Abstract
Exosomal microRNAs (miRNAs) critically regulate several major intracellular and metabolic activities, including cancer evolution. Currently, increasing evidence indicates that exosome harbor and transport these miRNAs from donor cells to neighboring and distantly related recipient cells, often in a cross-species manner. Several studies have reported that plant-based miRNAs can be absorbed into the serum of humans, where they hinder the expression of human disease-related genes. Moreover, few recent studies have demonstrated the role of these xenomiRs in cancer development and progression. However, the cross-kingdom gene regulation hypothesis remains highly debatable, and many follow up studies fail to reproduce the same. There are reports that show no effect of plant-derived miRNAs on mammalian cancers. The foremost cause of this controversy remains the lack of reproducibility of the results. Here, we reassess the latest developments in the field of cross-kingdom transference of miRNAs, emphasizing on the role of the diet-based xenomiRs on cancer progression.
Keywords: miRNA, Exosomes, Cross-kingdom gene regulation, Cancer
1. Cancer: A multifaceted disease
Cancer is a heterogeneous group of diseases and has become a prominent reason of death worldwide (Fitzmaurice et al., 2017). The global cancer burden amounts to 14.1 million worldwide, while ~ 9.6 million deaths occurred in 2018 due to cancer (WHO 2018). The number of expected cancer cases may reach 23.6 million by 2030 (Si et al., 2019). Cancer is a complex pathological condition characterized by altered gene expression, aberrant genetic mutations and genomic instability. Moreover, the tumor microenvironment (TME), comprising of cellular and non-cellular components of the tumoral niche, plays a crucial role in cancer development as well as metastasis (Wang et al., 2017a, Sun et al., 2018). Accumulating evidence suggests that TME greatly influences the metabolic rewiring of cancer cells via extracellular vehicles (EVs) (Sullivan et al., 2017).
EVs are spherical, bilayered, heterogeneous cell derived membranous organelles (from normal/cancerous cells) that package cellular material; thereby influencing neighboring as well as remote cells with its payload (Colombo et al., 2014). The components of the cargo varies highly and depends on the kind of cell it originated from. Cancer cells are known to secrete many-fold vesicles than normal dividing cells (Akers et al., 2013, Mao et al., 2018). Studies suggests that their content plays a critical function as the crosstalk between constituent cells of the TME through the transport of specific cargo molecules, such as growth factors, chemokines, and microRNAs (miRNAs) (Kahlert and Kalluri, 2013, Benmoussa et al., 2017). EVs secreted from cancer cells can remodel the microenvironment and promote cancer progression, metastasis, and drug resistance (Xie et al., 2019, Chiarugi and Cirri, 2016, Tkach and Thery, 2016).
Recent evidences have shown that EVs transport miRNAs that act as messengers between tumoral and stromal cells (Donnarumma et al., 2017). This interaction can affect the TME, influencing the extracellular matrix (ECM), epithelial-mesenchymal transition, angiogenesis, and reprograming of immune function (Ingenito et al., 2019). Altered miRNA expression in TME favors the acquisition of cancer hallmarks, leading to the development and progression of tumors. (Soon and Kiaris, 2013, Rupaimoole et al., 2016, Kuninty et al., 2016, Fanini and Fabbri, 2017, Curtale, 2018, Bandini et al., 2019).
miRNAs can circulate in the blood and may be absorbed by cells and tissues at distant sites (Hasegawa et al., 2017). Plant-based exosomal miRNA (which protects RNase) was shown to be delivered to humans in a cross-kingdom fashion (Zhang et al., 2012a), where they can mediate cancer metastasis (Zhou et al., 2014a, Zhou et al., 2014b, Zhou et al., 2014c). Further studies showed that plant exomiRs may regulate the expression of cancer-related genes and proteins (Pastrello et al., 2016, Chin et al., 2016). In this article, we have summarized and reviewed cross-kingdom gene regulation by plant-derived exomiRs with special emphasis on cancer progression.
2. Extracellular vesicles — Connecting kingdoms
Currently, EVs are known to play a pivotal function in diverse cell-to-cell signaling processes. Multivesicular bodies (MVBs), later revealed to be exosomes, were documented in the 1950s, first in algae (Sager and Palade, 1957) and later in mammalian cells (Sotelo et al., 1959). Simultaneously, outer membrane vesicles (OMVs) have been discovered in bacteria (De, 1959, Chatterjee et al., 1959). Soon after, MVBs were found in higher plants (Jensen, 1965) as well as in fungi (Takeo et al., 1973). At that time, the researchers could not realize the significance of this discovery. Two decades later, exosomes were found to function as garbage-dumping devices. It was assumed that their function was to simply “defenestrate” remnants instead of its degradation (Harding et al., 1983, Pan et al., 1985, Johnstone et al., 1987, Thery, 2011). This notion of waste disposal remained until 1996 when Raposo et al. (1996) discovered that EVs may influence antigen presentation in vivo. Since then, tremendous research on EVs has suggested their importance in cellular communication.
Presently, three different types of EVs are widely accepted according to the consensus nomenclature (Han et al 2019) (Table 1): apoptotic bodies, microvesicles, and exosomes (Fuhrmann et al., 2017, Mathieu et al., 2019). Apoptotic bodies (ApoBDs), the largest EVs (with a diameter of 1–5 μm), are discharged as membrane-surrounded fragments during apoptosis. ApoBDs can carry protein, lipid, DNA, RNA as well as cell organelles (van Niel et al., 2018). Microvesicles (ectosomes) are larger in size (0.1–1.0 μm) than exosomes, originating from outward budding and fission of the plasma membrane (Thery, 2011, Rutter and Innes, 2016, Zempleni, 2017, Raposo and Stoorvogel, 2013). Oncosomes are 100–400 nm, specialized vehicles transporting abnormal proteins that play crucial role in vascular growth, cancer evolution and metastasis (Minciacchi et al., 2015, Jaiswal and Sedger, 2019). Exosomes, the smallest (30–100 nm) among EVs, are membranous vesicles secreted by a plethora of cell types, including cancer cells; and are released extracellularly via fusion of MVBs with the cell membrane (Hessvik et al., 2013). These secreted EVs has been detected in virtually every kind of body fluids; such as blood (serum and plasma) (Caby et al., 2005, McDonald et al., 2013), seminal fluid (Hoog and Lotvall, 2015), urine (Pisitkun et al., 2004), saliva (Machida et al., 2015, Han et al., 2018), ascites (Keller et al., 2011), breast milk (Lasser et al., 2011, Wang, 2017), cerebrospinal fluid (Bachy et al., 2008), and amniotic fluid (Keller et al., 2011, Ebert and Rai, 2019).
Table 1.
Major subtypes of EVs and their representative features (Modified from Han et al 2019, Open Access article distributed under the terms of the Creative Commons Attribution 4.0 International License).
| EV subtype | Exosomes | Microvesicles (MVs)/ ectosomes/microparticles | Apoptotic bodies (ABs) |
|---|---|---|---|
| Subcellular origin | Multivesicular bodies (MVBs)/ endosomes | Plasma membrane | Apoptotic blebs |
| Regular diameter | 30–120 nm | 100–1000 nm | 0.8–5.0 μm |
| Sedimentation force | 100,000–120,000 × g | 10,000 g | 2000 g |
| Types of generation | Constitutive | Regulated | Regulated |
| Filtration | 20–200 nm | >200 nm | >1000 nm |
| Intracellular storage | Yes | No | No |
| Biogenetic mechanisms | Rab proteins (i.e. Rab7, Rab11, Rab27A/B, Rab35), NSMase, ESCRTs, syndecan, syntenin, ATG12, tetraspanins | ASMase, flippase, flippase and scramblase (TMEM16F), ARF6 | Annexin V, Caspase 3 |
| Organelles | No | No | Yes |
| Content | Proteins, cholesterol, ceramide, noncoding RNA, mRNA, miRNA, and cytosol | Proteins, phosphatidylserine, cholesterol, mRNA, miRNA, and cytosol | Proteins, phosphatidylserine, DNA, rRNA, and cytosol |
| Marker proteins | CD 9, CD63 and CD61, tetraspanins, HSP70, HSP90, Alix, Rab5a/b | TyA and C1a, ARF6 and VCAMP3, β1 integrins, selectins, CD40, MMP, lineage markers, and ezrin | Calreticulin, TSP and C3b, and histones |
| Impact on the immune system | Immunostimulators | Immunosuppressors | Immunosuppressors |
Exosomes packed with donor tumor cell-derived biologically-active oncogenic proteins and nucleic acids can affect the cellular functions of nearby and remote normal recipient cells (Whiteside 2016). Accumulating evidence reveals that tumor cell-derived exosomes have become a central candidate for promoting tumor cell proliferation, invasion, angiogenesis, distant metastasis and remodeling of the TME through transmitting onco-miRNAs (Rahbarghazi et al., 2019). These exosome-derived miRNAs can regulate intercellular signaling pathways (Tan et al., 2020); inducing functional alterations that impact on tumor progression and metastasis (Whiteside 2016). It has been demonstrated that tumor cell-released exosomal miRNAs such as miR-9, miR-23a, miR-92a, miR-103, miR-105, miR-126, miR-132 miR-135b, miR-210, miR-221 and cytokines (e.g., interleukins: IL-6 and IL-8, TNF-α, transforming growth factor β, FGF2, and VEGF) are proangiogenic factors to promote neovascularization and metastasis (Umezu et al., 2014, Zhou et al., 2014a, Zhou et al., 2014b, Zhou et al., 2014c, Martinelli, 2017, Mentkowski et al., 2018, Ma et al., 2018, Fortunato et al., 2019, Wortzel et al., 2019). It has also been suggested that exosomal miRNAs play a key role in surpassing endothelial cells, cells in the stroma and blood–brain barrier, important in the development of brain metastases (Tominaga et al., 2015, Zhou et al., 2014a, Zhou et al., 2014b, Zhou et al., 2014c). Considering these findings, exo-miRNAs might play an key function during the transformation of normal cells into malignant cells.
In comparison to intercellular communication displayed by low molecular weight compounds (e.g., hormones), EVs transport and deliver their payload in a highly concentrated form, unaltered by diffusion or dilution (Wolf and Casadevall, 2014). Exchange of information can be done either by intake of EV by receptor-mediated internalization, endocytosis, docking, and fusion to target cells or interaction through cell surface receptor (Van Niel et al., 2018, Malloci et al., 2019). Since EVs ferry nucleic acids, they participate in post-transcriptional gene regulation in the target cell.
3. Small RNAs — Key regulators in inter-kingdom communication
In 2007, researchers discovered that EVs can transport messenger RNA (mRNA) and small non-coding RNA (sRNA), allowing cells to trade genetic message; thus, renewing the understanding of cell–cell interaction (Valadi et al., 2007, Simons and Raposo, 2009, Vlassov et al., 2012, Otsuka et al., 2018). sRNAs (~22 nucleotides single-stranded RNA), including small interfering RNAs (siRNAs) and microRNAs (miRNAs), are crucial managers of several intracellular activities. The sRNA binds to its complementary mRNA, which results in post-transcriptional gene-silencing (Bartel, 2004). These sRNAs can influence several biological processes, including cell growth, differentiation, metabolism, and apoptosis (Bartel, 2004). Thus, sRNAs may act as key regulatory elements in EV-mediated (inter-kingdom) interactions (Record, 2013).
Two most critical parameters of bioavailability of sRNA are its stability and absorption. The vesicular envelope of EVs protect miRNAs from degradation by extracellular RNases enabling them to be more stable than free miRNAs and to be efficiently integrated by specific recipient cells (Kosaka et al., 2010a, Kosaka et al., 2010b, Zhang et al., 2010, Koga et al., 2011, Keller et al., 2011, Buck et al., 2014, Zempleni et al., 2015, Vautrot et al., 2019). This can be evident from the fact that processes such as ultrasonication, pasteurization and homogenization that disrupt the vesicular integrity of the exosomes, greatly reduces detectable levels of sRNA (Zempleni et al., 2017, Baier et al., 2014, Howard et al., 2015).
4. miRNA – A small RNA with diverse functions
miRNAs are endogenous, highly conserved, non-coding single stranded RNAs consisting of ~ 20–25 nucleotides that suppress translation or stimulate target mRNA degradation (Bartel, 2004). MiRNA was initially unearthed in Caenorhabditis elegans (C. elegans) by Ambros and coworkers who named it as lin-4 (Lee et al., 1993). In 2000, another miRNA (termed let-7) was detected in C. elegans (Reinhart et al., 2000). Subsequently, miRNAs have been isolated from almost all eukaryotes. Some miRNAs are highly conserved and highlight their evolutionarily functional relevance across species in post-transcriptional gene silencing. (Lagos-Quintana et al., 2001, Lau et al., 2001). Today, the expression of approximately 30–60% of protein-coding mRNAs is estimated to be controlled by miRNAs (Berezikov et al., 2005, Boffelli et al., 2003, Lewis et al., 2005, John et al., 2004).
