Abstract
Acinetobacter baumannii is one of the most difficult-to-treat pathogens worldwide, due to developed resistance. The aim of this study was to evaluate the use of widely prescribed antimicrobials and the respective resistance rates of A. baumannii, and to explore the relationship between antimicrobial use and the emergence of A. baumannii resistance in a tertiary care hospital. Monthly data on A. baumannii susceptibility rates and antimicrobial use, between January 2014 and December 2017, were analyzed using time series analysis (Autoregressive Integrated Moving Average (ARIMA) models) and dynamic regression models. Temporal correlations between meropenem, cefepime, and ciprofloxacin use and the corresponding rates of A. baumannii resistance were documented. The results of ARIMA models showed statistically significant correlation between meropenem use and the detection rate of meropenem-resistant A. baumannii with a lag of two months (p = 0.024). A positive association, with one month lag, was identified between cefepime use and cefepime-resistant A. baumannii (p = 0.028), as well as between ciprofloxacin use and its resistance (p < 0.001). The dynamic regression models offered explanation of variance for the resistance rates (R2 > 0.60). The magnitude of the effect on resistance for each antimicrobial agent differed significantly.
Keywords: acinetobacter, antibiotic resistance, time series analysis, dynamic regression models, antimicrobial stewardship, meropenem, cefepime, ciprofloxacin
1. Introduction
Acinetobacter species are Gram-negative bacteria associated with bacteremia and hospital-acquired pneumonia, including ventilator-associated pneumonia, surgical site infection, secondary meningitis, and urinary tract infections [1,2,3]. Acinetobacter baumannii has been reported as one of the most important and difficult-to-treat pathogens in the hospital setting [4,5]. Moreover, the number of A. baumannii infections is steadily increasing, while the optimal treatment of these infections has not yet been established [6]. The lack of new antimicrobial agents against A. baumannii and its ability to accumulate multiple antibiotic resistance genes has led to the current situation of multidrug-resistant (MDR) or extensively drug-resistant Acinetobacter isolates [7,8,9]. Multidrug resistance is considered to be non-susceptibility to at least one agent from three or more antibiotic classes that would otherwise serve as clinically effective treatments and has become a significant cause of increased morbidity and mortality in critically ill patients with severe sepsis [10,11].
Previous use of carbapenems, third- and fourth-generation cephalosporins, and fluoroquinolones are independent risk factors for acquisition of MDR-Acinetobacter [12,13]. Since carbapenems, especially meropenem, are frequently administered to treat A. baumannii, carbapenem-resistant A. baumannii (CRAB) strains have become a major therapeutic challenge in recent years [4,14,15]. CRAB is one of the critical priority pathogens on the World Health Organization priority list for antibiotic-resistant bacteria, which require effective drug development [16]. Cephalosporins could be another option for the treatment of A. baumannii, but cephalosporins are not usually considered suitable for the treatment of A. baumannii infections due to widespread high resistance rates [4]. High nosocomial A. baumannii resistance has also been observed for fluoroquinolones (e.g., ciprofloxacin) [17,18,19]. Recently, Butler et al. published a detailed study on the treatment options for MDR A. baumannii infections [20]. Despite the large number of studies on A. baumannii resistance, there are few data on the correlation between hospital antimicrobial use and resistance development, information that is critical for the development of an antimicrobial stewardship program (ASP) [21,22,23].
Numerous methods have been applied to investigate the above correlation; however, time series models and relevant analyzes, such as Autoregressive Integrated Moving Average models (ARIMA), dynamic regression models, are recognized as valuable tools for modeling antibiotic use-related resistance [24]. These methods offer the possibility of considering the influence of the timing of an intervention on this relationship, ensuring that the possible cause of antibiotic use precedes the effect of antibiotic resistance. These mathematical models also analyze the behavior of the dependent variable (antimicrobial resistance) as a function of its prior value, trends, and steep changes in the recent past.
Considering the above, the aim of this study was to use monthly hospital data to investigate the resistance rates of A. baumannii and the use of three broad-spectrum antimicrobial agents over a period of four years. In addition, the correlation of the above data was investigated by using time series analysis (ARIMA) and dynamic regression models.
2. Results
A. baumannii isolates were tested for susceptibility to all available antimicrobial agents and Clinical & Laboratory Standards Institute (CLSI) criteria were applied [25]. The evolution of mean yearly antimicrobial use expressed as DDD/100 patient days from January 2014 to December 2017 is shown in Table 1. It was observed that meropenem had the highest mean consumption of the antibiotics studied and its use increased significantly from 2014 to 2017. In addition, ciprofloxacin and the β-lactam combination piperacillin/tazobactam exhibited high levels of consumption per year, while imipenem use decreased gradually between 2014 and 2016 and eventually was practically annihilated in 2017.
Table 1.
