Abstract
The coronavirus disease 2019 (COVID-19) pandemic has caused the most devasting social and economic impact of this century. The current pandemic will end only after a safe, effective vaccine becomes available and protective herd immunity has been achieved through vaccination. The key parameter to gauge protective immunity is neutralizing antibody levels. Thus, reliable serology testing is essential to diagnose whether an individual has been previously infected, as a large proportion of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections is asymptomatic. For both naturally infected and vaccinated individuals, it is critical to monitor their neutralizing antibody titers over time. This is because, when neutralizing antibody levels wane below a threshold which remains to be determined, they become vulnerable to reinfection. Due to the importance of serology testing, academia and industry have developed different platforms for serological diagnosis, many of which have achieved the Food and Drug Administration (FDA) Emergency Use Authorizations (EUA). Here we summarize the status of COVID-19 serology testing, discuss challenges, and provide future directions for improvement.
Keywords: COVID-19, SARS-CoV-2, Serology testing, Binding antibodies, Neutralizing antibodies
Coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has swept across the world since December 2019 and continues to cause devastation (Huang et al., 2020; Zhu et al., 2020). As of December 2020, there have been over 82 million cases and close to 1.8 million deaths worldwide (https://coronavirus.jhu.edu/map.html). Communities, businesses, and governments across the world have implemented social distancing policies that have changed daily life and the economy. It has become widely accepted that the only solution to successfully end lockdowns and to resolve the pandemic is a safe, effective vaccine that can confer immunity to a critical majority of the population. For this to occur, not only must there be accurate serology testing to determine whether an individual has recent or prior infection, but also specifically the detection of neutralizing antibodies (nAb) which are believed to be primarily responsible for protective immunity. Thus, quantification of nAb levels is an integral part of vaccine development. Monitoring of individuals' nAb level is important to estimate the risk of re-infection. Understanding herd immunity is essential for public policymakers to safely open our communities. Here, we review the status and challenges of current SARS-CoV-2 serology testing. We also discuss some future directions to improve the serology testing of COVID-19.
There are two major types of virus detection: (i) diagnostic tests such as molecular RNA and protein antigen tests that detect the virus itself, and (ii) antibody tests that detect the host's adaptive immune response to the viral infection (Table 1 ). The former determines whether an individual is actively infected by the virus, while the latter indicates a recent or previous infection by the virus. Molecular diagnostic tests detect viral RNA using techniques such as Real Time-Polymerase Chain Reaction (RT-PCR) and Isothermal Amplification (Dao Thi et al., 2020). The Lucira™ COVID-19 All-In-One Test Kit (https://www.fda.gov/media/143808/download, n.d.), which uses reverse transcription loop-mediated isothermal amplification (RT-LAMP) technology to detect viral RNA of the N gene for SARS-CoV-2, is the first test with EUA that allows for individuals to swab, run, and see test results completely at home. Antigen tests detect specific viral proteins by employing antigen lateral flow with either instrument-mediated detection (e.g., Sofia SARS Antigen FIA by Quidel) or visual read (e.g., BinaxNOW™ COVID-19 Ag Card by Abbott).
Table 1.
Comparison of laboratory tests for COVID-19.