4.1. miRNA biogenesis
The biogenesis of miRNAs is complex (Breving and Esquela-Kerscher, 2012). miRNAs are transcribed in the nucleus by RNA polymerase II (although Pol III transcription has also been observed) to generate a precursor, primary (pri)-miRNA, which are often 5′ capped and 3′ polyadenylated. A pri-miRNA transcript can encode multiple miRNA genes, and each gene is processed into a ~70-nucleotide pre-miRNA hairpin precursor by the RNase III enzyme Drosha and its cofactor, DiGeorge syndrome critical region 8 (DGCR8). An independent subclass of pre-miRNAs, termed “mirtrons,” do not rely on Drosha processing and are generated from mRNA transcripts as by-products of exon splicing and intron disbranching phenomena (Chan and Slack, 2007).
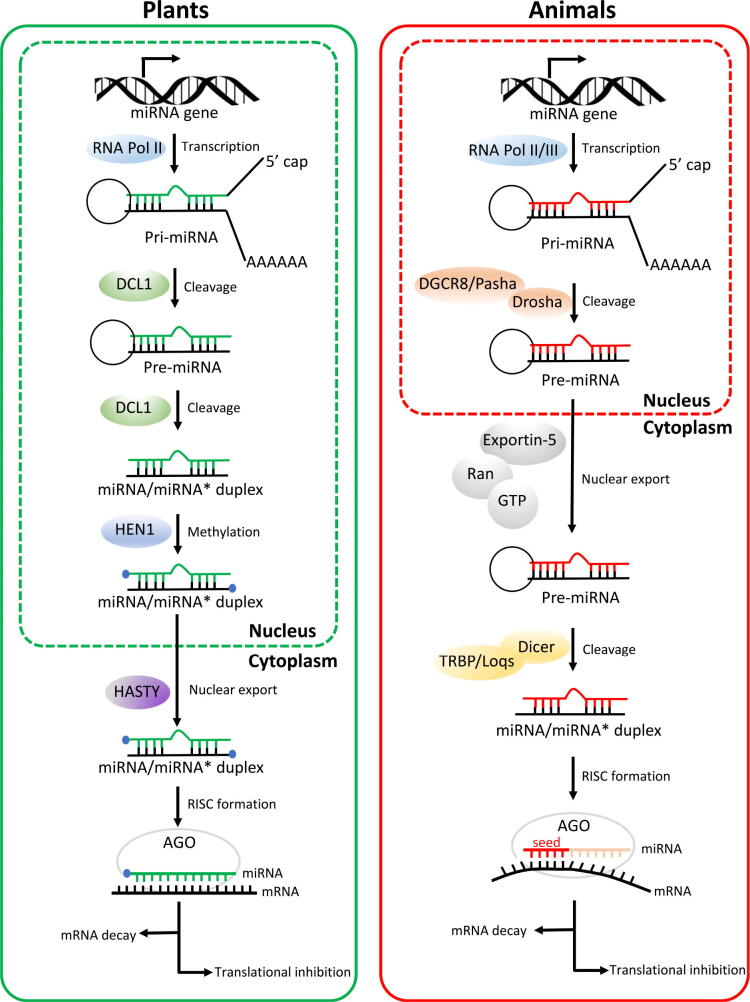
Pre-miRNAs are subsequently exported out of the nucleus by RAN-GTP and Exportin. Once in the cytoplasm, pre-miRNAs are processed further by the RNase III enzyme, Dicer, and its co-factor, TAR RNA-binding protein (TRBP) to generate a ~ 22-nucleotide double-stranded RNA duplex. One strand of the duplex attaches with a large multiprotein miRNA-associated RNA-induced silencing complex (miRISC). Argonaute (AGO) is the key catalytic component of this complex responsible for miRNA strand selection and mediating miRNA-based alterations of gene expression (Peng and Croce, 2016). The associated miRNA escorts the AGO/miRISC complex to the targeted site via miRNA binding to the complementary sequences within the mRNA transcript, and ultimately results in target mRNA degradation and/or blocked protein translation (Fig. 1).
Fig. 1.
Comparison of miRNA biogenesis and activity pathways in plants and animals. In both plants and animals, the biogenesis of miRNAs initiates within the nucleus. In plants, miRNA/miRNA* duplexes are cleaved from pri-miRNAs through the action of DCL1 endonuclease in two steps. DCL1 firstly cuts off the imperfectly folded ends of pri-miRNAs to generate pre-miRNAs with stem-loop hairpin secondary structures. The resulting pre-miRNAs are further excised by DCL1 to mature miRNA/miRNA* duplexes. Then the 3′-terminal of duplexes is methylated by HEN1. By contrast, in animals, pre-miRNAs are produced in the nucleus by the action of the Drosha enzyme, together with its DGCR8 protein (in mammals) or Pasha protein (in flies). Duplexes of miRNA/miRNA* are further processed after being exported from the nucleus to the cytoplasm, where pre-miRNAs are cleaved by Dicer and TRBP (in mammals) or Loqs (in flies). In plants, HASTY is responsible for the transport of miRNA/miRNA* duplexes from the nucleus to the cytoplasm, whereas in animals’ pre-miRNAs are recognized and then exported by Exportin-5 in a Ran-GTP-dependent manner. During RISC loading, one strand of the small RNA duplexes is selected as the guide strand (green in plants or red in animals) and incorporated into AGO to form a functional RISC, whereas the other strand is removed and degraded. In plants, miRNAs have near-perfect complementarity to their target mRNAs. By contrast, animal miRNAs often have targets with imperfect complementarity and the major determinant for the binding of animal miRNAs to their target mRNAs is a 6–8 nucleotide domain at the 5′ extremity or seed sequence. The arrows indicate the direction of the subsequent activity pathways. Both plant and animal miRNAs can regulate gene expression via mRNA decay and translational inhibition (Adapted from Li et al., 2018, an Open access article distributed under the terms of the Creative Commons Attribution 4.0 International License).
Human miRNAs binds to the 3′ untranslated regions (3′ UTR) of the target mRNA with imperfect complementarity; although miRNA binding within the 5′ UTR and coding regions of the mRNA target can also modulate gene expression. Perfect base pairing of a highly conserved “seed sequence” (nucleotides 2–8) in the miRNA is important for proper miRNA targeting (Doench and Sharp, 2004). A few notable exceptions exist where miRNAs activate target gene expression via epigenetic regulation of the enhancer regions in the nucleus or post-transcriptionally to induce protein translation (Valinezhad Orang et al., 2014, Xiao et al., 2017). Evidence show that a single miRNA can recognize up to 100 well-defined targets (Brennecke et al 2005), and therefore can regulate multiple non-overlapping biological pathways simultaneously. From a therapeutic standpoint, single miRNA therapy could be a powerful tool to treat cancer in patients that harbor an accumulation of genetic alterations and can be effective without identifying the key mutations that lead to tumor formation.
MiRNA gene silencing machinery is highly conserved in most classes of eukaryotes; however substantial differences have been identified between the plant and animal counterparts at the level of the biogenesis, maturation, and mode of action (Achkar et al., 2016). In plants, a ribonuclease III enzyme, Dicer-like 1 (DCL1), plays major role in processing miRNA (Fukudome et al., 2017). This mechanism of miRNA processing can be understood by dividing this process in two distinct steps. In the first step, DCL1 along with its accessory proteins, SERRATE (SE) and HYPONASTC LEAVES1 (HYL1), forms a ‘dicing complex’ inside the nucleus (Fang and Spector 2007), which catalyzes the conversion of pri-miRNA to pre-miRNA by cleaving the imperfectly-folded end of the pri-miRNA to create a stem-loop hairpin secondary structure of pre-miRNA (Dong et al 2008). In the second step, pre-miRNA is expurgated by DCL1 generating a nascent miRNA/miRNA* duplex (Wang et al., 2019). This second step also takes place in the nucleus at the nuclear processing centre termed as dicing bodies (D-bodies) (Fang and Spector, 2007, Song et al., 2007). In the animal counterparts, maturation of pri-miRNA into a mature miRNA involves two different proteins namely ‘Drosha’ and ‘Dicer’ which sequentially carry out the process in two different compartments of the cell i.e. nucleus and cytoplasm (O’Brien et al., 2018).
Unlike animals, nascent miRNA/miRNA* duplex generated in the nucleus of plant cells are universally methylated at their 3′ ends before being exported to the cytoplasm. Both the strands of nascent miRNA/miRNA* have 3′ nucleotide overhangs, phosphate group at 5′ ends, and two hydroxyl groups at the 3′ ends (2′-OH and 3′-OH). The 2′-hydroxyl group at 3′ end of nascent miRNA/miRNA* is methylated by HUA Enhancer (HEN1) which has been identified as a small RNA methyltransferase (Singh et al., 2018). This step prevents uridylation and degradation of the methylated miRNA/miRNA* (Li et al., 2005). Export of methylated miRNA/miRNA* duplex from nucleus to cytoplasm is performed by ‘Hasty’ (HST) protein which is a homologue of animal Exportin 5 (EXPO 5) (Wahid et al., 2010). Inside cytoplasm, guide strand of matured duplex is loaded onto Argonaute (AGO) protein and the complex so formed targets specific genes through base-pairing, in-turn mediating gene-silencing (Wang and Xu, 2015).
4.2. miRNA and cancer
The association between miRNA and cancer was demonstrated by Dr. Croce and colleagues while trying to recognize the tumor suppressor genes in the chromosomal region 13q14 in B-cell chronic lymphocytic leukemia (BLL) cells (Calin et al., 2002). They found that this region, often deleted or downregulated in BLL, codes for two miRNAs, namely miR-15a and miR-16–1. Further studies revealed the role of miR-15 and miR-16–1 as tumor-suppressor genes that repress Bcl-2, an anti-apoptotic protein known to overexpress in malignant non-dividing B cells, and several solid tumors (Cimmino et al., 2005, Calin et al., 2008). In later years, miRNA profiling and deep sequencing shed some direct evidence on relationship between miRNA expression and cancer dysregulation, providing more focus on its usage in tumor classification, diagnosis, and prognosis.
miRNAs extensively impact the progression of many human cancers (Calin and Croce, 2006a, Calin and Croce, 2006b, Berindan-Neagoe et al., 2014) acting both as tumor suppressors (Fang et al., 2012, Hatziapostolou et al., 2011) and promoters (Mi et al., 2017, Zhu et al., 2018). Abnormal upregulation or downregulation of miRNAs has been shown to play a major role in tumor proliferation, epithelial-mesenchymal transition (EMT), angiogenesis, and metastasis.
5. Cross-species sRNA transfer
Experimentations have unveiled that sRNAs are not confined to the cells, which produce them but exhibit inter-species/inter-kingdom communication within an organism or between hosts and pathogens, such as parasites (Choy et al., 2008, Sarkies and Miska, 2014, Colombo et al., 2014, Islam et al., 2017, Knip et al., 2014). Several studies have demonstrated inter-species translocation of sRNA molecules (Xia et al., 2008, Hansen et al., 2010, Knip et al., 2014, Philip et al., 2015). This makes the cross-species transfer of sRNAs a novel form of intra-species and inter-species cross-talk (Knip et al., 2014).
miRNAs have often been reported to be transferred across species, regulating key processess (Table 2). Several reports identify the unique phenomenon of transfer of miRNA from parasites into their vector and host species. Garcia-Silva et al., (2014) demonstrated the channelization of tRNA derived small RNAs (tsRNAs) along with distinctive proteins, to their mammalian host cells packaged as vesicular structures resembling Golgi complex in parasitic protozoan Trypanosoma cruzi. Another study shows miRNAs produced in Schistosoma mansoni, a blood fluke, to exist very stably in the tissues and body fluids of their host like mice and humans and being used as a bio-marker for the disease (Hoy et al., 2014). According to Buck et al., (2014), Heligmosomoides polygyrus, a gastrointestinal parasitic nematode, can manipulate the innate immunity of their hosts by secretion of exosomal vesicles containing miRNAs and parasite derived AGO proteins, thereby elucidating a novel mechanism of RNA transfer between species.
Table 2.
