Significance
Primary cilia (PC) are sensory organelles essential for the development and maintenance of adult tissues. Accordingly, dysfunction of PC causes human disorders called ciliopathies. Hence, a thorough understanding of the molecular regulation of PC is critical. Our findings highlight CSNK2A1 as a modulator of cilia trafficking and stability, tightly related to TTBK2 function. Enriched at the centrosome, CSNK2A1 prevents abnormal accumulation of key ciliary proteins, instability at the tip, and aberrant activation of the Sonic Hedgehog pathway. Furthermore, we establish that Csnk2a1 mutations associated with Okur-Chung neurodevelopmental disorder (OCNDS) alter cilia morphology. Thus, we report a potential linkage between CSNK2A1 ciliary function and OCNDS.
Keywords: primary cilia, CSNK2A1, TTBK2, OCNDS, actin modulators
Abstract
Cilia biogenesis is a complex, multistep process involving the coordination of multiple cellular trafficking pathways. Despite the importance of ciliogenesis in mediating the cellular response to cues from the microenvironment, we have only a limited understanding of the regulation of cilium assembly. We previously identified Tau tubulin kinase 2 (TTBK2) as a key regulator of ciliogenesis. Here, using CRISPR kinome and biotin identification screening, we identify the CK2 catalytic subunit CSNK2A1 as an important modulator of TTBK2 function in cilia trafficking. Superresolution microscopy reveals that CSNK2A1 is a centrosomal protein concentrated at the mother centriole and associated with the distal appendages. Csnk2a1 mutant cilia are longer than those of control cells, showing instability at the tip associated with ciliary actin cytoskeleton changes. These cilia also abnormally accumulate key cilia assembly and SHH-related proteins. De novo mutations of Csnk2a1 were recently linked to the human genetic disorder Okur-Chung neurodevelopmental syndrome (OCNDS). Consistent with the role of CSNK2A1 in cilium stability, we find that expression of OCNDS-associated Csnk2a1 variants in wild-type cells causes ciliary structural defects. Our findings provide insights into mechanisms involved in ciliary length regulation, trafficking, and stability that in turn shed light on the significance of cilia instability in human disease.
Primary cilia (PC) are microtubule-based cellular projections that function as vital cellular-signaling organelles, transducing signals through a variety of important developmental pathways. In addition to their roles in embryonic development, cilia also persist on the cells of most adult tissues (1–4). PC are dynamic organelles, and bidirectional trafficking of proteins and other materials within cilia by the process of intraflagellar transport (IFT) is vital for the maintenance of ciliary structure and signaling (5–7). However, little is known about the processes and pathways upstream of IFT that control the presence or absence of a cilium. In prior work, we identified a kinase, TTBK2, that is required for ciliogenesis. TTBK2 mediates both the recruitment of IFT proteins to the mother centriole and removal of centrosomal proteins that suppress cilium assembly (8). TTBK2 is also important for maintaining the stability of the ciliary axoneme (9), and both of these requirements are linked to a role for TTBK2 in a human neurodegenerative disease (4, 10). A limited number of interactors and effectors of TTBK2 in ciliogenesis have been identified, including the distal appendage proteins CEP164 (11, 12) and CEP83 (13), and a suppressor of ciliogenesis, M-phase phosphoprotein 9 (14). However, the pathway through which TTBK2 functions to mediate cilium assembly and stability is largely undefined.
Here, to uncover additional players in this pathway, we undertook two distinct unbiased approaches to identify previously unknown physical and functional interactors of TTBK2: proximity-dependent biotin identification (BioID) to identify TTBK2-proximate proteins, and a CRISPR-based screen to uncover genetic modifiers of a hypomorphic Ttbk2 phenotype (9). We identified Casein kinase II subunit alpha (CSNK2A1), the main catalytic subunit of casein kinase II (CK2) (15), in both screens, making it an attractive candidate as a modulator of TTBK2 function in cilia regulation.
CK2 is a widely expressed serine-threonine kinase involved in many cellular processes, including cell-cycle progression, circadian-clock regulation, and WNT signaling (16–20). Mutations in Csnk2a1 are associated with Okur-Chung neurodevelopmental syndrome (OCNDS), a dominantly inherited disorder characterized by dysmorphic facial features and neurological impairments (21–26). We show that CSNK2A1 acts in opposition to TTBK2 in establishing and maintaining ciliary structure, with knockout of Csnk2a1 partially rescuing ciliary phenotypes of Ttbk2 hypomorphic mutant cells. We also demonstrate that CSNK2A1 mainly localizes to the mother centriole and is required for cilia structure and stability. Csnk2a1-knockout cells have long cilia that exhibit a variety of defects in trafficking and actin dynamics. Additionally, overexpression of Csnk2a1 variants associated with OCNDS causes ciliary structural abnormalities. Thus, we reveal a previously undescribed role for CSNK2A1 in cilia homeostasis, define a kinase network that regulates the stability of PC, and identify links between cilia integrity and human disease.
Results
CSNK2A1 Is a Modulator of TTBK2 at the Mother Centriole.
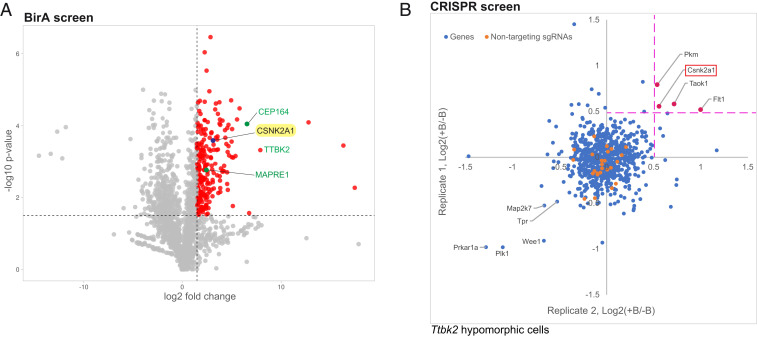
In order to identify additional components of the TTBK2-dependent pathways that regulate ciliary assembly and/or stability, we undertook two unbiased screens: A BioID proximity-labeling screen to find proteins that physically interact with TTBK2 and might, therefore, be effectors or upstream regulators of this crucial kinase (Fig. 1A); and a CRISPR loss-of-function screen to identify proteins that functionally interact with TTBK2 (Fig. 1B). In our proximity-labeling screen, we tagged TTBK2 at the N terminus with the biotin ligase BirA, mutated to be constitutively active (27). We verified that BirA*-tagged TTBK2 is active and able to perform its normal cellular function mediating cilium assembly (SI Appendix, Fig. S1A). We then inducibly expressed TTBK2-BirA* in HEK-293T cells, labeled cells with biotin, and carried out cell streptavidin affinity purification and mass spectrometry (SI Appendix, Fig. S1 B and C). Using this approach, we identified 370 proteins that were enriched by twofold or more in cells expressing TTBK2-BirA* compared with the GFP-BirA* negative control (Fig. 1A and Dataset S1). Of these, 351 were statistically significant at a false discovery rate of 0.01.
Fig. 1.
Identification of CSNK2A1 as a putative TTBK2-interacting protein. (A) Volcano plot of TTBK2-proximate proteins labeled by BioID. CEP164, TTBK2, and MAPRE1, known interactors of TTBK2, are shown in green. CSNK2A1, a candidate TTBK2-proximate protein is highlighted in yellow. The x-axis denotes the log2 fold change of TTBK2:GFP control. In the y-axis, the significance displays the negative log10-transformed P value for each protein. The red dots show the proteins considered as hits, delimited by the selected thresholds ≥ 1.5 for the log2 fold change and statistical significance. (B) The CRISPR screen plot for Ttbk2null/gt cells. The suppressor screen identifies potential negative modifiers of TTBK2, including CSNK2A1 (red). Each gene is plotted using the mean of medians values (defined as the two median ratios of sgRNAs counts, calculated as follows: +blasticidin(+SAG): −Blasticidin(+SAG); note that four sgRNAs represent each gene). Orange dots represent the nontargeting sgRNAs.
In parallel, we performed a CRISPR loss-of-function kinome screen to identify additional kinases that cooperate with TTBK2 in cilium assembly. We employed a recently described Hedgehog (HH) pathway-responsive reporter, in which 8× GLI response elements drive the expression of blasticidin resistance genes (28). We expressed the reporter and a mouse-kinome small guide RNA (sgRNA) library [no. 75316, Addgene; (29)] in wild-type (WT) mouse embryonic fibroblasts (MEFs), as well as MEFs derived from hypomorphic Ttbk2 mutant embryos [Ttbk2null/gt transheterozygous embryos (9) (SI Appendix, Fig. S2A)]. We defined a hit in our screen as any sgRNAs that became enriched within the population following blasticidin selection in the Ttbk2null/gt cells but not in the WT MEFs. In other words, hits were sgRNAs that specifically improve the responsiveness of cells to HH pathway activation when TTBK2 activity is impaired. This approach would predominantly identify negative regulators of TTBK2 function either in cilium initiation or stability or both (Fig. 1B, SI Appendix, Fig. S2B, and Dataset S2).
