Summary
Identifying lineage-specific markers is pivotal for understanding developmental processes and developing cell therapies. Here, we investigated the functioning of a cardiomyogenic cell-surface marker, latrophilin-2 (LPHN2), an adhesion G-protein-coupled receptor, in cardiac differentiation. LPHN2 was selectively expressed in cardiac progenitor cells (CPCs) and cardiomyocytes (CMCs) during mouse and human pluripotent stem cell (PSC) differentiation; cell sorting with an anti-LPHN2 antibody promoted the isolation of populations highly enriched in CPCs and CMCs. Lphn2 knockdown or knockout PSCs did not express cardiac genes. We used the Phospho Explorer Antibody Array, which encompasses nearly all known signaling pathways, to assess molecular mechanisms underlying LPHN2-induced cardiac differentiation. LPHN2-dependent phosphorylation was the strongest for cyclin-dependent kinase 5 (CDK5) at Tyr15. We identified CDK5, Src, and P38MAPK as key downstream molecules of LPHN2 signaling. These findings provide a valuable strategy for isolating CPCs and CMCs from PSCs and insights into the still-unknown cardiac differentiation mechanisms.
Keywords: cardiac differentiation, G-protein-coupled receptor, marker, stem cells, signaling
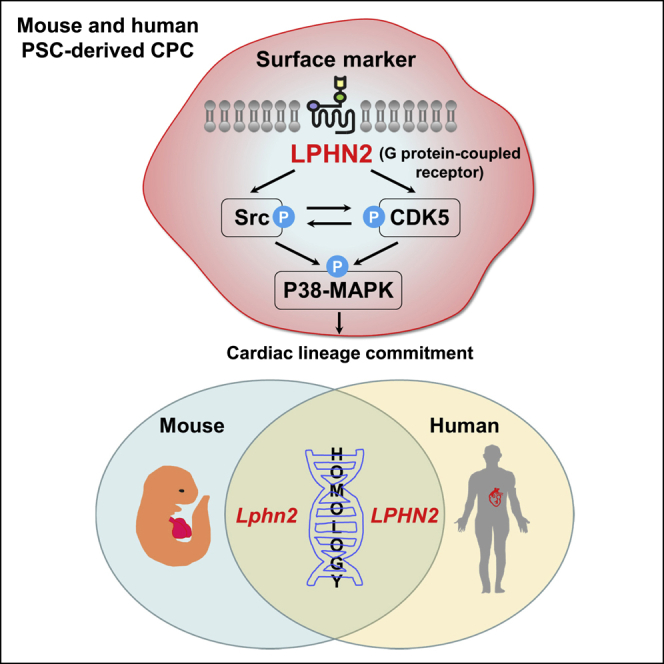
Graphical abstract
Highlights
-
•
LPHN2 is a cell-surface marker used to separate mouse PSC-derived CPCs and CMCs
-
•
CDK5 is a crucial LPHN2 signaling molecule and with Src induces P38MAPK phosphorylation
-
•
LPHN2+ cells differentiate to human multipotent CPCs
In this article, Kim and colleagues demonstrate latrophilin-2 (LPHN2) to be a functionally significant marker of mouse and human cardiac differentiation. Further analysis indicates that CDK5 is downstream of LPHN2 and interacts with Src kinase to induce P38MAPK phosphorylation, subsequently activating cardiac-specific gene transcription. LPHN2 can be pivotal to achieving cardiac lineage differentiation, facilitating the clinical application of stem cells in cardiovascular disease.
Introduction
To take advantage of the beneficial properties of embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs), the development of protocols for differentiating pluripotent stem cells (PSCs) into precursor and mature functional somatic cells is required. The demand for ESC- or iPSC-derived cardiomyocytes (CMCs) for use in cardiovascular disease research has increased in recent years (Fox et al., 2014; Oikonomopoulos et al., 2018). In addition, specific surface markers that enable monitoring of cell subtypes have been developed over the past several years to establish conditions that promote PSC differentiation into cardiac lineage cells (Oikonomopoulos et al., 2018).
A previous study showed that FLK-1 (also known as KDR or VEGFR-2) and PDGFR-α are co-expressed in the cardiac mesoderm (Kattman et al., 2011). These markers are expressed in both cardiac and vascular progenitor cells, but they appear transiently during development and require two-color flow cytometry for practical enrichment. Moreover, signal-regulatory protein α(SIRPα or CD172a) (Dubois et al., 2011) and podoplanin (PDPN) (Birket et al., 2015) have been used as surface markers for the selective enrichment and expansion of cardiac cell populations derived from human PSCs. Although several sets of cell-surface markers that distinguish cardiac progenitor cells (CPCs) from PSCs have been reported, the functional significance of these molecules remains elusive.
To develop a widely applicable strategy for the enrichment of PSC-derived cardiac cells, we conducted microarray screening to identify cell-surface markers specific to CPCs and focused on functional molecules, such as G-protein-coupled receptors (GPCRs) (Lee et al., 2019). GPCRs play an essential role in pathophysiology and represent attractive drug targets for the treatment of several diseases (Hilger et al., 2018). Recently, we discovered latrophilin-2 (LPHN2, Adgrl2) as a novel cell-surface marker for cardiomyogenic lineage cells during in vitro differentiation of mouse PSCs (Lee et al., 2019). LPHN2 is an adhesion GPCR characterized by large extracellular domains, and it has been reported to be ubiquitously expressed in multiple organ tissues of adult mice (Boucard et al., 2014). Another study showed that LPHN2 maintains synapse numbers through a postsynaptic mechanism in the mouse brain (Anderson et al., 2017). Although LPHN2 has been shown to play a role in the central nervous system, LPHN2 expression during cardiac differentiation and development and its clinical implication in heart disease are unclear.
Here, we demonstrate LPHN2 to be a functional marker for CPCs and CMCs during in vitro PSC differentiation. In addition, we investigate the underlying molecular mechanism of action of LPHN2 in cardiac differentiation. Our findings provide a vital strategy for achieving cardiomyogenic lineage cell differentiation.
Results
LPHN2 Is Expressed in CPCs and CMCs during Mouse PSC Differentiation
To optimize the conditions for cardiac lineage cell differentiation, we compared the spontaneous and directed differentiation of mouse PSCs. Moreover, we established a protocol for directed PSC differentiation into CMCs after exposing cells to various combinations of cytokines for different periods, based on the biology of embryonic development (Greber et al., 2010; Hudson et al., 2012; Kattman et al., 2011; Laflamme et al., 2007; Yang et al., 2008; Yu et al., 2011). For the directed differentiation of mouse iPSCs into the cardiac lineage cells, embryoid bodies (EBs) were generated in an AggreWell plate after culturing for a day in EB medium in the presence of bone morphogenetic protein 4 (BMP-4), with the subsequent addition of activin A and basic fibroblast growth factor (FGF2) for 3 additional days (Figure S1A). On day 4, EBs were transferred to the cardiac differentiation medium containing epithelial growth factor, FGF2, cardiotrophin-1, and vascular endothelial growth factor (Lee et al., 2019). On day 14, we observed a 4.5-fold increase in the number of beating foci in cells undergoing optimized differentiation compared with that in cells undergoing spontaneous differentiation (Figure S1B). Immunofluorescence (IF) analysis of beating cells revealed a strong expression of α-sarcomeric actinin (α-SA) (Figure S1C).
