Abstract
This current study presents, for the first time, the complete chloroplast genome of two Cleomaceae species: Dipterygium glaucum and Cleome chrysantha in order to evaluate the evolutionary relationship. The cp genome is 158,576 bp in length with 35.74% GC content in D. glaucum and 158,111 bp with 35.96% GC in C. chrysantha. Inverted repeats IR 26,209 bp, 26,251 bp each, LSC of 87,738 bp, 87,184 bp and SSC of 18,420 bp, 18,425 bp respectively. There are 136 genes in the genome, which includes 80 protein coding genes, 31 tRNA genes and four rRNA genes were observed in both chloroplast genomes. 117 genes are unique while the remaining 19 genes are duplicated in IR regions. The analysis of repeats shows that the cp genome includes all types of repeats with more frequent occurrences of palindromic; Also, this analysis indicates that the total number of simple sequence repeats (SSR) were 323 in D. glaucum, and 313 in C. chrysantha, of which the majority of the SSRs in these plastid genomes were mononucleotide repeats A/T which are located in the intergenic spacer. Moreover, the comparative analysis of the four cp sequences revealed four hotspot genes (atpF, rpoC2, rps19, and ycf1), these variable regions could be used as molecular makers for the species authentication as well as resources for inferring phylogenetic relationships of the species. All the relationships in the phylogenetic tree are with high support, this indicate that the complete chloroplast genome is a useful data for inferring phylogenetic relationship within the Cleomaceae and other families. The simple sequence repeats identified will be useful for identification, genetic diversity, and other evolutionary studies of the species. This study reported the first cp genome of the genus Dipterygium and Cleome. The finding of this study will be beneficial for biological disciplines such as evolutionary and genetic diversity studies of the species within the core Cleomaceae.
Keywords: Cleomaceae, Dipterygium glaucum, Cleome chrysantha, Chloroplast genome (cp), SSR
Abbreviations: DNA, Deoxyribonucleic acid; LSC, Large single copy region; SSC, Small single copy region; IR, Inverted repeat; SSR, Simple sequence repeats
1. Introduction
Cleomaceae Bercht. et J. Presl (1825) family include 18 genera and 150–200 species (Patchell et al., 2014), are distributed in the tropics and subtropics areas, and widely uses as ornamentals (Heywood et al., 2007, Fay and Christenhusez, 2010) and a hypothesized origin in central Asia (Feodorova et al., 2010). Cleomaceae are herbs or shrubs; leaves usually palmately compound; fruits capsules, nutlets, or schizocarps, absent septum; and seeds with a testa, usually not arillate (Hall et al., 2002, Iltis et al., 2011).
Until recently, the Cleomaceae has been thought to be closely related with Capparaceae based on the morphological and chemical data (Hall et al., 2002, Iltis, 1957, Rodman et al., 1993, Rodman et al., 1996, Rodman et al., 1998, Rollins, 1993). Some authors merge Brassicaceae, Capparaceae and Cleomaceae into Brassicaceae (Angiosperm Phylogeny Group, 1998, Angiosperm Phylogeny Group, 2003, Judd et al., 1994, Judd et al., 2007). Pax and Hoffmann, 1936, Melchior, 1964 classified the family Capparaceae into two major subfamilies, Capparoideae and Cleomoideae. Recently, the two subfamilies have been elevated to familial rank, which had been previously proposed and believed by earlier taxonomists (Airy Shaw, 1965, Hutchinson, 1967). The phylogeny analysis of the chloroplast genome data strongly supports that the Cleomaceae is a monophyletic family, also the chloroplast sequences data highly supports that Cleomaceae is sister to Brassicaceae (Hall et al., 2002, Hall et al., 2004, Simpson, 2006, Heywood et al., 2007, Hall, 2008, Martín-Bravo et al., 2009, Angiosperm Phylogeny Group, 2009, Angiosperm Phylogeny Group, 2016, Alzahrani et al., 2020). In this study, the Cleomaceae family is represented in two genera: Cleome (Cleome chrysantha) and Dipterygium (Dipterygium glaucum).
Dipterygium glaucum Decne. is a monotypic genus with one species, a medicinal herb, widespread in the tropical and subtropical regions such as Saudi Arabia, Egypt, Sudan, and Pakistan (Ahmad et al., 2014, Mehmood et al., 2010). It is one of the medicinal plants that has multiple uses, it is commonly used for its treatment of miss‑breathing troubles as trachea dilating agent (Moussa et al., 2012), to treat jaundice, blood purifier, psoriasis, and ringworm infestation and as an antiasthma drug (Ahmad et al., 2014, Rahman et al., 2004). It has been stated that the herb of D. glaucum contains significant antioxidant, cytotoxic, and antimicrobial activities (Shaheen et al. 2017). Previous studies demonstrated that D. glaucum plants includes vital phytochemical compounds such as alkaloids, cardiac glycoside, bound anthraquinones, saponins, terpenoids, and sterols (Mehmood et al., 2010, Abdel-Mogib et al., 2000). Hutchinson, 1967, placed genus Dipterygium in Brassicaceae, while Pax and Hoffmann 1936 and Hedge et al., 1980 sited the genus into the Capparaceae family. Based on the evedince of presence of methyl-glucosinolate, some authors have placed the genus Dipterygium within the Capparaceae family, subfamily Dipterygioideae (Hedge et al., 1980, Luning et al., 1992). Presence of six stamens of equal length and a short gynophore are the main floral features that located the genus Dipterygium in Cleomoideae. Patchell et al., 2014 reported Dipterygium belongs to Cleomaceae based on three cpDNA (ndhF, matK, ycf1), one mtDNA (rps3), and one nrDNA (ITS) regions. The result of the phylogenetic analysis in Alzahrani et al. 2020 showed D. glaucum and Tarenaia hassleriana from Cleomaceae in one clade, which confirms placement of D. glaucum in Cleomaceae and is sister to T. hassleriana.
Cleome L. is the largest genus of the subfamily Cleomoideae, Cleomaceae, including 180 to 200 species of herbaceous annual or perennial plants, widespread distribution worldwide in tropical and subtropical areas (Abdullah et al., 2016). Various studies have reported diverse pharmacological activities of plants of the genus Cleome including antidiabetic, anticancer, antibacterial, anti-inflammatory, analgesic, antidiarrheal and antimalarial, as a result of the chemical compounds present in different parts of the Cleome plants such as essential oils, terpenes, flavonoids, glucosinolates and alkaloids (Tripti et al., 2015, Abdullah et al., 2016).
The complete chloroplast genomic has provided large genetic information and molecular markers that are useful for resolve obscure phylogenetic relationships in seed plants (Luo et al., 2014). The majority of the chloroplast genomes of land plants range from 120 to 160 kb and possess their own genomes is rich of evolutionary and phylogenetic information (Raubeson and Jansen, 2005, Yap et al., 2015). At present, more than 3000 complete chloroplast genomes are available in the NCBI database (https://www.ncbi.nlm.nih.gov/genome/GenomesGroup.cgi?taxid = 2759&opt = plastid) (Li et al., 2019). However, there is only two sequence from the chloroplast genome of Cleomaceae species in GenBank.
In this study, we reported the characteristics of the complete chloroplast genome sequences of Dipterygium glaucum and Cleome chrysantha for the first time. We also compared the cp genomes of four Cleomaceae species to investigate the diversity among chloroplast genomes, and SSR was used as a tool to facilitate the assessment of molecular diversity and to identify related species. To understand the relationships of the D. glaucum and C. chrysantha with other species in related families, we constructed the phylogenetic tree using their fully sequenced chloroplast genome sequences.
2. Materials and methods
2.1. Plant material and DNA extraction
Fresh young leaf materials for D. glaucum and C. chrysantha were collected through field survey in Makkah region, Saudi Arabia. Total genomic DNA was extracted from the samples using Qiagen genomic DNA extraction kit according to the manufacturer’s protocols.
2.2. Library construction, sequencing and assembly
A total amount of 1.0μ g DNA was used as input material for the DNA sample preparations. Sequencing libraries were generated using NEBNext DNA Library Prep Kit following manufacturer’s recommendations and indices were added to each sample. The genomic DNA is randomly fragmented to a size of 350 bp by shearing, then DNA fragments were end polished, A-tailed, and ligated with the NEBNext adapter for Illumina sequencing and further PCR enriched by P5 and indexed P7 oligos. The PCR products were purified (AMPure XP system) and resulting libraries were analyzed for size distribution by Agilent 2100 Bio analyzer and quantified using real-time PCR. The qualified libraries are fed into Illumina sequencers after pooling according to its effective concentration and expected data volume. The raw reads were filtered to get the clean reads (5 Gb) using PRINSEQlite v0.20.4 (Schmieder and Edwards, 2011) and were subjected to de novo assembly using NOVOPlasty2.7.2 (Dierckxsens et al., 2017) with kmer (K-mer = 33) to assemble the complete chloroplast genome from the whole genome sequence. Finally, for each species one contig containing the complete chloroplast genome sequence was generated.
2.3. Gene annotation
Genes were annotated using DOGMA (Dual Organellar GenoMe Annotator, University of Texas at Austin, Austin, TX, USA) (Wyman et al., 2014). The positions of start and stop codon were adjusted manually. tRNA genes were identified by the trnAscan-SE server (http://lowelab.ucsc.edu/tRNAscan-SE/) (Schattner et al., 2005). The circular chloroplast genome maps were drawn using OGDRAW (Organellar Genome DRAW) (Lohse et al., 2007). The sequences of the chloroplast genome were deposited in the GenBank database: D. glaucum (MT041700) and C. chrysantha (MT948188).
2.4. Sequence analysis
The relative synonymous codon usage (RSCU) values, base composition and codon usage were analysed using MEGA 6.0 software. Potential RNA editing sites present in the protein coding genes were predicted by the PREP suite (Kurtz et al., 2001) with the cutoff value set to 0.8
2.5. Repeat analysis in chloroplast genome
The online software MIcroSAtellite (MISA) (Thiel et al., 2003) was used to identify Simple Sequence Repeats (SSRs) in the chloroplast genome with the following parameters: eight, five, four and three repeats units for mononucleotides, dinucleotides, trinucleotides and tetra, penta, hexa nucleotides SSR motifs respectively. The REPuter software (https://bibiserv.cebitec.uni-bielefeld.de/reputer) (Kurtz et al., 2001) was used with default settings to detect the size and location of the long repeats in the two Cleomaceae species.
2.6. Genome comparison
The chloroplast genome of D. glaucum, C. chrysantha, C. lutea (NC_049613) and T. hassleriana (NC_034354) were compared using mVISTA program (Mayor et al., 2000) and the annotation of D. glaucum used as reference in the Shuffle-LAGAN mode (Frazer et al., 2004). Comparison of the large single copy (LSC), inverted repeat (IR), small single copy (SSC), and inverted repeat (IR) boundaries among the four species of Capparaceae plastomes.
2.7. Characterization of substitution rate
The nonsynonymous (dN) and synonymous (dS) substitution rates were calculated using the DNAsp v5.10.01 (Librado and Rozas 2009). The plastome of D. glaucum was compared with the plastome of C. chrysantha, C. lutea and T. hassleriana to identify the genes that are under selective pressure. Geneious software v. 8.1.3 (Biomatters, Ltd., Auckland, New Zealand) was used to align the individual protein coding genes separately, the aligned sequences were then translated into protein sequence.