CSNK2A1, the catalytic subunit of CK2, was identified as a hit in both of these screens (Fig. 1 A and B). Because it was the only protein identified by both of our approaches, we prioritized CSNK2A1 for further analysis. To examine the relationship between CSNK2A1 and TTBK2, we verified that these two kinases associate during ciliogenesis. We coexpressed TTBK2-V5 with GFP or CSNK2A1-GFP in serum-starved HEK-293T cells. We detected an interaction between the two fusion proteins by co-immunoprecipitation, whereas no interaction was observed in the control pulldown (Fig. 2A). This suggests that these two kinases interact either directly, or as part of a complex with additional proteins.
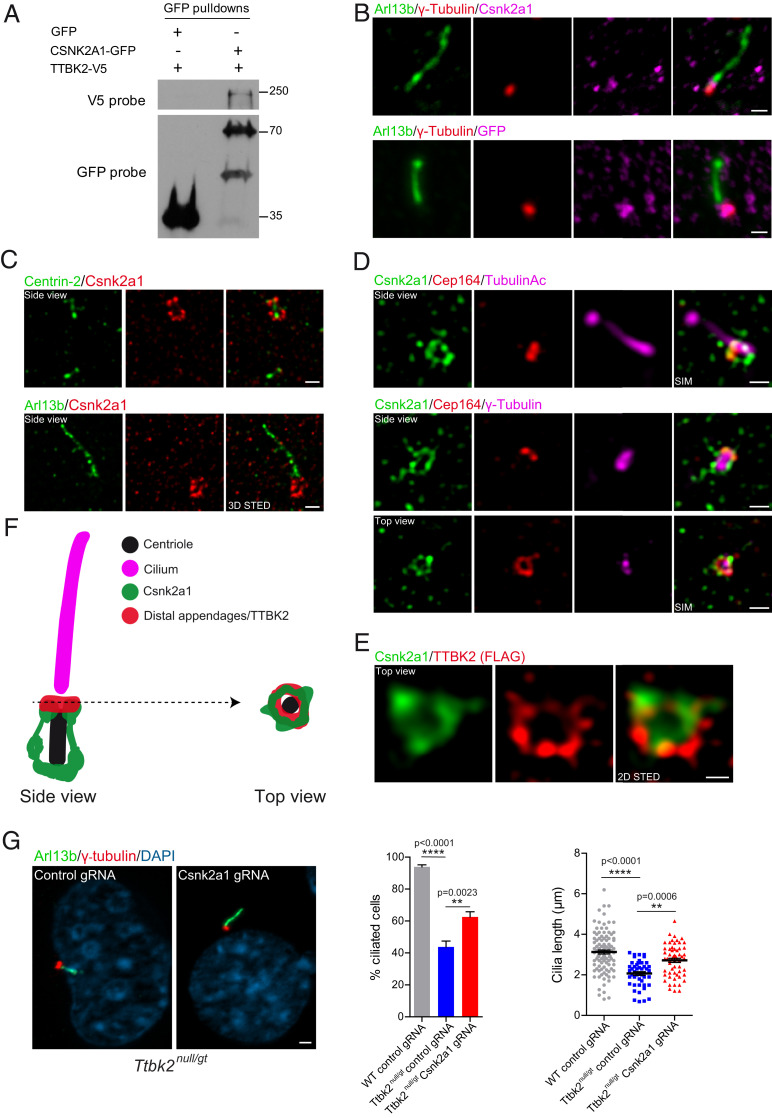
Fig. 2.
CSNK2A1 is a centrosomal protein that opposes TTBK2 function. (A) TTBK2-V5 was overexpressed with GFP or CSNK2A1-GFP in HEK293T cells. GFP pulldowns were performed on total cell lysates, and Western blots were probed with antibodies to V5 and GFP tags. (B) Endogenous (Upper) and GFP-tagged (Lower, stably expressed, both magenta) CSNK2A1, costained with the centrosomal protein γ-tubulin (red) and ciliary marker ARL13B (green) in WT MEFs. (Scale bars, 1 μm.) (C) 3D STED superresolution images show the endogenous CSNK2A1 (red) surrounding the centriole core (Centrin-2 green, Upper). Lower shows CSNK2A1 signal at the vicinity of the cilium. (Scale bars, 500 nm.) (D) SIM acquisitions show that CSNK2A1 exhibits a complex basal body-associated organization. (Upper) A side view of CSNK2A1 (green) at the base of the cilium labeled with acetylated alpha-tubulin (magenta) and partially overlapping with CEP164 (red). (Lower) CSNK2A1 tightly surrounds the pericentriolar material, labeled by γ-tubulin (magenta), partially overlapping with CEP164 (Upper and Lower). (Scale bars, 500 nm.) (E) Representative two-dimensional STED images from WT MEFs, exhibiting the top view of CSNK2A1 (green), in a ring-shaped structure and colocalizing with TTBK2 (red). (Scale bar, 250 nm.) (F) Cartoon illustrating the subcellular organization of CSNK2A1 at the basal body. (G) Cilia defects in Ttbk2null/gt cells are partially rescued by Csnk2a1 knockout. Representative immunofluorescence images from cells labeled with ARL13B (Green) and γ-tubulin (red). DNA was stained with DAPI. (Scale bar, 2 μm.) Graphs show the mean percentage of ciliated cells (left graph: WT control guide RNA (gRNA) n = 105; Ttbk2null/gt control gRNA n = 251; and Ttbk2null/gt Csnk2a1 gRNA n = 282 cells) and cilia length (right graph: WT control gRNA n = 105; Ttbk2null/gt control gRNA n = 48; and Ttbk2null/gt Csnk2a1 gRNA n = 58 cells). Error bars denote the SEM. Statistical comparison was performed by one-way ANOVA with Tukey’s multiple comparisons test. P values are shown on the graph.
TTBK2 localizes to the distal appendages of the mother centriole prior to the initiation of cilium assembly (8, 13, 30). To resolve the precise subciliary localization of CSNK2A1 and assess whether CSNK2A1 colocalizes with TTBK2 in that compartment, we performed conventional and superresolution immunofluorescence microscopy using antibodies against endogenous CSNK2A1, as well as a GFP fusion construct. With both approaches, we found that CSNK2A1 is specifically enriched at the centrosome, consistent with previous findings (31) (Fig. 2B and SI Appendix, Fig. S3). We then used three-dimensional stimulated emission depletion (STED) microscopy to examine endogenous CSNK2A1 localization relative to the centriolar core component Centrin-2 and the ciliary membrane protein ARL13B. This analysis revealed that CSNK2A1 predominantly localizes to the mother centriole at the vicinity of the axoneme (Fig. 2C). CSNK2A1 signal shows a distinctive localization pattern, wherein it surrounds the mother centriole in a barrel-like fashion (Fig. 2C). We distinguished two main loci of the CSNK2A1 signal. A symmetrical arc-like structure is found at the proximal end of the mother centriole. Fainter parallel linkers to centriolar sidewalls connect the proximal CSNK2A1 to a tighter distal locus (Fig. 2 C and F). In addition, we detect a weaker signal of the kinase at the daughter centriole (Fig. 2 C, Upper).
To further characterize the localization of CSNK2A1 at the distal mother centriole, we analyzed its positioning relative to the distal appendage protein CEP164 together with γ-tubulin and acetylated α-tubulin using the structured illumination microscopy (SIM) (Fig. 2D). In a side view, we observed an identical pattern to that obtained from colabeling with Centrin-2. In a top-down view, we noted a ring of CSNK2A1 partially overlapping with that of CEP164 (Fig. 2 D and F). Top-view acquisition costained with TTBK2 confirmed partial colocalization between CSNK2A1 and TTBK2 rings at the mother centriole (Fig. 2E) and indicates that CSNK2A1 is a centriolar distal appendage-associated protein (Fig. 2 D–F). These results show that CSNK2A1 and TTBK2 distal rings exhibit similar positioning at the distal appendages, with evidence of a more complex organization for CSNK2A1 (Fig. 2F).
We identified CSNK2A1 from a CRISPR screen for genes that modify the function of TTBK2, specifically as a gene that, when knocked out, increases the ability of hypomorphic Ttbk2null/gt cells to activate a HH signaling response (Fig. 1B). Ttbk2null/gt cells form cilia at a reduced frequency, and these cilia have defects in IFT protein localization as well as structural abnormalities (9). Therefore, we tested whether the loss of CSNK2A1 function is sufficient to rescue or partially rescue cilia defects seen in Ttbk2null/gt cells. To this end, we tested five different sgRNAs against Csnk2a1, from which we chose the most efficient sgRNA and selected individual clones to validate Csnk2a1 knockout by Western blot and sequencing (SI Appendix, Fig. S4A). Simultaneously, we isolated a control clonal cell line, generated by a nontargeting sgRNA. Consistent with the identification of Csnk2a1 in our CRISPR screen, sgRNA-mediated knockout of Csnk2a1 in Ttbk2null/gt cells partially rescued defects in cilia frequency as well as cilia length (Fig. 2G).
Loss of CSNK2A1 Perturbs Ciliary Trafficking.
