Significance
In natural environments, plants establish intimate interactions with a wide diversity of microbes. It is unknown, however, how microbiota composed of commensal bacteria colonize roots in the face of a sophisticated plant immune system that evolved to recognize microbial-associated molecular patterns. We investigate the interaction between plant immune system function and the root microbiota. We report that root-associated commensal bacteria actively suppress the host immune response in the context of a community. Suppressors and nonsuppressors co-occur in the root microbiome and the presence of the former can enhance the colonization ability of the latter. We highlight the role of a specific sector of the plant immune system and its suppression in gating microbial access to the roots.
Keywords: plant immunity, root microbiome, MAMP, SynCom, flg22
Abstract
Plants have an innate immune system to fight off potential invaders that is based on the perception of nonself or modified-self molecules. Microbe-associated molecular patterns (MAMPs) are evolutionarily conserved microbial molecules whose extracellular detection by specific cell surface receptors initiates an array of biochemical responses collectively known as MAMP-triggered immunity (MTI). Well-characterized MAMPs include chitin, peptidoglycan, and flg22, a 22-amino acid epitope found in the major building block of the bacterial flagellum, FliC. The importance of MAMP detection by the plant immune system is underscored by the large diversity of strategies used by pathogens to interfere with MTI and that failure to do so is often associated with loss of virulence. Yet, whether or how MTI functions beyond pathogenic interactions is not well understood. Here we demonstrate that a community of root commensal bacteria modulates a specific and evolutionarily conserved sector of the Arabidopsis immune system. We identify a set of robust, taxonomically diverse MTI suppressor strains that are efficient root colonizers and, notably, can enhance the colonization capacity of other tested commensal bacteria. We highlight the importance of extracellular strategies for MTI suppression by showing that the type 2, not the type 3, secretion system is required for the immunomodulatory activity of one robust MTI suppressor. Our findings reveal that root colonization by commensals is controlled by MTI, which, in turn, can be selectively modulated by specific members of a representative bacterial root microbiota.
Plants are inhabited by hundreds of species of commensals, many of which have beneficial effects on the host (1). These microbes often express the same immunogenic microbe-associated molecular patterns (MAMPs) that are found in pathogens, highlighting their potential to trigger immune responses in their hosts (2–4). How plants mount effective defenses against pathogens while allowing the colonization of commensals remains a mystery. Plant-associated microbial communities are much less diverse than those of the surrounding environment (1), indicating that the host exerts selection pressure over their microbiota and that some microbes are better adapted to colonize plant tissues than others. While multiple environmental and genetic factors likely orchestrate microbiota assembly and structure, recent research indicates that the plant immune system operates as a major gatekeeper. Arabidopsis thaliana plants (hereafter Arabidopsis) compromised in the signaling of the defense phytohormones salicylic acid and jasmonic acid harbor altered microbiota (5, 6). Similarly, mutants impaired in MAMP-triggered immunity (MTI) and in the MIN7-vesicle trafficking pathway carry altered endophytic phyllosphere microbiota and display leaf-tissue damage associated with dysbiosis under conditions of high humidity (7). Recent evidence demonstrates that perception of flg22 is usually low in most root cells, but up-regulated following tissue wounding associated with infection and potentially colonization (8, 9).
The ability to suppress the host immune response is a hallmark of successful pathogens (10). In animals, both specific and redundant immunomodulatory effects have been defined for taxonomically diverse commensal gut bacteria (11). Likewise, plant-associated commensals have been shown to modulate MTI (12–19). Nevertheless, studies in plants have so far focused on single microbes and the specific immunomodulatory effects of different strains have not been integrated in the context of complex communities. Furthermore, the significance of immunomodulation for community assembly remains unexplored. In this work, we investigated how a community of root-associated commensal bacteria interacts with the Arabidopsis immune system. We verified that the ability to suppress MTI is common and taxonomically widespread among these commensals. High-throughput gene-expression analyses of plants colonized by synthetic communities (SynComs) or by single microbes led to the identification of a set of defense-related genes that were commonly manipulated by phylogenetically distinct suppressor strains, highlighting a sector of the plant immune system that may control plant colonization by the microbiota. Notably, suppressors could promote the growth of other nonsuppressor strains, indicating that certain microbes may benefit from co-occurring suppressor strains. Furthermore, a genetic screening revealed that the type 2 secretion system (T2SS) is required for the robust suppressor Dyella japonica MF79 to interfere with root MTI, while the type 3 secretion system (T3SS) was dispensable. Our results expand our understanding of how the plant immune system functions in roots colonized by commensals, underscoring the role of MTI and of its suppression in microbiota assembly.
Results
A Community of Root Commensals Suppresses MTI.
To understand how MAMP-triggered immunity affects the composition of the root microbiome and whether root-associated bacteria can modulate this immune response in the context of a commensal community, we established a model system (SI Appendix, Fig. S1 A–E). Here, 7-d-old Arabidopsis seedlings were exposed for 12 d to 1 µM of the bacterial peptide MAMP flg22. Although MTI has been traditionally studied within minutes/hours of MAMP exposure (15, 20, 21), we validated that the continuous presence of flg22 in our system induces a defense-related transcriptional signature that is functionally overlapping with the one observed in “typical” systems (SI Appendix, Fig. S1F), indicating that flg22 signaling and immunogenicity persist for at least 12 d. Using this system, we concurrently inoculated plants with 1 µM flg22 or not and a 35-member bacterial synthetic community composed of a random consortium of Arabidopsis root commensals (SynCom35) (Dataset S1) that is phylogenetically representative of wild-soil root-associated microbiota (22). Interestingly, bacterial 16S profiles revealed only minor differences between the communities of bacteria colonizing flg22-treated roots and those colonizing untreated plants (Fig. 1A). Only two bacterial taxa had differential abundances in the roots of plants exposed to flg22 (SI Appendix, Fig. S1G and Dataset S2). Thus, overall community composition was not affected by the addition of the elicitor flg22, suggesting that SynCom35 interferes with the plant immune response to facilitate community assembly in the face of flg22-driven MTI.
Fig. 1.