2.8. Phylogenetic analysis
The analysis was conducted based on the complete chloroplast genome sequences of four Cleomaceae species (D. glaucum, C. chrysantha, C. lutea and T. hassleriana), four Capparaceae species, eight species of Brassicaceae and two species of Malvaceae that were used as outgroup. All of the sequences were aligned using MAFFT (Katoh and Standley 2013) with default settings. The phylogenetic trees were reconstructed based on Maximum Parsimony (MP) method using PAUP version 4.0b10 (Felsenstein 1978) and a using heuristic search strategy of 1000 random sequence addition replicates with tree bisection– reconnection (TBR) branch swapping, saving a maximum 100 trees pear replicate, with MulTrees on, gaps were treated as missing data. statistical support was assessed for clades with nonparametric bootstrap analysis using 1000 bootstrap replicates. A 50% majority-rule consensus tree was calculated from all the most parsimonious trees. Bayesian inference (BI) analyses were performed in MrBayes version 3.2.6 (Fredrik et al., 2012) the best models were selected using jModelTest version 3.7 (Posada 2008).
3. Results
3.1. Characteristics of two chloroplast genome
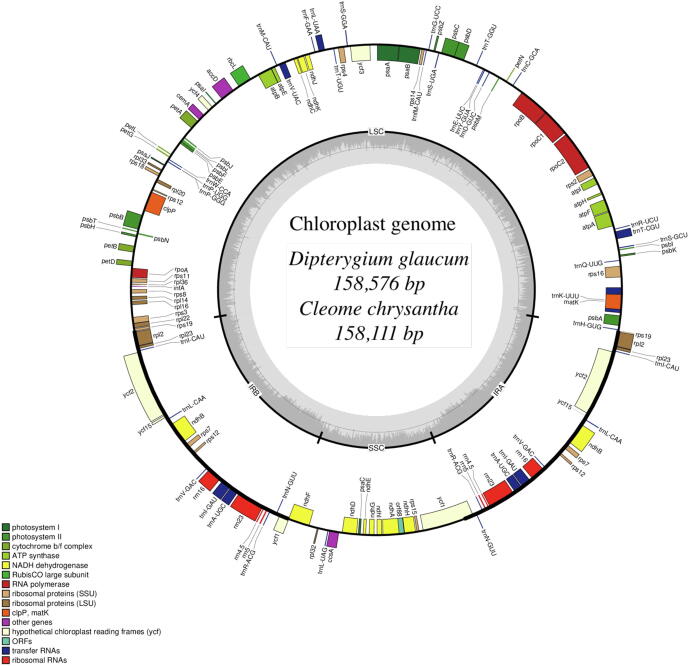
The complete plastome sequence has a circular and quadripartite structure, the total length of the D. glaucum genome is 158,576 bp (Alzahrani et al., 2020), 158,111 bp in C. chrysantha. The plastome has four distinct regions in D. glaucum and C. chrysantha which are Large Single Copy (LSC) length is 87,738 bp and 87,184 bp respectively, Small Single Copy (SSC) length is 18,420 bp and 18,425 bp respectively and a pair of Inverted repeats (IRa and IRb) length is 26,209 bp and 26,251 bp respectively which separates the LSC and SSC regions (Fig. 1; Table 1). The region coding for genes in D. glaucum and C. chrysantha respectively is 76, 598 bp – 76,905 bp in length which constitutes 48.30% − 48.63% of the genome, the remaining 72, 286 bp – 71,042 bp is the non-coding region which includes intron and intergenic spacer (45.58% − 44.93%). The plastome sequence has GC of 35.74% − 35.96% and AT content of 64.23% − 64.02% respectively (Table 1). The LSC regions possessed GC content of 33.27% − 33.59% and the SSC regions content of 28.76% − 28.97%, while the inverted repeats IRa and IRb have 42.34% − 42.32%. The chloroplast genome sequences were deposited in the GenBank: Accession Number for D. glaucum is (MT041700) and for C. chrysantha is (MT948188).
Fig. 1.
Chloroplast genome maps of the two Cleomaceae (A) D. glaucum and (B) C. chrysantha. Genes inside of the circles are transcribed clockwise, while genes outside circles are transcribed counterclockwise. The light grey inner circle and the dark grey corresponds to the AT and GC content respectively. The different colours represent the different functional groups of the genes.
Table 1.
Bases content in the D. glaucum and C. chrysantha chloroplast genomes.
| Species | D. glaucum | C. chrysantha |
|---|---|---|
| Genome size (bp) | 158,576 | 158,111 |
| IR (bp) | 26,209 | 26,251 |
| LSC (bp) | 87,738 | 87,184 |
| SSC (bp) | 18,420 | 18,425 |
| Total number of genes | 136 | 136 |
| rRNA | 4 | 4 |
| tRNA | 31 | 31 |
| Protein-coding genes | 80 | 80 |
| T (U) % | 32.57 | 32.48 |
| C % | 18.2 | 18.32 |
| A % | 31.66 | 31.54 |
| G % | 17.54 | 17.64 |
Result of the genes annotation revealed a total of 136 in the two species, 117 are unique; the remaining 19 are duplicated in the inverted region. The plastome contained 80 protein coding genes, 31 tRNA genes and four rRNA genes (Fig. 1 and Table 2). The inverted repeat region contained eight protein coding genes, seven tRNA in D. glaucum and eight tRNA in C. chrysantha and four rRNA in the single copy region; the LSC contained 61 protein coding genes and 23 tRNA genes, the remaining 13 protein coding genes and one tRNA are situated within the SSC region.
Table 2.
Gene contents in the chloroplast genomes of D. glaucum and C. chrysantha.
| Category | Group of genes | Name of genes |
|---|---|---|
| RNA genes | ribosomal RNA genes (rRNA) | rrn5, rrn4.5, rrn16, rrn23 |
| Transfer RNA genes (tRNA) | trnH-GUG, trnK-UUU+, trnQ-UUG, trnS-GCU, trnT-CGU+, trnR-UCU, trnC-GCA, trnD-GUC, trnY-GUA, trnE-UUC (A, B+,a), trnT-GGU, trnS-UGA, trnG-UCC, trnfM-CAU, trnS-GGA, trnT-UGU, trnL-UAA+, trnF-GAA, trnV-UAC+, trnM-CAU, trnW-CCA, trnP-UGG, trnP-GGG, trnI-CAUa, trnL-CAAa, trnV-GACa, trnI-GAU (A+,a,B), trnA-UGC+,a, trnR-ACGa, trnN-GUUa, trnL-UAG | |
| Ribosomal proteins | Small subunit of ribosome | rps2, rps3, rps4, rps7a, rps8, rps11, rps12a, rps14, rps15, rps16+, rps18, rps19 |
| Transcription | Large subunit of ribosome | rpl2+,a, rpl14, rpl16, rpl20, rpl22, rpl23a, rpl32, rpl33, rpl36 |
| DNA dependent RNA polymerase | rpoA, rpoB, rpoC1+, rpoC2 | |
| Translational initiation factor | infA | |
| Protein genes | Photosystem I | psaA, psaB, psaC, psaI, psaJ, ycf3++ |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Subunit of cytochrome | petA, petB, petD, petG, petL, petN | |
| Subunit of synthase | atpA, atpB, atpE, atpF+, atpH, atpI | |
| Large subunit of rubisco | rbcL | |
| NADH dehydrogenase | ndhA+, ndhB+,a, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| ATP dependent protease subunit P | clpP++ | |
| Chloroplast envelope membrabe protein | cemA | |
| Other genes | Maturase | matK |
| Subunit acetyl-coA carboxylase | accD | |
| C-type cytochrome systhesis | ccsA | |
| Hypothetical proteins | ycf2a, ycf4, ycf15 (A++,a,B) | |
| Component of TIC complex | ycf1a |
*(A) D. glaucum cp genome; (B) C. chrysantha cp genome.
Gene with one intron, ++ Gene with two intron and a Gene with copies.
The composition of A is 31.66% − 31.54%, C is 18.2% − 18.32%, T is 32.57% − 32.48% and G is 17.54% − 17.64% in D. glaucum and C. chrysantha respectively. Our results revealed that the majority of protein coding genes start with the typical ATG codon, which is responsible in the coding of methionine, while others begin with the codons ATC, GTG and ACG, as in most Angiosperms plant chloroplast genomes (Raman and Park, 2016, Park et al., 2017, Li et al., 2017). Intron is present in several of the protein coding and tRNA genes of the D. glaucum and C. chrysantha chloroplast genomes, similar to other chloroplast genomes of flowering plants (Raman and Park, 2016, Park et al., 2017).
There are 15 genes in D. glaucum and C. chrysantha that contain intron out of the total genes, among the 15 genes, nine are protein coding genes while the remaining six are tRNA genes. One gene has the intron namely ndhA located in SSC region, five genes namely rpl2, ycf15, ndhB, trnI-GAU and trnA-UGC are located in the IR region in D. glaucum and four genes namely rpl2, ndhB, trnE-GAU and trnA-UGC in C. chrysantha, while the remaining nine are located in the LSC region. Only three genes ycf3, clpP and ycf15 have two introns in D. glaucum and two genes ycf3 and clpP in C. chrysantha, the other 12 genes have only one intron. The tRNA, trnK-UUU has the longest intron of 2570 bp in D. glaucum and 2568 pb in C. chrysantha, this is as a result of the matK gene being located within the intron of the trnK.
3.2. Codon usage
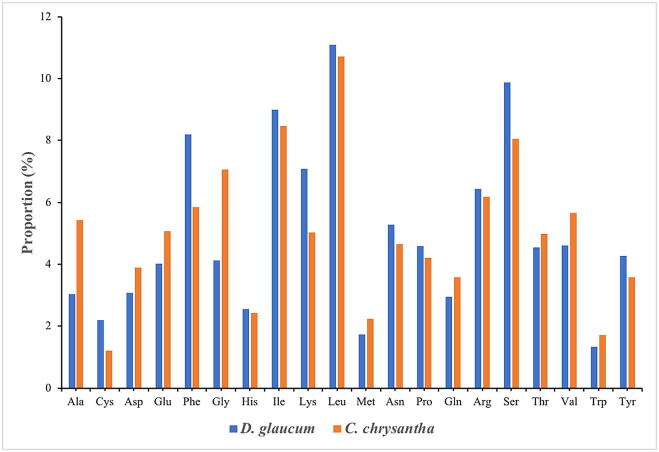
Codon usage is vital in the influence of the chloroplast genome evolution. Scientists have stated that the evolutionary phenomenon occurs as a result of bias in mutation (Li et al., 2017). The nucleotides of protein coding and tRNA genes were used in computing the codon usage bias of the plastome. The nucleotides sequences in D. glaucum: 94,112 bp and in C. chrysantha: 89,715 bp. Supplementary Table A1, Table A2 present the relative synonymous codon usage (RSCU) of each codon in the genome, the result indicated that all the genes in D. glaucum are encoded by 31,366 codons, while in C. chrysantha they encoded by 26,475 codons, coding for the amino acids Leucine are the most frequent codons, this has been stated in other flowering plant genomes (Liu et al., 2018); 2,772 (8.83%) in D. glaucum and 2,831 (10.69%) in C. chrysantha (Fig. 2), whereas codons coding are the least in the genome for Tryptophan 659 (2.10%) in D. glaucum and 452 (1.70%) in C. chrysantha (Fig. 2). A- and T- ending are discovered to be less frequent than their counterparts G- and C-. Appendices Table A1, Table A2 presented the result of the analysis that indicates the codon usage bias is low in the chloroplast genome of Cleomaceae species. The RSCU values of 28 codons were greater than 1 and all having A/T endings, whereas other 28 codons were less than 1 and all end with G/C. The RSCU values of Tryptophan and Methionine amino acids 1, therefore they have only one codon.
