Summary
The ability to genetically manipulate organisms has led to significant insights into functional genomics in many species. In birds, manipulation of the genome is hindered by the inaccessibility of the one-cell embryo. During embryonic development, avian primordial germ cells (PGCs) migrate through the bloodstream and reach the gonadal anlage, where they develop into mature germ cells. Here, we explored the use of PGCs to produce transgenic offspring in the zebra finch, which is a major animal model for sexual brain differentiation, vocal learning, and vocal communication. Zebra finch PGCs (zfPGCs) obtained from embryonic blood significantly proliferated when cultured in an optimized culture medium and conserved the expression of germ and stem cell markers. Transduction of cultured zfPGCs with lentiviral vectors was highly efficient, leading to strong expression of the enhanced green fluorescent protein. Transduced zfPGCs were injected into the host embryo and transgenic songbirds were successfully generated.
Keywords: primordial germ cell culture, lentiviral vector, eGFP, transgenic zebra finch, songbird
Graphical abstract

Highlights
-
•
Blood-derived zebra finch primordial germ cells (zfPGCs) can be expanded in vitro
-
•
Lentiviral vectors can efficiently transduce zfPGCs in vitro
-
•
Genetically modified zfPGCs can be used to generate transgenic zebra finches
In this article, Gessara and colleagues used zebra finch primordial germ cells obtained from embryonic blood for the genetic modification of a songbird species using lentiviral vectors.
Introduction
Songbirds are major animal models for studying the genetic and neural basis of vocal learning and communication (Mello, 2014; Mooney, 2020; Prather et al., 2017) as well as sex-hormone-dependent brain development (Balthazart et al., 2010; Gahr, 2007; McCarthy and Arnold, 2011) and adult neurogenesis (Goldman and Nottebohm, 1983; Paton and Nottebohm, 1984). For the zebra finch, transgenic models have been successfully developed (Abe et al., 2015; Agate et al., 2009; Liu et al., 2015). Agate et al. (2009) were the first to inject lentiviral vectors for GFP into the blastodisc of freshly laid zebra finch eggs in order to target the primordial germ cells (PGCs), the precursors of spermatocytes and oocytes. However, due to the inefficiency of the method, only two other transgenic models have been generated over the past 10 years using this method (Abe et al., 2015; Liu et al., 2015).
During avian development, PGCs are located in the central area of the blastodisc until Eyal-Giladi and Kochav (EGK) stage X (Eyal-Giladi et al., 1976; Kochav et al., 1980); from here, PGCs translocate to the germinal crescent (Ginsburg and Eyal-Giladi, 1986) and finally migrate through the developing vascular system to reach the gonadal anlage (Fujimoto et al., 1976; Jung et al., 2019; Nakamura et al., 1988). Avian PGCs are easily isolated from the circulatory system by blood aspiration between Hamburger and Hamilton (HH) stages 14 and 17 (Hamburger and Hamilton, 1951). In the domestic chicken, embryonic-blood-derived PGCs can be propagated in vitro for several months, genetically modified, and re-injected into early embryo surrogate hosts. Their ability to migrate to the gonadal anlage was unaffected by this treatment and the resulting hosts exhibited a high germline transmission rate (Macdonald et al., 2010; Van De Lavoir et al., 2006). For the zebra finch, it has been shown that PGCs of the embryonic gonads (zfgPGCs) can be cultured for up to 30 days on a feeder cell layer in the presence of relatively high concentrations of undefined fetal bovine serum, be genetically modified with transposons, and retain the ability to colonize embryonic host gonads (Jung et al., 2019). However, to the best of our knowledge the generation of transgenic songbirds using this approach has not yet been reported.
Here, we explored the use of blood-borne zebra finch PGCs (zfPGCs) to generate transgenic zebra finches using a feeder-free culture medium that contained fetal bovine serum at a minimal concentration to avoid precocious differentiation and using lentiviral transduction for genetic modification. Cultured zfPGCs were efficiently transduced with lentiviral vectors expressing enhanced green fluorescent protein (eGFP). After injection underneath the early zebra finch blastodisc of a host embryo (EGK stage X), genetically modified zfPGCs migrated to and colonized the gonadal anlage following incubation. These surrogate host embryos were successfully hatched and raised by foster parents and, when mated, efficiently transmitted the transgene to their offspring (Figure S1).
Results
Expansion of Embryonic-Blood-Derived zfPGCs in Culture
Blood samples (1–3 μL) extracted from zebra finch embryos at Murray stages 13–15 (Murray et al., 2013) contained a mixed population of erythrocytes and about 140 PGCs, with large individual differences in the number of PGCs observed between samples (40–500). Like in chicken (Macdonald et al., 2010), blood-derived zfPGCs were round, with a diameter of 16–20 μm, and showed tiny membrane protrusions visible under phase contrast, a prominent nucleus, and small cytoplasmic vesicles that enclosed polysaccharides, which could be stained by the periodic acid-Schiff (PAS) reaction (Figures 1A–1C).
Figure 1.
Characterization and Expansion of Cultured zfPGCs
(A) Phase-contrast image of an early blood cell culture with zfPGCs (white arrows) that contained polysaccharide vesicles and showed typical cellular protrusions (red arrow). The inset shows cellular protrusions at higher magnification. Note that PGCs were larger in size than other blood cells.
(B and C) PAS stains of blood smear samples taken from a zebra finch (B) and a chicken (C) embryo at embryonic stage 14, staining polysaccharide vesicles of PGCs in magenta. A cytoplasmic fragment of the zfPGC (red arrow) is shown in the inset at a higher magnification (B).
(D) ZfPGCs growing in loose clumps at 7 DIV conserve their native morphology.
(E) Blood samples from single embryos were split in two and cultured, one-half in the presence of activin and the other with bone morphogenetic protein 4 (BMP4). Cell numbers were counted at 1 and 15 DIV and numbers of cell divisions at 15 DIV are shown (mean ± SEM).
(F) Growth curve of zfPGCs cultured in medium containing BMP4. Different letters indicate statistically significant differences between cell numbers on different days (ANOVA; n = 32, p = 0.05). ZfPGCs proliferated significantly until 15 DIV. At 20 DIV a significant decrease in the cell number due to death was detectable.
(G) Boxplot showing fold increase in zfPGCs during a period of 30 days in culture compared with the number of zfPGCs at 1 DIV. Scale bars in (A––D) represent 20 μm.
See also Figure S2.
To culture zfPGCs, we used a feeder-layer-free culture and a defined culture medium (FAIcs; see Table S1) with low calcium concentration that we had developed for chicken PGCs (chPGCs) obtained from embryonic blood (Whyte et al., 2015), but in which we replaced the chicken serum with a low concentration of fetal bovine serum. In this culture medium, we found that a low concentration of calcium (0.3 mM) supported the proliferation of zfPGCs in loose, non-adherent clumps (Figures 1D and S2A). Higher concentrations of calcium (medium with 100% KnockOut DMEM containing 1.8 mM calcium) promoted stronger cell-cell interactions, leading to the formation of dense zfPGC clumps with indistinguishable cell boundaries. We also tested a culture medium previously developed for zfgPGCs (Jung et al., 2019) that contained higher serum concentrations. In this medium, zfPGCs extracted from embryonic blood attached to the bottom of the culture plate (Figure S2B), showed a lower growth rate, and exhibited signs of cell death.