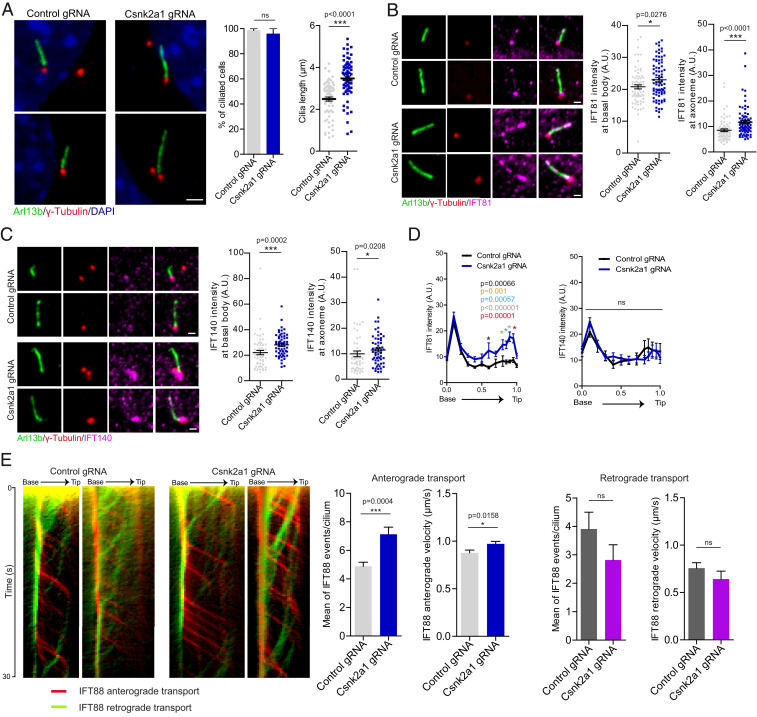
To further examine the requirements for CSNK2A1 in cilia assembly and function, we generated Csnk2a1-knockout cell lines in WT MEFs using the CRISPR-Cas9 system. Two clonal cell lines were isolated with distinct frameshift mutations within the Csnk2a1 gene (SI Appendix, Fig. S4B). The frequency of cilia formation was not affected in the absence of CSNK2A1. However, Csnk2a1-depleted cells formed longer cilia with a wide range of lengths, suggesting possible ciliary instability (Fig. 3A).
Fig. 3.
CSNK2A1 regulates cilia length and IFT trafficking. (A) Csnk2a1 knockout cells form longer cilia. Representative immunofluorescence images stained for to ARL13B (green) and γ-tubulin (red), from control and Csnk2a1 KO-1 cells as indicated. Graphs show the mean percentage of ciliated cells and the average cilia length ± SEM. Statistical comparison was performed using the nonparametric Mann–Whitney U test. P values are displayed on the graph (ns, not significant). The total number of cells analyzed for both graphs is 69 cells per condition. (Scale bar, 2 μm.) (B) IFT81 accumulates at the basal body and cilia in Csnk2a1 KO-1 cells. Representative immunofluorescence images labeled with ARL13B (green), γ-tubulin (red), and IFT81 (magenta). Graphs show the mean intensity of IFT81 ± SEM at the basal body (left graph) and the axoneme (right graph) in control and Csnk2a1 KO-1 cells. Statistical comparison was performed using the nonparametric Mann–Whitney U test. P values are shown on the graph. Cell numbers are as follows: Basal body, control gRNA n = 72 and Csnk2a1 gRNA n = 79 cells; axoneme, control gRNA n = 71 and Csnk2a1 gRNA n = 79 cells. (Scale bars, 1 μm.) (C) IFT140 accumulates at the basal body and cilia in Csnk2a1 KO-1 cells. Representative immunofluorescence of cilia stained with ARL13B (green), γ-tubulin (red), and IFT140 (magenta). Graphs show the mean intensity of IFT140 ± SEM at the basal body (left graph) and the axoneme (right graph) in control and Csnk2a1 KO-1 cells. Statistical comparison was performed using the nonparametric Mann–Whitney U test. P values are shown on the graph. Basal body, control gRNA n = 58 and Csnk2a1 gRNA n = 57 cells; axoneme, control gRNA n = 56 and Csnk2a1 gRNA n = 57 cells. (Scale bars, 1 μm.) (D) IFT81, but not IFT140, accumulates in the distal cilium of Csnk2a1 KO-1 cells. The mean intensity of IFT81 (left graph) and IFT140 (right graph) ± SEM is profiled along the cilium. All cilia lengths were normalized to 1 and displayed on the x-axis. Statistical comparison was executed by multiple t tests. P values are shown on the graph (ns, not significant). Left graph, control gRNA n = 28 and Csnk2a1 gRNA n = 29 cells; right graph, control gRNA n = 30 and Csnk2a1 gRNA n = 35 cells. (E) The anterograde trafficking of IFT88-GFP, but not the retrograde, is increased in Csnk2a1 KO-1 cells. Representative kymographs of IFT88-GFP for anterograde (red) and retrograde (green) trafficking are shown from the base to the ciliary tip for a duration of 30 s. Graphs exhibit the mean number of IFT88-GFP events per cilium and their velocities. Error bars denote the SEM. Statistical comparison was performed using the nonparametric Mann–Whitney U test. P values are shown on the graph (ns, not significant). Number of cilia quantified is as follows: anterograde transport, control gRNA n = 18 and Csnk2a1 gRNA n = 15 cilia; retrograde transport, control gRNA n = 15 and Csnk2a1 gRNA n = 13 cilia.
Cilia formation is a multistep process with TTBK2 acting upstream of IFT complexes to trigger the final steps required for the conversion of the mother centriole into a basal body (8). Since CSNK2A1 partially opposes TTBK2 function, we evaluated IFT recruitment and localization of Csnk2a1-knockout cilia. In contrast with Ttbk2 mutant cells, components of the IFT-B complex, IFT81 and IFT88, are seen at abnormally high levels within Csnk2a1-knockout cilia, especially within the more distal axoneme (Fig. 3 B and D and SI Appendix, Fig. S4 C and D). We also assessed the localization and intensity of IFT140, a component of the IFT-A complex, which is implicated in retrograde transport as well as in the trafficking of specialized ciliary membrane proteins and HH pathway components (32, 33). In WT cells, IFT140 accumulates primarily at the ciliary base. In Csnk2a1-knockout cilia, we do not observe substantial changes in localization pattern; however, there is an increase in IFT140 intensity at the ciliary base and the axoneme (Fig. 3 C and D).
We postulated that the changes we observed in IFT localization may point to defects in intraciliary transport. To examine the bidirectional IFT, we monitored IFT88-GFP within the cilium by live imaging and measured anterograde and retrograde dynamics (Fig. 3E). Mutant cilia had a greater number of IFT88 tracks moving toward the ciliary tip, with increased velocity. Retrograde measurements did not show a significant change compared to control cilia (Fig. 3E). Altogether, these results suggest that CSNK2A1 regulates IFT levels in PC.
Trafficking of HH Pathway Components and Signaling Are Increased in Csnk2a1-Knockout Cilia.
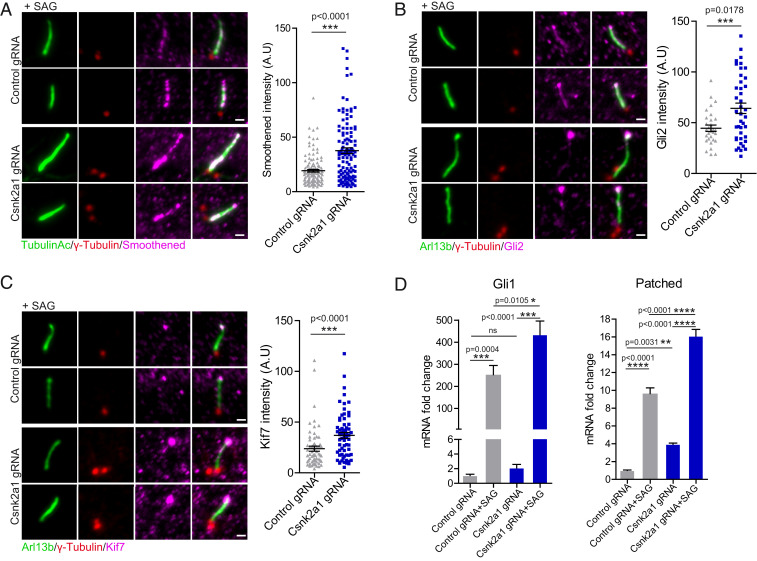
Given our observation of abnormal ciliary trafficking in Csnk2a1-knockout cells, we assessed whether the ciliary trafficking of key HH pathway components, including SMO, GLI2, and KIF7, is affected by the loss of Csnk2a1. Upon activation of HH signaling, the transmembrane protein SMO becomes enriched throughout the ciliary membrane (34). GLI2, a transcriptional activator of the pathway, becomes enriched at the distal tip of cilia (35), as does the atypical kinesin KIF7, which regulates the structure of the ciliary tip compartment and participates in HH signaling (36, 37). For each of these proteins, we observed that their accumulation upon treatment with SMO agonist (SAG) was increased in Csnk2a1-knockout cilia (Fig. 4 A–C).
Fig. 4.