A synthetic community comprised of 35 bacterial strains (SynCom35) modulates a sector of the Arabidopsis immune system. (A) Canonical analysis of principal coordinates (CAP) based on Bray–Curtis dissimilarities of 16S rDNA profiling reveals no significant effect of flg22 in the composition of root bacterial community. Significance of model terms was determined with a PERMANOVA analysis using 5,000 permutations (α = 0.05). (B) Number of differentially expressed genes (DEG) identified by RNA sequencing in roots treated with flg22 or not in the absence of bacteria, with heat-killed SynCom35 or with SynCom35 alive. (C) Overlap among the sets of flg22-responsive genes in the “No bacteria,” “Heat-killed SynCom35,” and “SynCom35” conditions. (D) Pearson correlation among flg22 responses in plants grown axenically (no bacteria) or in the presence of dead or living SynCom35. Note the low correlation between “SynCom35” and the other two conditions. (E) CAP coordinates shows that plants grown in the presence of SynCom35 display a unique transcriptional signature that is not differentiated by addition of flg22. (F) Hierarchical clustering of the 716 genes that responded to flg22 in at least one of the experimental conditions. SynCom35 triggers a transcriptional signature that is very similar to the flg22 response except for three sets of genes (Cluster SC1, Cluster SC7, and Cluster SC8), which display a remarkably different expression profile when compared to control plants. (G) Expression pattern of each of the clusters defined in the heatmap. The P values on the bottom refer to the comparison between groups (flgs22 vs. mock) for each condition using two-tailed t tests. (H) Gene ontology enrichment analyses showing enriched biological processes in each of the clusters. Cluster SC1, which is suppressed by SynCom35, contains defense genes that are activated by flg22. Cluster SC7, which is activated by SynCom35, is enriched in auxin metabolism. All results refer to plants exposed to the treatments for 12 d. HK, heat-killed SynCom35; FDR, false-discovery rate; NB, no bacteria; SC, SynCom35. The complete enrichment analysis and the associated statistics are shown in Dataset S3.
To investigate whether SynCom35 interferes with the flg22-induced immune response in roots, we cultivated plants in axenic conditions, in the presence of SynCom35, or in the presence of heat-killed SynCom35 (2 h at 100 °C) (SI Appendix, Fig. S1 C–E). Roots exposed to flg22 for 12 d without any bacteria or in the presence of the heat-killed SynCom35 displayed a clear transcriptional reprogramming, with 492 and 523 genes responding to the elicitor, respectively (Fig. 1B and Dataset S3). The flg22-responsive genes from these control treatments were not only highly overlapping (Fig. 1C), but also showed highly correlated expression levels (Fig. 1D), demonstrating that the dead bacterial community does not interfere with the plant response to flg22. In contrast, only seven genes responded to flg22 in plants inoculated with the live SynCom35 (Fig. 1B), and no correlation was observed between this condition and the two other controls (Fig. 1 C and D). Indeed, SynCom35 triggered a unique transcriptional signature that persisted in the presence of flg22 (Fig. 1E). Hierarchical clustering revealed a set of 281 genes (39% of the total flg22-responsive genes) that were clearly activated by flg22 in the roots of control plants, but strongly suppressed when SynCom35 was also present (cluster SC1 in Fig. 1 F and G). Cluster SC1 is strongly enriched in immune-related genes (Fig. 1H), demonstrating that this bacterial commensal community possesses immunomodulatory activity.
WRKY transcription factors, which include master regulators of plant immunity (23), were overrepresented in cluster SC1 (P = 8.8E-6) (SI Appendix, Fig. S2 A–E), which contained seven of the eight WRKY genes that were up-regulated by flg22. Consistent with a suppressed WRKY regulatory network, cluster SC1 was also enriched in predicted targets of these transcription factors (SI Appendix, Fig. S2F). SynCom35 also suppressed the activation of genes encoding receptor-like kinases (RLKs) (SI Appendix, Fig. S2G), including the MTI marker FRK1 (FLG22-INDUCED RECEPTOR-LIKE KINASE 1) and the flg22 receptor itself, FLS2 (FLAGELLIN-SENSITIVE 2). RLKs are receptors that form the first line of defense in the plant immune system by transducing the perception of extracellular molecules into intracellular signals that activate immunity (24). Genes involved in the biosynthesis of secondary metabolites were also prevalent in cluster SC1. In particular, the master regulator of indole glucosinolate biosynthesis MYB51 (25), along with three indole glucosinolate methyltransferases genes (IGMT2, IGMT3, and IGMT4) and various glutathione S-transferases and cytochrome P450s were not activated by flg22 in plants colonized by SynCom35 (SI Appendix, Fig. S2H). Root-exuded secondary metabolites act as chemical barriers against microbial invaders and suppression of their biosynthesis may favor colonization of microbial commensals (26). These results demonstrate that key components of the plant immune system, from receptors to transcription factors and biochemical executors, are modulated by SynCom35.
Since cluster SC1 overlaps significantly with a set of flg22-responsive genes that were suppressed by a different and independently derived SynCom from an independent study (SI Appendix, Fig. S3A and Dataset S3) (27), modulation of this sector of the plant immune system is likely to be a common strategy employed by root commensals. Cluster SC1 also overlaps significantly (84 of 281 genes; P = 3.27E-58) (Dataset S3) with a set of 868 genes that are flg22-regulated across four Brassicaceae species (28), indicating that SynCom35 has the potential to modulate an evolutionarily conserved sector of MTI. Although defense-related genes were not activated by flg22 in plants grown with SynCom35 at day 12, they were up-regulated on the first day of treatment (SI Appendix, Fig. S3 B–F and Dataset S3), indicating that SynCom35, once fully established, suppresses an ongoing immune response instead of preventing its activation.
The subset of flg22-activated genes that were not suppressed by SynCom35 at day 12 (clusters SC2 and SC4) (Fig. 1F) were enriched in root morphogenesis and differentiation processes (Fig. 1H). These genes were constitutively activated by SynCom35 even in the absence of flg22 (Fig. 1G). Impaired primary root elongation is a phenotype of plants exposed to flg22 (29), a phenotype that is also triggered by SynCom35 (SI Appendix, Fig. S4A). Arthrobacter strains present in SynCom35 affect root development, likely via auxin production (30). Consistent with this observation, auxin-responsive genes were highly activated in plants grown with SynCom35 (cluster SC7 in Fig. 1 F–H). Furthermore, the auxin-resistant mutant axr2-1 expressed a significantly reduced root growth-inhibition (RGI) phenotype in the presence of Arthrobacter MF161 (SI Appendix, Fig. S4B). These results demonstrate that the SynCom35 effect on root morphology is uncoupled from its immunomodulatory activity.