Fig. 2.
Amino acids frequencies in D. glaucum and C. chrysantha chloroplast genome protein coding sequences.
3.3. RNA editing sites
RNA editing sites feature a set of processes that include inserting, deleting or modifying nucleotides which alter the DNA-encoded sequence of a transcribed RNA (Mower 2009), which saves a way to create transcript and protein diversity (Bundschuh et al., 2011). Some chloroplast RNA editing site are preserved in plants (Zeng et al., 2007). The PREP suite program was used to predict the RNA editing sites in the two species chloroplast genomes, the first codon position of the first nucleotide used in all the analysis. The results (Appendices Table A3, Table A4) indicates that the amino acid Serine to Leucine are the majority of the conversion in the codon positions, this conversion is found to occur more frequently (Luo et al., 2014). In all, the programme revealed 41 editing sites in the genome distributed among 16 protein coding genes in D. glaucum and 35 editing sites distributed among 14 protein coding genes in C. chrysantha. As reported in previous researches (Wang et al., 2017, Kumbhar et al., 2018, Park et al., 2018) in D. glaucum, the ndhB gene have the highest number of editing site (10 sites) followed by ndhD (7 sites) and accD, psaB, psbE, psbF, rpoC2, rps14, rps16 have the least one site each; in C. chrysantha the ndhD gene have the highest number of editing site (seven sites) followed by ndhB (five sites) and the following genes: psbE, psbF, rpoC1, rpoC2, rps14, rps16 have the least, one site each. Certain RNA sites amidst all the conversion in the RNA editing (modification) site changed the amino acid from Proline to Serine, one site in D. glaucum and two sites in C. chrysantha. RNA predicting site in the first codon of the first nucleotides are not present in the following genes atpA, atpB, atpF, atpI, ccsA, petB, petD, petG, psaI, psbB, psbL, rpl2, rpl20, rpl23, rpoA, rps2, rps8 and ycf3 among others, except the genes ndhA, petL and psaB which are only found in C. chrysantha. This result indicated that the preservation of RNA editing is fundamental (Magdalena et al., 2009, Huang et al., 2013).
3.4. Repeat analysis
3.4.1. Long repeats
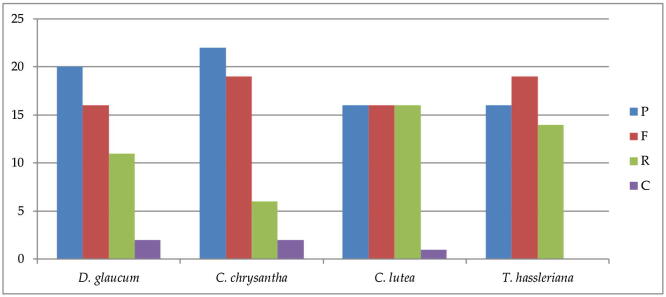
Repeats sequences in the chloroplast genomes of the four species were determined by the REPuter programme with default settings; obtained results clearly show that forward, reverse, palindrome, and complemented repeats were detected in the cp genomes. The long repeats analysis showed in Dipterygium glaucum, Cleome chrysantha, Cleomella lutea and Tarenaya hassleriana: 20–22-16–16 palindromic repeats, 16–19-16–19 forward repeats, 11–6-16–14 reverse repeats and 2–2-1–0 complement repeat respectively (Fig. 3). Majority of the repeats size respectively are: between 20 and 29 bp (81.63%-89.79%-91.83%-59.18%), followed 30–39 bp (12.24%-6.12%-6.12%-24.48%), whereas 40–49 bp (2.04%-2.04%- absent-14.28%), followed 50–59 bp (4.08%-2.04%- absent-2.04%) and 60–69 bp present only in C. lutea (2.04%) (Appendices Table A5, Table A6). In total, the chloroplast genome of the four species there are 49 repeats. In the first location the codon region harbored 61.22% of the repeats in D. glaucum and 67.34% in C. chrysantha, tRNA contained 3 repeats (6.12%) in D. glaucum and 6 repeats (12.24%) in C. chrysantha, the remaining are located in the protein coding genes 8 repeats (16.32%) in D. glaucum and 5 repeats (10.20%) in C. chrysantha. The length of repeated sequences in four chloroplast genomes ranged from 10 to 69 bp, analogous to the lengths in other angiosperm plants (Li et al., 2017, Greiner et al., 2008, Song et al., 2017).
Fig. 3.
Number of different repeats in the chloroplast genomes of the four Cleomaceae species. P = palindromic, F = forward, R = reverse and C = complement.
3.4.2. Simple sequence repeats (SSRs).
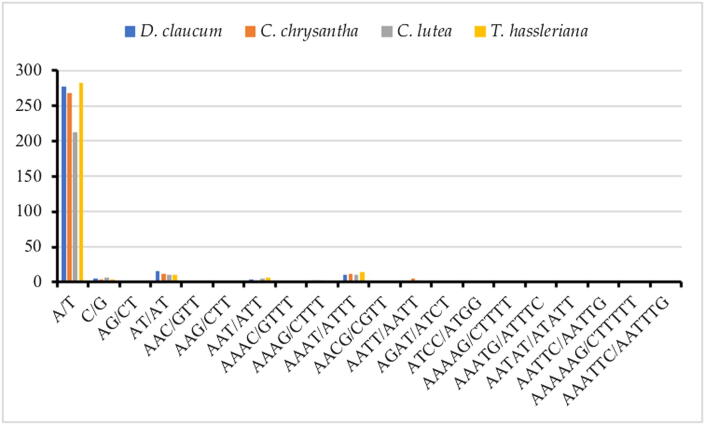
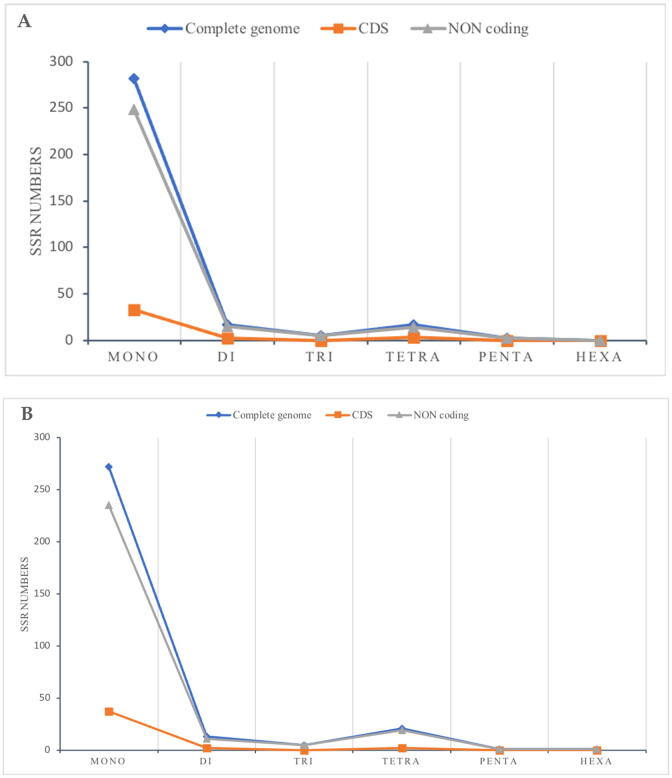
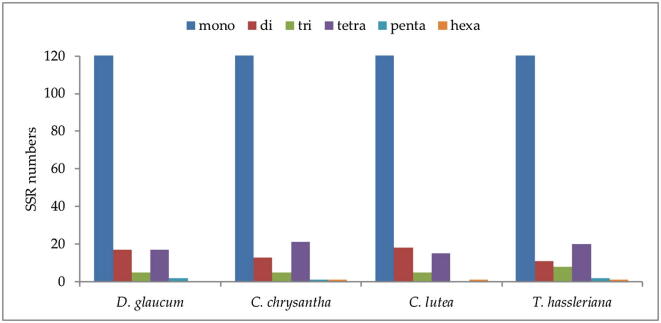
The SSRs or microsatellites are a group of short repeat sequences of nucleotide series (1–6 bp), which are used as a tool that facilitates the assessment of molecular diversity (Kaila et al., 2017). The genetic variation within and among species with the valuable molecular marker of the SSRs are extremely important for studying genetic heterogeneity, and contributes to species recognition (Bryan et al., 1999, Provan, 2000, Ebert and Peakall, 2009). In this study, the microsatellites were found in plastid genome of D. glaucum is 323, C. chrysantha is 313, C. lutea is 258 and of T. hassleriana is 328 (Table 3). Majority of SSRs in the cp genome in D. glaucum, C. chrysantha, C. lutea and T. hassleriana are mononucleotide (90.24%-86.93%-84.88%-87.19%) respectively of which most are poly T and A (Fig. 4) Poly T (polythymine) constituted (51.21%-48.24%-49.18%-55.61%) whereas poly A (polyadenine) (37.56%-36.68%-31.89%-30.61%) respectively. Only two poly C (polycytosine) (0.97%) in D. glaucum, three (1.50%) in C. chrysantha and (1.53%) in T. hassleriana and five (2.7%) in C. lutea and only a single poly G (polyguanine) in three species (0.48% in D. glaucum, 0.50% in C. chrysantha, 0.54% in C. lutea) and absent in T. hassleriana. The dinucleotide AT/AT is found in the all genomes. Reflecting series complementary, three trinucleotide AAC/GTT, AAG/CTT and AAT/ATT, seven tetranucleotide AAAC/GTTT, AAAG/CTTT, AAAT/ATTT, AACG/CGTT, AATT/AATT, AGAT/ATCT and ATCC/ATGG, four pentanucleotide AAATG/ATTTC, AAATG/ATTTC, AATAT/ATATT and AATTC/AATTG and two hexanucleotide were discovered in the genome (Fig. 4). High richness in mono nucleotides poly A and T has been observed in most flowering plants cp genome (Li et al., 2017). The density of microsatellite in the intergenic spacer regions are significantly more (88.29%-86.39%) than the coding regions (11.70%-13.60%) in D. glaucum and C. chrysantha respectively (Fig. 5).
Table 3.
The SSRs in four chloroplast genomes of Cleomaceae.
| SSR type | Repeat unit | Species |
|||
|---|---|---|---|---|---|
| D. glaucum | C. chrysantha | C. lutea | T. hassleriana | ||
| Mono | A/T | 277 | 268 | 212 | 282 |
| C/G | 5 | 4 | 7 | 4 | |
| Di | AG/CT | 1 | 1 | 1 | 0 |
| AT/AT | 16 | 12 | 17 | 11 | |
| Tri | AAC/GTT | 1 | 1 | 0 | 0 |
| AAG/CTT | 0 | 1 | 0 | 1 | |
| AAT/ATT | 4 | 3 | 5 | 7 | |
| Tetra | AAAC/GTTT | 0 | 0 | 0 | 1 |
| AAAG/CTTT | 2 | 2 | 0 | 1 | |
| AAAT/ATTT | 11 | 12 | 10 | 14 | |
| AACG/CGTT | 1 | 0 | 0 | 1 | |
| AATT/AATT | 1 | 5 | 3 | 1 | |
| AGAT/ATCT | 1 | 1 | 1 | 1 | |
| ATCC/ATGG | 1 | 1 | 1 | 1 | |
| Penta | AAAAG/CTTTT | 0 | 0 | 0 | 1 |
| AAATG/ATTTC | 1 | 0 | 0 | 0 | |
| AATAT/ATATT | 1 | 0 | 0 | 1 | |
| AATTC/AATTG | 0 | 1 | 0 | 0 | |
| Hexa | AAAAAG/CTTTTT | 0 | 0 | 1 | 0 |
| AAATTC/AATTTG | 0 | 1 | 0 | 1 | |
Fig. 4.