CSNK2A1-mutant cilia exhibit increased SHH response. (A) SMO accumulates in Csnk2a1-mutant cilia. Representative immunofluorescence images stained for acetylated tubulin (green), γ-tubulin (red), and SMO (magenta), from control and Csnk2a1 KO-1 cells. The graph shows the mean intensity of SMO ± SEM along the cilium in control and Csnk2a1 KO-1 cells. Statistical comparison was performed using the nonparametric Mann–Whitney U test. P value is displayed on the graph. Data collected from control gRNA n = 123, Csnk2a1 gRNA n = 120 cilia. (Scale bars, 1 μm.) (B) Csnk2a1-mutant cilia exhibit higher levels of GLI2 at the tip. Representative immunofluorescence images labeled with antibodies to ARL13B (green), γ-tubulin (red), and GLI2 (magenta), from control and Csnk2a1 KO-1 cells. Graph shows the mean intensity of GLI2 ± SEM at the ciliary tip in control and Csnk2a1 KO-1 cells. Statistical comparison was performed using the nonparametric Mann–Whitney U test. P value is displayed on the graph. Data points collected from control gRNA n = 30 cilia, Csnk2a1 gRNA n = 41 cilia. (Scale bars, 1 μm.) (C) Csnk2a1-mutant cilia show increased levels of KIF7 at the ciliary tip. Representative immunofluorescence images of cilia labeled with antibodies to ARL13B (green), γ-tubulin (red), and KIF7 (magenta), from control and Csnk2a1 KO-1 cells. Graph shows the mean intensity of KIF7 ± SEM. Statistical comparison was performed using the nonparametric Mann–Whitney U test. P value is shown on the graph. Examined data points were collected from control gRNA n = 63 cilia, Csnk2a1 gRNA n = 64 cilia. (Scale bars, 1 μm.) (D) Real-time qPCR shows a significant increase in messenger RNA (mRNA) fold change (relative to untreated control) ± SEM of Gli1 and patched transcripts in Csnk2a1 KO-2 cells. Graphs were produced from three replicates per condition from two independent experiments. Statistical comparison was performed by two-way analysis ANOVA with Tukey’s multiple comparisons test. P values are shown on the graph (ns, not significant).
Increased accumulation or trafficking of HH pathway components into the cilium is often associated with changes in the transcriptional output of the pathway (38, 39). To test whether this is the case for the Csnk2a1-knockout cells, we assessed the expression of two direct transcriptional targets of the HH pathway, Ptch and Gli1, using qPCR. Consistent with the increased accumulation of HH pathway components within the Csnk2a1-knockout cilia, we observed higher expression levels of both Ptch and Gli1 upon SAG treatment in Csnk2a1-knockout cell lines relative to WT (Fig. 4D and SI Appendix, Fig. S5A). In addition, we also noted that levels of Ptch1 are higher in the Csnk2a1-knockout cells relative to WT even in the absence of SAG, suggesting that the basal levels of HH might be higher in the absence of CSNK2A1 (Fig. 4D). Altogether, these results suggest that CSNK2A1 is a negative regulator of Sonic Hedgehog (SHH) pathway.
CSNK2A1 Is Required for Cilium Stability.
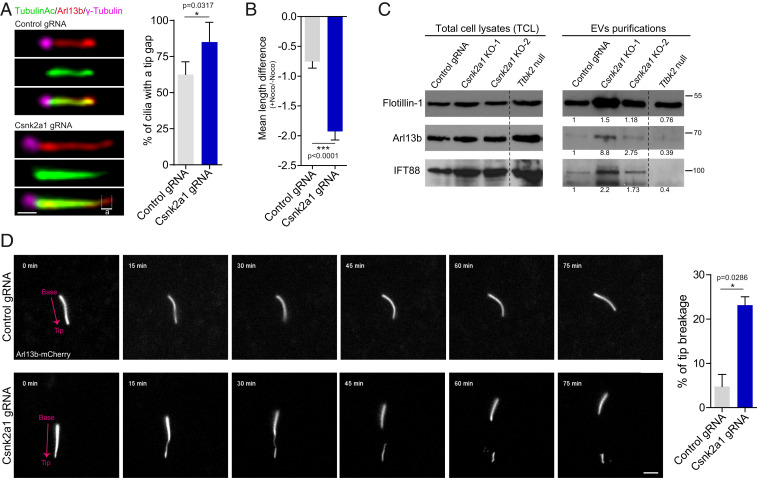
Because Csnk2a1-knockout cilia are elongated and exhibit a variety of trafficking defects, we further examined the stability of these cilia. Csnk2a1-depleted cells display a higher percentage of cilia with a tip gap that is positive for ARL13B but negative for acetylated α-tubulin (Fig. 5A). This gap, in addition to the accumulation of IFT81, GLI2, and KIF7 in mutant cilia, points to abnormalities in the ciliary tip compartment. To test whether defects at the ciliary tip were associated with decreased ciliary stability, we incubated the cells with nocodazole (Noco) to induce microtubule depolymerization and to challenge the axonemal integrity of the cilium. We then calculated the ciliary length difference between the Noco-treated and the Noco-untreated conditions and noted that mutant cilia are less stable than control cilia (Fig. 5B). Taken together, these data indicate that Csnk2a1-mutant cilia have an abnormal ciliary tip compartment and are unstable.
Fig. 5.
CSNK2A1 is essential for tip stability. (A) A tip gap is a tip positive for ARL13B and negative for acetylated alpha-tubulin (illustrated by ‘’a’’). (Scale bars, 1 μm.) Graph shows the mean percentage of cilia that display tip gaps ± SEM (47 control cilia and 49 mutant cilia). (B) Under Noco treatment, mutant cilia are less stable than control cilia. Cells were serum starved for 48 h and then treated with 10 μM Noco or dimethyl sulfoxide (DMSO) as a control for 35 min. Cilia length was collected and data were displayed as a mean difference of cilia length [+Noco]−[−Noco] ± SEM. Measurements were produced from 74 control cilia and 70 Csnk2a1 mutant cilia. (C) Csnk2a1-knockout cells shed more ciliary EVs. Western blots from total cell lysates (TCL, left blots) and EVs purifications (right blots) are probed with antibodies to FLOTILLIN-1, ARL13B, and IFT88. For each probe, the fold change value was calculated as follows: ratio of [EVs_Area under the curve]:[TCL_Area under the curve]. The fold change was normalized to the control value. Dashed line marks where the Western blot images were cut to remove experimental conditions that were not relevant to the current study. (D) Csnk2a1-knockout cilia are prone to tip breakage. Nonclonal control and Csnk2a1-depleted cells were generated in transgenic ARL13B-mCherry MEFs. Cilia were imaged every 15 min for 75 min. Breakage events were quantified and normalized to the total number of imaged cilia. The graph represents the mean percentage of tip breakage ± SEM for control (n = 51 cilia) and Csnk2a1-depleted cells (n = 63 cilia). (Scale bars, 3 μm.)
The ciliary tip is also a site of ectocytosis, a process by which vesicles are shed from the cilium. This process is linked to signaling from PC, and regulation of ectocytosis is important for controlling the ciliary length and for maintaining stability (40–44). To test whether ciliary scission and ectocytosis are affected in the absence of CSNK2A1, we purified extracellular vesicles (EVs) from supernatants collected from serum-starved control and Csnk2a1-knockout cells. Nonciliated cells can shed EVs (45), however, under our conditions, EVs are enriched for ciliary proteins, suggesting shedding through tip scission (41). We noted that among the supernatants, we observed higher levels of FLOTILLIN, a marker of EVs, as well as IFT88 and ARL13B in the Csnk2a1-knockout cells relative to control cells WT cells (Fig. 5C). Ttbk2 null cells, which do not form PC (8), have FLOTILLIN levels in the supernatant at levels comparable to WT cells; however, we observed a dramatic reduction of ciliary-associated proteins recovered in this fraction. Additionally, ciliary tip markers GLI2 and IFT81 were also found specifically enriched in supernatants from Csnk2a1-knockout cells (SI Appendix, Fig. S5B). Taken together, these data suggest that Csnk2a1-mutant cilia exhibit an increased amount of ciliary scission and/or EV formation.
To extend these findings and examine how the loss of CSNK2A1 might affect cilium stability through ciliary scission, we performed live imaging of cilia in MEFs derived from a transgenic mouse line expressing ARL13B-mCherry (46) and transduced with previously described lentiviruses coding for control or Csnk2a1 sgRNAs. After selection, we validated by immunofluorescence a 60% depletion of CSNK2A1. Csnk2a1-depleted cilia frequently showed a thinning of the distal axoneme followed by breakage of the ciliary tip. Compared to control cells, mutant cilia showed an average increase of ∼fivefold in the frequency of tip breakage/scission (Fig. 5D and Movies S1 and S2).
CSNK2A1 Regulates Actin Modulators and Ciliary Actin Dynamics.
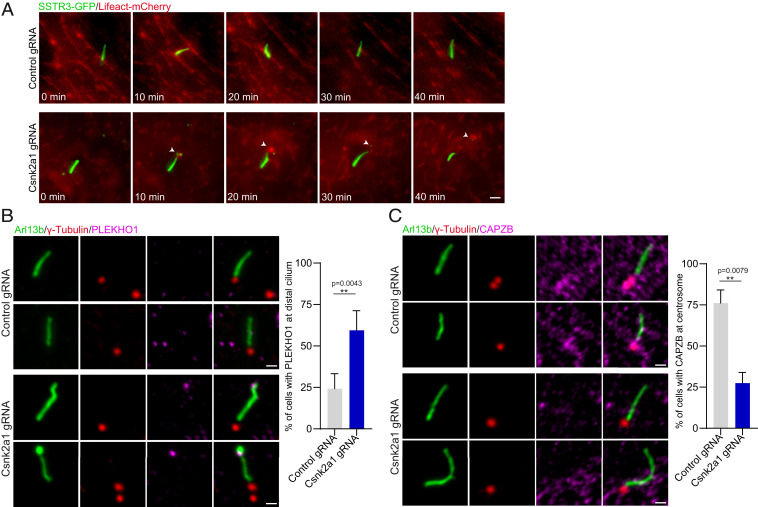
Previously, actin and actin-binding proteins have been linked to ciliary scission (41, 42). We therefore examined whether the ciliary breaks observed in the Csnk2a1-knockout cells were driven by changes in the actin cytoskeleton within the PC. Live imaging of cells expressing SSTR3-GFP to label cilia and Lifeact-mCherry (F-actin) showed that F-actin was transiently recruited to the breakage site in Csnk2a1-knockout cilia, prior to the excision event (Fig. 6A and Movies S3 and S4). Similarly, induction of cilia disassembly by serum readdition led to a distal enrichment of F-actin in Csnk2a1-knockout cilia (SI Appendix, Fig. S5C). These results indicate that CSNK2A1 is required for the integrity of the ciliary tip and that the protein depletion causes spontaneous ciliary tip excision, likely driven by changes in actin cytoskeleton dynamics.