Dissecting the Immunomodulatory Activity of Commensals.
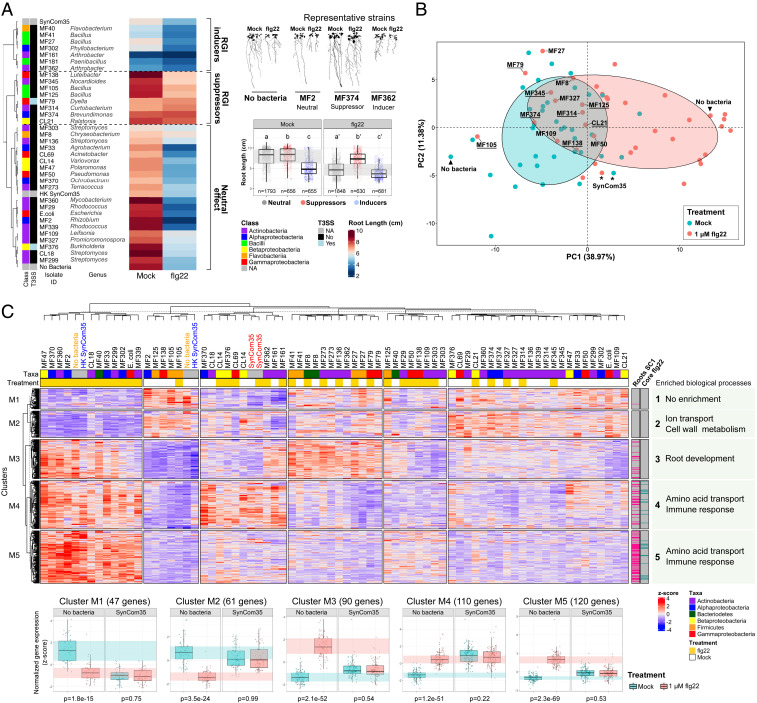
After establishing that SynCom35 had immunomodulatory activity on Arabidopsis roots, we performed monoassociation experiments to evaluate the interaction of individual strains with the plant immune system. Seven-day-old Arabidopsis seedlings were inoculated with each member of SynCom35 in the presence or absence of 1 µM flg22 (SI Appendix, Fig. S5). Evaluation of root lengths revealed that individual strains interacted with the plants in three different ways (Fig. 2A). Twenty strains did not alter the flg22 effect on root morphology. Seven strains caused RGI even in the absence of flg22 (RGI inducers), a phenotype reminiscent of the SynCom35 effect on the plant. Importantly, eight strains suppressed the RGI phenotype triggered by flg22 (RGI suppressors), supporting longer root lengths in the presence of the MAMP. Because plants grown in the presence of SynCom35 display short roots (Fig. 2A and SI Appendix, Fig. S4A), the RGI inducers in this consortium are functionally dominant over the suppressors for this trait.
Fig. 2.
Suppression of the flg22 response is a common feature among members of SynCom35. (A) Arabidopsis root length in the presence of each strain of SynCom35. While some strains have a neutral effect on the plant root elongation, others are able to suppress the flg22-induced RGI or induce the same phenotype even in the absence of flg22. The taxonomic class of each strain and the presence or not of a T3SS are annotated as colored boxes on the left of the heatmap. The images (Right) show the plant phenotype in the presence of a representative neutral (MF2), suppressor (MF374), and inducer strain (MF362). The boxplots summarize the effect of each group from the heatmap on root length (multiple comparisons were performed with ANOVA followed by a Tukey test, α = 0.05). (B) PCA based on 428 genes that responded to flg22 in axenic plants demonstrates that some, but not all, strains of SynCom35 interfere with the transcriptional regulation of the flg22 regulon. Ellipses show the parametric smallest area around the mean that contains 70% of the probability mass for each group (blue: mock; red: 1 μM flg22). Only controls (“no bacteria” and “SynCom35”) and suppressors of this gene set are labeled for clarity. Underlined labels indicate that the strain also suppressed the RGI phenotype triggered by flg22 shown in A. (C) Hierarchical clustering based on the expression of the same 428 genes in seedlings grown in the presence of each strain from SynCom35 along with controls (no bacteria, heat-killed SynCom35, and SynCom35). Five clusters were defined and their profiles in seedlings grown under axenic conditions or with SynCom35 are shown on the bottom. The P values refer to the comparison between groups (flg22 vs. mock) using two-tailed t tests. The boxes on the top of the heatmap indicate the taxonomic classification of the inoculated strain and treatment (orange) or not (white) with flg22. Lanes on the right of the heatmap indicate whether each gene was suppressed by SynCom35 in roots (cluster SC1) (Fig. 1F) or whether they belong to the set of 84 core flg22 marker genes (SI Appendix, Fig. S6). Representative biological processes that are enriched in each cluster are shown on the right. The complete enrichment analysis and the associated statistics are shown in Dataset S4.
We next asked whether strains that suppressed flg22-mediated RGI were also efficient suppressors of the flg22 regulon. A principal component analysis (PCA) based on the expression of 428 genes that define the flg22 regulon in axenic plants (Dataset S4) revealed suppression of this transcriptional response by various members of SynCom35 (Fig. 2B), including all strains that suppressed flg22-mediated RGI (Fig. 2A). Thus, efficient suppression of the flg22 regulon predicts suppression of the flg22 effect on root elongation. This conclusion was validated using an orthogonal approach. We generated a literature-based core set of 84 flg22 transcriptional markers, defined by both chronic and acute flg22 treatments of diverse Arabidopsis tissues (SI Appendix, Fig. S6 A and B and Dataset S5). These 84 genes contained 40 cluster SC1 genes (P = 2.82E-57) and 52 of the evolutionarily conserved flg22 regulon (28) (P = 9.34e-56). We clustered our data based on their expression (SI Appendix, Fig. S6C). The two resulting groups reflected the expression levels of these defense-related genes: Group I (SI Appendix, Fig. S6C, blue) displayed low expression of these marker genes and was mostly comprised of mock-treated plants; and group II (SI Appendix, Fig. S6C, red) presented higher expression of the same genes and was mostly comprised of flg22-treated plants. However, some strains prevented the activation of the flg22 core set when plants were exposed to the elicitor, while others activated the same genes even in the absence of flg22. These results show that the SynCom35 commensal consortium contains members that have opposing effects on the plant immune response to flg22.