Frequency of different SSR motifs in different repeat types in four chloroplast genomes of Cleomaceae.
Fig. 5.
SSR Number in complete genome, protein coding regions, and noncoding genes: (A) D. glaucum, (B) C. chrysantha.
The result of comparative analysis of the simple sequence repeat between the chloroplast genome sequences of the four Cleomaceae species (Fig. 6) indicated that the more frequent occurrences are the mononucleotide repeats. The highest number of mononucleotide in T. hassleriana is 286. Pentanucleotide is not present in C. lutea and hexanucleotide is not present in D. glaucum however they are present in the other species.
Fig. 6.
Number of different SSR types in the four-chloroplast genome of Cleomaceae.
3.5. Comparative analysis of the cp genome of the Cleomaceae species.
To analyse the DNA sequence divergence among the chloroplast genomes of the four Cleomaceae species: D. glaucum, C. chrysantha, C. lutea and T. hassleriana comparative analysis was done using mVISTA program to align the sequences. To understand the structural characteristics in the cp genomes, the annotation of D. glaucum used as reference. The alignment outcome reveals highly conserved genomes with few variations. As in many flowering plants cp genomes, the noncoding gene regions were less conserved than the coding regions (Fig. 7). Among the four cp genomes, the results showed that the following genes, rps16 trnQ, psbK-trnS, atpF-atpH, atpH-atpI, rpoB-trnC, psbM-trnD, trnD-trnY, trnE-trnT, trnT-psbD, trnT-trnL, trnM-atpE, rbcL-accD, petA-psbJ, psbE-petL, rbs16-rbs3, ndhF-rpl32 and trnV-rps12, were the most divergent non-coding regions. However, considerable slight variation was observed in four genes (atpF, rpoC2, rps19 and ycf1) of the four chloroplast genomes sequences.
Fig. 7.
Comparison of four chloroplast genomes in Cleomaceae family using mVISTA. Annotation of D. glaucum was used as reference. The top arrows above the alignment indicate gene orientation. Blue bars represent protein coding, pink bars represent noncoding sequences (CNS) and light green indicates tRNAs and rRNAs. The horizontal axis indicates the coordinates in the cp genome, the vertical scale indicates the percentage identity between 50 and 100%.
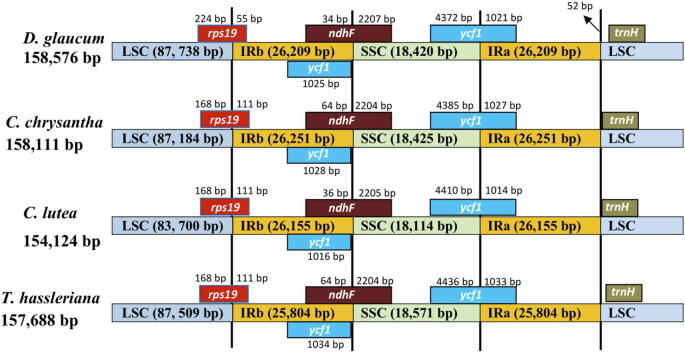
In this current study, the IR-LCS and IR-SSC boundaries of the four Cleomaceae species genomes were compared. Even though the result showed that there are similarities among the compared cp genomes of all four species (Fig. 8), C. lutea has the smallest chloroplast genome (154,124 bp), whereas D. glaucum has the largest chloroplast genome (158,576 bp). The smallest IR region in T. hassleriana (25,804 bp) and the largest in C. chrysantha (26,251 bp). Furthermore, the lengths of LSC regions varied among the four Cleomaceae species, it is 87,738 bp in D. glaucum, 87,184 bp in C. chrysantha, 83,700 bp in C. lutea, and 87,509 bp in T. hassleriana. Also, comparative analysis of the cp genome of the four Cleomaceae species revealed that the location of the rpsl9 gene is between the LSC and IRb regions. The ycf1 gene was located in IRb regions and crossed the SSC/IRa region and extends into the SSC region by different lengths depending on the genome (D. glaucum 4,372 bp; C. chrysantha 4,385 bp; C. lutea 4,410 bp and T. hassleriana 4,436 bp); the IRb region includes 1,021; 1,027; 1,014 and 1,033 bp respectively of the ycf1 gene. The ndhF was found in the IRb/SSC to have 34 bp in D. glaucum, 64 bp in C. chrysantha and T. hassleriana and 36 bp in C. lutea in the IRb region and extends into the SSC region 2,207 bp in D. glaucum, 224 bp in C. chrysantha and T. hassleriana and 2,205 bp in C. lutea.
Fig. 8.
Comparison of the IR, SSC and LSC junction positions among four chloroplast genomes of Cleomaceae.
3.6. Divergence of protein coding gene sequence
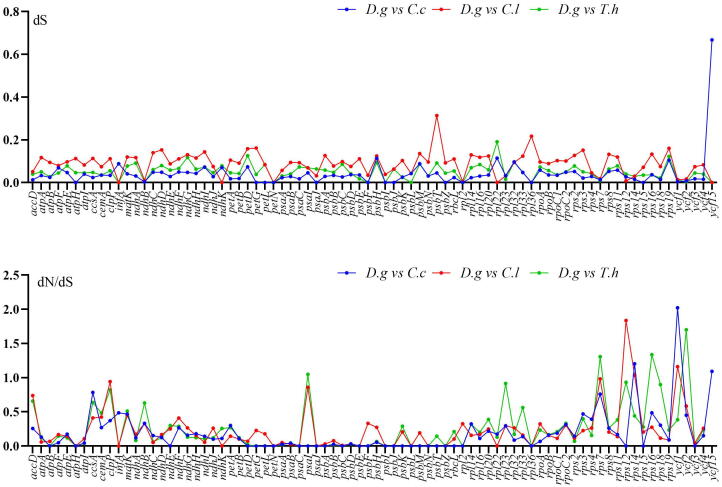
The rates of synonymous (dS) and nonsynonymous (dN) substitution and dN/dS ratio were calculated to detect the selective pressure among 80 protein coding genes in the cp genome of four Cleomaceae species. The results showed that the dN/dS ration is less than 1 in all of the paired genes except rps14, ycf1 and ycf15 in D. glaucum vs C. chrysantha having 1.19, 1 and 2.16 respectively, rps12, rps14 and ycf1 in D. glaucum vs C. lutea having 1.83, 1.03 and 1.16 respectively and psaI, rps7, rps16 and ycf2 in D. glaucum vs T. hassleriana having 1.04, 1.3, 1.33 and 1.7 respectively (Fig. 9). The synonymous (dS) values in all the genes ranges from 0 to 0.32 (Fig. 9).
Fig. 9.
The synonymous (dS) and dN/dS ration values of 80 protein coding genes from four Cleomaceae cp genomes.
3.7. Phylogenetic analysis
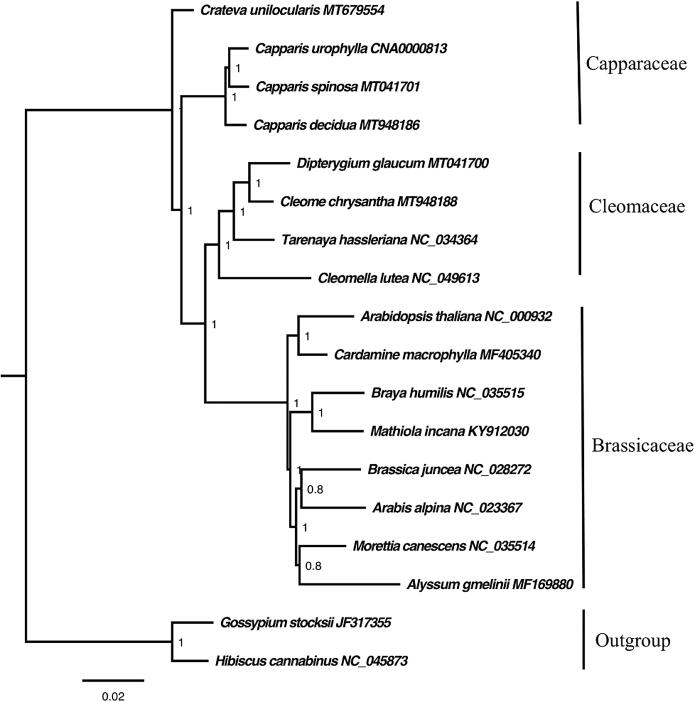
Phylogenetic relationships based on the Bayesian and Maximum Parsimony Analysis placed all samples into three main clades where the results match in the two analysis with strong support in all the nodes PP, 1.00, and MP, 100 (Fig. 10). The first clade contains species of Capparaceae family. The second clade comprised of Cleomaceae species while the third clade includes species from Brassicaceae family. The phylogenetic tree showed that the family Cleomaceae was separated from Capparaceae and becomes sister to Brassicaceae family which is consistent with some previous classifications of the order Brassicales (Angiosperm Phylogeny Group, 2009, Angiosperm Phylogeny Group, 2016).
Fig. 10.
Phylogenetic tree reconstruction based on the complete chloroplast genome of sixteen taxa inferred from Bayesian Inference (BI) methods showing relationship within Cleomaceae and other families. Numbers in the clade represent posterior probability (PP) values.
4. Discussion
Next Generation Sequencing (NGS) provide sufficient information for molecular genetic markers, species identification, relationships and evolution within and between different species (Powell et al., 1995, Grassi et al., 2002, Doorduin et al., 2011, Straub et al., 2012). The complete chloroplast genome has provided large genetic information and molecular markers that are valuable tools to solve obscure phylogenetic relationships among land plants (Luo et al., 2014).
The chloroplast genomes of D. glaucum and C. chrysantha have similar roots to chloroplast genome of angiosperms (Raman and Park, 2016, Park et al., 2017, Chen et al., 2018). This study presents for the first time the characterization of complete chloroplast genomes of C. chrysantha and D. glaucum sequenced. The chloroplast genome size of D. glaucum is 158,576 bp and 158,111 bp in C. chrysantha (Fig. 1). The plastome sequence of D. glaucum and C. chrysantha has GC of 35.74% − 35.96% and AT content of 64.23% − 64.02% respectively (Table 1). The GC content in the IR region is higher (42.34% − 42.32%) than that of the LSC (33.27% − 33.59%) and SSC regions (28.76% − 28.97%).