Fig. 6.
Actin and actin modifiers are perturbed in Csnk2a1-mutant cilia. (A) F-actin transiently localizes to mutant cilia prior to cilia excision. SSTR3-GFP and Lifeact-mCherry were overexpressed in control and Csnk2a1 KO-1 cell lines. Time-lapse imaging was performed on ciliated cells every 10 min for 40 min. (Scale bars, 2 μm.) (B) PLEKHO1 abnormally localizes to the distal domain of Csnk2a1-mutant cilia. Representative immunofluorescence images labeled with ARL13B (green), γ-tubulin (red), and PLEKHO1 (magenta). Graphs show the mean percentage of cells with PLEKHO1 at distal cilium in control and Csnk2a1 KO-1 cells. Statistical comparison was performed using the nonparametric Mann–Whitney U test. P values are shown on the graph. Cell number details: control gRNA n = 37 and Csnk2a1 gRNA n = 65 cells. (Scale bars, 1 μm.) (C) CAPZB centrosomal localization is disrupted in Csnk2a1-knockout cilia. Representative immunofluorescence images labeled ARL13B (green), γ-tubulin (red), and CAPZB (magenta). Graphs show the mean percentage of cells with CAPZB at the centrosome in control and Csnk2a1 KO-1 cells. Statistical comparison was performed using the nonparametric Mann–Whitney U test. P values are shown on the graph. Cell number details: control gRNA n = 37 and Csnk2a1 gRNA n = 65 cells. (Scale bars, 1 μm.)
Several studies have reported that CSNK2A1 regulates actin cytoskeleton through actin modulators such as PLEKHO1 (pleckstrin homology domain containing O1), cortactin, and capping proteins (47–50). To investigate how CSNK2A1 present at the distal appendages regulates the ciliary tip stability, we examined the localization of PLEKHO1 and CAPZB (F-actin-capping protein subunit beta) in control and Csnk2a1-knockout cilia. PLEKHO1 is a bona fide interactor of CK2 (49), and both proteins are reported to inhibit the activity of actin-capping proteins at the barbed ends, thereby promoting polymerization of branched actin (48). In control cilia, there was no detectable signal of PLEKHO1 in the axoneme. In contrast, a localized enrichment of the protein was detected specifically at the distal end of Csnk2a1-knockout cilia (Fig. 6B). Additionally, the capping-protein subunit CAPZB was enriched at the centrosome in control cells but was absent from this organelle in Csnk2a1-knockout cells (Fig. 6C). These changes correlate with a distally localized F-actin formation at the breakage sites of Csnk2a1-mutant cilia (Fig. 6A), supporting a model where CSNK2A1 regulates the ciliary tip stability through the modulation of actin cytoskeleton dynamics. To investigate whether CSNK2A1 kinase activity is implicated in the regulation of PLEKHO1 and CAPZB localization, we generated a stable MEF cell line expressing a kinase-dead variant of CSNK2A1 (CSNK2A1K68M) (51) and compared the localization of actin modulators in cilia with that of cilia from cells expressing CSNK2A1WT (SI Appendix, Fig. S5D). While no change was observed with CAPZB localization, PLEKHO1 was distally localized in cilia from cells expressing CSNK2A1K68M. This suggests a dominant-negative effect of the kinase-dead mutation on the localization of PLEKHO1 but not that of CAPZB (SI Appendix, Fig. S5D). Similarly, we looked at cells stably expressing CSNK2A1D156H, an OCNDS-associated mutation located at the active site (52), and did not observe any change in PLEKHO1 and CAPZB localizations (SI Appendix, Fig. S5D). In comparison to the kinase-dead mutation, the OCNDS mutation may not completely inactivate the kinase activity, which may explain the absence of the dominant-negative effect in this context.
OCNDS-Associated Csnk2a1 Mutations Cause Defects in Cilia Morphology.
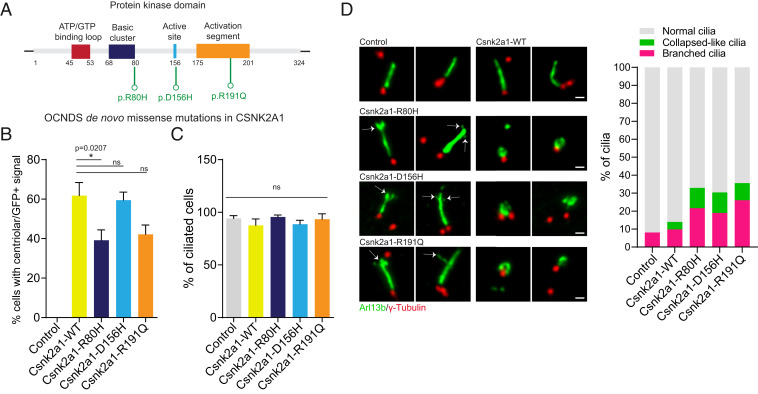
Dominant mutations in Csnk2a1 are associated with the human genetic disorder OCNDS (21–26). Because there is some phenotypic overlap between OCNDS and ciliopathies, and because we have shown that Csnk2a1 is required for normal ciliary trafficking and structure, we tested whether the OCNDS mutations are associated with ciliary defects. We stably expressed mouse CSNK2A1WT as well as three different OCNDS-associated variants: R80H, D156H, and R191Q (Fig. 7A) (22, 24, 52) in WT MEFs. GFP tagged-CSNK2A1WT localizes to the centrosome, similar to the endogenous protein (Fig. 2B). Most Csnk2a1 variants were still recruited to the centrosome, except for CSNK2A1R80H, which exhibited impaired centrosomal recruitment (Fig. 7B). We assessed cilia frequency and found no significant difference between these cell lines (Fig. 7C). Because the primary phenotypes of Csnk2a1-knockout cilia are related to cilia stability, we examined cilia morphology and consistently found between the three CSNK2A1 variants a higher frequency of cilia with morphological abnormalities compared to WT CSNK2A1 (Fig. 7D). About 70% of these abnormalities include a bifurcated ciliary tip and/or membranous branch-like protrusions from the distal axoneme. An additional 30% of these abnormal cilia showed a collapsed morphology, where ARL13B-positive cilia exhibited nonelongated bulgy and hollow-curled structures (Fig. 7D). Our results indicate that the OCNDS-associated mutations may alter the morphology and structure of PC. Our work, therefore, establishes a link between Csnk2a1 and cilia regulation and raises the possibility that OCNDS might be linked, at least partially, to disrupted cilia structure or function.
Fig. 7.
OCNDS-associated Csnk2a1 mutations cause ciliary structural defects. (A) The kinase domain illustration of CSNK2A1 and the OCNDS-associated mutations. OCNDS de novo missense mutations p.R80H, p.D156H, and p.R191Q are displayed in green. (B) Graphs show the mean percentage of cells with centrosomal GFP-positive signal in control and CSNK2A1-expressing cell lines. Statistical comparison was performed by one-way analysis ANOVA with Tukey’s multiple comparisons test. P values are shown on the graph (ns, not significant). Results analyzed from control cells n = 46; CSNK2A1-WT n = 83; CSNK2A1-R80H n = 64; CSNK2A1-D156H n = 96; and CSNK2A1-R191Q n = 78 cells. (C) Graphs show the mean percentage of cilia in control and CSNK2A1-expressing cell lines. Statistical comparison was performed by one-way analysis ANOVA with Tukey’s multiple comparisons test (ns, not significant). Each graph displays quantified from control cells n = 46; CSNK2A1-WT n = 83; CSNK2A1-R80H n = 64; CSNK2A1-D156H n = 96; and CSNK2A1-R191Q n = 78 cells. (D) WT MEFs stably expressing CSNK2A1-R80H-GFP, CSNK2A1-D156H-GFP, and CSNK2A1-R191Q-GFP exhibit a higher percentage of abnormal cilia. (Scale bars, 1 μm.) Control cells, n = 41; CSNK2A1-WT, n= 127; CSNK2A1-R80H, n = 60; CSNK2A1-D156H, n = 107; and CSNK2A1- R191Q, n = 85 cells. The percentage of each defect is detailed as follows: for the branching phenotype, control: 8.09%, CSNK2A1-WT: 9.8%, CSNK2A1-R80H: 21.64%, CSNK2A1-D156H: 19%, and CSNK2A1- R191Q: 26.03%; for the ciliary collapsed-like defect, control: 0%, CSNK2A1-WT: 4.19%, CSNK2A1-R80H: 11.28%, CSNK2A1-D156H: 11.4%, and CSNK2A1- R191Q: 9.52%. Statistical comparison was performed by one-way analysis ANOVA with Tukey’s multiple comparisons test on the mean percentage of abnormal cilia (Branched + collapsed-like phenotypes). P values are detailed as follows: CSNK2A1-WT versus CSNK2A1-R80H, P value = 0.0102; CSNK2A1-WT versus CSNK2A1-D156H, P value = 0.0334; and CSNK2A1-WT versus CSNK2A1-R191Q, P value = 0.0026.