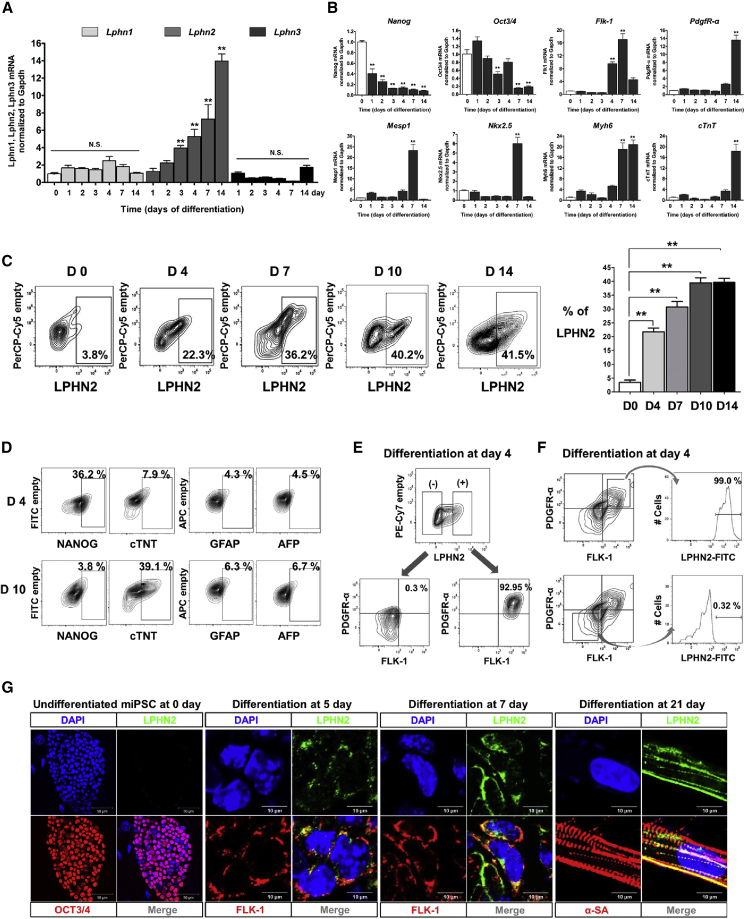
We investigated the expression patterns of all three LPHN-family genes during differentiation. Lphn2 expression gradually increased during cardiac differentiation and plateaued 14 days after induction of differentiation, whereas the expression levels of latrophilin-1 (Lphn1, Adgrl1) and latrophilin-3 (Lphn3, Adgrl3) remained unaltered (Figure 1A). Following induction of differentiation, the pluripotency genes Nanog and Oct3/4 (Pou5f1) (Cho et al., 2010) were found to be downregulated, whereas the mesoderm and CPC markers (Flk-1, Mesp1, and Nkx2.5) were expressed transiently 7 days postinduction (Figure 1B). Similarly, the CMC markers Myh6 and cTnT (Ieda et al., 2010) were highly expressed 14 days postinduction.
Figure 1.
Expression Pattern of LPHN2 during Cardiac Differentiation
(A) Gene expression analysis of the latrophilin family in mouse induced pluripotent stem cells (iPSCs) during serial differentiation stages. Cultures of iPSCs under differentiation were harvested at the indicated days, and gene expression in cells was analyzed by qPCR. Values are shown relative to day 0. ∗∗p < 0.01, N.S., not significant, one-way ANOVA and post hoc Bonferroni test, n = 3 independent replicates.
(B) mRNA expression levels of Nanog, Oct3/4, Flk-1, PdgfR-α, Mesp1, Nkx2.5, Myh6, and cTnT in mouse iPSC-derived cells at successive stages during cardiac differentiation. ∗∗p < 0.01, one-way ANOVA and post hoc Bonferroni test, n = 3 independent replicates.
(C) Flow cytometric analysis of LPHN2 expression during cardiac differentiation. (Left) LPHN2 expression in mouse iPSC-derived cardiac cells at various time points. (Right) Quantification of the plots shown on the left. ∗∗p < 0.01, one-way ANOVA and post hoc Bonferroni test, n = 4 independent replicates.
(D) Flow cytometric analysis of cells on consecutive days after cardiac differentiation. NANOG was used as a pluripotency marker, cTNT as a CMC marker, glial fibrillary acidic protein (GFAP) as an ectoderm marker, and α-fetoprotein (AFP) as an endoderm marker.
(E and F) Representative fluorescence-activated cell sorting (FACS) plots showing the correlation between LPHN2 and co-expression of FLK-1 and PDGFR-α. FACS analysis for LPHN2 expression in differentiated cells at day 4 is shown. (E) The LPHN2+ and LPHN2− cell populations were analyzed using anti-FLK-1 and anti-PDGFR-α antibodies. (F) The FLK-1+/PDGFR-α+ and FLK-1−/PDGFR-α− cell populations were divided, and LPHN2 expression was analyzed.
(G) Immunostaining for LPHN2 and OCT3/4, FLK-1, or α-SA in monolayer cultures in the early (day 7) and late (day 21) stages of cardiac differentiation. Blue, nuclear counterstaining with 4,6-diamidino-2-phenylindole (DAPI). PSC, pluripotent stem cell.
Flow cytometric analysis revealed LPHN2 expression on the surface of 41.5% of cells after 14 days of optimized cardiac differentiation (Figure 1C). Moreover, the expression of the pluripotency-related marker (NANOG) decreased during differentiation, and that of the cardiac lineage marker (cTNT) gradually increased (Figure 1D). The expression of the other lineage markers (glial fibrillary acidic protein and α-fetoprotein) did not change (Figure 1D).
To determine whether LPHN2 expression correlates with the expression of the CPC markers, we divided the cells into LPHN2-negative (LPHN2−) and LPHN2-positive (LPHN2+) subpopulations 4 days post differentiation. The LPHN2+ population overlapped with the FLK-1+PDGFR-α+ (F+P+) population by 92.95%, validating LPHN2 as a CPC marker (Figure 1E). In addition, 99% of the F+P+ subpopulation expressed LPHN2, whereas only 0.32% of the FLK-1−PDGFR-α− (F−P−) subpopulation expressed LPHN2 (Figure 1F). Furthermore, immunostaining analysis revealed an absence of LPHN2 in undifferentiated cells; its expression gradually increased following induction of differentiation (Figure 1G). Moreover, LPHN2 co-localized with FLK-1 on day 7 of differentiation and α-SA on day 21 (Figure 1G).
To examine the expression of LPHN2 during differentiation of mouse PSCs into non-CMC cell types, we induced endothelial cell (EC) differentiation, as described previously (Joo et al., 2012). When FLK-1+ mesodermal precursor cells were purified on day 4, Cd31 expression was found to increase gradually in the EC differentiation medium (Figure S1D). IF staining showed that the CD31+ EC colonies on day 21 did not express LPHN2 (Figure S1E), suggesting that LPHN2 was detected only in PSC-derived CPCs and CMCs from the mesoderm lineage.
To explore the broad-spectrum utility of LPHN2 as a cell-surface marker, we validated our findings by examining other mouse ESC (mESC) lines. mESC-derived CPCs and CMCs exhibited Lphn2 gene and LPHN2 protein expression levels similar to those noted for iPSC-derived CPCs and CMCs (Figures S2A–S2D).
Enrichment of Cardiac Lineage Cells by LPHN2
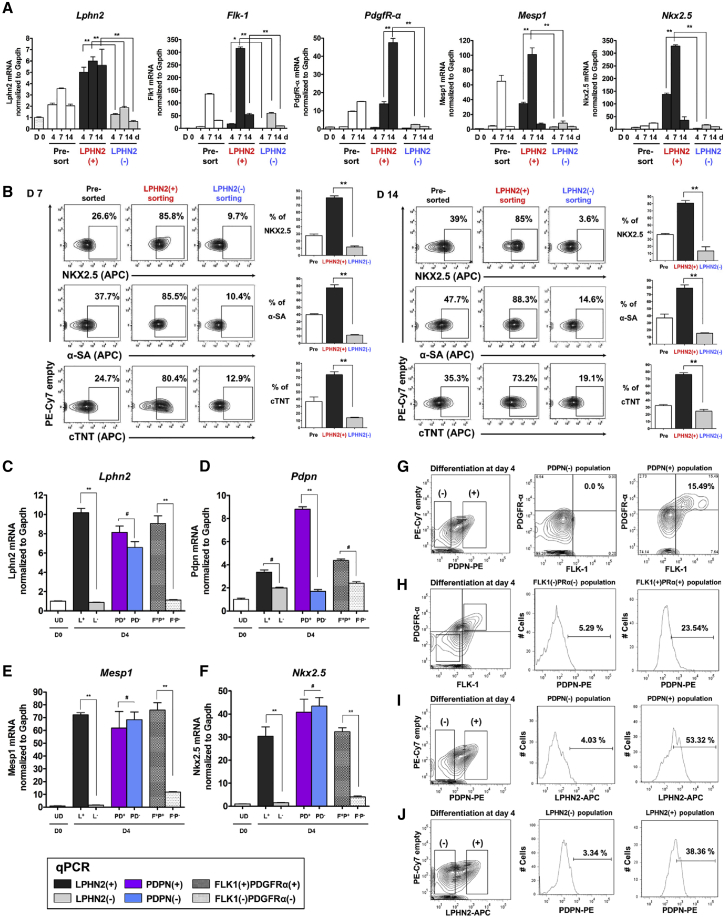
To determine whether LPHN2 can be used as a marker to select populations enriched in the cardiac lineage cells, LPHN2+ and LPHN2− fractions were isolated from mouse iPSC-derived cell populations after 4, 7, and 14 days of differentiation by cell sorting. The purities of the LPHN2+ and LPHN2− sorted populations are shown in Figure S3. Gene expression analyses revealed higher expression of Lphn2, Flk-1, PdgfR-α, Mesp1, and Nkx2.5 in LPHN2+ cells than in LPHN2− cells (Figure 2A). After cell sorting on days 7 and 14, the LPHN2+ fractions at both stages were dominantly enriched for NKX2.5, α-SA, and cTNT expression, representing CPCs and CMCs, compared with the LPHN2− fractions (Figure 2B). In addition, fluorescence-activated cell sorting (FACS)-based separation of various cell lines reproduced the significant enrichment of mESC-derived CMCs (Figure S4).
