Summary
Bipolar disorder (BD) is characterized by cyclical mood shifts. Studies indicate that BD patients have a peripheral pro-inflammatory state and alterations in glial populations in the brain. We utilized an in vitro model to study inflammation-related phenotypes of astrocytes derived from induced pluripotent stem cells (iPSCs) generated from BD patients and healthy controls. BD astrocytes showed changes in transcriptome and induced a reduction in neuronal activity when co-cultured with neurons. IL-1β-stimulated BD astrocytes displayed a unique inflammatory gene expression signature and increased secretion of IL-6. Conditioned medium from stimulated BD astrocytes reduced neuronal activity, and this effect was partially blocked by IL-6 inactivating antibody. Our results suggest that BD astrocytes are functionally less supportive of neuronal excitability and this effect is partially mediated by IL-6. We confirmed higher IL-6 in blood in a distinct cohort of BD patients, highlighting the potential role of astrocyte-mediated inflammatory signaling in BD neuropathology.
Keywords: iPSC, mood disorders, inflammation, glia, cytokine, IL-6, psychiatry, astrocytes, neuronal activity
Highlights
-
•
Bipolar disorder astrocytes are functionally less supportive of neuronal activity
-
•
Bipolar disorder astrocytes response to IL-1β is transcriptionally distinct
-
•
IL-6 secretion in bipolar disorder astrocytes reduces neuronal activity
-
•
Bipolar disorder patients show higher circulating levels of IL-6 in blood
In this article, Gage and collaborators show that astrocytes differentiated from induced pluripotent stem cells generated from bipolar disorder patients are functionally less supportive of neuronal activity. Bipolar disorder astrocytes' response to pro-inflammatory cytokines is characterized by a unique transcriptional response and increased IL-6 secretion that directly and negatively impacted on neuronal activity. Increased peripheral IL-6 was confirmed in a distinct clinical cohort highlighting the potential role of astrocyte-mediated inflammatory signaling in the neuropathology of bipolar disorder.
Introduction
Bipolar disorder (BD) is a chronic neuropsychiatric illness that usually starts in young adulthood and is characterized by recurrent mood states ranging from high energy and elation, known as mania, to low energy and sad or depressive episodes. Estimates show that 1% to 3% of the world population suffers from BD and it has heritability of up to 80%, suggesting not only a strong genetic component but also that the disorder is the result of a complex interplay with environmental factors (Craddock and Sklar, 2013; Hayden and Nurnberger, 2006). Several lines of evidence suggest a link between imbalanced inflammatory signaling and BD (Munkholm et al., 2013; Najjar et al., 2013). BD patients show a higher prevalence of comorbid diseases with an inflammatory component, such as cardiovascular disease, diabetes mellitus, and the presence of an immune-related “metabolic syndrome” (Cassidy et al., 1999; Leboyer et al., 2012; Osby et al., 2001; Weiner et al., 2011). Also, BD patients develop organ-specific autoimmunity such as thyroid failure and atrophic gastritis due to thyroperoxidase and proton pump H+/K+-ATPase antibodies, respectively (Vonk et al., 2007). There is evidence that BD patients have chronic and mild inflammation that is thought to act on processes that trigger atherosclerosis, hypertension, and diabetes (Drake et al., 2011; Goldstein et al., 2009; Hamdani et al., 2012; Ross, 1999). Meta-analyses of several human studies reported higher concentrations of circulating pro-inflammatory cytokines such as interleukin (IL)-6 and tumor necrosis factor (TNF)-α in BD patients (Goldstein et al., 2009; Modabbernia et al., 2013; Munkholm et al., 2013). These studies suggest that BD may be a multi-system disease with an inflammatory component, both peripherally and in the CNS (Pinto et al., 2017).
In the CNS, astrocytes are key mediators of immune responses downstream of inflammatory signals. They play crucial roles in sustaining neuronal function via regulation of processes such as synapse maintenance and elimination, energy balance, lipid processing, homeostasis of ions, glutamate, and secondary messengers (Vasile et al., 2017). Astrocytes are immunocompetent cells known to participate in the inflammatory cascade within the brain by being activated by pro-inflammatory cytokines such as IL-1β and in turn secreting cytokines that participate in the process of neuroinflammation (Colombo and Farina, 2016). Due to a growing understanding of the role of neuroinflammation in psychiatric disorders, we asked whether altered inflammation-driven signaling in astrocytes was associated with BD. We have previously developed a method for rapidly generating inflammation-responsive astrocytes from human induced pluripotent stem cells (iPSCs) via a glial precursor intermediate (Santos et al., 2017). iPSC technology has enabled the study of specific phenotypes and cellular subtypes in the context of psychiatric disorders (Brennand et al., 2011; Mertens et al., 2015; Soliman et al., 2017; Vadodaria et al., 2018, 2019a, 2019b). In this study, we searched for an inflammation signature in astrocytes generated from BD patients compared with healthy individuals’ iPSCs. Analysis of whole transcriptome showed hundreds of differential expressed genes in BD astrocytes but gene ontology analysis did not revealed an enrichment in inflammation-related genes. However, IL6 gene was upregulated in BD astrocytes compared with controls suggesting that this cytokine may contribute to BD defects. Confirming published studies, we observed significant higher IL-6 levels in the blood of BD patients in a separate cohort of more than a hundred individuals. The response of BD astrocytes to pro-inflammatory cytokines revealed a unique transcriptional response to inflammation, with increased IL-6 secretion that directly and negatively impacted on the activity of co-cultured neurons.