To establish improved culture conditions for zfPGCs, we adapted the FAIcs by altering the ligand of the TGF-β signaling pathway. Both growth factors activin A and bone morphogenetic protein (BMP) 4 have been shown to be sufficient for propagating chPGCs in vitro (Whyte et al., 2015). When zfPGCs were cultured in the presence of BMP4, we detected a significant increase in the cell number compared with control cultures without this growth factor (average ± SEM, 206 ± 36%; p = 0.05; n = 5) after 15 days in vitro (DIV). Because we found a trend toward higher numbers of cell divisions in the presence of BMP4 for zfPGCs (average ± SEM, 4.33 ± 0.88 for activin and 6.97 ± 1.87 for BMP4; n = 3; Figure 2E), we replaced activin with BMP4 for our germline transmission experiments.
Figure 2.
Conserved Expression of Germ and Stem Cell-Specific Markers in Cultured zfPGCs
Microphotographs of PGCs were taken after immunofluorescent staining for (A–D) stage-specific embryonic antigen 1 (SSEA-1) and (E–I) epithelial membrane antigen 1 (EMA-1). Staining was performed with blood smears obtained from zebra finch (A and E) and chicken (B and F) embryos as well as with zfPGCs cultured for 10 DIV (C, D, and G–I). Fluorescence images (immunostaining in red and nuclear staining with DAPI in blue) and phase contrast (C and G) images are shown. In (A) arrows point to a zfPGC (white arrow) and an erythrocyte (red arrow). Note that in the blood smear samples PGCs were surrounded by immunonegative blood cells. Scale bars represent 20 μm. In (I) zfPGCs growing in a cluster are shown.
(J) Photographs of agarose gels after electrophoretic separation and ethidium bromide staining of RT-PCR products. Cultured zfPGCs (PGCs; 10 DIV) continued to express germ cell markers such as DAZL (1, 2) and DDX4 (3, 4) and, interestingly, also stem cell markers such as POU5F1 (5) and NANOG (6). In contrast, cultured fibroblasts (EF) of zebra finch embryos did not express any of these marker genes. All cDNA samples from zebra finch blastodisc cells (Blast) that include early zfPGCs turned out to be PCR positive for the same marker genes. Detection of γ-actin (ACTG1) (7) expression was used to control all samples for equal cDNA quality. Sizes of molecular weight markers (left slots) and expected amplicons are indicated. For both DAZL and DDX4, two RT-PCRs were carried out using primer pairs that were reported by Mak et al. (2015) (DAZL(I), DDX4(I)) and newly designed for this study to be intron spanning (DAZL(II), DDX4(II)) (see Experimental Procedures).
When our optimized zfPGC culture medium was used (Table S1), proliferation of zfPGCs was detectable after 2 DIV, and loose cell clusters were observed forming after 5 DIV (Figure 1D). During the first 2 weeks of culture, zfPGCs divided two to nine times, reaching a maximum of 8,000 cells. On average, we obtained 1,331 zfPGCs at 10 DIV and 1,045 zfPGCs at 15 DIV per blood sample (n = 32; Figures 1F and 1G). Between 15 and 20 DIV, zfPGCs started to undergo cell death.
Expression of Cell-Type-Specific Markers by Cultured zfPGCs
To test the effect of in vitro propagation on PGC-specific gene expression, we assessed germ and stem cell marker expression by zfPGCs at 10 DIV by performing immunofluorescent staining and RT-PCR analyses. Stage-specific embryonic antigen 1 (SSEA-1), a carbohydrate epitope associated with cell adhesion and migration, is normally expressed by avian PGCs during migration to the gonadal anlage in turkeys (D'Costa and Petitte, 1999), as well as in zfgPGCs (Jung et al., 2019) and also by chPGCs (Macdonald et al., 2010) in vitro. Furthermore, epithelial membrane antigen 1 (EMA-1), a cell surface glycoprotein, is expressed by chPGCs at early embryonic stages and in vitro (Raucci et al., 2015; Urven et al., 1988). Correspondingly, migrating zfPGCs that were freshly isolated from the embryonic bloodstream at Murray stages 13–15, as well as zfPGCs at 10 DIV, were found to be immunopositive for both SSEA-1 (Figures 2A and 2D) and EMA-1 (Figures 2E, 2H, and 2I). In both antibody stainings, the cell surface of zfPGCs was strongly labeled (Figure 2D); in contrast, erythrocytes remained unstained (Figures 2A, 2B, 2E, and 2F).
In both chPGCs and zfPGCs, the germline-specific genes DAZL (Deleted in Azoospermia-like) and DDX4 (DEAD-Box Helicase 4), as well as the pluripotency markers POU5F1 (POU Class 5 Homeobox 1) and NANOG (Nanog Homeobox), are expressed (Jung et al., 2019; Macdonald et al., 2010; Van De Lavoir et al., 2006). Our RT-PCR analyses revealed that cultured zfPGCs harvested after 10 DIV expressed these stem cell markers (Figure 2J). In control samples, zebra finch blastodiscs from freshly laid eggs, which contain PGCs, showed a similar expression pattern. In contrast, cultured fibroblasts from zebra finch embryos did not express these markers (Figure 2J). Furthermore, we identified the PGC marker DAZL to be highly expressed in the transcriptome of both zfPGCs freshly extracted from blood and those cultured for 10 DIV.
Highly Efficient Transduction of Cultured zfPGCs with Lentiviral Vectors
Electroporation (BTX, Gemini System) and lipofection resulted in extensive cell mortality of cultured zfPGCs and low transfection efficiency (data not shown). To determine whether cultured zfPGCs could be genetically modified, we transduced cultured zfPGCs at 7 DIV with a lentiviral vector containing the eGFP gene under control of the human phosphoglycerate kinase (hPGK) promoter (Figure 3A) when they were growing in cell clumps. Two days post-transduction, the majority of the cells exhibited strong eGFP expression (Figures 3B–3D), while they continued to express the zfPGC marker EMA-1 (Figure 3E). In contrast to zfPGCs cultured from embryonic blood, zfgPGCs (Jung et al., 2019) showed a relatively low lentiviral transduction efficiency (Figures S2G and S2H). We next compared the eGFP expression in cultured zfPGCs after transduction with lentiviral vectors that contained different constitutive promoters, including the hUBC promoter, the human elongation factor 1α promoter, the cytomegalovirus (CMV) promoter, and the CMV enhancer fused to the chicken β-actin promoter. For all these lentiviral constructs the levels of eGFP expression after transduction turned out to be similar (Figure S3).
Figure 3.
Cultured zfPGCs Were Efficiently Transduced with a Lentiviral Vector for eGFP and Colonized Host Embryo Gonads
(A) The lentiviral vector rrl-hPGK-eGFP that was used to transduce zfPGCs in vitro and generate founder birds.
(B–F) Microphotographs of cultured zfPGCs that were transduced with rrl-hPGK-eGFP for 48 h. Phase-contrast (B) and eGFP expression (C) of zfPGCs are shown at low magnification. Note that almost all zfPGCs were expressing the reporter gene. In (D–F), fluorescence images of a PGC cluster show the same cells expressing eGFP (D), being immunopositive for the PGC marker EMA-1 (E), and after DAPI staining (F). The inset in (E) shows a merged eGFP- and anti-EMA-1 image of a subset of cells (indicated by the arrow). Note that eGFP-expressing cells were EMA-1 immunopositive.