Previous studies have shown that the plastomes of flowering plants are much conserved in structural organization and gene content; however, contraction and expansion do occur (Chang et al., 2006, Chen et al., 2018). There are 136 genes in the both genomes, which includes 80 protein coding genes, 31 tRNA genes and 4 rRNA genes. The IR region contained 8 protein coding genes, 7 tRNA in D. glaucum and 8 tRNA in C. chrysantha and 4 rRNA; the LSC contained 61 protein coding genes and 23 tRNA genes, the SSC region contained 13 protein coding genes and 1 tRNA. Intron is present in several of the protein coding and tRNA genes of the D. glaucum and C. chrysantha chloroplast genomes, similar to other chloroplast genomes of flowering plants (Raman and Park, 2016, Park et al., 2017, Chen et al., 2018). Out of the total genes in the cp genomes of both species, 15 genes are containing intron, nine of which are protein coding genes, whereas the remaining six genes are tRNA.
The results showed the genes in the plastome are encoded by 31,366 codons in D. glaucum and 26,475 codons in C. chrysantha, the most common codons are the coding for the amino acids Leucine, which has been previously stated in several cp genome of flowering plants (Liu et al., 2018). The Result of RNA editing sites revealed that most of the amino acid conversions in the codon positions were Serine to Leucine, presenting 41 editing sites in the genome distributed among 16 protein coding genes in D. glaucum and 35 editing sites distributed among 14 protein coding genes in C. chrysantha. The repeat sequence was identified in the chloroplast genomes of D. glaucum, C. chrysantha, C. lutea and T. hassleriana using default settings. The long repeats analysis showed 20–22-16–16 palindromic repeats, 16–19-16–19 forward repeats, 11–6-16–14 reverse repeats and 2–2-1–0 complement repeat respectively (Fig. 3). The length of repeated sequences in the four chloroplast genomes ranged from 10 to 69 bp, analogous to the lengths in other angiosperm plants (Greiner et al., 2008, Li et al., 2017, Song et al., 2017). Majority of SSRs in the cp genomes are mononucleotide, the largest number was found in D. glaucum of which most are poly T and A (Fig. 4) as in most flowering plants cp genome (Li et al., 2017). The highest constituted of Poly T (polythymine) in T. hassleriana whereas poly A (polyadenine) in D. glaucum. Only two poly C (polycytosine) in D. glaucum, three in C. chrysantha and T. hassleriana and five in C. lutea and only a single poly G (polyguanine) in three species (D. glaucum, C. chrysantha, C. lutea) and absent in T. hassleriana. The dinucleotide, trinucleotide, tetranucleotide, pentanucleotide and hexanucleotide are found in all four genomes (Fig. 4).
The degree of DNA sequence divergence was examined in the four-chloroplast genome. There were many different non-coding regions among the four cp genomes in addition to following genes atpF, rpoC2, rps19 and ycf1. Similarly, to the majority of the angiosperm cp genomes, the gene-coding regions are more highly conserved than those of their noncoding counterparts (Fig. 7). Among the four cp genomes, some divergent non-coding regions have been observed in addition to the following genes atpF, rpoC2, rps19 and ycf1. This study compared between IR-LCS and IR-SSC boundaries of the four cp genomes of Cleomaceae, the results showed that C. lutea has the smallest chloroplast genome from the four studied, while D. glaucum has the greatest chloroplast genome in size. The smallest IR region in T. hassleriana and the largest in C. chrysantha, the smallest LSC region in C. lutea and largest in D. glaucum and the smallest SSC region in C. lutea and largest in T. hassleriana. The results to rates of the selective pressure among 80 protein coding genes in the chloroplast genome of the four Cleomaceae species revealed that the dN/dS ration is less than 1 in all of the paired genes, yet there were a few exceptions (Fig. 7). The identical (dS) values in all of the protein coding genes ranges from 0 to 0.32 (Fig. 9). Phylogenetic relationships based on the Bayesian and Maximum Parsimony Analysis placed all samples into three main clades, where every family is in a separate clade (Fig. 10). The phylogenetic tree showed that the family Cleomaceae was separated from Capparaceae and becomes sister to Brassicaceae family which is consistent with some previous classifications of the order Brassicales (Angiosperm Phylogeny Group, 2009, Angiosperm Phylogeny Group, 2016).
5. Conclusion
This current study used the Illumina HiSeq 2500 platform to sequence the complete chloroplast genome of Dipterygium glaucum and Cleome chrysantha, which provided valuable plastid genomic resources for these medicinally important plants. We annotated the cp genome for both species, moreover, we identified the base composition, codon usage and RNA editing site, SSRs and Long repeat.
6. Data availability
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number (D. glaucum MT041700; C. chrysantha MT948188).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Peer review under responsibility of King Saud University.
Appendix.
.
Table A1.
Codon – anticodon recognition patterns and codon usage of D. glaucum chloroplast genome.
| Codon | Amino Acid | RSCU | tRNA | Codon | Amino Acid | RSCU | tRNA |
|---|---|---|---|---|---|---|---|
| UUU | Phe | 1.3 | trnF-GAA | UAU | Tyr | 1.45 | trnY-GUA |
| UUC | Phe | 0.7 | UAC | Tyr | 0.55 | ||
| UUA | Leu | 1.76 | trnL-UAA | UAA | Stop | 1.08 | |
| UUG | Leu | 1.15 | trnL-CAA | UAG | Stop | 0.7 | |
| CUU | Leu | 1.28 | trnL-UAG | CAU | His | 1.5 | trnH-GUG |
| CUC | Leu | 0.52 | CAC | His | 0.5 | ||
| CUA | Leu | 0.86 | CAA | Gln | 1.52 | trnQ-UUG | |
| CUG | Leu | 0.42 | CAG | Gln | 0.48 | ||
| AUU | Ile | 1.46 | trnI-GAU | AAU | Asn | 1.43 | trnN-GUU |
| AUC | Ile | 0.73 | AAC | Asn | 0.57 | ||
| AUA | Ile | 0.81 | trnI-CAU | AAA | Lys | 1.48 | trnK-UUU |
| AUG | Met | 1 | trnM-CAU | AAG | Lys | 0.52 | |
| GUU | Val | 1.45 | trnV-GAC | GAU | Asp | 1.51 | trnD-GUC |
| GUC | Val | 0.73 | GAC | Asp | 0.49 | ||
| GUA | Val | 1.27 | GAA | Glu | 1.45 | trnE-UUC | |
| GUG | Val | 0.54 | trnV-UAC | GAG | Glu | 0.55 | |
| UCU | Ser | 1.44 | trnS-GGA | UGU | Cys | 1.2 | trnC-GCA |
| UCC | Ser | 0.92 | UGC | Cys | 0.8 | ||
| UCA | Ser | 1.3 | UGA | Stop | 1.22 | ||
| UCG | Ser | 0.69 | trnS-UGA | UGG | Trp | 1 | trnW-CCA |
| CCU | Pro | 1.4 | trnP-UGG | CGU | Arg | 0.83 | trnR-ACG |
| CCC | Pro | 0.78 | CGC | Arg | 0.32 | trnR-UCU | |
| CCA | Pro | 1.19 | CGA | Arg | 1.12 | ||
| CCG | Pro | 0.63 | CGG | Arg | 0.49 | ||
| ACU | Thr | 1.29 | AGA | Arg | 0.99 | ||
| ACC | Thr | 0.86 | AGG | Arg | 0.65 | ||
| ACA | Thr | 1.23 | trnT-GGU | AGU | Ser | 2.07 | trnS-GCU |
| ACG | Thr | 0.61 | trnT-UGU | AGC | Ser | 1.18 | |
| GCU | Ala | 1.62 | trnA-UGC | GGU | Gly | 1.09 | |
| GCC | Ala | 0.65 | GGC | Gly | 0.51 | ||
| GCA | Ala | 1.24 | GGA | Gly | 1.54 | ||
| GCG | Ala | 0.49 | GGG | Gly | 0.86 | trnG-UCC |
Table A2.
Codon – anticodon recognition patterns and codon usage of C. chrysantha chloroplast genome.
| Codon | Amino Acid | RSCU | tRNA | Codon | Amino Acid | RSCU | tRNA |
|---|---|---|---|---|---|---|---|
| UUU | Phe | 1.31 | trnF-GAA | UAU | Tyr | 1.6 | trnY-GUA |
| UUC | Phe | 0.69 | UAC | Tyr | 0.4 | ||
| UUA | Leu | 1.88 | trnL-UAA | UAA | Stop | 0.86 | |
| UUG | Leu | 1.16 | trnL-CAA | UAG | Stop | 1.48 | |
| CUU | Leu | 1.25 | trnL-UAG | CAU | His | 1.51 | trnH-GUG |
| CUC | Leu | 0.46 | CAC | His | 0.49 | ||
| CUA | Leu | 0.84 | CAA | Gln | 1.55 | trnQ-UUG | |
| CUG | Leu | 0.41 | CAG | Gln | 0.45 | ||
| AUU | Ile | 1.48 | trnI-GAU | AAU | Asn | 1.54 | trnN-GUU |
| AUC | Ile | 0.61 | AAC | Asn | 0.46 | ||
| AUA | Ile | 0.92 | trnI-CAU | AAA | Lys | 1.49 | trnK-UUU |
| AUG | Met | 1 | trnM-CAU | AAG | Lys | 0.51 | |
| GUU | Val | 1.42 | trnV-GAC | GAU | Asp | 1.61 | trnD-GUC |
| GUC | Val | 0.55 | GAC | Asp | 0.39 | ||
| GUA | Val | 1.43 | GAA | Glu | 1.47 | trnE-UUC | |
| GUG | Val | 0.6 | trnV-UAC | GAG | Glu | 0.53 | |
| UCU | Ser | 1.7 | trnS-GGA | UGU | Cys | 1.45 | trnC-GCA |
| UCC | Ser | 0.92 | UGC | Cys | 0.55 | ||
| UCA | Ser | 1.21 | UGA | Stop | 0.67 | ||
| UCG | Ser | 0.58 | trnS-UGA | UGG | Trp | 1 | trnW-CCA |
| CCU | Pro | 1.58 | trnP-UGG | CGU | Arg | 1.33 | trnR-ACG |
| CCC | Pro | 0.78 | CGC | Arg | 0.4 | trnR-UCU | |
| CCA | Pro | 1.14 | CGA | Arg | 1.38 | ||
| CCG | Pro | 0.5 | CGG | Arg | 0.48 | ||
| ACU | Thr | 1.63 | AGA | Arg | 1.71 | ||
| ACC | Thr | 0.74 | AGG | Arg | 0.7 | ||
| ACA | Thr | 1.21 | trnT-GGU | AGU | Ser | 1.17 | trnS-GCU |
| ACG | Thr | 0.42 | trnT-UGU | AGC | Ser | 0.42 | |
| GCU | Ala | 1.85 | trnA-UGC | GGU | Gly | 1.3 | |
| GCC | Ala | 0.59 | GGC | Gly | 0.37 | ||
| GCA | Ala | 1.1 | GGA | Gly | 1.66 | ||
| GCG | Ala | 0.45 | GGG | Gly | 0.67 | trnG-UCC |
Table A3.