Mean monthly antimicrobial consumption data1 estimated between January 2014 and December 2017.
| Antimicrobial Agent | Year | |||
|---|---|---|---|---|
| 2014 | 2015 | 2016 | 2017 | |
| Imipenem | 1.29 | 1.10 | 0.41 | 0.02 |
| Meropenem | 4.28 | 8.49 | 7.77 | 6.42 |
| Ceftazidime | 0.51 | 0.31 | 0.62 | 0.48 |
| Cefepime | 0.88 | 2.75 | 1.69 | 2.08 |
| Ciprofloxacin | 4.41 | 4.89 | 4.86 | 4.44 |
| Piperacillin/Tazobactam | 3.59 | 3.88 | 4.45 | 3.84 |
| Tigecyclin | 0.91 | 1.72 | 2.08 | 2.69 |
1 Mean monthly use in Defined Daily Doses (DDD) [26] per 100 Patient Days (PD).
According to our data, a large number of A. baumannii strains were found to be multi-drug resistant, considering five classes of antibiotics that may be used against this pathogen (cephalosporins, fluoroquinolones, carbapenems, polymyxins and combinations of β-lactam/β-lactamase inhibitor). In particular, 15% of isolates were found to be resistant to all five classes, 42% were resistant to four classes, and 30% were resistant to three classes. Consequently, only 5.5% of the strains were sensitive to all the aforementioned antimicrobial classes. These findings are in accordance with previously reported data concerning the alarming resistance rates and resistance trends over time (1996–2017) of A. baumannii in Greek hospitals [27,28]. The distribution of isolates in terms of the biological sample and the hospital department is presented in Table 2.
Table 2.
Distribution of A. baumannii isolates gathered during the four-year study.
| Per Specimen | Percent of Isolates | Per Department | Percent of Isolates |
|---|---|---|---|
| Blood | 14.63% | Medical wards | 52.44% |
| Urine | 15.24% | Surgical wards | 25% |
| Broncho-Alveolar Lavage | 21.95% | Intensive Care Units | 16.46% |
| Sputum | 14.63% | Oncology/Hematological wards | 4.88% |
| Trauma | 11.59% | Mixed medical/surgical ward | 1.22% |
| Other | 21.95% |
Time series analysis and transfer function models were applied to all antimicrobial agents. The only antimicrobials that presented a correlation between their use with their A. baumannii resistance pattern were meropenem, cefepime, and ciprofloxacin (Table 3, Table 4 and Table 5). No correlation between prior antimicrobial use and pathogen resistance rates was revealed for tigecycline, piperacillin/tazobactam, imipenem, and ceftazidime (Table A1).
Table 3.
Autoregressive Integrated Moving Average (ARIMA) models for meropenem-resistant A. baumannii (A) and meropenem use (B). Dynamic regression model for the association between meropenem-resistant A. baumannii and hospital meropenem use (C).
| Estimate | Model Parameter | Standard Error | p-Value |
|---|---|---|---|
| A. A. baumannii resistance | |||
| ar1 | −0.517 | 0.118 | 0.000 |
| ar2 | −0.578 | 0.114 | <0.001 |
| AIC | 186.93 | ||
| R2 | 0.531 | ||
| B. Meropenem use (in DDD/100 PD) | |||
| ar1 | −0.831 | 0.122 | <0.001 |
| ar2 | −0.637 | 0.144 | <0.001 |
| ar3 | −0.569 | 0.117 | <0.001 |
| AIC | 242.91 | ||
| R2 | 0.638 | ||
| C. Impact of meropenem use on A. baumannii resistance | |||
| ar1 | −0.564 | 0.126 | <0.001 |
| ar2 | −0.610 | 0.124 | <0.001 |
| mer2 | 0.130 | 0.057 | 0.024 |
| AIC | 166.25 | ||
| R2 | 0.626 | ||
Key: AIC, the estimated Akaike Information Criterion value for the model; ar1, autoregression term with a lag of one month of the ARIMA model; ar2, autoregressive component with lag equal to two months of the ARIMA model; ar3, autoregressive component with lag equal to three months of the ARIMA model; mer2, meropenem use of two months with lag time of two months; R2, the coefficient of determination of the model; DDD, Defined Daily Dose; PD, Patient days.
Table 4.
ARIMA models for cefepime-resistant A. baumannii (A) and cefepime use (B). Dynamic regression model for the association between cefepime-resistant A. baumannii and hospital cefepime use (C).
| Estimate | Model Parameter | Standard Error | p-Value |
|---|---|---|---|
| A. A. baumannii resistance | |||
| ar1 | −0.677 | 0.129 | <0.001 |
| ar2 | −0.710 | 0.122 | <0.001 |
| AIC | 100.56 | ||
| R2 | 0.580 | ||
| B. Cefepime use (in DDD/100 PD) | |||
| ar1 | −0.463 | 0.137 | <0.001 |
| ar2 | −0.466 | 0.133 | <0.001 |
| AIC | 139.03 | ||
| R2 | 0.619 | ||
| C. Impact of cefepime use on A. baumannii resistance | |||
| ar1 | −0.576 | 0.210 | 0.006 |
| ar2 | −0.559 | 0.222 | 0.011 |
| cef1 | 0.865 | 0.395 | 0.028 |
| AIC | 85.53 | ||
| R2 | 0.660 | ||
Key: AIC, the estimated Akaike Information Criterion value for the model; ar1, autoregression term with a lag of one month of the ARIMA model; ar2, autoregressive component with lag equal to two months of the ARIMA model; cef1, cefepime use of one month with lag time of one month; R2, the coefficient of determination of the model, DDD, Defined Daily Dose; PD, Patient days.
Table 5.
