Abstract
Background:
Group B Streptococcus (GBS) is a leading cause of neonatal sepsis and meningitis and an important cause of invasive infections in pregnant and nonpregnant adults. Vaccines targeting capsule polysaccharides and common proteins are under development.
Methods:
Using whole genome sequencing (WGS), a validated bioinformatics pipeline, and targeted antimicrobial susceptibility testing, we characterized 6,340 invasive GBS isolates recovered during 2015–2017 through population-based Active Bacterial Core surveillance (ABCs) in eight states.
Results:
Six serotypes accounted for 98.4% of isolates (21.8% Ia, 17.6% V, 17.1% II, 15.6% III, 14.5% Ib, 11.8% IV). Most (> 98%) of these six serotypes were in seven clonal complexes (CCs) comprised of multilocus sequence types (MLSTs) identical or closely related to STs 1, 12, 17, 19, 22, 23, and 459. Fifty-four isolates (0.87%) had point mutations within pbp2x (31 different alleles) associated with reduced susceptibility to one or more β-lactam antibiotics. Genes conferring resistance to macrolides and/or lincosamides were found in 56% of isolates; 85.2% of isolates had tetracycline resistance genes. Two isolates carrying vanG were vancomycin-nonsusceptible (MIC 2μg/ml). Nearly all isolates possessed capsule genes, 1–2 of the three main pilus gene clusters (or islands), and one of four homologous Alpha/Rib family determinants. Presence of hvgA virulence gene was primarily restricted to serotype III/CC17 isolates (465 isolates), but 8 exceptions (7 IV/CC23 and 1 IV/CC17) were observed.
Conclusions:
This first comprehensive, population-based quantitation of strain features in the United States suggests current vaccine candidates should have good coverage. Beta-lactams remain appropriate for first line treatment and prophylaxis, but emergence of nonsusceptibility warrants ongoing monitoring.
Keywords: Capsular serotypes, surface protein distributions, resistance determinants, beta-lactam nonsusceptibility
summary
We describe demographic and genomic-based features of invasive GBS recovered from population-based surveillance capturing 9% of the US population in 2015–2017. Annotated documentation of capsule, surface proteins and antimicrobial resistance determinants inform vaccine development, prophylaxis, and treatment policies.
Background
Lancefield Group B Streptococcus (GBS), also referred to as Streptococcus agalactiae, is a major bacterial cause of sepsis and meningitis in neonates and infants [1, 2]. GBS has also been associated with high rates of invasive infections in adults, especially in patients >65 years and in people with underlying medical conditions [3]. Intrapartum antibiotic prophylaxis (IAP) to women at risk for GBS infection has dramatically lowered the burden of early-onset disease (<7 days of age; EOD) [4], but not of late-onset disease (7–89 days; LOD) [5]. The incidence rates in adults have been increasing [3].
Beta-lactam antibiotics are recommended as first-line therapy against GBS infections and for IAP because GBS are considered susceptible to these agents. However, GBS isolates with reduced or non-susceptibility to β-lactams have been detected from invasive and non-invasive infections [6–9]. Associations with mutations in pbp2x have been well documented [6–9] making this an excellent target for ongoing surveillance. In the case of penicillin allergy, GBS IAP guidelines suggest the use of cefazolin, or for women at high risk of anaphylaxis to a beta-lactam antibiotic, clindamycin (if the GBS is susceptible) or vancomycin [10]. Increased levels of erythromycin and clindamycin resistance among GBS have been reported in the USA and elsewhere [3, 5, 11].
Concerns about emerging antimicrobial resistance and the potential for microbiome dysbiosis resulting from IAP [12], as well as a need for prevention strategies appropriate for low income settings in Africa and Asia [1], and the potential for primary prevention among high risk adults [3], have motivated active efforts to develop vaccines against GBS [1].
Ten structurally and antigenically unique capsular polysaccharide types (Ia, Ib, II - IX) have been described (13). While GBS of all serotypes are capable of causing invasive infections, six serotypes (Ia, Ib, II, III, IV, and V) account for the majority of disease in neonates and adults (1, 3, 14–16). A 6 valent (Ia, Ib, II, III, IV, and V) conjugate vaccine is currently under development [17, 18].
Protein vaccines are also under development, targeting surface adhesins and virulence factors [19]. A fusion protein of the N-terminal domains of the alpha C protein and Rib (GBS-NN) was a promising vaccine in a mouse model [20] and has been tested in phase I clinical trials in pregnant women. Current phase I trials investigating the safety and immunogenicity of this vaccine have been performed [17]. A potential advantage of the GBS-NN vaccine is the expression of highly related Alpha family proteins in a high proportion of strains [21]. Similarly, a vaccine targeting three different pilus structures could potentially be broadly protective [22].
Here, we use WGS to describe a large population-based sampling of invasive GBS (iGBS) collected in the United State from 2015–2017. We quantitate distributions of potential vaccine targets (capsule polysaccharides, surface proteins) and their associations with major antibiotic resistance markers within well-recognized clonal lineages.
MATERIALS AND METHODS
Invasive GBS Isolates
ABCs conducts active, population- and laboratory-based surveillance for iGBS disease in select counties of 10 states across the United States. The areas under surveillance, methods and key surveillance data through 2017 are described at https://www.cdc.gov/abcs/methodology/surv-pop.html [23]. All available GBS isolates (n=6,340 from 7,114 cases; 89.1%) recovered from eight of the 10 sites during 2015–2017 were characterized. Isolates were from individuals of all ages for all sites except for isolates from NY which were limited to invasive infections in infants <90 days old.
Whole genome sequencing and bioinformatics pipeline
Chromosomal DNA extraction, genomic libraries and WGS was performed as previously described [9]. Strain features with isolate identifiers and genome accession numbers are listed in sTable 1. Serotypes, multilocus sequence types (MLST), antibiotic resistance determinants, and predicted minimum inhibitory concentrations (MICs) were determined using a previously described GBS bioinformatics pipeline [9]. The ARG-ANNOT and ResFinder databases were incorporated to detect additional resistance determinants [24, 25]. Conventional broth microdilution results were available for all year 2015 isolates [9] and years 2016–2017 isolates recovered from Minnesota. Additional strain features that were extracted from the genomic data included presence/absence of surface protein genes encoding the hypervirulent GBS adhesin (hvga) [26], serine-rich repeat (srr) proteins [27], alpha protein family (Alpha, Rib, Alp2/3, Alp1) [19] and pilus islands (PI1, PI2A and PI2B) [28] (https://github.com/BenJamesMetcalf) (sTable 2). For each of these sequence queries, ≥ 95% identity was required for a positive result.
MLST data
The STs were grouped via the eBURST program [29] into clonal complexes (CCs) whose members shared at least five of the seven MLST loci; otherwise, an ST was considered an outlier. The relationship between STs and different parameters of GBS isolates were illustrated by the minimum spanning tree (PHYLOVIZ software version 2.0; PHYLOViZ team, Lisbon, Portugal).
RESULTS
Isolates.
Among the 6,340 iGBS strains identified over the 3-year period, 5,778 (91.1%) were from adult patients (≥18 years), with 56 isolates from individuals 90 days – 17 years (sFigure 1) and 47 from pregnant women. There were 506 isolates from infants < 3 months of age, with 232 (3.7%) and 274 (4.3%) from EOD and LOD, respectively (Fig. 1A). Most (86.8%) iGBS isolates were recovered from blood; 72 (1.1%) were from CSF and remaining 12.1% from other body fluids (not shown). The largest number of isolates over the 3-year period were from Maryland (1,692; 26.7%) and Minnesota (1,508, 23.8%) (sFigure 2).
Figure 1.