| Classification | Example (FDA authorized) | Intended Use: detection of | TAT1 | Strength | Weakness |
|---|---|---|---|---|---|
| NAAT2, high throughput platform | Hologic Fusion SARS-CoV-2 Assay | Active infection | 1100 samples/24 h | High sensitivity, specificity, and throughput | Long TAT, high instrument cost |
| NAAT, sample-to-answer platform | Abbott ID NOW COVID-19 | Active infection | 15 min/sample | Short TAT | Each instrument can only run one test at a time |
| Antigen LFA3, instrument read | Quidel Sofia 2 SARS Antigen FIA | Active infection | 15 min/sample | POCT4, short TAT | Less sensitive or specific than NAAT |
| Antigen LFA, visual read | Abbott BinaxNOW COVID-19 Ag Card | Active infection | 15 min/sample | POCT, short TAT, no instrument requirement, low cost | Less sensitive or specific than NAAT, subjective to readout |
| Serology, EIA5, high throughput | Ortho Vitros Anti-SARS-CoV-2 IgG | Recent or prior infection, bAb6 | 1000 samples/24 h | High throughput, can be semi-quantitative | Qualitative, high instrument cost |
| Serology, EIA, medium throughput | Euroimmun Anti-SARS-CoV-2 ELISA (IgG) | Recent or prior infection, bAb | 4 h/96-well plate | Can potentially be quantitative | Qualitative |
| Serology, LFA | Premier Biotech RightSign COVID-19 IgG/IgM Rapid Test Cassette | Recent or prior infection, bAb | 15 min/sample | POCT, short TAT, no instrument requirement, low cost | Less sensitive or specific than EIA, subjective readout |
| Serology, total neutralizing antibodies | GenScript cPass SARS-CoV-2 Neutralization Antibody Detection Kit | Recent or prior infection, nAb7 | 4 h/96-well plate | Can potentially quantify neutralizing activity | Qualitative test, cross-reactive to SARS-CoV |
| PRNT8 | – | Recent or prior infection, nAb titer | 4 days | Gold standard to quantify nAb, highly specific | Requires highly trained staff, BSL-39 containment, resource-intensive, slow TAT, not commercially available |
| Reporter virus neutralization test | None | Recent or prior infection, nAb titer | 5–20 h | Quantifies nAb & correlate well with PRNT titer, highly specific | BSL-3 containment, not commercially available |
| Pseudotype virus neutralization test | None | Recent or prior infection, nAb titer | 1 day | No BSL-3 requirement | Only spike protein present on the pseudotype virus, not commercially available |
TAT: Turnaround time.
NAAT: Nucleic acid amplification test.
LFA: Lateral Flow Assay.
POCT: Point-of-Care Test.
EIA: Enzyme Immunoassay.
bAb: binding antibody.
nAb: neutralization antibody.
PRNT: Plaque reduction neutralization test.
BSL-3: Biosafety Level 3.
Antibody tests, or serology tests, use a variety of techniques to detect antibodies from the blood, which will be reviewed in the next section. Upon SARS-CoV-2 infection, our immune system produces antibodies, but not all antibodies can block viral infection. This is because some antibodies bind to viral antigens at epitopes that are not important for viral infection, these antibodies cannot neutralize the virus; other antibodies can bind and neutralize the virus but at different potencies. Thus, it is critical to measure individuals' neutralizing antibody levels for vaccine clinical trials, research studies, and disease prevention. Currently, it is not known what minimal neutralizing antibody level is required to protect one from infection. It is also not known how long the protective immunity can last after natural infection or vaccination (Mulligan et al., 2020; Sahin et al., 2020; Walsh et al., 2020). Answers to these questions, all of which depend on reliable serology testing, are essential for disease control and prevention.
In February 2020, in response to the emergence of COVID-19, the United States Food and Drug Administration (FDA) started to issue Emergency Use Authorizations (EUA) for diagnosis, treatment, and prevention of SARS-CoV-2. As of December 2020, there are 57 assays with EUA for serology testing for COVID-19 (https://www.fda.gov/emergency-preparedness-and-response/mcm-legal-regulatory-and-policy-framework/emergency-use-authorization, n.d.). All but one approved assays detect binding antibodies (bAb) against viral proteins (e.g., spike and/or nucleocapsid proteins), with the intended use to identify whether an individual was recently or previously infected by the virus. These serology tests can be broadly categorized by their readout platforms used to detect SARS-CoV-2 antibodies (Table 1). Enzyme