Exogenous miRNA studies of cross-kingdom regulation (Adapted with permission from Zhang et al 2019).
| miRNA | Source | Target cell or animal | Target gene | Function | N/P | Reference |
|---|---|---|---|---|---|---|
| miR-451 | Human | Plasmodium falciparum | PKA-R | Inhibits parasite growth | P | Choy et al., 2008 |
| miR-BART5 | Epstein-Barr virus | Human NPC cells | PUMA | Facilitates the establishment of latent infection by promoting host cell survival | P | Xia et al., 2008 |
| miR-K12-6, miR-K12-11 | Kaposi sarcoma herpesvirus | Lymphatic endothelial cells/blood vessel endothelial cells | MAF | Influences the differentiation status of infected cells and contribute to KSHV-induced oncogenesis | P | Hansen et al., 2010 |
| miR-BHRF 1–3 | Epstein-Barr virus | Two BL early-passage cell lines (BL-5 and BL-8) | CXCL-11 | Cancer immunosurveillance | P | Snow et al., 2013 |
| miR-172 | Brassica oleracea | Mice | — | Absorbed | P | Liang et al., 2014, Hou et al., 2018) |
| mol-miR168a | Moringa oleifera | Human hepatoma cell line G2 (HEPG2) cells | SIRT1 | Absorbed | P | (Pirrò et al., 2016, LaMonte et al., 2012 |
| miR166a | Arabidopsis thaliana | Human breast milk exosomes | — | Absorbed | P | Lukasik & Zielenkiewicz 2014 |
| miR159 | Arabidopsis thaliana | Mice | TCF7 | Suppresses the growth of xenograft breast tumors | P | Chin et al., 2016, Luo et al., 2017 |
| miR162a | Bee pollen | Drosophila | dmTOR | Delays the development and decreases the body and ovary size in bees | P | Zhu et al., 2017 |
| zma-miR164a-5p | Maize | Pigs | OTX1, PLAGL2, CSPG4 | Absorbed | Luo et al., 2017, Philip et al., 2015 |
Alternatively, it has been seen that miRNAs may be transferred from host cells into the parasites as a means of building resistance against the pathogen. The anomalous production of two miRNAs (miR-451 and let7i) in erythrocytes having alleles for sickle-cell disease, led to impairment of translational machinery inside Plasmodium falciparum, thereby disturbing the intra-erythrocytic life-cycle of the parasite and providing resistance against the disease (LaMonte et al., 2012). This non-canonical RNAi mechanism act most likely by forming chimeric fusions miRNA transfer from host to its parasite and may act as a mechanism of selection and regulation of parasitic population. The gut-microbiota consisting of various species of bacteria has also been shown to be controlled by the host. This is done through host miRNA mediated regulation of bacterial genes resulting in their growth perturbations (Liu et al., 2016). Altogether, these reports suggest that cross-species/cross-kingdom transfer of miRNA is not uncommon.
6. Plants and bidirectional cross-kingdom sRNA transport
Plants also exhibit cross-kingdom transfer of double-stranded siRNAs (Pirrò et al., 2016, Hou et al., 2018). This strategy known as plant-mediated RNA interference (PM-RNAi), is used by plants to suppress the growth of harmful insects. A study by Baum et al. (2005) found out that the ingestion of plant dsRNAs, through feeding or topical application, can lead to larval stunting and morbidity in several coleopteran insects. They suggested that the RNAi pathway can be exploited to control insect pests via in-planta expression of a particular dsRNA. Similarly, plants like cotton have been shown to export plant-specific miRNAs into their fungal pathogens to induce cross-kingdom gene silencing and confer disease resistance. This study conducted by Zhang et al. (2016), showed that two miRNAs (miR166 and miR159) produced in cotton plants are exported to the hyphae of Verticillium dahliae (pathogenic fungus causing wilt disease in crops). These miRNAs specifically target two V. dahliae genes viz. a Ca2+-dependent cysteine protease (Clp-1) and an isotrichodermin C-15 hydroxylase (HiC-15) that are essential for fungal virulence.
A reveling study by Weiberg et al (2013) exemplifies cross-kingdom RNAi as a virulence mechanism of Botrytis cinerea, a fungal pathogen responsible for gray mould disease in many plant species. sRNAs of this pathogen (Bc-sRNAs) vandalize host’s immune response by selectively paralysing the host RNAi machinery and building infection. Compared to the sRNA transfer from pathogen to host plants, the functional movement of sRNAs from parasitic plants to host plants has rarely been reported. However, recent cases of miRNA movements, where double-stranded small RNA molecules (~22 nucleotide long) move into host plants from their parasitic-plant counterparts like Cuscuta campestris, have revealed robust roles of miRNA species in host-parasite interaction and regulation through siRNA mediated target gene silencing (Shahid et al., 2018).
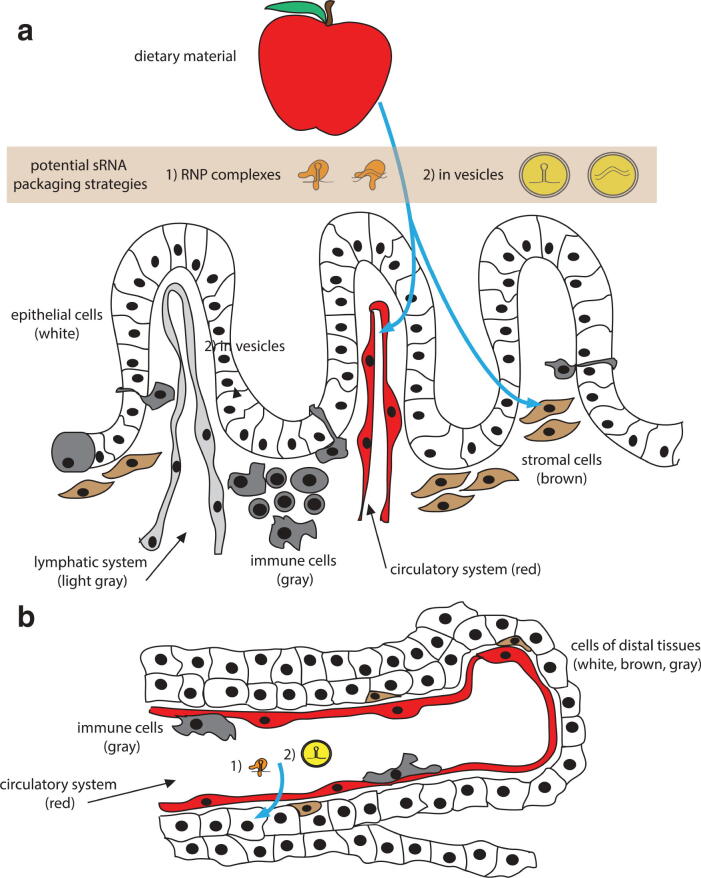
A highly debatable and unresolved issue is whether a cross-kingdom transmission of miRNA from plants to humans can occur. Growing experimental evidence has suggested that the regular usage of plant-based food substances is beneficial against metabolic disease and cancer (Willett, 2002, Liu, 2004). Since daily food intake of humans is considerable, it is intriguing to speculate that exogenous plant-based miRNA (termed xenomiRs) can affect human health and disease pathology (Motti and Meccariello, 2018). Foods, including vegetables, fruits and whole grains, have been shown to contain miRNAs that can survive digestion, translocate into the human blood, and exhibit vital role in regulating gene expression in a cross-kingdom fashion (Witwer, 2012, Chin et al., 2016, Zhang et al., 2012a, Zhou et al., 2015) (Fig. 2). This natural uptake of diet-derived xenomiRs is reported to be biologically active in vast variety of lower eukaryotes or invertebrates (Li et al., 2018). Recent studies has proven that xenomiRs can be effectively packaged into exosomal-like nanoparticles that provides resistance against harsh conditions (RNase and extreme pH) and can be relocated into adjacent/distant cells where they regulate gene function (Luo et al., 2017). However, the actual underlying mechanism still needs to be elucidated.
Fig. 2.
Model for the uptake of dietary sRNA from the digestive tract. To carry RNAi regulatory activity on gene expression in an ingesting organism, (a) sRNAs from the diet (potentially packaged in (1) ribonucleoprotein (RNP) complexes or (2) in vesicles) should cross the epithelial cell (white) barrier via transcellular or paracellular mechanisms or conveyance by immune cells (gray). They should then be absorbed by proximal cells, such as stromal cells (brown) or must gain access to the circulatory (red) or lymphatic system (light gray) for systemic dissemination. (b) Subsequently, after exiting from the circulatory system (red), uptake of sRNAs is ensued by the cells of various tissues and organs (gray, brown, and yellow). None of these putative steps is understood at the level of the molecular mechanism (Adapted and modified from Chan and Snow, 2017, Open Access article distributed under the terms of the Creative Commons Attribution 4.0 International License).
Several reports have suggested the prospect of cross-kingdom gene regulation by plant-derived miRNAs (Table 3). A breakthrough (yet controversial) study by Zhang et al. (2012a) presented high levels of plant-based miRNAs in human sera. They demonstrated that the miR-168a enriched in rice (Oryza sativa), may be absorbed through intestines, where it inhibits the expression of its target low-density lipoprotein receptor 1 (LDLRAP1), in liver (Zhang et al., 2012a). Researchers proposed that ingested plant-based miRNAs can be selectively loaded into microvesicles (exosomes), subsequently being absorbed by the gastrointestinal epithelial cells, which repackage them into EVs and unfetter them in blood. The study was challenged by Dickinson et al (2013) that showed no evidence of intake of ingested plant miRNAs by mice nor any reduction in LDLRAP1 levels in liver. To address this, Chen et al., (2013) proposed that the alteration in results could be due to the nutritional disparities in the diet constituents. They also argued that the sequencing strategy employed by Dickinson et al (2013) may not be able to accurately measure the plant miRNAs.
Table 3.
Supporting evidence of cross-kingdom communication by diet/plant-derived miRNAs (Adapted with permission from Li et al., 2018, an Open access article distributed under the terms of the Creative Commons Attribution 4.0 International License).