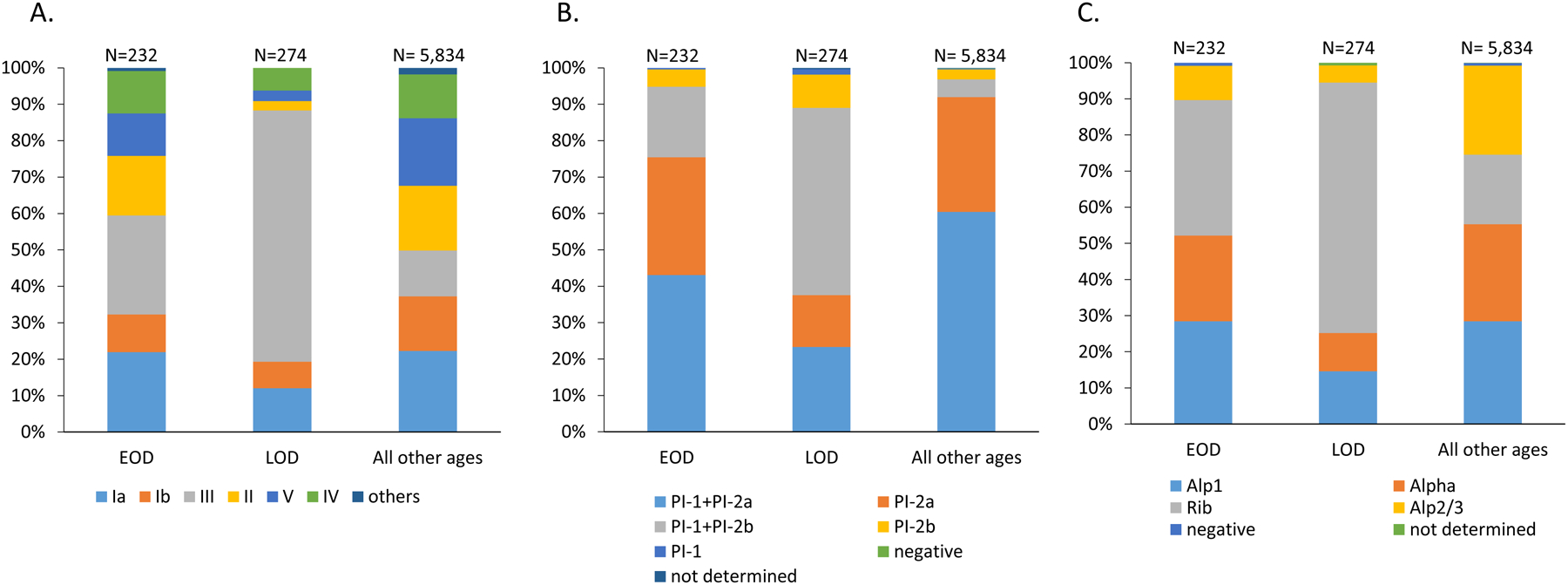
A. Distribution of invasive GBS serotypes among isolates recovered from young infants (EOD and LOD) and from all other ages. B. Distribution of invasive GBS pilus backbone protein types among isolates recovered from young infants (EOD and LOD) and from all other ages. C. Distribution of invasive GBS alpha family protein types among isolates recovered from young infants (EOD and LOD) and from all other ages.
Vaccine candidate distributions.
Serotypes were predicted for all 10 capsular types with few non-typeable isolates (13/6,340; 0.2%). Overall predicted serotype distribution was similar when comparing serotypes across each of the eight states (sFigure 2). Serotypes Ia (1,384; 21.8%), V (1,116; 17.6%), II (1,082; 17.1%), III (987; 15.6%), Ib (919; 14.5%) and IV (748; 11.8%) accounted for 98.4% of the isolates (Figure 1A, sTable 3). Serotypes VI, VII, VIII and IX were rare (<2% of isolates). The predominant serotype for both EOD (63/232, 27.2%) and LOD (189/274, 69.0%) isolates was serotype III; serotype III accounted for 12.6% of the 5,834 remaining isolates (Figure 1A). Serotypes Ia, Ib, and III represented 138/232 (59.5%) of EOD isolates, 242/274 (88.3%) of LOD isolates, and 46% of the remaining isolates (Figure 1A). Five serotypes (Ia, Ib, III, II, V) accounted for 203/232 (87.5%) of EOD isolates, 257/274 (93.8%) of LOD isolates, and 5,028/5,834 (86.2%) of the remaining isolates. Serotype IV accounted for 44/506 (8.7%) of the combined isolates from young infants (27/232, 11.6% of EOD and 17/274, 6.2% of LOD). There were two serotype VI EOD isolates (not shown). In summary, the top five serotypes represented 91.3% (462/506) of cases in young infants, with the addition of serotype IV increasing this proportion to 99.6% (504/506).
At least one of the three pilus backbone determinants, corresponding to three distinct pili and pathogenicity islands, were detected in almost all (98.5%) GBS isolates (Figure 1B, sTable 3). The most frequently identified “pilus type” was the combination PI-1 and PI-2a (n = 3,691, 58.3%) followed by PI-2a alone (n = 1,953, 30.8%), PI-1 + PI-2b (n = 469, 7.4%), PI-2b (n = 203, 3.2%) and PI-1 (n = 16, 0.3%). The distributions of the pilus protein types among isolates recovered from neonatal disease (EOD and LOD) varied markedly from those isolated from older ages, due to the high proportion of isolates from young infants that were positive for both PI-1 and PI-2b (186/506, 36.8%), nearly all of which (183/186) were serotype III.
Nearly all (n = 6,289, 99.2%) GBS isolates shared a 522 bp sequence with ≥ 95% sequence identity to one of the four homologous alpha protein family gene queries (alpha, rib, alp2/3, alp1) (sTable 3). These four alpha family DNA sequence queries, corresponding to the 174 residue N-terminal domains used for the GBS-NN vaccine [20] share approximately 68–75% sequence identity between them (not shown). The frequencies of the alpha family protein determinants were similar: alp1 (28.1%), alpha (26.3%), alp2/3 (23.4%) and rib (22.2%) (Figure 1C), however rib was much more common among isolates from young infants. Of the 1,399 rib-positive isolates, 937 (67%) isolates were of serotype III. These rib- positive isolates included 277 (54.7%) of the 506 isolates from young infants (87 EOD, 190 LOD), of which 247 (89.2%) were serotype III. Most (1,105/1,766, 62.3%) alp1-positive isolates were of serotype Ia and 864/1,473 (58.7%) of alp2/3-positive isolates were serotype V (sTable 3).
Distributions of serotypes and protein vaccine candidate distributions among clonal complexes.
MLST data was available for 6,336 GBS isolates and 389 STs were identified (sTable 4). These STs were grouped into seven major CCs: CC23 (24.3%), CC1 (22.1%), CC19 (14.3%), CC12 (12.8%), CC459 (9.4%), CC22 (8.7%) and CC17 (7.3%) (Table 1 and Figures 2A–2B). Each CC was represented by a dominant serotype: CC585/41 (V; 100%), CC17 (III; 99.8%), CC459 (IV; 99.7%), CC22 (II; 99.6), CC26/1087 (V; 96.7%), CC23 (Ia; 87.4%), CC12 (Ib; 82.8%), CC328 (V; 77.8%), CC1 (V; 62.3%) and CC19 (III; 53.2%) (Table 1 and Figures 3A–3B). There were 28 STs (sTable 5) that were represented by multiple serotypes.
Table 1.