Results
Characterization and Stimulation of BD Patient iPSC-Derived Astrocytes
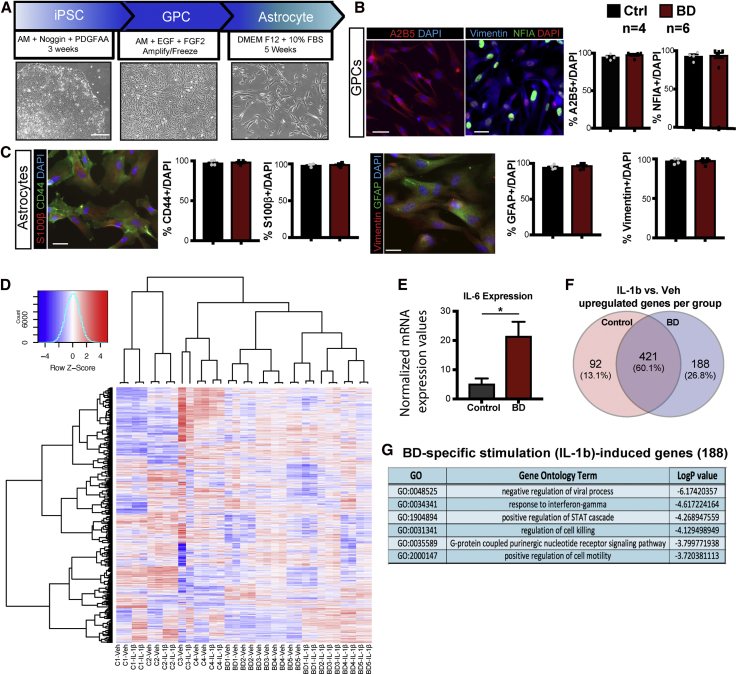
We have previously described a method to efficiently generate glial progenitor cells (GPCs) from human iPSCs such that they can be propagated, expanded, frozen, and thawed as proliferating intermediates (Santos et al., 2017). Using this method, free-floating embryoid bodies were generated from six patient and four control iPSC lines, giving rise to GPCs and then differentiated to astrocytes (Figure 1A). Embryoid bodies were cultured in astrocyte induction media in the presence of noggin and platelet-derived growth factor (PDGF)AA (Bonni et al., 1997; Chambers et al., 2009; Dell'Albani et al., 1998; Fan et al., 2005), following which GPCs were maintained in media containing epithelial growth factor (EGF) and fibroblast growth factor 2 (FGF2). The glial identity of GPCs derived from all control and BD individuals was assessed by immunostaining and quantifying the percentage of cells expressing the cell surface ganglioside antigen A2B5 and nuclear factor I A (NFIA), a glial transcription factor (Deneen et al., 2006). A vast majority (over 80%) of GPCs expressed both markers in control and BD groups (Figure 1B). To differentiate GPCs from astrocytes, cells were cultured for an additional 5 weeks in media containing 10% fetal bovine serum (FBS). We observed changes in the morphology of differentiating cells, as they became flatter with more surface area, resembling astrocytes (Figure 1A). We next confirmed in iPSC-derived astrocytes from all individuals that they expressed markers such as the intermediate filament proteins Vimentin and glial fibrillary acidic protein (GFAP), the calcium binding protein S100β, and the cell surface glycoprotein CD44. We found that nearly all (90% to 100%) differentiated astrocytes expressed these astrocytic markers, with no significant differences between control and BD groups (Figure 1C). We next performed whole transcriptome analyses on control and patient-derived astrocytes and found that there were differences between groups. Hierarchical clustering analyses showed that controls and BD patient lines separated with 817 genes significantly dysregulated in BD versus controls (Figure S1). We further examined the families of genes dysregulated and gene ontology (GO) analysis showed that a most significant categories were “cell division” and “cell cycle” related (Figure S1). These suggest that there were baseline differences between BD and control astrocytes. Interestingly, inflammation and related categories were not among the most significant GO categories (Figure S1). However, given the links between inflammation and BD, we especially sought to explore how BD astrocytes responded to inflammatory stimuli. Using RNA-sequencing as well as a flow cytometry assay, we had previously shown that iPSC-derived astrocytes robustly responded to inflammatory cytokines such as IL-1β (Santos et al., 2017). Astrocytes from BD patients and controls were stimulated with IL-1β for 5 h. Using whole transcriptome analysis, we asked whether inflammation-driven pathways were differentially regulated between groups. Unsupervised hierarchical clustering of unstimulated and stimulated controls and BD we made two key observations: (1) control and BD astrocytes clustered separately, highlighting some baseline differences and (2) stimulated BD astrocytes clustered distinctly from control-stimulated astrocytes and from unstimulated BD astrocytes, suggesting that BD astrocytes had the most distinct response to IL-1β (Figure 1D). Baseline differentially expressed genes (DEGs) between control and BD (unstimulated) did not contribute to the IL-1β upregulated genes in stimulated versus vehicle-treated BD astrocytes (Figure S1). In fact, only two genes contributed to the IL-1β upregulated genes (Figures 1F and S1). Interestingly however, even under baseline conditions (unstimulated) the inflammatory cytokine IL-6 was significantly higher in BD astrocytes as compared with controls (Figure 1E). In examining IL-1β-induced genes we observed that a majority of the IL-1β-induced genes were common to both the control and BD groups (Figure 1F), as expected. However, a separate set of 188 genes was upregulated only in BD astrocytes, and GO analysis of those 188 unique genes revealed GO terms such as “negative regulation of viral process” and “response to interferon-gamma” (Figure 1G, Tables S2 and S3). Only two of the 188 genes were DEG between BD and control unstimulated astrocytes (Figure S1). There were fewer (28 genes) uniquely downregulated in IL-1β-treated BD astrocytes enriching in GO terms not directly related to inflammation (Figure S1). Together, these results suggested that BD astrocytes have a different transcriptional profile compared with control astrocytes and that in addition to a generalized response to inflammation, BD astrocytes displayed a unique inflammatory signature that might contribute to the pathology of the disorder.
Figure 1.
Characterization and Validation of BD Patient iPSC-Derived Astrocytes through a Glial Precursor Intermediate
(A) Summary of an 8- to 10-week cell culture timeline to differentiate human astrocytes from iPSCs. Bright field images show cell types: patient fibroblast-derived iPSCs, GPCs, and 5-week-old astrocytes (scale bar, 300 μm).
(B) Representative images and quantification of GPCs immunostained with early glial fate markers: A2B5, Vimentin and NFIA. Scale bar, 50 μm.
(C) Representative images and quantification of 4- to 5-week-old astrocytes immunostained with astrocytic markers: S100β, CD44, and GFAP. Scale bar, 50 μm. Data are expressed as mean ± SEM of percent of stained cells over total number detected by DAPI. Experiments in triplicate with n = 4 controls and n = 6 BD patients.
(D) Transcriptome of BD astrocytes shows inflammatory response to IL-1β: unsupervised hierarchical clustering of top expressed genes from whole transcriptome analysis (increased, red; decreased, blue) in vehicle and IL-1β-treated astrocytes from four control and five BD individuals (in duplicate).
(E) Normalized expression values for IL-6 in unstimulated (veh) BD versus control astrocytes.
(F) Venn diagram showing the number of upregulated genes in IL-1β treatment versus vehicle in controls (92) and BD (188), with 421 overlapping genes.
(G) List of significantly enriched GO terms for 188 uniquely upregulated genes in IL-1β-treated BD astrocytes.
