Abstract
The ongoing coronavirus disease 2019 pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has posed a serious threat to global public health and social stability. There is an urgent need for understanding the nature and infection mechanism of the virus. Owing to its high infectivity and pathogenicity and lack of effective treatments, live SARS-CoV-2 has to be handled in biosafety level 3 laboratories, which has impeded research into SARS-CoV-2 and the development of vaccines and therapeutics. Pseudotyped viruses that lack certain gene sequences of the virulent virus are safer and can be investigated in biosafety level 2 laboratories, providing a useful virological tool for the study of SARS-CoV-2. In this review, we will discuss the construction of SARS-CoV-2 pseudoviruses based on different packaging systems, current applications, limitations, and further explorations.
Keywords: COVID-19, SARS-CoV-2, pseudovirus, spike protein, packaging system
Introduction
The outbreak of coronavirus disease 2019 (COVID-19) has caused major global human health and social upheaval. According to the World Health Organization (WHO), as of February 23, 2021, more than 111 million cases and 2.4 million deaths have been confirmed worldwide, with numbers continuing to increase on a daily basis (https://covid19.who.int/). Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a new member of the genus Betacoronaviruses and a close relative of SARS-CoV, has been identified as the cause of the global pandemic1-3. Studies indicate that both SARS-CoV-2 and SARS-CoV utilize angiotensin-converting enzyme 2 (ACE2) as the host cell surface receptor to gain entry into target cells4. SARS-CoV-2 is a positive-strand RNA virus with a 30 kb genome5. It has a round or elliptic morphology, with a diameter of approximately 60-140 nm, and is characterized by a crown-like appearance under electron microscopy observation owing to the presence of spike glycoproteins on the envelope. The first two-thirds of the genome, named the ORF1ab region, encodes 16 non-structural proteins. The rest of the genome encodes several accessory proteins and four structural proteins, including spike (S) glycoprotein, envelope (E), matrix (M), and nucleocapsid (N) proteins, which are important for maintaining the structural integrity of the enveloped SARS-CoV-2 virion6-8.
Studies of live SARS-CoV-2 are restricted to biosafety level 3 laboratories, which has made SARS-CoV-2 research inaccessible to the majority of research laboratories around the world, which would seriously delay the development of vaccines and therapeutics against SARS-CoV-2 if there were no alternative solution. Pseudoviruses are a kind of recombinant virus with their core or backbone and surface proteins derived from different viruses9. Genes inside pseudoviruses are usually altered or modified to abolish native surface protein expression. An additional plasmid is then used to express alternative surface proteins, producing a pseudovirus that can infect susceptible host cells but can only replicate intracellularly for a single round10, 11. As viral surface proteins play pivotal roles in gaining entry into host cells, the conformational structures of pseudoviral surface proteins have high similarity to that of the native viral proteins; however, pseudoviruses have attenuated virulence compared with wild-type (WT) viruses, allowing them to be safely handled in biosafety level 2 laboratories12. As a result, pseudoviruses are widely used for the study of cellular tropism13, receptor recognition14, drug discovery15, and for developing and evaluating antibodies and vaccines16, 17. In this review, we discuss the current development and applications of SARS-CoV-2 pseudotyped viruses, their limitations, and further development directions.
Packaging systems for SARS-CoV-2 pseudoviruses
The human immunodeficiency virus (HIV-1)-based lentiviral packaging system
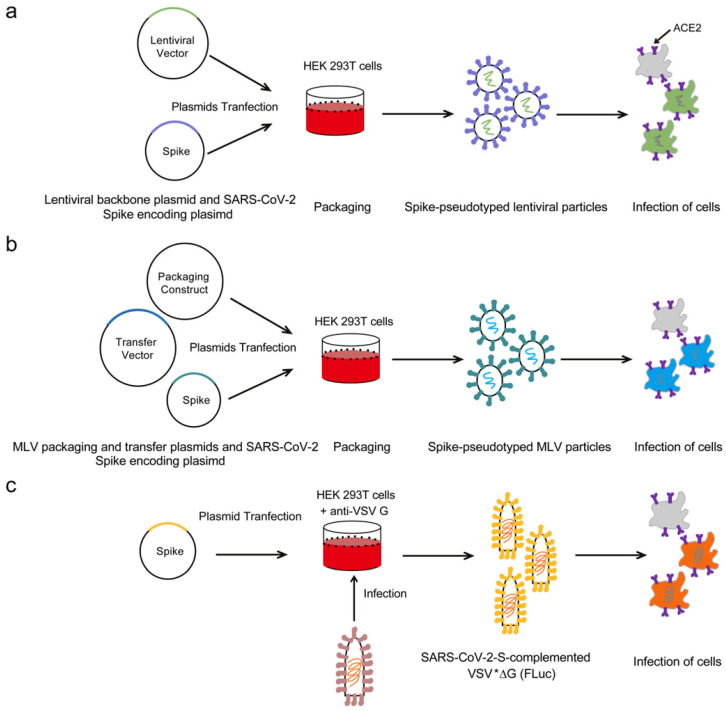