(G and H) In microphotographs of cryosections produced from an adult founder ovary that showed eGFP expression and blue nuclear counterstain with DAPI are presented at low (G) and high (H) magnification. Note that in the ovary eGFP-positive germ cells of different sizes display various stages of differentiation. Scale bars represent 100 μm (C, G, and H) and 20 μm (D–F).
(I–K) Photographs of agarose gels after electrophoretic separation and ethidium bromide staining of molecular weight markers (left lanes) and PCR products. PCR products that were obtained using the lentiviral vector plasmid (lane 1 in I and J) are shown as positive controls. Genomic DNA (gDNA) was isolated from a founder ovary (I, lane 2) and testis (I, lane 3), and from blood samples of three F1 birds, two males (J, lanes 2 and 3) and one female (J, lane 4). The right arrowhead in (I and J) points to the hPGK-promoter-eGFP amplicon (602 bp). To verify expression of the transgene by RT-PCR (K), RNA was isolated from the brain of a wild-type bird (K, lane 1) as well as brain (K, lanes 2 and 3) and liver (K, lanes 4 and 5) samples of different F1 birds. Although transgene expression could not be detected in skeletal muscle of transgenic birds by RT-PCR, the transgene was detectable in skeletal muscle gDNA of transgenic birds (K, lanes 7 and 8), in contrast to the wild type (K, lane 6). The right arrowhead in (K) points to the eGFP-WPRE amplicon (894 bp).
Because viral titer is a crucial factor for a successful lentiviral vector transduction, we also assayed reporter gene expression in cultured zfPGCs when using increasingly diluted viral particle suspensions. Using the lentiviral vector for rrl-hPGK-eGFP, we found the highest eGFP expression at a final concentration of 2 × 108 transducing units (TU)/mL and greater (Figure S4). For gene editing studies in which Cas9, CRISPR guide RNAs (gRNAs), and a selection marker are used at the same time, it would be necessary to simultaneously transduce zfPGCs with two different lentiviral vectors. To test this we treated the same cells with lentiviral vectors for both UBC-eGFP and CMV-Tomato at a titer of 2 × 108 TU/mL each. Two days after transduction, most zfPGCs expressed both reporter genes at comparable strength (Figure S5). Together, these findings demonstrated that lentiviral vectors provide an efficient and promising way to introduce transgenes into cultured zfPGCs.
Upregulation of LDLR Gene Family Members in Cultured zfPGCs
The viral vectors used to transduce zfPGCs in this study were vesicular stomatitis virus glycoprotein G (VSV-G) pseudotyped lentivirus for which the low-density lipoprotein receptor (LDLR) family constitutes the main cell surface receptors (Finkelshtein et al., 2013; Nikolic et al., 2018). Transcriptome analysis of zfPGCs that were freshly extracted from embryonic blood and cultured for 10 DIV revealed the expression of the LDLR gene family members LR11, LRP1B, and LRP3 to be significantly upregulated after 10 DIV. The expression of other gene family members, such as LRP2, LRP6, LRP8, and LRP1, remained unchanged and was found to be downregulated for VLDLR and LRP4 (Table 1). Absence of the prototypic LDLR gene in the zfPGC transcriptome was in line with the role of LDLR in steroidogenesis by somatic cells of chicken ovarian follicles (Hummel et al., 2003).
Table 1.
Expression Profile of Low-Density Lipoprotein Receptor Family in zfPGCs Cultured for 10 Days
| Gene Symbol | Gene Name | Zebra Finch Ensembl Gene ID | Fold Change | Adjusted p |
|---|---|---|---|---|
| LR11 (SORL1) | low-density lipoprotein receptor relative with 11 ligand-binding repeats | ENSTGUG00000000462 | 2.816 | 0.05 |
| LRP1B | low-density lipoprotein receptor-related protein 1b | ENSTGUG00000011849 | 2.602 | 0.043 |
| LRP3 | low-density lipoprotein receptor-related protein 3 | ENSTGUG00000009422 | 1.8494 | 0.008 |
| LRP2 (Megalin) | low-density lipoprotein receptor-related protein 2 | ENSTGUG00000007663 | 1.172 | 0.175 |
| LRP6 | low-density lipoprotein receptor-related protein 6 | ENSTGUG00000012717 | 0.412 | 0.698 |
| LRP8 (APOER2) | low-density lipoprotein receptor-related protein 8 | ENSTGUG00000009223 | −0.008 | 0.994 |
| LRP1 | low-density lipoprotein receptor-related protein 1 | ENSTGUG00000015747 | −0.560 | 0.620 |
| VLDLR | very low-density lipoprotein receptor | ENSTGUG00000005386 | −1.558 | 0.017 |
| LRP4 (MEGF7) | low-density lipoprotein receptor-related protein 4 | ENSTGUG00000010533 | −2.688 | 0.004 |
Cultured zfPGCs Colonized the Gonadal Anlage of Host Embryos after Re-injection Subgerminally
Cultured zfPGC clumps expressing eGFP under the control of the hPGK promoter were pooled from 5–10 cultures and dissociated to single cells after digestion with papain. Around 500 cells were injected under the blastodisc of freshly laid zebra finch eggs. The eggs were sealed and incubated for 48 h before being transferred into the nests of foster parents for further incubation. From 22 injected eggs, 10 founder birds (45.4%) were hatched and raised, 6 females and 4 males. Using histological sections of founder gonads, we found that cultured and re-injected zfPGCs migrated to the gonadal anlage of the host embryo and differentiated into germinal cells (Figures 3G and 3H). Furthermore, integration of the hPGK-eGFP construct in genomic DNA (gDNA) extracted from founder gonads was verified by PCR in both ovaries and testes (Figure 3I). All founders that were examined contained hPGK-eGFP-positive zfPGCs in their gonads.
Songbird Transgenesis
Founders were crossed with wild-type birds, and gDNA from blood samples of the offspring (F1 birds) was analyzed by PCR for the presence of the hPGK-eGFP construct (Figure 3J). F1 birds that were hPGK-eGFP positive by PCR were sacrificed to produce histological sections and confirm eGFP protein expression by anti-GFP immunofluorescence staining. All F1 birds that were found to be hPGK-eGFP-positive by PCR turned out to be GFP immunopositive as well. In addition, RNA samples were collected from liver, brain, and skeletal muscle of gDNA-PCR-positive F1 birds. In RT-PCR analyses, we found eGFP to be expressed in brain and liver (Figure 3K, lanes 2–5), but not in the skeletal muscle of the transgenic birds. However, genomic integration of the hPGK-eGFP construct was detectable also in skeletal muscle by gDNA PCR (Figure 3K, lanes 7 and 8). In summary, all 10 founder birds produced transgenic offspring showing a germline transmission rate between 4% and 22% (Table 2). In the transgenic birds, we observed eGFP expression in kidney, heart, lung, liver, ovary, and brain (Figures 3 and 4), including forebrain song control nuclei like the high vocal center (HVC) (Figures 5A–5C) and the robust nucleus of the arcopallium (RA) (Figures 5D–5F). The percentage of eGFP-expressing cells varied from 1% to 70% depending on the organ and the individual (Table S2). All transgenic birds generated were phenotypically normal and did not present any pathologies (Figure 5G).
Table 2.