Functional Impact of Stimulated BD Astrocytes on Neuronal Activity
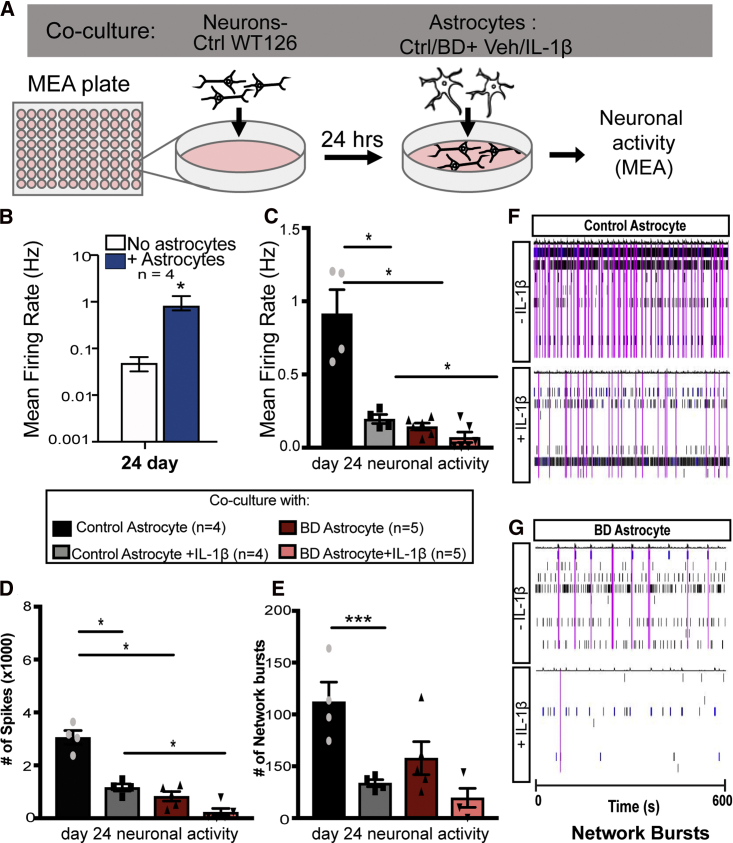
Since BD astrocytes displayed differential inflammatory responses to IL-1β stimulation, we next asked whether this feature had functional consequences. To examine the functional impact of the altered inflammatory response of BD astrocytes on neurons, we performed an astrocyte-neuron co-culture experiment and neuronal activity was measured with a multi-electrode array (MEA) system. For this experiment, neurons derived from one healthy individual (WT126; unrelated to the cohort used to differentiate astrocytes) were plated onto an MEA plate and astrocytes derived from four healthy controls or five BD patients were plated on top (Figure 2A). This experiment helped dissect the astrocyte-specific contribution to neuronal activity since neurons were differentiated from neural precursor cells of a single iPSC line. We first performed a time course experiment to examine how neuronal activity changed with and without co-cultured astrocytes and over a period of 3 to 4 weeks. We observed that the mean firing rate of neurons increased with time from day 0 to day 24, and that it was significantly higher in the presence of astrocytes at day 24 post-differentiation. Therefore, we used day 24 time point to examine neural activity in the presence of IL-1β-stimulated and non-stimulated astrocytes (5 h before co-culture) derived from controls and BD patients (Figures 2B–2G). IL-1β treatment in control astrocytes significantly impacted the activity of co-cultured neurons, causing a reduction in three key parameters: mean firing rate, number of spikes, and network bursts. Neurons co-cultured with non-stimulated BD astrocytes also displayed significant reduction in these parameters as compared with non-stimulated control astrocytes. Stimulation of BD astrocytes with IL-1β impacted activity of co-cultured neurons, further reducing the average mean firing rate and number of spikes, as compared with stimulated control astrocytes (Figures 2C–2E). A raster plot example shows the comparative network burst activity between control and BD astrocyte groups (Figures 2F and 2G). These results revealed the negative functional impact of BD astrocytes and inflammation on activity of co-cultured neurons.
Figure 2.
Decreased Activity of Neurons Co-cultured with IL-1β-Stimulated BD Patient Astrocytes
(A) Control (WT126) neurons were differentiated as a monolayer on a MEA and co-cultured with different groups of patient-derived astrocytes, and neuronal activity was measured.
(B) Twenty-four days after differentiation of WT126-neurons on the MEA plate, mean firing rate of cultured neurons was measured either with or without co-cultured control astrocytes.
(C–G) Neurons were co-cultured with astrocytes from different groups pretreated with either vehicle or IL-1β for 5 h. At 24 days of neural differentiation, electrical parameters were measured in the MEA and the results are graphed as mean ± SEM for (C) mean firing rate, (D) number of spikes, and (E) number of network bursts. (F and G) Example raster plot showing spikes (black lines) and network bursts (pink lines) in the MEA for neurons cultured with (E) control astrocytes or (F) BD astrocytes. Individual electrodes are plotted (y axis) over time in seconds (x axis). Statistical significance was determined using the unpaired non-parametric (A) Mann-Whitney two-tailed test, and (B–D) ordinary one-way ANOVA, with multiple comparisons between groups. n = 4 per group. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. ∗∗∗∗p < 0.0001.
Stimulation-Induced Cytokines in BD Astrocytes
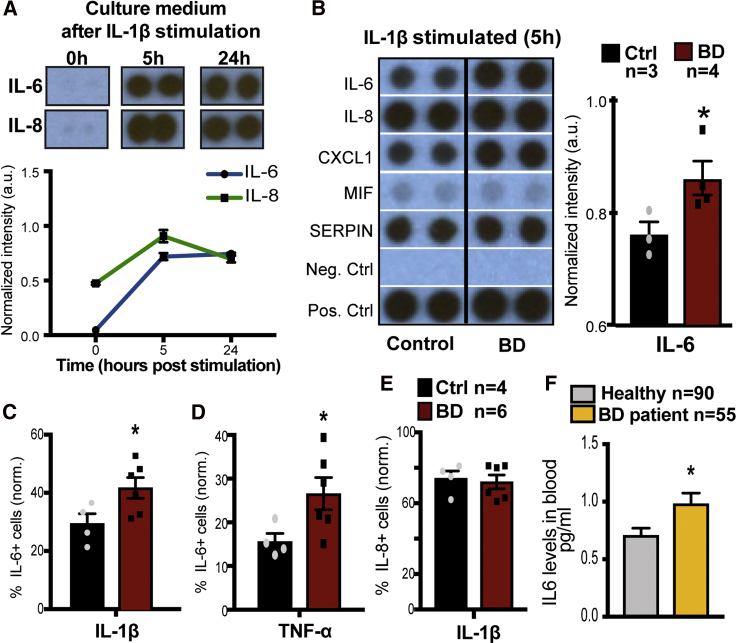
Given the striking effect of stimulated BD astrocytes on neuronal activity, we sought to examine whether the observed effects were potentially downstream of stimulation-induced secretion of cytokines. To test this hypothesis, astrocytes were cultured for 3 to 4 weeks and stimulated with IL-1β following which culture medium was collected for analysis of the secreted cytokines (blot array). We first performed a time course experiment to determine the peak of cytokine release in astrocyte culture medium following treatment with IL-1β for 5 and 24 h (Figure 3A). Culture medium was collected from astrocytes at baseline (0), and 5 and 24 h after stimulation. Using human cytokine blot arrays, we observed that key downstream cytokine targets such as IL-6 and IL-8 peaked at 5 h after stimulation and levels plateaued by 24 h (Figure 3A). We next examined the profile of multiple secreted cytokines 5 h after stimulation of astrocytes (Figure 3B). We observed a significant induction of several cytokines, such as IL-6, IL-8, CXCL1, MIF, and SERPIN, following stimulation with IL-1β in both control and BD groups (Figure 3B). While a majority of secreted cytokines were at comparable levels between controls and BD, IL-6 was significantly higher in the culture medium of stimulated BD astrocytes (Figure 3B), suggesting that stimulated BD astrocyte-specific effects might lie downstream of IL-6. To further understand the IL-6 response, we examined IL-6 and IL-8 protein expression in BD astrocytes using flow cytometry (Figures 3D–3F). In this assay, astrocytes were treated for 5 h with IL-1β along with monensin and brefeldin A compounds, which inhibit Golgi-mediated protein transport, resulting in the intracellular accumulation of proteins, including cytokines that normally would be secreted. Five hours after stimulation, astrocytes were fixed and permeabilized for detection of IL-6 and IL-8, followed by a flow cytometry assay to compare percentages of immunopositive cells across different groups. We observed that, following IL-1β treatment, BD astrocytes had a significantly higher percentage of IL-6-positive cells compared with controls (Figure 3D). Next, we asked if the effect observed was IL-1β specific or was also observed downstream of another pro-inflammatory cytokine, such as TNF-α. Here we also observed a significantly higher percentage of IL-6-positive cells with TNF-α stimulation (Figure 3E). On the other hand, IL-8-expressing astrocytes were not significantly different between control and BD groups with IL-1β-mediated stimulation (Figure 3F). These results were in agreement with the cytokine blot array data, suggesting a specific upregulation of IL-6 in BD astrocytes. Lithium is a commonly prescribed medication for BD, and the BD-cohort utilized in our study has been previously characterized with patients being either lithium responsive or non-responsive. This led us to examine whether being clinically responsive or refractory to lithium treatment correlated with differential inflammatory responses. Using the fluorescence-activated cell sorting (FACS) assay following stimulation, we did not find significant group differences among BD patients that therapeutically responded to lithium versus those who did not (Figure S2). Next, we tested whether lithium treatment in vitro impacted the heightened inflammatory response in BD astrocytes using the FACS and did not find significant differences between groups (Figure S2).