Discussion
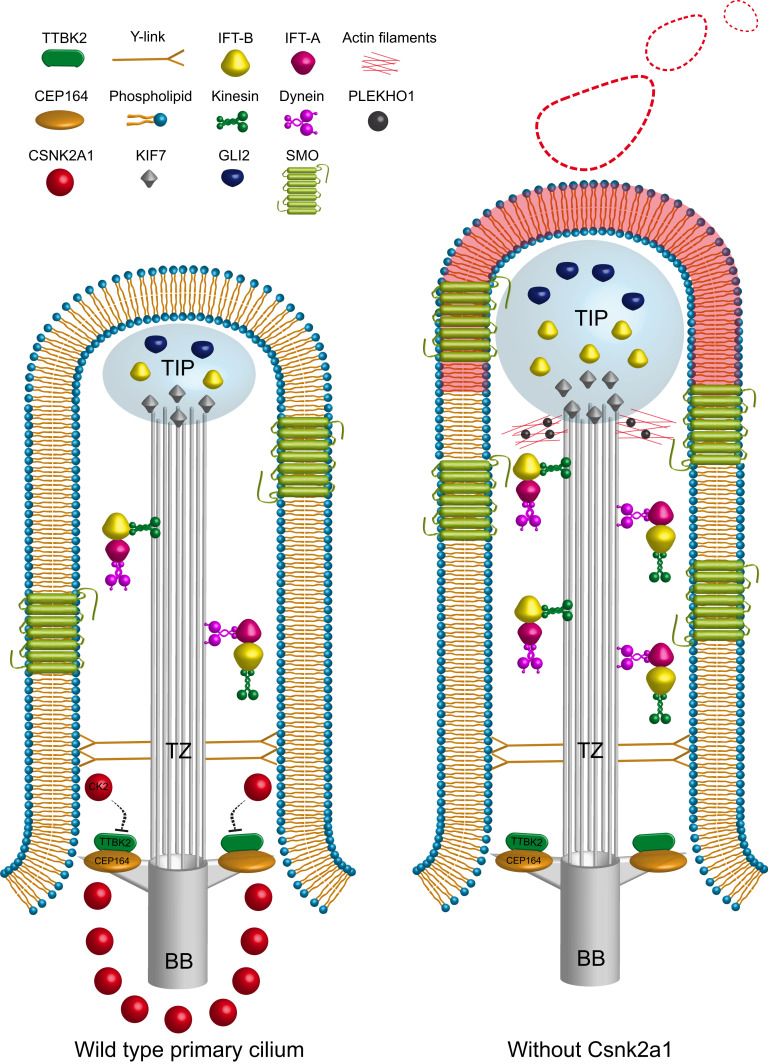
Our present work identifies the CK2 catalytic subunit CSNK2A1 as a negative regulator of TTBK2 and the SHH pathway. CSNK2A1 opposes TTBK2 function mediating cilia structure and trafficking. Our results reveal that CSNK2A1 and TTBK2 physically and functionally interact and colocalize at the distal appendages. In the absence of CSNK2A1, mutant cilia are longer and exhibit instability at the ciliary tip with significant changes in actin cytoskeleton dynamics within the cilium. These defects are accompanied by increased anterograde trafficking and accumulation of IFT and SHH-related proteins within the cilium (Fig. 8). In addition, OCNDS-associated mutations to CSNK2A1 cause ciliary abnormalities, suggesting this human disorder may be linked to ciliary dysfunction.
Fig. 8.
Working model of CSNK2A1 role in cilia homeostasis and function. CSNK2A1 acts as a negative regulator of TTBK2 and SHH signaling. It is differentially enriched at the mother centriole and the distal appendages. The kinase knockout leads to the aberrant trafficking of several critical regulators of cilia assembly, SHH pathway, and actin dynamics. The red shading on the ciliary membrane illustrates the part that will be decapitated.
CK2 plays a key role in numerous cellular processes, including signal transduction, cell-cycle progression, and apoptosis (53–57). The CK2 holoenzyme is generally comprised of two catalytic subunits (CSNK2A1 and/or CSNK2A2) and two regulatory CSNK2B subunits (15, 58, 59). It is not yet clear whether the role we have identified for CSNK2A1 in ciliary stability and trafficking is an independent function of that subunit or whether it represents a requirement for the CK2 holoenzyme. Several reports showed that CK2 and the catalytic subunit CSNK2A1 can localize differentially. For example, CK2 was found associated with the Golgi complex and the endoplasmic reticulum (60), whereas CSNK2A1 (61) and CSNK2A2, but not CSNK2B, were detected at the centrosome (31). CSNK2A1 can also autonomously interact and/or phosphorylate an additional set of proteins, including PP2A, CALMODULIN, and MDM2 (62–65). On the other hand, both Csnk2a1 and Csnk2b were identified as potential negative regulators of the SHH pathway in a cell-based screen (66). Thus, the role of the regulatory subunit warrants further study in the context of PC function.
Our imaging reveals a complex localization pattern for CSNK2A1 at the centrosome, in particular, the cage-like organization observed from side views. A comparable structure was observed with the secreted metalloprotease ADAMTS9 (67), which is required for ciliogenesis. The ADAMTS9 signal is similar but not identical to that of CSNK2A1, with a larger and less-organized structure surrounding the centrosome (67). CSNK2A1 also forms a ring structure at the distal appendages, which is a more common centriolar structure similar to and colocalizing with TTBK2 and CEP164 (Fig. 2 D and E). A fainter signal of CSNK2A1 was also detected at the daughter centriole. Altogether, CSNK2A1 barrel-like localization needs further examination to uncover its role in ciliary function.
Several studies reported that CK2 regulates actin cytoskeleton dynamics (47, 48, 68, 69). Our data showed that CSNK2A1 mediates the ciliary tip stability, likely by regulating the localization of actin modulators such as PLEKHO1 and CAPZB, respectively, at the cilium and the centrosome (Fig. 6 B and C). PLEKHO1 becomes enriched at the ciliary tip in Csnk2a1-knockout cilia (Fig. 6B). Expression of PLEKHO1 is reportedly sufficient to increase F-actin levels (48). Thus, the presence of PLEKHO1 at the distal tip of mutant cilia could trigger the formation of transient F-actin structures leading to ciliary excision (Fig. 8). Moreover, PLEKHO1 interacts specifically with the CSNK2A1 subunit, and this interaction modulates the subcellular localization of PLEKHO1 and its regulation of actin (48, 70). In addition, the capping subunit CAPZB that blocks actin polymerization (71) was enriched at the centrosome in control cells but absent in Csnk2a1-knockout cells. Consistent with our findings, recent evidence collected by cryoelectron tomography has emerged showing that PC do contain actin filaments around the ciliary axoneme (72). The centrosome itself also appears to function as an actin-organizing center capable of promoting the assembly of actin filaments at the vicinity of the cilium (73). Additionally, the kinase-dead mutation of CSNK2A1 displayed a dominant-negative effect on the ciliary localization of PLEKHO1 but not that of CAPZB, which indicates a possible implication of CSNK2A1 kinase activity on regulating PLEKHO1 through phosphorylation. Altogether, our findings suggest that CSNK2A1 controls the dynamics of actin modulators within the cilium-centrosome compartment in order to inhibit the aberrant formation of ciliary F-actin (Fig. 8).
In addition to knockout of Csnk2a1 causing ciliary defects, we show that stable expression of variants associated with the human disorder OCNDS is sufficient to perturb cilia morphology and structure. OCNDS-associated missense mutations occur throughout the kinase domain and are predicted to be deleterious to the kinase activity (9). Our data show that the overexpression of the OCNDS mutation located at the active site is not sufficient to affect either PLEKHO1 or CAPZB localization, contrary to the kinase-dead mutation. This suggests that the human mutation may not lead to complete loss of the kinase activity. Most of the CSNK2A1-mutated variants that we examined still localize to the centrosome (Fig. 7B). Thus, these may interfere and/or compete with the endogenous CSNK2A1WT, affecting its overall activity and/or function. An additional possibility is that mutations to CSNK2A1 could alter its protein–protein interaction networks, leading to branched and collapsed ciliary phenotypes we observed.
In summary, our present work identifies a distal appendage-associated kinase module that regulates cilia stability and signaling. CSNK2A1 acts as a negative modifier of TTBK2 function and modulates the ciliary actin cytoskeleton, mediating PC trafficking and stability. Thus, our work proposes a failsafe mechanism for ciliary tip stability mediated by CSNK2A1 by which it prevents aberrant F-actin polymerization within the cilium. Our findings also expand our knowledge of a potential implication of ciliary dysfunction in OCNDS. Further molecular dissection will be important to define the ciliary mechanisms/changes through which OCNDS mutations trigger clinical features and neurodevelopmental delay in patients. Examining this link in detail and in vivo is therefore of high interest to us moving forward.
Materials and Methods
Cell Culture and Cell Lines.