The HIV-1 lentiviral packaging system is currently the most widely used SARS-CoV-2 pseudoviral packaging system. Figure 1a shows the basic strategies to generate the SARS-CoV-2 pseudoviruses based on the HIV-1-derived lentiviral particle. In general, two or three plasmids are co-transfected into the cells to produce the pseudoviruses. The two-plasmid packaging system is the most widely used SARS-CoV-2 pseudoviral packaging system, which includes a plasmid to express the SARS-CoV-2 S protein and another HIV-1 backbone plasmid to express the packaging proteins and signals but with the envelope gene deleted. Several HIV-1 lentivial backbone plasmids are used in the two-plasmid packaging system, such as pNL4-3-kfs18 and pNL4-3.Luc.R-E-19-21. The HIV-1 three-plasmid packaging system is usually comprised of a packaging plasmid, a transfer plasmid containing the reporter gene, and a SARS-CoV-2 S protein-expressing plasmid. Specifically, this system involves splitting the HIV‐1 backbone into separate packing and transfer plasmids. The packaging plasmid expresses the Gag and Pol proteins, while the transfer plasmid contains the cis‐regulatory elements needed for HIV-1 reverse transcription, integration, and packaging, as well as multiple cloning sites and a reporter gene under the control of a heterogeneous promoter22, 23. The SARS-CoV-2 S protein-expressing plasmid is made of a vector carrying the S gene driven by a cytomegalovirus (CMV) promoter. Several independent groups have reported that co-transfection of pNL4-3.Luc.R-E- and a SARS-CoV-2 S protein-expressing plasmid can produce SARS-CoV-2 pseudoviruses15, 19-21. Our group produced pseudotyped viruses through co-transfection of pNL4-3-kfs and a SARS-CoV-2 S protein-expressing plasmid (unpublished). Ou et al co-transfected a SARS-CoV-2 S protein-expressing plasmid, psPAX2, and pLenti-GFP into HEK 293T cells to produce SARS-CoV-2 pseudoviruses23. Yan et al co-transfected a pCDH plasmid harboring SARS-CoV-2 specific genes, psPAX, and pMD2G into the HEK 293T cells to successfully produce SARS-CoV-2 pseudoviruses24.
Figure 1.
The strategies to acquire SARS-CoV-2 pseudoviruses based on different packaging systems. (a) HEK 293T cells were transfected with a plasmid encoding lentiviral backbone and a plasmid expressing SARS-CoV-2 Spike. The transfected cells produced Spike-pseudotyped lentiviral particles and these viral particles can infect cells that express the ACE2 receptor. (b) HEK 293T cells were co-transfected with an Spike encoding-plasmid, an MLV Gag-Pol packaging construct and the MLV transfer vector encoding a luciferase reporter. The transfected cells produced Spike-pseudotyped MLV particles and these viral particles can infect cells that express the ACE2 receptor. (c) HEK 293T cells were transfected with SARS-CoV-2 Spike expression plasmid, after 24 h post-transfection, the cells were inoculated with VSV*∆G (Fluc) encoding firefly luciferase. After an incubation period of 1h at 37 ℃, the inoculum was removed and cells were washed with PBS before medium supplemented with anti VSV-G antibody was added in order to neutralize residual input virus. Spike-pseudotyped particles were harvested 20 h postinoculation and could infect cells that express the ACE2 receptor.
The murine leukemia virus (MLV)-based packaging system
MLVs are typical simple retroviruses, they are enveloped and contain positive-strand RNA bearing three genes (gag, pol and env) that encode viral capsid proteins, viral enzymes and envelope proteins, respectively25. Early work by Witte and colleagues showed that when vesicular stomatitis virus (VSV) is used to infect cells in which MLV is packaged, pseudoviruses can be harvested for use in neutralization antibody assays, indicating its potential as a packaging system for the production of pseudotyped viruses26. Figure 1b shows the strategies to generate the SARS-CoV-2 pseudoviruses based on the MLV packaging system. Recently, Zheng et al co-transfected MLV packaging system containing three plasmids into HEK 293FT cells to generate SARS-CoV-2 pseudovirion particles: the first plasmid was the packaging construct, SV-Psi--Env--MLV; the second plasmid was L-LUC-SN, a minimal retroviral transfer vector encoding the luciferase reporter gene; and the third plasmid was an expression construct encoding the SARS-CoV-2 S protein27. Walls et al co-transfected an MLV Gag-Pol packaging construct, a MLV transfer vector encoding a luciferase reporter, and an S protein-encoding plasmid into the HEK 293T cells to successfully produce SARS-CoV-2 pseudoviruses28.