| Year | miRNAs involved | Contents | Source origin | miRNA levels | Potential mechanism | Function | Detection methods | Conclusion | Reference |
|---|---|---|---|---|---|---|---|---|---|
| 2012 | miR168a | Plant miRNAs were present in human and animal sera and organs. miR168a regulated mouse | Rice | fM Level | Associated with AGO2 complex and packaged in MVs | LDLRAP1 expression and consequently decreased LDL removal from mouse plasma. | HTS, RT-qPCR, Bioinformatics, NB, WB, AGO2 immuno-precipitation | Exogenous plant miRNAs in food could regulate the expression of target genes in mammals. | Zhang et al., 2012a |
| 2014 | miR172 | miR172 from cabbage (Brassica oleracea) was detected in blood, spleen, liver and kidney of mice after feeding with plant extract. | Cabbage | Stomach contained about 4.5–0.4% (2–24 h after feeding), intestines 2.4–0.2% (2–36 h), blood 1.3–0.2% (2–72 h) and spleen 0.38–0.04% (2–72 h) of the miR172 orally administered. | sRNA could survive for>36 h in blood and fecal samples | No phenotypic changes were found in all the mice fed with the foreign RNA. | RT-qPCR, Electrophoresis | Exogenous plant miRNAs could survive in the murine GI tract, enter peripheral blood and continue to access other organs. | Liang et al., 2014 |
| 2015 | 18 plant miRNAs (miR156a, miR157a, miR158a, etc.) | Plant miRNAs were detectable in human plasma of volunteers after drinking juice. | Watermelon juice and mixed fruits | fM Level | Largely encapsulated in MVs | Not mentioned | RT-qPCR, NB | Plant miRNAs in human plasma could be efficiently detected and reliably compared by RT-qPCR. Provided a SOP for measuring plant miRNAs in human and animal plasma. | Liang et al; 2015 |
| 2015 | miR166, miR167, miR168 | Even after an extensive pretreatment, plant-derived miRNA delivered by typical dietary ingestion remained bioavailable for uptake during early digestion. | Soybean and rice | In vitro methods | Not mentioned | Not mentioned | RT-qPCR | Storage, processing and cooking did not abolish plant miRNAs in food. | Philip et al., 2015 |
| 2015 | miR2911 | miR2911 was highly stable in honeysuckle decoction, and continuous drinking or gavage feeding of honeysuckle decoction significantly elevated miR2911 levels in mouse blood and lung. | Honeysuckle | fM Level | A unique sequence and high GC content, MVs mediated pathway | miR2911 could directly target multiple viral genes and suppress viral infections. | HTS, RT-qPCR, NB, Fluorescent labeled tracing assay, Luciferase reporter assay, Ago2 Immunoprecipitation | Provided evidence of physiological function of exogenous plant miRNAs in human and animals. | Zhou et al., 2015 |
| 2015 | miR2911, miR168a | Using a chow diet containing honeysuckle plant-based sRNAs could be detected in sera and urine of mice | Honeysuckle | fM Level | Consumers of particular diets and/or with increased intestinal permeability | Altered or damaged guts lining could enhance dietary miRNA uptake | RT-qPCR, droplet digital PCR | Dietary sRNAs could survive circulation and are excreted in urine | Yang et al., 2015a |
| 2015 | miR2911 | miR2911 was detectable in sera and urine of the honeysuckle decoction consuming mice. | Dried herbs or flowers | fM Level | Circulating miR2911 was not bound by AGO2, but due to high GC content. | Not mentioned | RT-qPCR, AGO2 immunoprecipitation | The uptake of miR2911 might be a more commonplace phenomenon that could occur when eating a variety of plant-based foods. | Yang et al., 2015b |
| 2016 | miR2911 | Plant-based miR2911 was measured 7 days after feeding in animals | Plants | fM Level | Circulating miR2911 was not associated with exosomes, but possibly with a protein | Not mentioned | RT-qPCR | Mice consuming diets rich in vegetables displayed enhanced serum levels of plant specific miR2911. | Yang et al., 2016 |
| 2017 | miR2911 | Plant-derived miR2911 was detectable in sera of mice fed with various vegetables. | Cabbage | Arabidopsis | miR2911 was detectable while other plant-based miRNAs failed to detect. Increased levels of miR2911 correlated with the degradation of plant foods and rRNAs. | Not mentioned | RT-qPCR, Dual-luciferase reporter assay, Bioinformatics | Provided insights into the atypical bioavailability of miR2911 and offered engineering strategies for plant-based sRNA therapeutics. | Yang et al., 2017 |
| 2015 | miR34a, miR143, miR145 | Orally administered tumor suppressor miRNAs reduced tumor burden in ApcMin/+ mice and were detectable in intestinal tissue | Synthesized methylated miRNAs | Intestinal miR34a was at a detectable level; detection of miR143 and miR145 in mouse intestines were failed | Not mentioned | Reduced tumor burden in the well-established ApcMin/+ mouse model of colon cancer. | RT-qPCR, Dissecting microscope | Tumor suppressor miRNAs designed to mimic sRNAs produced in plants were taken up by the digestive tract of ApcMin/+ mice upon ingestion. | Mlotshwa et al., 2015 |
| 2016 | miR159 | Plant miR159 could be detected in human sera and tumor tissues and was associated with breast cancer progression. | Synthesized methylated miRNAs | fM Level | Predominantly present in MVs | The miR159 in human serum was capable of inhibiting cell proliferation. | RT-qPCR, HTS, Dual-luciferase reporter assay, WB, In situ hybridization, Immunohistochemistry, | The feasibility of using synthetic forms of plant miRNAs as dietary supplements in the treatment of human cancers, including those outside of the GI track. | Chin et al., 2016 |
| 2016 | FvmiR168 | Strawberry fruit FvmiR168 affected properties of dendritic cells and their ability to respond to inflammatory stimuli | Strawberry fruit | biologically relevant amount | The immunomodulatory effect of plant miRNA was not sequence or plant specific | Plant-based miRNAs modified dendritic cells ability to respond to inflammatory agents by limiting T cell proliferation | RT-qPCR, Flow cytometry, Fluorescence microscopy | A potential for therapeutic use of plant miRNAs in the prevention of chronic inflammation related diseases | Cavalieri et al., 2016 |
| 2017 | miR451 miR144 | Ingestion of wild type blood increased the levels of miR451 and miR144 in peripheral blood of miR144/451-null mice | Wild type mice blood | At very low level but biologically relevant amount | Exosomes | Exogenous miR451 existing in miR144/451 knockout mice enhanced antioxidant activity in vivo via increasing the activity of Foxo3 pathway | Two different RT-qPCR, Dual-luciferase reporter assay, WB, FACS | miRNAs in foods or dietary supplements could affect the functions of the consumer | Wang et al., 2017b |
MVs microvesicles, HTS high-throughput sequencing; RT-qPCR quantitative real time polymerase chain reaction, NB Northern blot, WB Western blot, ELISA enzyme-linked immunosorbent assay, FACS Fluorescent
In another study, Liang et al. (2014) administered cruciferous miRNAs extracted from Brassica oleracea (a member of the cabbage family) in food or oral gavage to mice (Liang et al., 2014). B. oleracea miR-172 was found to be resistant to the gastrointestinal tract degradation, and this miRNA was recovered in the blood, stomach, intestines, spleen, and feces of treated animals up to 72 h after feeding. This study is particularly intriguing from the perspective of nutritional therapeutics because epidemiological data support the cancer-protective role of cruciferous vegetables such as broccoli (Higdon et al., 2007). Additional studies indicate that miRNAs from Zea mays (corn), Solanum lycopersicum (tomato), Vitis vinifera (grapes), and Glycine max (soybean) were found to be present in the blood of animals when they were fed on diets containing these plants (Wang et al., 2012, Yang et al., 2015a, Yang et al., 2015b).
Similarly, Zhou et al., (2015) showed that a plant-based miRNA from honeysuckle inhibits the replication of influenza A. In a similar study, significantly higher levels of a plant specific miRNA were found to be abundantly present in the serum of mice consuming diets rich in vegetables (Yang et al., 2016). They further studied the abnormal abundance of miR2911 and its distinctive origin. The sRNA was found to be closely associated with the mammalian miRISC (Yang et al., 2017). In a recent study, Luo et al., (2017) demonstrated that the dietary maize miRNAs were able to cross the pig gastrointestinal tract and can be detected in porcine tissues and serum. Therefore, dietary intake of exogenous plant miRNAs may be more widespread than initially appreciated and can contribute to recipient gene expression and human health.
On the other hand, there have been many contrasting views regarding presence of foreign miRNAs in mammalian specimens that were absorbed through dietary sources (Table 4), especially plants (Tosar et al., 2014, Snow et al., 2013, Witwer et al., 2013, Dickinson et al., 2013, Chen et al., 2013). Although several previous studies have reported the detection of miRNAs from exogenous organisms in human serum (Wang et al., 2012) and breast milk (Lukasik and Zielenkiewicz, 2014); however, other set of reports have not been able to identify foreign miRNAs after exposure to various food types (Snow et al., 2013, Witwer et al., 2013). There are arguments regarding this contrasting set of opinions, largely based of methodological aspects of the later studies, where a selective set of miRNAs were focused for the study rather than sequencing and screening a complete dataset of all known plant miRNAs. Another group of studies done by a relatively smaller number of researchers reveal another aspect of plant miRNAs in mammals, where miRNAs have been shown to have potential functions in regulating mammalian cells, however several aspects in this area need further exploration (Zhou et al., 2015, Liang et al., 2014, Liang et al., 2015 Yang et al., 2015b).
Table 4.
Contradicting evidence of cross-kingdom communication by diet/plant-derived miRNAs (Adapted with permission from Li et al., 2018, an Open access article distributed under the terms of the Creative Commons Attribution 4.0 International License).
| Year | Contents | miRNAs involved | Source origin | miRNA levels | Refuting points | Detectionmethods | Conclusion | Reference |
|---|---|---|---|---|---|---|---|---|
| 2013 | Little or no plant miRNAs or miR168a were detected in blood or liver of mice fed with rice-containing diets | miR168a | Rice | Unmeasurable | The observed changes in LDL levels might be due to the release of endogenous cholesterol stores in response to negligible dietary cholesterol intake in mice fed with only rice | HTS, RT-qPCR, ELISA | Dietary exposure to miR168a did not affect plasma LDL levels. Plasma LDL changes reported by Zhang resulted from nutritional imbalances between test and control groups rather than an RNAi mediated effect of consuming miR168a in rice | Dickinson et al., 2013 |
| 2013 | Plant miRNAs were not detectable in the plasma from healthy human subjects after intake of a western diet containing fruits. | miR156a miR159a miR169a |
Plant material | Undetectable | Low measurable uptake | RT-qPCR | Horizontal delivery of miRNAs via oral ingestion of a typical diet was neither a frequent nor a prevalent event across multiple recipient animal organisms | Snow et al., 2013 |
| Negligible expression of miR21 in plasma or organ tissue in miR21 knockout mice after oral diets replete with endogenous miR21 | miR21 | Animal lard diet replete With miR21 | Undetectable in plasma; less than one copy per cell in the liver, lungs, kidneys and stomach | |||||
| Negligible expression of miR156a, miR159a and miR169a in plasma or organs in mice after diets replete with these miRNAs. | miR156a miR159a miR169a |
Vegetariandiets replete with these miRNAs | miR156a: far less than one copy of miRNA per cell in liver, lungs, kidneys and stomach; miR159a and miR169a: undetectable in either plasma and/or organs. | |||||
| Negligible expression of plant-derived miRNAs in recipient honeybee tissues. | miR156a miR159a miR169a |
Plant derived miRNA | Only miR156a but not miR159a or miR169a, was detected in abdominal tissue derived from nurses and foragers, but again at exceptionally low levels. | |||||
| 2012 | Predominant monocot miR168 sequence was present as a result of contamination from a non-plant source | miR168a | Plant | Not available | Contamination | HTS, NB | The observed plant miRNAs in animal sRNA datasets could originate in the process of sequencing, and accumulation of plant miRNAs via dietary exposure was not universal in animals. | Zhang et al., 2012b |
| 2014 | Cross-contamination during library preparation was a source of exogenous RNAs | miR168a miR156a miR167a |
Plant | Not available | Contamination | HTS | Variable amounts of plant miRNAs were found in publicly available sRNA-seq data sets of human tissues. | Tosar et al., 2014 |
| 2014 | Failed to observe a postprandial increase in the brassica-specific miR824 or miR167a in broccoli sprouts feeding study | miR167a miR824 |
Broccoli sprouts | Below detection limit | Low measurable uptake | RT-qPCR | Skeptical of the bioavailability and biologic activity of plant-borne miRNAs | Baier et al., 2014 |
| 2013 | Nonhuman primates failed to uptake dietary plant miRNAs | miR156 miR160 miR166 miR167 miR168 miR172 |
Fruit | Not available | The concentrations were too low to be specific and reliable. | RT-qPCR, Droplet digital PCR | The level of miRNAs was too low to be true and/or amplification was non–specific | Witwer et al., 2013 |
| 2018 | Corn miRNA was extensively degraded in the GI tract and that the uptake into circulation and tissues was minimal | miR156a miR164a miR167a |
Corn | No corn miRNAs could be detected in whole blood, fecal or liver of animals | Significant degradation of corn miRNAs occurred during digestion | — | No evidence of increased levels of corn miRNAs in whole blood or tissues after supplementation of corn miRNAs in the diet was observed in a mouse model. | Huang et al., 2018 |
MVs microvesicles, HTS high-throughput sequencing, NB Northern blot, WB Western blot, ELISA enzyme-linked immunosorbent assay, LDL low-density lipoprotein, sRNA small RNA
The debate has, since then, continued in the form of follow-up studies that have provided evidences either for or against the notion concerning stability and biological function of plant miRNAs consumed by mammals through various dietary sources. A study by Dickinson et al (2013) forms a major cornerstone of this extended discussion, where miR168a derived from rice, exhibited no cross-kingdom modulation in the levels of a LDLRAP1 found in mouse liver, as discussed earlier. Another study on mice showed negligible evidences for the presence of miRNAs in the blood plasma after ingestion of plant-based diet (Snow et al., 2013). Further, some reports on macaque species have concluded that there has been no detection of dietary miRNAs in blood samples when investigated using droplet-digital PCR (Witwer et al., 2013). Huang et al., (2018) recovered less than 1% of corn miRNAs in the gastrointestinal tract of animals fed with plant-based diet.
Arguments putting up questions against such studies generally root from the notion concerning construction and sequencing of plant sRNA libraries. It has been identified that as compared to animal miRNA libraries, plant miRNA libraries are not constructed with the same precision due to presence of 2′-O-methylated 3′ end modification (Yu et al., 2005); which has a negative influence on the efficiency of adapter ligation, thereby underestimating the quantification of plant-based miRNAs (Munafo & Robb, 2010). Studies showing contrasting results due to methodological dissimilarities have strengthened the ground for such debates. For example, Zhang et al (2012a) utilized oxidized deep sequencing to retrieve miRNAs of plant origin in human and mice blood samples, whereas Dickinson et al., (2013) could not detect any such small RNAs in mice samples, demonstrating a bias for sequencing. An additional dispute involved in the debate for existence of cross-kingdom miRNAs in human fluids is that nucleic acids are common contaminants in experiments involving genomic library preparations and sequencing, thereby showing false positive results (Tosar et al., 2014). Hence, when extremely low quantities of miRNAs are detected, the first assumption is whether any contamination or background noise exists in the apparatus. Although, these experimental issues can be resolved through cross-validation of results using multiple assay platforms including Chip-seq, RT-PCR and deep sequencing techniques.