Figure 3.
IL-6 Release Is Increased in BD Astrocytes in Response to Inflammatory Stimulus
(A) Representative images of time course of secreted IL-6 and IL-8 in the culture medium of control astrocytes stimulated with IL-1β at baseline (0), or 5 or 24 h. Blot intensity was quantified and plotted over time (arbitrary units, a.u.). Graphs show mean ± SEM for n = 3.
(B) Representative images showing the levels of cytokines in the culture medium of control and BD astrocytes 5 h after IL-1β stimulation. Quantification of IL-6 from cytokine array blots (normalized intensity, arbitrary units) in control and BD groups.
(C and D) (C) Quantification of percentage of IL-6-positive cells normalized to vehicle-stimulated cells (by subtraction) following a 5-h pretreatment with IL-1β or (D) TNF-α.
(E) Quantification of percentage of IL-8-positive cells (normalized to vehicle-stimulated cells) following a 5-h pretreatment with IL-1β. Graphs show mean ± SEM for control (n = 4) and BD (n = 6) repeated in duplicate. Significance was determined using the unpaired non-parametric Mann-Whitney one-tailed test, ∗p < 0.05.
(F) IL-6 concentration measured in the blood of healthy controls (n = 90) or BD patients (n = 55). Significance was determined using Welsh's two-tailed test not assuming equal distribution, ∗p < 0.05.
To understand the implications of our data in a clinical context, we asked whether BD patients have an increased inflammatory drive with altered IL-6 levels. To this end, we examined blood IL-6 levels from a large but distinct cohort of BD patients (n = 55) versus age-matched healthy controls (n = 90). Remarkably, IL-6 levels, averaged over three blood draws, were significantly higher in BD patients (Figure 3G). Taken together, these results indicate that BD patients may have a general propensity for increased IL-6 and that secreted IL-6 in inflammation-primed BD astrocytes may contribute to the functional consequences on the activity of co-cultured neurons.
Secreted IL-6 from BD Astrocytes Regulates Neuronal Activity
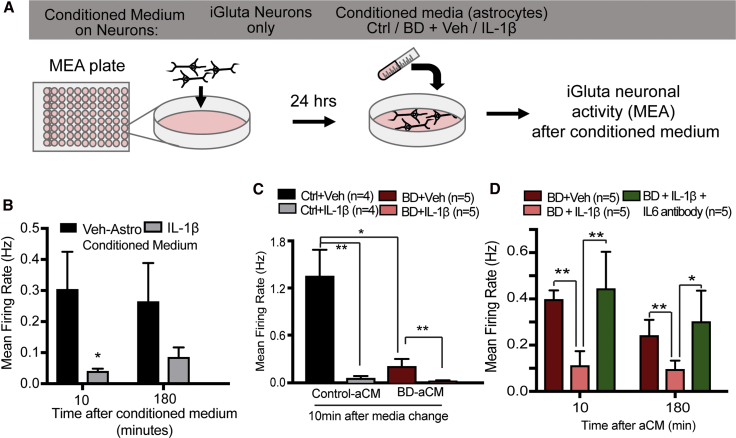
Our results led us to hypothesize that some of the observed effects of stimulated BD astrocytes on neuronal activity were mediated via increased secretion of IL-6. To test this hypothesis, we examined the effect of astrocyte-conditioned neuronal medium (aCM) from control or BD groups on the activity on MEA of iGluta neurons (commercially available) in culture (Figure 4A). Astrocyte-conditioned neuronal medium was obtained from each healthy (n = 3) and BD patient astrocyte (n = 4) line pretreated with vehicle or IL-1β (4 groups in total). First, we performed a time course experiment over 12 h, examining the activity of neurons when incubated with aCM from control astrocytes. We observed the maximum increase in neuronal activity immediately after (10 min) incubation with aCM with a gradual decrease by 6 to 12 h (Figure S3). These results indicated that astrocyte-secreted factors played a supportive role in promoting neuronal activity. Next, we asked whether secreted factors in IL-1β-stimulated astrocytes impacted neuronal activity. For this experiment, aCM was collected from IL-1β-stimulated control astrocytes and incubated with neurons for activity measurements at 10 min and 180 min after incubation. aCM from stimulated control astrocytes substantially reduced neuronal activity within 10 min. We observed the greatest negative impact within 10 min after medium change to aCM from control astrocytes, which continued up to 3 h (Figure 4B), gradually normalizing by 12 h (Figure S3). Note that the data are shown in minutes in the figures.
Figure 4.
IL-1β-Stimulated BD Astrocyte-Conditioned Medium Decreases Neuronal Activity via IL-6
(A) Control neurons plated on an MEA plate were incubated with conditioned neuronal media from different groups and neural activity was measured. Differentiated control (four individuals) or BD astrocytes (five individuals) were treated with vehicle or IL-1β for 5 h, following which they were incubated with neuronal media; 18 h later, astrocyte-conditioned neuronal medium was collected for treating iGluta commercially derived glutamatergic neurons.
(B) Time course measurement of activity of iGluta neurons with aCM 10 min and 180 min after exposure, from vehicle or IL-1β-stimulated astrocytes.
(C) Neuronal activity of iGluta neurons (10 min) after media change to aCM from control (n = 4 individuals) and BD astrocytes (n = 5 individuals) that were either vehicle or IL-1β treated.
(D) Neuronal activity after incubation with conditioned medium (10 min and 180 min) from BD astrocytes that were IL-1β treated or IL-1β + IL-6 antibody treated. Graphs show mean ± SEM for each group. Significance was determined using (B) paired two-tailed t test, (C) ordinary two-way ANOVA, and (D) unpaired one-tailed t test. ∗p < 0.05, ∗∗p < 0.01.