To explore strain-specific effects on the plant immune response in more detail, the 428 flg22-responsive genes that were defined in axenic plants were subjected to hierarchical clustering based on their expression levels in the context of monoassociations (Fig. 2C). Eleven of the 35 strains (31%) had no effect on the plant response to flg22; seedlings grown in the presence of these bacteria responded to the MAMP similarly to seedlings grown without bacteria or with the heat-killed SynCom35 (Fig. 2C). The remaining 24 strains (69%) suppressed the plant transcriptional response to flg22 either partially or completely. Of the three clusters containing genes activated by flg22 (clusters M3, M4, and M5) (Fig. 2C), one was enriched in root development (cluster M3) (Fig. 2C and Dataset S4) and two were enriched in defense-related genes (clusters M4 and M5) (Fig. 2C and Dataset S4). Importantly, cluster M5 was the only set of flg22-responsive genes whose activation by the MAMP was consistently suppressed by all 24 strains, indicating that modulation of these defense-related genes is a common feature shared among phylogenetically diverse commensal bacteria. Cluster M5 is also highly overlapping (76 of 120 genes) with the cluster SC1 genes suppressed by SynCom35 in Arabidopsis roots (Figs. 1F and 2C, column labeled “Roots SC1”).
Evaluation of the Arabidopsis transcriptome in response to each member of SynCom35 in the absence of exogenous MAMPs complemented the analyses focused on the flg22 response (SI Appendix, Fig. S7 A and B). Most strains (22 of 35) triggered the activation of defense-related genes to some extent (SI Appendix, Fig. S7C). However, activation of defense-related genes was undetectable in plants grown in the presence of the other, taxonomically diverse, 13 strains (SI Appendix, Fig. S7C). Eleven of these strains also suppressed cluster M5 of flg22-responsive genes in plants treated with flg22 (Fig. 2C). Thus, these results extend our observations and demonstrate that phylogenetically diverse commensals can modulate immune responses triggered by endogenous MAMPs.
We assessed the modulation of MTI by root commensals using an independent and conceptually distinct experimental system. Because flg22 triggers transcriptional responses in plant cells within minutes and over hours of its perception (15, 20, 21), we evaluated whether members of SynCom35 also interfere with the acute response that is triggered only 5 h after root exposure to flg22 using the same set-up that was previously employed to characterize MTI in the roots (19). For this, we utilized an Arabidopsis line carrying the GUS reporter gene under control of the flg22-responsive promoter pCYP71A12, which is activated by flg22 specifically in the root elongation zone (19). Plants treated with 100 nM flg22 under axenic conditions activated the pCYP71A12 promoter in roots within 5 h of treatment. This response was absent in the control treatments [i.e., plants treated with 10 µM MeJA or with the Pseudomonas simiae strain WCS417 (19)] but was observed in plants inoculated with most of the members of SynCom35 (SI Appendix, Fig. S6D). Importantly, seven strains suppressed pCYP71A12 activation by flg22 in this assay, indicating that these commensals can also interfere with the plant acute response to this MAMP.
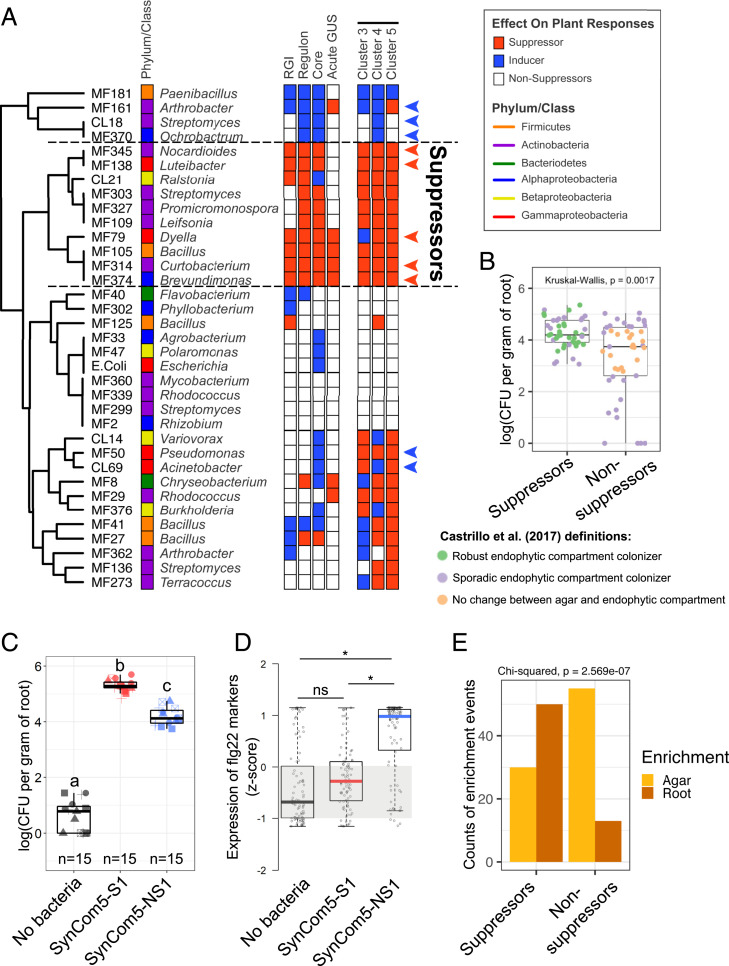
By collating the results from the independent and conceptually different monoassociation experiments, a set of 10 robust MTI suppressors emerged within the members of SynCom35 (Fig. 3A). Although some strains of this community remained neutral or even activated the plant immune system in some assays, a total of 23 strains (65%) suppressed the plant response to flg22 in at least one assay (Fig. 3A). These results confirm that the ability to suppress the plant immune system is common and taxonomically widespread among root commensals. Interestingly, no strain among six tested robust suppressors modulated root immunity by lowering the extracellular pH (SI Appendix, Fig. S8A), a strategy recently described for commensal Pseudomonas (12). Interestingly, some suppressor strains interfered with the plant response to other peptide ligands and prevented RGI phenotypes induced by the damage-associated pep1 and developmental regulator clv3 peptides, while failing to suppress the RGI phenotype induced by hormones methyl jasmonate or auxin (SI Appendix, Fig. S8B). Thus, a diversity of extracellular MTI-suppression molecular mechanisms likely evolved among plant-associated bacteria in order to interfere with plant immunity.