ARIMA models for ciprofloxacin-resistant A. baumannii (A) and ciprofloxacin use (B). Dynamic regression model for the association between ciprofloxacin-resistant A. baumannii and hospital ciprofloxacin use (C).
| Estimate | Model Parameter | Standard Error | p-Value |
|---|---|---|---|
| A. A. baumannii resistance | |||
| ma1 | −0.900 | 0.181 | <0.001 |
| AIC | 99.38 | ||
| R2 | 0.486 | ||
| B. Ciprofloxacin use (in DDD/100 PD) | |||
| ar1 | −0.527 | 0.147 | <0.001 |
| ar3 | −0.299 | 0.152 | 0.004 |
| AIC | 77.76 | ||
| R2 | 0.550 | ||
| C. Impact of ciprofloxacin use on A. baumannii resistance | |||
| cip1 | 0.733 | 0.081 | <0.001 |
| AIC | 77.52 | ||
| R2 | 0.617 | ||
Key: AIC, the estimated Akaike Information Criterion value for the model; ar1, autoregression term with a lag of one month of the ARIMA model; ar3, autoregressive component with lag equal to three months of the ARIMA model; ma1, moving average component with lag equal to one month of the ARIMA model; cip1, ciprofloxacin use of one month with lag time of one month; R2, the coefficient of determination of the model; DDD, Defined Daily Dose; PD, Patient days.
2.1. Meropenem
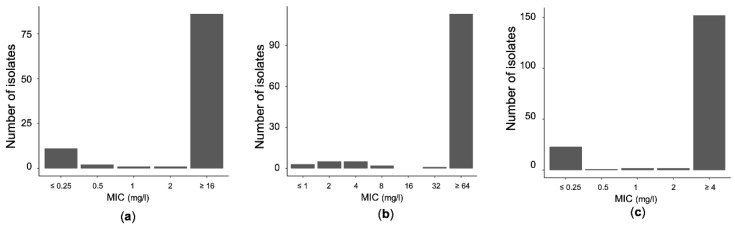
Between January 2014 and December 2017, the observed mean detection rate of meropenem-resistant A. baumannii was 1.89 isolates per month; Figure 1a shows the meropenem Minimum Inhibitory Concentration (MIC) distribution of strains collected during the study period.
Figure 1.
Distribution of meropenem (a), cefepime (b), and ciprofloxacin (c) minimum inhibitory concentrations (MICs) from A. baumannii blood isolates during the four-year period of the study.
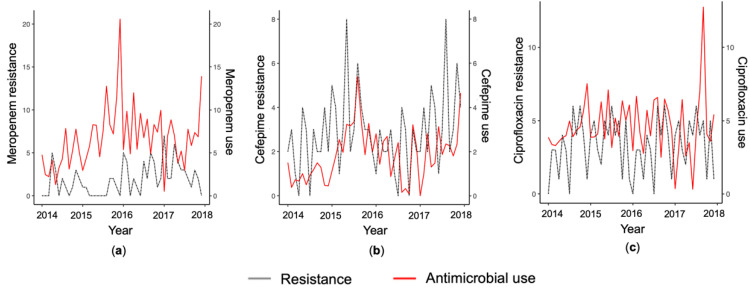
The monthly mean use of meropenem was 6.74 Defined Daily Doses (DDDs) per 100 Patient Days (PDs). Smoothed data of meropenem use and meropenem-resistant A. baumannii are depicted in Figure 2a.
Figure 2.
Smoothed monthly resistance detection rate of A. baumannii and hospital use of meropenem (a), cefepime (b), and ciprofloxacin (c). Usage is expressed in defined daily doses per 100 patient days.
The ARIMA models and their corresponding parameters are shown in Table 3A,B for the meropenem-resistant A. baumannii and meropenem consumption series, respectively. Prior to these models, time series became stationary through differencing. Two significant autoregressive terms of orders (lags) of 1 and 2 months were identified for meropenem resistance. The coefficient of determination (R2) was 0.531 and the Akaike Information Criterion (AIC) was 186.73. In addition, three significant autoregressive terms of orders of 1, 2, and 3 months were identified for meropenem use; the R2 was 0.638 and the AIC was 242.91. Autocorrelation function (ACF) and partial autocorrelation function (PACF) plots of both model residuals confirmed that the series residuals corresponded to white noise.
To investigate a possible relationship between meropenem-resistant A. baumannii detection rate and meropenem use, we built a dynamic regression model via the linear transfer function (Table 3C). Examination of the cross-correlation function of the residuals of the two previous ARIMA models showed only one significant correlation with lag of 2 months. Therefore, two-month lags in the meropenem use time series were introduced in the transfer function model. The R2 of the transfer function model was 0.626 and the AIC was 166.25. The transfer function model could also be presented by the following equation:
| R(t) = −0.564 R(t − 1) − 0.61 R(t − 2) + 0.13 U(t − 2) + e(t) | (1) |
where R is the detection rate of meropenem resistance observed at t months, U is the hospital meropenem use, and e(t) represents the residual error.
2.2. Cefepime
The mean monthly detection rate of cefepime-resistant A. baumannii was 2.37 isolates during the study period. The observed mean monthly cefepime consumption was 1.85 DDD/100 patient days. The distribution of MICs of cefepime is shown in Figure 1b. Smoothed data on cefepime use and cefepime-resistant A. baumannii are shown in Figure 2b.