The VSV packaging system
VSV is a negative-strand RNA virus that possesses a single type envelope glycoprotein (G) mediating its entry29. Previous studies have shown that G-deleted VSV can be complemented in trans with the envelope glycoproteins of even unrelated viruses, including coronaviruses30, 31. Stillman et al cloned the VSV genome into a plasmid to make stable VSV, which was subsequently used to generate pseudoviruses carrying heterogeneous glycoproteins32. Various reporter genes were successively inserted into this plasmid to facilitate the detection of infected cells in a reasonable period of time33. Figure 1c shows the outlines to generate the SARS-CoV-2 pseudoviruses based on the VSV packaging system. Briefly, HEK 293T cells were grown in cell culture dishes and transfected with SARS-CoV-2 spike protein expression plasmid, after 24 h post-transfection, the cells were washed and inoculated with VSV*∆G (Fluc) encoding firefly luciferase. After an incubation period of 1h at 37 ℃, the inoculum was removed and cells were washed with PBS before medium supplemented with anti VSV-G antibody was added in order to neutralize residual input virus. Pseudotyped particles were harvested 20 h postinoculation. Recently, Zettl et al co-transfected a pseudotyped VSV*∆G (FLuc), a G-deleted VSV encoding both GFP and firefly luciferase, with the SARS-CoV-2 S protein and used it for the detection of SARS-CoV-2 neutralizing antibodies in the sera of convalescent COVID-19 patients17. Letko et al used VSV∆G-luc bearing SARS-spike chimeras to study cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses34. Nie et al successfully constructed a pseudovirus neutralization assay for SARS-CoV-2, consisting of a pseudotyped VSV bearing the full-length spike protein of SARS-CoV-235. Notably, when the VSV packaging system is used to make the SARS-CoV-2 pseudovirus, there may be residual VSV virus mixed with the pseudovirus, which may interfere with the neutralization assay by producing false-positive results. Preferably, the amount of VSV should be minimized; however, if excess VSV is believed to be interfering with a pseudovirus-based assay, treatment of the pseudovirus preparation with a VSV neutralizing antibody could be considered36.
Other packaging systems
Besides the aforementioned SARS-CoV-2 pseudovirus packaging systems, researchers have also produced SARS-CoV-2 pseudoviruses with other packaging systems. Yan et al used a phage vector to produce a SARS-CoV-2 pseudovirus. Specifically, an MS2 vector was used to insert SARS-CoV-2 genes for N, E, and ORF1ab24. In general, while packaging 500 bp of RNA is efficient using MS2, packaging 1.5 kb of RNA does not work well37. Although MS2-based armored L-RNA technology can package >2 kb RNA sequences38, the packaging capacity of MS2 is still not as good as lentiviruses. Owing to this limitation, genes for N, E, and nine parts of ORF1ab gene were inserted into the MS2 vector separately, and different MS2-based pseudoviruses were mixed together to form quality control materials. Researchers also used a NS1-deleted influenza A virus as the vector to express SARS-CoV-2 Spike protein, and this influenza virus-based pseudovirus is now being tested as candidate vaccine in a clinical trial39. The adenovirus vector was used to express SARS-CoV-2 S protein and being evaluated as candidate vaccine against SARS-CoV-240-42.
Applications of SARS-CoV-2 pseudoviruses
Mechanistic study of viral infection and entry
Pseudoviruses have been widely used for conducting in vitro studies on the interaction between viruses and host cells43. They have also proven to be very useful for in vivo studies, particularly on the mechanism of viral infection and biodistribution44. Reporter genes are usually incorporated into pseudoviruses to easily perform quantitative analyses. These mainly include luciferase and fluorescent protein-encoding genes, with each option displaying particular advantages and disadvantages. For example, bioluminescence assays usually have lower background noise and are more sensitive, but data acquisition and analysis can time-consuming and expensive. In contrast, assays using a fluorescent protein as the indicator are much cheaper and easier to operate; however, they may face higher background noise issues and are less sensitive45-47. Along with the aforementioned study in Letko et al, Hoffmann and colleagues used pseudotyped SARS-CoV-2 particles to show that SARS-CoV-2 infection depends on the host cell factors ACE2 and transmembrane protease, serine 2 (TMPRSS2)36. Li et al recovered rVSV-eGFP-SARS-CoV-2 particles and confirmed that the pseudovirus displayed entry characteristics similar to that of the WT virus, such as cell tropism and pH-dependence48. Ou et al used a lentiviral pseudotype system to determine cell type susceptibility, virus receptor, and entry pathway for SARS-CoV-223. Yi et al utilized a spike-containing SARS-CoV-2 pseudovirus to infect neural layers within human embryonic stem cell-derived brain organoids and monolayer cortical neurons, revealing that mature neurons express ACE2 at the soma and are susceptible to invasion of the spike-pseudotyped SARS-CoV-2, establishing an in vitro model to study the impact of SARS-CoV-2 on the human brain49. Some examples of the applications of SARS-CoV-2 pseudoviruses are listed in Table 1.