Fig. 3.
Microbiota members that suppress the plant immune system are better colonizers of Arabidopsis roots either in monoassociation or in the context of SynComs. (A) Summary of the assays performed to define suppression of the flg22 response in this study. Strains are clustered based on their response to a total of five assays that are illustrated as rows of boxes to the right. RGI: the ability of a strain to suppress flg22-induced RGI or induce this phenotype even in the absence of the MAMP (Fig. 2A). Regulon: the ability to interfere with the overall expression signature of 428 flg22-responsive genes defined in control (no bacteria) seedlings (Fig. 2B). Core: the ability of each strain to interfere specifically with a set of 84 defense-related genes that are consistently induced by flg22 under diverse experimental conditions (SI Appendix, Fig. S6C). Acute GUS: the ability of each strain to interfere with the acute response to flg22 (5 h treatment) specifically in roots based on the pCYP71A12::GUS reporter line (SI Appendix, Fig. S6D). Clusters 3, 4, and 5: the suppression ability of each strain on specific transcriptional response sectors of the flg22 regulon (Fig. 2C). Members of SynCom5-S1 and SynCom5-NS1 are marked with red and blue arrowheads, respectively. (B) Suppressors are better colonizers of Arabidopsis roots in monoassociation experiments. The growth of five suppressors (members of SynCom5-S1) and taxonomically matching nonsuppressors (members of SynCom5-NS1) were individually assayed in Arabidopsis roots after 12 d of colonization in three biological replicates. Each dot represents an individual strain/replicate. Strains are colored based on their behavior in the context of SynCom35 in a previous study (22). Note that all robust endophytic compartment colonizers are suppressor strains. A Kruskal–Wallis test was performed on the CFU counts from three independent experiments each with three technical reps. n = 9 for each strain and n = 45 for each group being compared. (C) A SynCom made of five suppressors (SynCom5-S1) colonizes Arabidopsis roots better than a SynCom made of five taxonomically matching nonsuppressors (SynCom5-NS1). Different symbols represent independent experimental repetitions. Letters indicate significantly different groups based on an ANOVA followed by a Tukey test (α = 0.05). (D) Seedlings grown with SynCom5-NS1 activate the expression of flg22 marker genes while seedlings inoculated with SynCom5-S1 do not. Statistical significance was determined using a permutation approach, where the actual group mean differences were compared to the group mean differences of 10,000 permutations of the gene counts. The group mean differences that were greater than 95% of the group mean differences calculated in the permutations were considered significant (asterisks). n.s. = not significant. (E) Suppressor (SynCom5-S1), but not nonsuppressor (SynCom5-NS1), strains are enriched in the endophytic compartment of Arabidopsis roots relative to agar in the context of a synthetic community. Suppressors n = 80, nonsuppressors n = 68.
MTI can Control Colonization by Commensals.
We then tested the hypothesis that the ability to suppress the plant immune response is advantageous to commensal bacteria and, therefore, has ecological and evolutionary significance. We first evaluated the colonization ability of five robust suppressors and taxonomically matched strains from SynCom35 that did not suppress all measured outputs (Fig. 3A and Dataset S1). Monoassociation assays demonstrated that most suppressors grew to higher titers in Arabidopsis roots than nonsuppressors (Fig. 3B). This conclusion was supported by experiments using two smaller SynComs made of the five suppressors (SynCom5-S1) and five taxonomically matched nonsuppressors (SynCom5-NS1). In agreement with the monoassociation assays, 10-fold more bacterial colonies were recovered from Arabidopsis roots inoculated with SynCom5-S1 than with SynCom5-NS1 (Fig. 3C). The same result was obtained in an independent experiment that used different SynComs (SynCom5-S2 and SynCom5-NS2) (SI Appendix, Fig. S9 and Dataset S1). As predicted, seedlings inoculated with SynCom5-NS1 showed stronger activation of defense-related genes in comparison to seedlings inoculated with SynCom5-S1 (Fig. 3D, SI Appendix, Fig. S9 B–E, and Dataset S6). These genes encompass well-characterized components of the plant immune system, including those involved in the biosynthesis of glucosinolates, as well as RLKs and WRKYs (SI Appendix, Fig. S9D). Many of these genes were also found in clusters SC1 (Fig. 1F) and M5 (Fig. 2C). Thus, efficient suppression of MTI by commensals correlates with enhanced root colonization. Supporting this conclusion, suppressors were detected at higher relative abundances more often than nonsuppressors in roots colonized with SynCom35 (Fig. 3E). Furthermore, strains that we defined as robust suppressors were also classified as robust endophytic compartment colonizers in a previous study using SynCom35 (22), while strains that we classified as nonsuppressors were not (Fig. 3B).