Table 4 shows ARIMA and transfer function models for estimating the detection rate of cefepime resistance among A. baumannii isolates. The series were stationary in variance and mean after log transformation and simple differencing. Specifically, Table 4A shows an ARIMA model for cefepime resistance rate among A. baumannii that identified two significant autoregressive terms of order of 1 and 2 months. The AIC value was 100.56 and the R2 was 0.58. The ARIMA model of cefepime use included two significant autoregressive terms of order of 1 and 2 months (Table 4B). In this case, the AIC was 139.03 and the R2 was 0.619. ACF and PACF plots verified that the series residuals for the above models corresponded to white noise.
The influence of cefepime use in cefepime-resistant A. baumannii was analyzed using dynamic regression models, i.e., using the transfer function (Table 4C). A lag of one month was identified from the cross-correlation function between the residuals of the two previous ARIMA models. Accordingly, we introduced a one-month lag of cefepime use in the transfer function model and two significant autoregressive terms of order of 1 and 2 months of the residuals. The R2 was 0.66 and the AIC was 85.53.
2.3. Ciprofloxacin
The monthly mean of ciprofloxacin-resistant A. baumannii isolates was 3.16 isolates and the mean of ciprofloxacin use was 4.65 DDDs/100 patient days during the study period. Figure 1c presents the distribution of ciprofloxacin MICs and Figure 2c shows smoothed monthly series of ciprofloxacin use and ciprofloxacin-resistant A. baumannii.
ARIMA and transfer function models were built to estimate the detection rate of ciprofloxacin resistance among A. baumannii isolates (Table 5).
The series were first differentiated to achieve stationarity. The ARIMA model of ciprofloxacin resistance rate of A. baumannii showed a significant first order moving average term (Table 5A). The AIC value was 99.38 and the R2 was 0.486. In addition, the identified model of the ciprofloxacin use series contained two significant autoregressive terms of order of 1 and 3 months (Table 5B). In this case, the AIC was 77.76 and the R2 was 0.55. Combining the above models, we developed a dynamic regression model of the ciprofloxacin-resistant A. baumannii detection rate and ciprofloxacin use by introducing a lag of 1 month in the ciprofloxacin use series (Table 5C). This lag was found by cross-correlating the residuals of the ARIMA models for ciprofloxacin use and resistance. The R2 was 0.617 and the AIC was 77.52.
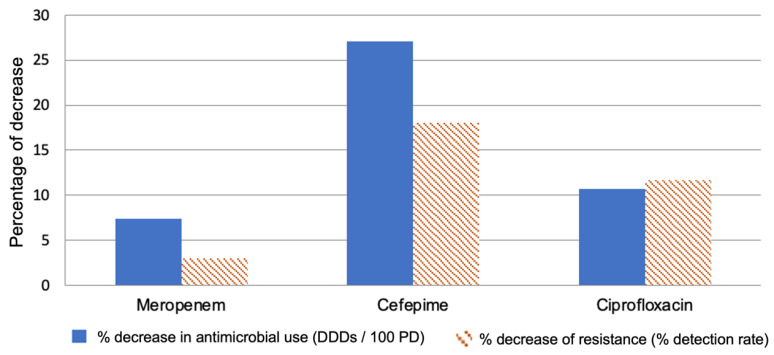
Furthermore, in Figure 3 we can observe the possible impact of a 0.5 DDD/100 patient days on each of the antimicrobial agents. In the case of meropenem, the reduction is approximately 7.5% and could probably be achieved by 3% reduction in the detection rate. On the contrary, for ciprofloxacin the same reduction would mean a 10.75% decrease in use that could result in a significant resistance decrease (11.7%).
Figure 3.
The predicted effect of a 0.5 Defined Daily Doses (DDD) per 100 patient days (PD) reduction for each antibiotic to the corresponding resistance.
3. Discussion
In recent years, A. baumannii has developed resistance to virtually all known antibiotics [4,7]. Combination therapies of high-dose, long-term infusions of sulbactam, meropenem, ceftazidime/avibactam, and meropenem/varobactam with polymyxins, minocycline, aminoglycosides, and fosfomycin are being considered for the treatment of MDR A. baumannii infections [20,29,30]. Taconelli et al. proposed a priority list for research and development of new antibiotics for antibiotic-resistant bacteria and concluded that CRAB is a critical priority pathogen [16]. As a result, efforts must be made to maintain the susceptibility of this pathogen to currently available antimicrobials, especially ASPs. Although data on the correlation between antimicrobial use and resistance rates are required for effective work by ASP, we still have limited relevant data on A. baumannii.