immunoassays include the medium-throughput Enzyme-Linked Immunosorbent Assay (ELISA) and Sandwich Enzyme Immunoassay with Final Fluorescence Detection (FEIA) techniques; and the high-throughput Chemiluminescence Immunoassay (CLIA), Chemiluminescent Microparticle Immunoassay (CMIA), and Electrochemiluminescence Immunoassay (ECLIA) techniques. For these immunoassays, a SARS-CoV-2 antigen is first exposed to patient serum and any patient antibodies against epitopes on that antigen will bind. Next, secondary anti-human IgG, IgM, or IgA antibodies conjugated to a detection platform are used to indicate the presence of bound SARS-CoV-2 antibodies in enzymatic immunoassays. The detection platform determines how quickly a sample can be processed, with medium-throughput systems taking about 4 h per 96-well plate, while high-throughput systems have a turnaround time of about 45 min (e.g., 45 min for the first specimen followed by 1 min per specimen for the next 1000 samples when using the Abbott SARS-CoV-2 IgG assay) (Charlton et al., 2020). Testing of six major enzyme immunoassays (SARS-CoV-2 IgG assay by Abbott, Novel Coronavirus COVID-19 IgG/IgM ELISA by Affinity, Platelia SARS-CoV-2 Total Ab by Bio-Rad, SARS-CoV-2 S1/S2 IgG assay by DiaSorin, Anti-SARS-CoV-2 ELISA IgG by Euroimmun, and Elecsys Anti-SARS-CoV-2 by Roche) found a sensitivity ranging from 50 to 100% and specificity of 92–100% in patients with greater than 21 days of symptom onset (Charlton et al., 2020). Four of the six immunoassays (Abbott, Affinity, Bio-Rad, and Euroimmun) showed >95% sensitivity after 21 days of symptom onset. A separate study found that four high-throughput commercial SARS-CoV-2 IgG tests with EUA (SARS-CoV-2 IgG assay by Abbott, SARS-CoV-2 S1/S2 IgG assay by DiaSorin, Vitros anti-SARS-CoV-2 IgG by Ortho, and Anti-SARS-CoV-2 ELISA (IgG) by Euroimmun), had excellent inter-test agreements and low false-positive rates despite differences in assay techniques (Prince et al., 2020).
The second category of serology tests is the lateral flow assay (LFA), which utilizes a cassette through which patient sample is injected, and a band that appears as positive or negative for bAb detection. Patient antibodies, if present, will attach to virus antigen bound to gold nanoparticles or another detection system. The complex migrates along the membrane to reach the test line, which contains a secondary antibody against the immune complex, causing the test line to develop a change in color that the human eye can detect. LFAs are the basis of point-of-care serology testing, taking only 15 min per sample. A study of six LFAs (BTNX, Biolidics, Deep Blue, Genrui, Getein BioTech, and Innovita) found that four had >95% sensitivity after 21 days of symptom onset. Specificities of the six assays ranged from 96 to 100% (Charlton et al., 2020). Similarly, a separate study of ten immunochromatographic LFAs (Biomedomics, Bioperfectus, DecomBio, DeepBlue, Innovita, Premier, Sure-Bio, UCP Bioscienc., VivaChek, and WondFo) agreed that test specificity peaked at greater than 20 days after symptom onset with specificities of 84.3% to 100% (Whitman et al., 2020). In addition, using a combination of IgM and IgG detection was found to be most specific (Whitman et al., 2020). Authors from both studies noted that positive/negative bands in LFAs were often hard to read and that reader training is required for reliable testing.
Other laboratory techniques used for serology testing with EUA's include fluorescent microsphere immunoassays (i.e., antibody attaches to antigen-coated microbeads and the antibody/antigen microbeads are detected by flow cytometry), photonic ring immunoassays (i.e., detection of the change in resonance wavelength when antigen complexes to antigens bound to a silicon chip), and photometric immunoassays (i.e., machine detects optical changes when silver nucleates around bound gold nanoparticles of a secondary antibody conjugate).
Vaccination has begun in the US, Europe, and many other countries. The assays that target spike protein, such as Liaison SARS-CoV-2 S1/S2 IgG, Vitros anti-SARS-CoV-2 IgG, and Euroimmun Anti-SARS-CoV-2 ELISA, will not be able to distinguish antibody responses to vaccination from responses to natural viral infection. However, the assays targeting nucleocapsid protein, such as Architect SARS-CoV-2 IgG, Platelia SARS-CoV-2 Total Ab, and Elecsys Anti-SARS-CoV-2, may only detect responses to viral infection but not to vaccination.
