Significance
The transcription factor IRF4 controls activation and differentiation of CD8+ T cells; however, its role in memory T cells is unclear. Here, we use a mouse model that allows removal of IRF4 from CD8+ T cells after their activation. We show that IRF4 is differentially required for long-term survival of CD8+ memory T cell subsets. In particular, maintenance of tissue-resident memory T cells appears to depend on IRF4. Upon reactivation, IRF4-deficient CD8+ memory T cells are strongly impaired in their proliferation and cytokine response. Thus, IRF4 is essential for the function CD8+ memory T cells.
Keywords: memory T cells, CD8+ T cells, interferon regulatory factor 4, Listeria monocytogenes
Abstract
The transcription factor IRF4 is required for CD8+ T cell activation, proliferation, and differentiation to effector cells and thus is essential for robust CD8+ T cell responses. The function of IRF4 in memory CD8+ T cells yet needs to be explored. To investigate the role of IRF4 for maintaining differentiation state and survival of CD8+ memory T cells, we used a mouse model with tamoxifen-inducible Irf4 knockout to preclude effects due to inefficient memory cell differentiation in absence of IRF4. We infected mice with ovalbumin-recombinant listeria and induced Irf4 knockout after clearance of the pathogen. Loss of IRF4 resulted in phenotypical changes of CD8+ memory T cells but did not cause a reduction of the total memory T cell population. However, upon reencounter of the pathogen, CD8+ memory T cells showed impaired expansion and acquisition of effector functions. When compared to CD8+ effector memory T cells, CD8+ tissue-resident memory T cells (TRM cells) expressed higher IRF4 levels. Mice with constitutive Irf4 knockout had diminished CD8+ TRM-cell populations, and tamoxifen-induced Irf4 deletion caused a reduction of this cell population. In conclusion, our results demonstrate that IRF4 is required for effective reactivation but not for general survival of CD8+ memory T cells. Formation and maintenance of CD8+ TRM cells, in contrast, appear to depend on IRF4.
The transcription factor interferon regulatory factor 4 (IRF4) is expressed in T cells and various other hematopoietic cells (1, 2). In T cells, IRF4 controls expression of a large number of genes, including genes coding for T cell effector proteins, for enzymes in central metabolism pathways, and for transcription factors and regulatory proteins involved in proliferation and differentiation processes. IRF4 is induced after T cell receptor (TCR) stimulation, and the level of its expression correlates with the strength of TCR signal. Thereby, IRF4 can act as a sensor that translates the quality of the TCR signal into distinct transcriptional programs. Consequently, IRF4 is considered as a central regulator of T cell differentiation (3–7).
IRF family members are characterized by their conserved DNA binding domains which recognize the interferon-stimulated response element. For IRF4, cooperative DNA binding at Ets-IRF composite elements (EICE) and AP-1-IRF composite elements (AICE) has been described. IRF4 interacts with the Ets family transcription factor PU.1 and SpiB in binding to EICE motifs or with heterodimers of BATF or BATF3 and Jun family members in binding to AICE motifs. Most T cell subsets express only low levels of PU.1, indicating that IRF4 binding to EICE is only of minor relevance for these cells. In contrast, T cells express BATF, BATF3, and various Jun family members, and the IRF4–AICE interaction could be demonstrated for CD8+ T cells and several CD4+ T cell subsets (8–10). Further cooperation partners of IRF4 include members of the signal transducer and activator of transcription and nuclear factor of activated T cells (NFAT) families of transcription factors (11–14). Binding often occurs at regulatory sites distant from the promotor region. It was therefore proposed that IRF4 can act as a pioneering factor enhancing gene accessibility for other transcription factors (6, 11).
Cytotoxic CD8+ T cells are central for the response against intracellular pathogens. After clearance of infection, a subpopulation of activated CD8+ T cells remains as memory T cells, able to act rapidly upon secondary encounter with the pathogen. Already during the acute T cell response, surface markers allow to distinguish between short-lived effector cells (KLRG1hi CD127lo) and long-lived memory precursor cells (KLRGlo CD127hi) (15). In CD8+ T cells, the T-box transcription factors TBX21 (T-bet) and EOMES regulate the development of effector and memory cells. Both factors cooperate in initial activation of CD8+ T cells. At later stages, strong TBX21 expression is associated with effector functions whereas high levels of EOMES correlate with memory cell formation but also with T cell exhaustion (15, 16). T cell longevity is further controlled by the stem cell transcription factor TCF-7 (T cell factor 7) (17).
IRF4 deficiency in CD8+ T cells results in impaired clonal expansion and diminished effector cell differentiation. In consequence, IRF4-deficient mice fail to mount CD8+ T cell responses to pathogens such as Listeria monocytogenes (3, 18), influenza virus (3–5), and lymphocytic choriomeningitis virus (LCMV) (3, 5, 19, 20) and are also compromised in the formation of pathogen-specific memory T cells (3, 18, 20, 21). During persisting infections, chronic T cell stimulation can result in T cell exhaustion. Exhaustion is defined by surface expression of inhibitory molecules, like the checkpoint inhibitor PD-1, limited effector functions, and metabolic impairment (16). In mice with chronic LCMV infection, exhausted CD8+ T cells express high levels of IRF4, and in cooperation with BATF and NFATc1, IRF4 controls the expression of genes associated with the exhaustion state (12). Interestingly, loss of one Irf4 allele protects CD8+ T cells from exhaustion and allows development of TCF-7+ memory T cells (3, 22). In human chimeric antigen receptor T cells, high levels of JUN disrupt the IRF4–BATF/BATF3–JUNB complex at AICE sites and thereby limit development of T cell exhaustion (23). In summary, results on IRF4 so far indicate that the amount and duration of IRF4 expression is decisive for the fate of CD8+ T cell differentiation. IRF4 expression is required for T cell activation and high IRF4 levels promote formation of effector T cells. However, during chronic infection, constitutively high IRF4 levels also result in T cell exhaustion.
The role of IRF4 in memory T cells is unclear. Low numbers of memory-like CD8+ T cells are found in IRF4-deficient mice and are dysfunctional upon activation (18, 21). However, these cells were generated in the absence of IRF4, thus, their functional state could reflect an impaired initial activation. In the current study, we examined the role of IRF4 in CD8+ memory T cells. We applied a mouse model with a tamoxifen-inducible Cre recombinase and floxed Irf4 alleles that allowed deletion of Irf4 at different time points of the CD8+ T cell response. In this model, we tested the function of IRF4 in CD8+ memory T cells in vitro and in the L. monocytogenes infection model in vivo.
Results
Deletion of Irf4 after Activation Does Not Impair Survival and Cytokine Production of CD8+ T Cells In Vitro.
The transcription factor IRF4 is required for effective activation of CD8+ T cells and their differentiation to effector T cells (3, 4, 18, 20, 24). Impaired activation in the absence of IRF4 might also influence the development of CD8+ memory T cells. To determine the role of IRF4 after T cells had been activated, we used an approach that allowed inducible deletion of the Irf4 gene. Mice with floxed Irf4 alleles (25) were crossed with CreERT2 mice (26). In Irf4fl/fl mice, Cre-mediated recombination results in the loss of exons I and II of Irf4 and the constitutive expression of green fluorescent protein (GFP) (25). Since GFP expression in Irf4fl/fl mice does not allow differentiation of cells with heterozygous or homozygous Irf4 mutation, we used Irf4fl/+×CreERT2 and Irf4fl/−×CreERT2 mice in which the floxed Irf4fl allele was combined with a wild-type Irf4+ or a mutant Irf4− allele (19), respectively (Fig. 1A). In the former mouse line, GFP expression indicates a switch from the Irf4 wild type to the heterozygous genotype, and in the latter, a switch from the heterozygous to the homozygous mutant genotype. In addition, Irf4fl/+×CreERT2 mice were on a CD90.1+ CD90.2+ background which allowed discrimination of their CD8+ T cells from those of Irf4fl/−×CreERT2 mice (CD90.1− CD90.2+) in coculture or cotransfer experiments.