In this study, ARIMA and transfer function models were identified to estimate the correlations between the detection rate of resistant A. baumannii isolates, previous resistance rates, and previous antimicrobial use. Our study provided significant results for three broad-spectrum antimicrobials, i.e., meropenem, ciprofloxacin, and cefepime. We identified an ARIMA model for the detection rate of meropenem-resistant A. baumannii; the rate was negatively related to the detection rate of the same resistance observed one and two months ago (Table 3A). The R2 was 0.531, meaning that 53% of the variance in this resistance series is anticipated by the model. Similarly, the ARIMA model for meropenem use showed a negative relationship with the same use series before one, two, and three months (Table 3B), where the R2 of the model was 0.638. Then, the influence of meropenem use in meropenem-resistant A. baumannii was analyzed using a dynamic regression model; according to it, the current rate of meropenem resistance was negatively related to resistance one and two months before and positively related to meropenem use two months before (Table 3C). An increase of 1 DDD/100 patient days for meropenem two months prior results in an increase of 0.13 in the meropenem resistance rate after accounting for meropenem use. In addition, ARIMA and transfer function models were estimated for the detection rate of cefepime resistance among A. baumannii isolates. These rates were negatively associated with the same rate one and two months ago (Table 4A), and the R2 of the model was 0.58. Cefepime use was also associated with the same use one and two months ago (Table 4B), and the R2 of the model was 0.619. The dynamic regression model implied that the current rate of cefepime resistance was negatively associated with resistance one and two months ago and positively associated with cefepime use one month ago (Table 4C). An increase of 1 DDD/100 patient days for cefepime two months prior results in an increase of 0.856 in the cefepime detection rate of resistant isolates after accounting for cefepime use. Finally, an ARIMA model for ciprofloxacin-resistant A. baumannii showed a negative association with the same series one month earlier (Table 5A). Ciprofloxacin use was negatively related to the same use one and three months ago and the coefficient of determination, R2, was 0.55 (Table 5B). The dynamic regression model showed that an increase in ciprofloxacin use by 1 DDD/100 patient days corresponded with an increase in the detection rate of ciprofloxacin-resistant isolates by 0.73 (Table 5C). These results are consistent with similar literature reports also indicating time delays between antimicrobial use and increased resistance [31,32,33]. For four other broad-spectrum antibiotics studied, i.e., tigecycline, piperacillin/tazobactam, imipenem, and ceftazidime, no correlation was found between antimicrobial use and corresponding resistance (Table A1).
It is worth noting that in all three models, the introduction of prior antimicrobial use significantly improved the models; the R2 increased to 0.6, meaning that more than 60% of the variance in the detection rate of resistant strains can be predicted by the described models. Figure 3 shows what a reduction of 0.5 DDD/100 PD would give for each of the antimicrobial agents studied; in relation to meropenem, this reduction, which is about 7.5%, could probably be achieved by an ASP but would give poor results, i.e., a 3% reduction in the detection rate. In contrast, the same reduction for ciprofloxacin would correspond to a 10.75% reduction in use, a still achievable target that could lead to a remarkable 11.7% reduction in resistance. This observation is an indication that ASPs should be planned based on local or regional epidemiological data, with pragmatic targets adapted to the results of the respective prediction models.
One of the limitations of this study is that the findings, on the association between antibiotic use and resistance, were examined on aggregate hospital data rather than at the individual patient level. In addition, molecular data were not available for the study period because genotypic testing is not routine in this hospital. Instead, data from phenotypic testing were used to inform antimicrobial treatment decisions. The benefits of molecular test information should be acknowledged, as this type of data would help in distinguishing between the different sources of resistance, e.g., horizontal genetic transfer or acquired resistance by mutation. The only limiting factor in the use of genotype data is its cost [34]. Despite the additional useful information that molecular data can provide, the phenotype data used in this study are adequate to fulfill the purpose of this study, which is to quantify the prior antimicrobial use and prior resistance rate on the observed resistance rate of A. baumannii [35]. It should also be recognized that mathematical models, despite their usefulness, have certain limitations, as they are based only on the mathematical properties of the series and not on the dynamics of infectious disease transmission [36]. Therefore, although the current results offered a reasonable level of reliability, future research may incorporate genotype data to gain better insight into the process of resistance of hospital-acquired infections.
Comparing our results with the literature, the complexity of the resistance phenomenon becomes clear; for example, with regard to meropenem, the results of this analysis are in agreement with previous studies, but no significant correlation of resistance was found with ceftazidime, while a strong positive correlation was found with the use of ciprofloxacin [23,37]. These observations contrast with the results of a recent study conducted in Serbia [21]. In addition, no effect of the rather limited imipenem use on resistance was found, which contrasts with the results of a Chinese study [38]. Such differences may be attributed to methodological reasons or may be due to the different epidemiological conditions in the hospital setting. Finally, another limitation of this study is that possible cross-correlations were not examined, such as the effect of ciprofloxacin use on meropenem resistance rates.
4. Materials and Methods
4.1. Clinical Setting
A retrospective study was performed from January 2014 to December 2017 in the General Military Hospital of Athens, a tertiary care hospital with medical and surgical wards as well as two Intensive Care Units (ICUs). This study research was approved by the hospital scientific committee. Informed consent was waived by the ethics review board. As to the hospital restriction policy, all prescriptions for broad spectrum antibiotics need composed endorsement by an infectious diseases specialist before administration; no adjustment in the limitation strategy or other significant change occurred during the investigation time frame.
4.2. Antibiotic Consumption
Monthly data of antimicrobial consumption were obtained for the study period from the hospital pharmacy database and were converted into defined daily doses (DDD). Antibiotic consumption was finally expressed as the number of defined daily doses/100 patient days according to the 2020 version of the ATC/DDD classification (World Health Organization Collaborating Centre for Drug Statistics Methodology, ATC/DDD index 2020) [26].
4.3. Microbiological Data
The results of susceptibility tests were obtained from the Clinical Microbiology Department of the hospital for clinical A. baumannii isolates for the study period. Interpretation of the results was performed according the CLSI criteria [25]. All clinical isolates of A. baumannii from every biological sample from all wards and ICUs were included in the analysis. The isolates with intermediate susceptibility were grouped with the resistant ones, forming the non-susceptible group. Detection rate of resistance was examined per month and was expressed as the number of multidrug-resistant A. baumannii isolates. Duplicate isolates were defined on the basis of the patient identity and the antibiotic phenotype.