When aCM from three controls and four BD astrocytes was incubated with iGluta neurons we observed a significant reduction in neuronal activity in IL-1β-stimulated control and BD astrocytes compared with the non-stimulated astrocytes (Figure 4C). These results corroborated our observations with the BD astrocytes/neurons co-culture experiment. They indicated that secreted factors from astrocytes directly regulated neuronal activity and that secreted factors from BD astrocytes, both stimulated and non-stimulated, reduced neuronal activity. Based on our results showing higher IL-6 levels in the culture medium of IL-1β-stimulated BD astrocytes, we hypothesized that IL-6 might mediate the negative impact of stimulated BD astrocytes on neuronal activity. To test this hypothesis, we performed an experiment using an IL-6 blocking antibody to antagonize the downstream effects of IL-6 on neurons. For this experiment, aCM from each BD astrocyte line (n = 4) stimulated with IL-1β was collected and split into two groups, with or without IL-6 blocking antibody. iGluta neurons on an MEA plate were treated with aCM from BD astrocytes, IL-1β-stimulated BD astrocytes, or IL-1β-stimulated BD astrocytes with IL-6 blocking antibody. Confirming previous results, we observed that neurons treated with aCM from stimulated BD astrocytes displayed lower activity compared with the non-stimulated astrocytes (Figure 4D). Furthermore, aCM from stimulated BD astrocytes with IL-6 blocking antibody had significantly higher levels of activity, mean firing rate (Figure 4D), spike number, but not network bursts (data not shown) compared with stimulated BD astrocytes (Figure 4D). It is likely that IL-6 mediates its impact on neuronal activity via the soluble IL-6 receptor, as human iPSC-derived neurons expressed significantly higher levels of the soluble IL-6 receptor as compared with the membrane-bound receptor (Figure S3). These results suggest that secreted IL-6 may partially mediate the negative impact of stimulated BD astrocytes on neuronal activity.
Discussion
In this in vitro study, we derived immune-responsive astrocytes from control and BD patient iPSCs and showed that BD astrocytes mounted a unique response to inflammatory stimuli. BD astrocytes are transcriptionally distinct from controls and are less supportive for neuronal activity, even without stimulation. Inflammatory stimulation further exacerbated these phenotypes and increased IL-6. Neuronal activity decreased following co-culture with IL-1β-stimulated BD astrocytes. Compared with control astrocytes, BD astrocytes secreted more IL-6 following inflammation, with a greater percentage of cells being immunopositive for IL-6. Conditioned culture medium of stimulated BD astrocytes was sufficient to decrease neuronal activity, and this effect was rescued in the presence of an IL-6 blocking antibody. These results suggest that secreted factors from astrocytes play a role in regulating neuronal activity and that, in the case of BD, IL-6 at least in part mediated the effects of inflammation-primed astrocytes on neuronal activity.
In addition to microglia and neurons, astrocytes are a source of IL-6 production following CNS injury and neuroinflammatory response. Elevated levels of IL-1β and IL-6 have been found in the blood and cerebrospinal fluid of patients with psychiatric disorders, including BD, and particularly during manic episodes (O'Brien et al., 2006). While mild inflammation can be beneficial for many neural processes, the overproduction of IL-6 may worsen BD symptoms and may be an important therapeutic target. As a key target of inflammation, IL-6 has been studied in various systems revealing pleiotropic effects. Acutely in vitro, it serves as a neuroprotectant, but chronic treatment is associated with neuronal cell injury and death (Xia et al., 2015). Overexpression of IL-6 in mice has a significant effect on dendrite arborization and death of distinct neural subtypes (Campbell et al., 1993). Our results suggest that overproduction of IL-6 has a negative impact on neuronal activity; however, given that our experiments are all short time exposure, the direct impact of IL-6 must be interpreted with caution. The high dose of IL-1β used in our studies was based on our previous studies; however, the physiological relevance of this dose needs to be determined. Given the short time scales in which we observed changes in neuronal activity following aCM and substantially higher expression of the soluble IL-6 receptor in neurons, it is possible that the effects are mediated by altered synaptic transmission rather than loss of synapses. BD and other CNS diseases are often accompanied by chronic inflammation, and our experiments mimic an acute model of inflammation. Given, also, that many cell types are involved in immune response in the CNS, adding complexity to our culture system via the study of microglia and a chronic treatment paradigm would be useful to provide a clearer picture of this nuanced signaling pathway.
While our results are robust across our patient-derived astrocytes, the current sample size is relatively small given the variability that is typically observed in cell culture models. Due to the low throughput nature of patient-stem cell work, it is currently difficult to extend the study to include a large enough sample set run together, as batch effects of lines generated independently can be significant. The role of IL-6 on astrocytes' impact on neuronal activity must be interpreted with caution, as it remains unclear whether IL-6 can directly regulate neuronal activity in our culture system and for the antibody blocking experiment whether a control immunoglobulin (Ig)G could also impact neuronal activity. It is also worth recognizing that iPSC-derived astrocytes are relatively immature compared with those in the brains of bipolar patients, and we do not yet have reliable biomarkers for pinpointing exact developmental age. Hence, direct extrapolation to in vivo phenotypes and understanding the compound impact on patient brains remains challenging. Future studies addressing these points could prove meaningful. Despite these limitations, our findings elucidate aspects of the understudied role of astrocytes in neuroinflammation in psychiatric disorders, with relevance for altered IL-6 and inflammatory signaling in BD patient astrocytes.
Experimental procedures
Differentiation of Astrocytes from iPSCs
All subjects were white males and provided written informed consent. Six of these were patients with BD type I who participated in a drug response clinical trial at the University of California, San Diego (Veteran's Study and Pharmacogenomics of Bipolar Disorder Study) and were described previously (Mertens et al., 2015), and there were four healthy controls (Supplemental Table S1). All procedures were approved by the University of California, San Diego (UCSD), the Salk Institute Institutional Review Board and the Embryonic Stem Cell Research Oversight Committee (IRB protocol #09-0003).
Fibroblasts obtained from sterile skin biopsies from healthy controls and BD patients were reprogrammed to iPSCs using the Cyto-Tune Sendai kit (Invitrogen) as previously described (Mertens et al., 2015). iPSCs were cultured on matrigel-coated plates in mTeSR1 medium (STEMCELL Technologies) and differentiation into astrocytes was performed as described (Santos et al., 2017). Embryoid bodies were generated from confluent iPSC cultures by mechanical dissociation with 1 mg/mL collagenase IV (Gibco) and then plated onto low-adherence plates in mTeSR1 medium containing 10 μM ROCK inhibitor (STEMCELL Technologies) and incubated overnight while shaking. The day after, the medium was changed to Astrocyte Medium (AM, ScienCell) with 500 ng/mL noggin (R&D Systems) and 10 ng/mL PDGFAA (PeproTech) for 14 days and was supplemented with only PDGFAA for an additional 7 days with agitation. Embryoid bodies were dissociated with papain (Papain dissociation system, Worthington), and the resulting GPCs were grown on 10 μg/mL poly-L-ornithine (poly-O, Sigma)- and 5 μg/mL laminin (Invitrogen)-coated plates in AM with 20 ng/mL FGF-2 (Joint Protein Central) and 20 ng/mL EGF (Humanzyme). Low-density GPC cultures were used for differentiation to astrocytes in DMEM/F-12 Glutamax, 2% B27 without vitamin A, 1% N2 (N2B27 media; all from Thermo Fisher Scientific) plus 10% FBS (Biowest). Human cerebellar fetal primary astrocytes (ScienCell) were cultured in AM ScienCell medium and used as a positive control for human astrocyte markers.