Eleven clonal complexes and their serotype distributions among the study isolates (2015–2017)a
| Clonal Complex | Ia | Ib | II | III | IV | V | VI | VII | VIII | IX | NT | No. isolates (% total) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC23 | 1344 | 2 | 17 | 18 | 144 | 9 | 0 | 0 | 0 | 0 | 4 | 1538 (24.3) |
| CC1 | 15 | 235 | 186 | 5 | 8 | 873 | 61 | 4 | 7 | 0 | 4 | 1398 (22.1) |
| CC19 | 15 | 2 | 210 | 482 | 0 | 182 | 0 | 1 | 10 | 0 | 2 | 904 (14.3) |
| CC12 | 2 | 674 | 118 | 12 | 0 | 7 | 0 | 0 | 0 | 0 | 1 | 814 (12.8) |
| CC459 | 0 | 1 | 0 | 0 | 591 | 0 | 0 | 0 | 0 | 0 | 1 | 593 (9.4) |
| CC22 | 1 | 0 | 547 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 549 (8.7) |
| CC17 | 0 | 0 | 0 | 461 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 462(7.3) |
| CC26/1087 | 0 | 0 | 0 | 1 | 0 | 29 | 0 | 0 | 0 | 0 | 0 | 30 (0.5) |
| CC3 | 6 | 4 | 1 | 1 | 4 | 5 | 0 | 0 | 0 | 0 | 0 | 21 (0.5) |
| CC328 | 0 | 0 | 0 | 7 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 9 (0.1) |
| CC585/41 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 4 (0.06) |
| Singletons | 0 | 1 | 2 | 0 | 0 | 3 | 0 | 0 | 0 | 8 | 0 | 14 (0.2) |
| Total | 1383 | 919 | 1081 | 987 | 748 | 1114 | 61 | 5 | 17 | 8 | 13 | 6336 (100) |
The number of isolates within the major serotype of a given CC is indicated in bold. Fifteen isolates (singletons) were unrelated to any of the 11 CCs in that their multilocus sequence types (MLSTs) did not share 5 of the 7 MLST locus sequences with members of the 11 different CCs.
Figure 2.
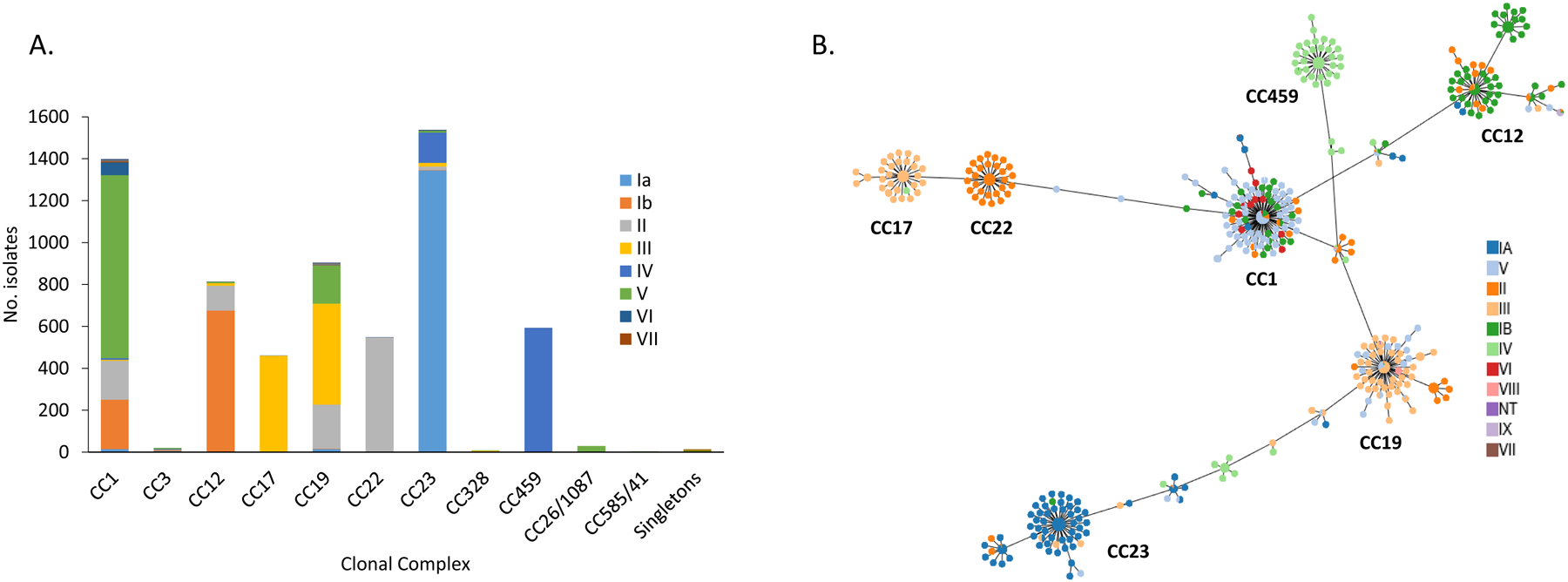
A. Serotype associations within individual clonal complexes. B. Phyloviz diagram depicting individual MLST and associations with the 10 different serotypes.
(Note: In the minimum spanning tree, each ST is displayed as a circle and serotypes are represented by different colors. The size of circles are all the same and do not represent the prevalence of each ST. Major clonal complexes (CCs) are indicated)
Figures 3.
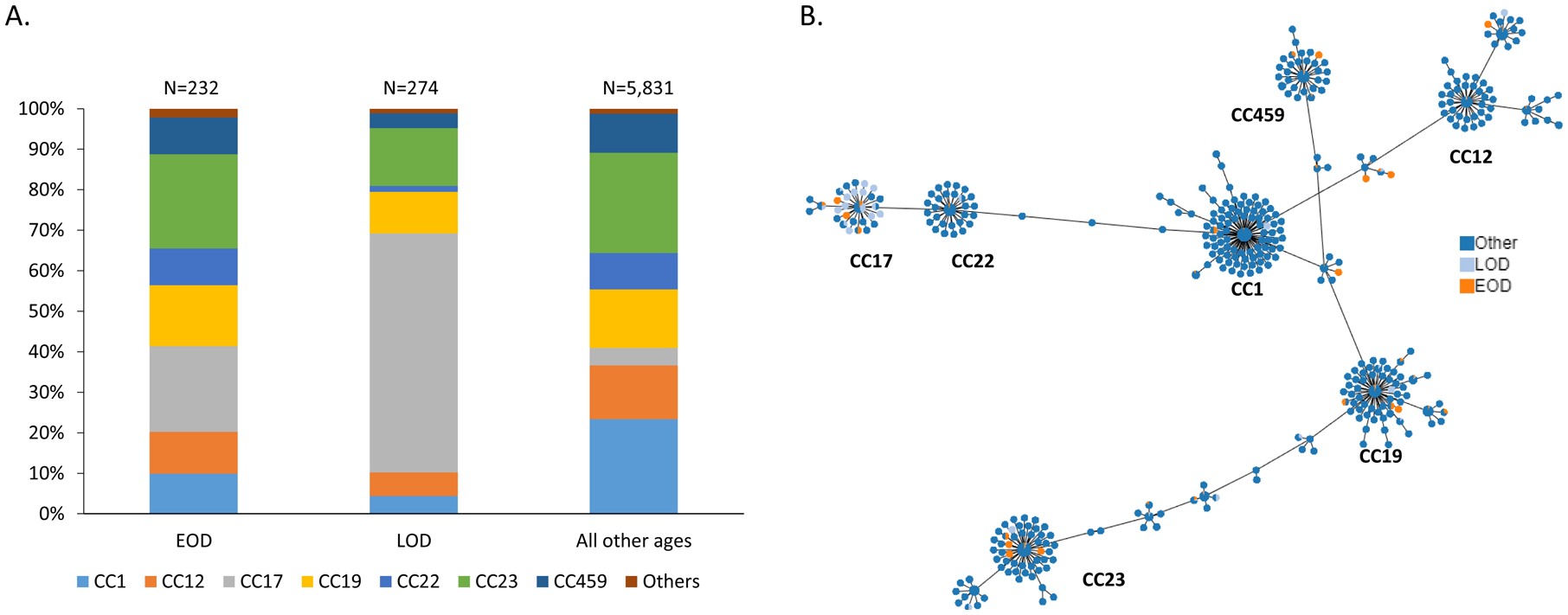
A. Clonal complex distributions among GBS causing EOD, LOD, and disease among all other ages. B. Phyloviz diagram depicting individual MLST and associations with EOD, LOD, and disease among all other age groups. (Note: In the minimum spanning tree, each ST is displayed as a circle and serotypes are represented by different colors. The size of circles are all the same and do not represent the prevalence of each ST. Major clonal complexes (CCs) are indicated)
The distributions of serotypes and CCs among the different age groups from individuals > 90 days of age were similar (not shown). Most EOD and LOD serotype III isolates corresponded to CC17 (49/63; 77.8% and 161/186; 86.6%, respectively), which in all ages is comprised almost entirely of serotype III isolates (Figures 3A and 3B). Most serotype III isolates (444/735, 60.4%) from other age groups were within CC19 (data not shown).