Table 1.
SARS-CoV-2 pseudoviruses
| Virus Protein | Packaging System | Application | Reference |
|---|---|---|---|
| SARS-CoV-2 Spike | pCMVdeltaR8.91, pLAS2w.RFP-C.Pneo, pMD.G-S | Neutralization antibody assay | 22 |
| SARS-CoV-2 Spike | HDM-IDTSpike-fixK, HDM-Hgpm2, HDM-tat1b, pRC-CMV-Rev1b, pHAGE-CMV-Luc2-IRES-ZsGreen-W | Neutralization antibody assay, Main protease inhibitors evaluation | 50, 51 |
| SARS-CoV-2 Spike | psPAX2, pLenti-GFP, pCMV14-3×Flag-S | Virus entry and immune cross-reactivity, identifying monoclonal antibodies | 23, 52 |
| SARS-CoV-2 Spike (D614/G614) | pHAGE-fullEF1α-ZsGreen-IRES-Puro(R), Lentiviral proteins encoding plasmids (Tat, Rev and Gag/Pol), pS (D614/G614) | MEK inhibitors evaluation | 53 |
| SARS-CoV-2 Spike | pcDNA3.1-SARS2-Spike, psPAX2, pUltra-hot | Brain organoids infection | 49 |
| SARS-CoV-2 Spike | pNL4-3.Luc.R-E-, pcDNA3.1-SARS-CoV-2-S or pCMV3-SARS-CoV-2-S | Neutralization antibody assay and entry inhibition test, entry inhibitors identification, membrane fusion inhibitors evaluation | 15, 19-21 |
| SARS-CoV-2 Spike | pNL4-3/KFS, pSARS-CoV-2-S | Virus assembly | 18 |
| SARS-CoV-2 Spike | pCMV-MLVgag-pol, pTG-Luc, pCAGGS-SARS-CoV-2-S | Virus entry and cross-neutralizing antibody assay | 28, 54 |
| SARS-CoV-2 Spike | SV-Psi--Env--MLV, L-LUC-SN, pSARS-CoV-2-S | Neutralization assay | 27 |
| SARS-CoV-2 Spike | rVSV∆G-G, pSARS-CoV-2-S | Entry and neutralization assay, vaccine | 17, 35, 36, 48, 55, 56 |
| SARS-CoV-2 ORF1ab, N, E | psPAX, PCDH-ORF1ab-N-E, pMD2G; MS2-ORF1ab, MS2-N, MS2-E | Comparing commercial SARS-CoV-2 nucleic acid detection reagents | 24 |
| SARS-CoV-2 Spike | NS1-deleted influenza A virus, pSARS-CoV-2-S | vaccine | 39 |
| SARS-CoV-2 Spike | adenovirus type 5, adenovirus type 26, ChAdOx, SARS-CoV-2-S | vaccine | 40-42 |
Quantification of neutralizing antibodies
Currently, the spike-containing SARS-CoV-2 pseudovirus is the mostly developed pseudoviral system. As various studies have verified that SARS-CoV-2 uses ACE2 as the host cell surface receptor, and the interaction between the S protein and ACE2 mainly mediates its entry to the target cells4, 28, thus, antibody neutralization assays target S protein based on SARS-CoV-2 pseudoviruses have been widely investigated. Compared with traditional assays, the reported pseudovirus-based assays have demonstrated good correlation with WT virus-based assays, while also typically being higher throughput and requiring less sample serum57. Zettl et al reported a VSV-based SARS-CoV-2 pseudovirus and used it to determine neutralizing antibody titers in convalescent COVID-19 patients, showing that most patients had only low titers of neutralizing antibodies (50% neutralizing dose < 320)17. Nie et al evaluated neutralizing antibodies against SARS-CoV-2 in biosafety level 2 facilities by using a VSV pseudovirus production system35. Crawford et al used pseudotyped lentiviral particles to measure the neutralizing activity of human sera or plasma against SARS-CoV-2 in a luciferase-based assay, providing a valuable complement to enzyme-linked immunosorbent assay-based methods that measure antibody binding rather than neutralization50.
Vaccines approach
Pseudovirus-based candidate vaccines against SARS-CoV-2 are also under development. These candidate vaccines with features of live attenuated vaccine which induces robust immune responses including mucosal immunity. The adenoviral vectors based pseudoviruses are the most commonly used vaccines against SARS-CoV-2, such as adenovirus type 540, adenovirus type 2641, and ChAdOx42. All of these candidate vaccines are now currently being tested in phase III clinical trials. The researchers of University of Hong Kong used a influenza A virus-based pseudovirus to develop SARS-CoV-2 vaccine, which is now in a clinical trial39. Case et al reported a live attenuated, replication-competent, VSV vector based vaccine protected against SARS-CoV-2-mediated pathogenesis in mice56. Researchers of Shenzhen Geno-Immune Medical Institute engineered the dendritic cells (DC) with the lentiviral vector expressing the SARS-CoV-2 structural proteins and protease to develop the candidate vaccine, which named LV-SMENP-DC39. In addition, measles virus expressing SARS-CoV-2 S was generated and used as a live attenuated vaccine, while not all immunized animals develop neutralizing activity detectable in the assay58.