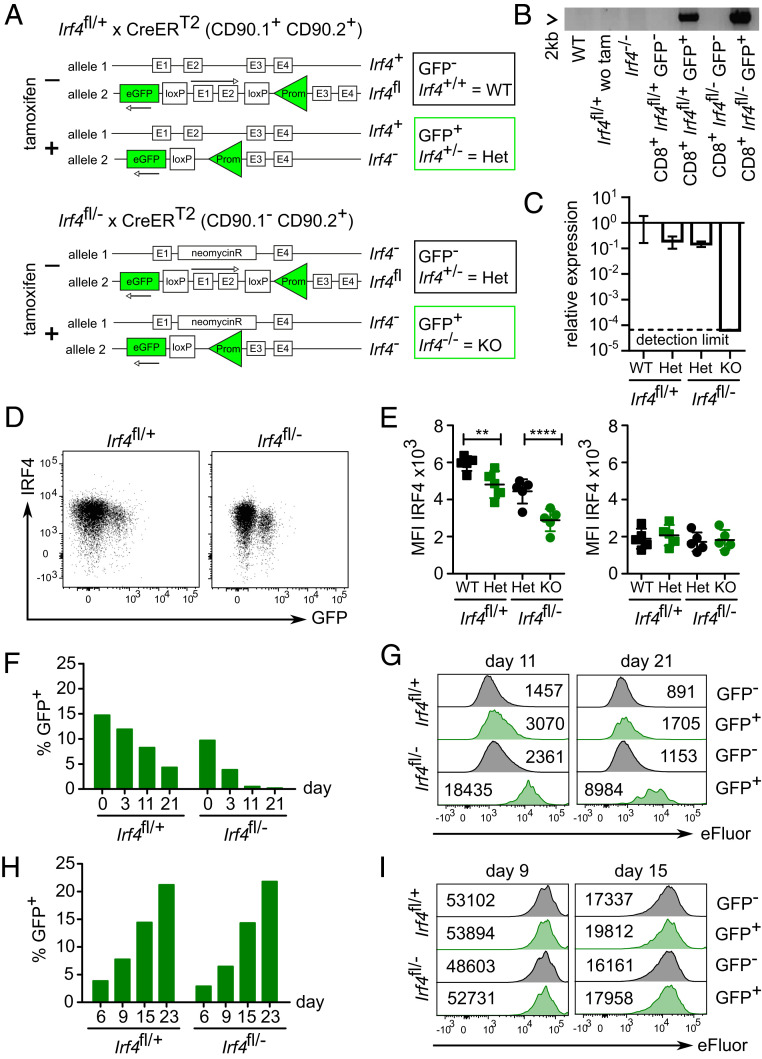
Fig. 1.
IRF4 is not required for maintenance of CD8+ memory T cells in vitro. (A) Scheme of genetic modifications of tamoxifen-inducible Irf4 knockout. (B–E) Irf4fl/+×CreERT2 and Irf4fl/−×CreERT2 mice were treated on 5 consecutive d with tamoxifen. Following 1 wk after the last treatment, spleen cells were analyzed. (B) Deletion of exons 1 and 2 of Irf4 in sorted CD8+ T cells. Primers are located upstream of the 5′ loxP site and within exon 3 and amplify a 2 kb fragment in the Cre-modified gene locus of the Irf4fl allele. (C) Expression of Irf4 exon 2 coding mRNA in sorted CD8+ T cells quantified by RT-qPCR. Normalized to 18S rRNA and to the Irf4 expression of GFP− Irf4fl/+×CreERT2 cells. (D) IRF4 expression in CD8+ T cells. Spleen cells were stimulated with PMA/ionomycin and after anti-IRF4 staining, analyzed with FACS. Representative FACS plots for activated CD8+ T cells. (E) Mean fluorescence intensity (MFI) values for stimulated (Left) and nonstimulated cells (Right; green symbols: GFP+ cells). Mean ± SEM, n = 5 per group, paired t test. **P ≤ 0.01, ***P ≤ 0.001. (F and G) CD8+ T cells purified from three to four pooled spleens from either Irf4fl/+×CreERT2 or Irf4fl/−×CreERT2 mice after tamoxifen treatment were stimulated for 3 d with anti-CD3 mAb, anti-CD28 mAb, and IL-2 and then eFluor670 labeled and incubated with IL-7 (SI Appendix, Fig. S1A). (F) % GFP+ of CD8+ T cells at indicated time points. (G) eFluor670 staining. The numbers give the MFI. (H and I) CD8+ T cells purified from three to four pooled spleens from either Irf4fl/+×CreERT2 or Irf4fl/−×CreERT2 mice were stimulated for 3 d with anti-CD3 mAb, anti-CD28 mAb, and IL-7, and then incubated with 4-hydroxytamoxifen and IL-7 (SI Appendix, Fig. S1C). After 6 d, cells were washed, labeled with eFluor670, and incubated for indicated time periods with IL-7. (H) % GFP+ of CD8+ T cells. (I) eFluor670 staining. (B) Result of one experiment with two pooled spleens of Irf4fl/+×CreERT2 and Irf4fl/−×CreERT2 mice, respectively. (C) Result of one experiment with three Irf4fl/+×CreERT2 or Irf4fl/−×CreERT2 mice. (D–I) Representative result of two or three independent experiments.
Irf4fl/+×CreERT2 and Irf4fl/−×CreERT2 mice were treated on 5 consecutive d with tamoxifen. After 1 wk following the last treatment, we consistently observed that between 10 and 15% of CD8+ T cells in spleens of both mouse lines had become GFP+. After polyclonal stimulation, PCR with primers specific for the Cre-modified gene locus amplified the expected 2 kb product only in sorted GFP+ but not in GFP− CD8+ T cells, indicating that GFP expression closely correlated with the Cre-mediated deletion of the Irf4fl locus (Fig. 1B). Quantitative PCR of sorted GFP+ CD8+ T cells also showed a pronounced reduction of messenger RNA (mRNA) coding for the deleted exon II of Irf4 in GFP+ CD8+ T cells from Irf4fl/−×CreERT2 mice (Fig. 1C). After polyclonal stimulation, Irf4-deficient CD8+ T cells (GFP+ Irf4fl/−) failed to up-regulate IRF4 and Irf4 heterozygous CD8+ T cells (GFP+ Irf4fl/+ and GFP− Irf4fl/−) had lower IRF4 expression than wild-type CD8+ T cells (GFP− Irf4fl/+) (Fig. 1 D and E).
To analyze memory formation and survival in vitro, spleen cells from tamoxifen-treated mice were activated with anti-CD3 and anti-CD28 mAb. After 3 d, cells were labeled with eFluor670 and cultured with IL-7 for up to 3 wk (SI Appendix, Fig. S1A) (27). In cultures of Irf4fl/−×CreERT2 CD8+ T cells, the frequency of GFP+ cells rapidly declined, and GFP+ cells showed only limited proliferation as indicated by the marginal loss of eFluor670 labeling (Fig. 1 F and G). We also observed a reduction of GFP+ cells in cultures of Irf4fl/+×CreERT2 CD8+ T cells, and CD8+ T cells heterozygous for a mutant allele (GFP+ Irf4fl/+×CreERT2 or GFP− Irf4fl/−×CreERT2) showed slightly higher levels of eFluor670 labeling at day 11 and 21. The latter result is consistent with the IRF4 dosage effect that has been described before for Irf4 heterozygous T cells (3, 5, 12, 22). Overall, results were consistent with the impaired response of Irf4+/− and Irf4−/− CD8+ T cells in coculture experiments (SI Appendix, Fig. S1B) and thus also confirm the genotype of GFP+ CD8+ cells.
In an alternative approach, CD8+ T cells from Irf4fl/+×CreERT2 and Irf4fl/−×CreERT2 mice were activated for 3 d and subsequently incubated with 4-hydroxytamoxifen and IL-7. After further 3 d, cells were eFluor670 labeled and incubated with IL-7 only (SI Appendix, Fig. S1C). Using this approach, we observed a steady increase of GFP+ cells in both Irf4fl/+×CreERT2 and Irf4fl/−×CreERT2 CD8+ T cells, possibly due to sustained recombinase activity and/or delayed GFP expression (Fig. 1H). eFluor670 staining revealed similar proliferation of CD8+ T cells from both mouse lines and for GFP+ and GFP− cells (Fig. 1I). Of note, due to the labeling at day six, we measured mainly IL-7 driven proliferation which was less pronounced than proliferation after initial TCR stimulation. After 3 wk, CD8+ T cells of all genotypes showed equal CD44 expression (SI Appendix, Fig. S1D). At this time point, CD8+ T cells from Irf4fl/−×CreERT2 mice had lower frequencies of IFN-γ+ cells as compared to CD8+ T cells from Irf4fl/+×CreERT2 mice following stimulation. However, in CD8+ T cells from both mouse lines, GFP+ cells were not impaired in cytokine production (SI Appendix, Fig. S1E). This response differed to that of CD8+ T cells from complete IRF4-deficient mice (SI Appendix, Fig. S1 F and G). Irf4−/− CD8+ T cells showed initial up-regulation of CD44. After 3 wk, remaining Irf4−/− CD8+ T cells had partially lost CD44 expression, and a reduced number of cells produced IFN-γ upon stimulation. In conclusion, Irf4 deletion after T cell activation did not substantially affect T cell proliferation and cytokine production in vitro.
Deletion of Irf4 after Activation Does Not Generally Impair Survival of CD8+ Memory T Cells In Vivo.