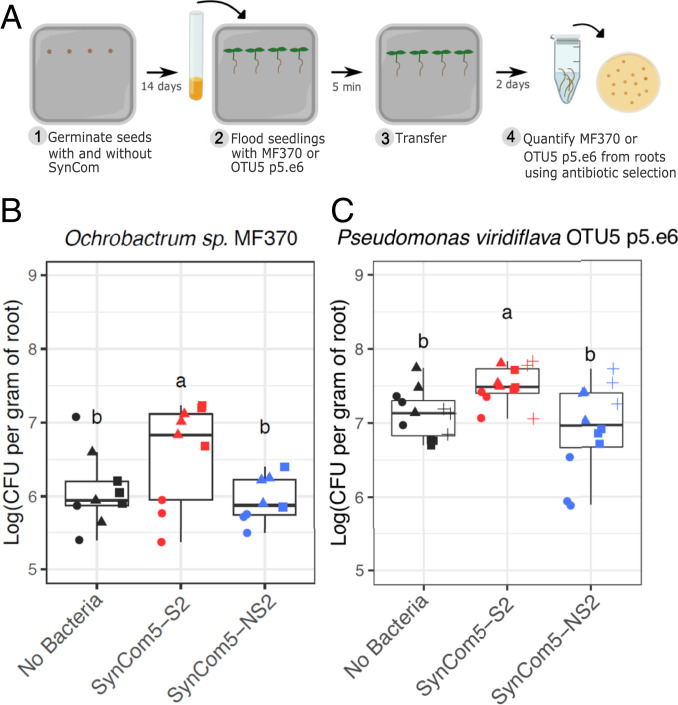
Since suppressor and nonsuppressor strains co-occur in the root microbiota, we evaluated whether modulation of MTI by suppressors enhances the colonization ability of other commensal strains (Fig. 4A). For this, Arabidopsis seedlings were initially grown for 14 d in the presence of synthetic communities comprised of five suppressors (SynCom5-S2) or five nonsuppressors (SynCom5-NS2) (Dataset S1). Subsequently, plants were flood-inoculated for 5 min with the inducer Ochrobactrum sp. MF370 and grown for 48 h. More Ochrobactrum sp. MF370 cells were recovered from roots colonized with SynCom5-S2 compared to those inoculated with SynCom5-NS2 or grown axenically (Fig. 4B). The same result was observed for a second commensal microbe, Pseudomonas viridiflava OTU5 strain p5.e6 (Fig. 4C), which can also be an opportunistic plant pathogen (31). Thus, suppressor strains can enhance the root colonization capacities of other microbes, indicating that MTI, and its suppression, can contribute to the control of microbial loads in the roots.
Fig. 4.
Immunomodulatory bacteria enhance the root colonization ability of other commensal strains. (A) Cartoon representation of the experimental system employed. Arabidopsis seedlings were grown axenically, in the presence of a community of five suppressors (SynCom5-S2) or in the presence of five nonsuppressors (SynCom5-NS2) and then inoculated with one of the commensal strains. (B) Quantification of Ochrobactrum sp. MF370 cells (kanamycin resistant) from Arabidopsis roots (n = 9). (C) Quantification of P. viridiflava OTU5 p5.e6 cells (gentamycin resistant) from Arabidopsis roots (n = 12). Different symbols represent independent experimental repetitions. Multiple comparisons were performed with ANOVA followed by a Tukey test (α = 0.05).
Immunomodulatory Strategies in Commensals.
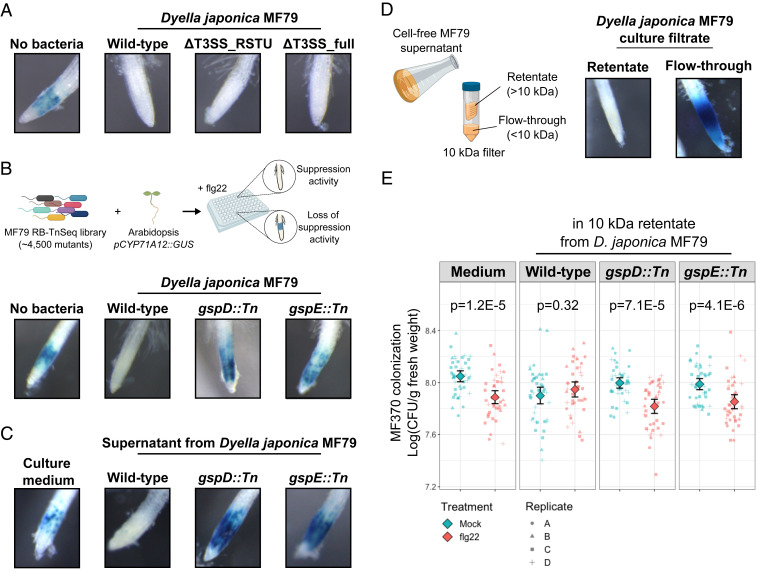
Finally, we sought to identify the mechanism of suppression by one of the robust suppressor strains, D. japonica MF79 (Fig. 3A). This strain carries a T3SS, a molecular syringe that is widely employed by bacterial pathogens to inject virulence effectors into the cells of their hosts to suppress MTI (32). Thus, we tested the hypothesis that this commensal relies on the T3SS to interfere with plant immunity. We constructed two T3SS deletion strains and evaluated their ability to modulate the root response to flg22 using the pCYP71A12::GUS reporter line. The first mutant (MF79 ΔT3SS_RSTU) deletes the T3SS inner membrane components (SctRSTU), whereas the second mutant (MF79 ΔT3SS_full) deletes the full T3SS locus (SI Appendix, Fig. S10A). Interestingly, neither of these mutants were impaired in their ability to suppress the root response to flg22 (Fig. 5A). This demonstrates that the T3SS in this strain is dispensable for immune suppression.
Fig. 5.
D. japonica MF79 requires the T2SS to suppress the root response to flg22. (A) The T3SS is not required for MTI suppression by D. japonica MF79. Two independent mutants (ΔT3SS_RSTU and ΔT3SS_full) retained the ability to prevent activation of the pCYP71A12::GUS reporter in roots exposed to 100 nM flg22 for 5 h. (B) The screening of a transposon insertion library revealed two major components of the bacterial T2SS (GspD and GspE) as required for suppression of the root response to flg22. The diagram (Upper) summarizes the strategy employed in the screening. Additional mutants and replicates are shown in SI Appendix, Fig. S10B. (C) The cell-free supernatant of wild-type D. japonica MF79, but not of T2SS mutants, is sufficient for the suppression of the root response to flg22. (D) The molecule responsible for the suppression activity is larger than 10 kDa. The diagram (Left) illustrates the preparation of 10-kDa retentate and flow-through from bacterial cultures. (E) The 10-kDa retentate of wild-type D. japonica MF79 prevents the flg22-mediated reduction of Ochrobactrum sp. MF370 colonization. Four independent experiments were performed (n = 42 per condition). Diamonds represent means with two times SE. P values were determined with the Wilcoxon test. Root images were captured at 43× magnification on a Leica M205 FA fluorescence stereo microscope.