Inhibitor screening
Screening of small‐molecule inhibitors against SARS-CoV-2 has also been performed using SARS-CoV-2 pseudoviruses. Yang et al used pseudotyped SARS-CoV-2 to screen an approved drug library of 1,800 small molecular drugs for SARS-CoV-2 virus entry inhibitors. Fifteen active drugs were identified as specific SARS-CoV-2 S pseudovirus entry inhibitors, and further antiviral tests using native SARS-CoV-2 virus in Vero E6 cells confirmed that seven of these drugs significantly inhibited SARS-CoV-2 replication, reducing supernatant viral RNA load with a promising level of activity15. Puhl et al repurposed Ebola and Marburg virus inhibitors tilorone, quinacrine, and pyronaridine for SARS-CoV-2 based on the SARS-CoV-2 pseudovirus59. Zhu et al used a lentiviral-based pseudotyped SARS-CoV-2 particle to evaluate potent membrane fusion inhibitors20. Besides the aforementioned SARS-CoV-2 entry small-molecule inhibitors screenings, efforts have been made into evaluating SARS-CoV-2 main protease and MEK inhibitors51, 53.
Other applications
Guo et al used a pseudovirus with the SARS-CoV-2 S protein as a model and confirmed that plasma-activated water (PAW) effectively inhibited pseudovirus infection through S protein inactivation60. Yan et al compared seven commercial SARS-CoV-2 nucleic acid detection reagents with pseudovirus as the quality control material and found that a pCDH-based pseudovirus was more similar to SARS-CoV-2 than an MS2-based pseudovirus, and was therefore more suitable for evaluating SARS-CoV-2 nucleic acid test kits24. Our group also made a lentiviral vector-based SARS-CoV-2 pseudovirus, combining it with a fluorescence labeling technique for real-time imaging of single SARS-CoV-2 entry in respiratory epithelial cells (unpublished).
Limitations and weakness
There are several issues that should be considered with SARS-CoV-2 pseudoviral systems. SARS-CoV-2 pseudoviruses typically only contain the S protein of WT SARS-CoV-2, which can largely mediate viral entry in a fashion similar to that of the WT virus. Because SARS-CoV-2 pseudoviruses can only replicate for a single round and may not always induce pathogenesis as their WT counterparts do, results from assays using pseudotyped viruses should be compared and validated against WT virus-based assays that remains the gold standard61. Additionally, the virus shape may influence its suitability for constructing a corresponding pseudotyping virus. For example, HIV-1 and MLV are spherical viruses, whereas VSV is bullet-shaped. Therefore, the pattern of S protein distribution/conformation/density on a pseudotyped virus may not reflect the “natural” state of S proteins on SARS-CoV-2. It is necessary to use various packaging systems to simultaneously prepare pseudoviruses62, 63. Currently, the S incorporated pseudotyped viruses were the mainly developed SARS-CoV-2 pseudoviruses, which were unable to conduct researches on functions and processes related to other viral proteins. In addition, to develop pseudoviruses-based candidate vaccines, researchers should consider the pre-existing immunity to the viral vectors. A phase I trial of an Ad5-nCoV demonstrated the result of diminishing vaccine efficiency in individuals with high pre-existing Ad5 immunity40. Moreover, the pseudoviruses-based candidate vaccines worked as live attenuated viruses may face the issues about genome instability which may lead to a back mutation recovering their virulence.
Further explorations
The currently developed SARS-CoV-2 pseudoviruses only contain the SARS-CoV-2 S protein, which restrict their applications; a pseudovirus with multiple viral components of SARS-CoV-2 will partly solve the issues, our group is now trying to alter or modify the viral genome to make a SARS-CoV-2 pseudovirus which contains multiple viral components. Genetic code expansion technology has been used to generate a live but replication-incompetent influenza A virus64, and such strategies could be considered for the construction of SARS-CoV-2 pseudoviruses.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (grant number 2020YFC0861100, grant number 2017YFA0205503), National Natural Science Foundation (21890743), Strategic Priority Research Program of the Chinese Academy of Sciences, China (grant number XDB29050100).