Frequency of Germline Transmission Detected by PCR and anti-GFP Immunostaining
| Founders |
F1 birds |
||||
|---|---|---|---|---|---|
| ID | Sex | Number | hPGK-eGFP Positive by PCR | GFP Immunopositive | Germline Transmission (%) |
| 1 | Male | 18 | 4 | 4 | 22 |
| 2 | Male | 8 | 1 | 1 | 12 |
| 3 | Female | 9 | 1 | 1 | 11 |
| 4 | Female | 11 | 1 | 1 | 9 |
| 5 | Male | 23 | 2 | 2 | 8 |
| 6 | Male | 13 | 1 | 1 | 7 |
| 7 | Female | 17 | 1 | 1 | 5 |
| 8 | Female | 19 | 1 | 1 | 5 |
| 9 | Female | 20 | 1 | 1 | 5 |
| 10 | Female | 23 | 1 | 1 | 4 |
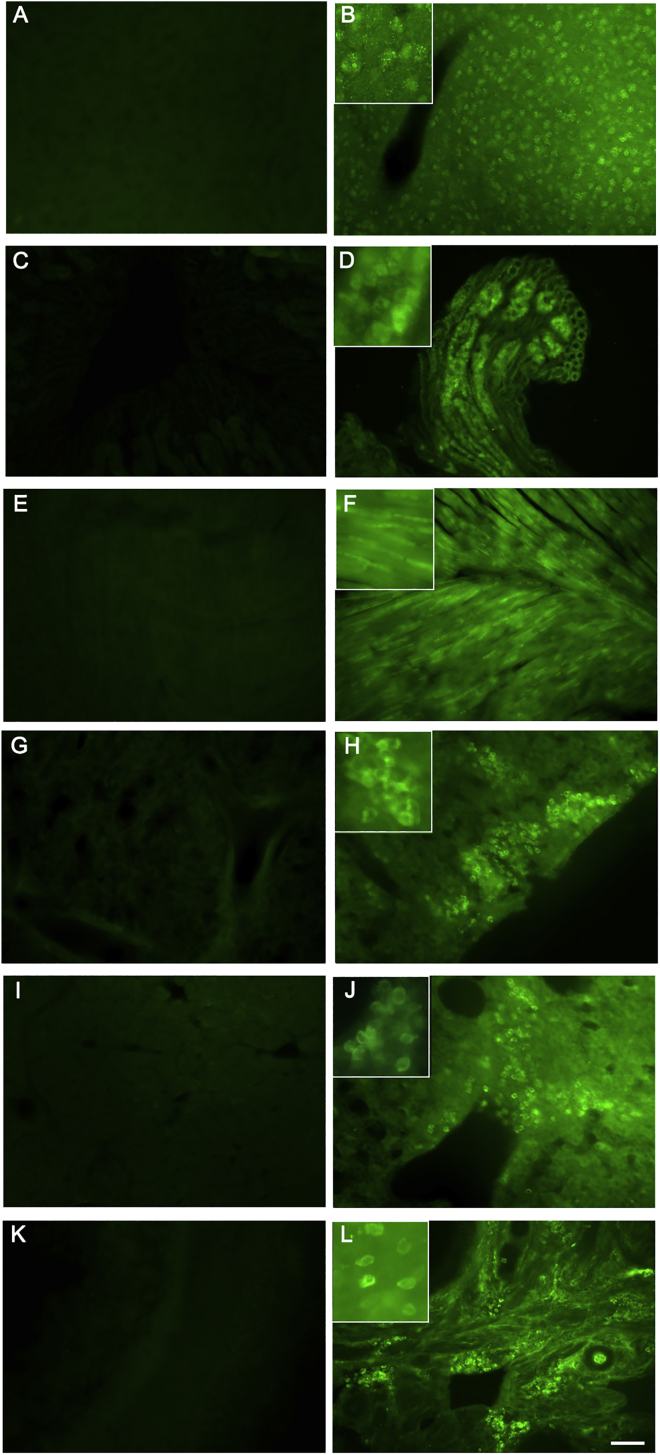
Figure 4.
EGFP Expression in Transgenic Birds
Anti-GFP immunofluorescence staining is shown for different organs of adult control (left) and eGFP-transgenic (right) birds: brain (A and B), kidney (C and D), heart (E and F), lung (G and H), liver (I and J), and ovary (K and L). Insets show eGFP-immunopositive cells at higher magnification. The scale bar represents 100 μm.
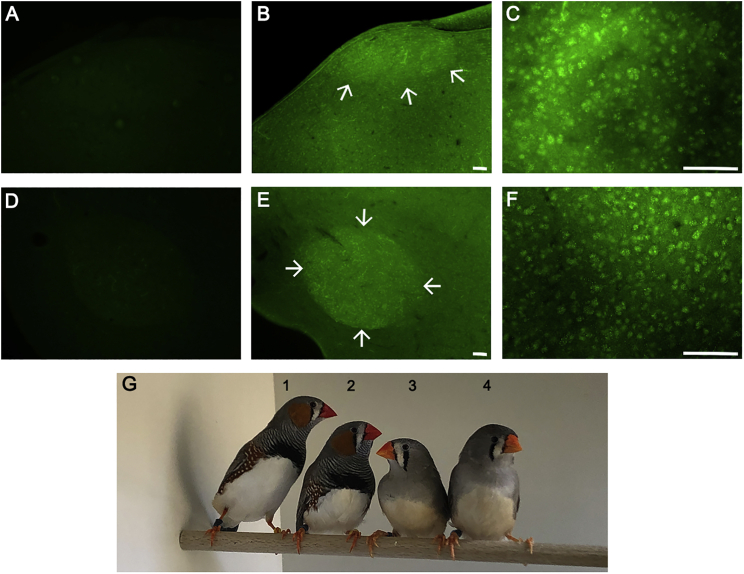
Figure 5.
EGFP Expression in the Song Control Nuclei HVC and RA of Transgenic Birds
Anti GFP-immunofluorescence staining is shown for adult male forebrain sections of control (A and D) and eGFP-transgenic birds (B, C, E, and F) that included the song control nuclei HVC (A–C) and RA (D–F) at low (left and middle) and high (right) magnification. At low magnification the location of the song control nucleus is indicated by arrows.
(G) Photograph of transgenic (1 and 3) and wild-type (2 and 4) adult zebra finches. The scale bars represent 100 μm.
Discussion
Songbirds, especially the zebra finch, are prominent animal models for studying neural circuit development, vocal communication, and vocal learning, among other topics (Bolhuis and Gahr, 2006; Konopka and Roberts, 2016). However, the wider use of the zebra finch in mechanistic research is hampered by the lack of transgenic technology, which is available for most standard animal model organisms (Ménoret et al., 2017). In chicken, long-term cultures of PGCs turned out to be extraordinarily useful for facilitating the production of transgenic chicken lines (Macdonald et al., 2010; Van De Lavoir et al., 2006). The ability to culture chPGCs almost indefinitely made it possible to modify the chicken genome in vitro and inject large numbers of genetically modified chPGCs into host embryos, substantially increasing the germline transmission rate in these birds (Sid and Schusser, 2018; Schusser et al., 2013, 2016; Dimitrov et al., 2016; Taylor et al., 2017; Oishi et al., 2016, 2018). Here, we adapted such an approach for the zebra finch to allow for fast and efficient generation of transgenic songbirds.