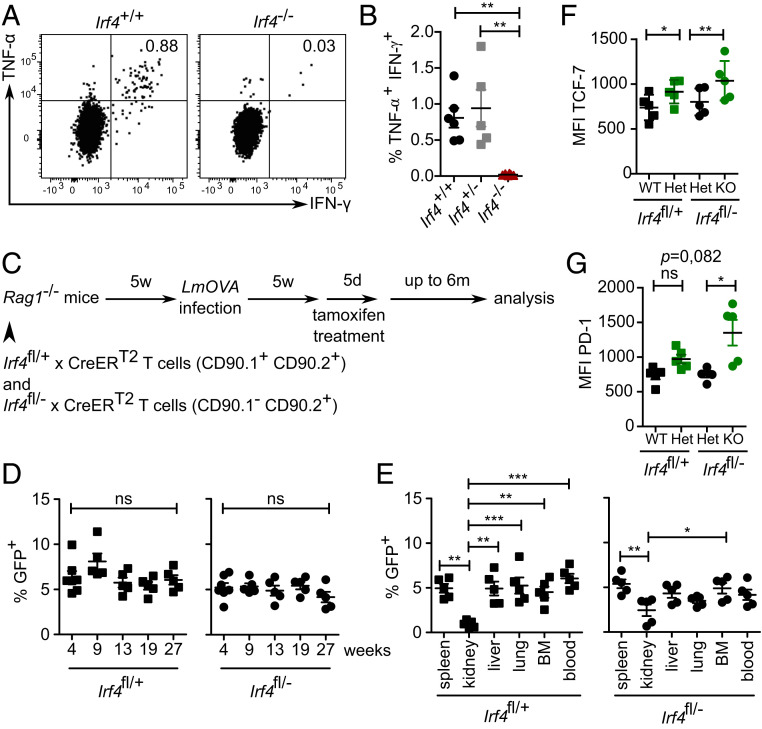
Irf4+/+, Irf4+/−, and Irf4−/− mice were infected with ovalbumin-recombinant listeria (LmOVA). After 6 wk, spleen cells were stimulated with OVA257–264 peptide and intracellular cytokines were determined. In contrast to Irf4+/+ and Irf4+/− mice, we did not detect ovalbumin-specific CD8+ T cells in Irf4−/− mice (Fig. 2 A and B). This result indicates that IRF4 deficiency largely prevented CD8+ memory T cell formation and confirms published results (3, 18, 20, 21). To analyze the role of IRF4 in CD8+ memory T cells, we reconstituted CD45.1+ Rag1−/− mice with T cells from naïve Irf4fl/+×CreERT2 and Irf4fl/−×CreERT2 mice (both CD45.2+). T cells from Irf4fl/+×CreERT2 mice expressed CD90.1, which allowed identification of different donor T cells after cotransfer (Fig. 2C and SI Appendix, Fig. S2A). Since it was shown before that heterozygous mutation of Irf4 could already alter the function of CD8+ T cells (3, 5, 12, 22), we used the cotransfer approach, which allowed analysis of the impact of a switch from Irf4 wild type to heterozygous and from heterozygous to homozygous mutant CD8+ T cells in the same recipient. After reconstitution, recipient mice were infected with LmOVA. After 5 wk, following recovery from infection, mice were treated with tamoxifen and analyzed as described before. With this approach, we restricted the Cre-mediated recombination to peripheral T cells, including CD8+ memory T cells generated during infection of recipient mice. Analysis of IRF4 in GFP− and GFP+ Irf4fl/+×CreERT2 T cells as well as of GFP− and GFP+ Irf4fl/−×CreERT2 CD8+ T cells revealed the graded expression in cells with two, one, or no functional Irf4 alleles and confirmed the accuracy of the GFP expression in this setting (SI Appendix, Fig. S3). After LmOVA infection and tamoxifen treatment, we followed the CD8+ T cells in the peripheral blood for up to 6 mo. Frequencies of GFP+ cells among Irf4fl/+ and among Irf4fl/− CD44+ CD8+ memory T cells remained stable over the whole observation period (Fig. 2D). After 6 mo, T cells were isolated from different tissues, and the frequencies of GFP+ cells were determined in Irf4fl/+×CreERT2 and Irf4fl/−×CreERT2 CD44+ CD8+ T cell subsets (Fig. 2E). Tissues differed slightly in frequencies of GFP+ cells, with lower frequencies in kidneys; however, overall frequencies were largely similar and did not substantially diverge between Irf4fl/+×CreERT2 and Irf4fl/−×CreERT2 CD44+ CD8+ T cells. Phenotypical analysis of CD44+ CD8+ T cell populations revealed that GFP+ T cells from both donor lines expressed higher levels of the transcription factor TCF-7 (Fig. 2F), and particularly GFP+ T cells from Irf4fl/−×CreERT2 mice had higher surface expression of PD-1 (Fig. 2G). In conclusion, acquisition of an Irf4 heterozygous or—more importantly—of an Irf4-deficient genotype by CD8+ T cells after infection did not generally impair long-term persistence of these cell populations.
Fig. 2.
Survival of CD8+ T cells in the absence of IRF4. (A and B) Mice were infected with LmOVA. After 5 wk, spleen cells were stimulated with OVA257–261 peptide for 4 h, and the production of IFN-γ and TNF-α in CD8+ T cells was determined by FACS. (A) Representative dot plots for CD8-gated cells and (B) %-values for CD8+ T cells from Irf4+/+, Irf4+/− and Irf4−/− mice. Pooled results from two independent experiments with five or six mice/group. Mean ±SEM, ANOVA, and Bonferroni’s post-test. (C–G) Rag1−/− mice were reconstituted with T cells from naïve Irf4fl/+×CreERT2 and Irf4fl/−×CreERT2 mice. Recipient mice were infected with LmOVA. After 5 wk, mice were treated with tamoxifen for 5 d. Mice were analyzed at the indicated time points. (For the gating strategy see SI Appendix, Fig. S2A). (D) Percentage of GFP+ Irf4fl/+ and Irf4fl/− CD44+ CD8+ T cells in blood at the indicated weeks after tamoxifen treatment. (E) Percentage of GFP+ cells of CD44+ CD8+ T cells in different tissues of recipients 6 mo after tamoxifen treatment. (D and E) Representative result of two independent experiments with five or six mice/experiment. Mean ± SEM, ANOVA, and Bonferroni’s post-test. (F and G) Mean fluorescence intensity of TCF-7 and PD-1 expression of GFP+ and GFP− CD8+ T cells from Irf4fl/+×CreERT2 and Irf4fl/−×CreERT2 donors 6 mo after tamoxifen treatment. Representative result of two independent experiments with five or six mice/experiment). Mean ± SEM, paired t test. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
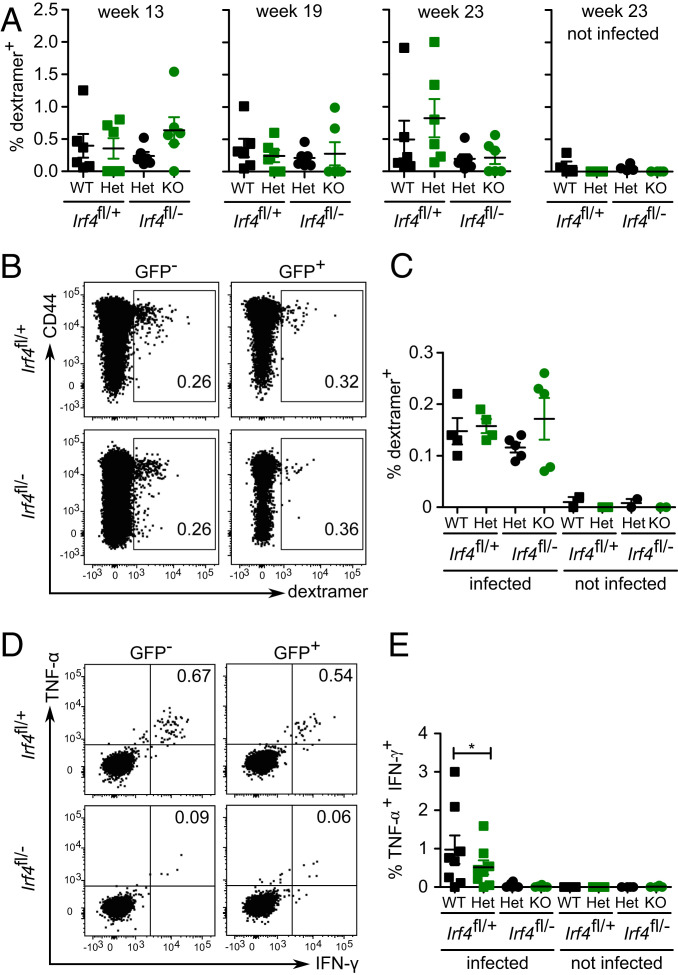
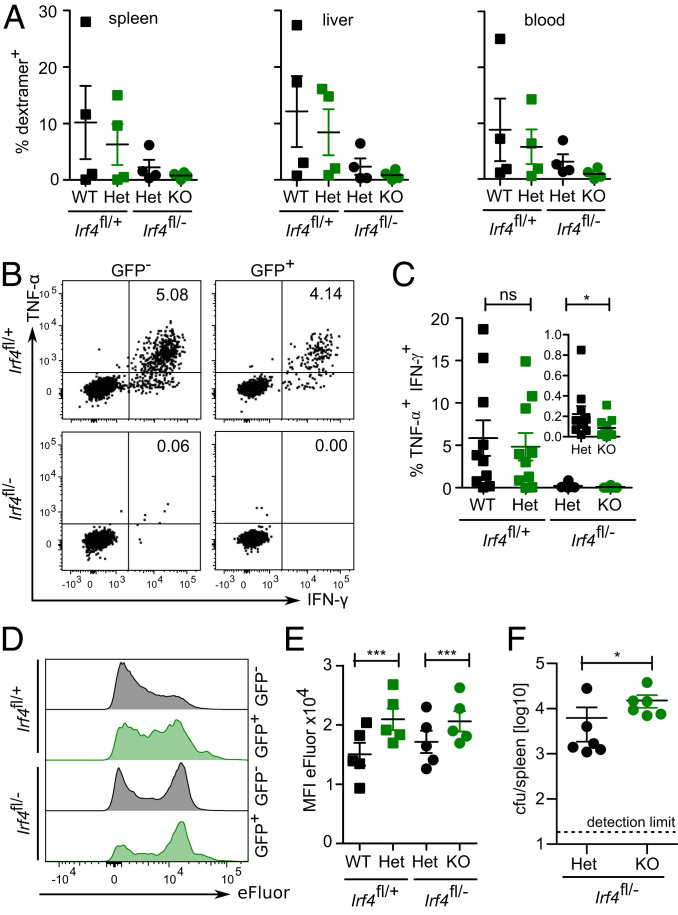
Frequencies of ovalbumin-specific CD8+ memory T cells were determined with OVA257–264 H-2Kb dextramers in GFP+ and GFP− CD8+ T cells from both donor cells populations. In the peripheral blood, dextramer+ cells were detected in low but similar frequencies in all four CD8+ T cell subpopulations and remained relatively stable over the entire observation period (Fig. 3A). After 6 mo, analysis of CD8+ T cells from spleens of mice confirmed this result, since we detected similar frequencies of dextramer+ cells in GFP+ and GFP− CD8+ T cells from both donor populations (Fig. 3 B and C). Spleen cells were also stimulated with OVA257–264 peptide, and IFN-γ and TNF-α were determined (Fig. 3 D and E). We detected only very low frequencies of IFN-γ+ TNF-α+ cells in GFP+ and GFP− Irf4fl/−×CreERT2 CD8+ T cells. CD8+ T cells derived from Irf4fl/+×CreERT2 donors responded to peptide, but here, GFP+ cells showed a weaker response than GFP− cells. In summary, loss of functional Irf4 alleles after memory formation did not cause reduced survival of ovalbumin-specific CD8+ memory T cells but impaired their ability to produce IFN-γ and TNF-α.