References
- 1.Huang CL, Wang YM, Li XW, Ren LL, Zhao JP, Hu Y. et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu RJ, Zhao X, Li J, Niu PH, Yang B, Wu HL. et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–74. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N, Zhang DY, Wang WL, Li XW, Yang B, Song JD. et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. New Engl J Med. 2020;382:727–33. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–3. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG. et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–9. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan JFW, Kok KH, Zhu Z, Chu H, To KKW, Yuan S. et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infec. 2020;9:221–36. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, Liu QY, Guo DY. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J Med Virol. 2020;92:418–23. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu AP, Peng YS, Huang BY, Ding X, Wang XY, Niu PH. et al. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe. 2020;27:325–8. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanders DA. No false start for novel pseudotyped vectors. Curr Opin Biotech. 2002;13:437–42. doi: 10.1016/s0958-1669(02)00374-9. [DOI] [PubMed] [Google Scholar]
- 10.Ory DS, Neugeboren BA, Mulligan RC. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. P Natl Acad Sci USA. 1996;93:11400–6. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L, Li QQ, Liu Q, Huang WJ, Nie JH, Wang YC. A bioluminescent imaging mouse model for Marburg virus based on a pseudovirus system. Hum Vacc Immunother. 2017;13:1811–7. doi: 10.1080/21645515.2017.1325050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Welch SR, Guerrero LW, Chakrabarti AK, McMullan LK, Flint M, Bluemling GR. et al. Lassa and Ebola virus inhibitors identified using minigenome and recombinant virus reporter systems. Antivir Res. 2016;136:9–18. doi: 10.1016/j.antiviral.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 13.Bartosch B, Dubuisson J, Cosset FL. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J Exp Med. 2003;197:633–42. doi: 10.1084/jem.20021756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Radoshitzky SR, Abraham J, Spiropoulou CF, Kuhn JH, Nguyen D, Li WH. et al. Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature. 2007;446:92–6. doi: 10.1038/nature05539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang L, Pei RJ, Li H, Ma XN, Zhou Y, Zhu FH, Identification of SARS-CoV-2 entry inhibitors among already approved drugs. Acta Pharmacol Sin. 2020. [DOI] [PMC free article] [PubMed]
- 16.Nie JH, Wu XH, Ma J, Cao SC, Huang WJ, Liu Q. et al. Development of in vitro and in vivo rabies virus neutralization assays based on a high-titer pseudovirus system. Sci Rep-Uk. 2017;7:42769. doi: 10.1038/srep42769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zettl F, Meister TL, Vollmer T, Fischer B, Steinmann J, Krawczyk A. et al. Rapid Quantification of SARS-CoV-2-Neutralizing Antibodies Using Propagation-Defective Vesicular Stomatitis Virus Pseudotypes. Vaccines-Basel. 2020;8:386. doi: 10.3390/vaccines8030386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He S, Waheed AA, Hetrick B, Dabbagh D, Akhrymuk IV, Kehn-Hall K. et al. PSGL-1 Inhibits the Incorporation of SARS-CoV and SARS-CoV-2 Spike Glycoproteins into Pseudovirions and Impairs Pseudovirus Attachment and Infectivity. Viruses. 2020;13:46. doi: 10.3390/v13010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang R, Huang B, A R, Li W, Wang W, Deng Y. et al. Development and effectiveness of pseudotyped SARS-CoV-2 system as determined by neutralizing efficiency and entry inhibition test in vitro. Biosaf Health. 2020;2:226–31. doi: 10.1016/j.bsheal.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu Y, Yu D, Yan H, Chong H, He Y. Design of Potent Membrane Fusion Inhibitors against SARS-CoV-2, an Emerging Coronavirus with High Fusogenic Activity. J Virol. 2020;94:e00635–20. doi: 10.1128/JVI.00635-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wan JK, Xing SH, Ding LF, Wang YH, Gu CJ, Wu YL. et al. Human-IgG-Neutralizing Monoclonal Antibodies Block the SARS-CoV-2 Infection. Cell Rep. 2020;32:107918. doi: 10.1016/j.celrep.2020.107918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang SW, Tai CH, Hsu YM, Cheng DN, Hung SJ, Chai KM. et al. Assessing the application of a pseudovirus system for emerging SARS-CoV-2 and re-emerging avian influenza virus H5 subtypes in vaccine development. Biomed J. 2020;43:375–87. doi: 10.1016/j.bj.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ou X, Liu Y, Lei X, Li P, Mi D, Ren L. et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan Y, Chang L, Luo W, Liu J, Guo F, Wang L. Comparison of Seven Commercial Severe Acute Respiratory Syndrome Coronavirus 2 Nucleic Acid Detection Reagents with Pseudovirus as Quality Control Material. J Mol Diagn. 2021;23:300–9. doi: 10.1016/j.jmoldx.2020.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan H. Leukemogenesis by Moloney murine leukemia virus: A multistep process. Trends Microbiol. 1997;5:74–82. doi: 10.1016/S0966-842X(96)10076-7. [DOI] [PubMed] [Google Scholar]