We developed a new method for the propagation of zfPGCs from early embryonic blood cultures. When grown in cell clumps after 7 DIV, zfPGCs exhibited high transduction rates using lentiviral vectors such that almost every zfPGC expressed the reporter gene. Previously, the transduction and transmission efficiency of lentiviral vectors after injection at the blastodermal stages was low (Agate et al., 2009). In particular, lentiviral vectors transduced zfPGCs much less efficiently in ovo compared with cultured zfPGCs growing in clumps. Factors like inactivation of lentiviral vectors in ovo and increased cellular uptake in vitro might be responsible for this difference. Thus, our approach constitutes a major improvement for the genomic modification of songbirds. Upregulation of LDLR gene family members like LR11, LRP1B, and LRP3 by zfPGCs may have contributed to the increased uptake of the VSV-G pseudotyped lentiviral vectors by zfPGCs growing in clumps. Interestingly, LR11 also promotes the cell adhesion process of hematopoietic stem cells (Nishii et al., 2013). We observed that the growth of zfPGCs in clumps was associated with an enhanced uptake of lentiviral vectors. Lower transduction rates were found in single zfPGCs obtained from cultured cell clumps after digestion with papain or when prepared from embryonic gonads (Jung et al., 2019). The highly efficient transduction of cultured zfPGCs with lentiviral vectors did not compromise the ability of the germ cells to differentiate in the host gonad and form functional gametes. All founder birds (100%) of both sexes that were generated produced transgenic offspring, which is astonishing considering that the injected PGCs were derived from a mixture of male and female embryos. An additional factor that contributed to the high efficiency we achieved was the relatively high hatching success (45.4%) of the injected host embryos. In comparison, injections of lentiviral vectors into blastoderm resulted in a hatching rate of 13.2% (Agate et al., 2009). We posit that the transfer of genetically modified zfPGCs into a host embryo by a single injection was less harmful to the embryo than the multiple vector injections required by the previous protocol used for the production of transgenic zebra finches (Agate et al., 2009). In conclusion, expansion of embryonic-blood-derived zfPGCs in vitro followed by transduction with lentiviral vectors constitutes an extremely efficient strategy for producing transgenic zebra finches.
We selected lentiviral constructs containing the hPGK promoter to generate transgenic zebra finches because they generated consistently strong eGFP expression in cultured zfPGCs. In our transgenic hPGK-eGFP F1 birds, eGFP was not ubiquitously expressed in all zebra finch tissues, but was detectable after immunostaining in most brain areas, including the song control system, and in the kidney, heart, lung, liver, and ovary. The heterogeneous and selective expression pattern of eGFP might be the result of a cell-type-specific activity or silencing of the hPGK promoter in the zebra finch. For cell-type-directed transgene expression, additional promoter constructs need to be tested to match the transgene expression pattern to the specific requirements that are defined by the various experimental questions for a transgenic songbird model.
The use of cultured zfPGCs that were transduced with lentiviral vectors appears to be an efficient transgenic approach for the overexpression of a transgene and, possibly, downregulation of an endogenous gene following random transgene insertion into the genome. The successful simultaneous transduction of cultured zfPGCs with two different lentiviral vectors suggested that in future CRISPR-Cas experiments, the use of high-titer lentiviral preparations for Cas proteins under the control of a PGK promoter and gRNAs under control of a U6 promoter will be a promising approach. We expect a culturing period of 2 weeks to be sufficiently long for the successful application of targeted gene editing techniques on zfPGCs, such as gene knockout by non-homologous end-joining repair of CRISPR-Cas-induced DNA breaks (Sid and Schusser, 2018). For more complex genome editing techniques that involve a homology-directed repair, culture conditions that permit a more extended growth of zfPGCs in vitro and clonal selection of genetically modified zfPGCs will need to be developed. Nevertheless, for transgenic applications in neuroethological studies, the method of generating transgenic songbird models reported here represents a fast, efficient, and straightforward procedure.
Experimental procedures
Ethics Statement
Animal handling was carried out in accordance with the European Communities Council Directive 2010/63 EU and legislation of the state of Upper Bavaria.
Culture of Embryonic-Blood-Derived zfPGCs
Freshly laid zebra finch eggs were incubated for 60 h at 37°C and relative humidity of 75% until Murray stage 13–15. Then a window was opened above the air chamber of the egg, and 1–3 μL of blood was extracted from the vasculature system using a pulled-glass needle. Blood samples were cultured separately in a 96 well plate (Falcon) with 150 μL of the zfPGC culture medium in each well (Table S1). Cells were maintained in culture for up to 30 days, and 80 μL of the medium was replaced every other day. Gonadal PGCs were obtained and cultured as described by Jung et al. (2019).
To quantify the number of cell divisions in cultures with activin or BMP4, the number of zfPGCs was scored in a minimum of eight fields under a 20× objective of an inverted microscope (Nikon Eclipse Ts2) at 1 and 15 DIV, and the fold change between the two culture periods was calculated.
Histological Staining
For PAS stain of blood smears, samples were extracted from zebra finch and chicken embryos at HH stage 14, dried on glass slides, and fixed with 70% ethanol for 15 min. For staining, a PAS kit (Sigma-Aldrich) was used according to the manufacturer's instructions, and cells were counterstained with hematoxylin solution Gill No. 3 (Sigma-Aldrich). For the immunofluorescence staining of freshly extracted zfPGCs, dried blood smears were fixed with 4% buffered formaldehyde for 10 min at room temperature. Slides were washed three times with washing buffer (0.2% Triton X-100 in phosphate-buffered saline [PBS]) and blocked for 1 h with a blocking solution of 10% pre-immune goat serum in washing buffer. After being washed, the samples were incubated for 12 h at 4°C with a primary antibody against SSEA-1 (1 μg/mL; D. Solter/B.B. Knowles; DSHB Hybridoma Bank) or the PGC surface marker EMA-1 (5 μg/mL; M. Eddy/A. Hahnel; DSHB Hybridoma Bank) diluted in blocking solution. Thereafter, the slides were washed three times with washing buffer and incubated for 3 h at room temperature with the secondary antibody (Alexa Fluor 594 goat-anti-mouse [IgM]; Life Technologies) diluted 1:300 in blocking solution. Following two washing steps with washing buffer, the slides were incubated in 0.1 μg/mL DAPI diluted in water for nuclear staining, washed with PBS, and mounted with 50% glycerol in PBS.
For cultured PGCs, the immunostaining was done in suspension with cells that we harvested after 10 DIV. Cells were centrifuged for 5 min at 2,500g, resuspended in 4% paraformaldehyde, incubated for 10 min at room temperature, and washed twice with the washing buffer. Subsequently, washed cell pellets were resuspended and incubated in blocking solution for 1 h at 20°C. After another washing step, the cells were resuspended in blocking solution containing the primary antibody and incubated for 12 h at 4°C. Then, the cells were washed again and incubated with the secondary antibody for 3 h at 20°C. Finally, the cells were washed, stained with DAPI, washed again, and resuspended in 50% glycerol in PBS. For immunostaining of living PGCs, the fixation step was omitted. Images were obtained by performing epifluorescence microscopy.
RT-PCR Analyses
Total RNA was isolated from embryonic tissues using the RNeasy mini kit (Qiagen) and from cultured zfPGCs using the Power SYBR Green Cells-to-CtT kit (Thermo Fisher), following the manufacturer's instructions. For cDNA synthesis, 2–3 μg of total RNA was denatured in 10 μL distilled water in the presence of 1 μL random hexamers (50 ng/μL) and 1 μL deoxyribonucleotide triphosphates (dNTP mix; 10 mM each) for 5 min at 65°C. After chilling the solution on ice for 1 min, we added 2 μL 10× reverse-transcriptase (RT) buffer, 4 μL 25 mM MgCl2, 2 μL 0.1 M 1,4-dithio-D-threitol, 1 μL RNaseOUT (40 U/μL; Thermo Fisher), and 1 μL RT SuperScript III (200 U/μL; Thermo Fisher). Reverse transcription was carried out in a thermocycler for 10 min at 25°C, followed by 50 min at 50°C and, finally, 5 min at 85°C. Next, the reaction mix was placed on ice for 5 min and, after 1 μL RNase H (2 U/μL; Thermo Fisher) was added, incubated for 20 min at 37°C.