Figure 2.
Enrichment of iPSC-Derived Cardiac Progenitor Cells and Cardiomyocytes by Cell Sorting Based on LPHN2 Expression
(A) qPCR analysis of cardiac lineage cell markers in pre-sorting and sorted (LPHN2+ and LPHN2−) cells at various stages of cardiac differentiation of iPSCs. Values are shown relative to day 0. ∗p < 0.05, ∗∗p < 0.01, one-way ANOVA and post hoc Bonferroni test; n = 3 independent replicates.
(B) Pre-sorting and sorted (LPHN2+ and LPHN2−) fractions at each time point were analyzed for NKX2.5, α-SA, and cTNT expression by intracellular flow cytometry. Quantification of the plots is shown on the right. ∗∗p < 0.01, one-way ANOVA and post hoc Bonferroni test; n = 3 independent replicates.
(C and D) qPCR analysis of (C) Lphn2 and (D) Pdpn expression in the sorted cell populations on day 4 of differentiation. ∗∗p < 0.01, #not significant, one-way ANOVA and post hoc Bonferroni test; n = 3 independent replicates. UD, undifferentiated.
(E and F) Differences in the mRNA expression of (E) Mesp1 and (F) Nkx2.5 in the sorted cell populations on day 4 of differentiation. Values are shown relative to day 0. ∗∗p < 0.01, #not significant, one-way ANOVA and post hoc Bonferroni test; n = 3 independent replicates. UD, undifferentiated.
(G and H) Representative flow cytometric plots showing correlation in the expression of the cardiac progenitor cell markers PDPN, FLK-1, and PDGFR-α on day 4 of differentiation.
(I and J) Representative flow cytometric plots showing correlation between LPHN2 and PDPN expression on day 4 of differentiation.
To investigate the feasibility of using LPHN2 as a cardiac-specific cell-surface marker, we assessed the advantages of using LPHN2+ CPCs over cells expressing other known markers (Birket et al., 2015; Dubois et al., 2011; Kattman et al., 2011). Interestingly, quantitative real-time polymerase chain reaction (qPCR) analysis revealed that F+P+ cells expressed higher levels of Lphn2 than F−P− cells, similar to LPHN2+ cells (Figure 2C). Conversely, the difference in Lphn2 expression between PDPN+ and PDPN− populations was not significant. Moreover, Pdpn expression was not significantly higher in LPHN2+ and F+P+ cells than in LPHN2− and F−P− cells, respectively (Figure 2D). The expression of Mesp1 and Nkx2.5, which encode CPC markers, was higher in LPHN2+ and F+P+ cells than in LPHN2− and F−P− cells, respectively (Figures 2E and 2F). However, the expression of CPC markers in the PDPN+ cells was not higher than that in the PDPN− cells. Moreover, flow cytometric analysis demonstrated higher expression of F+P+ in PDPN+ cells than in PDPN− cells; however, the expression was lower than that in the LPHN2+ cells (Figure 2G). Conversely, F+P+ cells expressed a marginally higher level of PDPN than F−P− cells, suggesting that PDPN is not a functional receptor for mouse CPC differentiation (Figure 2H). The correlation between PDPN and LPHN2 was examined, and 53.32% of the PDPN+ population was found to express LPHN2 (Figure 2I). In addition, 38.36% of the LPHN2+ population expressed PDPN, indicating a low correlation between the LPHN2 and the PDPN markers (Figure 2J).
Taken together, these cell sorting analyses demonstrated that LPHN2 expression distinguished cardiac lineage cells during differentiation of mouse iPSCs and ESCs, and cell sorting with the anti-LPHN2 antibody allowed isolation of populations highly enriched in CPCs and CMCs.
Functional Significance of LPHN2 in Cardiac Differentiation
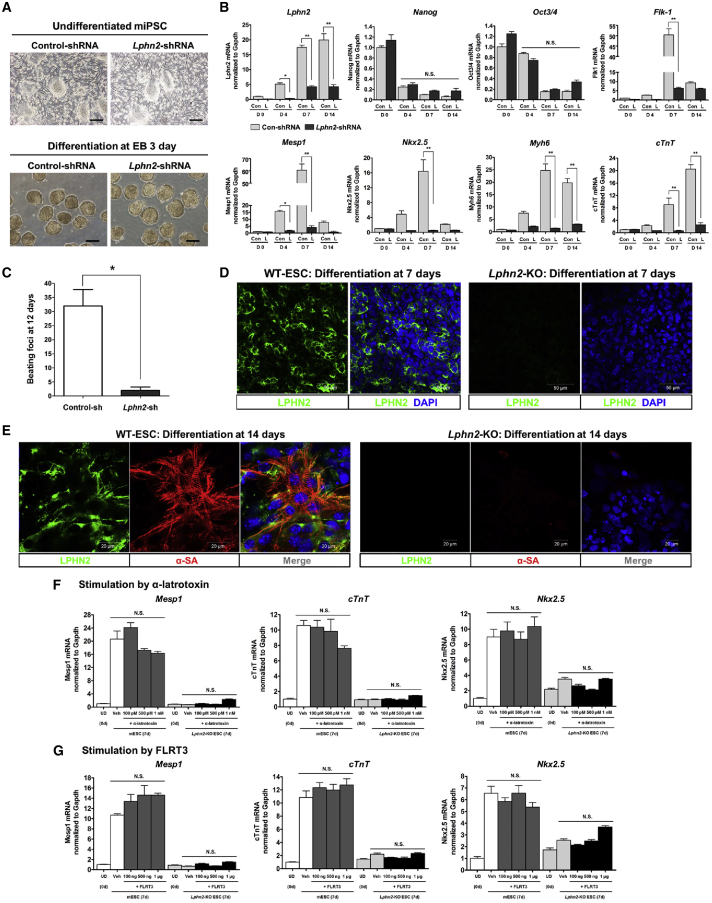
To examine the functional significance of LPHN2 in vitro, we used a short-hairpin RNA (shRNA) against Lphn2 mRNA to knock down (KD) Lphn2 expression in mouse iPSCs, achieving 70% reduction in Lphn2 expression (Figures 3A and 3B). No differences were observed between control-iPSCs and Lphn2-KD cells in the expression of pluripotency-related genes (Nanog and Oct3/4) or the gross morphology of EBs formed during differentiation (Figures 3A and 3B). However, Lphn2-KD cells did not show complete cardiac differentiation in terms of cardiac gene expression and did not produce beating cells (Figures 3B and 3C, Videos S1 and S2).
Figure 3.
Functional Significance of LPHN2 during In Vitro Cardiac Differentiation
(A) Gross morphology of control induced pluripotent stem cells (iPSCs) and Lphn2-knockdown (KD) iPSCs transduced by control-shRNA and Lphn2-shRNA, respectively. No differences were observed in the colony formation or EB formation. Scale bar, 200 μm. EB, embryoid body.
(B) Effects of Lphn2 KD on cardiac differentiation. Transcriptional profiling of Lphn2, Nanog, Oct3/4, Flk-1, Mesp1, Nkx2.5, Myh6, and cTnT in control iPSC- and Lphn2-KD iPSC-derived cells by qPCR. ∗p < 0.05, ∗∗p < 0.01, N.S., not significant, one-way ANOVA and post hoc Bonferroni test; n = 3 independent replicates.
(C) Quantification of spontaneous beating foci in control and Lphn2-KD cells on day 12 of cardiac differentiation. ∗p < 0.05, Mann-Whitney U test; n = 4 independent replicates.
(D) Immunostaining for LPHN2 (green) expression in wild-type embryonic stem cell (WT-ESC) and Lphn2-knockout (KO) ESC-derived cells on day 7 of cardiac differentiation. Blue, nuclear counterstaining with DAPI. Scale bar, 50 μm.