WT, Ttbk2null/gt, Ttbk2null, and CRISPR-engineered mouse MEFs, and HEK293T cells were grown in high glucose Gibco Dulbecco's modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS) in a humidified 5% CO2 incubator at 37 °C. Cilia formation was induced by serum starving the cells using media with 0.5% FBS. WT cell lines stably expressing TTBK2-FLAG-EGFP, CSNK2A1WT-FLAG-EGFP, CSNK2A1R80H-FLAG-EGFP, CSNK2A1D156H-FLAG-EGFP, CSNK2A1R191Q, and CSNK2A1K68M-FLAG-EGFP were maintained in growth media with 500 µg/mL of Geneticin.
Constructs.
We employed the Gateway destination vector pFLAP-Dest-EGFP-3xFLAG, where we introduced Csnk2a1WT in the C terminus of EGFP, separated by a linker of 67 amino acids. The site-directed mutagenesis method was utilized to generate pFLAP-DEST- CSNK2A1R80H-EGFP, CSNK2A1D156H-EGFP, CSNK2A1R191Q-EGFP, and CSNK2A1K68M-EGFP. The following plasmids were also used in this study: pmCherry-pmCherry-Lifeact (Plasmid no. 54491, Addgene), pEGFPN3-SSTR3 (Plasmid no. 35623, Addgene), pCAGGS-mCherry-ARL13B (Goetz laboratory), and pCAGGS-EGFP-IFT88 (Goetz laboratory). BioID constructs for TTBK2 and GFP were generated using Gateway cloning into pDEST 5′ BirA*-FLAG-pcDNA5-FRT-TO (gift of Anne-Claude Gingras, Lunenfeld-Tanenbaum Research Institute at Mount Sinai Hospital, Canada).
Live Imaging of Cilia.
Cells were seeded in 8-well Nunc Lab-Tek II chambered cover glass and serum starved for 24 h. At all times, cells were kept in a humidified chamber at 37 °C. The live imaging was performed using a Zeiss Axio Observer-Z1 widefield microscope. The Zeiss Plan-Apochromat 63×/1.4 Oil objective with the Axiocam 506 monochrome camera collected the fluorescent signal. The Zeiss definite focus was employed to maintain cilia on focus. Time-lapse series were generated using the Fiji software.
IFT88 Transport Measurements.
Cells were cotransfected with pCAGGS-mCherry-ARL13B and pCAGGS-EGFP-IFT88 and serum starved for 24 h. Cilia were live imaged every 250 ms for 30 s to generate fast time-lapse series. These image sequences were converted into an 8-bit format. The unspecific background was removed with the correction of the unwanted cilia movement. IFT88 bidirectional transport along the cilium was displayed using kymographs that were generated using a Fiji macro toolset called KymographClear 2.0 (74).
Ciliation Rate and Fluorescence Intensity Quantification.
Measurements of cilia rate and fluorescence intensity were performed from at least five randomly selected fields of cells for each condition. The fluorescence intensity of centrosomal and ciliary markers was quantified using the Fiji software. The polygon tool was chosen to select the positive pixels of the immunostained protein. We subsequently measured the mean fluorescence intensity, which integrates the total fluorescence intensity by the selected surface area.
RT-qPCR.
Control and Csnk2a1 knockout cells were plated in a 6-well plate. Cells were serum starved for 48h and treated or not with 200 nM of SAG. Total RNA was purified using the RNeasy kit (Qiagen). From each condition, a total of 600 ng RNA was reverse transcripted to complementary DNA (cDNA) using the iScript Reverse Transcription Supermix for RT-qPCR (no. 1708840). Real-time quantitative PCRs were performed following the PowerUp SYBR Green (ThermoFisher) manufacturer instructions. We used the following primers: GLI1 forward 5′-TTATGGAGCAGCCAGAGAGA-3′, reverse 5′- GAGCCCGCTTCTTTGTTAAT-3′; PTCH1 forward 5′- TGACAAAGCCGACTACATGC-3′, reverse 5′- AGCGTACTCGATGGGCTCT3′; and ACTIN forward 5′- TGGCTCCTAGCACCATGA-3′, reverse 5′- CCACCGATCCACACAGAG-3′.
Statistical Analysis.
Data are reported as arithmetic means ± SEM. Statistical analyses were performed using the nonparametric Mann–Whitney U test with GraphPad Prism 8 software. For multiple comparisons, we executed a one- or two-way analysis ANOVA with Tukey’s multiple comparisons test. To analyze the intensity profile data, we implemented multiple t tests (one per row), found in the grouped analyses tab in GraphPad Prism 8 software. P ≤ 0.05 was used as the cutoff for statistical significance.
Additional materials and methods are described in detail in SI Appendix.
Supplementary Material
Acknowledgments
We thank Drs. Kris Wood and Kevin Lin for their guidance with the CRISPR screen. We are grateful to Drs. David Breslow, Anne-Claude Gingras, Laurence Pelletier, and Saikat Mukhopadhyay for generously sharing reagents. We also thank Dr. Erik Soderblom and the Duke Proteomics and Metabolomics Core for their help with the TTBK2 BioID. We thank Dr. Oliver Tress and Kathryn Schallhorn for acquiring the SIM data. This work was supported by the NIH (R01 HD099784 to S.C.G.) and the Regeneration Next Initiative at Duke University (RNI Postdoctoral Fellowship to A.L.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2018740118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Bishop G. A., Berbari N. F., Lewis J., Mykytyn K., Type III adenylyl cyclase localizes to primary cilia throughout the adult mouse brain. J. Comp. Neurol. 505, 562–571 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Tong C. K., et al., Primary cilia are required in a unique subpopulation of neural progenitors. Proc. Natl. Acad. Sci. U.S.A. 111, 12438–12443 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ford M. J., et al., A cell/cilia cycle biosensor for single-cell kinetics reveals persistence of cilia after G1/S transition is a general property in cells and mice. Dev. Cell 47, 509–523.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowie E., Goetz S. C., TTBK2 and primary cilia are essential for the connectivity and survival of cerebellar Purkinje neurons. eLife 9, e51166 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kozminski K. G., Johnson K. A., Forscher P., Rosenbaum J. L., A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl. Acad. Sci. U.S.A. 90, 5519–5523 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pazour G. J., et al., Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J. Cell Biol. 151, 709–718 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye F., Nager A. R., Nachury M. V., BBSome trains remove activated GPCRs from cilia by enabling passage through the transition zone. J. Cell Biol. 217, 1847–1868 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goetz S. C., Liem K. F. Jr, Anderson K. V., The spinocerebellar ataxia-associated gene Tau tubulin kinase 2 controls the initiation of ciliogenesis. Cell 151, 847–858 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowie E., Norris R., Anderson K. V., Goetz S. C., Spinocerebellar ataxia type 11-associated alleles of Ttbk2 dominantly interfere with ciliogenesis and cilium stability. PLoS Genet. 14, e1007844 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Houlden H., et al., Mutations in TTBK2, encoding a kinase implicated in tau phosphorylation, segregate with spinocerebellar ataxia type 11. Nat. Genet. 39, 1434–1436 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Čajánek L., Nigg E. A., Cep164 triggers ciliogenesis by recruiting Tau tubulin kinase 2 to the mother centriole. Proc. Natl. Acad. Sci. U.S.A. 111, E2841–E2850 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oda T., Chiba S., Nagai T., Mizuno K., Binding to Cep164, but not EB1, is essential for centriolar localization of TTBK2 and its function in ciliogenesis. Genes Cells 19, 927–940 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Lo C.-H., et al., Phosphorylation of CEP83 by TTBK2 is necessary for cilia initiation. J. Cell Biol. 218, 3489–3505 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang N., et al., M-Phase Phosphoprotein 9 regulates ciliogenesis by modulating CP110-CEP97 complex localization at the mother centriole. Nat. Commun. 9, 4511 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lozeman F. J., et al., Isolation and characterization of human cDNA clones encoding the alpha and the alpha’ subunits of casein kinase II. Biochemistry 29, 8436–8447 (1990). [DOI] [PubMed] [Google Scholar]
- 16.Homma M. K., Homma Y., Regulatory role of CK2 during the progression of cell cycle. Mol. Cell. Biochem. 274, 47–52 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Maier B., et al., A large-scale functional RNAi screen reveals a role for CK2 in the mammalian circadian clock. Genes Dev. 23, 708–718 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang S., Jones K. A., CK2 controls the recruitment of Wnt regulators to target genes in vivo. Curr. Biol. 16, 2239–2244 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Song D. H., Sussman D. J., Seldin D. C., Endogenous protein kinase CK2 participates in Wnt signaling in mammary epithelial cells. J. Biol. Chem. 275, 23790–23797 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Seldin D. C., et al., CK2 as a positive regulator of Wnt signalling and tumourigenesis. Mol. Cell. Biochem. 274, 63–67 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Okur V., et al., De novo mutations in CSNK2A1 are associated with neurodevelopmental abnormalities and dysmorphic features. Hum. Genet. 135, 699–705 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Chiu A. T. G., et al., Okur-Chung neurodevelopmental syndrome: Eight additional cases with implications on phenotype and genotype expansion. Clin. Genet. 93, 880–890 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Akahira-Azuma M., Tsurusaki Y., Enomoto Y., Mitsui J., Kurosawa K., Refining the clinical phenotype of Okur-Chung neurodevelopmental syndrome. Hum. Genome Var. 5, 18011 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Owen C. I.et al.; Deciphering Developmental Disorders Study , Extending the phenotype associated with the CSNK2A1-related Okur-Chung syndrome-A clinical study of 11 individuals. Am. J. Med. Genet. A. 176, 1108–1114 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Duan H. L., et al., [A case of Okur-Chung syndrome caused by CSNK2A1 gene variation and review of literature]. Zhonghua Er Ke Za Zhi 57, 368–372 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Wu R., et al., [Identification of a novel de novo variant of CSNK2A1 gene in a boy with Okur-Chung neurodevelopmental syndrome]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 37, 641–644 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Roux K. J., Kim D. I., Raida M., Burke B., A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breslow D. K., et al., A CRISPR-based screen for Hedgehog signaling provides insights into ciliary function and ciliopathies. Nat. Genet. 