PCR was carried out with the HOT FIREPol DNA polymerase in buffer B1 (both Solis BioDyne). A 20 μL reaction mixture included 12.5 μL H2O, 2 μL 10× buffer B1, 1.6 μL MgCl2 (25 mM), 1 μL cDNA, 2 μL primer pair (10 μM), 0.4 μL dNTP mix, and 0.5 μL HOT FIREPol. The thermocycling conditions were as follows: denaturation at 95°C for 10 min, 35 amplification cycles (95°C for 30 s, 60°C for 40 s, and 72°C for 30 s), and a final extension at 72°C for 5 min. The amplified products were resolved by gel electrophoresis in 2% ROTIGarose (high-resolution agarose; Roth) in 0.5× UltraPure Tris-borate-EDTA buffer (Thermo Fisher). Non-intron-spanning primers for DAZL (DAZL(I)), DDX4 (DDX4(I)), NANOG, SOX3, and POU5F1 were the same as in Mak et al. (2015). We designed the following intron-spanning primer pairs for zebra finch DAZL (DAZL(II): forward, 5′-GAAACCCAGCACTCAAACGC-3′; reverse, 5′-AAGACGCTCCGAATTTCAGC-3′) and DDX4 (DDX4(II): forward, 5′-CTGGAAGCCTACTCCAGTGC-3′; reverse, 5′-TCCCTCATCATTTGGGCCAC-3′). The primer pair designed for ACGT1 was forward, 5′-AACCGGACTGTTTCCAACAC-3′; reverse, 5′-CACCTTCACCGTTCCAGTTT-3′.
Lentiviral Transduction and Injection of Cultured zfPGCs
The lentiviral vector for eGFP under control of the human EF1α promoter was purchased from SignaGen Laboratories (Gaithersburg, MD, USA). All other lentiviral vectors were made in the department of Prof. Dr. Alexander Pfeifer (University of Bonn). For the production of lentiviral vectors, vector plasmids as well as the packaging plasmids pMDLg/pRRE, RSV-rev, and pMD2.G were co-transfected into HEK293T cells seeded on poly-L-lysine-coated dishes. The supernatant was collected and centrifuged in an ultracentrifuge with an SW32 Ti rotor at 61,700g at 17°C for 2 h. Virus suspensions were concentrated by ultracentrifugation over a 20% (w/v) sucrose cushion in an SW55 Ti rotor at 53,500g at 17°C for 2 h. Viral titer was quantified using RT enzyme-linked immunosorbent assay.
To test for germline transmission, we used the vector for hPGK-eGFP. After 7 DIV, zfPGCs growing in clumps were pooled from 5–10 embryos without being separated by sex to reach a density of 1–3 × 103 cells in 50 μL of culture medium, and lentiviral vectors were added to a final concentration of 2 × 108 TU/mL. The culture medium was replaced after 12 h and the cells were cultured for an additional 2 days. To dissociate zfPGC clumps, the cells were washed with PBS 48 h after viral transduction and digested for 30 min at 37°C with a papain (Worthington, LS003119) solution (2 mg/mL KnockOut DMEM without calcium) that had been sterilized using a 0.2 μm syringe filter. Then the zfPGCs were washed with PBS, centrifuged for 5 min at 2,500g, and resuspended in culture medium to give a final concentration of 500 cells/μL for injection into a host embryo.
To inject transduced cultured zfPGCs, freshly laid zebra finch eggs were incubated for 4 h at 38°C and then placed on a silicone surface with the blunt end facing upward. With a light source that illuminated the egg from below, we were able to observe the blastodisc through the eggshell. Using a scalpel, we opened a 0.5–1 mm window in the eggshell above the blastodisc. Care was taken not to disturb the inner and outer egg membranes, as doing so would reduce the survival of the embryo. Using a pulled-glass needle connected to a microinjector, we injected 300–500 transduced cultured zfPGCs into the subgerminal cavity of the host embryo. The window in the eggshell of the host embryo was sealed with chicken egg membrane and closed with zebra finch eggshell that was glued to the host eggshell with zebra finch egg white. After 72 h of incubation at 38°C, injected eggs that showed host embryo survival and ongoing development were taken to nests of foster parents for further egg incubation, hatching, and development until sexual maturity.
Generation and Screening of Transgenic Zebra Finches
Founder birds were raised by foster parents until sexual maturity and paired with wild-type birds. Fertilized eggs produced by the founders were incubated by foster parents that raised the hatchlings until maturity. To test for the presence of the eGFP gene in the offspring (F1 birds), blood samples were collected from each hatchling, and gDNA was extracted using the DNeasy Blood & Tissue Kit (Qiagen) following the manufacturer's instructions to perform PCR analyses. The primers used for the hPGK-eGFP-PCR produced an amplicon (602 bp) that included sequences of both the hPGK promotor and the eGFP gene: the forward primer was 5′-CACTAGTACCCTCGCAGACG-3′, the reverse primer was 5′-TCTTGTAGTTGCCGTCGTCC-3′. For each PCR 50–100 ng gDNA was used in a 20 μL reaction mixture that contained 12.5 μL H2O, 2 μL primer mix (10 μM), 0.4 μL dNTPs (10 μM), as well as 2 μL buffer B1 (10×), 1.6 μL MgCl2 (25 mM), and 0.5 μL FIREPol DNA polymerase (all three from Solis BioDyne). Thermocycling conditions were the following: initial denaturation at 95°C for 5 min; 35 cycles of 95°C for 30 s, 65°C for 30 s, and 72°C for 30 s; followed by 72°C for 5 min. The amplified products were resolved by electrophoresis in a 2% Tris-acetate-EDTA agarose gel. All steps were performed in a lab free of eGFP usage or in a UV-decontaminated PCR box. To confirm eGFP expression by RT-PCR the forward primer was 5′-GGGCACAAGCTGGAGTACAA-3′ and the revers primer was 5′-AAGGGAGATCCGACTCGTCT-3′. Thermocycling conditions were the following: initial denaturation at 95°C for 5 min; 40 cycles of 95°C for 30 s, 68°C for 1 min, and 72°C for 10 s; followed by 72°C for 5 min.
To confirm eGFP expression by immunostaining, F1 birds that were PCR positive for hPGK-eGFP were sacrificed and transcardially perfused with PBS followed by 4% paraformaldehyde in PBS. Tissues were post-fixed in 4% paraformaldehyde, cryoprotected in 10% and 30% sucrose in PBS, and sectioned at a freezing microtome. Free-floating tissue sections were washed three times for 10 min with PBS and then incubated for 2 h in a PBS blocking solution containing 0.5% saponin (Sigma-Aldrich 84510) and 10% pre-immune goat serum (Thermo Fisher). For anti-GFP immunostaining, sections were incubated for 36 h at 4°C with a chicken anti-GFP antibody (Aves GFP-1020) at a concentration of 1:1,000 in PBS blocking solution. After three subsequent washes with PBS, the samples were incubated for 3 h at room temperature with the goat anti-chicken antibody conjugated with Alexa Fluor 488 (Abcam 150169) at a concentration of 1:500 in PBS blocking buffer. Finally, the sections were washed three times with PBS and mounted on a glass slide with VECTASHIELD (Vector Laboratories). Images were obtained by performing epifluorescence microscopy. All birds that were PCR positive for hPGK-eGFP and GFP immunopositive were considered to be transgenic (Table 2). Gonads of founders were used fresh for DNA extraction followed by PCR for hPGK-eGFP and after fixation for anti-GFP immunostaining.