(E) WT-ESCs express LPHN2 (green) and α-SA (red) with striations at day 14 after differentiation, indicating cardiomyocyte differentiation, but Lphn2-KO ESCs did not differentiate into cardiomyocytes. Blue, nuclear counterstaining with DAPI. Scale bar, 20 μm.
(F and G) qPCR analysis of gene expression of cardiac-specific Mesp1, Nkx2.5, and cTnT after stimulation with presumptive ligands. Treatment with α-latrotoxin (F) and FLRT3 (G) did not increase the expression of cardiac lineage genes in mESCs or Lphn2-KO ESCs after 7 days under cardiac differentiation conditions. Values are shown relative to the expression in WT-mESCs on day 0. N.S., not significant, one-way ANOVA and post hoc Bonferroni test; n = 3 independent replicates. UD, undifferentiated; Veh, vehicle.
On day 10 of cardiac differentiation, control-shRNA-transduced iPSCs exhibited robust beating.
On day 10 of cardiac differentiation, Lphn2-shRNA-transduced iPSCs did not beat spontaneously.
To confirm the importance of LPHN2 in cardiac differentiation, we further established mouse Lphn2-knockout (KO) ESCs (Lee et al., 2019). IF analysis showed no LPHN2 expression in Lphn2-KO ESC-derived cells (Figure 3D). From day 10 to 14, wild-type ESC-derived cells began to contract (Video S3) and expressed α-SA with striations (Figure 3E). Lphn2-KO cells, however, did not generate contracting cardiac cells (Video S4).
On day 10 of cardiac differentiation, there were several spontaneous beating foci in wild-type ESCs.
On day 10 of cardiac differentiation, there were no spontaneous beating foci in Lphn2-KO ESCs
As LPHN2 is a GPCR, we investigated its potential ligands. Two potential ligands have been reported for the LPHN family, particularly LPHN1 and LPHN3: α-latrotoxin (α-LTX) and fibronectin leucine-rich repeat transmembrane 3 (FLRT3), respectively (Langenhan et al., 2013; Lelianova et al., 1997; O'Sullivan et al., 2012). Cardiac differentiation was not significantly altered when cells were stimulated with α-LTX or FLRT3 (Figures 3F and 3G). Nonetheless, further research is required to determine the native ligands involved in LPHN2-mediated cardiac differentiation and develop artificial agonists.
Mechanisms Underlying LPHN2-mediated Cardiac Differentiation
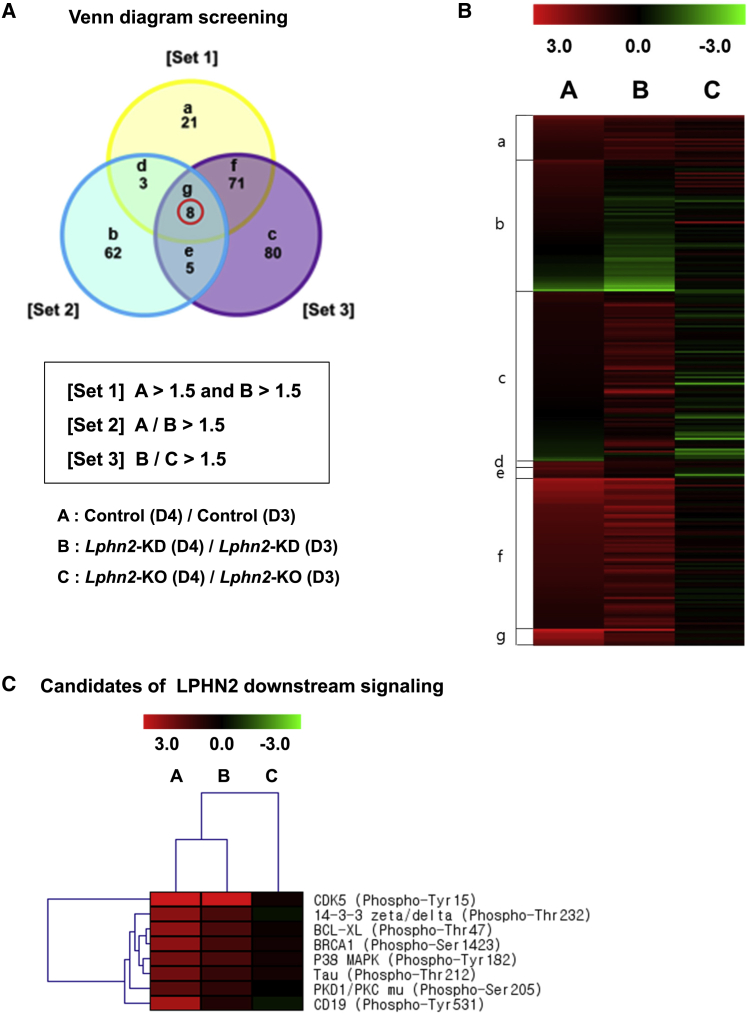
To investigate the molecular mechanisms underlying the induction of cardiac differentiation by LPHN2, we used the Phospho Explorer Antibody Array, which encompasses nearly all known signaling pathways. From the array data, we first calculated the degree of increased phosphorylation on differentiation day 4 relative to the phosphorylation on day 3 (phosphorylation ratio D4/D3) in three different groups: A, control-shRNA-transfected cells differentiating from mESCs; B, Lphn2-shRNA-transfected cells differentiating from mESCs; and C, Lphn2-KO cells differentiating from KO mESCs (Figure S5). Based on the phosphorylation ratios obtained, we delineated three different sets to construct a Venn diagram as follows. Set 1 included proteins with a phosphorylation ratio >1.5 for both groups A and B; set 2 included proteins with a >1.5-fold higher phosphorylation ratio for group A than for group B; and set 3 included proteins with a >1.5-fold higher phosphorylation ratio for group B than for group C (Figure 4A). The purpose of defining sets 1, 2, and 3 was to classify proteins according to a progressive increase in phosphorylation (from group C to A). The heatmap constructed from the data shown in the Venn diagram shows the differential phosphorylation ratios (Figure 4B). The proteins with specific phosphorylation sites in the intersecting region of the Venn diagram are shown in Figure 4C. LPHN2-dependent phosphorylation was the strongest for cyclin-dependent kinase 5 (CDK5) at Tyr15 (Figure 4C).
Figure 4.
Screening of LPHN2-mediated Cardiac Differentiation Mechanisms Based on the Phospho Explorer Antibody Array
(A) A Venn diagram showing the intersection of sets 1, 2, and 3, with the most highly phosphorylated proteins in group A during differentiation of control-shRNA pluripotent stem cells. Lphn2-KO, Lphn2-knockout; Lphn2-KD, Lphn2-knockdown.
(B) Heatmap constructed from the data shown in (A) shows the differential phosphorylation ratios in groups A, B, and C.
(C) Phosphorylation sites of the most highly phosphorylated proteins in group A were considered as candidates for LPHN2 downstream signaling.