Predicted RNA editing site in the D. glaucum chloroplast genome.
| Gene | Nucleotide Position | Amino Acid Position | Codon | Amino Acid | Score |
|---|---|---|---|---|---|
| accD | 791 | 264 | TCG => TTG | S => L | 0.8 |
| clpP | 464 | 155 | GCT => GTT | A => V | 1 |
| 493 | 165 | CAT => TAT | H => Y | 1 | |
| matK | 706 | 236 | CAT => TAT | H => Y | 1 |
| 1250 | 417 | TCA => TTA | S => L | 0.86 | |
| 1309 | 437 | CAC => TAC | H => Y | 1 | |
| ndhA | 125 | 42 | ACA => ATA | T => I | 0.8 |
| 341 | 114 | TCA => TTA | S => L | 1 | |
| ndhB | 149 | 50 | TCA => TTA | S => L | 1 |
| 467 | 156 | CCA => CTA | P => L | 1 | |
| 586 | 196 | CAT => TAT | H => Y | 1 | |
| 611 | 204 | TCA => TTA | S => L | 0.8 | |
| 746 | 249 | TCT => TTT | S => F | 1 | |
| 830 | 277 | TCA => TTA | S => L | 1 | |
| 836 | 279 | TCA => TTA | S => L | 1 | |
| 1255 | 419 | CAT => TAT | H => Y | 1 | |
| 1481 | 494 | CCA => CTA | P => L | 1 | |
| 1526 | 509 | CCT => CTT | P => L | 1 | |
| ndhD | 65 | 22 | TCT => TTT | S => F | 0.8 |
| 401 | 134 | TCA => TTA | S => L | 1 | |
| 692 | 231 | TCG => TTG | S => L | 1 | |
| 896 | 299 | TCA => TTA | S => L | 1 | |
| 905 | 302 | CCC => CTC | P => L | 1 | |
| 1328 | 443 | TCA => TTA | S => L | 0.8 | |
| 1423 | 475 | CTT => TTT | L => F | 0.8 | |
| ndhF | 205 | 69 | CAT => TAT | H => Y | 0.8 |
| 290 | 97 | TCA => TTA | S => L | 1 | |
| 586 | 196 | CTT => TTT | L => F | 0.8 | |
| ndhG | 166 | 56 | CAT => TAT | H => Y | 0.8 |
| 314 | 105 | ACA => ATA | T => I | 0.8 | |
| psaB | 452 | 151 | ACA => ATA | T => I | 1 |
| psbE | 214 | 72 | CCT => TCT | P => S | 1 |
| psbF | 77 | 26 | TCT => TTT | S => F | 1 |
| rpoB | 338 | 113 | TCT => TTT | S => F | 1 |
| 551 | 184 | TCA => TTA | S => L | 1 | |
| 2432 | 811 | TCA => TTA | S => L | 0.86 | |
| rpoC1 | 41 | 14 | TCA => TTA | S => L | 1 |
| 1943 | 648 | ACT => ATT | T => I | 0.86 | |
| rpoC2 | 2335 | 779 | GCC => GTC | A => V | 0.86 |
| rps14 | 80 | 27 | TCA => TTA | S => L | 1 |
| rps16 | 176 | 59 | TCA => TTA | S => L | 0.83 |
Table A4.
Predicted RNA editing site in the C. chrysantha chloroplast genome.
| Gene | Nucleotide Position | Amino Acid Position | Codon | Amino Acid | Score |
|---|---|---|---|---|---|
| accD | 791 | 264 | TCG => TTG | S => L | 0.8 |
| 1400 | 467 | CCT => CTT | P => L | 1 | |
| clpP | 167 | 56 | GCT => GTT | A => V | 1 |
| 196 | 66 | CAT => TAT | H => Y | 1 | |
| matK | 709 | 237 | CAT => TAT | H => Y | 1 |
| 1253 | 418 | TCA => TTA | S => L | 0.86 | |
| 1312 | 438 | CAC => TAC | H => Y | 1 | |
| ndhB | 149 | 50 | TCA => TTA | S => L | 1 |
| 467 | 156 | CCA => CTA | P => L | 1 | |
| 586 | 196 | CAT => TAT | H => Y | 1 | |
| 611 | 204 | TCA => TTA | S => L | 0.8 | |
| 746 | 249 | TCT => TTT | S => F | 1 | |
| ndhD | 20 | 7 | ACG => ATG | T => M | 1 |
| 401 | 134 | TCA => TTA | S => L | 1 | |
| 692 | 231 | TCA => TTA | S => L | 1 | |
| 896 | 299 | TCA => TTA | S => L | 1 | |
| 905 | 302 | CCC => CTC | P => L | 1 | |
| 1328 | 443 | TCA => TTA | S => L | 0.8 | |
| 1423 | 475 | CTT => TTT | L => F | 0.8 | |
| ndhF | 205 | 69 | CAT => TAT | H => Y | 0.8 |
| 290 | 97 | TCA => TTA | S => L | 1 | |
| 586 | 196 | CTT => TTT | L => F | 0.8 | |
| ndhG | 166 | 56 | CAT => TAT | H => Y | 0.8 |
| 314 | 105 | ACA => ATA | T => I | 0.8 | |
| psbE | 214 | 72 | CCT => TCT | P => S | 1 |
| psbF | 77 | 26 | TCT => TTT | S => F | 1 |
| rpoB | 338 | 113 | TCT => TTT | S => F | 1 |
| 551 | 184 | TCA => TTA | S => L | 1 | |
| 566 | 189 | TCG => TTG | S => L | 1 | |
| 1981 | 561 | CCC => TCC | P => S | 0.86 | |
| 2434 | 812 | TCA => TTA | S => L | 0.86 | |
| rpoC1 | 1517 | 506 | ACT => ATT | T => I | 0.86 |
| rpoC2 | 2342 | 781 | GCC => GTC | A => V | 0.86 |
| rps14 | 80 | 27 | TCA => TTA | S => L | 1 |
| rps16 | 176 | 59 | TCA => TTA | S => L | 0.83 |
Table A5.
Repeat sequences present in the D. glaucum chloroplast genome.
| SN | Repeat Size | Repeat Position 1 | Repeat Type | Repeat Location | Repeat Position 2 | Repeat Location 2 | E-Value |
|---|---|---|---|---|---|---|---|
| 1 | 55 | 0 | P | IGS | 87,683 | rps19 | 5.45E-24 |
| 2 | 52 | 30,403 | P | IGS | 30,403 | IGS | 3.49E-22 |
| 3 | 47 | 40,642 | F | psaB | 42,866 | psaA | 3.57E-19 |
| 4 | 39 | 45,750 | F | ycf3 Intron | 102,076 | IGS | 2.34E-14 |
| 5 | 39 | 45,750 | P | ycf3 Intron | 144,199 | IGS | 2.34E-14 |
| 6 | 34 | 9240 | R | IGS | 9240 | IGS | 2.40E-11 |
| 7 | 34 | 44,864 | P | ycf3 Intron | 44,864 | ycf3 Intron | 2.40E-11 |
| 8 | 31 | 9248 | R | IGS | 9248 | IGS | 1.53E-09 |
| 9 | 30 | 8750 | P | IGS | 8750 | IGS | 6.13E-09 |
| 10 | 29 | 30,292 | F | IGS | 30,321 | IGS | 2.45E-08 |
| 11 | 29 | 95,068 | F | ycf2 | 95,086 | ycf2 | 2.45E-08 |
| 12 | 29 | 95,068 | P | ycf2 | 151,199 | ycf2 | 2.45E-08 |
| 13 | 29 | 95,086 | P | ycf2 | 151,217 | ycf2 | 2.45E-08 |
| 14 | 29 | 151,199 | F | ycf2 | 151,217 | ycf2 | 2.45E-08 |
| 15 | 28 | 8507 | P | trnS-GCU | 46,871 | trnS-GGA | 9.81E-08 |
| 16 | 26 | 9240 | F | IGS | 9253 | IGS | 1.57E-06 |
| 17 | 26 | 10,325 | P | IGS | 10,357 | IGS | 1.57E-06 |
| 18 | 25 | 38,275 | R | IGS | 38,275 | IGS | 6.28E-06 |
| 19 | 25 | 128,760 | R | ycf1 | 128,760 | ycf1 | 6.28E-06 |
| 20 | 24 | 8760 | R | IGS | 8760 | IGS | 2.51E-05 |
| 21 | 24 | 32,513 | P | IGS | 32,513 | IGS | 2.51E-05 |
| 22 | 24 | 44,474 | P | IGS | 73,687 | clpP Intron | 2.51E-05 |
| 23 | 24 | 127,804 | P | IGS | 127,804 | IGS | 2.51E-05 |
| 24 | 23 | 3434 | P | matK | 91,728 | ycf2 | 1.01E-04 |
| 25 | 23 | 3434 | F | matK | 154,563 | ycf2 | 1.01E-04 |
| 26 | 23 | 8872 | F | IGS | 8895 | IGS | 1.01E-04 |
| 27 | 23 | 10,643 | R | IGS | 10,643 | IGS | 1.01E-04 |
| 28 | 23 | 30,247 | F | IGS | 30,268 | IGS | 1.01E-04 |
| 29 | 23 | 44,479 | R | IGS | 44,479 | IGS | 1.01E-04 |
| 30 | 23 | 92,588 | F | ycf2 | 92,612 | ycf2 | 1.01E-04 |
| 31 | 23 | 92,588 | P | ycf2 | 153,679 | ycf2 | 1.01E-04 |
| 32 | 23 | 92,612 | P | ycf2 | 153,703 | ycf2 | 1.01E-04 |
| 33 | 23 | 153,679 | F | ycf2 | 153,703 | ycf2 | 1.01E-04 |
| 34 | 22 | 10,745 | F | IGS | 10,767 | IGS | 4.02E-04 |
| 35 | 22 | 15,273 | C | IGS | 28,813 | IGS | 4.02E-04 |
| 36 | 22 | 43,907 | P | IGS | 96,899 | ycf2 | 4.02E-04 |
| 37 | 22 | 43,907 | F | IGS | 149,393 | IGS | 4.02E-04 |
| 38 | 22 | 44,898 | P | ycf3 Intron | 44,898 | ycf3 Intron | 4.02E-04 |
| 39 | 22 | 48,054 | F | IGS | 48,074 | IGS | 4.02E-04 |
| 40 | 22 | 63,409 | P | IGS | 63,409 | IGS | 4.02E-04 |
| 41 | 22 | 103,201 | R | IGS | 103,201 | IGS | 4.02E-04 |
| 42 | 22 | 103,201 | C | IGS | 143,091 | IGS | 4.02E-04 |
| 43 | 22 | 113,539 | P | ycf1 | 113,539 | ycf1 | 4.02E-04 |
| 44 | 22 | 113,539 | F | ycf1 | 132,753 | ycf1 | 4.02E-04 |
| 45 | 22 | 132,753 | P | ycf1 | 132,753 | ycf1 | 4.02E-04 |
| 46 | 22 | 143,091 | R | IGS | 143,091 | IGS | 4.02E-04 |
| 47 | 21 | 8511 | F | trnS-GCU | 37,322 | trnS-UGA | 1.61E-03 |
| 48 | 21 | 8723 | R | IGS | 8723 | IGS | 1.61E-03 |
| 49 | 21 | 9240 | R | IGS | 9240 | IGS | 1.61E-03 |
Table A6.