To identify novel D. japonica MF79 genes required for the suppression activity, ∼4,500 strains from a transposon insertion library (33) were screened on the pCYP71A12::GUS reporter line in a high-throughput 96-well format. We found six mutant strains that were unable to suppress the activation of the pCYP71A12::GUS reporter by flg22 (Fig. 5B). Remarkably, all six transposon insertions mapped to the T2SS: one to the intracellular component gspE and five (four independent transposon hits, one duplicate) to the outer membrane component gspD (SI Appendix, Fig. S10B and Dataset S7). Thus, the T2SS is required for the secretion of substrates with immune suppressive activity to the extracellular space. Wild-type and mutant strains grew at the same rate in vitro (SI Appendix, Fig. S10C), suggesting that the loss of suppression ability in the gspD and gspE mutants is not merely due to growth defects. This was confirmed by experiments in which supernatants of bacteria grown in plant-free medium were used. Although no bacterial growth differences were observed in vitro, supernatant from wild-type D. japonica MF79 suppressed the root response to flg22, while neither the gspD nor gspE mutant supernatants had suppressive activity (Fig. 5C). The suppressive activity from wild-type D. japonica MF79 was retained by the filter of a 10-kDa protein concentrator, but not in the flow-through (Fig. 5D). Importantly, the 10-kDa retentate from wild-type D. japonica MF79 prevented the flg22-mediated reduction in root colonization of the nonsuppressor strain Ochrobactrum sp. MF370, and this effect required type 2 secretion components (Fig. 5E). Both the gspD and the gspE mutants were slightly poorer colonizers of Arabidopsis roots than the wild-type strain in monoassociation assays (SI Appendix, Fig. S10D). Taken together, these results suggest that suppression of the plant response to flg22 by D. japonica MF79 is mediated by at least one T2SS-secreted protein that is larger than 10 kDa and that the suppressive ability correlates with enhanced colonization of Arabidopsis roots.
Discussion
MTI is a well-known plant immune response to invading microbes, and its modulation by disease-causing pathogens has been extensively reported (10). Recent evidence suggests that the ability to suppress MTI has also evolved in commensal microbes (2). While previous studies focused on single microbes and on relatively simple MTI readouts, we demonstrate that a complex bacterial community of Arabidopsis root commensals interferes with a specific sector of the plant transcriptional response to the MAMP flg22 (cluster SC1) (Fig. 1 F and G). The suppression of these genes thus defines the set of host immune response genes that are likely modulated to facilitate successful commensal colonization. Suppression of subsets of MAMP-responsive genes has been observed in Arabidopsis roots colonized by the single commensal strains P. simiae WCS417 and Bacillus subtilis FB17 (15, 34). Although these studies did not investigate the effect of the microbes on the immune response elicited with exogenous MAMPs, they support our conclusion that diverse commensal bacteria interfere with specific sectors of the plant immune system. Thus, similar to what has been established for pathogenic microbes, suppression of the plant immune system is a hallmark of root colonization by commensals. Importantly, our findings agree with and significantly extend the proposal that the ability to suppress the host immune system is common and taxonomically widespread among root-associated bacteria (12, 13), a concept that has also emerged for human gut commensals (11). Recent evidence suggests that root commensals may avoid host cell damage, a signal that enhances MTI in root tissues, to prevent the activation of plant immunity during colonization (8). Since we observed a clear response to flg22 in our control conditions (no bacteria and heat-killed SynCom35), suppression of defense-related genes in roots colonized by SynCom35 is not merely due to damage avoidance. It is noteworthy that the suppression of immune responses is a dominant trait in our SynCom.
A distinctive feature of our study is the dissection of the immunomodulatory capacity of 35 individual commensal strains and the integration of this information with the outcome observed in a community context. Monoassociation experiments revealed that root commensals trigger both shared and unique responses in Arabidopsis, without any obvious taxonomic signature (Fig. 2 and SI Appendix, Fig. S6). Interestingly, a common set of defense-related genes was suppressed by most strains (cluster M5) (Fig. 2C), indicating that phylogenetically diverse bacteria likely employ multiple mechanisms to exert a redundant immunomodulatory effect on the host. Many of these genes were also suppressed by a different bacterial community characterized in an independent study (SI Appendix, Fig. S3A) (27), supporting the hypothesis that root commensals manipulate a core set of biological processes in their hosts. Members of the WRKY family of transcription factors (e.g., WRKY28, -30, and -33) were consistently activated by flg22 in the roots, but suppressed by our strains and by those reported by Ma et al. (27) (SI Appendix, Fig. S3A). These WRKYs might constitute key regulators of the plant immune response to commensals, possibly playing important roles in the homeostasis of immune responses during commensal community assembly. Importantly, a robust, widespread, and redundant immunomodulatory capacity among commensals could buffer plant-associated communities against perturbations and maintain homeostasis when some members are lost or when external stimuli are present, as demonstrated for SynCom35 (Fig. 1A). Furthermore, the unique effect of each strain on the host may account for specific features, such as the ability to prime the plant immune system or to trigger induced systemic resistance (35).
One hypothesis supported by our findings is that the presence of suppressor strains might benefit other nonsuppressors in the context of plant-associated communities. Although suppressors were often more efficient root colonizers than nonsuppressor strains, the latter (including D. japonica MF79 T2SS mutants) could still be recovered from Arabidopsis roots, either in monoassociation or SynCom experiments. This indicates that suppression of the plant immune system likely facilitates bacterial growth within plant tissues but is unlikely to be a requirement for the colonization by most commensal strains. Interestingly, root colonization of nonsuppressive bacteria was enhanced by the presence of a suppressor SynCom (Fig. 4). We predict that this effect might involve specific, rather than general, microbial combinations. Niche occupancy and specific microbe–microbe interactions are important factors contributing to microbial colonization of plant tissues, resulting in different community contexts and complex phenotypes. It is interesting to note, however, that cell-free supernatant from wild-type D. japonica MF79, but not from T2SS mutants, prevented the flg22-mediated reduction in root colonization of the nonsuppressor strain Ochrobactrum sp. MF370 (Fig. 5E). This indicates that nonsuppressor bacteria can indeed benefit from the immunomodulatory activity of other community members under specific circumstances.