7. Plant-derived xenomiRs affect cancer progression
Exosomal miRNAs delivered through the diet may mediate cellular effects in the human body (Fig. 2), including development of cancer. Wolf et al., (2015) showed that bovine exosomes are transported into human colon carcinoma Caco-2 cells in culture, resulting in elevated miR-29b and miR-200c levels. Interestingly, both miRNAs are shown to be incriminated in human cancers (Kwon et al., 2018, Kumar et al., 2015). These studies may open a new arena of treatment of human diseases (including cancer) exploiting plant-based miRNAs.
Although the source and scope of plant miRNAs detected in mammalian specimens remain debatable, Mlotshwa et al., (2015) described that mammalian miRNAs that were 2′-O-methylated like plant miRNAs (Yu et al 2005), could reduce mouse intestinal tumor burden when administered orally for longer durations. They also unraveled that further reduction in the tumor load is possible if tumor suppressor miRNAs are administered together with the plant miRNAs in the similar subjects (Mlotshwa et al., 2015). This study, however, concluded that a larger sample size may be required to confirm the therapeutic role of plant-based xenomiRs ingested by mammals. Moreover, it is still a subject of investigation, whether these cross-kingdom miRNAs have any beneficiary therapeutic role in treating cancers outside of the GI tract.
Recently, an inverse association between the abundance of plant miRNA and breast cancer incidence and progression was reported (Chin et al., 2016). The authors detected the presence of a plant originating miRNA, miR159 in the sera of human EVs that exhibited resistance to sodium periodate oxidation, which may be due to the 2′-O-methylation. They also found that the concentration of orally ingested miRNA was inversely related to the chances and progression of the cancer. Through administration of synthetic mimic of miR159 in animal model, a transcription factor encoded by TCF7 gene was identified as the target gene for RNAi mediated silencing. This study concluded that miR159 can significantly reduce the growth of breast tumors in mammals, becoming the first report showing the role of a plant-based miRNA in mammalian cancer. More recently, a plant-based mir167e-5p was found to be inhibiting the proliferation of Caco-2 cancer cell lines dose- and time-dependently in a epigenetic manner by targeting β-catenin pathway (Li et al., 2019). However, another study showed no such effect of plant derived miRNAs on mammalian cancers (Xiao et al., 2020). Using deep sequencing analysis, the researchers identified ~750 miRNA from sprouts of B. rapa sylvestris (commonly called broccoletti) and predicted their human targets using bioinformatics techniques. Two new miRNAs were identified from this database. It was observed that the overexpression of selected miRNAs and their administration, did not affect any major metabolic activity of pancreatic cancer cells. This study creates scientific evidence against the perception of regulation of human genes from plant-derived miRNAs present in food (Xiao et al., 2020).
8. Conclusion
The role of mobile miRNAs in cross-kingdom RNAi is intriguing; yet, it remains skeptical how these sRNAs ‘traverse’ long distances surviving diverse cellular boundaries between plants and their target entities. Notably, this cross-kingdom miRNA transfer may not be due to chance, rather it may occur through a selective transport of functional miRNAs. miRNAs present in diets are now being detected in the sera of the consumer where they function to regulate various biological processes including cancer. Recent evidence suggests that plant xenomiRs may hinder the expression of human disease-related genes (Zhou et al., 2015, Chin et al., 2016, Sharma et al., 2016, Tian et al., 2016, Choi et al., 2017). However, our understanding of the cross-kingdom gene regulation remains inconclusive (Mar-Aguilar et al., 2020), and it is too early to make a final stand in favor or against of the notion that the plant-based xenomiRs regulate gene expression in humans.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
The author would like to thank the Deanship of Scientific Research at Majmaah University for supporting this work under Project Number No. R-2021-3.
Funding
This work did not receive any grant from any funding agencies.
Footnotes
Peer review under responsibility of King Saud University.
References
- Achkar N.P., Cambiagno D.A., Manavella P.A. miRNA biogenesis: a dynamic pathway. Trends Plant Sci. 2016;21(12):1034–1044. doi: 10.1016/j.tplants.2016.09.003. [DOI] [PubMed] [Google Scholar]
- Akers J.C., Gonda D., Kim R., Carter B.S., Chen C.C. Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J. Neurooncol. 2013;113:1–11. doi: 10.1007/s11060-013-1084-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachy I., Kozyraki R., Wassef M. The particles of the embryonic cerebrospinal fluid: How could they influence brain development? Brain Res Bull. 2008;75:289–294. doi: 10.1016/j.brainresbull.2007.10.010. [DOI] [PubMed] [Google Scholar]
- Baier S.R., Nguyen C., Xie F., Wood J.R., Zempleni J. MicroRNAs are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow milk and affect gene expression in peripheral blood mononuclear cells, HEK-293 kidney cell cultures, and mouse livers. J. Nutr. 2014;144:1495–1500. doi: 10.3945/jn.114.196436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandini E., Rossi T., Gallerani G., Fabbri F. Adipocytes and microRNAs crosstalk: A key tile in the mosaic of breast cancer microenvironment. Cancers (Basel). 2019;11:1451. doi: 10.3390/cancers11101451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Baum J.A., Bogaert T., Clinton W., Heck G.R., Feldmann P., Ilagan O., Johnson S., Plaetinck G., Munyikwa T., Pleau M., Vaughn T., Roberts J. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 2005;25:1322–1326. doi: 10.1038/nbt1359. [DOI] [PubMed] [Google Scholar]
- Benmoussa A., Ly S., Shan S., Laugier J., Boilard E., Gilbert C., Provost P. A subset of extracellular vesicles carries the bulk of microRNAs in commercial dairy cow's milk. J. Extracell. Vesicles. 2017;6:1401897. doi: 10.1080/20013078.2017.1401897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E., Guryev V., van de Belt J., Wienholds E., Plasterk R.H., Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Berindan-Neagoe I., Monroig Pdel.C., Pasculli B., Calin G.A. MicroRNAome genome: a treasure for cancer diagnosis and therapy. CA Cancer J. Clin. 2014;64:311–336. doi: 10.3322/caac.21244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffelli D., McAuliffe J., Ovcharenko D., Lewis K.D., Ovcharenko I., Pachter L., Rubin E.M. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science. 2003;299:1391–1394. doi: 10.1126/science.1081331. [DOI] [PubMed] [Google Scholar]
- Brennecke J., Stark A., Russell R.B., Cohen S.M. Principles of microRNA–Target recognition. PLoS Biol. 2005;3(3) doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breving K., Esquela-Kerscher A. The complexities of microRNA regulation: mirandering around the rules. Int. J. Biochem. Cell Biol. 2010;42:1316–1329. doi: 10.1016/j.biocel.2009.09.016. [DOI] [PubMed] [Google Scholar]
- Buck A.H., Coakley G., Simbari F., McSorley H.J., Quintana J.F., Bihan T.L., Kumar S., Abreu-Goodger C., Lear M., Harcus Y., Ceroni A., Babayan S.A., Blaxter M., Ivens A., Maizels R.M. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat. Commun. 2014;5:5488. doi: 10.1038/ncomms6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caby M.P., Lankar D., Vincendeau-Scherrer C., Raposo G., Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int. Immunol. 2005;17:879–887. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- Calin G.A., Cimmino A., Fabbri M., Ferracin M., Wojcik S.E., Shimizu M., Taccioli C., Zanesi N., Garzon R., Aqeilan R.I., Alder H., Volinia S., Rassenti L., Liu X., Liu C.G., Kipps T.J., Negrini M., Croce C.M. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc. Natl. Acad. Sci. U.S.A. 2008;105:5166–5171. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Calin G.A., Croce C.M. MicroRNAs and chromosomal abnormalities in cancer cells. Oncogene. 2006;25:6202–6210. doi: 10.1038/sj.onc.1209910. [DOI] [PubMed] [Google Scholar]
- Calin G.A., Dumitru C.D., Shimizu M., Bichi R., Zupo S., Noch E., Aldler H., Rattan S., Keating M., Rai K., Rassenti L., Kipps T., Negrini M., Bullrich F., Croce C.M. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U.S.A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalieri D., Rizzetto L., Tocci N., Rivero D., Asquini E., Si-Ammour A., Bonechi E., Ballerini C., Viola R. Plant microRNAs as novel immunomodulatory agents. Sci. Rep. 2016;6:25761. doi: 10.1038/srep25761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S.Y., Snow J.W. Formidable challenges to the notion of biologically important roles for dietary small RNAs in ingesting mammals. Genes Nutr. 2017;12:13(2017). doi: 10.1186/s12263-017-0561-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S.P., Slack F.J. And now introducing mammalian mirtrons. Dev. Cell. 2007;13:605–607. doi: 10.1016/j.devcel.2007.10.010. [DOI] [PubMed] [Google Scholar]
- Chatterjee K.R., Das Gupta N.N., De M.L. Electron microscopic observations on the morphology of Mycobacterium leprae. Exp. Cell Res. 1959;18:521–527. doi: 10.1016/0014-4827(59)90317-9. [DOI] [PubMed] [Google Scholar]
- Chen X., Zen K., Zhang C.Y. Reply to Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat. Biotechnol. 2013;31:967–969. doi: 10.1038/nbt.2741. [DOI] [PubMed] [Google Scholar]
- Chiarugi P., Cirri P. Metabolic exchanges within tumor microenvironment. Cancer Lett. 2016;380:272–280. doi: 10.1016/j.canlet.2015.10.027. [DOI] [PubMed] [Google Scholar]
- Chin A.R., Fong M.Y., Somlo G., Wu J., Swiderski P., Wu X., Wang S.E. Cross kingdom inhibition of breast cancer growth by plant miR159. Cell Res. 2016;26:217–228. doi: 10.1038/cr.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.J., Lin C.P., Risso D., Chen S., Kim T.A., Tan M.H., Li J.B., Wu Y., Chen C., Xuan Z., Macfarlan T., Peng W., Lloyd K.C., Kim S.Y., Speed T.P., He L. Deficiency of microRNA miR-34a expands cell fate potential in pluripotent stem cells. Science. 2017;355:eaag1927. doi: 10.1126/science.aag1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy Y.W., Siu K.L., Kok K.H., Lung W.M., Chi M.T., To K.F., Kwong L.W., Tsao S.W., Jin D.Y. An Epstein-Barr virus–encoded microRNA targets PUMA to promote host cell survival. J. Exp. Med. 2008;205:2551–2560. doi: 10.1084/jem.20072581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino A., Calin G.A., Fabbri M., Iorio M.V., Ferracin M., Shimizu M., Wojcik S.E., Aqeilan R.I., Zupo S., Dono M., Rassenti L., Alder H., Volinia S., Liu C.G., Kipps T.J., Negrini M., Croce C.M. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. U.S.A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M., Raposo G., Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- Curtale G. MiRNAs at the Crossroads between innate immunity and cancer: focus on macrophages. Cells. 2018;7:12. doi: 10.3390/cells7020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De S.N. Enterotoxicity of bacteria-free culture-filtrate of Vibrio cholerae. Nature. 1959;183:1533–1534. doi: 10.1038/1831533a0. [DOI] [PubMed] [Google Scholar]
- Dickinson B., Zhang Y., Petrick J.S., Heck G., Ivashuta S., Marshall W.S. Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat. Biotechnol. 2013;31:965–967. doi: 10.1038/nbt.2737. [DOI] [PubMed] [Google Scholar]
- Doench J.G., Sharp P.A. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z., Han M.-H., Fedoroff N. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. PNAS. 2008;105(29):9970–9975. doi: 10.1073/pnas.0803356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnarumma E., Fiore D., Nappa M., Roscigno G., Adamo A., Iaboni M., Russo V., Affinito A., Puoti I., Quintavalle C., Rienzo A., Piscuoglio S., Thomas R., Condorelli G. Cancer-associated fibroblasts release exosomal microRNAs that dictate an aggressive phenotype in breast cancer. Oncotarget. 2017;8:19592–19608. doi: 10.18632/oncotarget.14752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert B., Rai A.J. Isolation and characterization of amniotic fluid-derived extracellular vesicles for biomarker discovery. Methods Mol. Biol. 2019;1885:287–294. doi: 10.1007/978-1-4939-8889-1_19. [DOI] [PubMed] [Google Scholar]
- Fang Y., Spector D.L. Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr. Biol. 2007;17:818–823. doi: 10.1016/j.cub.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Xue J.L., Shen Q., Chen J., Tian L. MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatol. 2012;55:1852–1862. doi: 10.1002/hep.25576. [DOI] [PubMed] [Google Scholar]