Figures s1, 4 – 5 show the distributions of three different vaccine candidate categories (capsular polysaccharides, pilus backbone proteins, alpha family proteins) among the different CCs. Of the seven major CCs, CCs 17, 22, and 459 were each represented almost entirely by a single serotype (Figure 2A and 2B), pilus type (Figures 4A and 4B), and Alpha family protein (Figures 5A and 5B). Other CCs showed much less type uniformity for each of the three potential vaccine classes (Figures 2, 4–5).
Figure 4.

A. Pilus protein determinant associations with individual clonal complexes. B. Phyloviz diagram depicting individual MLST and associations with the 5 different pilus profiles. (Note: In the minimum spanning tree, each ST is displayed as a circle and serotypes are represented by different colors. The size of circles are all the same and do not represent the prevalence of each ST. Major clonal complexes (CCs) are indicated)
Figure 5.
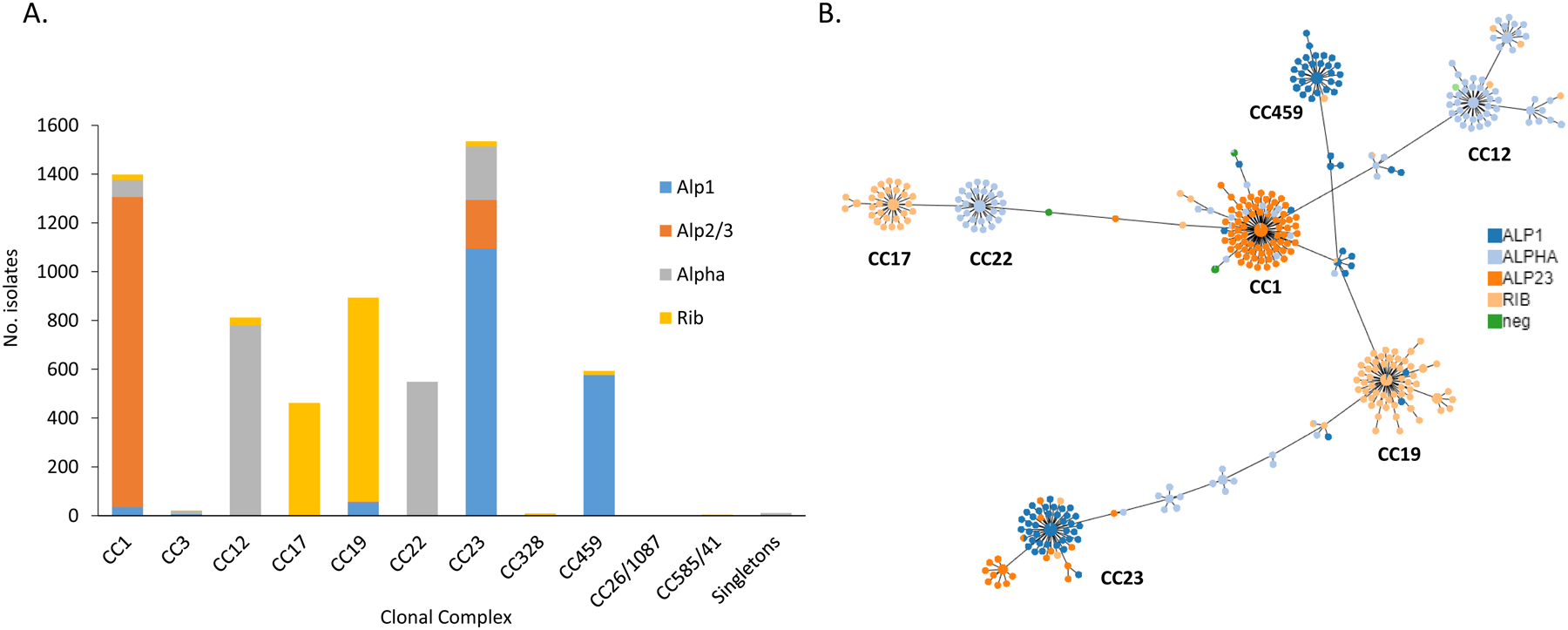
A. Alpha family protein associations with individual clonal complexes. B. Phyloviz diagram depicting individual MLST and associations with the 4 different alpha family protein types. (Note: In the minimum spanning tree, each ST is displayed as a circle and serotypes are represented by different colors. The size of circles are all the same and do not represent the prevalence of each ST. Major clonal complexes (CCs) are indicated)
Hvga virulence protein.
All 465 serotype III/CC17 GBS were positive for the hvgA determinant encoding the major GBS adhesin/invasion, including 161 LOD and 49 EOD isolates. The remaining eight hvgA-positive isolates were serotype IV (CC17 = 1; CC23 =7) from adults ranging in age from early 40s to 90 years of age (sTable 3).
Srr1 and Srr2.
Serine-rich repeat glycoprotein determinants (srr1 or srr2) were identified in most of the iGBS isolates (n=6,113, 96.4%) (sTable 3), with srr1 more predominant (n = 5,507, 90.1%). Srr2 was associated with only two CCs: CC17 (n = 460, 76%) and CC23 (n = 145, 24%). Isolates negative for srr genes were primarily of the ST28 lineage (80%).
Resistance determinants
Of the 6,340 isolates subjected to WGS, 87 (1.4%) did not yield PBP2X types due to assembly errors. These 87 isolates were subjected to conventional MIC testing and found to be susceptible to β-lactam antibiotics. There were 79 PBP2x types (sTable 1), defined as any amino acid sequence difference from the major allele (3,615/6,235, 58%). There were only 54 isolates (0.87%) among the 6,235 examined with one of 31 point mutant alleles of pbp2x that were determined through phenotypic testing to have decreased susceptibility to one or more β-lactam antibiotics based on previously described cutoffs [9] (Table 2, Figure 6). Of these 54 isolates, 37 (68.5%) were also macrolide-resistant (compared to 55% for all isolates combined). These pbp2x point mutants were distributed over the three years (14, 23, and 17 over years 2015, 2016, and 2017, respectively). Of the 31 point mutant pbp2x alleles, eight (corresponding to PBP2X types 17, 24, 26, 32, 42, 47, 59, and 68) accounted for two or more of the 54 isolates recovered during at least two of the three years. Six of these eight types were over-represented by a major strain complex (PBP2x-17, 3/6 Ib/CC12; PBP2x-24, 4/4 Ia/ST23; PBP2x-26, 5/5 Ia/ST26; PBP2x-42, 2/2 Ia/ST23; PBP2x-47, 3/3 V/ST1; PBP2x-59, 2/2 V/ST1).
Table 2.