50, 460–471 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doench J. G., et al., Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang T. T., et al., Super-resolution architecture of mammalian centriole distal appendages reveals distinct blade and matrix functional components. Nat. Commun. 9, 2023 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faust M., Gänther J., Morgenstern E., Montenarh M., Götz C., Specific localization of the catalytic subunits of protein kinase CK2 at the centrosomes. Cell. Mol. Life Sci. 59, 2155–2164 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mukhopadhyay S., et al., TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes Dev. 24, 2180–2193 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fu W., Wang L., Kim S., Li J., Dynlacht B. D., Role for the IFT-A complex in selective transport to the primary cilium. Cell Rep. 17, 1505–1517 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corbit K. C., et al., Vertebrate Smoothened functions at the primary cilium. Nature 437, 1018–1021 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Haycraft C. J., et al., Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 1, e53 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liem K. F. Jr, He M., Ocbina P. J. R., Anderson K. V., Mouse Kif7/Costal2 is a cilia-associated protein that regulates Sonic hedgehog signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 13377–13382 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He M., et al., The kinesin-4 protein Kif7 regulates mammalian Hedgehog signalling by organizing the cilium tip compartment. Nat. Cell Biol. 16, 663–672 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Svärd J., et al., Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev. Cell 10, 187–197 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Zhang Q., Seo S., Bugge K., Stone E. M., Sheffield V. C., BBS proteins interact genetically with the IFT pathway to influence SHH-related phenotypes. Hum. Mol. Genet. 21, 1945–1953 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Long H., et al., Comparative analysis of ciliary membranes and ectosomes. Curr. Biol. 26, 3327–3335 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nager A. R., et al., An actin network dispatches ciliary GPCRs into extracellular vesicles to modulate signaling. Cell 168, 252–263.e14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phua S. C., et al., Dynamic remodeling of membrane composition drives cell cycle through primary cilia excision. Cell 168, 264–279.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoang-Minh L. B., Dutra-Clarke M., Breunig J. J., Sarkisian M. R., Glioma cell proliferation is enhanced in the presence of tumor-derived cilia vesicles. Cilia 7, 6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang G., et al., Rab7 regulates primary cilia disassembly through cilia excision. J. Cell Biol. 218, 4030–4041 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Niel G., D’Angelo G., Raposo G., Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 19, 213–228 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Bangs F. K., Schrode N., Hadjantonakis A.-K., Anderson K. V., Lineage specificity of primary cilia in the mouse embryo. Nat. Cell Biol. 17, 113–122 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Markwell S. M., et al., Cortactin phosphorylation by casein kinase 2 regulates actin-related protein 2/3 complex activity, invadopodia function, and tumor cell invasion. Mol. Cancer Res. 17, 987–1001 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Canton D. A., et al., The pleckstrin homology domain-containing protein CKIP-1 is involved in regulation of cell morphology and the actin cytoskeleton and interaction with actin capping protein. Mol. Cell. Biol. 25, 3519–3534 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bosc D. G., et al., Identification and characterization of CKIP-1, a novel pleckstrin homology domain-containing protein that interacts with protein kinase CK2. J. Biol. Chem. 275, 14295–14306 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Olsten M. E. K., Canton D. A., Zhang C., Walton P. A., Litchfield D. W., The Pleckstrin homology domain of CK2 interacting protein-1 is required for interactions and recruitment of protein kinase CK2 to the plasma membrane. J. Biol. Chem. 279, 42114–42127 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Penner C. G., Wang Z., Litchfield D. W., Expression and localization of epitope-tagged protein kinase CK2. J. Cell. Biochem. 64, 525–537 (1997). [PubMed] [Google Scholar]
- 52.Trinh J., et al., A novel de novo mutation in CSNK2A1: Reinforcing the link to neurodevelopmental abnormalities and dysmorphic features. J. Hum. Genet. 62, 1005–1006 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Sayed M., Pelech S., Wong C., Marotta A., Salh B., Protein kinase CK2 is involved in G2 arrest and apoptosis following spindle damage in epithelial cells. Oncogene 20, 6994–7005 (2001). [DOI] [PubMed] [Google Scholar]
- 54.Shin S., et al., Caspase-2 primes cancer cells for TRAIL-mediated apoptosis by processing procaspase-8. EMBO J. 24, 3532–3542 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bae J. S., et al., CK2α phosphorylates DBC1 and is involved in the progression of gastric carcinoma and predicts poor survival of gastric carcinoma patients. Int. J. Cancer 136, 797–809 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Lüscher B., Kuenzel E. A., Krebs E. G., Eisenman R. N., Myc oncoproteins are phosphorylated by casein kinase II. EMBO J. 8, 1111–1119 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keller D. M., et al., A DNA damage-induced p53 serine 392 kinase complex contains CK2, hSpt16, and SSRP1. Mol. Cell 7, 283–292 (2001). [DOI] [PubMed] [Google Scholar]
- 58.Pinna L. A., Casein kinase 2: An ‘eminence grise’ in cellular regulation? Biochim. Biophys. Acta 1054, 267–284 (1990). [DOI] [PubMed] [Google Scholar]
- 59.Litchfield D. W., Dobrowolska G., Krebs E. G., Regulation of casein kinase II by growth factors: A reevaluation. Cell. Mol. Biol. Res. 40, 373–381 (1994). [PubMed] [Google Scholar]
- 60.Faust M., Jung M., Günther J., Zimmermann R., Montenarh M., Localization of individual subunits of protein kinase CK2 to the endoplasmic reticulum and to the Golgi apparatus. Mol. Cell. Biochem. 227, 73–80 (2001). [PubMed] [Google Scholar]
- 61.McKendrick L., Milne D., Meek D., Protein kinase CK2-dependent regulation of p53 function: Evidence that the phosphorylation status of the serine 386 (CK2) site of p53 is constitutive and stable. Mol. Cell. Biochem. 191, 187–199 (1999). [PubMed] [Google Scholar]
- 62.Bidwai A. P., Reed J. C., Glover C. V., Phosphorylation of calmodulin by the catalytic subunit of casein kinase II is inhibited by the regulatory subunit. Arch. Biochem. Biophys. 300, 265–270 (1993). [DOI] [PubMed] [Google Scholar]
- 63.Li D., Dobrowolska G., Krebs E. G., The physical association of casein kinase 2 with nucleolin. J. Biol. Chem. 271, 15662–15668 (1996). [DOI] [PubMed] [Google Scholar]
- 64.Hériché J. K., et al., Regulation of protein phosphatase 2A by direct interaction with casein kinase 2alpha. Science 276, 952–955 (1997). [DOI] [PubMed] [Google Scholar]
- 65.Guerra B., Götz C., Wagner P., Montenarh M., Issinger O.-G., The carboxy terminus of p53 mimics the polylysine effect of protein kinase CK2-catalyzed MDM2 phosphorylation. Oncogene 14, 2683–2688 (1997). [DOI] [PubMed] [Google Scholar]
- 66.Pusapati G. V., et al., CRISPR screens uncover genes that regulate target cell sensitivity to the morphogen sonic hedgehog. Dev. Cell 44, 113–129.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nandadasa S., et al., Secreted metalloproteases ADAMTS9 and ADAMTS20 have a non-canonical role in ciliary vesicle growth during ciliogenesis. Nat. Commun. 10, 953 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yue J., et al., Cutaneous human papillomavirus type 38 E7 regulates actin cytoskeleton structure for increasing cell proliferation through CK2 and the eukaryotic elongation factor 1A. J. Virol. 85, 8477–8494 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xavier C.-P., et al., Phosphorylation of CRN2 by CK2 regulates F-actin and Arp2/3 interaction and inhibits cell migration. Sci. Rep. 2, 241 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Canton D. A., Olsten M. E. K., Niederstrasser H., Cooper J. A., Litchfield D. W., The role of CKIP-1 in cell morphology depends on its interaction with actin-capping protein. J. Biol. Chem. 281, 36347–36359 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Edwards M., et al., Capping protein regulators fine-tune actin assembly dynamics. Nat. Rev. Mol. Cell Biol. 15, 677–689 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kiesel P., et al., The molecular structure of mammalian primary cilia revealed by cryo-electron tomography. Nat. Struct. Mol. Biol. 27, 1115–1124 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Farina F., et al., The centrosome is an actin-organizing centre. Nat. Cell Biol. 18, 65–75 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mangeol P., Prevo B., Peterman E. J. G., KymographClear and KymographDirect: Two tools for the automated quantitative analysis of molecular and cellular dynamics using kymographs. Mol. Biol. Cell 27, 1948–1957 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.