Fig. 3.
Survival of CD8+ memory T cells in the absence of IRF4. Rag1−/− mice were reconstituted, infected, and tamoxifen treated as described in Fig. 2. GFP+ and GFP− CD8+ T cells from Irf4fl/+×CreERT2 and Irf4fl/−×CreERT2 donors were analyzed at the indicated time points. (A) Frequencies of ovalbumin-specific CD8+ T cells in peripheral blood were determined with OVA257–264 dextramers at 13, 19, and 23 wk after tamoxifen treatment. (B and C) Dextramer+ CD8+ T cells in spleens 6 mo after tamoxifen treatment. (B) Representative dot plot and (C) %-values of dextramer+ CD8+ T cells. (A–C) Representative result of two independent experiments with five or six mice in infected groups and two or four mice in not infected groups. Mean ± SEM, paired t test. (D and E) Following 3 wk after tamoxifen treatment, spleen cells were stimulated with OVA257–264 peptide for 4 h, and the production of IFN-γ and TNF-α in CD8+ T cells was determined by FACS. (D) Representative dot plots for CD8-gated cells and (E) %-values of cytokine-positive CD8+ T cells. (D and E) Representative results of three independent experiments with five to eight mice in infected groups and four or five mice in not infected groups. Mean ± SEM, paired t test. *P ≤ 0.05.
Impaired Reactivation of IRF4-Deficient CD8+ Memory T Cells.
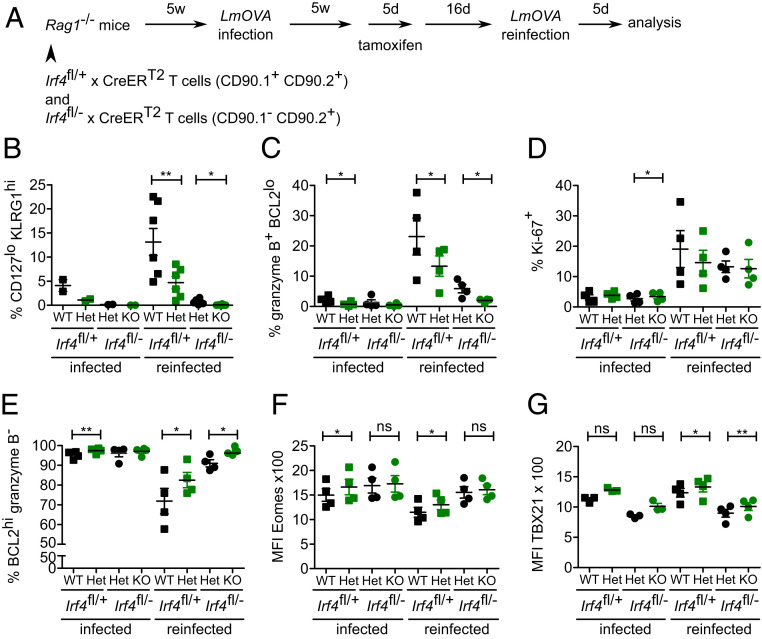
To test whether depletion of Irf4 in CD8+ memory T cells affected reactivation of these cells, Rag1−/− mice were reconstituted with Irf4fl/+×CreERT2 and Irf4fl/−×CreERT2 T cells, infected with LmOVA, and after recovery from infection treated with tamoxifen. Following 16 d after tamoxifen treatment, mice were reinfected with LmOVA, and after further 5 d, the T cell response was analyzed (Fig. 4A). Reinfection of mice caused an expansion of the CD44hiCD62Llo effector/effector memory (Eff/EM) cell subset (SI Appendix, Fig. S4 A and B) and particularly of the CD44hiCD62LloCD127loKLRG1hi short-lived effector cell (SLEC) subset in GFP− Irf4fl/+×CreERT2 CD8+ T cells (Fig. 4B and SI Appendix, Fig. S4C). Loss of one functional Irf4 allele in GFP+ Irf4fl/+×CreERT2 CD8+ T cells significantly reduced the expansion of both T cell subsets. GFP− Irf4fl/−×CreERT2 CD8+ T cells showed only marginal formation of Eff/EM and SLEC subsets. GFP+ Irf4fl/−×CreERT2 CD8+ T cells completely failed to generate these effector subsets. In line with their maturation to SLEC, a subpopulation of wild-type CD8+ T cells (GFP− Irf4fl/+×CreERT2) up-regulated expression of granzyme B. Up-regulation was less pronounced in Irf4 heterozygous CD8+ T cells (GFP+ Irf4fl/+×CreERT2 and GFP− Irf4fl/−×CreERT2) and almost absent in Irf4 homozygous mutant (GFP+ Irf4fl/−×CreERT2) T cells (Fig. 4C and SI Appendix, Fig. S4D). Without reinfection, only few CD8+ T cells proliferated, as indicated by nuclear Ki-67 staining (Fig. 4D and SI Appendix, Fig. S4E). Reinfection caused up-regulation of Ki-67 in all CD8+ T cells independent of their genotype. Without reinfection, almost all CD8+ T cells were BCL2+ (Fig. 4E and SI Appendix, Fig. S4D). Following reinfection, Irf4 wild-type CD8+ T cells down-regulated BCL2, but reduction was less prominent in Irf4 heterozygous CD8+ T cells and absent in Irf4 deficient cells (GFP+ Irf4fl/−×CreERT2). In summary, mutation of already one Irf4 allele reduced the formation of a CD8+ effector T cell population upon reinfection, and in Irf4 deficient cells, effector formation was completely blocked despite Ki-67 up-regulation.
Fig. 4.
IRF4 is required for reactivation of CD8+ T cells. Rag1−/− mice were reconstituted, infected, and tamoxifen treated as described in Fig. 2. Following 2 wk after tamoxifen treatment, mice were reinfected with LmOVA. After five d, CD8+ T cells were analyzed. (A) The experimental scheme. (B) Frequencies of CD44+ CD62L− KLRG+ CD127− short-lived CD8+ effector T cells (C) of granzyme B+ BCL2lo CD8+ T cells, (D) Ki-67+ CD8+ T cells, and (E) BCL2hi granzyme B− CD8+ T cells in spleens of mice with LmOVA reinfection and of mice similarly treated but without reinfection (infected). Mean fluorescence intensity of staining for EOMES (F) and TBX21 (G) in CD8+ T cells from spleens of mice. (B–G) Representative results of three independent experiments with four to six mice in reinfected groups and two to four mice in infected groups. Mean ± SEM, paired t test. *P ≤ 0.05, **P ≤ 0.01.