To confirm the integration of eGFP in the genome, genome walking was performed using the Universal GenomeWalker (TAKARA 636406) and following the manufacturer’s instructions.
Separation of Cells by Density Gradient Centrifugation
To separate zfPGCs from red blood cells, two Ficoll 400 (Sigma F2637) solutions (16% and 6.3%) were prepared in cell culture medium. Embryonic blood samples were pooled from five individuals and centrifuged for 5 min at 2,500g. The cell pellet was resuspended in 100 μL of culture medium, mixed gently with 900 μL of the 16% Ficoll solution, placed underneath 200 μL of the 6.3% Ficoll solution, and centrifuged for 30 min at 3,400g. ZfPGCs could be collected from the most superficial layer (200 μL) of the gradient. After 10 μL was removed for cell counting, the rest of the cell suspension was diluted with 300 μL of culture medium and centrifuged for 5 min at 2,500g. After the supernatant was carefully removed, the cell pellet was resuspended in the amount of culture medium necessary for RNA extraction.
RNA Extraction and Next-Generation Sequencing
Four samples with 400 zfPGCs each were collected for each group (1 and 10 DIV), and RNA was extracted using the SMART-Seq v.4 Ultra Low Input RNA kit for sequencing (TAKARA) following the manufacturer’s instructions. Illumina high-output sequencing was done by the NGS group at the Max Planck Institute for Molecular Genetics.
Statistical Analysis
For statistical comparison of zfPGC numbers obtained in cultures with BMP4 and activin we performed a one-way ANOVA with p < 0.05. To analyze the growth of zfPGCs in vitro over time we performed a one-way ANOVA followed by a post hoc test with the p value being adjusted for multiple comparisons.
The Galaxy web platform (https://usegalaxy.eu/) was used for the analysis of the sequencing data (Afgan et al., 2018). Quality control of the FASTQ files was performed with FASTQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Nextera adapters were removed with Trimmomatic (Bolger et al., 2014). The reads were mapped with STAR (Dobin et al., 2013) and counted with featureCounts (Liao et al., 2014). Quality control of the mapped sequences was done with MultiQC (Ewels et al., 2016). Differentially expressed genes between the two groups were identified with DESeq2 (Love et al., 2014).
Data and Code Availability
The raw data fastq.gz files were deposited in the SRA database under accession no. PRJNA637305.
Author contributions
I.G., F.D., M.H., M.M., and M.G. conceived the study. M.G., M.M., and A.P provided instruments, materials, and reagents. I.G. performed the experiments. S.H. produced the lentiviral vectors. C.F.V. contributed to the analysis of the sequencing data. I.G. wrote the manuscript and all coauthors contributed to manuscript revision. I.G. is a member of the International Max Planck Research School for Organismal Biology and the work was funded by the Max Planck Society.
Acknowledgments
We are grateful to Anja Lohrentz, Christina Reusch, Judith Kammerlander, Antje Bakker, Sunil Nandi, and Lorna Taylor for their excellent technical support and Bernd Timmermann for performing the next-generation sequencing of our samples. We want to thank David Witkowski, Frances Weigl, and Frank Lehmann for their outstanding care of the birds in this study. Thanks go to Luisana Carballo for comments on the manuscript draft. I.G. also thanks the International Max Planck Research School for training and support. The study was funded by the Max Planck Society.
Published: March 18, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.02.015.
Supplemental information
References
- Abe K., Matsui S., Watanabe D. Transgenic songbirds with suppressed or enhanced activity of CREB transcription factor. Proc. Natl. Acad. Sci. U S A. 2015;112:7599–7604. doi: 10.1073/pnas.1413484112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afgan E., Baker D., Batut B., Van den Beek M., Bouvier D., Čech M., Chilton J., Clements D., Coraor N., Grüning B. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46:W537–W544. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agate R.J., Scott B.B., Haripal B., Lois C., Nottebohm F. Transgenic songbirds offer an opportunity to develop a genetic model for vocal learning. Proc. Natl. Acad. Sci. U S A. 2009;106:17963–17967. doi: 10.1073/pnas.0909139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balthazart J., Charlier T.D., Barker J.M., Yamamura T., Ball G.F. Sex steroid-induced neuroplasticity and behavioral activation in birds. Eur. J. Neurosci. 2010;32:2116–2132. doi: 10.1111/j.1460-9568.2010.07518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolhuis J.J., Gahr M. Song system Neural mechanisms of birdsong memory. Nat. Rev. Neurosci. 2006;7:347–357. doi: 10.1038/nrn1904. [DOI] [PubMed] [Google Scholar]
- D’Costa S., Petitte J.N. Characterization of stage-specific embryonic antigen-1 (SSEA-1) expression during early development of the Turkey embryo. Int. J. Dev. Biol. 1999;43:349–356. [PubMed] [Google Scholar]
- Dimitrov L., Pedersen D., Ching K.H., Yi H., Collarini E.J., Izquierdo S., Van de Lavoir M.C., Leighton P.A. Germline gene editing in chickens by efficient CRISPR-mediated homologous recombination in primordial germ cells. PLoS One. 2016;11:e0154303. doi: 10.1371/journal.pone.0154303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels P., Magnusson M., Lundin S., Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyal-Giladi H., Kochav S., Menashi M.K. On the origin of primordial germ cells in the chick embryo. Differentiation. 1976;6:13–16. doi: 10.1111/j.1432-0436.1976.tb01462.x. [DOI] [PubMed] [Google Scholar]
- Finkelshtein D., Werman A., Novick D., Barak S., Rubinstein M. LDL receptor and its family members serve as the cellular receptors for vesicular stomatitis virus. Proc. Natl. Acad. Sci. U S A. 2013;110:7306-7311. doi: 10.1073/pnas.1214441110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto T., Ukeshima A., Kiyofuji R. The origin, migration and morphology of the primordial germ cells in the chick embryo. Anat. Rec. 1976;185:139–153. doi: 10.1002/ar.1091850203. [DOI] [PubMed] [Google Scholar]
- Gahr M. Sexual differentiation of the vocal control system of birds. Adv. Genet. 2007;59:67–105. doi: 10.1016/S0065-2660(07)59003-6. [DOI] [PubMed] [Google Scholar]
- Ginsburg M., Eyal-Giladi H. n nmj and spatial aspects of the gradual migration of primordial germ cells from the epiblast into the germinal crescent in the avian embryo. Development. 1986;95:53–71. [PubMed] [Google Scholar]
- Goldman S.A., Nottebohm F. Neuronal production, migration, and differentiation in a vocal control nucleus of the adult female canary brain. Proc. Natl. Acad. Sci. U S A. 1983;80:2390–2394. doi: 10.1073/pnas.80.8.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V., Hamilton H.L. A series of normal stages in the development of the chick embryo. Dev. Dyn. 1951;88:49–92. [PubMed] [Google Scholar]