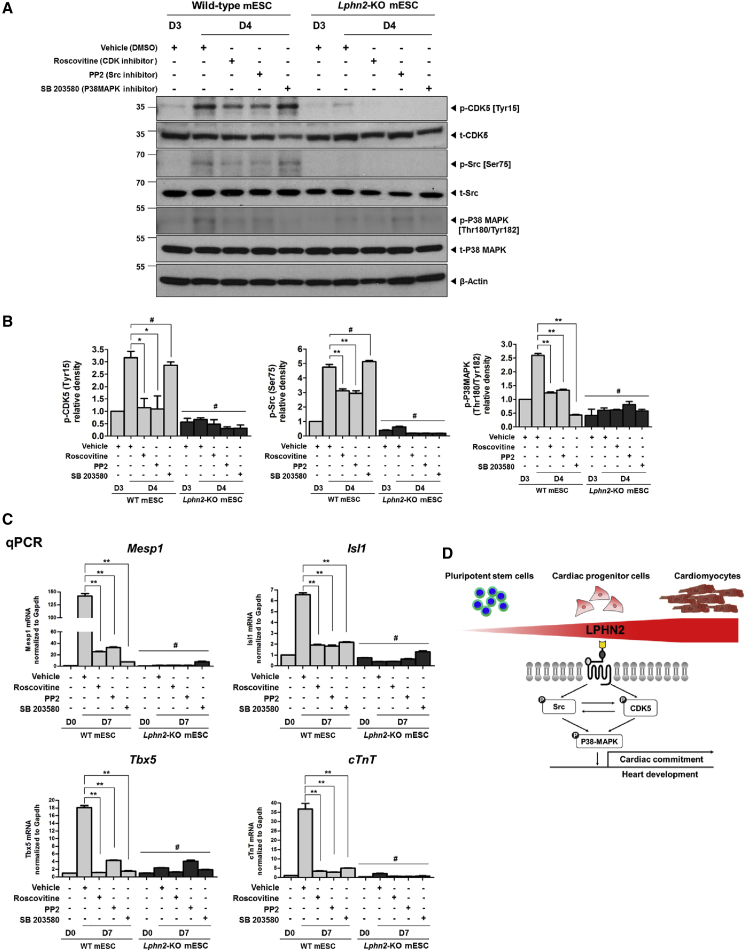
To confirm this finding, we performed western blot analysis with antibodies against phosphorylated CDK5 (Tyr15), as well as Src (Ser75) and P38MAPK (Tyr182), which are presumably downstream of CDK5. In wild-type mESCs, cardiac differentiation following exposure to cytokines significantly increased the phosphorylation of CDK5, Src, and P38MAPK on day 4 compared with that on day 3, whereas this phosphorylation was not observed in Lphn2-KO mESCs (Figures 5A and 5B). Treatment with roscovitine (a CDK5 inhibitor) or PP2 (a Src inhibitor) resulted in decreased phosphorylation of CDK5, Src, and P38MAPK. In contrast, upon application of SB 203580 (a P38MAPK inhibitor), neither CDK5 nor Src phosphorylation was inhibited; however, P38MAPK phosphorylation was decreased (Figures 5A and 5B). In Lphn2-KO mESCs, none of these inhibitors affected the level of phosphorylation, suggesting that LPHN2 signaling involved CDK5, Src, and P38MAPK activation.
Figure 5.
CDK5 Activated P38MAPK via Src and Induced Differentiation of Pluripotent Stem Cells into Cardiomyocytes
(A) Phosphorylation levels of CDK5, Src, and P38MAPK in wild-type mouse ESCs (mESCs) and Lphn2-knockout (KO) mESCs treated with the respective inhibitors during cardiac differentiation, as detected by western blotting.
(B) Quantification of the phosphorylation ratios of CDK5, Src, and P38MAPK in wild-type mESCs (WT mESCs) and Lphn2-KO mESCs. Values are shown relative to the phosphorylation ratio for the vehicle group of WT mESCs on day 3 of differentiation. ∗p < 0.05, ∗∗p < 0.01, #not significant, one-way ANOVA and post hoc Bonferroni test; n = 3 independent replicates.
(C) Gene expression analysis of Mesp1, Isl1, Tbx5, and cTnT in WT mESCs and Lphn2-KO mESCs treated with inhibitors of CDK5, Src, and P38MAPK on day 7 of cardiac differentiation. Values are shown relative to the expression in WT mESCs on day 0. ∗∗p < 0.01, #not significant, one-way ANOVA and post hoc Bonferroni test; n = 3 independent replicates.
(D) Schematic illustration of the molecular mechanism of LPHN2-mediated cardiac differentiation via the CDK5-Src-P38MAPK signaling cascade.
qPCR analysis further confirmed that each inhibitor (roscovitine, PP2, and SB 203580) downregulated cardiac-specific gene expression (Mesp1, Isl1, Tbx5, and cTnT) in wild-type mESCs, but not in Lphn2-KO mESCs (Figure 5C). Therefore, our results suggested that CDK5 and Src interacted in parallel downstream of LPHN2 to induce cardiac differentiation, and P38MAPK, which is located downstream of CDK5 and Src, was also involved (Figure 5D). We concluded that LPHN2 was critical in the differentiation of PSCs into CPCs and CMCs, and CDK5, Src, and P38MAPK were key downstream molecules of LPHN2 signaling.
LPHN2 Expression in Human PSC Differentiation
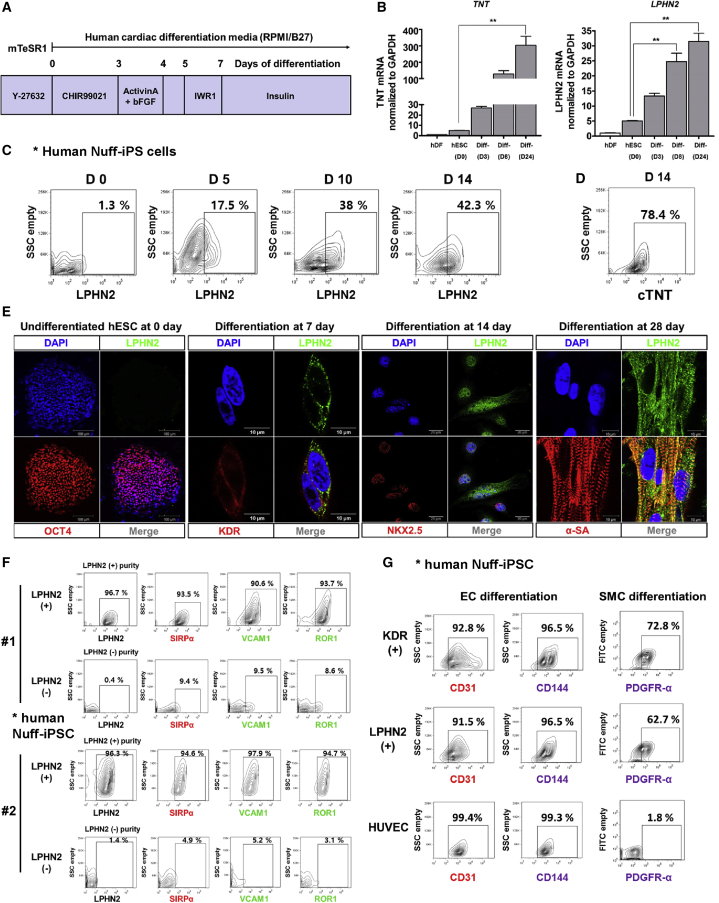
Next, we analyzed the expression of LPHN2 (ADGRL2) in human PSCs. To differentiate human PSCs into CMCs efficiently, we optimized the previously developed protocols (Figure 6A). Similar to our findings in murine cells, TNT and LPHN2 were not expressed in human dermal fibroblasts or undifferentiated human ESCs, including HUVECs (Figures 6B and S6A). As expected, TNT and LPHN2 expression gradually increased throughout the process of differentiation of human ESCs into CPCs and CMCs (Figure 6B). We examined the time course of LPHN2 expression during CMC differentiation using flow cytometry. LPHN2 expression gradually increased to 42.3%, and the cTNT expression level reached approximately 78.4% on day 14 (Figures 6C and 6D). Immunostaining analysis did not reveal the expression of LPHN2 in undifferentiated ESCs or HUVECs (Figures 6E and S6B). LPHN2, however, co-localized with KDR at day 7 of differentiation, and LPHN2+ cells expressed NKX2.5 at day 14 (Figure 6E). Sequentially, IF staining confirmed that LPHN2 was expressed in beating CMCs expressing α-SA characterized by a striated morphology at day 28 post differentiation (Figure 6E).
Figure 6.
LPHN2 Expression and Enriched CPC Potential in Human Pluripotent Stem Cells
(A) A schematic timeline demonstrating the major steps of the culturing procedure used for the directed differentiation of human pluripotent stem cells into cardiomyocytes.
(B) qPCR analysis of TNT and LPHN2 expression in human embryonic stem cell (ESC)-derived cardiomyocytes. Expression values are shown relative to that in hDFs. ∗∗p < 0.01, one-way ANOVA and post hoc Bonferroni test; n = 3 independent replicates. hDF, human dermal fibroblast cell; hESC, human embryonic stem cell; Diff, differentiated.
(C) FACS plots for LPHN2 expression in undifferentiated human Nuff-iPS cells and during a time course of cardiac differentiation.
(D) FACS analysis of the cTNT expression in human Nuff-iPS cells during cardiac differentiation at day 14.
(E) Immunostaining for LPHN2 (green), OCT4 (red), KDR (red), NKX2.5 (red), and α-SA (red) expression in human ESCs and ESC-derived cardiomyocytes. Blue, nuclear counterstaining with DAPI.
(F) Flow cytometric analysis showing expression of CPC markers in sorted LPHN2+ and LPHN2− populations at day 5 of differentiation.