PBP2x substitutions associated with reduced susceptibility to β-lactam antibiotics among Group B streptococci from invasive disease, 2015–2017
| PBP2X type | PBP2x Substitutionsa | No. isolates | Isolate (serotype, ST) | Ampicillinc | Cefotaximec | Cefoxitinc | Cefazolinc | Penicillinc | Ceftizoximec |
|---|---|---|---|---|---|---|---|---|---|
| 10 | L534S | 1 | 20155117 (V,1) | 0.12 | 0.12 | 16 | 0.5 | 0.12 | 4 |
| 17 | A400V | 6 | 20154959 (Ib,8) | 0.25 | 0.12 | 16 | 0.5 | 0.12 | 1 |
| 20153631 (Ib,1) | 0.25 | 0.12 | 16 | 0.5 | 0.12 | 1 | |||
| 20165790 (Ib/10) | 0.25 | 0.12 | 16 | 1 | 0.12 | 1 | |||
| 20173788 (Ib,8) | 0.25 | 0.12 | 16 | 0.5 | 0.12 | 1 | |||
| 20175374 (V/1) | 0.25 | 0.12 | 16 | 1 | 0.12 | 1 | |||
| 20175554 (V/1) | 0.25 | 0.12 | 16 | 0.5 | 0.12 | 1 | |||
| 24 | I377V,V510I,V525I | 4 | 20155719 (Ia,23) | 0.25 | 0.12 | 16 | 0.5 | 0.12 | 1 |
| 20165538 (Ia,88) | 0.25 | 0.06 | 8 | 0.5 | 0.12 | 1 | |||
| 20166195 (Ia,23) | 0.25 | 0.12 | 16 | 0.5 | 0.12 | 1 | |||
| 20175104 (Ia,23) | 0.25 | 0.12 | 16 | 0.5 | 0.12 | 1 | |||
| 26 | I377V,G415E,V510I | 10 | 20156531 (Ia,23) | 0.25 | 0.12 | 16 | 0.5 | 0.12 | 0.5 |
| 20162445 (Ia,23) | 0.25 | 0.12 | 16 | 0.5 | 0.12 | 0.5 | |||
| 20164013 (Ia,23) | 0.25 | 0.12 | 16 | 0.5 | 0.12 | 0.5 | |||
| 20172717 (Ia,23) | 0.25 | 0.12 | 16 | 0.5 | 0.12 | 0.5 | |||
| 20174129 (Ia,23) | 0.25 | 0.12 | 16 | 0.5 | 0.12 | 1 | |||
| 20164903 (Ia,1031)b | 0.12 | 0.06 | 16 | 0.25 | 0.06 | 0.5 | |||
| 20166529 (Ia,23) | 0.25 | 0.12 | 16 | 0.5 | 0.12 | 0.5 | |||
| 20171112 (Ia,23) | 0.25 | 0.12 | 16 | 0.5 | 0.12 | 0.5 | |||
| 20171215 (Ia,23)b | 0.25 | 0.12 | 16 | 0.5 | 0.12 | 0.25 | |||
| 20176075 (Ia,23) | 0.25 | 0.12 | 16 | 0.5 | 0.12 | 0.5 | |||
| 29 | 521-Y-522 insertion | 1 | 20160409 (V,1) | 0.25 | 0.12 | 16 | 0.25 | 0.12 | 2 |
| 32 | T394I | 2 | 20161556 (IV,459) | 0.25 | 0.12 | 8 | 0.25 | 0.12 | 2 |
| 20162626 (V,1) | 0.25 | 0.25 | 16 | 0.5 | 0.12 | 4 | |||
| 33 | R433C | 1 | 20161558 (III,19) | 0.12 | 0.25 | 8 | 0.5 | 0.12 | 0.5 |
| 34 | I377V,G406D | 1 | 20161102 (II,22) | 0.5 | 0.25 | 16 | 1 | 0.25 | 16 |
| 35 | I377V,A400V,V510I | 1 | 20161114 (IV,452) | 0.25 | 0.12 | 16 | 0.5 | 0.12 | 1 |
| 37 | I377V,A400T | 1 | 20162216 (II,22) | 0.25 | 0.12 | 16 | 0.5 | 0.12 | 1 |
| 39 | I377V,G398A,G627V | 1 | 20175390 (III/109)b | 0.5 | 0.25 | 16 | 0.5 | 0.25 | 1 |
| 42 | I377V,A400T,V510I | 2 | 20162746 (Ia,23) | 0.25 | 0.12 | 8 | 0.25 | 0.12 | 1 |
| 20164794 (Ia,23) | 0.25 | 0.12 | 16 | 0.5 | 0.12 | 1 | |||
| 44 | A400T,Q412K | 1 | 20165260 (V/1) | 0.25 | 0.25 | 16 | 0.5 | 0.12 | 4 |
| 47 | F399V | 3 | 20166135 V/1) | 0.25 | 0.12 | 8 | 0.5 | 0.12 | 4 |
| 20176687 V/1 | 0.5 | 0.12 | 16 | 0.5 | 0.12 | 4 | |||
| 20163262 (V/1) | 0.25 | 0.12 | 8 | 0.5 | 0.12 | 2 | |||
| 48 | I378V,V510I,A514G,N575K | 1 | 20166262 (Ia/23) | 0.5 | 0.12 | 4 | 0.25 | 0.12 | 0.25 |
| 55 | I377V,T394I | 1 | 20164062, (II,22) | 0.25 | 0.12 | 16 | 0.5 | 0.12 | 2 |
| 58 | I377V,Q412R | 1 | 20166357 (Ia,22) | 0.25 | 0.12 | 16 | 0.25 | 0.12 | 0.5 |
| 59 | G406D | 2 | 20170772 (V/1) | 0.5 | 0.25 | 16 | 1 | 0.25 | 32 |
| 20183967 (V/1) | 0.25 | 0.12 | 8 | 1 | 0.12 | 16 | |||
| 60 | Del K358 | 1 | 20171010 (III,19) | 0.12 | 0.06 | 16 | 0.25 | 0.06 | 2 |
| 63 | I377V,V510I,Q557E | 1 | 20172108 (Ia/ST23) | 0.25 | 0.25 | 16 | 1 | 0.25 | 2 |
| 66 | I377V, G406D,I510V | 1 | 20172365 (Ia/ST23) | 0.5 | 0.12 | 16 | 1 | 0.25 | 16 |
| 68 | T394A | 2 | 20172646 (Ib,8) | 0.12 | 0.12 | 8 | 0.25 | 0.06 | 2 |
| 20181729 (IV,459) | 0.12 | 0.12 | 8 | 0.25 | 0.06 | 2 | |||
| 70 | I377V, A400V | 1 | 20173768 (III,17) | 0.25 | 0.12 | 16 | 0.5 | 0.12 | 1 |
| 80 | I377V,V510I,A514T | 1 | 20174532 (Ia,23) | 0.25 | 0.12 | 32 | 0.5 | 0.12 | 0.5 |
| 81 | A400V, V525I | 1 | 20176362 (V,1) | 0.25 | 0.12 | 16 | 0.5 | 0.12 | 8 |
| 83 | A550V | 1 | 20175389 (IV/821) | 0.12 | 0.12 | 16 | 0.5 | 0.06 | 2 |
| 85 | I377V,V510I,A616V | 1 | 20177021 (Ia,23) | 0.25 | 0.12 | 16 | 0.5 | 0.12 | 0.5 |
| 87 | T432I | 1 | 20180100 (III,19) | 0.12 | 0.12 | 16 | 0.5 | 0.12 | 2 |
| 91 | F395C | 1 | 20181122 (III,19) | 0.12 | 0.12 | 4 | 0.5 | 0.12 | 1 |
| 94 | I377V; L534S; G627V | 1 | 20182381 (III/828) | 0.25 | 0.25 | 16 | 0.5 | 0.25 | 8 |
| 95 | F486L;V510L | 1 | 20183394 (III,23) | 0.25 | 0.12 | 16 | 0.5 | 0.12 | 0.5 |
Residues in bold appear to be associated with decreased β-lactam susceptibility.
Isolates 20164903 (EOD), 20171215 (LOD), 20175390 (EOD) were associated with EOD and LOD cases. All other isolates were from adult disease cases.
Values in bold indicate elevated MICs.
Figure 6.