Immunocytochemistry
Cells were fixed with 4% paraformaldehyde solution for 15 min at 4°C. Antigen blocking and cell permeabilization were done using 10% horse serum and 0.1% Triton X-100 in PBS for 1 h at room temperature. Primary antibodies prepared in blocking solution were incubated overnight at 4°C, and the next day secondary antibodies (1:250, Jackson Laboratories) were incubated for 1 h at room temperature after washes. The primary antibodies used were mouse anti-Nestin (1:500; EMD Millipore; MAB5326), rabbit anti-S100β (1:1,000; Dako; Z0311), mouse anti-GFAP monoclonal (1:250; EMD Millipore; IF03L), rat anti-CD44 (1:100; BD Pharmingen; 550538), rabbit anti-NFIA (1:250; Novus Bio; NBP1-81406), mouse anti-ALDH1L1 (1:100; EMD Millipore; MABN495), and mouse immunoglobulin M anti-A2B5 (1:50; EMD Millipore; MAB312). Fluorophore-conjugated secondary antibodies (donkey) were used (Jackson Labs). Cells were counterstained with DAPI for visualizing cell nuclei.
Flow Cytometry Assay for Cytokine Stimulation
Astrocytes from four controls and six BD patients were differentiated for 5 weeks and stimulated with 10 ng/mL recombinant human IL-1β (R&D) or with 50 ng/mL recombinant human TNF-α (R&D) or PBS (vehicle) for 5 h. Protein transport inhibitors (1:1,000 BD GolgiPlug and BD GolgiStop; BD Biosciences) were added to all samples, including non-stimulated controls. Five hours after stimulation, cells were dissociated at room temperature for 1 min in a 1:1 mixture of enzymes accutase (STEMCELL) and papain (Papain dissociation system, Worthington). Cells were washed and stained with the viability dye Zombie UV fixable kit (BioLegend). Cells were fixed and permeabilized using the BD Cytofix/Cytoperm and BD Perm/Wash (BD Biosciences). IL-6 or IL-8 cytokines were detected following incubation in BD Perm/Wash containing PerCP conjugated anti-IL-8 (BioLegend BH0814) and APC conjugated anti-IL-6 (BioLegend MQ213A5) antibodies for 20 min at 4°C. Quantification was done on the BD Canto II cytometer (BD Biosciences) and analysis was performed on FlowJo software (TreeStar). Negative gating controls for anti-IL-6 were done with rat IgG1-APC (BioLegend RTK2071) and for anti-IL-8 with mouse IgG2b-PercP (BioLegend MPC-11). Also, negative gates were determined for each experiment using non-stimulated controls, and normalized by subtraction from positive cells (data not shown).
Cytokine Array Blots
Five-week-old astrocytes were washed in PBS and treated with 10 ng/mL recombinant human IL-1β (R&D) or PBS (vehicle) in astrocyte differentiation medium. Five hours after stimulation, 1 mL of medium was collected and flash frozen in liquid nitrogen for cytokine blot arrays. The remaining cells were detached as described above and counted to confirm a similar number across lines and experimental conditions. Three control (C1, C2, C3) and four BD astrocyte lines (BD1, BD2, BD4, BD6) were used for this experiment. The level of cytokines released in the medium was determined using Proteome Profiler Human Cytokine Array from R&D Systems according to the manufacturer's instructions. Comparative quantification of image intensity of cytokines was done using FIJI software (26). The intensity of cytokine dots (in stimulation group) was quantified using FIJI software and normalized per blot using positive controls provided by the manufacturer.
Library Preparation and RNA-Sequencing
RNA-sequencing (RNA-Seq) analysis was performed on 5-week-old astrocytes, either non-stimulated (PBS) or stimulated with 10 ng/mL IL-1β (without protein transport inhibitors). After 5 h, the cells were collected in RNABee solution (Tel test, Inc) and total RNA was extracted using the DNA-Free RNA Kit (Zymo Research) according to the manufacturer's instructions. RNA quality was assayed using Agilent Technologies 2200 TapeStation and only samples with high RNA quality (RIN >8) were used for library preparation. Stranded mRNA-Seq libraries were prepared using the Illumina TruSeq Stranded mRNA Library Prep Kit according to the manufacturer's instructions. Sequenced reads were quality-tested using FASTQC (27) and aligned to the hg19 (28) human genome using the STAR aligner (29) version 2.4.0k. Mapping was carried out using default parameters (up to 10 mismatches per read, and up to nine multi-mapping locations per read). The genome index was constructed using the gene annotation supplied with the hg19 iGenomes collection (Illumina) and overhang value of 100. Raw gene expression was quantified across all gene exons (RNA-Seq) using the top-expressed isoform as proxy for gene expression. Differential expression was performed using DESeq2 (30) package version 1.20.0. DEGs were defined as having a false discovery rate (FDR) < 0.05 and a log2 fold change >1. Hierarchical clustering was performed using the R language (v.3.3.2) with Ward's hierarchical agglomerative clustering method and 1-correlation as a distance metric. For the RNA-Seq experiment, astrocytes from four healthy controls and five BD patients (listed in Figure 1) were differentiated in duplicates and sequenced (Table S1, Figure 1).
BD Patient IL-6 Blood Measurements
A separate sample of people with and without BD was recruited from outpatient clinics, community settings, and other research studies for a longitudinal study of cognition and inflammation; data from year one of the study are presented. BD had DSM-IV diagnoses of Bipolar I or II Disorder, as assessed using a modified structured clinical interview that was also used for diagnosis of the skin sample donor patients. Exclusion criteria were acute illness or pregnancy, a recent vaccination, history of various health conditions (dementia, seizures, Parkinson's, stroke, or head trauma), cancer treatment in past, diabetes/hypertension that is not controlled, among others. Healthy comparison (HC) participants were excluded who had a history of DSM-IV Axis I disorders, previous use of psychotropic medications, as well as having a first-degree relative with history of depression, BD, or schizophrenia. This study was approved by the UCSD Institutional Review Board; all participants completed informed consent prior to study involvement. Smokers were excluded, as smoking correlated with increased IL-6. Three study visits occurred in year one over a 2-week period, each a week apart. Clinical and demographic data were collected at the first visit; non-fasting blood (15 mL) was drawn by a licensed phlebotomist at all three visits, at a similar time of day. Samples were immediately centrifuged for 10 min at 500 × g. Plasma was extracted and stored at −80°C until assay. Samples from 55 BD and 90 HC participants were processed in the laboratory of Cristian Achim, PhD, in the UCSD Department of Psychiatry. Plasma aliquots from all participants were tested using a sandwich immunoassay with MSD Human V-PLEX Custom Human Cytokine kits (Meso Scale Discovery, Rockville, MD) and analyzed on a SECTOR Imager 2400 instrument (Rockville, MD). The laboratory technician performing the assays was “blind” to the subject's diagnosis and samples were run in duplicate. No sample showed biomarker levels below the detection limits. Three concentration ranges of quality controls were used to evaluate assay accuracy and precision in order to ensure reliable and accurate results. Intra-assay variability was less than 5% for all assays. We log-transformed IL-6 values and averaged the results across (up to) three blood draws for analyses.