- Fanini F., Fabbri M. Cancer-derived exosomic microRNAs shape the immune system within the tumor microenvironment: State of the art. Semin. Cell Dev. Biol. 2017;67:23–28. doi: 10.1016/j.semcdb.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzmaurice C., Allen C., Barber R.M., Barregard L., Bhutta Z.A., Brenner H., Dicker D.J., Chimed-Orchir O., Dandona R., Dandona L., Fleming T., Forouzanfar M.H., Hancock J., Hay R.J., Hunter-Merrill R., Huynh C., Hosgood H.D., Johnson C.O., Jonas J.B. Global, regional, and National cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: A systematic analysis for the global burden of disease study. JAMA Oncol. 2017;3:524–548. doi: 10.1001/jamaoncol.2016.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortunato O., Gasparini P., Boeri M., Sozzi G. Exo-miRNAs as a new tool for liquid biopsy in lung cancer. Cancers. 2019;11(6):888. doi: 10.3390/cancers11060888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann G., Neuer A.L., Herrmann I.K. Extracellular vesicles - a promising avenue for the detection and treatment of infectious diseases? Eur. J. Pharm. Biopharm. 2017;118:56–61. doi: 10.1016/j.ejpb.2017.04.005. [DOI] [PubMed] [Google Scholar]
- Fukudome A., Fukuhara T. Plant dicer-like proteins: double-stranded RNA-cleaving enzymes for small RNA biogenesis. J. Plant Res. 2017;130:33–44. doi: 10.1007/s10265-016-0877-1. [DOI] [PubMed] [Google Scholar]
- Garcia-Silva M.R., das Neves R.F., Cabrera-Cabrera F., Sanguinetti J., Medeiros L.C., Robello C., Naya H., Fernandez-Calero T., Souto-Padron T., de Souza W., Cayota A. Extracellular vesicles shed by Trypanosoma cruzi are linked to small RNA pathways, life cycle regulation, and susceptibility to infection of mammalian cells. Parasitol. Res. 2014;113:285–304. doi: 10.1007/s00436-013-3655-1. [DOI] [PubMed] [Google Scholar]
- Han L., Lam E.W., Sun Y. Extracellular vesicles in the tumor microenvironment: old stories, but new tales. Mol. Cancer. 2019;18:59. doi: 10.1186/s12943-019-0980-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Jia L., Zheng Y., Li W. Salivary exosomes: emerging roles in systemic disease. Int. J. Biol. Sci. 2018;14:633–643. doi: 10.7150/ijbs.25018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen A., Henderson S., Lagos D., Nikitenko L., Coulter E., Roberts S., Gratrix F., Plaisance K., Renne R., Bower M., Kellam P., Boshoff C. KSHV-encoded miRNAs target MAF to induce endothelial cell reprogramming. Genes Dev. 2010;24:195–205. doi: 10.1101/gad.553410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding C., Heuser J., Stahl P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J. Cell Biol. 1983;97:329–339. doi: 10.1083/jcb.97.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa T., Lewis H., Esquela-Kerscher A. The role of noncoding RNAs in prostate cancer. In: Laurence J., editor. Translating microRNAs to the clinic. Academic Press; Boston, Massachusetts: 2017. pp. 329–369. [Google Scholar]
- Hatziapostolou M., Polytarchou C., Aggelidou E., Drakaki A., Poultsides G.A., Jaeger S.A., Ogata H., Karin M., Struhl K., Hadzopoulou-Cladaras M., Iliopoulos D. An HNF4a-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell. 2011;147:1233–1247. doi: 10.1016/j.cell.2011.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessvik N.P., Sandvig K., Llorente A. Exosomal miRNAs as biomarkers for prostate cancer. Front. Genet. 2013;4:36. doi: 10.3389/fgene.2013.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higdon J.V., Delage B., Williams D.E., Dashwood R.H. Cruciferous vegetables and human cancer risk: epidemiologic evidence and mechanistic basis. Pharmacol. Res. 2007;55:224–236. doi: 10.1016/j.phrs.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoog J.L., Lotvall J. Diversity of extracellular vesicles in human ejaculates revealed by cryo-electron microscopy. J. Extracell. Vesicles. 2015;4:28680. doi: 10.3402/jev.v4.28680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou D., He F., Ma L., Cao M., Zhou Z., Wei Z., Xue Y., Sang X., Chong H., Tian C., Zheng S., Li J., Zen K., Chen X., Hong Z., Zhang C.Y., Jiang X. The potential atheroprotective role of plant MIR156a as a repressor of monocyte recruitment on inflamed human endothelial cells. J. Nutr. Biochem. 2018;57:197–205. doi: 10.1016/j.jnutbio.2018.03.026. [DOI] [PubMed] [Google Scholar]
- Howard K.M., Jati Kusuma R., Baier S.R., Friemel T., Markham L., Vanamala J., Zempleni J. Loss of miRNAs during processing and storage of cow's (Bos taurus) milk. J. Agric. Food Chem. 2015;63:588–592. doi: 10.1021/jf505526w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoy A.M., Lundie R.J., Ivens A., Quintana J.F., Nausch N., Forster T., Jones F., Kabatereine N.B., Dunne D.W., Mutapi F., Macdonald A.S., Buck A.H. Parasite-derived microRNAs in host serum as novel biomarkers of helminth infection. PLoS Negl. Trop. Dis. 2014;8 doi: 10.1371/journal.pntd.0002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Davis C.D., Wang T.T. Extensive degradation and low bioavailability of orally consumed corn miRNAs in mice. Nutrients. 2018;10:215. doi: 10.3390/nu10020215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingenito F., Roscigno G., Affinito A., Nuzzo S., Scognamiglio I., Quintavalle C., Condorelli G. The role of exo-miRNAs in cancer: a focus on therapeutic and diagnostic applications. Int. J. Mol. Sci. 2019;20:4687. doi: 10.3390/ijms20194687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam W., Islam S.U., Qasim M., Wang L. Host-pathogen interactions modulated by small RNAs. RNA Biol. 2017;14:891–904. doi: 10.1080/15476286.2017.1318009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal R., Sedger L.M. Intercellular vesicular transfer by exosomes, microparticles and oncosomes - Implications for cancer biology and treatments. Front. Oncol. 2019;9:125. doi: 10.3389/fonc.2019.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen W.A. The composition and ultrastructure of the nucleus in cotton. J. Ultrastruct. Res. 1965;13:112–128. [Google Scholar]
- John B., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S. Human MicroRNA targets. PLoS Biol. 2004;2 doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone R.M., Adam M., Hammond J.R., Orr L., Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) J. Biol. Chem. 1987;262:9412–9420. [PubMed] [Google Scholar]
- Kahlert C., Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J. Mol. Med. (Berl) 2013;91:431–437. doi: 10.1007/s00109-013-1020-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller S., Ridinger J., Rupp A.K., Janssen J.W., Altevogt P. Body fluid derived exosomes as a novel template for clinical diagnostics. J. Transl. Med. 2011;9:86. doi: 10.1186/1479-5876-9-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knip M., Constantin M.E., Thordal-Christensen H. Trans-kingdom cross-talk: small RNAs on the move. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga Y., Yasunaga M., Moriya Y., Akasu T., Fujita S., Yamamoto S., Matsumura Y. Exosome can prevent RNase from degrading microRNA in feces. J. Gastrointest. Oncol. 2011;2:215–222. doi: 10.3978/j.issn.2078-6891.2011.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka N., Iguchi H., Yoshioka Y., Takeshita F., Matsuki Y., Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 2010;285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka N., Izumi H., Sekine K., Ochiya T. MicroRNA as a new immune-regulatory agent in breast milk. Silence. 2010;1:7. doi: 10.1186/1758-907X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Nag A., Mandal C.C. A comprehensive review on miR-200c, a promising cancer biomarker with therapeutic potential. Curr. Drug Targets. 2015;16(12):1381–1403. doi: 10.2174/1389450116666150325231419. [DOI] [PubMed] [Google Scholar]
- Kuninty P.R., Schnittert J., Storm G., Prakash J. MicroRNA targeting to modulate tumor microenvironment. Front. Oncol. 2016;6:3. doi: 10.3389/fonc.2016.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon J.J., Factora T.D., Dey S., Kota J. A systematic review of miR-29 in cancer. Mol. Ther. Oncolytics. 2018;12:173–194. doi: 10.1016/j.omto.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- LaMonte G., Philip N., Reardon J., Lacsina J.R., Majoros W., Chapman L., Thornburg C.D., Telen M.J., Ohler U., Nicchitta C.V., Haystead T., Chi J.T. Translocation of sickle cell erythrocyte microRNAs into Plasmodium falciparum inhibits parasite translation and contributes to malaria resistance. Cell Host Microbe. 2012;12:187–199. doi: 10.1016/j.chom.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasser C., Alikhani V.S., Ekström K., Eldh M., Paredes P.T., Bossios A., Sjöstrand M., Gabrielsson S., Lötvall J., Valadi H. Human saliva, plasma and breast milk exosomes contain RNA: Uptake by macrophages. J. Transl. Med. 2011;9:9. doi: 10.1186/1479-5876-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li J., Yang Z., Yu B., Liu J., Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Chen T., He J.J., Wu J.-H., Luo J.-Y., Ye R.-S., Xie M.-Y. Plant MIR167e-5p inhibits enterocyte proliferation by targeting β-catenin. Cells. 2019;8(11):1385. doi: 10.3390/cells8111385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Xu R., Li N. MicroRNAs from plants to animals, do they define a new messenger for communication? Nutr. Metab. 2018;Lond). 15:68. doi: 10.1186/s12986-018-0305-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Zhu Y., Sun B., Shao Y., Jing A., Wang J., Xiao Z. Assessing the survival of exogenous plant microRNA in mice. Food Sci. Nutr. 2014;2:380–388. doi: 10.1002/fsn3.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H., Zhang S., Fu Z., Wang Y., Wang N., Liu Y., Zhao C., Wu J., Hu Y., Zhang J., Chen X., Zen K., Zhang C.Y. Effective detection and quantification of dietetically absorbed plant microRNAs in human plasma. J. Nutr. Biochem. 2015;26:505–512. doi: 10.1016/j.jnutbio.2014.12.002. [DOI] [PubMed] [Google Scholar]
- Liu R.H. Potential synergy of phytochemicals in cancer prevention: mechanism of action. J. Nutr. 2004;134:3479S–3485S. doi: 10.1093/jn/134.12.3479S. [DOI] [PubMed] [Google Scholar]
- Liu S., da Cunha A.P., Rezende R.M., Cialic R., Wei Z., Bry L., Comstock L.E., Gandhi R., Weiner H.L. The host shapes the gut microbiota via fecal microRNA. Cell Host Microbe. 2016;19:32–43. doi: 10.1016/j.chom.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukasik A., Zielenkiewicz P. In silico identification of plant miRNAs in mammalian breast milk exosomes–a small step forward? PLoS One. 2014;9 doi: 10.1371/journal.pone.0099963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Wang P., Wang X., Wang Y., Mu Z., Li Q., Fu Y., Xiao J., Li G., Ma Y., Gu Y., Jin L., Ma J., Tang Q., Jiang A., Li X., Li M. Detection of dietetically absorbed maize-derived microRNAs in pigs. Sci. Rep. 2017;7:645. doi: 10.1038/s41598-017-00488-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T., Chen Y., Chen Y. MicroRNA-132, delivered by mesenchymal stem cell-derived exosomes, promote angiogenesis in myocardial infarction. Stem Cells Int. 2018:1–18. doi: 10.1155/2018/3290372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida T., Tomofuji T., Ekuni D., Maruyama T., Yoneda T., Kawabata Y., Mizuno H., Miyai H., Kunitomo M., Morita M. MicroRNAs in salivary exosome as potential biomarkers of aging. Int. J. Mol. Sci. 2015;16:21294–21309. doi: 10.3390/ijms160921294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malloci M., Perdomo L., Veerasamy M., Andriantsitohaina R., Simard G., Martinez M.C. Extracellular vesicles: mechanisms in human health and disease. Antioxid. Redox Signal. 2019;30:813–856. doi: 10.1089/ars.2017.7265. [DOI] [PubMed] [Google Scholar]
- Mao L., Li X., Gong S., Yuan H., Jiang Y., Huang W., Sun X., Dang X. Serum exosomes contain ECRG4 mRNA that suppresses tumor growth via inhibition of genes involved in inflammation, cell proliferation, and angiogenesis. Cancer Gene Ther. 2018;25:248–259. doi: 10.1038/s41417-018-0032-3. [DOI] [PubMed] [Google Scholar]
- Mar-Aguilar F., Arreola-Triana A., Mata-Cardona D., Gonzalez-Villasana V., Rodríguez-Padilla C., Reséndez-Pérez D. Evidence of transfer of miRNAs from the diet to the blood still inconclusive. PeerJ. 2020;8 doi: 10.7717/peerj.9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli, C., 2017. Exosomes: new biomarkers for targeted cancer therapy in Molecular Oncology: Underlying Mechanisms and Translational Advancements. Farooqi AA, İsmail M. (Eds). Springer International Publishing, 129–157.