4.4. Data Analysis
Prior to statistical analysis, any patients’ recognizable information was changed into anonymous information by the principal investigator of the study. A time series analysis of the monthly detection rate of A. baumannii resistance isolates and the monthly consumption of the respective broad-spectrum antimicrobials was performed. ARIMA models were used to analyze the temporal behavior of each variable in relation to its previous values, its trends, and any sudden changes. To identify an ARIMA model from the observed time series, it was first investigated whether the time series was stationary using the Augmented Dickey-Fuller test for Unit Roots. Then, the required assumptions of the Box and Jenkins methods were checked [39]. From the autocorrelation function and partial autocorrelation function, the appropriate order of the autoregressive and moving average terms of the model was selected. The parameters of the identified model were estimated using the maximum likelihood function or the unconditional least squares function. Goodness-of-fit criteria were estimated, such as the Akaike information criterion and the coefficient of determination, which corresponds to the percentage of variance in the observed time series explained by the model. Diagnostic checks, statistical significance of the parameters, and the ACF and PACF residuals of the model corresponded to white noise and were estimated to select the appropriate model.
Once the basic ARIMA models were established, dynamic time series modeling techniques were used to assess the relationships between antimicrobial use series (DDD/100 patient days) and resistance series (detection of meropenem, cefepime, and ciprofloxacin Acinetobacter resistance isolates per 1000 patient days). In particular, the linear transfer function method proposed by Haugh (1976) was used to estimate these relationships [25]. The cross-correlation function between the residuals of these ARIMA models was calculated to identify the adequate lags in the antimicrobial use time series to be further introduced into the transfer function model. Cross-correlation of the resistance and use series was performed with lags of up to one year, and backward selection was applied to eliminate non-significant correlations. If no correlations were found, no lag order was selected. In addition, a diagnostic check, goodness-of-fit calculation, AIC, and R2 were implemented. The above techniques were analyzed using R software version 3.6.1 (the R project for statistical computing; http://www.r-project.org accessed on 15 April 2021).
5. Conclusions
In the present study, the association between the use of broad-spectrum antimicrobials (meropenem, cefepime, and ciprofloxacin) and resistant A. baumannii isolates in a tertiary hospital was investigated using time series analysis. Statistically significant associations were found between the use of these antimicrobials (meropenem, cefepime, ciprofloxacin) and corresponding resistance in A. baumannii. For each antimicrobial agent, a different pattern of correlation with resistance was revealed in terms of time lag and magnitude of effect. These results have important implications for strategies to contain resistance and highlight the need for antimicrobial stewardship programs to adjust their targets in accordance with locally developed predictive models.
Appendix A
Table A1.
The association in univariate time series analysis between antimicrobial use and the corresponding antibiotic resistance of A. baumannii.
| Antimicrobial Agent/Class | Order 1 | p-Value 2 | AIC 3 | R2 4 |
|---|---|---|---|---|
| Imipenem | 0 | 0.058 | 155.46 | 0.286 |
| Ceftazidime | 0 | 0.305 | 174.69 | 0.042 |
| Piperacillin/Tazobactam | 0 | 0.386 | 162.17 | 0.22 |
| Tigecyclin | 0 | 0.201 | 180.3 | 0.149 |
1 Delay before effect is observed (months).2 p-value for the association between antimicrobial use and A. baumannii resistance of the ARIMA model.3 Akaike information criterion.4 Coefficient of determination of the model.
Author Contributions
Conceptualization, C.A. and V.K.; methodology, R.K. and V.K.; software, R.K..; validation, C.A., G.I., and V.K.; formal analysis, R.K.; investigation, R.K.; resources, C.A., K.L., and O.I.; data curation, R.K. and G.I.; writing—original draft preparation, R.K.; writing—review & editing, R.K., C.A., G.I., and V.K.; visualization, R.K. and V.K.; supervision, V.K.; project administration, V.K.; funding acquisition, R.K. All authors have read and agreed to the published version of the manuscript.
Funding
R.K. was supported by the Hellenic Foundation for Research and Innovation (HFRI) under the HFRI PhD Fellowship grant (Fellowship Number: 261).
Institutional Review Board Statement
Ethical review and approval were waived for this study, due to the retrospective character of the study and the fact that no personal information was required for this study.
Informed Consent Statement
Patient consent was waived due to the retrospective character of the study and the fact that no personal information was required for this study.