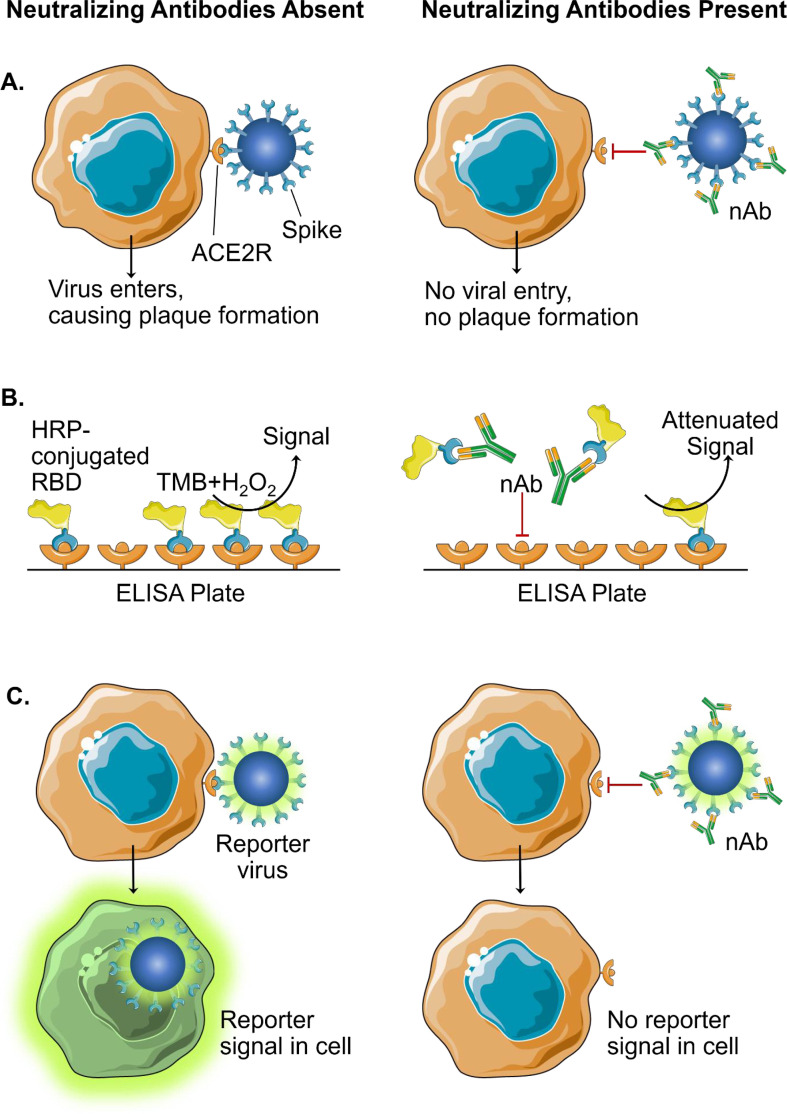
The major limitation to all the afore-mentioned serology tests is that they all detect bAb against SARS-CoV-2 antigens. bAb-detection is only useful for sero-surveillance of the population to gain insight into COVID-19 epidemiology such as seroprevalence. The results from the bAb-assays do not directly indicate whether an individual is immune to SARS-CoV-2 infection. Though antibodies against many epitopes of the SARS-CoV-2 spike protein inhibit virions from infecting the host cell, one cannot be entirely sure of the neutralizing potential unless a neutralizing test is used. The gold standard for detection of nAb is the Plaque Reduction Neutralization Test (PRNT), which requires patient serum to be serially diluted and incubated with authentic live virus followed by infection of cells (Fig. 1A). nAbs prevent the virus from infecting cells and creating plaques, which are counted after a two- to three-day incubation. PRNT is labor and resource-intensive, has a turnaround time of at least 4 days for SARS-CoV-2, and requires bio-safety level 3 (BSL-3) containment. A microneutralization assay has been established to conduct PRNT in a 96-well format, allowing for medium-throughput capacity (Amanat et al., 2020; Manenti et al., 2020), but it is still limited by a 4-day turnaround time and requires BSL-3 laboratories.
Fig. 1.