- Mathieu M., Martin-Jaular L., Lavieu G., Théry C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell. Biol. 2019;21:9–17. doi: 10.1038/s41556-018-0250-9. [DOI] [PubMed] [Google Scholar]
- McDonald M.K., Capasso K.E., Ajit S.K. Purification and microRNA profiling of exosomes derived from blood and culture media. J. Vis. Exp. 2013;76 doi: 10.3791/50294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentkowski K.I., Snitzer J.D., Rusnak S., Lang J.K. Therapeutic potential of engineered extracellular vesicles. AAPS J. 2018;20(3):50. doi: 10.1208/s12248-018-0211-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi Y., Zhang D., Jiang W., Weng J., Zhou C., Huang K., Tang H., Yu Y., Liu X., Cui W. miR-181a-5p promotes the progression of gastric cancer via RASSF6-mediated MAPK signaling activation. Cancer Lett. 2017;389:11–22. doi: 10.1016/j.canlet.2016.12.033. [DOI] [PubMed] [Google Scholar]
- Minciacchi V.R., You S., Spinelli C., Morley S., Zandian M., Aspuria P., Cavallini L., Ciardiello C., Sobreiro M., Morello M., Kharmate G., Jang S., Chul Kim D. Large oncosomes contain distinct protein cargo and represent a separate functional class of tumor-derived extracellular vesicles. Oncotarget. 2015;6:11327–11341. doi: 10.18632/oncotarget.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlotshwa S., Pruss G.J., MacArthur J.L., Endres M.W., Davis C., Hofseth L.J., Pena M.M., Vance V.A. A novel chemopreventive strategy based on therapeutic microRNAs produced in plants. Cell Res. 2015;25:521–524. doi: 10.1038/cr.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motti M.L., Meccariello D.A.S.R. MicroRNAs, cancer, and diet: facts and new exciting perspectives. Curr. Mol. Pharmacol. 2018;11:90–96. doi: 10.2174/1874467210666171013123733. [DOI] [PubMed] [Google Scholar]
- Munafo D.B., Robb G.B. Optimization of enzymatic reaction conditions for generating representative pools of cDNA from small RNA. RNA. 2010;16:2537–2552. doi: 10.1261/rna.2242610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien J., Hayder H., Zayed Y., Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka K., Yamamoto Y., Matsuoka R., Ochiya T. Maintaining good miRNAs in the body keeps the doctor away: perspectives on the relationship between food-derived natural products and microRNAs in relation to exosomes/extracellular vesicles. Mol. Nutr. Food Res. 2018;62(1):1700080. doi: 10.1002/mnfr.201700080. [DOI] [PubMed] [Google Scholar]
- Pan B.T., Teng K., Wu C., Adam M., Johnstone R.M. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J. Cell Biol. 1985;101:942–948. doi: 10.1083/jcb.101.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastrello C., Tsay M., McQuaid R., Abovsky M., Pasini E., Shirdel E., Angeli M., Tokar T., Jamnik J., Kotlyar M., Jurisicova A., Kotsopoulos J., El-Sohemy A., Jurisica I. Circulating plant miRNAs can regulate human gene expression in vitro. Sci. Rep. 2016;6:32773. doi: 10.1038/srep32773. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Peng Y., Croce C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target Ther. 2016;1:15004. doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip A., Ferro V.A., Tate R.J. Determination of the potential bioavailability of plant microRNAs using a simulated human digestion process. Mol. Nutr. Food Res. 2015;59:1962–1972. doi: 10.1002/mnfr.201500137. [DOI] [PubMed] [Google Scholar]
- Pirrò S., Minutolo A., Galgani A., Potestà M., Colizzi V., Montesano C. Bioinformatics prediction and experimental validation of microRNAs involved in cross-kingdom interaction. J. Comput. Biol. 2016;23:976–989. doi: 10.1089/cmb.2016.0059. [DOI] [PubMed] [Google Scholar]
- Pisitkun T., Shen R.F., Knepper M.A. Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. U.S.A. 2004;101:13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahbarghazi R., Jabbari N., Sani N.A., Asghari R., Salimi L., Kalashani S.A., Feghhi M., Etemadi T., Akbariazar E., Mahmoudi M., Rezaie J. Tumor-derived extracellular vesicles: Reliable tools for cancer diagnosis and clinical applications. Cell Commun. Signal. 2019;17(1):73. doi: 10.1186/s12964-019-0390-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G., Nijman H.W., Stoorvogel W., Liejendekker R., Harding C.V., Melief C.J., Geuze H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996;183:1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G., Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Record M. Exosome-like nanoparticles from food: Protective nanoshuttles for bioactive cargo. Mol. Ther. 2013;21:1294–1296. doi: 10.1038/mt.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Rupaimoole R., Calin G.A., Lopez-Berestein G., Sood A.K. miRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov. 2016;6:235–246. doi: 10.1158/2159-8290.CD-15-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter B.D., Innes R.W. Extracellular vesicles isolated from the leaf apoplast carry stress-response proteins. Plant Physiol. 2016;175:728–741. doi: 10.1104/pp.16.01253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R., Palade G.E. Structure and development of the chloroplast in Chlamydomonas. I. The normal green cell. J. Biophys. Biochem. Cytol. 1957;3:463–488. doi: 10.1083/jcb.3.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkies P., Miska E.A. Small RNAs break out: the molecular cell biology of mobile small RNAs. Nat. Rev. Mol. Cell Biol. 2014;15:525–535. doi: 10.1038/nrm3840. [DOI] [PubMed] [Google Scholar]
- Shahid S., Kim G., Johnson N.R., Wafula E., Wang F., Coruh C., Bernal-Galeano V., Phifer T., dePamphilis C.W., Westwood J.H., Axtellet M.J. MicroRNAs from the parasitic plant Cuscuta campestris target host messenger RNAs. Nature. 2018;553:82–85. doi: 10.1038/nature25027. [DOI] [PubMed] [Google Scholar]
- Sharma A., Sahu S., Kumari P., Gopi S.R., Malhotra R., Biswas S. Genome-wide identification and functional annotation of miRNAs in anti-inflammatory plant and their cross-kingdom regulation in Homo sapiens. J. Biomol. Struct. Dyn. 2016;35:1389–1400. doi: 10.1080/07391102.2016.1185381. [DOI] [PubMed] [Google Scholar]
- Si W., Shen J., Zheng H., Fan W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin. Epigenet. 2019;11:25. doi: 10.1186/s13148-018-0587-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M., Raposo G. Exosome—Vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 2009;21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Singh A., Gautam V., Singh S., Das S.S., Verma S., Mishra V., Mukherjee S., Sarkar A.K. Plant small RNAs: advancement in the understanding of biogenesis and role in plant development. Planta. 2018;248(3):545–558. doi: 10.1007/s00425-018-2927-5. [DOI] [PubMed] [Google Scholar]
- Snow J.W., Hale A.E., Isaacs S.K., Baggish A.L., Chan S.Y. Ineffective delivery of diet-derived microRNAs to recipient animal organisms. RNA Biol. 2013;10:1107–1116. doi: 10.4161/rna.24909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Han M.H., Lesicka J., Fedoroff N. Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc. Natl. Acad. Sci. U.S.A. 2007;104:5437–5442. doi: 10.1073/pnas.0701061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soon P., Kiaris H. MicroRNAs in the tumour microenvironment: big role for small players. Endocr. Relat. Cancer. 2013;20:R257–R267. doi: 10.1530/ERC-13-0119. [DOI] [PubMed] [Google Scholar]
- Sotelo J.R., Porter K.R. An electron microscope study of the rat ovum. J. Biophys. Biochem. Cytol. 1959;5:327–342. doi: 10.1083/jcb.5.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan R., Maresh G., Zhang X., Salomon C., Hooper J., Margolin D., Li L. The Emerging roles of extracellular vesicles as communication vehicles within the tumor microenvironment and beyond. Front. Endocrinol. 2017;8:194. doi: 10.3389/fendo.2017.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Shi K., Yang S., Liu J., Zhou Q., Wang G., Song J., Li Z., Zhang Z., Yuan W. Effect of exosomal miRNA on cancer biology and clinical applications. Mol. Cancer. 2018;17:147. doi: 10.1186/s12943-018-0897-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeo K., Uesaka I., Uehira K., Nishiura M. Fine structure of Cryptococcus neoformans grown in vitro as observed by freeze-etching. J. Bacteriol. 1973;113:1442–1448. doi: 10.1128/jb.113.3.1442-1448.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S., Xia L., Yi P. Exosomal miRNAs in tumor microenvironment. J. Exp. Clin. Cancer Res. 2020;39:67. doi: 10.1186/s13046-020-01570-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery C. Exosomes: Secreted vesicles and intercellular communications. F1000 Biol. Rep. 2011;3:15. doi: 10.3410/B3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y., Cai L., Tian Y., Tu Y., Qiu H., Xie G., Huang D., Zheng R., Zhang W. miR156a mimic represses the epithelial-mesenchymal transition of human nasopharyngeal cancer cells by targeting junctional adhesion molecule A. PLoS One. 2016;11 doi: 10.1371/journal.pone.0157686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkach M., Thery C. Communication by extracellular vesicles: Where we are and where we need to go. Cell. 2016;164:1226–1232. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- Tominaga N., Kosaka N., Ono M., Katsuda T., Yoshioka Y., Tamura K. Brain metastatic cancer cells release microRNA-181c-containing extracellular vesicles capable of destructing blood–brain barrier. Nat. Commun. 2015;6:6716. doi: 10.1038/ncomms7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosar J.P., Rovira C., Naya H., Cayota A. Mining of public sequencing databases supports a non-dietary origin for putative foreign miRNAs: underestimated effects of contamination in NGS. RNA. 2014;20:754–757. doi: 10.1261/rna.044263.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezu T., Tadokoro H., Azuma K., Yoshizawa S., Ohyashiki K., Ohyashiki J.H. Exosomal miR-135b shed from hypoxic multiple myeloma cells enhances angiogenesis by targeting factor-inhibiting HIF-1. Blood. 2014;124(25):3748–3757. doi: 10.1182/blood-2014-05-576116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi H., Ekstrom K., Bossios A., Sjostrand M., Lee J.J., Lotvall J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- Valinezhad Orang A., Safaralizadeh R., Kazemzadeh-Bavili M. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int. J. Genomics. 2014;2014 doi: 10.1155/2014/970607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Niel G., D’Angelo G., Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018;19:213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- Vautrot V., Chanteloup G., Elmallah M., Cordonnier M., Aubin F., Garrido C., Gobbo J. Exosomal miRNA: Small molecules, big impact in colorectal cancer. J. Oncol. 2019;2019:8585276. doi: 10.1155/2019/8585276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlassov A.V., Magdaleno S., Setterquist R., Conrad R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. BBA Gen. Subj. 2012;1820:940–948. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- Wahid F., Shehzad A., Khan T., Kim Y.Y. MicroRNAs: Synthesis, mechanism, function, and recent clinical trials. Biochimica et Biophysica Acta (BBA) – Mol. Cell Res. 2010;1803(11):1231–1243. doi: 10.1016/j.bbamcr.2010.06.013. [DOI] [PubMed] [Google Scholar]
- Wang J., Mei J., Ren G. Plant microRNAs: Biogenesis, homeostasis, and degradation. Frontiers Plant Sci. 2019;10:360. doi: 10.3389/fpls.2019.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Li H., Yuan Y., Etheridge A., Zhou Y., Huang D., Wilmes P., Galas D. The complex exogenous RNA spectra in human plasma: an interface with human gut biota? PLoS One. 2012;7 doi: 10.1371/journal.pone.0051009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Zhao J., Zhang L., Wei F., Lian Y., Wu Y., Gong Z., Zhang S., Zhou J., Cao K., Li X., Xiong W., Li G., Zeng Z., Guo C. Role of tumor microenvironment in tumorigenesis. J. Cancer. 2017;8:761–773. doi: 10.7150/jca.17648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Hang C., Zhang Y., Chen M., Meng X., Cao Q., Song N., Itkow J., Shen F., Yu D. Dietary miR-451 protects erythroid cells from oxidative stress via increasing the activity of Foxo3 pathway. Oncotarget. 2017;8:107109–107124. doi: 10.18632/oncotarget.22346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. Isolation of extracellular vesicles from breast milk. Methods Mol. Biol. 2017;1660:351–353. doi: 10.1007/978-1-4939-7253-1_28. [DOI] [PubMed] [Google Scholar]
- Wang Z.H., Xu C.J. Research progress of microRNA in early detection of ovarian cancer. Chin. Med. J. 2015;128:3363–3370. doi: 10.4103/0366-6999.171459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiberg A., Wang M., Lin F.M., Zhao H., Zhang Z., Kaloshian I., Huang H.D., Jin H. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science. 2013;342:118–123. doi: 10.1126/science.1239705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteside T.L. Tumor-derived exosomes and their role in cancer progression. Adv. Clin. Chem. 2016;74:103–141. doi: 10.1016/bs.acc.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Fact Sheet, 2018. Available at https://www.who.int/news-room/fact-sheets/detail/cancer (accessed 1 December 2020).
