xu et al., 2018 | N/A |
| FP8v1-rTTHC-EN-nanoparticle | This study | N/A |
| Critical commercial assays | ||
| Turbo293 Transfection Kit | ThermoFisher Scientific Inc. | Cat# A14525 |
| BirA biotin-protein ligase bulk reaction kit | Avidity | BirA500 |
| Experimental models: Cell lines | ||
| Expi293F cells | ThermoFisher Scientific Inc | Cat# A14527 |
| FreeStyle 293-F cells | ThermoFisher Scientific Inc | Cat# R79007 |
| Experimental models: Organisms/strains | ||
| Indian origin rhesus macaque | This paper | N/A |
| Recombinant DNA | ||
| pVRC8400 vector | https://www.addgene.org | Cat# 63160 |
| pVRC8400-RM19R plasmid | This paper | N/A |
| pVRC8400-RM20A2 plasmid | This paper | N/A |
| pVRC8400-RM19B1 plasmid | This paper | N/A |
| pVRC8400-RM20G plasmid | This paper | N/A |
| Software and algorithms | ||
| GraphPad Prism Software | GraphPad Prism Software, Inc. | N/A |
| The PyMol Molecular Graphics System, v2 | Schrödinger, LLC | https://pymol.org/2/ |
| BLI Acquisition & Analysis Software | ForteBio | https://www.sartorius.com/en/products/protein-analysis/octet-systems-software |
| EPU | ThermoFisher Scientific | https://www.thermofisher.com/us/en/home/electron-microscopy/products/software-em-3d-vis/epu-software.html |
| EMAN2.1 | Tang et al., 2007 | https://blake.bcm.edu/emanwiki/EMAN2 |
| RELION | Scheres, 2012 | https://www3.mrc-lmb.cam.ac.uk/relion/index.php/Main_Page |
| SPIDER | Shaikh et al., 2008 | https://spider.wadsworth.org/spider_doc/spider/docs/spider.html |
| UCSF Chimera | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera/ |
| FREALIGN | Grigorieff, 2007, Lyumkis et al., 2013 | https://grigoriefflab.umassmed.edu/frealign |
Highlights.
Devise methods to quantify antibody responses targeting the base of HIV-1 Env trimers
Fusion-peptide (FP) priming reduces anti-base responses upon HIV Env trimer boost
Lower percentage of anti-base responses correlates with improved neutralization breadth
ACKNOWLEDGMENTS
We thank members of the Non-human Primate Immunogenicity Core at the VRC for assistance with the NHP studies; J. Noor and E. McCarthy of the Translational Research Program and Bioqual veterinary technical staff; K. Saunders and B.F. Haynes for discussions on CH505; Brenda Hartman for assistance with figures; and members of the Virology Laboratory, Vaccine Research Center, for discussions and comments on the manuscript. Support for this work was provided by the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health, and by the International AIDS Vaccine Initiative’s (IAVI’s) Neutralizing Antibody Consortium. This project has also been funded in part with federal funds from the Frederick National Laboratory for Cancer Research, NIH, under contract HHSN261200800001E (to T.S. and Y.T.).
Footnotes
DECLARATION OF INTERESTS
P.D.K., J.R.M., L.O., Y.T., K.X., and B.Z. are inventors on U.S. Patent Application 62/735,188 filed March 26, 2020, entitled “HIV-1 ENV fusion peptide immunogens and their use.” The other authors declare no competing interests.
CONSORTIA
The VRC Production Program includes Nadia Amharref, Nathan Barefoot, Christopher Barry, Elizabeth Carey, Ria Caringal, Kevin Carlton, Naga Chalamalsetty, Adam Charlton, Rajoshi Chaudhuri, Mingzhong Chen, Peifeng Chen, Nicole Cibelli, Jonathan W. Cooper, Hussain Dahodwala, Marianna Fleischman, Julia C. Frederick, Haley Fuller, Jason Gall, Isaac Godfroy, Daniel Gowetski, Krishana Gulla, Vera Ivleva, Lisa Kueltzo, Q. Paula Lei, Yile Li, Venkata Mangalampalli, Sarah O’Connell, Aakash Patel, Erwin Rosales-Zavala, Elizabeth Scheideman, Nicole A. Schneck, Zachary Schneiderman, Andrew Shaddeau, William Shadrick, Alison Vinitsky, Sara Witter, Yanhong Yang, and Yaqiu Zhang.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.108937.