The transcription factors TBX21 (T-bet) and EOMES are both induced during CD8+ T cell activation. Effector cells are characterized by high expression levels of TBX21, whereas memory precursors and memory T cells but also exhausted CD8+ T cells express enhanced EOMES levels (16). Without reinfection, loss of one or both functional Irf4 alleles was associated with slightly higher expression of EOMES in CD8+ T cells (Fig. 4F and SI Appendix, Fig. S4F). Reinfection of mice caused down-regulation of EOMES in Irf4 wild-type CD8+ T cells; however, reduction was less pronounced in CD8+ T cells lacking one or both functional Irf4 alleles, in line with observation of increased EOMES expression in CD8+ T cells of Irf4−/− mice (3, 12, 18). CD8+ T cells derived from Irf4fl/−×CreERT2 mice showed generally lower TBX21 expression; however, in both Irf4fl/+×CreERT2 and Irf4fl/−×CreERT2 cell populations, GFP+ CD8+ T cells had slightly higher TBX21 expression than GFP− cells. TBX21 expression only marginally changed in all CD8+ T cell populations upon reinfection (Fig. 4G and SI Appendix, Fig. S4F). In conclusion, induced Irf4 deficiency prevented down-regulation of EOMES in CD8+ T cells but had only limited impact on TBX21 expression upon reinfection.
OVA257–264 dextramer staining revealed expansion of ovalbumin-specific CD8+ T cells in the GFP− and GFP+ Irf4fl/+×CreERT2 cell populations in spleen, liver, and blood of reinfected mice (Fig. 5A and SI Appendix, Fig. S2B). We observed large variation in the strength of response between individual mice. Irf4 heterozygous GFP− Irf4fl/−×CreERT2 CD8+ T cells contained lower frequencies of dextramer+ cells, and in all analyzed mice, dextramer+ T cells remained at very low levels in the IRF4-deficient GFP+ Irf4fl/−×CreERT2 T cell population. Stimulation of spleen cells with OVA257–264 peptide induced a cytokine response in CD8+ T cells from the GFP− and GFP+ Irf4fl/+×CreERT2 cell populations (Fig. 5 B and C). In contrast, only marginal frequencies of TNF-α+ IFN-γ+ cells were detected among GFP− Irf4fl/−×CreERT2 CD8+ T cells, and cytokine-positive cells were absent in the GFP+ Irf4fl/−×CreERT2 T cell population. In vitro proliferation assays using OVA257–264 peptide stimulation largely confirmed these observations (Fig. 5 D and E). We observed strong proliferation of CD8+ T cells with two functional Irf4 alleles, reduced proliferation in Irf4-heterozygous CD8+ T cells, and only marginal proliferation in Irf4-deficient CD8+ T cells.
Fig. 5.
IRF4 is required for reactivation of CD8+ memory T cells. Rag1−/− mice were reconstituted and treated as described in Fig. 4. (A) Frequencies of OVA257–264 dextramer+ CD8+ T cells in tissues of reinfected mice (see SI Appendix, Fig. S2B for representative dot plots). Representative result of three independent experiments with four to six mice. Mean ± SEM, paired t test. (B and C) Spleen cells of reinfected mice were stimulated with OVA257–264 peptide and frequencies of IFN-γ+ and TNF-α+ CD8+ T cells were determined. (B) Representative plots and (C) frequencies of IFN-γ+ TNF-α+ T cells (Insert: results for Irf4fl/− cells with magnified y axis). Pooled results from two independent experiments with four and six mice. Mean ± SEM, paired t test. (D and E) Spleen cells from reinfected mice were labeled with eFluor670 and cultivated for 4 d with OVA257–264 peptide. (D) Representative histograms of CD8-gated T cells and (E) mean fluorescence intensity of eFluor670 staining of CD8+ T cells. Representative result of two independent experiments with four and five mice. Mean ± SEM, paired t test. (F) Irf4fl/−×CreERT2 mice were infected with LmOVA and after 3 wk, treated with tamoxifen. Following 9 d after the tamoxifen treatment, 2 × 105 sorted GFP− or GFP+ CD8+ T cells were transferred into Rag1−/− mice. Recipients were infected with LmOVA and after 3 d, listeria titers in spleens were determined. One experiment with six recipient mice/group. Median, Mann–Whitney test. *P ≤ 0.05, ***P ≤ 0.001.
In order to test the protective function of CD8+ memory T cells in which Irf4 was deleted after the primary activation, Irf4fl/−×CreERT2 mice were infected with LmOVA and, after recovery from infection, treated with tamoxifen. After 9 d, sorted GFP+ and GFP− CD8+ T cells were transferred into Rag1−/− mice and recipients were infected with LmOVA (Fig. 5F). After 3 d postinfection, we observed significantly reduced listeria titers in recipients of Irf4-heterozygous GFP− CD8+ T cells when compared to recipients of Irf4-deficient GFP+ CD8+ T cells. Overall, these results indicate that loss of Irf4 in memory CD8+ T cells impaired their response upon reactivation.
IRF4 Supports Survival of CD8+ TRM Cells.
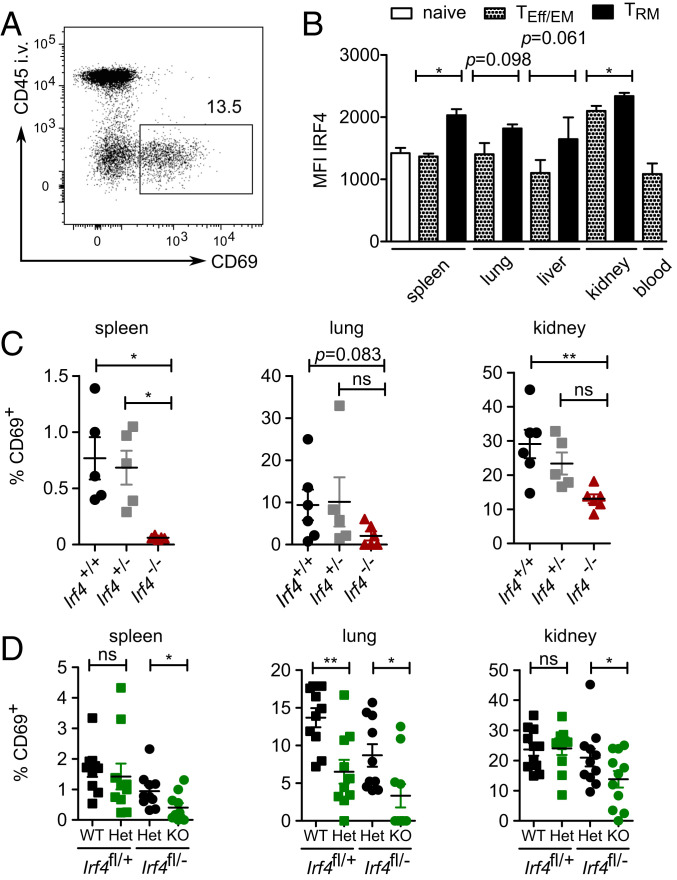
CD8+ TRM cells constitutively express Irf4 mRNA (28, 29), indicating that IRF4 might be required for the function of these cells. We analyzed tissues from mice for CD8+ T cells with the CD69+ CD44+ CD62L− phenotype of TRM cells. Nonvascular cells in tissues were identified by intravenous (IV) injection of fluorochrome-conjugated anti-CD45 mAb 3 min prior to T cell isolation and subsequently gating on CD45-negative (CD45IV−) cells (Fig. 6A). After 5 wk post-LmOVA infection, CD45IV− CD69+ CD8+ TRM cells from spleen, lung, and kidney of wild-type mice expressed higher baseline IRF4 levels than corresponding CD69− CD8+ TEff/EM cells or naïve CD8+ T cells (Fig. 6B). Compared to Irf4+/+ mice, Irf4−/− mice had lower frequencies of CD8+ T cells with a TRM-cell phenotype in spleen, lung, and kidney (Fig. 6C).
Fig. 6.
IRF4 supports survival of CD8+ tissue-resident memory T cells. (A) Representative gating strategy for CD45IV− CD69+ CD8+ TRM cells in lung. (B) Irf4+/+ mice were infected with LmOVA. After 5 wk, tissues were analyzed for CD8+ T cells with a TRM phenotype. Mean fluorescence intensity of IRF4 staining of CD45IV− CD44+ CD62L− CD69− CD8+ T effector/effector memory (TEff/EM) cells, of CD45IV− CD44+ CD62L− CD69+ CD8+ TRM cells and corresponding CD45IV+ CD8+ TEff/EM cells in blood, and of CD44− CD62L+ naïve CD8+ T cells. Representative result of two independent experiments, each with three individually analyzed mice. Mean ± SEM, paired t test. (C) Irf4+/+, Irf4+/−, and Irf4−/− mice were infected with LmOVA. After 5 wk, tissues were analyzed. Frequencies of CD8+ T cells with a TRM-cell phenotype among CD45IV− CD8+ T cells. Pooled results from two independent experiments with five or six mice/group. Mean ± SEM, ANOVA, and Bonferroni’s post-test. (D) Rag1−/− mice were reconstituted with T cells from naïve Irf4fl/+×CreERT2 and Irf4fl/−×CreERT2 mice. Recipient mice were infected with LmOVA. After 5 wk, mice were treated with tamoxifen for 5 d. After 6 mo, tissues were analyzed for CD8+ T cells with a TRM phenotype among CD45IV− CD8+ T cells. Pooled results from two independent experiments with five and six mice. Mean ± SEM, paired t test. *P ≤ 0.05, **P ≤ 0.01.