(G) FACS measurements of CD31, CD144 after EC differentiation, and PDGFR-α after SMC differentiation of sorted populations.
When we sorted cells depending on the LPHN2 expression after 5 days of differentiation of human iPSCs toward CPCs, we observed significant enrichment of cardiac lineage cells in the LPHN2-positive cell population and could exclude cardiac lineage cells from the LPHN2-negative cell population (Figure 6F). Consistent with our findings in murine cells, human LPHN2+ CPCs expressed other well-known CPC markers, such as SIRPα (Dubois et al., 2011), VCAM1 (Uosaki et al., 2011), and ROR1 (Halloin et al., 2019), in amounts higher than that expressed by LPHN2− cells. Under differentiation conditions, LPHN2+ cells yielded SIRPα+, VCAM1+, and ROR1+ CPCs with 90% efficiency, but LPHN2− cells failed to show CPC marker expression (10% efficiency) (Figure 6F).
To evaluate the multipotent potential, we purified KDR+ or LPHN2+ cells at day 5 by FACS analysis and re-cultured the purified cell populations for 14 days under EC or smooth muscle cell (SMC) differentiation conditions. Under the EC culture conditions, the KDR+ and LPHN2+ cell populations almost completely differentiated into CD31+ or CD144+ ECs with >90% efficiency (Figure 6G). In addition, the KDR+ and LPHN2+ cell populations predominantly differentiated into PDGFR-α+ SMCs (Figure 6G). Immunostaining also showed that most LPHN2+ cells at day 14 differentiated into VE-Cadherin+ or CD31+ ECs under differentiation conditions, similar to KDR+ cells (Figures S6C and S6D).
Taken together, these data indicated that LPHN2 was highly expressed on the surface of human CPCs and CMCs. In conclusion, LPHN2 expression during human cardiac differentiation was identical to that in murine cardiac differentiation.
Discussion
Although clinical trials have been performed for applying several types of stem cells in the treatment of acute myocardial infarction, CMCs derived from stem cells are not ready for clinical use. Extensive studies on pre-clinical and clinical cell therapies for heart diseases have employed several types of cells for cardiac repair. However, owing to the low efficiency of cardiac differentiation by various types of stem cells, effective methods to generate homogeneous cardiac cells in amounts sufficient for clinical applications are still lacking. Therefore, practical methods for generating homogeneous CMCs are a prerequisite for their eventual clinical application. Although CPCs have been identified using multiple markers (Chen and Wu, 2016), it is very challenging to isolate PSC-derived CPCs and CMCs in vitro because most cardiac-specific markers are intracellular molecules (Elliott et al., 2011) or transcription factors (Bu et al., 2009). Thus, cell-surface markers are required to purify CPCs and CMCs from heterogeneous cell populations during stem cell differentiation.
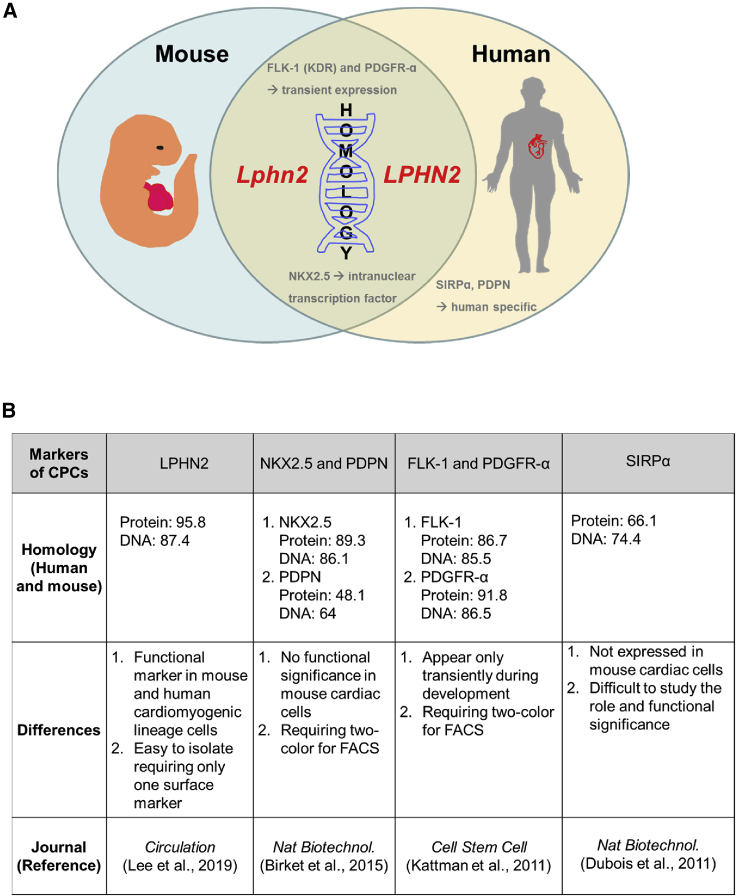
In this study, we demonstrated that a cardiac lineage-specific cell-surface marker, LPHN2, which is highly specific to CPCs and CMCs, possesses functional significance in mice and humans. Intriguingly, the mouse Lphn2 gene is highly homologous to its human ortholog, with 95.8% identity. Recent studies have utilized SIRPα (Dubois et al., 2011), VCAM1 (Uosaki et al., 2011), ROR1 (Halloin et al., 2019), and PDPN (Birket et al., 2015) as cell-surface markers to purify human cardiac cell populations or enrich CMCs. However, SIRPα is not expressed in mouse cardiac cells, making it difficult to study its role and functional significance in KO or genetically modified models. In addition, cell sorting with anti-PDPN antibody was less useful in isolating mouse CPCs because of the differences in the PDPN protein homology between mice and humans. Herein, we reported several notable differences between LPHN2 and other CPC markers (Figures 7A and 7B).
Figure 7.
A Schematic Illustration and Summary Indicating the Applicability of LPHN2 as a Marker of Cardiac Differentiation
(A) Schematic illustration showing the differences in cardiac progenitor cell markers based on homology in mice and humans.
(B) Notable differences between LPHN2 and other reported cardiac progenitor cell markers for cardiac differentiation.
The LPHN family includes three adhesion GPCRs (LPHN1–LPHN3) that mediate synaptic exocytosis caused by α-LTX in the venom of black widow spiders, which are an invertebrate species (Lelianova et al., 1997). However, their endogenous ligands and functions remain unclear (Ichtchenko et al., 1999). Similarly, there are three forms of LPHN present in vertebrates, each of which is expressed in different tissues. LPHN1 was first thought to be brain specific, but RNA blotting techniques revealed ubiquitous expression of the protein in all tissues at low levels (Boucard et al., 2014). In contrast, LPHN3 is expressed only in the brain. However, the potential mechanism underlying LPHN-mediated exocytosis remains unknown. As LPHN proteins are GPCRs, it might be expected that α-LTX triggers an intracellular G-protein-linked second-messenger cascade that ultimately leads to cardiac differentiation. Nevertheless, when we stimulated cells with α-LTX and another known LPHN3 ligand, FLRT3 (O'Sullivan et al., 2012), cardiac differentiation was not induced. These observations should therefore prompt further studies on GPCR-mediated signaling as well as innate and synthetic ligands for LPHN2-promoted cardiac differentiation.
We used the Phospho Explorer Antibody Array to investigate the molecular mechanism of LPHN2 in cardiac differentiation. The antibody array analysis showed that CDK5 phosphorylation at Tyr15 increased the most during cardiac differentiation from mESCs. Previous reports have shown that CDK5 is involved in myogenesis (Lazaro et al., 1997), whereas Src and P38MAPK are involved in ESC differentiation (Li et al., 2011) and heart function (Engel et al., 2006). In terms of a molecular hierarchy, Src is phosphorylated at S75 by CDK5 (Pan et al., 2011), and it has been shown to regulate P38MAPK activation. Thus, we first postulated that LPHN2 activated the downstream signaling cascade from CDK5 to Src and eventually to P38MAPK. LPHN2 activates P38MAPK via CDK5 and Src, which possibly affects the epithelial-mesenchymal transition events during differentiation to allow cardiac mesoderm formation (Graichen et al., 2008). However, our inhibitory experiments suggested that CDK5 and Src might act independently, rather than synergistically, during cardiac differentiation. In addition, it is interesting that CDK5, and not P38MAPK, appeared at the top of the molecular hierarchy, as Src and P38MAPK are better known for their functions in cardiac differentiation. CDK5 is known to be involved in myogenesis, but not in cardiac differentiation as such. Therefore, it is surprising that CDK5 phosphorylation was found to be critical for LPHN2 signaling during cardiac differentiation from PSCs.