- Hummel S., Lynn E.G., Osanger A., Hirayama S., Nimpf J., Schneider W.J. Molecular characterization of the first avian LDL receptor: role in sterol metabolism of ovarian follicular cells. J. Lipid Res. 2003;44:1633–1642. doi: 10.1194/jlr.M300014-JLR200. [DOI] [PubMed] [Google Scholar]
- Jung K.M., Kim Y.M., Keyte A.L., Biegler M.T., Rengaraj D., Lee H.J., Mello C.V., Velho T.A.F., Fedrigo O., Haase B. Identification and characterization of primordial germ cells in a vocal learning Neoaves species, the zebra finch. FASEB J. 2019;33:13825–13836. doi: 10.1096/fj.201900760RR. [DOI] [PubMed] [Google Scholar]
- Kochav S., Ginsburg M., Eyal-Giladi H. From cleavage to primitive streak formation: a complementary normal table and a new look at the first stages of the development of the chick: II. Microscopic anatomy and cell population dynamics. Dev. Biol. 1980;79:296–308. doi: 10.1016/0012-1606(80)90117-7. [DOI] [PubMed] [Google Scholar]
- Konopka G., Roberts T.F. Insights into the neural and genetic basis of vocal communication. Cell. 2016;164:1269–1276. doi: 10.1016/j.cell.2016.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Liu W.C., Kohn J., Szwed S.K., Pariser E., Sepe S., Haripal B., Oshimori N., Marsala M., Miyanohara A., Lee R. Human mutant huntingtin disrupts vocal learning in transgenic songbirds. Nat. Neurosci. 2015;18:1617–1622. doi: 10.1038/nn.4133. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald J., Glover J.D., Taylor L., Sang H.M., McGrew M.J. Characterisation and germline transmission of cultured avian primordial germ cells. PLoS One. 2010;5:e15518. doi: 10.1371/journal.pone.0015518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak S., Alev C., Nagai H., Wrabel A., Matsuoka Y., Honda A., Sheng G., Ladher R.K. Characterization of the finch embryo supports evolutionary conservation of the naive stage of development in amniotes. eLife. 2015;4:e07178. doi: 10.7554/eLife.07178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy M.M., Arnold A.P. Reframing sexual differentiation of the brain. Nat. Neurosci. 2011;14:677–683. doi: 10.1038/nn.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C.V. The zebra finch, Taeniopygia guttata: an avian model for investigating the neurobiological basis of vocal learning. Cold Spring Harb Protoc. 2014;2014:1237–1242. doi: 10.1101/pdb.emo084574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménoret S., Tesson L., Remy S., Usal C., Ouisse L.H., Brusselle L., Chenouard V., Anegon I. Advances in transgenic animal models and techniques. Transgenic Res. 2017;26:703–708. doi: 10.1007/s11248-017-0038-x. [DOI] [PubMed] [Google Scholar]
- Mooney The neurobiology of innate and learned vocalizations in rodents and songbirds. Curr. Opin. Neurobiol. 2020;2020:24–31. doi: 10.1016/j.conb.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J.R., Varian-Ramos C.W., Welch Z.S., Saha M.S. Embryological staging of the zebra finch, Taeniopygia guttata. J. Morphol. 2013;274:1090–1110. doi: 10.1002/jmor.20165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M., Kuwana T., Miyayama Y., Fujimoto T. Extragonadal distribution of primordial germ cells in the early chick embryo. Anat. Rec. 1988;222:90–94. doi: 10.1002/ar.1092220113. [DOI] [PubMed] [Google Scholar]
- Nikolic J., Belot L., Raux H., Legrand P., Gaudin Y., Albertini A. Structural basis for the recognition of LDL-receptor family members by VSV glycoprotein. Nat. Commun. 2018;9:1029. doi: 10.1038/s41467-018-03432-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishii K., Nakaseko C., Jiang M., Shimizu N., Takeuchi M., Schneider W., Bujo H. The soluble form of LR11 protein is a regulator of hypoxia-induced, urokinase-type plasminogen activator receptor (uPAR)-mediated adhesion of immature hematological cells. J. Biol. Chem. 2013;288:11877–11886. doi: 10.1074/jbc.M112.442491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi I., Yoshii K., Miyahara D., Kagami H., Tagami T. Targeted mutagenesis in chicken using CRISPR/Cas9 system. Sci. Rep. 2016;6:23980. doi: 10.1038/srep23980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi I., Yoshii K., Miyahara D., Tagami T. Efficient production of human interferon beta in the white of eggs from ovalbumin gene-targeted hens. Sci. Rep. 2018;8:10203. doi: 10.1038/s41598-018-28438-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton J.A., Nottebohm F.N. Neurons generated in the adult brain are recruited into functional circuits. Science. 1984;225:1046–1048. doi: 10.1126/science.6474166. [DOI] [PubMed] [Google Scholar]
- Prather J.F., Okanoya K., Bolhuis J.J. Brains for birds and babies: neural parallels between birdsong and speech acquisition. Neurosci. Biobehav Rev. 2017;81:225–237. doi: 10.1016/j.neubiorev.2016.12.035. [DOI] [PubMed] [Google Scholar]
- Raucci F., Fuet A., Pain B. In vitro generation and characterization of chicken long-term germ cells from different embryonic origins. Theriogenology. 2015;84:732–742. doi: 10.1016/j.theriogenology.2015.04.032. [DOI] [PubMed] [Google Scholar]
- Schusser B., Collarini E.J., Pedersen D., Yi H., Ching K., Izquierdo S., Thoma T., Lettmann S., Kaspers B., Etches R.J. Expression of heavy chain-only antibodies can support B-cell development in light chain knockout chickens. Eur. J. Immunol. 2016;46:2137–2148. doi: 10.1002/eji.201546171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schusser B., Collarini E.J., Yi H., Izquierdo S.M., Fesler J., Pedersen D., Klasing K.C., Kaspers B., Harriman W.D., van de Lavoir M.C. Immunoglobulin knockout chickens via efficient homologous recombination in primordial germ cells. Proc. Natl. Acad. Sci. U S A. 2013;110:20170–20175. doi: 10.1073/pnas.1317106110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sid H., Schusser B. Applications of gene editing in chickens: a new era is on the horizon. Front Genet. 2018;9:456. doi: 10.3389/fgene.2018.00456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor L., Carlson D.F., Nandi S., Sherman A., Fahrenkrug S.C., McGrew M.J. Efficient TALEN-mediated gene targeting of chicken primordial germ cells. Development. 2017;144:928–934. doi: 10.1242/dev.145367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urven L.E., Erickson C.A., Abbott U.K., McCarrey J.R. Analysis of germ line development in the chick embryo using an anti-mouse EC cell antibody. Development. 1988;103:299–304. doi: 10.1242/dev.103.2.299. [DOI] [PubMed] [Google Scholar]
- Van De Lavoir M.C., Diamond J.H., Leighton P.A., Mather-Love C., Heyer B.S., Bradshaw R., Kerchner A., Hooi L.T., Gessaro T.M., Swanberg S.E. Germline transmission of genetically modified primordial germ cells. Nature. 2006;441:766–769. doi: 10.1038/nature04831. [DOI] [PubMed] [Google Scholar]
- Whyte J., Glover J.D., Woodcock M., Brzeszczynska J., Taylor L., Sherman A., Kaiser P., McGrew M.J. FGF, insulin, and SMAD signaling cooperate for avian primordial germ cell self-renewal. Stem Cell Rep. 2015;5:1171–1182. doi: 10.1016/j.stemcr.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data fastq.gz files were deposited in the SRA database under accession no. PRJNA637305.