Data Availability Statement
Data is contained within the article.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fournier P.E., Richet H. The epidemiology and control of A. baumannii in health care facilities. Clin. Infect. Dis. 2006;42:692–699. doi: 10.1086/500202. [DOI] [PubMed] [Google Scholar]
- 2.Peleg A.Y., Seifert H., Paterson D.L. A. baumannii: Emergence of a successful pathogen. Clin. Microbiol. Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergogne-Berezin E., Towner K.J. Acinetobacter spp. as nosocomial pathogens: Microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 1996;9:148–165. doi: 10.1128/CMR.9.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans B., Hamouda A., Amyes S. The rise of carbapenem-resistant A. baumannii. Curr. Pharm. Des. 2013;19:223–238. doi: 10.2174/138161213804070285. [DOI] [PubMed] [Google Scholar]
- 5.Dijkshoorn L., Nemec A., Seifert H. An increasing threat in hospitals: Multidrug-resistant A. baumannii. Nat. Rev. Microbiol. 2007;5:939–951. doi: 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- 6.Vila J., Pachon J. Therapeutic options for A. baumannii infections. Expert Opin. Pharmacother. 2008;9:587–599. doi: 10.1517/14656566.9.4.587. [DOI] [PubMed] [Google Scholar]
- 7.Baron S., Hadjadj L., Rolain J.M., Olaitan A.O. Molecular mechanisms of polymyxin resistance: Knowns and unknowns. Int. J. Antimicrob. Agents. 2016;48:583–591. doi: 10.1016/j.ijantimicag.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 8.Paterson D.L. The epidemiological profile of infections with multidrug-resistant Pseudomonas aeruginosa and Acinetobacter species. Clin. Inf. Dis. 2006;43:S43–S48. doi: 10.1086/504476. [DOI] [PubMed] [Google Scholar]
- 9.Hsueh P.R., Teng L.J., Chen C.Y., Chen W.H., Ho S.W., Luh K.T. Pandrug-resistant A. baumannii causing nosocomial infections in a university hospital, Taiwan. Emerg. Infect. Dis. 2002;8:827–832. doi: 10.3201/eid0805.020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magiorakos A.P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C.G., Paterson D.L. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 11.Kanafani Z., Kanj S. Acinetobacter Infection: Treatment and Prevention. UpToDate. [(accessed on 28 February 2021)];2015 Available online: https://www.uptodate.com/contents/acinetobacter-infection-treatment-and-prevention.
- 12.Ye J.J., Huang C.T., Shie S.S., Huang P.Y., Su L.H., Chiu C.H., Chiang P.C. Multidrug resistant A. baumannii: Risk factors for appearance of imipenem resistant strains on patients formerly with susceptible strains. PLoS ONE. 2010;5:e9947. doi: 10.1371/journal.pone.0009947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falagas M.E., Kopterides P. Risk factors for the isolation of multi-drug resistant A. baumannii and Pseudomonas aeruginosa: A systematic review of the literature. J. Hosp. Infect. 2006;64:7–15. doi: 10.1016/j.jhin.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 14.Kim Y.J., Kim S.I., Hong K.W., Kim Y.R., Park Y.J., Kang M.W. Risk factors for mortality in patients with carbapenem-resistant A. baumannii bacteremia: Impact of appropriate antimicrobial therapy. J. Korean Med. Sci. 2012;27:471–475. doi: 10.3346/jkms.2012.27.5.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meric M., Kasap M., Gacar G., Budak F., Dundar D., Kolayli F., Vahaboglu H. Emergence and spread of carbapenem-resistant A. baumannii in a tertiary care hospital in Turkey. FEMS Microbiol. Lett. 2008;282:214–218. doi: 10.1111/j.1574-6968.2008.01129.x. [DOI] [PubMed] [Google Scholar]
- 16.Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D.L., Ouellette M. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018;18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 17.Coyne S., Rosenfeld N., Lambert T., Courvalin P., Perichon B. Overexpression of Resistance-Nodulation-Cell Division Pump AdeFGH Confers Multidrug Resistance in A. baumannii. Antimicrob. Agents Chemother. 2010;54:4389–4393. doi: 10.1128/AAC.00155-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacoby G.A., Strahilevitz J., Hooper D.C. Plasmid-mediated quinolone resistance. Microbiol. Spectrum. 2014;2:10.1128. doi: 10.1128/microbiolspec.PLAS-0006-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamed S.M., Elkhatib W.F., El-Mahallawy H.A., Helmy M.M., Ashour M.S., Aboshanab K.M. Multiple mechanisms contributing to ciprofloxacin resistance among Gram negative bacteria causing infections to cancer patients. Sci. Rep. 2018;8:1–10. doi: 10.1038/s41598-018-30756-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Butler D.A., Biagi M., Tan X., Qasmieh S., Bulman Z.P., Wenzler E. Multidrug Resistant A. baumannii: Resistance by Any Other Name Would Still be Hard to Treat. Curr. Infect. Dis. Rep. 2019;21:46. doi: 10.1007/s11908-019-0706-5. [DOI] [PubMed] [Google Scholar]
- 21.Pavlovic R.R., Jankovic S.M. Inverse correlation of Acinetobacter spp. resistance rate and ciprofloxacin utilization. J. Antib. 2014;67:273–275. doi: 10.1038/ja.2013.123. [DOI] [PubMed] [Google Scholar]