To exclude that reduced frequencies of CD8+ TRM cells were due to impaired T cell activation in the absence of IRF4, we applied the T cell transfer assays as described before (Fig. 2C). T cells from Irf4fl/+×CreERT2 and Irf4fl/−×CreERT2 mice were cotransferred into Rag1−/− mice. Recipients were infected with LmOVA and after recovery from infection treated with tamoxifen. After 6 mo, frequencies of GFP+ CD8+ TRM cells were determined in spleen, lung, and kidney (Fig. 6D). In all tissues, CD8+ T cells derived from Irf4fl/−×CreERT2 donors comprised lower frequencies of TRM cells, and among GFP+ Irf4fl/−×CreERT2 CD8+ T cells, frequencies of TRM cells were further reduced. In the lung, we also observed a reduction of TRM cells in the GFP+ Irf4fl/+×CreERT2 CD8+ T cell population. Thus, absence of IRF4 impaired the maintenance of TRM cells.
Discussion
IRF4 has an essential role in the activation and differentiation of CD8+ T cells; however, its function in memory T cells is less clear. Studies in mice with constitutive IRF4-deficient T cells are problematic since absence of IRF4 during activation could affect the function of the few memory T cells generated in these mice (18, 21). To circumvent this problem, we used CreERT2-mediated Irf4 deletion after the induction of the CD8+ T cell response. We detected Irf4 deletion in only 10 to 15% of CD8+ T cells in Irf4fl/+×CreERT2 and Irf4fl/−×CreERT2 mice and in Rag1−/− mice reconstituted with cells from these mice. Reasons for the limited rate of recombination in our model might be a low expression level and the only transient nuclear localization of Cre. Because of the low recombination efficacy, we used cells in which only one Irf4 allele was targeted by the tamoxifen treatment to generate heterozygous and homozygous Irf4 mutant cells from Irf4fl/+×CreERT2 and Irf4fl/−×CreERT2 T cells, respectively. Subsequent PCR analysis confirmed that the GFP expression reliably indicated the Cre-induced modification of the Irf4 allele in CD8+ T cells, and analysis of IRF4 by flow cytometry revealed a graded expression in cells with two, one, or no functional Irf4 allele. Reliability of the tamoxifen-induced Irf4 deletion was further confirmed by in vitro experiments in which the response of GFP+ Irf4fl/+×CreERT2 and Irf4fl/−×CreERT2 CD8+ T cells resembled those of Irf4+/− and Irf4−/− CD8+ T cells, respectively.
In the T cell transfer model, in which the peripheral T cell pool is not continuously replenished with naïve cells from the thymus, frequencies of peripheral GFP+ CD44+ CD8+ memory T cells, including cells with a switch from wild type to Irf4 heterozygous and from Irf4 heterozygous to homozygous mutant mice, were stable for up to 6 mo. Ovalbumin-specific CD8+ memory T cells were also maintained in the GFP+ cell populations when Irf4 alleles were deleted after recovery from LmOVA infection. These observations are in contrast to the profound loss of LCMV-specific CD8+ T cells when Irf4 was deleted in the late phase of the acute LCMV response using Cre expression under control of Gzmb gene-regulatory elements (3) or during chromic LCMV infection by tamoxifen activation of CreERT2 (12). Results are rather consistent with the relative stability of the few CD8+ memory T cells generated in mice with constitutive Irf4 deletion in T cells (18, 21) and indicate that IRF4 is not required anymore for survival of CD8+ T cells after formation of the memory state. Interestingly, despite persistence of ovalbumin-specific CD8+ memory T cells as determined with major histocompatibility complex class I multimers, we failed to detect IFN- γ and TNF-α production after short-term stimulation of cells with ovalbumin peptide in GFP− and GFP+ Irf4fl/−×CreERT2 CD8+ T cells, and loss of one allele caused reduction of the cytokine response in GFP+ Irf4fl/+×CreERT2 CD8+ T cells. Thus, high IRF4 expression is required for the formation of memory T cells that can rapidly produce IFN-γ and TNF-α, and loss of one allele in the memory state of cells reduces their capacity for immediate secretion of these cytokines. These results further indicate that although CD8+ memory T cells do not require IRF4 for survival, their rapid effector response highly depends on the transcription factor.
Loss of one functional allele in Irf4 wild-type cells and of the remaining allele in Irf4-heterozygous cells caused phenotypical changes in these CD8+ T cell populations already under steady-state conditions, despite only low Irf4 expression in CD8+ memory T cells (SI Appendix, Fig. S5, based on the ImmGen consortium database) (30). Particularly, the switch from wild-type to heterozygous genotype resulted in the reduction of the CD44hiCD62Llo CD8+ effector T cell population and the loss of the KLRG1hiCD127lo CD8+ short-lived effector T cells. In addition, GFP+ cells showed up-regulation of TCF-7, PD-1, and EOMES. Thus, IRF4 appears to be required for the survival of effector and, in particular, of short-lived effector CD8+ T cells. Suppression of EOMES and TCF-7 expression by IRF4 has been described before (3, 5, 12, 24). However, higher TCF-7 expression could also reflect a reduction of TCF-7lo effector cells in favor of TCF-7+ CD8+ memory T cell subsets (31). High PD-1 expression in complete Irf4 mutant mice was unexpected since high PD-1 expression is associated with activation but particularly with chronic activation and exhaustion of CD8+ T cells and is, at least in the latter situation, driven by high IRF4 levels (3, 12). This phenotype needs to be analyzed in further studies to elucidate the exact role of IRF4 in this setting.
Functional impairment of memory T cells that had lost Irf4 alleles after recovery from infection was more prominent upon in vivo reactivation of cells. Transferred GFP+ CD8+ T cells from Irf4fl/−×CreERT2 mice provided less protection to recipients than their GFP− counterparts with a still-functional Irf4 allele. Removal of functional Irf4 alleles in CD8+ T cells resulted in the diminished formation of BCL2lo granzyme B+ CD8+ effector T cells and prevented down-regulation of EOMES upon reinfection of mice. The higher TBX21 expression in GFP+ cells was unexpected since constitutive IRF4-deficient CD8+ T cells show impaired up-regulation of Tbx21 (4, 18), and enhanced TBX21 expression has been associated with effector cell differentiation (32) which was suppressed in CD8+ T cells after induced knockout of Irf4 alleles. Analysis of ovalbumin-specific T cells after reinfection was consistent with an impaired re-expansion, particularly of the GFP+ Irf4fl/−×CreERT2 CD8+ T cell population which had lost the remaining functional Irf4 allele. Thus, similar to naïve CD8+ T cells, CD8+ memory T cells appear to require high levels of IRF4 for robust expansion and acquisition of effector functions, and the loss of both functional Irf4 alleles completely prevents this response. Unexpectedly, CD8+ T cells of all genotypes had become Ki-67+ despite the failure to form an effector cell population, particularly in GFP+ Irf4fl/−×CreERT2 T cells. Ki-67+ cells in Irf4 heterozygous or homozygous mutant T cells could represent memory or even naïve T cells that had become activated but were stalled in the cell cycle in a Ki-67 positive stage. Ki-67 up-regulation in Irf4 mutant T cells could also indicate proliferation in response to mitogenic cytokines provided by IRF4-sufficient cells in recipient mice during reinfection. This proliferation without acquisition of an effector phenotype would be consistent with the observation that Irf4−/− CD8+ memory T cells demonstrate regular homeostatic proliferation in lymphopenic hosts (21).
Although GFP+ Irf4fl/+×CreERT2 and GFP− Irf4fl/−×CreERT2 CD8+ memory T cells share the Irf4 heterozygous genotype, cells differ in phenotype and function. Irf4fl/+×CreERT2 CD8+ T cells had two functional alleles during primary infection and one allele was deleted thereafter in the GFP+ subset. In contrast, GFP− Irf4fl/−×CreERT2 CD8+ T cells were activated with only one functional allele. Following CD8+ T cell activation, the level of IRF4 expression determines the strength of the T cell response, and a gene dosage effect for Irf4 heterozygous CD8+ T cells has been described (3, 5, 12, 22). Thus, the initial activation of CD8+ T cells with either one or two functional Irf4 alleles could well explain the differences in the two Irf4 heterozygous memory T cell populations.