In summary, we demonstrated that LPHN2 is a unique cell-surface marker of cardiac muscle progenitor cells and a functionally significant marker of cardiac differentiation. Analysis of the LPHN2 signaling pathway indicated that CDK5 is downstream of LPHN2 and interacts with Src kinase to induce P38MAPK phosphorylation, subsequently activating cardiac-specific gene transcription. The specific expression pattern of LPHN2 in PSC-derived cardiac lineage cells in mice and humans suggests that this receptor plays a pivotal and functional role across all strata of the cardiomyogenic lineage. Our findings, therefore, provide a strategy for achieving cardiac lineage cell differentiation that facilitates clinical application of stem cells in cardiovascular disease treatment.
Experimental procedures
Additional details on experimental procedures can be found in the Supplemental Experimental Procedures.
Ethics Statement
All experiments with human products were conducted with informed consent and were approved by the Institutional Review Board of Seoul National University Hospital (IRB no. H-0908-036-290).
Cell Lines and Maintenance
Cell culture was performed as previously described with slight modifications (Cho et al., 2010). C57BL/6-background mESCs (C57-mESCs; American Type Culture Collection [ATCC], cat. no. SCRC-1002) and iPSCs generated by FVB-background skin fibroblasts were cultured on a mitomycin C (Sigma-Aldrich)-treated STO (ATCC, cat. no. CRL-1503) or MEF (ATCC, cat. no. SCRC-1040) feeder layer in 0.1% gelatin (Sigma-Aldrich)-coated tissue culture dishes at 37°C in a 5% CO2 atmosphere. All cell lines used in the study were regularly tested for mycoplasma, and no contamination was observed. The mESC culture medium was changed daily and composed of Dulbecco's modified Eagle’s medium (DMEM; Gibco) with 2 mM L-glutamine, 10% fetal bovine serum (FBS; Gibco), 0.1 mM β-mercaptoethanol (Sigma-Aldrich, filter sterilized), 1% nonessential amino acids (Gibco), 50 IU/mL penicillin (Gibco), 50 mg/mL streptomycin (Gibco), and 2000 U/mL (20 ng/mL) recombinant leukemia inhibitory factor (LIF). The human ESC line ESI-049 was purchased from BioTime (cat. no. ES-702). Human ESCs were cultured with DMEM/F12 Glutamax (cat. no. 10565-018, Gibco) on an STO feeder layer. To generate human iPSCs from Nuff (newborn foreskin fibroblast) cells (cat. no. AMS.GSC-3006G, AMS Biotechnology), we introduced the reprogramming factors OCT4, SOX2, KLF4, and cMYC using lentiviruses (Takahashi et al., 2007). Human iPSCs were maintained on STO feeder cells with mTESR-1 medium (cat. no. 85851, STEMCELL Technologies).
EB Formation and Cardiac Differentiation
To assess the in vitro spontaneous differentiation potential, mESCs and iPSCs (Cho et al., 2010) were passaged in 0.1% gelatin-coated tissue culture dishes without feeder layers. Next, 1 × 106 cells were cultured by suspension in 100-mm Petri dishes containing EB medium (ESC medium without LIF). After 7 days, aggregated cells (EBs) were plated onto 0.1% gelatin-coated tissue culture dishes and cultured for another 7 or 14 days. Spontaneously contracting EBs were filmed with Olympus IX71 and Olympus DP71 digital cameras (15 fps/s at 680 × 512; Olympus). For cardiac differentiation of mouse PSCs, EBs were generated using AggreWell plates (STEMCELL Technologies) for 1 day and ultra-low attachment plates (Corning) for 3 days in the EB medium with activin A (10 ng/mL), BMP-4 (10 ng/mL), and bFGF (10 ng/mL). On day 4, EBs were attached to 0.1% gelatin-coated plates in the cardiac differentiation medium (35% Iscove’s modified Dulbecco’s medium, 65% DMEM/F12, 1% FBS, 2% B27, 2 mM L-glutamine, 0.1 mM 2-mercaptoethanol, and antibiotics in the presence of 20 ng/mL EGF, 20 ng/mL bFGF, 40 ng/mL cardiotrophin-1, and 5 ng/mL VEGF). For cardiac differentiation of human ESCs and iPSCs, we optimized the previously described protocols (Lian et al., 2013). Briefly, human PSC colonies were detached by dispase (cat. no. 17105-041, Gibco) and dissociated into single cells and seeded in Matrigel (cat. no. 354277, Corning)-coated 35-mm dishes containing mTeSR1 medium (cat. no. 85851, STEMCELL Technologies) supplemented with 5 μM Y-27632 (cat. no. 72302, STEMCELL Technologies). When human PSCs reached 100% confluency, cardiac differentiation was induced in a monolayer supplemented with a mixture of cytokines that was changed sequentially. The cytokine treatments used were as follows: CHIR99021 (cat. no. 252917-06-9, Cayman) for 3 days, activin A (cat. no. 338-AC, R&D Systems) and bFGF (cat. no. 13256029, Invitrogen) for a day, and IWR1 (cat. no. I0161, Sigma-Aldrich) for 2 days. The medium was replaced once every 2 days with the human cardiac differentiation medium. The human cardiac differentiation medium was supplemented with B27 supplement in RPMI 1640 medium (cat. no. 11875-085, Gibco).
Statistics
All experiments were performed independently at least three times. For all cell types, multiple experiments were performed independently to verify the reproducibility of results. The number of samples (n) used for each experiment is indicated in the figures and their legends. The results are presented as the mean ± standard error of the mean (SEM). SPSS version 18.0 (SPSS) was used for all statistical analyses. Statistical analyses between two groups were conducted using the unpaired Student’s t test or the Mann-Whitney U test, as appropriate. Comparison of more than two groups was performed using a one-way ANOVA, and post hoc comparisons were performed with the Bonferroni test. Statistical significance was defined at p < 0.05 and indicated as ∗p < 0.05 and ∗∗p < 0.01.
Author contributions
C.-S.L. designed and performed the majority of the experiments, conducted data analysis, prepared figures, and wrote the initial manuscript draft. H.-J.C. designed the experiments and contributed to manuscript writing. J.-W.L. assisted in conduction of the experiments. H.J.S. and J.C. assisted in performing the human PSC experiments. H.-S.K. designed the research, coordinated the research team, and finalized the manuscript.
Declaration of interests
The authors declare no competing interests.
Acknowledgments
This study was supported by grants from the Korea Health Technology Research and Development Project “Strategic Center of Cell and Bio Therapy” (grant HI17C2085) and “Korea Research-Driven Hospital” (grant HI14C1277) through the Korea Health Industry Development Institute, funded by the Ministry of Health and Welfare, Korea.
Published: April 1, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.03.003.