Assays for the detection of neutralizing antibodies (nAb). A. Plaque-reduction neutralization test (PRNT) is the current gold standard for detection of nAb. When nAb are absent, spike proteins of SARS-CoV-2 interact with ACE2 receptors (ACE2) to allow viral entry, replication, and subsequent plaque formation. Presence of nAb inhibits viral entry and the formation of plaques. B. A surrogate neutralization assay using a blocking ELISA: horseradish peroxidase (HRP) conjugated to the receptor-binding domain (RBD) of the SARS-CoV2 spike protein is pre-incubated on ACE2R-coated ELISA plate. If nAb are present, HRP-conjugated RBD is blocked from binding, resulting in an attenuated signal when 3,3′,5,5′-tetramethylbenzidine (TMB) substrate is provided. C. Reporter virus neutralization assays are based on the expression level of reporter signal in infected cells. Reporter signal will occur when there is viral entry into cells. When nAb are present, inhibition of entry causes decreased reporter signal from cells. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
There is currently a singular assay with EUA that detects nAb: the cPass SARS-CoV-2 Neutralizing Antibody Detection Kit by GenScript USA. It is well-established that coronaviruses, including SARS-CoV-2, enter host tissues through an interaction with the viral spike protein and host Angiotensin Converting Enzyme-2 (ACE2) receptors (Shang et al., 2020). The GenScript kit detects nAb against the receptor-binding domain (RBD) of SARS-CoV-2 spike protein through a blocking ELISA: ACE2 receptors are conjugated to the ELISA plate and preincubated with horseradish peroxidase (HRP)-conjugated RBD. When patient sample is added, if nAb are present, they displace HRP-RBD from binding to ACE2 receptors. This translates to a decreased signal when HRP substrate is added (Fig. 1B). The GenScript assay is currently approved as a qualitative test, with a signal inhibition above the threshold of 30% resulting in a positive test (i.e., nAb detected). It has had a 100% positive percent agreement and 100% negative percent agreement with viral infection-based PRNT50 and PRNT90 (antibody titers needed to reduce plaque formation by 50% and 90%, respectively) in clinical study. Because this assay does not use live virus, it can be performed in regular BSL-2 laboratories and is much higher throughput than PRNT. In addition, the assay is isotype-independent, thus eliminating the need for separate IgG, IgM, and IgA tests (Perera et al., 2020; Tan et al., 2020). Although the current EUA-approved version uses a single serum dilution and is not quantitative, the assay was reported to be capable of quantifying nAb when specimens were tested in a serially diluted fashion, as evidenced by a strong correlation between the neutralization titers derived from this assay and the PRNT assay (Perera et al., 2020).
Other promising nAb assays include the use of reporter viruses: modified SARS-CoV-2 containing a reporter gene that allows for detection of virus entry into cells and replication (Fig. 1C). Replacement of the sequence of SARS-CoV-2 open-reading-frame 7 (ORF7) with the mNeonGreen gene has successfully created a genetically stable reporter virus that replicates similarly to the wild-type (WT) virus (Muruato et al., 2020). Similarly, replacement of ORF7 with the Nanoluciferase gene demonstrated an even more robust reporter virus with a greater dynamic range and higher sensitivity than the fluorescent mNeonGreen (Xie et al., 2020a). Reporter viruses can be incubated with ACE2 receptor-bearing cells along with patient serum. nAb in the patient serum would prevent the reporter virus from infecting cells. The nAb titers can be determined by comparing the degree of attenuation of reporter signal after an incubation time of 4 h in the case of Nanoluciferase SARS-CoV-2; thus, the turnaround time for the neutralization assay could thus be as short as 5 h. Because the readout is a simple plate reader detection of the signal, the reporter virus assays could be conducted in a 96-well to 384- or 1536-well format, allowing much higher throughput than PRNT while maintaining the gold standard of serology test. These assays have also demonstrated high specificity, without cross-reactivity to sera from patients with different pathogen infections (Muruato et al., 2020). The reporter virus assays have also been shown to be useful for screening antivirals (Xie et al., 2020a) and for evaluating vaccine candidates (Muruato et al., 2020). The current limitation with these reporter virus systems is the requirement of BSL-3 containment. This challenge could be overcome by using an attenuated version of SARS-CoV-2. Attenuated SARS-CoV-2 could be generated by using the reverse genetic system of the virus (Xie et al., 2020b). However, a thorough validation of the attenuation of the mutant virus must be performed in both cell cultures and animal models before the biosafety regulatory committee/agency approves its downgrade to BSL-2.