- 26.Witte ON, Baltimore D. Mechanism of Formation of Pseudotypes between Vesicular Stomatitis-Virus and Murine Leukemia-Virus. Cell. 1977;11:505–11. doi: 10.1016/0092-8674(77)90068-x. [DOI] [PubMed] [Google Scholar]
- 27.Zheng Y, Larragoite ET, Williams ESCP, Lama J, Cisneros I, Delgado JC. et al. Neutralization assay with SARS-CoV-1 and SARS-CoV-2 spike pseudotyped murine leukemia virions. Virol J. 2021;18:1. doi: 10.1186/s12985-020-01472-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–92. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lichty BD, Power AT, Stojdl DF, Bell JC. Vesicular stomatitis virus: re-inventing the bullet. Trends in Molecular Medicine. 2004;10:210–6. doi: 10.1016/j.molmed.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 30.Huang AS, Palma EL, Hewlett N, Roizman B. Pseudotype Formation between Enveloped Rna and DNA Viruses. Nature. 1974;252:743–5. doi: 10.1038/252743a0. [DOI] [PubMed] [Google Scholar]
- 31.Fukushi S, Watanabe R, Taguchi F. Pseudotyped vesicular stomatitis virus for analysis of virus entry mediated by SARS coronavirus spike proteins. Methods Mol Biol. 2008;454:331–8. doi: 10.1007/978-1-59745-181-9_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stillman EA, Rose JK, Whitt MA. Replication and amplification of novel vesicular stomatitis virus minigenomes encoding viral structural proteins. J Virol. 1995;69:2946–53. doi: 10.1128/jvi.69.5.2946-2953.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moeschler S, Locher S, Conzelmann KK, Kramer B, Zimmer G. Quantification of Lyssavirus-Neutralizing Antibodies Using Vesicular Stomatitis Virus Pseudotype Particles. Viruses-Basel. 2016;8:254. doi: 10.3390/v8090254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5:562–9. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nie JH, Li QQ, Wu JJ, Zhao CY, Hao H, Liu H. et al. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg Microbes Infec. 2020;9:680–6. doi: 10.1080/22221751.2020.1743767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S. et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–80. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pasloske BL, Walkerpeach CR, Obermoeller RD, Winkler M, DuBois DB. Armored RNA technology for production of ribonuclease-resistant viral RNA controls and standards. J Clin Microbiol. 1998;36:3590–4. doi: 10.1128/jcm.36.12.3590-3594.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei Y, Yang C, Wei B, Huang J, Wang L, Meng S. et al. RNase-resistant virus-like particles containing long chimeric RNA sequences produced by two-plasmid coexpression system. J Clin Microbiol. 2008;46:1734–40. doi: 10.1128/JCM.02248-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaur SP, Gupta V. COVID-19 Vaccine: A comprehensive status report. Virus Res. 2020;288:198114. doi: 10.1016/j.virusres.2020.198114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu FC, Li YH, Guan XH, Hou LH, Wang WJ, Li JX. et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet. 2020;395:1845–54. doi: 10.1016/S0140-6736(20)31208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mercado NB, Zahn R, Wegmann F, Loos C, Chandrashekar A, Yu J. et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020;586:583–8. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Doremalen N, Lambe T, Spencer A, Belij-Rammerstorfer S, Purushotham JN, Port JR. et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature. 2020;586:578–82. doi: 10.1038/s41586-020-2608-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steffen I, Simmons G. Pseudotyping Viral Vectors With Emerging Virus Envelope Proteins. Curr Gene Ther. 2016;16:47–55. doi: 10.2174/1566523216666160119093948. [DOI] [PubMed] [Google Scholar]
- 44.Hutchens M, Luker GD. Applications of bioluminescence imaging to the study of infectious diseases. Cell Microbiol. 2007;9:2315–22. doi: 10.1111/j.1462-5822.2007.00995.x. [DOI] [PubMed] [Google Scholar]
- 45.Chan E, Heilek-Snyder G, Cammack N, Sankuratri S, Ji C. Development of a Moloney murine leukemia virus-based pseudotype anti-HIV assay suitable for accurate and rapid evaluation of HIV entry inhibitors. J Biomol Screen. 2006;11:652–63. doi: 10.1177/1087057106288881. [DOI] [PubMed] [Google Scholar]