- 22.Yang P., Chen Y., Jiang S., Shen P., Lu X., Xiao Y. Association between antibiotic consumption and the rate of carbapenem-resistant Gram-negative bacteria from China based on 153 tertiary hospitals data in 2014. Antimicrob. Resist. Infect. Control. 2018;7:137. doi: 10.1186/s13756-018-0430-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoon Y.K., Yang K.S., Lee S.E., Kim H.J., Sohn J.W., Kim M.J. Effects of Group 1 versus Group 2 carbapenems on the susceptibility of Acinetobacter baumannii to carbapenems: A before and after intervention study of carbapenem-use stewardship. PLoS ONE. 2014;9:e99101. doi: 10.1371/journal.pone.0099101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.López-Lozano J.M., Lawes T., Nebot C., Beyaert A., Bertrand X., Hocquet D., Aldeyab M., Scott M., Conlon-Bingham G., Farren D., et al. THRESHOLDS study group. A nonlinear time-series analysis approach to identify thresholds in associations between population antibiotic use and rates of resistance. Nat. Microbiol. 2019;4:1160–1172. doi: 10.1038/s41564-019-0410-0. [DOI] [PubMed] [Google Scholar]
- 25.Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing: 30th Informational Supplement. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2020. [(accessed on 6 April 2021)]. CLSI Document M100. Available online: https://clsi.org/media/3481/m100ed30_sample.pdf. [Google Scholar]
- 26.WHO Collaborating Centre for Drug Statistics Methodology, ATC Classification Index with DDDs, 2021. Norwegian Institute of Public Health; Oslo, Norway: 2020. [Google Scholar]
- 27.Falagas M.E., Mourtzoukou E.G., Polemis M., The Greek System for Surveillance of Antimicrobial Resistance. Vatopoulos A.C. Trends in antimicrobial resistance of Acinetobacter baumannii clinical isolates from hospitalised patients in Greece and treatment implications. Clin. Microbiol. Infect. 2007;13:816–819. doi: 10.1111/j.1469-0691.2007.01761.x. [DOI] [PubMed] [Google Scholar]
- 28.Polemis M., Tryfinopoulou K., Giakkoupi P., WHONET-Greece Study Group. Vatopoulos A. Eight-year trends in the relative isolation frequency and antimicrobial susceptibility among bloodstream isolates from Greek hospitals: Data from the Greek Electronic System for the Surveillance of Antimicrobial Resistance—WHONET-Greece, 2010 to 2017. Eurosurveillance. 2020;25:1900516. doi: 10.2807/1560-7917.ES.2020.25.34.1900516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karaiskos I., Lagou S., Pontikis K., Rapti V., Poulakou G. The “Old” and the “New” Antibiotics for MDR Gram-Negative Pathogens: For Whom, When, and How. Front Public Health. 2019;7:151. doi: 10.3389/fpubh.2019.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kengkla K., Kongpakwattana K., Saokaew S., Apisarnthanarak A., Chaiyakunapruk N. Comparative efficacy and safety of treatment options for MDR and XDR Acinetobacter baumannii infections: A systematic review and network meta-analysis. Antimicrob. Chemother. 2018;73:22–32. doi: 10.1093/jac/dkx368. [DOI] [PubMed] [Google Scholar]
- 31.Tansarli G.S., Papaparaskevas J., Balaska M., Samarkos M., Pantazatou A., Markogiannakis A., Daikos G.L. Colistin resistance in carbapenemase-producing Klebsiella pneumoniae bloodstream isolates: Evolution over 15 years and temporal association with colistin use by time series analysis. Int. J. Antimicrob. Agents. 2018;52:397–403. doi: 10.1016/j.ijantimicag.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 32.Monnet D.L., López-Lozano J.M., Campillos P., Burgos A., Yagüe A., Gonzalo N. Making sense of antimicrobial use and resistance surveillance data: Application of ARIMA and transfer function models. Clin. Microbiol. Infect. 2001;7:29–36. doi: 10.1046/j.1469-0691.2001.00071.x. [DOI] [PubMed] [Google Scholar]
- 33.López-Lozano J.M., Monnet D.L., Yagüe A., Burgos A., Gonzalo N., Campillos P., Saez M. Modelling and forecasting antimicrobial resistance and its dynamic relationship to antimicrobial use: A time series analysis. Int. J. Antimicrob. Agents. 2000;14:21–31. doi: 10.1016/S0924-8579(99)00135-1. [DOI] [PubMed] [Google Scholar]
- 34.World Health Organization Global Antimicrobial Resistance Surveillance System (GLASS): Molecular Methods for Antimicrobial Resistance (AMR) Diagnostics to Enhance the Global Antimicrobial Resistance Surveillance System. [(accessed on 6 April 2021)];2019 WHO/WSI/AMR/2019.1. Available online: https://apps.who.int/iris/handle/10665/310993.
- 35.Bard J.D., Lee F. Why Can’t We Just Use PCR? The Role of Genotypic versus Phenotypic Testing for Antimicrobial Resistance Testing. Clin. Microbiol. Newsl. 2018;40:87–95. doi: 10.1016/j.clinmicnews.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allard R. Use of time-series analysis in infectious disease surveillance. Bull. World Health Organ. 1998;76:327–333. [PMC free article] [PubMed] [Google Scholar]
- 37.Mozes J., Ebrahimi F., Goracz O., Miszti C., Kardos G. Effect of carbapenem consumption patterns on the molecular epidemiology and carbapenem resistance of Acinetobacter baumannii. J. Med. Microbiol. 2014;63:1654–1662. doi: 10.1099/jmm.0.082818-0. [DOI] [PubMed] [Google Scholar]
- 38.Guo W., Sun F., Liu F., Cao L., Yang J., Chen Y. Antimicrobial resistance surveillance and prediction of Gram-negative bacteria based on antimicrobial consumption in a hospital setting: A 15-year retrospective study. Medicine. 2019;98:e17157. doi: 10.1097/MD.0000000000017157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Box G., Jenkins G.M. Time Series Analysis: Forecasting and Control. 4th ed. John Wiley & Sons; Hoboken, NJ, USA: 2008. pp. 21–192. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data is contained within the article.