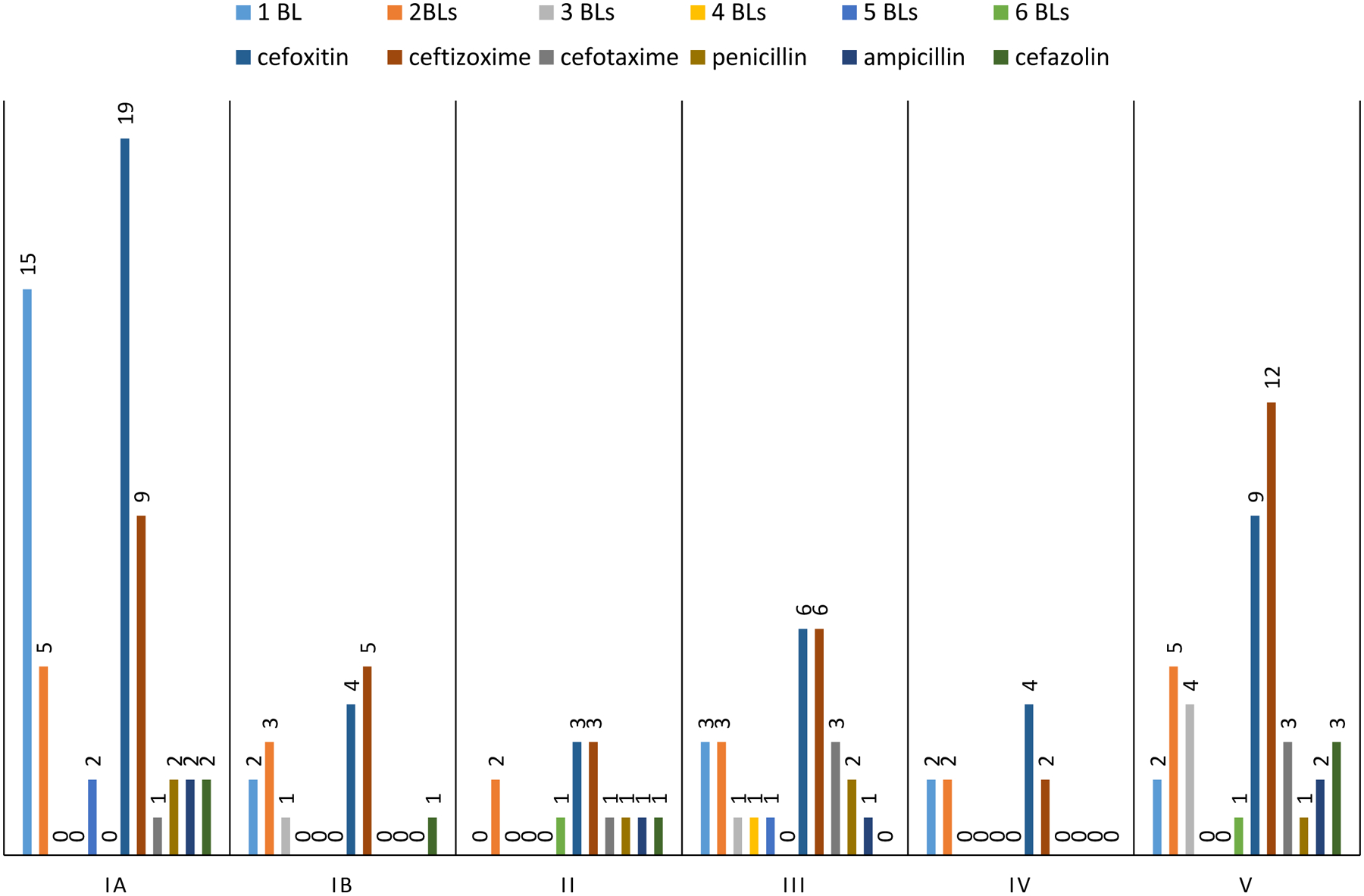
Serotype distribution of the 54 GBS isolates from this study with reduced susceptibility or low-level nonsusceptibility of up to 6 different β-lactam (BL) antibiotics (also shown in Table 2), including data for reduced or low-level susceptibility for each of the 6 β-lactams used. The number of isolates are shown above each of the 10 columns. The total number of isolates represented by each serotype is the sum of the first six columns.
Only nine isolates within eight PBP2X types 34, 39, 47, 48, 59, 63, 66 and 94 had MICs above the susceptible breakpoints for ampicillin and/or penicillin (≥0.25 mg/L and ≥0.12 mg/L, respectively) (Table 2). Of these eight PBP2X types, types 34, 39, 59, and 94 also revealed elevated MICs for the third generation cephalosporins cefotaxime (0.25 mg/L) and ceftizoxime (1–32 mg/L). The relevant substitutions within these PBP2X types included G406D (n=4), G398A (n=1), F399V (n=1), A514G+N575K (n=1), L354S (n=1) and Q557E (n=1), which generally map close to the three conserved catalytic motifs within PBP2X: 344-STMK-347, 402-SSN-404, and 552-KSG-554. PBP2X type 59 (G406D), found within two serotype V/ST1 isolates, was unique in that it was the only one of these eight PBP2X types observed in more than one isolate (once in each of years 2016 and 2017), and was also associated with above-normal MICs for all six of the β-lactam antibiotics tested. In total, there were 14 isolates of one of 12 different PBP2X types with elevated MICs for 1–4 of the clinically relevant β-lactams (ampicillin, penicillin, cefotaxime, cefazolin) used in this study. Thirteen isolates contained a substitution (V or T) at the position A400 (PBP2X type 17, 35, 37, 42, 44, 70, 81). The PBP2X G415E substitution was found in 10 serotype Ia/CC23 isolates. There was no evidence of contributing mutations within the PBP genes pbp1a (sTable 1) and pbp2b (data not shown). The exception was PBP1a-62 in association with PBP2X-83, with a D370N substitution. Elevated MICs for cefoxitin and/or ceftizoxime were the most common, with 78 instances compared to 26 instances for the four other β lactams combined (Figure 6). It follows that there were only 12 isolates that with reduced susceptibility to three or more of the six β-lactams (Figure 6).
A very small proportion of isolates were uniformly susceptible to the various antimicrobials assessed (Figure 7). Erythromycin-resistance (in combination with clindamycin-resistance or alone) within the six major serotypes ranged from 39.2% (serotype III) to 77.7% (serotype IV) (Figure 7). There were several different genes conferring resistance to one or more of the antibiotic classes including macrolides, lincosamides and streptogramins (sTable 6). Overall, resistance to erythromycin and clindamycin was predicted to be 55.2% (n= 3,497) and 43.9% (n= 2,783), respectively. The erm determinants (ermB, ermTR, ermT, ermA) that confer resistance to macrolides, lincosamide and streptogramin B (MLSB) antibiotics were found alone or in combination with other determinants in 2,690 strains and represented most (99.3%) of the combined resistance to erythromycin and clindamycin. The remaining isolates (18/2,710; 0.7%) contained mef/msrD (conferring erythromycin resistance) and either the lnu (lnuB or lnuC) and/or lsa (lsaA, lsaC, or lsaE) determinants known to confer resistance to lincosamides [9, 30]. The M phenotype (erythromycin resistance and clindamycin susceptibility) was predicted in 779 mef/msrD–positive isolates. One isolate (20161487) contained an apparent mef homologue (designated mefSL1) with 54.5%–53.5% sequence identity over the entire 398 codon structural gene with the mefA-10 determinant of the same length in ResFinder and mefB within ArgAnnot (sTable 6, footnote d). There were 67 isolates with predicted resistance to clindamycin only; 59 isolates contained lsaC, seven with both lnuB and lsaE, and one with lnuC alone. Tetracycline resistance was predicted in 85.2% (5,399) of isolates, primarily conferred by tetM (n = 5,011, 92.8%) and tetO (4.6%) (sTable 7). There were 103 (1.62%) GBS isolates with predicted MICs of ≥4 mg/L for levofloxacin due to known amino acid substitutions (sTable 8). The GyrA S81L combined with ParC S79F/Y were the most common substitutions (n= 69, 67%) associated with fluoroquinolone resistance. Two GBS isolates (20166174 and 20170296), both serotype V/ST1 and from adult patients in Maryland, contained vanG elements and had MICs of 2 mg/L to vancomycin (sTable 9). The catQ gene conferring chloramphenicol resistance was detected in 10 isolates. Eighteen isolates had substitutions within the RpoB regions associated with rifampin resistance. Two isolates carried the dfrG determinant that confers high-level trimethoprim resistance. Determinants found to be associated with gentamicin (aac6-aph2) and other aminoglycosides (aph3-III or ant6-Ia) were detected in 17 and 238 isolates, respectively.