- 46.Nie J, Liu Y, Huang W, Wang Y. Development of a Triple-Color Pseudovirion-Based Assay to Detect Neutralizing Antibodies against Human Papillomavirus. Viruses. 2016;8:107. doi: 10.3390/v8040107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Q, Liu Q, Huang W, Li X, Wang Y. Current status on the development of pseudoviruses for enveloped viruses. Rev Med Virol. 2018;28:e1963. doi: 10.1002/rmv.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li HY, Zhao CY, Zhang YH, Yuan F, Zhang Q, Shi XL. et al. Establishment of replication-competent vesicular stomatitis virus-based recombinant viruses suitable for SARS-CoV-2 entry and neutralization assays. Emerg Microbes Infec. 2020;9:2269–77. doi: 10.1080/22221751.2020.1830715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yi SA, Nam KH, Yun J, Gim DM, Joe D, Kim YH. et al. Infection of Brain Organoids and 2D Cortical Neurons with SARS-CoV-2 Pseudovirus. Viruses-Basel. 2020;12:1004. doi: 10.3390/v12091004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crawford KHD, Eguia R, Dingens AS, Loes AN, Malone KD, Wolf CR. et al. Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses. 2020;12:513. doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu Y, Ma C, Szeto T, Hurst B, Tarbet B, Wang J. Boceprevir, calpain inhibitors II and XII, and GC-376 have broad-spectrum antiviral activity against coronaviruses in cell culture. bioRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 52.Chen X, Li R, Pan Z, Qian C, Yang Y, You R. et al. Human monoclonal antibodies block the binding of SARS-CoV-2 spike protein to angiotensin converting enzyme 2 receptor. Cell Mol Immunol. 2020;17:647–9. doi: 10.1038/s41423-020-0426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou L, Huntington K, Zhang S, Carlsen L, So EY, Parker C, Natural Killer cell activation, reduced ACE2, TMPRSS2, cytokines G-CSF, M-CSF and SARS-CoV-2-S pseudovirus infectivity by MEK inhibitor treatment of human cells. bioRxiv. 2020.
- 54.Pinto D, Park YJ, Beltramello M, Walls AC, Tortorici MA, Bianchi S, Structural and functional analysis of a potent sarbecovirus neutralizing antibody. bioRxiv. 2020.
- 55.Xiong HL, Wu YT, Cao JL, Yang R, Liu YX, Ma J. et al. Robust neutralization assay based on SARS-CoV-2 S-protein-bearing vesicular stomatitis virus (VSV) pseudovirus and ACE2-overexpressing BHK21 cells. Emerg Microbes Infec. 2020;9:2105–13. doi: 10.1080/22221751.2020.1815589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Case JB, Rothlauf PW, Chen RE, Kafai NM, Fox JM, Smith BK. et al. Replication-Competent Vesicular Stomatitis Virus Vaccine Vector Protects against SARS-CoV-2-Mediated Pathogenesis in Mice. Cell Host Microbe. 2020;28:465–74. doi: 10.1016/j.chom.2020.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bentley EM, Mather ST, Temperton NJ. The use of pseudotypes to study viruses, virus sero-epidemiology and vaccination. Vaccine. 2015;33:2955–62. doi: 10.1016/j.vaccine.2015.04.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Horner C, Schurmann C, Auste A, Ebenig A, Muraleedharan S, Dinnon KH 3rd. et al. A highly immunogenic and effective measles virus-based Th1-biased COVID-19 vaccine. Proc Natl Acad Sci U S A. 2020;117:32657–66. doi: 10.1073/pnas.2014468117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Puhl AC, Fritch EJ, Lane TR, Tse LV, Yount BL, Sacramento CQ, Repurposing the Ebola and Marburg Virus Inhibitors Tilorone, Quinacrine and Pyronaridine: In vitro Activity Against SARS-CoV-2 and Potential Mechanisms. bioRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 60.Guo L, Yao Z, Yang L, Zhang H, Qi Y, Gou L, Plasma-activated water: An alternative disinfectant for S protein inactivation to prevent SARS-CoV-2 infection. Chem Eng J. 2020: 127742. [DOI] [PMC free article] [PubMed]
- 61.Tamin A, Harcourt BH, Lo MK, Roth JA, Wolf MC, Lee B. et al. Development of a neutralization assay for Nipah virus using pseudotype particles. Journal of Virological Methods. 2009;160:1–6. doi: 10.1016/j.jviromet.2009.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zeilfelder U, Bosch V. Properties of wild-type, C-terminally truncated, and chimeric maedi-visna virus glycoprotein and putative pseudotyping of retroviral vector particles. Journal of Virology. 2001;75:548–55. doi: 10.1128/JVI.75.1.548-555.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen Y, Ott CJ, Townsend K, Subbaiah P, Aiyar A, Miller WM. Cholesterol supplementation during production increases the infectivity of retroviral and lentiviral vectors pseudotyped with the vesicular stomatitis virus glycoprotein (VSV-G) Biochem Eng J. 2009;44:199–207. doi: 10.1016/j.bej.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Si LL, Xu H, Zhou XY, Zhang ZW, Tian ZY, Wang Y. et al. Generation of influenza A viruses as live but replication-incompetent virus vaccines. Science. 2016;354:1170–3. doi: 10.1126/science.aah5869. [DOI] [PubMed] [Google Scholar]