Another avenue being pursued for overcoming BSL-3 containment is the engineering of a pseudotype virus, which expresses and presents SARS-CoV-2 spike protein on the surface of a non-coronavirus. Ideally, the resulting pseudotype virus has the same machinery as the authentic SARS-CoV-2 spike necessary for entering cells and include all the epitopes that nAb would recognize. Almost all pseudotype virus systems only allow one round of viral infection and can be performed in BSL-2 laboratories. Currently, there are SARS-CoV-2 pseudotype viruses using Vesicular Stomatitis Virus (VSV) (Almahboub et al., 2020; Case et al., 2020; Xiong et al., 2020; Zettl et al., 2020) and Human Immunodeficiency Virus (HIV) (Hu et al., 2020; Yang et al., 2020). One potential disadvantage of the pseudotype virus is that the spike protein is displayed in the absence of other SARS-CoV-2 structural proteins (including membrane, envelope, and nucleocapsid proteins). Thus, caution must be taken to ensure that the entry machinery of pseudotype viruses mimics the authentic SARS-CoV-2. For example, discrepant results were obtained when comparing the function of a spike mutation D614G pseudotyped virus and authentic SARS-CoV-2: the D614G mutation was shown to alter the spike protein cleavage and shedding on the pseudotyped virus (Daniloski et al., 2020; Zhang et al., 2020), whereas no such effect was observed on SARS-CoV-2 (Plante et al., 2020). This example underscores the importance of using authentic SARS-CoV-2 to measure neutralizing antibodies.
In summary, enormous efforts from both academia and industry have been made to develop COVID-19 diagnostic tests at an unprecedented speed. Table 1 summarizes one representative assay in each category and highlights the strengths and weaknesses of each assay platform. A reliable serology test to rapidly quantify neutralizing antibody levels in a high-throughput manner is essential for diagnosis, vaccine development, and antiviral development–especially once a minimal threshold of nAb has been defined for disease prevention in the near future. Even in a post-vaccination era, serology tests will remain to be critical for studying both individual and the community's protective immunity to safeguard public health around the world.
References
- Almahboub S.A., Algaissi A., Alfaleh M.A., ElAssouli M.Z., Hashem A.M. Evaluation of neutralizing antibodies against highly pathogenic coronaviruses: a detailed protocol for a rapid evaluation of neutralizing antibodies using vesicular stomatitis virus pseudovirus-based assay. Front Microbiol. 2020:11. doi: 10.3389/fmicb.2020.02020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F., White K.M., Miorin L., Strohmeier S., McMahon M., Meade P., Liu W.-C., Albrecht R.A., Simon V., Martinez-Sobrido L., Moran T., Garcia-Sastre A., Krammer F. An in vitro microneutralization assay for SARS-CoV-2 serology and drug screening. Curr Protoc Microbiol. 2020;e108:58. doi: 10.1002/cpmc.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case J.B., Rothlauf P.W., Chen R.E., Liu Z., Zhao H., Kim A.S., Bloyet L.-M., Zeng Q., Tahan S., Droit L., Ilagan M.X.G., Tartell M.A., Amarasinghe G., Henderson J.P., Miersch S., Ustav M., Sidhu S., Virgin H.W., Wang D., Ding S., Corti D., Theel E.S., Fremont D.H., Diamond M.S., Whelan S.P.J. Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2. Cell Host Microbe. 2020;28:475–485. doi: 10.1016/j.chom.2020.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton C.L., Kanji J.N., Johal K., Bailey A., Plitt S.S., MacDonald C., Kunst A., Buss E., Burnes L.E., Fonseca K., Berenger B.M., Schnabl K., Hu J., Stokes W., Zelyas N., Tipples G. Evaluation of six commercial mid- to high-volume antibody and six point-of-care lateral flow assays for detection of SARS-CoV-2 antibodies. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.01361-20. e01361–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniloski Z., Jordan T.X., Ilmain J.K., Guo X., Bhabha G., Oever B.R., Sanjana N.E. The spike D614G mutation increases SARS-CoV-2 infection of multiple human cell types. bioRxiv. 