CD8+ TRM cells express enhanced levels of Irf4 mRNA (28, 29) and we also observed higher IRF4 protein levels in these cells. TRM cells have a sentinel function in tissues and can rapidly respond upon re-encounter of their antigen (33, 34). Enhanced steady-state levels of IRF4 in TRM cells could support the fast production of large amounts of cytokines and other effector proteins required in this situation. Interestingly, Irf4−/− mice harbored reduced levels of CD8+ T cells with a tissue-resident phenotype, and in the transfer model, deletion of the remaining allele in Irf4fl/−×CreERT2 CD8+ T cells caused a significant reduction of TRM cells in this populations. Unfortunately, the frequencies of ovalbumin-specific CD8+ TRM cells in the transfer model were too low to reliably evaluate impact of induced Irf4 mutations in established ovalbumin-specific TRM-cell populations. Still, our results clearly indicate that constitutive IRF4 expression is required for survival of these cells. These results would also imply that CD8+ TRM cells profoundly differ in their requirement for IRF4 from other CD8+ memory T cell subsets. The signals that maintain higher levels of IRF4 in CD8+ TRM cells, as well as the function and target genes of IRF4 in these cells, are currently unknown and require further investigation.
In conclusion, our results demonstrate that IRF4 is not required for survival of conventional CD8+ memory T cells once they have established this status. However, IRF4 becomes essential as soon as these cells are reactivated. In contrast, IRF4 appears to be essential for CD8+ TRM cell maintenance.
Materials and Methods
Mice.
Irf4−/− mice (B6.129P2-Irf4tm1Mak/J) (19), Irf4fl/fl mice (25), CreERT2 mice (26), Rag1−/− mice (B6.129S7-Rag1tm1Mom/J) (35), CD45.1 congenic mice (B6.SJL-Ptprca Pepcb/BoyJ), and CD90.1 congenic mice (B6.PL-Thy1a/CyJ) were on the C57BL/6 background. All other mice used in this study were derived by intercrosses of these mouse strains. Genotypes of mice were determined by PCR as described previously (19, 25) and by flow cytometry using anti-CD90.1 and CD90.2 mAb. All mice were housed in the animal facility of the University Medical Center Hamburg-Eppendorf under specific pathogen free conditions with standard food and water ad libitum. During experiments, mice were controlled daily, and mice with signs of severe disease were eliminated to minimize suffering. In all experiments, age and sex matched groups of mice were used. Animal experiments were conducted in agreement with the German animal protection law, and experimental protocols were approved by the local committee for animal experiments of the City of Hamburg (registration number: 17/17).
T cell Transfer and Infection of Mice.
T cells from spleens of Irf4fl/+×CreERT2 mice (CD90.1+ CD90.2+) and Irf4fl/−×CreERT2 mice (CD90.1− CD90.2+) were purified by negative selection (EasySep Mouse T cell Isolation Kit, Stemcell Technologies) according to the manufacturer’s protocol. T cells were mixed to a 1 : 1 ratio and 8 × 106 T cells per mouse were transferred into CD45.1+ Rag1−/− mice by tail vein injection. Transferred Irf4fl/+×CreERT2 and Irf4fl/−×CreERT2 T cells could be distinguished by CD90.1 staining. For activation of the Cre recombinase in mice, 2 mg of tamoxifen (Sigma Aldrich) per day was applied intraperitoneally on 5 consecutive d. Analyses were performed at least 7 d after the last tamoxifen injection (26). Tamoxifen (Sigma Aldrich) was dissolved in corn oil (Sigma Aldrich).
Mice were IV infected with 1 × 104 colony forming units (CFU) of a L. monocytogenes strain recombinant for LmOVA (36). For reinfection, mice received 1 × 105 CFU. Bacterial inocula were controlled by plating serial dilutions on tryptic soy broth agar plates and colonies were counted after 2 d of incubation at room temperature (RT). For the protection assay, Irf4fl/−×CreERT2 mice were infected with 5 × 103 CFU of LmOVA. After 3 wk, mice were treated on 5 consecutive d with tamoxifen. After 9 d, fluorescence-activated cell sorting (FACS)-sorted GFP+ and GFP− CD8+ T cells from spleens and lymph nodes were IV transferred into Rag1−/− mice, which were IV infected with 1 × 105 CFU of LmOVA at the same day. After 3 d, bacterial titers in spleens were determined by plating serial dilution of tissue suspensions.
T cell Isolation and Analysis.
To protect T cells from NAD+-induced cell death during purification, mice received intraperitoneal 50 µg of an ART2A blocking nanobody (S+16) 45 min before harvesting the organs (37). We IV injected 2.5 µg of PerCP-conjugated anti-CD45 mAb (clone 30F-11, BioLegend) 3 min before the mice were killed to stain intravascular cells (38). Peripheral heparin blood was collected by heart puncture. Erythrocytes were lysed with lysis buffer (155 mM NH4Cl, 10 mM KHCO3, 100 μM ethylenediaminetetraacetic acid (EDTA), pH 7.2) for 3 min. Spleens were forced through a 70 μm cell strainer and erythrocytes were lysed with lysis buffer. Mesenteric lymph nodes were forced through a 30 µm cell strainer. Bone marrow was harvested from the femur and erythrocytes were lysed as described before. After removing of the gall bladder, the liver was forced through a 100 µm cell strainer. Cells were washed and the cell suspension was layered over a 40% Percoll solution (GE Healthcare). After centrifugation, cells in the interphase were collected and remaining erythrocytes were lysed. Lungs and kidneys were minced and further dissociated for 45 min in Iscove's Modified Dulbecco's Medium (IMDM) containing DNase 1 (1 µl/mL, Sigma Aldrich) and collagenase D (0,25 mg/mL, Roche Diagnostics) with a gentleMACS Octo Dissociator (Miltenyi Biotec). Cells were washed and lymphocytes were enriched with Percoll gradient centrifugation and erythrocyte lysis, as described before.
T cell Culture.
Spleens were isolated as described before. For some experiments, CD8+ T cells were labeled with biotinylated anti-CD8β mAb (clone Ly3.2, BD Bioscience) and purified by positive selection (EasySep Mouse Biotin Selection Kit, Stemcell Technologies) according to the manufacturer’s protocol. T cells were cultured in 96-well round-bottom plates in IMDM supplemented with 5% fetal calf serum (FCS), glutamine, gentamicin, and 2-mercaptoethanol, at 37 °C and 5% CO2. For activation, 2 × 105 T cells per well were incubated in wells coated with anti-CD3ε mAb (2μg/mL phosphate-buffered saline (PBS) overnight at 4 °C; clone 145-2C11, Biolegend). Medium-contained anti-CD28 mAb (1 µg/mL; clone 37.51, Biolegend) and cultures with purified CD8+ T cells were additionally supplemented with recombinant human IL-2 (100 U/mL, Proleukin S, Novartis). After 3 d, cells were washed and 5 × 104 cells per well were incubated with medium containing recombinant mouse IL-7 (10 ng/mL, Peprotech). To induce Cre recombination in vitro, T cells from Irf4fl/+×CreERT2 and Irf4fl/−×CreERT2 mice were incubated with 4-hydroxytamoxifen (2 μM, Sigma Aldrich) for 3 d.
For polyclonal stimulation, cells were transferred into 5 mL round bottom tubes and incubated in medium with phorbol 12 myristate 13 acetate (PMA) (50 ng/mL, Sigma Aldrich) and ionomycin (1 µM, Sigma Aldrich) for 4 h at 37 °C. For antigen-specific stimulation, cells were incubated with ovalbumin257–264 peptide (SIINFEKL, 10−6M, JPT). Brefeldin A (10 µg/mL, Sigma Aldrich) was added to the cultures to prevent cytokine secretion. In controls, medium only contained brefeldin A.
Proliferation was determined by loss of eFluor670 labeling. Spleen cells (1 × 107 cells/mL) were incubated in PBS with 2µM eFluor670 (eBioscience) for 10 min at 37 °C. Labeling was stopped by adding the fivefold volume of IMDM 10% FCS.
Antibody Staining and Flow Cytometry.
After culture or after isolation from tissue, cells were incubated on ice in PBS containing 1% rat serum and anti–Fc-receptor mAb (10 µg/mL, clone 2.4G2, BioXCell). For extracellular staining, fluorochrome-conjugated antibodies and a fixable dead cell stain (Pacific Orange succinimidyl ester, Life Technologies) were added. Cells were incubated for 15 min on ice. Intracellular antibody staining was conducted with the Foxp3/Transcription Factor Staining Buffer Set (eBioscience) according to the manufacturer’s protocol. For intracellular staining of Irf4fl/+×CreERT2 and Irf4fl/−×CreERT2 T cells, we fixed cells with 3.7% formaldehyde for 20 min at RT. Cells were washed with PBS 1% FCS, then permeabilized with 0.1% Igepal CA-630 (Sigma Aldrich) for 5 min at RT. Cells were washed with PBS 1% FCS and incubated with antibodies for intracellular staining in PBS 1% FCS for 20 min at RT.
Cells were sorted with a FACS AriaFusion or acquired with a BD Canto II or BD Celesta (BD Bioscience), and data were analyzed with the FlowJO 10 software (BD Bioscience).