Astrocyte-neuron Co-culture and MEA
Differentiation of a single control iPSC line from an independent cohort (WT126) was utilized for co-culture MEA experiments (described below). WT126 iPSCs were differentiated into neural progenitor cells (NPCs) utilizing a pan-neuronal differentiation protocol (Marchetto et al., 2010). For MEA analysis, 10,000 NPCs from WT126 iPSCs were plated for neuronal differentiation in a 96-well poly-O (Sigma, 100 μg/mL)- and laminin- (Invitrogen, 100 μg/mL) coated MEA plates. Three weeks after neuronal differentiation, 4-week-old astrocytes from different groups/treatments were plated on top of differentiating neurons in the MEA plate. Cells were fed three times a week with co-culture medium: N2B27 media, 0.2 μM ascorbic acid (Sigma), 20 ng/mL glial cell-derived neurotrophic factor (PeproTech), 20 ng/mL brain-derived neurotrophic factor (PeproTech), 500 μg/mL cyclic AMP (cAMP, TOCRIS), 1% FBS, and 1% penicillin streptomycin (ScienCell). Measurements on the MEA system were taken before media changes. Recordings were performed in a Maestro MEA system and AxIS software (Axion Biosystems) using a bandwidth with a filter for 200 Hz to 3 kHz cutoff frequencies. Spike detection was performed using an adaptive threshold set to six times the standard deviation of the estimated noise on each electrode. Each plate was acclimatized for 10 min in the Maestro Instrument and recorded for 10 min for quantification. Multi-electrode data analysis was performed using the Axion Biosystems Neural Metrics Tool. An electrode was considered active at a threshold of five spikes per minute. Bursts were identified in the data recorded from each individual electrode using an adaptive Poisson surprise algorithm. Network bursts were identified for each well using a non-adaptive algorithm requiring a minimum of 10 spikes with a maximum inter-spike interval of 100 ms. Only the wells that exhibited bursting activity were included in this analysis. In graphs, the x axes represent time and the y axes represent the multiple parameters of neuronal activity recorded per well.
Conditioned Media Experiments
iGluta neurons (100,000 to 150,000 cells; Fujifilm Cellular Dynamics, Inc.) were plated on each well of a 96-well MEA plate (Axion Biosystems) and cultured in medium according to manufacturer's instructions. The baseline activity of the plate with iGluta neurons was recorded for 1 week. In parallel, 4-week-old astrocytes from three neurotypical controls (C1, C2, C4) and four BD patients (BD2, BD4, BD5, BD6) stimulated with IL-1β or vehicle (PBS) for 5 h were washed and culture medium was changed to neuronal medium (iGluta medium); 1% FBS was added to the astrocytes to generate conditioned medium; 24 h later, this astrocyte-conditioned medium was removed from astrocyte cultures and added to the neurons already in culture for 1 week on MEA plates. Neuronal activity was measured on the MEA before and immediately after changing to conditioned medium.
For IL-6 blocking antibody experiments, conditioned medium was collected as described and split into two sets, with 50 ng/mL IL-6 blocking antibody (R&D) or without blocking antibody. Neuronal activity was measured on the MEA immediately before and after changing to conditioned medium with/without IL-6 blocking antibody. Conditioned medium components were 100 mL of neuronal medium (BrainPhys Neuronal Media, STEMCELL Technologies), 2 mL of iCell Neural Supplement B (CDI), 1 mL of iCell Nervous System Supplement (CDI), 1 mL N2 (Invitrogen), 100 μL of 1 μg/mL laminin (Invitrogen), 1% penicillin streptomycin (ScienCell), and 1% FBS (Biowest).
Data Analysis and Statistics
For FACS, cytokine, neuronal co-culture, and conditioned medium experiments, three control and six BD patient-derived astrocytes were used. Experiments were run in triplicate and averaged for each line. Normalized averages across multiple experiments were used to generate graphs of mean from each individual. Over multiple of experiments, occasionally a line does not grow well due to technical reasons. To be on the conservative side of interpretation, we did not factor into our analyses lines that did not grow well in each experiment. Yet all experiments have at least four BD patients, although the lines vary. The average for each individual is represented as a single dot in column graphs in most figures. Statistical n was performed solely on number of individuals per group (control n = 3–4 and BD n = 4–6, detailed in each figure and figure legend). Statistical comparisons were performed by comparing average values from individuals per group with non-parametric statistical tests. Statistical tests, n values, and p values for the experiments are stated in the figure legends, and p < 0.05 was considered statistically significant. For RNA-Seq experiments, four control and five BD patient-derived astrocytes (run in duplicate) were used, and the DESeq2 statistical package was used.
Data and Code Availability
RNA-Seq data has been uploaded to NIH. Publicly accessible via accession number: GSE157509.
Author contributions
K.C.V., R.S., and C.M.M. designed the study, performed experiments, analyzed the data, interpreted the results, and wrote the manuscript. A.P.D.M., A.M., V.P., and K.J.H performed experiments and analyzed the data. R.O. performed experiments. G.E., M.N.S., and K.C.V. performed whole transcriptome data analysis. M.J., L.E., and J.R.K. provided clinical data analysis and contributed with materials. F.H.G. contributed to the design of the study and interpretation of the results and edited the manuscript.
Conflicts of interests
All authors declare no conflicts of interest.
Acknowledgments
This research was supported by the Robert and Mary Jane Engman Foundation and Lynn and Edward Streim. The authors acknowledge support from the Paul G. Allen Family Foundation, Bob and Mary Jane Engman, the Leona M. and Harry B. Helmsley Charitable Trust grant #2012-PG-MED002, Annette C. Merle-Smith, R01 MH095741 (F.H.G.), U19MH106434 (F.H.G.) and by The G. Harold & Leila Y. Mathers Foundation. Grants/contracts to J.R.K. were from the National Institute of Mental Health (U01 MH92758) supporting the Pharmacogenomics of Bipolar Disorder Study and from the Department of Veterans Affairs (5I01CX000363), and NIH grant R01MH103318 (L.E.). We thank the patients that participated in the study. The authors also thank M.L. Gage for editorial comments.
Published: March 4, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.02.004.
Contributor Information
Renata Santos, Email: renata.santos@inserm.fr.
Maria C. Marchetto, Email: marchetto@salk.edu.
Fred H. Gage, Email: gage@salk.edu.