Figure 7.

Proportions of each GBS serotype resistant to the individual antimicrobial classes shown and the proportion susceptible to all the antimicrobials assessed in this study (includes those susceptible to the drugs shown and additionally susceptible to clindamycin alone, vancomycin, rifampin, chloramphenicol, and aminoglycosides).
Discussion
During the past 70 years GBS emerged as the most common bacterial cause of newborn sepsis in the US and globally. Efforts are now ongoing towards development of effective vaccines to be implemented in pregnancy, and there is also more recognition of the increasing scope of iGBS disease within adults.
GBS has an extensive and variable array of surface structures, some of which have been documented as promising vaccine candidates [19]. A hexa-valent polysaccharide formulation would target nearly all combined EOD and LOD cases. Even in view of this encouraging data, the concept of universal protein-based vaccines that could target all GBS is attractive. Our data are consistent with past work indicating that a vaccine containing the 3 pilus protein constituents could conceivably target virtually all GBS [31, 32]. The GBS-NN vaccine, consisting of a protein that exactly correlates to the rib and alpha queries used for this survey, could also possibly protect against GBS strains, since nearly all isolates carry and potentially express highly homologous Alpha family proteins [21]. There is uncertainty in this prediction, since N-terminal regions lack antigenic cross-reactivity and published data indicating cross-protection of GBS-NN against strains expressing Alp1 and Alp2/3 is lacking [20, 33]. Strains within the established serotype III MLST type 17 (ST17) lineage, that uniformly express the hypervirulent GBS adhesin HvgA, are disproportionally associated with neonatal meningitis and sepsis [26] and as shown in this work are uniformly positive for the Rib protein that comprises half of the GBS-NN vaccine.
Within the four major CCs it appears likely that there has been frequent recombinational switching of genetic determinants encoding capsular serotype, pilus types, and alpha family protein type. It is theoretically possible that one or more of the three rarely occurring serotypes (1.64%) could emerge as a major cause of iGBS disease, possibly facilitated through expansion of a serotype-switch variant within a major virulent clonal complex. It should also be noted that the primary CC of each of these three rarely occurring serotypes in invasive disease are highly associated with an alpha family determinant type and one or more pilus types.
Although there have been several reports of decreased susceptibility to β-lactam antibiotics during the past 11 years [6–8, 34] our data from 3 years of surveillance indicates that this phenotype remains rare within the United States and that impactful emergence of such strains has not occurred. Nonetheless, the small number of these rare mutant alleles within members of the same major strain complexes within multiple years suggests that in each of these strains a level of fitness has been attained that compensates for its altered Ppb2X protein that is an essential component of the peptidoglycan synthetic apparatus.
We performed conventional testing for > 2,000 of the isolates described here that were recovered during 2015 and as other studies have described [8, 35, 36], we found that the detection of unusual PBP2X substitutions within its transpeptidase domain was effective in flagging all GBS with unusually elevated MICs to β-lactam antibiotics.
Recently we identified two independent vancomycin-nonsusceptible serotype II/ST22 invasive isolates (each carrying a distinct vanG element) [37]. In the current study we identified and phenotypically confirmed two vanG-positive, serotype V/ST1 isolates. The vanG elements detected in GBS to date show near identity to elements found within Enterococcus, suggestive of a common gastrointestinal interspecies reservoir for accessory element resistance determinants. Although GBS non-susceptibility to clinically relevant antibiotics such as β-lactams and vancomycin remains very rare, continued vigilance is warranted. Continued monitoring of diverse resistance features is essential in view of the rapidly growing proportion of iGBS that is resistant to macrolides and lincosamides [3, 5] due to an increasing variety of genetic determinants [9, 30].
Conclusions
We present population-based analysis of more than 6,300 iGBS isolates from the US. Annotated features from each isolate are linked to a publicly available genomic sequence. These data establish an important baseline for monitoring this rapidly evolving pathogen and evaluating the impact of future vaccine candidates. Current capsular serotype and surface protein distributions indicate that these potential vaccine targets would provide excellent coverage, especially in young infants.
Supplementary Material
sFigure 1 Serotype distribution of 6,340 GBS isolates (2015–2017) by 7 different age groups
sFigure 2. Serotype distribution of 6,340 GBS isolates (2015–2017) by state
(CA: California (3 counties); CO: Colorado (5 counties); GA: Georgia (20 counties); MD: Maryland (state-wide); MN: Minnesota (state-wide); NM: New Mexico (state-wide); NY: New York (15 counties); OR: Oregon (3 counties). Isolates were for all ages for all sites except NY which included only <90 day olds)
Acknowledgements
We thank all persons in the Active Bacterial Core Surveillance areas who are involved with surveillance and maintenance of the system. We also thank the laboratory staff at the 10 sites who isolate ABCs pathogens and make it possible to track these infections. We acknowledge the CDC ABCs Team and the Streptococcus Laboratory for isolate characterization and the Minnesota Department of Public Health laboratory for performing phenotypic antimicrobial susceptibility testing of all isolates from Minnesota. We would like to acknowledge the following members of the ABCs team at the study sites: California (Art Reingold, Mirasol Apostol, Herschel Kirk), Colorado (Rachel Herlihy and Shelli Marks), Georgia (Amy Tunali, Stephanie Thomas, Melissa Tobin-D’Angelo, Ashley Moore), Minnesota (Corinne Holtzman, Kathy Como-Sabetti, Anita Glennan), New Mexico (Lisa Onischuk and Nicole Espinoza), Oregon (Tasha Poissant, Heather Jamieson), New York (Kari Burzlaff, Glenda Smith, Nancy Spina, Rachel Wester).
This publication made use of the Streptococcus agalactiae MLST website (https://pubmlst.org/sagalactiae/) sited at the University of Oxford (Maiden 2010); development of this site has been funded by the Wellcome Trust.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Funding.
This work was supported by CDC funding for the Active Bacterial Core surveillance program through cooperative agreements with the Active Bacterial Core surveillance sites. The whole genome sequencing of GBS isolates was supported in part by CDC’s Advanced Molecular Detection (AMD) program.
Footnotes
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the CDC.
Conflict of Interest
The authors declare no potential conflicts of interest.