2020 doi: 10.1101/2020.06.14.151357. Published online June 15, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao Thi V.L., Herbst K., Boerner K., Meurer M., Kremer L.P.M., Kirrmaier D., Freistaedter A., Papagiannidis D., Galmozzi C., Stanifer M.L., Boulant S., Klein S., Chlanda P., Khalid D., Miranda I.B., Schnitzler P., Krausslich H.-G., Knop M., Anders S. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci Transl Med. 2020;12 doi: 10.1126/scitranslmed.abc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- https://www.fda.gov/emergency-preparedness-and-response/mcm-legal-regulatory-and-policy-framework/emergency-use-authorization
- https://www.fda.gov/media/143808/download
- Hu J., Gao Q., He C., Huang A., Tang N., Wang K. Development of cell-based pseudovirus entry assay to identify potential viral entry inhibitors and neutralizing antibodies against SARS-CoV-2. Genes Dis. 2020 doi: 10.1016/j.gendis.2020.07.006. Published online July 13, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manenti A., Maggetti M., Casa E., Martinuzzzzi D., Torelli A., Trombetta C.M., Marchi S., Montomoli E. Evaluation of SARS-CoV-2 neutralizing antibodies using a CPE-based colorimetric live virus micro-neutralization assay in human serum samples. J. Med. Virol. 2020;92:2096–2104. doi: 10.1002/jmv.25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan M.J., Lyke K.E., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Raabe V., Bailey R., Swanson K.A., Li P., Koury K., Kalina W., Cooper D., Fontes-Garfias C., Shi P.-Y., Türeci Ö., Tompkins K.R., Walsh E.E., Frenck R., Falsey A.R., Dormitzer P.R., Gruber W.C., Sahin U., Jansen K.U. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586:589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- Muruato A.E., Fontes-Garfias C.R., Ren P., Garcia-Blanco M.A., Menachery V.D., Xie X., Shi P.-Y. A high-throughput neutralizing antibody assay for COVID-19 diagnosis and vaccine evaluation. Nat. Commun. 2020;11:4059. doi: 10.1038/s41467-020-17892-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera R.A.P.M., Ko R., Tsang O.T.Y., Hui D.S.C., Kwan M.Y.M., Brackman C.J., To EMW, Yen H.-L., Keung K., Chen S.M.S., Chan K.H., Chan K.C.K., Li K.-C., Saif L., Barrs V.R., Wu J.T., Sit T.H.C., Poon L.L.M., Peiris M. Evaluation of a SARS-CoV-2 surrogate virus neutralization test for detection of antibody in human, canine, cat and hamster sera. J Clin Microbiol. 2020 doi: 10.1128/JCM.02504-20. Published online November 2, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., Mirchandani D., Scharton D., Bilello J.P., Ku Z., An Z., Kalveram B., Freiberg A.N., Menachery V.D., Xie X., Plante K.S., Weaver S.C., Shi P.-Y. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2020 doi: 10.1038/s41586-020-2895-3. Published online October 26, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince H.E., Givens T.S., Lape-Nixon M., Clarke N.J., Schwab N.J., Batterman H.J., Jones R.S., Meyer W.A., III, Kapoor H., Rowland C.M., Haji-Sheikhi F., Marlowe E.M. Detection of SARS-CoV-2 IgG targeting nucleocapsid or spike protein by four high-throughput immunoassays authorized for emergency use. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.01742-20. e01742–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U., Muik A., Derhovanessian E., Vogler V., Kranz L.M., Vormehr M., Baum A., Pascal K., Quandt J., Maurus D., Brachtendorf S., Lörks V., Sikorski J., Hilker R., Becker D., Eller A.-K., Grützner J., Boesler C., Rosenbaum C., Kühnle M.-C., Luxemburger U., Kemmer-Brück A., Langer D., Bexon M., Bolte S., Karikó K., Palanche T., Fischer B., Schultz A., Shi P.-Y., Fontes-Garfias C., Perez J.L., Swanson K.A., Loschko J., Scully I.L., Cutler M., Kalina W., Kyratsous C.A., Cooper D., Dormitzer P.R., Jansen K.U., Türeci Ö. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C.W., Chia W.N., Qin X., Liu P., Chen M.I., Tiu C., Hu Z., Chen V.C., Young B.E., Sia W.R., Tan Y.-J., Foo R., Yi Y., Lye D.C., Anderson D.E., Wang L.-F. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2–spike protein–protein interaction. Nat. Biotechnol. 2020;38:1073–1078. doi: 10.1038/s41587-020-0631-z. [DOI] [PubMed] [Google Scholar]