REFERENCES
- Aida Y, and Pabst MJ (1990). Removal of endotoxin from protein solutions by phase separation using Triton X-114. J. Immunol. Methods 132, 191–195. [DOI] [PubMed] [Google Scholar]
- Antanasijevic A, Ueda G, Brouwer PJM, Copps J, Huang D, Allen JD, Cottrell CA, Yasmeen A, Sewall LM, Bontjer I, et al. (2020). Structural and functional evaluation of de novo-designed, two-component nanoparticle carriers for HIV Env trimer immunogens. PLoS Pathog. 16, e1008665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi M, Turner HL, Nogal B, Cottrell CA, Oyen D, Pauthner M, Bastidas R, Nedellec R, McCoy LE, Wilson IA, et al. (2018). Electron-microscopy-based epitope mapping defines specificities of polyclonal antibodies elicited during HIV-1 BG505 envelope trimer immunization. Immunity 49, 288–300.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Xu K, Kong R, Chuang GY, Corrigan AR, Geng H, Hill KR, Jafari AJ, O’Dell S, Ou L, et al. (2019). Consistent elicitation of cross-clade HIV-neutralizing responses achieved in guinea pigs after fusion peptide priming by repetitive envelope trimer boosting. PLoS ONE 14, e0215163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Duan H, Xu K, Chuang GY, Corrigan AR, Geng H, O’Dell S, Ou L, Chambers M, Changela A, et al. ; VRC Production Program (2020). Immune monitoring reveals fusion peptide priming to imprint cross-clade HIV-neutralizing responses with a characteristic early B cell signature. Cell Rep. 32, 107981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottrell CA, van Schooten J, Bowman CA, Yuan M, Oyen D, Shin M, Morpurgo R, van der Woude P, van Breemen M, Torres JL, et al. (2020). Mapping the immunogenic landscape of near-native HIV-1 envelope trimers in non-human primates. PLoS Pathog. 16, e1008753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doores KJ, Kong L, Krumm SA, Le KM, Sok D, Laserson U, Garces F, Poignard P, Wilson IA, and Burton DR (2015). Two classes of broadly neutralizing antibodies within a single lineage directed to the high-mannose patch of HIV envelope. J. Virol. 89, 1105–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkowska E, Le KM, Ramos A, Doores KJ, Lee JH, Blattner C, Ramirez A, Derking R, van Gils MJ, Liang CH, et al. (2014). Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity 40, 657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorieff N (2007). FREALIGN: high-resolution refinement of single particle structures. J. Struct. Biol. 157, 117–125. [DOI] [PubMed] [Google Scholar]
- Hioe CE, Wrin T, Seaman MS, Yu X, Wood B, Self S, Williams C, Gorny MK, and Zolla-Pazner S (2010). Anti-V3 monoclonal antibodies display broad neutralizing activities against multiple HIV-1 subtypes. PLoS ONE 5, e10254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hraber P, Seaman MS, Bailer RT, Mascola JR, Montefiori DC, and Korber BT (2014). Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS 28, 163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JK, Crampton JC, Cupo A, Ketas T, van Gils MJ, Sliepen K, de Taeye SW, Sok D, Ozorowski G, Deresa I, et al. (2015). Murine antibody responses to cleaved soluble HIV-1 envelope trimers are highly restricted in specificity. J. Virol. 89, 10383–10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Kang BH, Ishida E, Zhou T, Griesman T, Sheng Z, Wu F, Doria-Rose NA, Zhang B, McKee K, et al. (2016). Identification of a CD4-binding-site antibody to HIV that evolved near-pan neutralization breadth. Immunity 45, 1108–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Burke V, Totrov M, Williams C, Cardozo T, Gorny MK, Zolla-Pazner S, and Kong XP (2010). Conserved structural elements in the V3 crown of HIV-1 gp120. Nat. Struct. Mol. Biol. 17, 955–961. [DOI] [PubMed] [Google Scholar]
- Kong R, Xu K, Zhou T, Acharya P, Lemmin T, Liu K, Ozorowski G, Soto C, Taft JD, Bailer RT, et al. (2016). Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody. Science 352, 828–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong R, Duan H, Sheng Z, Xu K, Acharya P, Chen X, Cheng C, Dingens AS, Gorman J, Sastry M, et al. ; NISC Comparative Sequencing Program (2019). Antibody lineages with vaccine-induced antigen-binding hotspots develop broad HIV neutralization. Cell 178, 567–584.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp DW, Steichen JM, Pauthner M, Hu X, Schiffner T, Liguori A, Cottrell CA, Havenar-Daughton C, Ozorowski G, Georgeson E, et al. (2017). Structure-based design of native-like HIV-1 envelope trimers to silence non-neutralizing epitopes and eliminate CD4 binding. Nat. Commun. 8, 1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YD, Pancera M, Acharya P, Georgiev IS, Crooks ET, Gorman J, Joyce MG, Guttman M, Ma X, Narpala S, et al. (2015). Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat. Struct. Mol. Biol. 22, 522–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Ozorowski G, and Ward AB (2016). Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 351, 1043–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Andrabi R, Su C-Y, Yasmeen A, Julien J-P, Kong L, Wu NC, McBride R, Sok D, Pauthner M, et al. (2017). A broadly neutralizing antibody targets the dynamic HIV envelope trimer apex via a long, rigidified, and anionic β-hairpin structure. Immunity 46, 690–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Migueles SA, Welcher B, Svehla K, Phogat A, Louder MK, Wu X, Shaw GM, Connors M, Wyatt RT, and Mascola JR (2007). Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat. Med. 13, 1032–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyumkis D, Brilot AF, Theobald DL, and Grigorieff N (2013). Likelihood-based classification of cryo-EM images using FREALIGN. J. Struct. Biol. 183, 377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JT, Cottrell CA, Antanasijevic A, Carnathan DG, Cossette BJ, Enemuo CA, Gebru EH, Choe Y, Viviano F, Tokatlian T, et al. (2020). Targeting HIV Env immunogens to B cell follicles in non-human primates through immune complex or protein nanoparticle formulations. NPJ Vaccines 2, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer TJ, Kato Y, Abraham W, Chang JYH, Kulp DW, Watson N, Turner HL, Menis S, Abbott RK, Bhiman JN, et al. (2020). Engineered immunogen binding to alum adjuvant enhances humoral immunity. Nat. Med. 26, 430–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogal B, Bianchi M, Cottrell CA, Kirchdoerfer RN, Sewall LM, Turner HL, Zhao F, Sok D, Burton DR, Hangartner L, and Ward AB (2020). Mapping polyclonal antibody responses in non-human primates vaccinated with HIV Env trimer subunit vaccines. Cell Rep. 30, 3755–3765.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou L, Kong WP, Chuang GY, Ghosh M, Gulla K, O’Dell S, Varriale J, Barefoot N, Changela A, Chao CW, et al. ; VRC Production Program (2020). Preclinical development of a fusion peptide conjugate as an HIV vaccine immunogen. Sci. Rep. 10, 3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, et al. (2014). Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Pintilie GD, Zhang J, Goddard TD, Chiu W, and Gossard DC (2010). Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J. Struct. Biol. 170, 427–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, and Moore JP (2017). Native-like Env trimers as a platform for HIV-1 vaccine design. Immunol. Rev. 275, 161–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Peña AT, Korzun J, et al. (2013). A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 9, e1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH (2012). RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh TR, Gao H, Baxter WT, Asturias FJ, Boisset N, Leith A, and Frank J (2008). SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat. Protoc. 3, 1941–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simek MD, Rida W, Priddy FH, Pung P, Carrow E, Laufer DS, Lehrman JK, Boaz M, Tarragona-Fiol T, Miiro G, et al. (2009). Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J. Virol. 83, 7337–7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Jones GB, Soto C, Lemmin T, Chuang GY, Druz A, Kong R, Thomas PV, Wagh K, Zhou T, Behrens AJ, et al. (2016). Trimeric HIV-1-Env structures define glycan shields from clades A, B, and G. Cell 165, 813–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, and Ludtke SJ (2007). EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46. [DOI] [PubMed] [Google Scholar]
- Tong T, Crooks ET, Osawa K, and Binley JM (2012). HIV-1 virus-like particles bearing pure env trimers expose neutralizing epitopes but occlude non-neutralizing epitopes. J. Virol. 86, 3574–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gils MJ, van den Kerkhof TL, Ozorowski G, Cottrell CA, Sok D, Pauthner M, Pallesen J, de Val N, Yasmeen A, de Taeye SW, et al. (2016). An HIV-1 antibody from an elite neutralizer implicates the fusion peptide as a site of vulnerability. Nat. Microbiol. 2, 16199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Simek MD, Priddy F, Gach JS, Wagner D, Zwick MB, Phogat SK, Poignard P, and Burton DR (2010). A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog. 6, e1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, et al. ; Protocol G Principal Investigators (2011). Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477, 466–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, et al. (2003). Antibody neutralization and escape by HIV-1. Nature 422, 307–312. [DOI] [PubMed] [Google Scholar]
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, et al. (2010). Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Acharya P, Kong R, Cheng C, Chuang GY, Liu K, Louder MK, O’Dell S, Rawi R, Sastry M, et al. (2018). Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1. Nat. Med. 24, 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakeri B, Fierer JO, Celik E, Chittock EC, Schwarz-Linek U, Moy VT, and Howarth M (2012). Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. USA 109, E690–E697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Doria-Rose NA, Cheng C, Stewart-Jones GBE, Chuang GY, Chambers M, Druz A, Geng H, McKee K, Kwon YD, et al. (2017). Quantification of the impact of the HIV-1-glycan shield on antibody elicitation. Cell Rep. 19, 719–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all data generated or analyzed during this study. This study did not generate new code.