Fluorochrome-conjugated antibodies against murine TCRβ (clone H57-597; BV605), CD4 (clone RM4-5; AF700, PerCP), CD8α (clone 53.6-7; BV650, PerCP), CD11b (clone M1/70; V500), CD19 (clone 6D5; V500), CD44 (clone IM7; PECy7), CD45 (clone 30F-11; PerCP), CD45.1 (clone A20; V500), CD62L (clone MEL-14; APCCy7), CD69 (clone H1.2F3; V450), CD90.1 (clone HIS51; V450), CD90.2 (clone 53-2.1; PE), CD127 (clone A7R34; PE), PD-1 (clone J43, PE), KLRG1 (clone 2F1; allophycocyanin [APC]), Ki-67 (clone SolA15, V450), Bcl-2 (clone 10C4; PECy7), Granzyme B (clone GB12; PE), EOMES (clone Dan11mag; APC), T-bet (clone 4B10; BV785), IRF4 (clone 3E4; PECy7), IFN-γ (clone XMG1.2; APCCy7), and TNF-α (clone MP6-XT22; V450) were obtained from Biolegend, eBioscience, Thermo Fisher or BD Bioscience. APC-conjugated H-2Kb/SIINFEKL dextramers were obtained from ImmuDex (Copenhagen, Denmark).
PCR for Modified Irf4 Alleles.
CreERT2-induced deletion of exon I and II was determined on FACS sorted GFP+ and GFP− CD8+ T cells from Irf4fl/+×CreERT2 and Irf4fl/−×CreERT2 mice after tamoxifen treatment. Genomic DNA was PCR amplified with primers directed against a region 5′ of Exon I (TGC CTT TGG GAC GGA TGC TC) and within Exon III (CAG AGC ACA TCG TAA TCT TGT CTT CC). Total RNA from sorted cells was isolated with the NucleoSpin Kit (Macharey-Nagel). RNA was reverse transcribed with the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher). Real-time qPCR using the SYBR green method was conducted with specific primers for exon II of Irf4 (fwd: GCA ATG GGA AAC TCC GAC AGT, rev: CAG CGT CCT CCT CAC GAT TGT) or for 18S ribosomal RNA (rRNA) (fwd: CAC GGC CGG TAC AGT GAA AC, rev: AGA GGA GCG AGC GAC CAA A). The measurement was performed on a StepOnePlus Real-Time PCR system (Thermo Fisher). Results of qPCR are given as values normalized to 18S rRNA and to the Irf4 expression of GFP− Irf4fl/+×CreERT2 cells.
Statistical Analyses.
Statistical analyses were performed with Prism software (GraphPad Software Inc). Results were analyzed with the tests indicated in the figure legends. In T cell transfer experiments with subsequent deletion of Irf4 alleles, we detected GFP+ cells and GFP− cells with deleted and nonmodified alleles, respectively. In these experiments, GFP+ and GFP− CD8+ T cells in individual mice were matched and results of groups of mice were analyzed with the paired t test. A P value of <0.05 was considered significant (*P < 0.05; **P < 0.01; ***P < 0.001; ns not significant).
Supplementary Material
Acknowledgments
We thank Dr. Antonia Zapf (Hamburg, Germany) for her advice on statistical analysis of complex data sets. We furthermore thank Drs. Dietmar Zehn (Freising, Germany), Hao Shen (Philadelphia, PA), Tobias Bopp (Mainz, Germany), Ulf Klein (Leeds, UK), and Hanna Taipaleenmäki (Hamburg, Germany) for providing bacteria strains and mouse lines. We are grateful to the animal facility of the University Medical Center Hamburg-Eppendorf for taking good care of our mice. This work benefitted from data assembled by the ImmGen consortium. This study was supported by grants from the Deutsche Forschungsgemeinschaft: GRK 841 to A.H. and C.S., SFB 841 and MI 471/7 to H.-W.M, and RA 2893/2 to F.R.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2014553118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Lohoff M., Mak T. W., Roles of interferon-regulatory factors in T-helper-cell differentiation. Nat. Rev. Immunol. 5, 125–135 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Huber M., Lohoff M., IRF4 at the crossroads of effector T cell fate decision. Eur. J. Immunol. 44, 1886–1895 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Man K., et al., The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat. Immunol. 14, 1155–1165 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Yao S., et al., Interferon regulatory factor 4 sustains CD8(+) T cell expansion and effector differentiation. Immunity 39, 833–845 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nayar R., et al., Graded levels of IRF4 regulate CD8+ T cell differentiation and expansion, but not attrition, in response to acute virus infection. J. Immunol. 192, 5881–5893 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Man K., Kallies A., Synchronizing transcriptional control of T cell metabolism and function. Nat. Rev. Immunol. 15, 574–584 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Krishnamoorthy V., et al., The IRF4 gene regulatory module functions as a read-write integrator to dynamically coordinate T helper cell fate. Immunity 47, 481–497.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glasmacher E., Agrawal S., Chang A. B.et al., A genomic regulatory element that directs assembly and function of immune-specific AP-1 - IRF complexes. Science 338, 975–980 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li P., et al., BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature 490, 543–546 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwata A., et al., Quality of TCR signaling determined by differential affinities of enhancers for the composite BATF-IRF4 transcription factor complex. Nat. Immunol. 18, 563–572 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciofani M., et al., A validated regulatory network for Th17 cell specification. Cell 151, 289–303 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Man K., et al., Transcription factor IRF4 promotes CD8+ T cell exhaustion and limits the development of memory-like T cells during chronic infection. Immunity 47, 1129–1141.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Kwon H., et al., Analysis of interleukin-21-induced Prdm1 gene regulation reveals functional cooperation of STAT3 and IRF4 transcription factors. Immunity 31, 941–952 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rengarajan J., et al., Interferon regulatory factor 4 (IRF4) interacts with NFATc2 to modulate interleukin 4 gene expression. J. Exp. Med. 195, 1003–1012 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaech S. M., Cui W., Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 12, 749–761 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wherry E. J., Kurachi M., Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 15, 486–499 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou X., et al., Differentiation and persistence of memory CD8(+) T cells depend on T cell factor 1. Immunity 33, 229–240 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raczkowski F., et al., The transcription factor interferon regulatory factor 4 is required for the generation of protective effector CD8+ T cells. Proc. Natl. Acad. Sci. U.S.A. 110, 15019–15024 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mittrücker H.-W., Matsuyama T., Grossman A.et al., Requirement for the Transcription Factor LSIRF/IRF4 for Mature B and T Lymphocyte Function. Science 275, 540–543 (1997). [DOI] [PubMed] [Google Scholar]
- 20.Grusdat M., et al., IRF4 and BATF are critical for CD8+ T cell function following infection with LCMV. Cell Death Differ. 21, 1050–1060 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyakoda M., et al., Differential requirements for IRF4 in the clonal expansion and homeostatic proliferation of naive and memory murine CD8+ T cells. Eur. J. Immunol. 48, 1319–1328 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Nayar R., et al., IRF4 regulates the ratio of T-bet to eomesodermin in CD8+ T cells responding to persistent LCMV infection. PLoS One 10, e0144826 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lynn R. C., et al., c-Jun overexpression in CAR T cells induces exhaustion resistance. Nature 576, 293–300 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nayar R., et al., TCR signaling via Tec kinase ITK and interferon regulatory factor 4 (IRF4) regulates CD8+ T cell differentiation. Proc. Natl. Acad. Sci. U.S.A. 109, E2794–E2802 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein U., et al., Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat. Immunol. 7, 773–782 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Hameyer D., et al., Toxicity of ligand-dependent Cre recombinases and generation of a conditional Cre deleter mouse allowing mosaic recombination in peripheral tissues. Physiol. Genomics 31, 32–41 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Salerno F., Guislain A., Cansever D., Wolkers M. C., TLR-mediated innate production of IFN-γ by CD8+ T cells is independent of glycolysis. J. Immunol. 196, 3695–3705 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Hombrink P., et al., Programs for the persistence, vigilance and control of human CD8+ lung-resident memory T cells. Nat. Immunol. 17, 1467–1478 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Mackay L. K., Minnich M., Kragten N. A. M.et al., Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 352, 459–463 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Heng T. S. P., Painter M. W.; Immunological Genome Project Consortium , The immunological genome project: Networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Zhao D.-M., Yu S., Zhou X.et al., Constitutive activation of wnt signaling favors generation of memory CD8 T cells. J Immunol. 184, 1191–1199 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Intlekofer A. M., et al., Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 6, 1236–1244 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Schenkel J. M., et al., T cell memory. Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science 346, 98–101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ariotti S., et al., T cell memory. Skin-resident memory CD8+ T cells trigger a state of tissue-wide pathogen alert. Science 346, 101–105 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Mombaerts P., et al., RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68, 869–877 (1992). [DOI] [PubMed] [Google Scholar]
- 36.Foulds K. E., et al., Cutting edge: CD4 and CD8 T cells are intrinsically different in their proliferative responses. J. Immunol. 168, 1528–1532 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Rissiek B., et al., In vivo blockade of murine ARTC2.2 during cell preparation preserves the vitality and function of liver tissue-resident memory T cells. Front. Immunol. 9, 1580 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anderson K. G., et al., Intravascular staining for discrimination of vascular and tissue leukocytes. Nat. Protoc. 9, 209–222 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.