- Willett W.C. Balancing life-style and genomics research for disease prevention. Science. 2002;296:695–698. doi: 10.1126/science.1071055. [DOI] [PubMed] [Google Scholar]
- Witwer K.W. XenomiRs and miRNA homeostasis in health and disease: evidence that diet and dietary miRNAs directly and indirectly influence circulating miRNA profiles. RNA Biol. 2012;9:1147–1154. doi: 10.4161/rna.21619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer K.W., McAlexander M.A., Queen S.E., Adams R.J. Real-time quantitative PCR and droplet digital PCR for plant miRNAs in mammalian blood provide little evidence for general uptake of dietary miRNAs: limited evidence for general uptake of dietary plant xenomiRs. RNA Biol. 2013;10:1080–1086. doi: 10.4161/rna.25246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf J.M., Casadevall A. Challenges posed by extracellular vesicles from eukaryotic microbes. Curr. Opin. Microbiol. 2014;22:73–78. doi: 10.1016/j.mib.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf T., Baier S.R., Zempleni J. The intestinal transport of bovine milk exosomes is mediated by endocytosis in human colon carcinoma Caco-2 cells and rat small intestinal IEC-6 cells. J. Nutr. 2015;145:2201–2206. doi: 10.3945/jn.115.218586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortzel I., Dror S., Kenific C.M., Lyden D. Exosome-mediated metastasis: communication from a distance. Dev. Cell. 2019;49(3):347–360. doi: 10.1016/j.devcel.2019.04.011. [DOI] [PubMed] [Google Scholar]
- Xia T., O’Hara A., Araujo I., Barreto J., Carvalho E., Sapucaia J.B., Ramos J.C., Luz E., Pedroso C., Manrique M., Toomey N.L., Brites C., Dittmer D.P., Harrington W.J., Jr. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 2008;68:1436–1442. doi: 10.1158/0008-5472.CAN-07-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M., Li J., Li W., Wang Y., Wu F., Xi Y., Zhang L., Ding C., Luo H., Li Y., Peng L., Zhao L., Peng S., Xiao Y., Dong S., Cao J., Yu W. MicroRNAs activate gene transcription epigenetically as an enhancer trigger. RNA Biol. 2017;14:1326–1334. doi: 10.1080/15476286.2015.1112487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Sticht C., Yin L., Liu L., Karakhanova S., Yin Y., Georgikou C., Gladkich J., Gross W., Gretz N., Herr I. Novel plant microRNAs from broccoletti sprouts do not showcross-kingdom regulation of pancreatic cancer. Oncotarget. 2020;11:1203–1217. doi: 10.18632/oncotarget.27527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F., Zhou X., Fang M., Li H., Su P., Tu Y., Zhang L., Zhou F. Extracellular vesicles in cancer immune microenvironment and cancer immunotherapy Adv. Sci. 2019;6:1901779. doi: 10.1002/advs.201901779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Farmer L.M., Agyekum A.A., Elbaz-Younes I., Hirschi K.D. Detection of an abundant plant-based small RNA in healthy consumers. PLoS One. 2015;10 doi: 10.1371/journal.pone.0137516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Farmer L.M., Agyekum A.A., Hirschi K.D. Detection of dietary plant-based small RNAs in animals. Cell Res. 2015;25:517–520. doi: 10.1038/cr.2015.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Hotz T., Broadnax L., Yarmarkovich M., Elbaz-Younes I., Hirschi K.D. Anomalous uptake and circulatory characteristics of the plant-based small RNA MIR2911. Sci. Rep. 2016;6:26834. doi: 10.1038/srep26834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Kongchan N., Primo, Planta C., Neilson J.R., Hirschi K.D. The atypical genesis and bioavailability of the plant-based small RNA MIR2911: bulking up while breaking down. Mol. Nutr. Food Res. 2017;61:1600974. doi: 10.1002/mnfr.201600974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Yang Z., Li J., Minakhina S., Yang M., Padgett R.W., Steward R., Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zempleni J. Milk exosomes: beyond dietary microRNAs. Genes Nutr. 2017;12:12. doi: 10.1186/s12263-017-0562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zempleni J., Aguilar-Lozano A., Sadri M., Sukreet S., Manca S., Wu D., Zhou F., Mutai E. Biological activities of extracellular vesicles and their cargos from bovine and human milk in humans and implications for infants. J. Nutr. 2017;147:3–10. doi: 10.3945/jn.116.238949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zempleni J., Baier S.R., Howard K.M., Cui J. Gene regulation by dietary microRNAs. Can. J. Physiol Pharmacol. 2015;93:1097–1102. doi: 10.1139/cjpp-2014-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Chen T., Yin Y., Zhang C.-Y., Zhang Y.-L. Dietary microRNA—a novel functional component of food. Adv. Nutr. 2019;10:711–721. doi: 10.1093/advances/nmy127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Hou D., Chen X., Li D., Zhu L., Zhang Y., Li J., Bian Z., Liang X., Cai X., Yin Y., Wang C., Zhang T., Zhu D., Zhang D., Xu J., Chen Q., Ba Y., Liu J., Wang Q. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross kingdom regulation by microRNA. Cell Res. 2012;22:107. doi: 10.1038/cr.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Zhao Y.L., Zhao J.H., Wang S., Jin Y., Chen Z.Q., Fang Y.Y., Hua C.L., Ding S.W., Guo H.S. Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat. Plants. 2016;2:16153. doi: 10.1038/nplants.2016.153. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu D., Chen X., Li J., Li L., Bian Z., Sun F., Lu J., Yin Y., Cai X., Sun Q., Wang K., Ba Y., Wang Q., Wang D., Yang J., Liu P., Xu T., Yan Q., Zhang J. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol. Cell. 2010;39:133–144. doi: 10.1016/j.molcel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wiggins B.E., Lawrence C., Petrick J., Ivashuta S., Heck G. Analysis of plant-derived miRNAs in animal small RNA datasets. BMC Genomics. 2012;13:381. doi: 10.1186/1471-2164-13-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Fong M.Y., Min Y., Somlo G., Liu L., Palomares M.R., Yu Y., Chow A., O'Connor S.T., Chin A.R., Yen Y., Wang Y., Marcusson E.G., Chu P., Wu J., Wu X., Li A.X., Li Z., Gao H., Ren X. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25:501–515. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Fong M.Y., Min Y. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25(4):501–515. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Fong M.Y., Min Y., Somlo G., Liu L., Palomares M.R. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25(4):501–515. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Li X., Liu J., Dong L., Chen Q., Liu J., Kong H., Zhang Q., Qi X., Hou D., Zhang L., Zhang G., Liu Y., Zhang Y., Li J., Wang J., Chen X., Wang H., Zhang J., Chen H. Honeysuckle-encoded atypical microRNA2911 directly targets influenza a viruses. Cell Res. 2015;25:39–49. doi: 10.1038/cr.2014.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K., Liu M., Fu Z., Zhou Z., Kong Y., Liang H., Lin Z., Luo J., Zheng H., Wan P., Zhang J., Zen K., Chen J., Hu F., Zhang C.Y., Ren J., Chen X. Plant microRNAs in larval food regulate honeybee caste development. PLoS Genet. 2017;13 doi: 10.1371/journal.pgen.1006946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Gu J., Li Y., Peng C., Shi M., Wang X., Wei G., Ge O., Wang D., Zhang B., Wu J., Zhong Y., Shen B., Chen H. miR-17-5p enhances pancreatic cancer proliferation by altering cell cycle profiles via disruption of RBL2/E2F4-repressing complexes. Cancer Lett. 2018;412:59–68. doi: 10.1016/j.canlet.2017.09.044. [DOI] [PubMed] [Google Scholar]
Further Reading
- Arroyo J.D., Chevillet J.R., Kroh E.M., Ruf I.K., Pritchard C.C., Gibson D.F., Mitchell P.S., Bennett C.F., Pogosova-Agadjanyan E.L., Stirewalt D.L., Tait J.F., Tewari M. Argonaute2 complexes carry a population of circulating -microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. U.S.A. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach A., Vyas G., Li A., Halushka M., Witwer K. Uptake of dietary milk miRNAs by adult humans: a validation study. 2016;F1000Res. 5:721. doi: 10.12688/f1000research.8548.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N., Baulcombe D. Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T.C., Wentzel E.A., Kent O.A., Ramachandran K., Mullendore M., Lee K.H., Kent O.A., Feldmann G., Yamakuchi M., Ferlito M., Lowenstein C.J., Arking D.E., Beer M.A., Maitra A., Mendell J.T. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. Small RNAs in development - insights from plants. Curr. Opin. Genet. Dev. 2012;22:361–367. doi: 10.1016/j.gde.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng G., Luo R., Hu C., Cao J., Jin Y. Deep sequencing-based identification of pathogen-specific microRNAs in the plasma of rabbits infected with Schistosoma japonicum. Parasitol. 2013;140:1751–1761. doi: 10.1017/S0031182013000917. [DOI] [PubMed] [Google Scholar]
- Fazi F., Racanicchi S., Zardo G., Starnes L.M., Mancini M., Travaglini L., Diverio D., Ammatuna E., Cimino G., Lo-Coco F., Grignani F., Nervi C. Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell. 2007;12:457–466. doi: 10.1016/j.ccr.2007.09.020. [DOI] [PubMed] [Google Scholar]
- Kurihara Y., Watanabe Y. Arabidopsis micro-RNA biogenesis through dicer like 1 protein functions. Proc. Natl. Acad. Sci. U.S.A. 2004;101:12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill M.J., Bourre L., Melgar S., O'Driscoll C.M. Intestinal delivery of non-viral gene therapeutics: physiological barriers and preclinical models. Drug Discov. Today. 2011;16:203–218. doi: 10.1016/j.drudis.2011.01.003. [DOI] [PubMed] [Google Scholar]
- Park M.Y., Wu G., Gonzalez-Sulser A., Vaucheret H., Poethig R.S. Nuclear processing and export of microRNAs in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2005;102:3691–3696. doi: 10.1073/pnas.0405570102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrick J.S., Brower-Toland B., Jackson A.L., Kier L.D. Safety assessment of food and feed from biotechnology-derived crops employing RNA-mediated gene regulation to achieve desired traits: A scientific review. Regul. Toxicol. Pharmacol. 2013;66:167–176. doi: 10.1016/j.yrtph.2013.03.008. [DOI] [PubMed] [Google Scholar]
- Qi Y., Denli A.M., Hannon G.J. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell. 2005;19:421–428. doi: 10.1016/j.molcel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Raver-Shapira N., Marciano E., Meiri E., Spector Y., Rosenfeld N., Moskovits N., Bentwich Z., Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol. Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Shukla N., Shukla V., Saxena S. Plant miRNAs and phytomolecules as anticancer therapeutics. In: Akhtar M., Swamy M., editors. Anticancer plants: Mechanisms and molecular interactions. Springer; Singapore: 2018. pp. 27–41. [Google Scholar]
- Tagawa H., Seto M. A microRNA cluster as a target of genomic amplification in malignant lymphoma. Leukemia. 2005;19:2013–2016. doi: 10.1038/sj.leu.2403942. [DOI] [PubMed] [Google Scholar]
- Vaucheret H. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 2006;20:759–771. doi: 10.1101/gad.1410506. [DOI] [PubMed] [Google Scholar]
- Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- Westwood J.H., Roney J.K., Khatibi P.A. Stromberg VK RNA translocation between parasitic plants and their hosts. Pest Manag Sci. 2009;65:533–539. doi: 10.1002/ps.1727. [DOI] [PubMed] [Google Scholar]
- Xoconostle-Cazares B., Xiang Y., Ruiz-Medrano R., Wang H.L., Monzer J., Yoo B.C., McFarland K.C., Franceschi V.R., Lucas W.J. Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science. 1999;283:94–98. doi: 10.1126/science.283.5398.94. [DOI] [PubMed] [Google Scholar]