Repeat sequences present in the C. chrysantha chloroplast genome.
| SN | Repeat Size | Repeat Position 1 | Repeat Type | Repeat Location | Repeat Position 2 | Repeat Location 2 | E-Value |
|---|---|---|---|---|---|---|---|
| 1 | 52 | 30,002 | P | IGS | 30,002 | IGS | 3.47E-22 |
| 2 | 47 | 40,405 | F | psaB | 42,629 | psaA | 3.55E-19 |
| 3 | 38 | 29,603 | P | IGS | 29,603 | IGS | 9.31E-14 |
| 4 | 34 | 32,963 | P | IGS | 32,963 | IGS | 2.38E-11 |
| 5 | 33 | 117,125 | F | IGS | 117,141 | IGS | 9.53E-11 |
| 6 | 29 | 92,074 | F | ycf2 | 92,098 | ycf2 | 2.44E-08 |
| 7 | 29 | 92,074 | P | ycf2 | 153,124 | ycf2 | 2.44E-08 |
| 8 | 29 | 92,098 | P | ycf2 | 153,148 | ycf2 | 2.44E-08 |
| 9 | 29 | 118,905 | F | IGS | 118,921 | IGS | 2.44E-08 |
| 10 | 29 | 153,124 | F | ycf2 | 153,148 | ycf2 | 2.44E-08 |
| 11 | 28 | 8401 | P | trnS-GCU | 46,575 | trnS-GGA | 9.76E-08 |
| 12 | 27 | 37,820 | R | IGS | 37,820 | IGS | 3.90E-07 |
| 13 | 26 | 79 | P | IGS | 79 | IGS | 1.56E-06 |
| 14 | 26 | 9949 | P | IGS | 9981 | IGS | 1.56E-06 |
| 15 | 26 | 116,265 | P | IGS | 116,265 | IGS | 1.56E-06 |
| 16 | 24 | 28,548 | P | IGS | 28,548 | IGS | 2.50E-05 |
| 17 | 24 | 32,078 | P | IGS | 32,078 | IGS | 2.50E-05 |
| 18 | 24 | 58,529 | P | IGS | 58,558 | IGS | 2.50E-05 |
| 19 | 24 | 76,983 | P | IGS | 77,008 | IGS | 2.50E-05 |
| 20 | 24 | 116,687 | F | IGS | 116,710 | IGS | 2.50E-05 |
| 21 | 23 | 4642 | F | IGS | 4708 | IGS | 9.99E-05 |
| 22 | 23 | 46,656 | F | IGS | 46,676 | IGS | 9.99E-05 |
| 23 | 23 | 97,249 | F | IGS | 97,271 | IGS | 9.99E-05 |
| 24 | 23 | 97,249 | P | IGS | 147,957 | IGS | 9.99E-05 |
| 25 | 23 | 97,271 | P | IGS | 147,979 | IGS | 9.99E-05 |
| 26 | 23 | 147,957 | F | IGS | 147,979 | IGS | 9.99E-05 |
| 27 | 22 | 38,011 | P | IGS | 38,011 | IGS | 4.00E-04 |
| 28 | 22 | 67,702 | F | IGS | 67,724 | IGS | 4.00E-04 |
| 29 | 22 | 113,002 | P | ycf1 | 113,002 | ycf1 | 4.00E-04 |
| 30 | 22 | 113,002 | F | ycf1 | 132,227 | ycf1 | 4.00E-04 |
| 31 | 22 | 117,470 | R | IGS | 117,470 | IGS | 4.00E-04 |
| 32 | 22 | 132,227 | P | ycf1 | 132,227 | ycf1 | 4.00E-04 |
| 33 | 21 | 314 | P | IGS | 363 | IGS | 1.60E-03 |
| 34 | 21 | 4878 | R | IGS | 4878 | IGS | 1.60E-03 |
| 35 | 21 | 5114 | C | IGS | 32,753 | IGS | 1.60E-03 |
| 36 | 21 | 5141 | F | IGS | 5166 | IGS | 1.60E-03 |
| 37 | 21 | 8068 | C | IGS | 30,261 | IGS | 1.60E-03 |
| 38 | 21 | 8405 | F | trnS-GCU | 37,002 | trnS-UGA | 1.60E-03 |
| 39 | 21 | 37,002 | P | trnS-UGA | 46,578 | trnS-GGA | 1.60E-03 |
| 40 | 21 | 38,404 | F | trnfM-CAU | 69,049 | trnP-GGG - trnP-UGG | 1.60E-03 |
| 41 | 21 | 56,602 | F | IGS | 56,623 | IGS | 1.60E-03 |
| 42 | 21 | 74,400 | R | IGS | 74,400 | IGS | 1.60E-03 |
| 43 | 21 | 117,116 | R | IGS | 117,116 | IGS | 1.60E-03 |
| 44 | 21 | 119,109 | R | IGS | 119,109 | IGS | 1.60E-03 |
| 45 | 20 | 160 | F | IGS | 8149 | psbI | 6.39E-03 |
| 46 | 20 | 4063 | F | trnK-UUU Intron | 23,615 | rpoC1 Intron | 6.39E-03 |
| 47 | 20 | 4063 | P | trnK-UUU Intron | 28,441 | IGS | 6.39E-03 |
| 48 | 20 | 4064 | F | trnK-UUU Intron | 111,518 | IGS | 6.39E-03 |
| 49 | 20 | 4064 | P | trnK-UUU Intron | 133,713 | IGS | 6.39E-03 |
References
- Abdel-Mogib M., Ezmirly S.T., Basaif S.A. Phytochemistry of Dipterygium glaucum and Capparis decidua. J. Saudi Chem. Soc. 2000;4:103–108. [Google Scholar]
- Abdullah W., Elsayed W.M., Abdelshafeek K.A., Nazif N.M., Singab N.B. Chemical Constituents and Biological Activities of Cleome Genus: A Brief Review. Inter. J. Pharm. Phyt. Res. 2016;8:777–787. [Google Scholar]
- Ahmad S., Wariss H.M., Alam K., Anjum S., Mukhtar M. Ethnobotanical studies of plant resources of Cholistan desert Pakistan. Int. J. Sci. Res. 2014;3:1782–1788. [Google Scholar]
- Airy Shaw, H.K. Diagnoses of new families, newnames, etc., for the seventh edition of Willis’s ‘Dictionary.’ Kew Bull. 1965, 18, 249–273.
- Alzahrani D., Albokhari E., Yaradua S., Abba A. Complete plastome genome of Dipterygium glaucum, Dipterygieae Cleomaceae. Mitochondrial DNA part B. 2020;5:1872–1873. doi: 10.1080/23802359.2019.1710291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- APG I (Angiosperm Phylogeny Group). An ordinal classification for the families of flowering plants. Ann. Missouri Bot. Gard. 1998, 85, 531–553.
- APG II (Angiosperm Phylogeny Group). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants. Bot. J. Linn. Soc. 2003, 141, 399-436
- APG III (Angiosperm Phylogeny Group). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants. Bot. J. Linn. Soc. 2009, 161, 105–121
- APG IV (Angiosperm Phylogeny Group). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants. Bot. J. Linn. Soc. 2016, 181, 1–20
- Bundschuh R., Altmuller J., Becker C., Nurnberg P., Gott J.M. Complete characterization of the edited transcriptome of the mitochondrion of Physarum polycephalum using deep sequencing of RNA. Nucleic Acids Res. 2011;39:6044–6055. doi: 10.1093/nar/gkr180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan G.J., McNicol J.W., Meyer R.C., Ramsay G., De Jong W.S. Polymorphic simple sequence repeat markers in chloroplast genomes of Solanaceous plants. Theor. Appl. Genet. 1999;99:859–867. [Google Scholar]
- Chang C.C., Lin H.C., Lin I.P., Chow T.Y., Chen H.H., Chen W.H., Cheng C.H., Lin C.Y., Liu S.M., Chang C.C., Chaw S.M. The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): Comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol. Biol. Evol. 2006;23:279–291. doi: 10.1093/molbev/msj029. [DOI] [PubMed] [Google Scholar]
- Chen H., Shao J., Zhang H., Jiang M., Huang L., Zhang Z., Yang D., He M., Ronaghi M., Luo X., Sun B., Wu W., Liu C. Sequencing and analysis of Strobilanthes cusia (Nees) Kuntze chloroplast Genome revealed the rare simultaneous contraction and expansion of the inverted repeat region in Angiosperm. Front. Plant Sci. 2018;9:324. doi: 10.3389/fpls.2018.00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierckxsens N., Mardulyn P., Smits G. Novoplasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017;45 doi: 10.1093/nar/gkw955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doorduin L., Gravendeel B., Lammers Y., Ariyurek T., Chin-A-Woeng T., Vrieling K. The complete chloroplast genome of 17 individuals of pest species Jacobaea vulgaris: SNPs, microsatellites and barcoding markers for population and phylogenetic studies. DNA Res. 2011;18:93–105. doi: 10.1093/dnares/dsr002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert D., Peakall R. Chloroplast simple sequence repeats (cpSSRs): Technical resources and recommendations for expanding cpSSR discovery and applications to a wide array of plant species. Mol. Ecol. Resour. 2009;9:673–690. doi: 10.1111/j.1755-0998.2008.02319.x. [DOI] [PubMed] [Google Scholar]
- Fay M.F., Christenhusez M.M. Brassicales - An order of plants characterised by shared chemistry. Curtis’s Bot. Mag. 2010;27:165–196. [Google Scholar]
- Felsenstein J. Cases in which parsimony or compatibility methods will be positively misleading. Syst. Zool. 1978;27:401–410. [Google Scholar]
- Feodorova T.A., Voznesenskaya E.V., Edwards G.E., Roalson E.H. Biogeographic patterns of diversification and the origins of C-4 in Cleome (Cleomaceae) Syst. Bot. 2010;35:811–826. [Google Scholar]
- Frazer K.A., Pachter L., Poliakov A., Rubin E.M., Dubchak I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004;32:273–279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredrik R., Maxim T., Paul V.M., Daniel L.A., Aaron D., Sebastian H., Bret L., Liang L., Mar A.S., John P.H. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Systematic. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassi F., Labra M., Scienza A., Imazio S. Chloroplast SSR markers to assess DNA diversity in wild and cultivated grapevines. Vitis. 2002;41:157–158. [Google Scholar]
- Greiner S., Wang X., Rauwolf U., Silber M.V., Mayer K., Meurer J., Haberer G., Herrmann R.G. The complete nucleotide sequences of the five genetically distinct plastid genomes of Oenothera, subsection Oenothera: I. sequence evaluation and plastome evolution. Nucleic Acids Res. 2008;36:2366–2378. doi: 10.1093/nar/gkn081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J.C. Systematics of Capparaceae and Cleomaceae: An evaluation of the generic delimitations of Capparis and Cleome using plastid DNA sequence data. Botany. 2008;86:682–696. [Google Scholar]
- Hall J.C., Sytsma K.J., Iltis H.H. Phylogeny of Capparaceae and Brassicaceae based on chloroplast sequence data. Am. J. Bot. 2002;89:1826–1842. doi: 10.3732/ajb.89.11.1826. [DOI] [PubMed] [Google Scholar]
- Hall J.C., Iltis H.H., Sytsma K.J. Molecular phylogenetics of core Brassicales, placement of orphan genera Emblingia, Forchhammeria, Tirania, and character evolution. Syst. Bot. 2004;29:654–669. [Google Scholar]
- Hedge I.C., Kjaer A., Malver O. Dipterygium—Cruciferae or Capparaceae? Notes from the Royal Botanical Garden. Edinburgh. 1980;38:247–250. [Google Scholar]
- Heywood V.H., Brummitt R.K., Culham A., Seberg O. Royal Botanic Gardens; Kew: 2007. Flowering Plant Families of the World. [Google Scholar]
- Huang Y.Y., Antonius J.M.M., Matzke M. Complete sequence and comparative analysis of the chloroplast Genome of Coconut Palm (Cocos nucifera) Plos One. 2013;8 doi: 10.1371/journal.pone.0074736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson J. Clarendon Press; Oxford: 1967. The genera of flowering plants. [Google Scholar]