References
- 1.Madrid L, Seale AC, Kohli-Lynch M, et al. Infant Group B streptococcal disease incidence and serotypes worldwide: systematic review and meta-analyses. Clin Infect Dis. 2017; 65(suppl2): S160–S172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furfaro LL, Chang BJ, Payne MS. Perinatal Streptococcus agalactiae epidemiology and surveillance targets. Clin Microbiol Rev. 2018; 31(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Francois Watkins LK, McGee L, Schrag SJ et al. Epidemiology of invasive Group B Streptococcal infections among nonpregnant adults in the United States, 2008–2016. JAMA Intern Med. 2019; February 18. doi: 10.1001/jamainternmed.2018.7269. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schrag SJ, Verani JR. Intrapartum. antibiotic prophylaxis for the prevention of perinatal Group B streptococcal disease: experience in the United States and implications for a potential group B streptococcal vaccine. Vaccine, 2013; 31 Suppl 4:D20±6. Epub 2012/12/12. [DOI] [PubMed] [Google Scholar]
- 5.Nanduri SA, Petit S, Smelser C. Epidemiology of invasive early-onset and late-onset Group B Streptococcal disease in the United States, 2006 to 2015: Multistate laboratory and population-based surveillance. JAMA Pediatr, 2019; January 14. doi: 10.1001/jamapediatrics.2018.4826. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kimura K, Suzuki S, Wachino J, et al. First molecular characterization of Group B streptococci with reduced penicillin susceptibility. Antimicrob Agents Chemother. 2008; 52: 2890–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dahesh S, Hensler ME, Van Sorge NM et al. Point mutation in the Group B streptococcal pbp2x gene conferring decreased susceptibility to beta-lactam antibiotics. Antimicrob Agents Chemother, 2008; 52:2195–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimura K, Nagano N, Arakawa Y. Classification of Group B streptococci with reduced β-lactam susceptibility (GBS-RBS) based on the amino acid substitutions in PBPs. Antimicrob Agents Chemother, 2015; 70: 1601–1603. [DOI] [PubMed] [Google Scholar]
- 9.Metcalf BJ, Chochua S, Gertz RE Jr., et al. Short-read whole genome sequencing for determination of antimicrobial resistance mechanisms and capsular serotypes of current invasive Streptococcus agalactiae recovered in the USA. Clin Microbiol Infect. 2017; 23(8):574.e7–574.e14. doi: 10.1016/j.cmi.2017.02.021. [DOI] [PubMed] [Google Scholar]
- 10.American College of Obstetricians and Gynecologists. Prevention of early-onset Group B Streptococcal disease in newborns. 2019. No. 782 (https://www.acog.org/GBS).
- 11.Wang P, Tong JJ, Ma XH. Serotypes, antibiotic susceptibilities, and multi-locus sequence type profiles of Streptococcus agalactiae isolates circulating in Beijing, China. PLoS One, 2015; 17: 10(3): e0120035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corvaglia L, Tonti G, Martini S, et al. Influence of intrapartum antibiotic prophylaxis for Group B streptococcus on gut microbiota in the first month of life. J Pediatr Gastroenterol Nutr. 2016; 62:304–308. [DOI] [PubMed] [Google Scholar]
- 13.Cieslewicz MJ, Chaffin D, Glusman G. Structural and genetic diversity of Group B Streptococcus capsular polysaccharides. Infect Immun, 2005; 73: 3096–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiely RA, Cotter L, Mollaghan AM, Cryan B, Coffey A, Lucey B. Emergence of Group B Streptococcus serotype IV in women of child-bearing age in Ireland. Epidemiol Infect, 2011; 139:236–8. [DOI] [PubMed] [Google Scholar]
- 15.Ferrieri P, Lynfield R, Creti R, Flores AE. Serotype IV and invasive Group B Streptococcus disease in neonates, Minnesota, USA, 2000–2010. Emerg Infect Dis, 2013; 19:551–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teatero S, McGeer A, Li A, et al. Population structure and antimicrobial resistance of invasive serotype IV Group B Streptococcus, Toronto, Ontario, Canada. Emerg Infect Dis, 2015; 21:585–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vekiemans J, Crofts J, Baker CJ, et al. The role of immune correlates of protection on the pathway to licensure, policy decision and use of Group B Streptococcus vaccines for maternal immunization: considerations from World Health Organization consultations. Vaccine, 2019; 37:3190–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buurman ET, Timofeyeva Y, Gu J, et al. A novel hexavalent capsular polysaccharide conjugate vaccine (GBS6) for the prevention of neonatal Group B streptococcal infections by maternal Immunization. J Infect Dis. 2019; 220:105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paoletti LC, Kasper DL. Surface structures of Group B Streptococcus important in human immunity. Microbiol Spectr, 2019; 7(2). doi: 10.1128/microbiolspec.GPP3-0001-2017. [DOI] [PubMed] [Google Scholar]
- 20.Stålhammar-Carlemalm M, Waldemarsson J, Johnsson E, Areschoug T, Lindahl G. Nonimmunodominant regions are effective as building blocks in a streptococcal fusion protein vaccine. Cell Host Microbe, 2007; 2:427–34. [DOI] [PubMed] [Google Scholar]
- 21.Maeland JA, Afset JE, Lyng RV, Radtke A. Survey of immunological features of the Alpha-like proteins of Streptococcus agalactiae. Clin Vaccine Immunol, 2015; 22:153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maione D, Margarit I, Rinaudo CD, et al. Identification of a universal Group B Streptococcus vaccine by multiple genome screen. Science 2005; 309:148–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langley G, Schaffner W, Farley MM, et al. Twenty years of Active Bacterial Core Surveillance. Emerg Infect Dis 2015; 9:1520–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta SK, Padmanabhan BR, Diene SM, et al. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother, 2014; 58:212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zankari E, Hasman H, Cosentino S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother, 2012; 67:2640–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tazi A, Disson O, Bellais S, et al. The surface protein HvgA mediates Group B streptococcus hypervirulence and meningeal tropism in neonates. J Exp Med, 2010; 207:2313–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Sorge NM, Quach D, Gurney MA, Sullam PM, Nizet V, Doran KS. The Group B streptococcal serine-rich repeat 1 glycoprotein mediates penetration of the blood-brain barrier. J Infect Dis, 2009; 199:1479–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosini R, Rinaudo CD, Soriani M, et al. Identification of novel genomic islands coding for antigenic pilus-like structures in Streptococcus agalactiae. Mol Microbiol. 2006; 61:126–41. [DOI] [PubMed] [Google Scholar]
- 29.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol, 2004; 186:1518–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hawkins PA, Law CS, Metcalf BJ, et al. Cross-resistance to lincosamides, streptogramins A and pleuromutilins in Streptococcus agalactiae isolates from the USA. J Antimicrob Chemother, 2017; 72:1886–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Margarit I, Rinaudo CD, Galeotti CL, et al. Preventing bacterial infections with pilus-based vaccines: the Group B streptococcus paradigm. J Infect Dis, 2009; 199:108–15. [DOI] [PubMed] [Google Scholar]
- 32.Nuccitelli A, Cozzi R, Gourlay LJ, et al. Structure-based approach to rationally design a chimeric protein for an effective vaccine against Group B Streptococcus infections. Proc Natl Acad Sci U S A, 2011; 108:10278–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larsson C, Stalhammar-Carlemalm M, Lindahl G. Experimental vaccination against Group B streptococcus, an encapsulated bacterium, with highly purified preparations of cell surface proteins Rib and alpha. Infect Immun, 1996; 64:3518–3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yi A, Kim CK, Kimura K, et al. First Case in Korea of Group B Streptococcus With Reduced Penicillin Susceptibility Harboring Amino Acid Substitutions in Penicillin-Binding Protein 2X. Ann Lab Med, 2019; 39:414–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimura K, Wachino J, Kurokawa H, et al. High cephalosporin resistance due to amino acid substitutions in PBP1A and PBP2X in a clinical isolate of Group B Streptococcus. J Antimicrob Chemother, 2013; 68:1533–6. [DOI] [PubMed] [Google Scholar]
- 36.Moroi H, Kimura K, Kotani T, Tsuda H, Banno H, Jin W, Wachino JI et al. Isolation of Group B Streptococcus with reduced β-lactam susceptibility from pregnant women. Emerg Microbes Infect, 2019;8:2–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Srinivasan V, Metcalf BJ, Knipe KM, et al. vanG element insertions within a conserved chromosomal site conferring vancomycin resistance to Streptococcus agalactiae and Streptococcus anginosus. Mbio, 2014; 5:e01386–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
sFigure 1 Serotype distribution of 6,340 GBS isolates (2015–2017) by 7 different age groups
sFigure 2. Serotype distribution of 6,340 GBS isolates (2015–2017) by state
(CA: California (3 counties); CO: Colorado (5 counties); GA: Georgia (20 counties); MD: Maryland (state-wide); MN: Minnesota (state-wide); NM: New Mexico (state-wide); NY: New York (15 counties); OR: Oregon (3 counties). Isolates were for all ages for all sites except NY which included only <90 day olds)