- Walsh E.E., Frenck R.W., Falsey A.R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M.J., Bailey R., Swanson K.A., Li P., Koury K., Kalina W., Cooper D., Fontes-Garfias C., Shi P.-Y., Türeci Ö., Tompkins K.R., Lyke K.E., Raabe V., Dormitzer P.R., Jansen K.U., Şahin U., Gruber W.C. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N Engl J Med. Published online October. 2020;14:2020. doi: 10.1056/nejmoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman J.D., Hiatt J., Mowery C.T., Shy B.R., Yu R., Yamamoto T.N., Rathore U., Goldgof G.M., Whitty C., Woo J.M., Gallman A.E., Miller T.E., Levine A.G., Nguyen D.N., Bapat S.P., Balcerek J., Bylsma S.A., Lyons A.M., Li S., Wong A.W., Gillis-Buck E.M., Steinhart Z.B., Lee Y., Apathy R., Lipke M.J., Smith J.A., Zheng T., Boothby I.C., Isaza E., Chan J., Acenas D.D., II, Lee J., Macrae T.A., Kyaw T.S., Wu D., Ng D.L., Gu W., York V.A., Eskandarian H.A., Callaway P.C., Warrier L., Moreno M.E., Levan J., Torres L., Farrington L.A., Loudermilk R.P., Koshal K., Zorn K.C., Garcia-Beltran W.F., Yang D., Astudillo M.G., Bernstein B.E., Gelfand J.A., Ryan E.T., Charles R.C., John Iafrate A., Lennerz J.K., Miller S., Chiu C.Y., Stramer S.L., Wilson M.R., Manglik A., Ye C.J., Krogan N.J., Anderson S.M., Cyster J.G., Ernst J.D., Wu A.H.B., Lynch K.L., Bern C., Hsu P.D., Marson A. Evaluation of SARS-CoV-2 serology assays reveals a range of test performance. Nat. Biotechnol. 2020;38:1174–1183. doi: 10.1038/s41587-020-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Muruato A.E., Zhang X., Lokugamage K.G., Fontes-Garfias C.R., Zou J., Liu J., Ren P., Balakrishnan M., Cihlar T., Tsen C.-T.K., Makino S., Menachery V.D., Bilello J.P., Shi P.-Y. A nanoluciferase SARS-CoV-2 for rapid neutralization testing and screening of anti-infective drugs for COVID-19. Nat. Commun. 2020;11:5214. doi: 10.1038/s41467-020-19055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Muruato A., Lokugamage K.G., Narayanan K., Zhang X., Zou J., Liu J., Schindewolf C., Bopp N.E., Aguilar P.V., Plante K.S., Weaver S.C., Makino S., LeDuc J.W., Menachery V.D., Shi P.-Y. An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe. 2020;27:841–848. doi: 10.1016/j.chom.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H.L., Wu Y.T., Cao J.-L., Yang R., Liu Y.-X., Ma J., Qiao X.-Y., Yao X.-Y., Zhang B.-H., Zhang Y.-L., Hou W.-H., Shi Y., Xu J.-J., Zhang L., Wang S.-J., Fu B.-R., Yang T., Ge S.-X., Zhang J., Yuan Q., Huang B.-Y., Li Z.-Y., Zhang T.-Y., Xia N.-S. Robust neutralization assay based on SARS-CoV-2 S-protein-bearing vesicular stomatitis virus (VSV) pseudovirus and ACE2-overexpressing BHK21 cells. Emerg Microbes Infect. 2020;9:2105–2113. doi: 10.1080/22221751.2020.1815589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Huang B., Li W., Wang W., Deng Y., Tan W. Development and effectiveness of pseudotyped SARS-CoV-2 system as determined by neutralizing efficiency and entry inhibition test in vitro. Biosaf Heal. 2020 doi: 10.1016/j.bsheal.2020.08.004. Published online August 21, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zettl F., Meister T.L., Vollmer T., Fischer B., Steinmann J., Krawczyk A., V’kovski P., Todt D., Steinmann E., Pfaender S., Zimmer G. Rapid quantification of SARS-CoV-2-neutralizing antibodies using propagation-defective vesicular stomatitis virus pseudotypes. Vaccines (Basel) 2020;8:386. doi: 10.3390/vaccines8030386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Jackson C.B., Mou H., Ojha A., Peng H., Quinlan B.D., Rangarajan E.S., Pan A., Vanderheiden A., Suthar M.S., Li W., Izard T., Rader C., Farzan M., Choe H. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat Commun. 2020;11:6013. doi: 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/nejmoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]