- Kaila T., Chaduvla P.K., Rawal H.C., Saxena S., Tyagi A., Mithra S.V., Solanke A.U., Kalia P., Sharma T.R., Singh N.K., Gaikwad K. Chloroplast Genome sequence of Clusterbean (Cyamopsis tetragonoloba L.): Genome structure and comparative analysis. Genes. 2017;8:212. doi: 10.3390/genes8090212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumbhar F., Nie X., Xing G., Zhao X., Lin Y., Wang S., Weining S. Identification and characterisation of rna editing sites in chloroplast transcripts of einkorn wheat (Triticummonococcum) Ann. Appl. Biol. 2018;172:197–207. [Google Scholar]
- Kurtz S., Choudhuri J.V., Ohlebusch E., Schleiermacher C., Stoye J., Giegerich R. Reputer: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iltis H.H. Studies in Capparidaceae. III. Evolution and phylogeny of the Western North American Cleomoideae. Ann. Missouri Bot. Gard. 1957;44:77–119. [Google Scholar]
- Iltis H.H., Hall J.C., Cochrane T.S., Sytsma K.J. Studies in the Cleomaceae I: On the separate recognition of Capparaceae, Cleomaceae, and Brassicaceae. Ann. Missouri Bot. Gard. 2011;98:28–36. [Google Scholar]
- Judd W.S., Sanders R.W., Donoghue M.J. Angiosperm family pairs: preliminary phylogenetic analyses. Harv. Pap. Bot. 1994;5:1–51. [Google Scholar]
- Judd W.S., Campbell C.S., Kellogg E.A., Stevens P.F., Donoghue M.J. third ed. Sinauer Associates; Sunderland, Massachusetts: 2007. Plant Systematics: A Phylogenetic Approach. [Google Scholar]
- Li, B., Lin, F., Huang, P., Guo, W., Zheng, Y. Complete chloroplast genome sequence of Decaisnea insignis: Genome organization, genomic resources and comparative analysis. Sci. Rep. vol. 7. 2017. [DOI] [PMC free article] [PubMed]
- Li X., Tan W., Sun J., Du J., Zheng C., Tian X., Zheng M., Xiang B., Wang Y. Comparison of Four Complete Chloroplast Genomes of Medicinal and Ornamental Meconopsis Species: Genome Organization and Species Discrimination. Sci. Rep. 2019;9:10567. doi: 10.1038/s41598-019-47008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P., Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Liu H.J., Ding C.H., He J., Cheng J., Pei L.Y., Xie L. Complete chloroplast genomes of Archiclematis, Naravelia and Clematis (Ranunculaceae), and their phylogenetic implications. Phytotaxa. 2018;343:214–226. [Google Scholar]
- Lohse M., Drechsel O., Bock R. Organellar Genome DRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- Luo J., Hou B.W., Niu Z.T., Liu W., Xue Q.Y., Ding X.Y. Comparative chloroplast genomes of photosynthetic orchids: insights into evolution of the Orchidaceae and development of molecular markers for phylogenetic capplications. PLoS One. 2014;9 doi: 10.1371/journal.pone.0099016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luning B., Kers L.E., Seffers P. Methyl glycosinolate confirmed in Puccionia and Dhofaria (Capparidaceae) Biochem. Syst. Ecol. 1992;29:394. [Google Scholar]
- Magdalena G.N., Ewa F., Wojciech P. Cucumber, melon, pumpkin, and squash: Are rules of editing in flowering plants chloroplast genes so well known indeed? Gene. 2009;434 doi: 10.1016/j.gene.2008.12.017. [DOI] [PubMed] [Google Scholar]
- Martín-Bravo S., Vargas P., Luceno M. Is Oligomeris (Resedaceae) indigenous to North America? Molecular evidence for a natural colonization from the old world. Am. J. Bot. 2009;96:507–518. doi: 10.3732/ajb.0800216. [DOI] [PubMed] [Google Scholar]
- Mayor C., Brudno M., Schwartz J.R., Poliakov A., Rubin E.M., Frazer K.A., Pachter L.S., Dubchak I. VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- Mehmood K., Mehmood S., Ramzan M., Arshad M., Yasmeen F. Biochemical and phytochemical analysis of Dipterygium glaucum collected from Cholistan desert. J. Sci. Res. 2010;40:13–18. [Google Scholar]
- Melchior H. twelth ed. Angiospermae; Bomtraeger, Berlin: 1964. Syllabus der Pflanzenfamilien. [Google Scholar]
- Moussa S.A., Taia W.K., Al-Ghamdy F.G. Acclimation of Dipterygium glaucum Decne. Grown in the Western Coastal part of Saudi Arabia to different water supplies. Int. J. Res. Chem. Environ. 2012;2:301–309. [Google Scholar]
- Mower J.P. The PREP suite: predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res. 2009;37:253–259. doi: 10.1093/nar/gkp337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M., Park H., Lee H., Lee B.-H., Lee J. The complete plastome sequence of anantarctic bryophyte Sanioniauncinata (Hedw.) loeske. Int. J. Mol. Sci. 2018;709:no. 19. doi: 10.3390/ijms19030709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, I., Kim, W.J., Yeo, S.-M., Choi, G., Kang, Y.-M., Piao, R., Moon, B.C. The complete chloroplast genome sequences of Fritillaria us suriensis maxim. In addition, Fritillaria cirrhosa D. Don, and comparative analysis with other Fritillaria species. Molecules vol. 282, no. 22. 2017. [DOI] [PMC free article] [PubMed]
- Patchell M.J., Roalson E.H., Hall J.C. Resolved phylogeny of Cleomaceae based on all three genomes. Taxon. 2014;63(2):315–328. [Google Scholar]
- Pax F., Hoffmann K. Capparidaceae. In: Engler A., Prantl K., editors. Die Nat€urlichen Pflanzenfamilien. 2nd ed. Wilhelm Engelmann; Leipzig: 1936. pp. 146–223. [Google Scholar]
- Powell W., Morgante M., McDevitt R., Vendramin G.G., Rafalski J.A. Polymorphic simple sequence repeat regions in chloroplast genomes: applications to the population genetics of pines. Proc. Natl. Acad. Sci. 1995;92:7759–7763. doi: 10.1073/pnas.92.17.7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provan J. Novel chloroplast microsatellites reveal cytoplasmic variation in Arabidopsis thaliana. Mol. Ecol. 2000;9:2183–2185. [PubMed] [Google Scholar]
- Raman, G., Park, S. The complete chloroplast genome sequence of Ampelopsis: Gene organization, comparative analysis, and phylogenetic relationships to other angiosperms. Front. Plant Sci. vol.341, no. 7. 2016. [DOI] [PMC free article] [PubMed]
- Raubeson L.A., Jansen R.K. Chloroplast genomes of plants. In: Henry R., editor. Diversity and evolution of plants-genotypic variation in higher plants. CABI Publishing; Oxfordshire: 2005. pp. 45–68. [Google Scholar]
- Rodman J.E., Price R.A., Karol K., Conti E., Sytsma K.J., Palmer J.D. Nucleotide sequences of the rbcL gene indicate monophyly of mustard oil plants. Ann. Missouri Bot. Gard. 1993;80:686–699. [Google Scholar]
- Rodman J.E., Karol K.G., Price R.A., Sytsma K.J. Molecules, morphology, and Dahlgren’s expanded order Capparales. Syst. Bot. 1996;21:289–307. [Google Scholar]
- Rodman J.E., Soltis P.S., Soltis D.E., Sytsma K.J., Karol K.G. Parallel evolution of glucosinolate biosynthesis inferred from congruent nuclear and plastid gene phylogenies. Am. J. Bot. 1998;85:997–1006. [PubMed] [Google Scholar]
- Rahman M.A., Mossa J.S., Al-Said M.S., Al-Yahya M.A. Medicinal plant diversity in the flora of Saudi Arabia 1: a report on seven plant families. Fitoterapia. 2004;75(2):149–161. doi: 10.1016/j.fitote.2003.12.012. [DOI] [PubMed] [Google Scholar]
- Rollins R.C. Stanford University Press; Stanford, California, USA: 1993. The Cruciferae of continental North America: systematics of the mustard family from the Arctic to Panama. [Google Scholar]
- Schattner P., Brooks A.N., Lowe T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33:686–689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R., Edwards R. “Quality control and preprocessing of metagenomic datasets,”. Bioinformatics. 2011;27(6):863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen U., Shoeib N., Temraz A., Abdelhady M.S. Flavonoidal constituents, antioxidant, antimicrobial, and cytotoxic activities of Dipterygium glaucum grown in Kingdom of Saudi Arabia. Phcog. Mag. 2017;13(51):484. doi: 10.4103/pm.pm_44_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson M.G. Elsevier Academic Press; Burlington, Massachusetts: 2006. Plant Systematics. [Google Scholar]
- Song Y., Wang S., Ding Y., Li M.F., Zhu S., Chen N. Chloroplast Genomic Resource of Paris for Species Discrimination. Sci. Rep. 2017;7:3427. doi: 10.1038/s41598-017-02083-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub S.C.K., Parks M., Weitemier K., Fishbein M., Cronn R.C., Liston A. Navigating the tip of the genomic iceberg: next-generation sequencing for plant systematics. Am. J. Bot. 2012;99:349–364. doi: 10.3732/ajb.1100335. [DOI] [PubMed] [Google Scholar]
- Thiel T., Michalek W., Varshney R., Graner A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.) Theor.l Appl. Genet. 2003;106:411–422. doi: 10.1007/s00122-002-1031-0. [DOI] [PubMed] [Google Scholar]
- Tripti J., Neeraj K., Preeti K. A Review on Cleome viscosa: An endogenous Herb of Uttarakhand. Inter. J. Pharma Res. & Rev. 2015;4:25–31. [Google Scholar]
- Yap J.Y., Rohner T., Greenfield A., Van Der Merwe M., McPherson H., Glenn W., Kornfeld G., Marendy E., Pan A.Y., Wilton A., Wilkins M.R., Rossetto M., Delaney S.K. Complete chloroplast genome of the Wollemi pine (Wollemia nobilis): structure and evolution. PLoS ONE. 2015;106 doi: 10.1371/journal.pone.0128126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., Yu, H., Wang, J., Lei, W., Gao, J., Qiu, X., Wang, J. The complete chloroplast genome sequences of the medicinal plant Forsythia suspense (oleaceae). Int. J. Mol. Sci. vol. 2288, no. 18. 2017. [DOI] [PMC free article] [PubMed]
- Wyman S., Jansen R., Boore J. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2014;20(17):3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- Zeng W.H., Liao S.C., Chang C.C. Identification of RNA editing sites in chloroplast transcripts of Phalaenopsis aphrodite and comparative analysis with those of other seed plants. Plant Cell Physiol. 2007;48:362–368. doi: 10.1093/pcp/pcl058. [DOI] [PubMed] [Google Scholar]
- Posada D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 2008;25:1253–1259. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number (D. glaucum MT041700; C. chrysantha MT948188).