Supplemental information
References
- Bonni A., Sun Y., Nadal-Vicens M., Bhatt A., Frank D.A., Rozovsky I., Stahl N., Yancopoulos G.D., Greenberg M.E. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science. 1997;278:477–483. doi: 10.1126/science.278.5337.477. [DOI] [PubMed] [Google Scholar]
- Brennand K.J., Simone A., Jou J., Gelboin-Burkhart C., Tran N., Sangar S., Li Y., Mu Y., Chen G., Yu D. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell I.L., Abraham C.R., Masliah E., Kemper P., Inglis J.D., Oldstone M.B., Mucke L. Neurologic disease induced in transgenic mice by cerebral overexpression of interleukin 6. Proc. Natl. Acad. Sci. U S A. 1993;90:10061–10065. doi: 10.1073/pnas.90.21.10061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy F., Ahearn E., Carroll B.J. Elevated frequency of diabetes mellitus in hospitalized manic-depressive patients. Am. J. Psychiatry. 1999;156:1417–1420. doi: 10.1176/ajp.156.9.1417. [DOI] [PubMed] [Google Scholar]
- Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo E., Farina C. Astrocytes: key regulators of neuroinflammation. Trends Immunol. 2016;37:608–620. doi: 10.1016/j.it.2016.06.006. [DOI] [PubMed] [Google Scholar]
- Craddock N., Sklar P. Genetics of bipolar disorder. Lancet. 2013;381:1654–1662. doi: 10.1016/S0140-6736(13)60855-7. [DOI] [PubMed] [Google Scholar]
- Dell'Albani P., Kahn M.A., Cole R., Condorelli D.F., Giuffrida-Stella A.M., de Vellis J. Oligodendroglial survival factors, PDGF-AA and CNTF, activate similar JAK/STAT signaling pathways. J. Neurosci. Res. 1998;54:191–205. doi: 10.1002/(SICI)1097-4547(19981015)54:2<191::AID-JNR7>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Deneen B., Ho R., Lukaszewicz A., Hochstim C.J., Gronostajski R.M., Anderson D.J. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron. 2006;52:953–968. doi: 10.1016/j.neuron.2006.11.019. [DOI] [PubMed] [Google Scholar]
- Drake C., Boutin H., Jones M.S., Denes A., McColl B.W., Selvarajah J.R., Hulme S., Georgiou R.F., Hinz R., Gerhard A. Brain inflammation is induced by co-morbidities and risk factors for stroke. Brain Behav. Immun. 2011;25:1113–1122. doi: 10.1016/j.bbi.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G., Martinowich K., Chin M.H., He F., Fouse S.D., Hutnick L., Hattori D., Ge W., Shen Y., Wu H. DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development. 2005;132:3345–3356. doi: 10.1242/dev.01912. [DOI] [PubMed] [Google Scholar]
- Goldstein B.I., Kemp D.E., Soczynska J.K., McIntyre R.S. Inflammation and the phenomenology, pathophysiology, comorbidity, and treatment of bipolar disorder: a systematic review of the literature. J. Clin. Psychiatry. 2009;70:1078–1090. doi: 10.4088/JCP.08r04505. [DOI] [PubMed] [Google Scholar]
- Hamdani N., Tamouza R., Leboyer M. Immuno- inflammatory markers of bipolar disorder: a review of evidence. Front. Biosci. (Elite Ed.) 2012;4:2170–2182. doi: 10.2741/e534. [DOI] [PubMed] [Google Scholar]
- Hayden E.P., Nurnberger J.I., Jr. Molecular genetics of bipolar disorder. Genes Brain Behav. 2006;5:85–95. doi: 10.1111/j.1601-183X.2005.00138.x. [DOI] [PubMed] [Google Scholar]
- Leboyer M., Soreca I., Scott J., Frye M., Henry C., Tamouza R., Kupfer D.J. Can bipolar disorder be viewed as a multi-system inflammatory disease? J. Affect. Disord. 2012;141:1–10. doi: 10.1016/j.jad.2011.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto M.C., Carromeu C., Acab A., Yu D., Yeo G.W., Mu Y., Chen G., Gage F.H., Muotri A.R. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens J., Wang Q.W., Kim Y., Yu D.X., Pham S., Yang B., Zheng Y., Diffenderfer K.E., Zhang J., Soltani S. Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature. 2015;527:95–99. doi: 10.1038/nature15526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modabbernia A., Taslimi S., Brietzke E., Ashrafi M. Cytokine alterations in bipolar disorder: a meta-analysis of 30 studies. Biol. Psychiatry. 2013;74:15–25. doi: 10.1016/j.biopsych.2013.01.007. [DOI] [PubMed] [Google Scholar]
- Munkholm K., Vinberg M., Vedel Kessing L. Cytokines in bipolar disorder: a systematic review and meta-analysis. J. Affect. Disord. 2013;144:16–27. doi: 10.1016/j.jad.2012.06.010. [DOI] [PubMed] [Google Scholar]
- Najjar S., Pearlman D.M., Alper K., Najjar A., Devinsky O. Neuroinflammation and psychiatric illness. J. Neuroinflammation. 2013;10:43. doi: 10.1186/1742-2094-10-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien S.M., Scully P., Scott L.V., Dinan T.G. Cytokine profiles in bipolar affective disorder: focus on acutely ill patients. J. Affect. Disord. 2006;90:263–267. doi: 10.1016/j.jad.2005.11.015. [DOI] [PubMed] [Google Scholar]
- Osby U., Brandt L., Correia N., Ekbom A., Sparen P. Excess mortality in bipolar and unipolar disorder in Sweden. Arch. Gen. Psychiatry. 2001;58:844–850. doi: 10.1001/archpsyc.58.9.844. [DOI] [PubMed] [Google Scholar]
- Pinto J.V., Moulin T.C., Amaral O.B. On the transdiagnostic nature of peripheral biomarkers in major psychiatric disorders: a systematic review. Neurosci. Biobehav. Rev. 2017;83:97–108. doi: 10.1016/j.neubiorev.2017.10.001. [DOI] [PubMed] [Google Scholar]
- Ross R. Atherosclerosis--an inflammatory disease. N. Engl. J. Med. 1999;340:115–126. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- Santos R., Vadodaria K.C., Jaeger B.N., Mei A., Lefcochilos-Fogelquist S., Mendes A.P.D., Erikson G., Shokhirev M., Randolph-Moore L., Fredlender C. Differentiation of inflammation-responsive astrocytes from glial progenitors generated from human induced pluripotent stem cells. Stem Cell Reports. 2017;8:1757–1769. doi: 10.1016/j.stemcr.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliman M.A., Aboharb F., Zeltner N., Studer L. Pluripotent stem cells in neuropsychiatric disorders. Mol. Psychiatry. 2017;9:1241–1249. doi: 10.1038/mp.2017.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadodaria K.C., Amatya D.N., Marchetto M.C., Gage F.H. Modeling psychiatric disorders using patient stem cell-derived neurons: a way forward. Genome Med. 2018;10:1. doi: 10.1186/s13073-017-0512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadodaria K.C., Ji Y., Skime M., Paquola A., Nelson T., Hall-Flavin D., Fredlender C., Heard K.J., Deng Y., Le A.T. Serotonin-induced hyperactivity in SSRI-resistant major depressive disorder patient-derived neurons. Mol. Psychiatry. 2019;24:795–807. doi: 10.1038/s41380-019-0363-y. [DOI] [PubMed] [Google Scholar]
- Vadodaria K.C., Ji Y., Skime M., Paquola A.C., Nelson T., Hall-Flavin D., Heard K.J., Fredlender C., Deng Y., Elkins J. Altered serotonergic circuitry in SSRI-resistant major depressive disorder patient-derived neurons. Mol. Psychiatry. 2019;24:808–818. doi: 10.1038/s41380-019-0377-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasile F., Dossi E., Rouach N. Human astrocytes: structure and functions in the healthy brain. Brain Struct. Funct. 2017;222:2017–2029. doi: 10.1007/s00429-017-1383-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonk R., van der Schot A.C., Kahn R.S., Nolen W.A., Drexhage H.A. Is autoimmune thyroiditis part of the genetic vulnerability (or an endophenotype) for bipolar disorder? Biol. Psychiatry. 2007;62:135–140. doi: 10.1016/j.biopsych.2006.08.041. [DOI] [PubMed] [Google Scholar]
- Weiner M., Warren L., Fiedorowicz J.G. Cardiovascular morbidity and mortality in bipolar disorder. Ann. Clin. Psychiatry. 2011;23:40–47. [PMC free article] [PubMed] [Google Scholar]
- Xia W., Peng G.Y., Sheng J.T., Zhu F.F., Guo J.F., Chen W.Q. Neuroprotective effect of interleukin-6 regulation of voltage-gated Na(+) channels of cortical neurons is time- and dose-dependent. Neural Regen. Res. 2015;10:610–617. doi: 10.4103/1673-5374.155436. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq data has been uploaded to NIH. Publicly accessible via accession number: GSE157509.