Supplemental information
References
- Anderson G.R., Maxeiner S., Sando R., Tsetsenis T., Malenka R.C., Sudhof T.C. Postsynaptic adhesion GPCR latrophilin-2 mediates target recognition in entorhinal-hippocampal synapse assembly. J. Cell Biol. 2017;216:3831–3846. doi: 10.1083/jcb.201703042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birket M.J., Ribeiro M.C., Verkerk A.O., Ward D., Leitoguinho A.R., den Hartogh S.C., Orlova V.V., Devalla H.D., Schwach V., Bellin M. Expansion and patterning of cardiovascular progenitors derived from human pluripotent stem cells. Nat. Biotechnol. 2015;33:970–979. doi: 10.1038/nbt.3271. [DOI] [PubMed] [Google Scholar]
- Boucard A.A., Maxeiner S., Sudhof T.C. Latrophilins function as heterophilic cell-adhesion molecules by binding to teneurins: regulation by alternative splicing. J. Biol. Chem. 2014;289:387–402. doi: 10.1074/jbc.M113.504779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu L., Jiang X., Martin-Puig S., Caron L., Zhu S., Shao Y., Roberts D.J., Huang P.L., Domian I.J., Chien K.R. Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature. 2009;460:113–117. doi: 10.1038/nature08191. [DOI] [PubMed] [Google Scholar]
- Chen I.Y., Wu J.C. Finding expandable induced cardiovascular progenitor cells. Circ. Res. 2016;119:16–20. doi: 10.1161/CIRCRESAHA.116.308679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H.J., Lee C.S., Kwon Y.W., Paek J.S., Lee S.H., Hur J., Lee E.J., Roh T.Y., Chu I.S., Leem S.H. Induction of pluripotent stem cells from adult somatic cells by protein-based reprogramming without genetic manipulation. Blood. 2010;116:386–395. doi: 10.1182/blood-2010-02-269589. [DOI] [PubMed] [Google Scholar]
- Dubois N.C., Craft A.M., Sharma P., Elliott D.A., Stanley E.G., Elefanty A.G., Gramolini A., Keller G. SIRPA is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells. Nat. Biotechnol. 2011;29:1011–1018. doi: 10.1038/nbt.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott D.A., Braam S.R., Koutsis K., Ng E.S., Jenny R., Lagerqvist E.L., Biben C., Hatzistavrou T., Hirst C.E., Yu Q.C. NKX2-5(eGFP/w) hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat. Methods. 2011;8:1037–1040. doi: 10.1038/nmeth.1740. [DOI] [PubMed] [Google Scholar]
- Engel F.B., Hsieh P.C., Lee R.T., Keating M.T. FGF1/p38 MAP kinase inhibitor therapy induces cardiomyocyte mitosis, reduces scarring, and rescues function after myocardial infarction. Proc. Natl. Acad. Sci. U S A. 2006;103:15546–15551. doi: 10.1073/pnas.0607382103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox I.J., Daley G.Q., Goldman S.A., Huard J., Kamp T.J., Trucco M. Stem cell therapy. Use of differentiated pluripotent stem cells as replacement therapy for treating disease. Science. 2014;345:1247391. doi: 10.1126/science.1247391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graichen R., Xu X., Braam S.R., Balakrishnan T., Norfiza S., Sieh S., Soo S.Y., Tham S.C., Mummery C., Colman A. Enhanced cardiomyogenesis of human embryonic stem cells by a small molecular inhibitor of p38 MAPK. Differentiation. 2008;76:357–370. doi: 10.1111/j.1432-0436.2007.00236.x. [DOI] [PubMed] [Google Scholar]
- Greber B., Wu G., Bernemann C., Joo J.Y., Han D.W., Ko K., Tapia N., Sabour D., Sterneckert J., Tesar P. Conserved and divergent roles of FGF signaling in mouse epiblast stem cells and human embryonic stem cells. Cell Stem Cell. 2010;6:215–226. doi: 10.1016/j.stem.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Halloin C., Schwanke K., Lobel W., Franke A., Szepes M., Biswanath S., Wunderlich S., Merkert S., Weber N., Osten F. Continuous WNT control enables advanced hPSC cardiac processing and prognostic surface marker identification in chemically defined suspension culture. Stem Cell Rep. 2019;13:775. doi: 10.1016/j.stemcr.2019.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilger D., Masureel M., Kobilka B.K. Structure and dynamics of GPCR signaling complexes. Nat. Struct. Mol. Biol. 2018;25:4–12. doi: 10.1038/s41594-017-0011-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson J., Titmarsh D., Hidalgo A., Wolvetang E., Cooper-White J. Primitive cardiac cells from human embryonic stem cells. Stem Cells Dev. 2012;21:1513–1523. doi: 10.1089/scd.2011.0254. [DOI] [PubMed] [Google Scholar]
- Ichtchenko K., Bittner M.A., Krasnoperov V., Little A.R., Chepurny O., Holz R.W., Petrenko A.G. A novel ubiquitously expressed alpha-latrotoxin receptor is a member of the CIRL family of G-protein-coupled receptors. J. Biol. Chem. 1999;274:5491–5498. doi: 10.1074/jbc.274.9.5491. [DOI] [PubMed] [Google Scholar]
- Ieda M., Fu J.D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B.G., Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo H.J., Choi D.K., Lim J.S., Park J.S., Lee S.H., Song S., Shin J.H., Lim D.S., Kim I., Hwang K.C. ROCK suppression promotes differentiation and expansion of endothelial cells from embryonic stem cell-derived Flk1(+) mesodermal precursor cells. Blood. 2012;120:2733–2744. doi: 10.1182/blood-2012-04-421610. [DOI] [PubMed] [Google Scholar]
- Kattman S.J., Witty A.D., Gagliardi M., Dubois N.C., Niapour M., Hotta A., Ellis J., Keller G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Laflamme M.A., Chen K.Y., Naumova A.V., Muskheli V., Fugate J.A., Dupras S.K., Reinecke H., Xu C., Hassanipour M., Police S. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat. Biotechnol. 2007;25:1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- Langenhan T., Aust G., Hamann J. Sticky signaling--adhesion class G protein-coupled receptors take the stage. Sci. Signal. 2013;6:re3. doi: 10.1126/scisignal.2003825. [DOI] [PubMed] [Google Scholar]
- Lazaro J.B., Kitzmann M., Poul M.A., Vandromme M., Lamb N.J., Fernandez A. Cyclin dependent kinase 5, cdk5, is a positive regulator of myogenesis in mouse C2 cells. J. Cell Sci. 1997;110:1251–1260. doi: 10.1242/jcs.110.10.1251. [DOI] [PubMed] [Google Scholar]
- Lee C.S., Cho H.J., Lee J.W., Lee J., Kwon Y.W., Son T., Park H., Kim J., Kim H.S. Identification of latrophilin-2 as a novel cell-surface marker for the cardiomyogenic lineage and its functional significance in heart development. Circulation. 2019;139:2910–2912. doi: 10.1161/CIRCULATIONAHA.119.040826. [DOI] [PubMed] [Google Scholar]
- Lelianova V.G., Davletov B.A., Sterling A., Rahman M.A., Grishin E.V., Totty N.F., Ushkaryov Y.A. Alpha-latrotoxin receptor, latrophilin, is a novel member of the secretin family of G protein-coupled receptors. J. Biol. Chem. 1997;272:21504–21508. doi: 10.1074/jbc.272.34.21504. [DOI] [PubMed] [Google Scholar]
- Li X., Zhu L., Yang A., Lin J., Tang F., Jin S., Wei Z., Li J., Jin Y. Calcineurin-NFAT signaling critically regulates early lineage specification in mouse embryonic stem cells and embryos. Cell Stem Cell. 2011;8:46–58. doi: 10.1016/j.stem.2010.11.027. [DOI] [PubMed] [Google Scholar]
- Lian X., Zhang J., Azarin S.M., Zhu K., Hazeltine L.B., Bao X., Hsiao C., Kamp T.J., Palecek S.P. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nat. Protoc. 2013;8:162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan M.L., de Wit J., Savas J.N., Comoletti D., Otto-Hitt S., Yates J.R., 3rd, Ghosh A. FLRT proteins are endogenous latrophilin ligands and regulate excitatory synapse development. Neuron. 2012;73:903–910. doi: 10.1016/j.neuron.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikonomopoulos A., Kitani T., Wu J.C. Pluripotent stem cell-derived cardiomyocytes as a platform for cell therapy applications: progress and hurdles for clinical translation. Mol. Ther. 2018;26:1624–1634. doi: 10.1016/j.ymthe.2018.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q., Qiao F., Gao C., Norman B., Optican L., Zelenka P.S. Cdk5 targets active Src for ubiquitin-dependent degradation by phosphorylating Src(S75) Cell. Mol. Life Sci. 2011;68:3425–3436. doi: 10.1007/s00018-011-0638-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Uosaki H., Fukushima H., Takeuchi A., Matsuoka S., Nakatsuji N., Yamanaka S., Yamashita J.K. Efficient and scalable purification of cardiomyocytes from human embryonic and induced pluripotent stem cells by VCAM1 surface expression. PLoS One. 2011;6:e23657. doi: 10.1371/journal.pone.0023657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Soonpaa M.H., Adler E.D., Roepke T.K., Kattman S.J., Kennedy M., Henckaerts E., Bonham K., Abbott G.W., Linden R.M. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- Yu P., Pan G., Yu J., Thomson J.A. FGF2 sustains NANOG and switches the outcome of BMP4-induced human embryonic stem cell differentiation. Cell Stem Cell. 2011;8:326–334. doi: 10.1016/j.stem.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
On day 10 of cardiac differentiation, control-shRNA-transduced iPSCs exhibited robust beating.
On day 10 of cardiac differentiation, Lphn2-shRNA-transduced iPSCs did not beat spontaneously.
On day 10 of cardiac differentiation, there were several spontaneous beating foci in wild-type ESCs.
On day 10 of cardiac differentiation, there were no spontaneous beating foci in Lphn2-KO ESCs