Our data suggest that the ability to suppress the plant immune system has evolved several independent times in plant commensals, likely resulting in numerous immunosuppressive mechanisms (2, 4). In contrast to most pathogens or mutualist microbes, root-associated commensals seem to display a much lower degree of host specialization. As a consequence, commensals may interfere with MTI mostly through nonspecific extracellular strategies. In turn, pathogens and specialized symbionts, such as nodule-forming rhizobia, often rely on highly specialized effector proteins that are injected into the host cell through the T3SS (36). Interestingly, D. japonica MF79 is the only suppressor commensal in SynCom35 to carry genes for the T3SS. Nevertheless, D. japonica MF79 ΔT3SS mutants retained their ability to suppress the flg22 response in Arabidopsis roots. Our transposon library screening resulted in six mutants in the T2SS, but no hits in other genes encoding putative T2SS substrates. This suggests that multiple proteins are secreted by D. japonica MF79 to the extracellular space and act redundantly to suppress MTI, consistent with previous demonstrations that T2SS can be required for pathogen virulence (37). Interestingly, only 3 of the 10 robust suppressors of the flg22 response (Fig. 3A) are predicted to carry a T2SS (Dataset S1), indicating the T2SS is dispensable for the suppression activity of most of these strains. We expect that new research will reveal a large number of novel strategies employed by commensals to modulate the plant immune system. Future studies should also elucidate the role of plant genes, specifically those in cluster SC1 (Fig. 1 F and G), in gating the assembly of microbial communities.
Materials and Methods
Details of the methods used in this work, including the preparation of bacterial synthetic communities, flg22 treatments, root length measurements, RNA extraction, RNA-sequencing analysis, DNA extraction, 16S amplicon sequencing, quantification of bacteria colonization of roots, GUS histochemical assays, genetic manipulation of bacteria, and mutant screenings, are described in SI Appendix, SI Text, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Ka-Wai Ma and Dr. Paul Schulze-Lefert for providing the gentamycin-resistant Pseudomonas viridiflava strain OTU5 p5.e6 and for sharing their bacteria colonization protocol and unpublished transcriptome data; Schulze-Lefert laboratory members for comments on the manuscript; the J.L.D. laboratory microbiome group for useful discussions; and Prof. Sarah Grant for comments and suggestions on the manuscript. This work was supported by NSF Grant IOS-1917270 and by Office of Science (Biological and Environmental Research), US Department of Energy Grant DE-SC0014395 (to J.L.D.) J.L.D is an Investigator of the Howard Hughes Medical Institute (HHMI), supported by the HHMI. P.J.P.L.T. was supported by The Pew Latin American Fellows Program in the Biomedical Sciences. N.R.C. was supported by NIH Training Grant T32GM135123.
Footnotes
Competing interest statement: J.L.D. is a cofounder of, and shareholder in, AgBiome LLC, a corporation whose goal is to use plant-associated microbes to improve plant productivity.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2100678118/-/DCSupplemental.
Data Availability
The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE156426).
References
- 1.Fitzpatrick C. R., et al., The plant microbiome: From ecology to reductionism and beyond. Annu. Rev. Microbiol. 74, 81–100 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Teixeira P. J. P., Colaianni N. R., Fitzpatrick C. R., Dangl J. L., Beyond pathogens: Microbiota interactions with the plant immune system. Curr. Opin. Microbiol. 49, 7–17 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Yu K., Pieterse C. M. J., Bakker P. A. H. M., Berendsen R. L., Beneficial microbes going underground of root immunity. Plant Cell Environ. 42, 2860–2870 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hacquard S., Spaepen S., Garrido-Oter R., Schulze-Lefert P., Interplay between innate immunity and the plant microbiota. Annu. Rev. Phytopathol. 55, 565–589 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Lebeis S. L., et al., PLANT MICROBIOME. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349, 860–864 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Kniskern J. M., Traw M. B., Bergelson J., Salicylic acid and jasmonic acid signaling defense pathways reduce natural bacterial diversity on Arabidopsis thaliana. Mol. Plant Microbe Interact. 20, 1512–1522 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Chen T., et al., A plant genetic network for preventing dysbiosis in the phyllosphere. Nature 580, 653–657 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou F., et al., Co-incidence of damage and microbial patterns controls localized immune responses in roots. Cell 180, 440–453.e18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emonet A., et al., Spatially Restricted Immune Responses Are Required for Maintaining Root Meristematic Activity upon Detection of Bacteria. Curr Biol 31, 1012–1028.e7, 10.1016/j.cub.2020.12.048 (2021). [DOI] [PubMed] [Google Scholar]
- 10.da Cunha L., Sreerekha M.-V., Mackey D., Defense suppression by virulence effectors of bacterial phytopathogens. Curr. Opin. Plant Biol. 10, 349–357 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Geva-Zatorsky N., et al., Mining the human gut microbiota for immunomodulatory organisms. Cell 168, 928–943.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu K., et al., Rhizosphere-associated Pseudomonas suppress local root immune responses by gluconic acid-mediated lowering of environmental pH. Curr. Biol. 29, 3913–3920.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Garrido-Oter R.et al.; AgBiome Team , Modular traits of the Rhizobiales root microbiota and their evolutionary relationship with symbiotic Rhizobia. Cell Host Microbe 24, 155–167.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Z., et al., A genome-wide screen identifies genes in rhizosphere-associated Pseudomonas required to evade plant defenses. MBio 9, e00433-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stringlis I. A., et al., Root transcriptional dynamics induced by beneficial rhizobacteria and microbial immune elicitors reveal signatures of adaptation to mutualists. Plant J. 93, 166–180 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plett J. M., et al., Effector MiSSP7 of the mutualistic fungus Laccaria bicolor stabilizes the Populus JAZ6 protein and represses jasmonic acid (JA) responsive genes. Proc. Natl. Acad. Sci. U.S.A. 111, 8299–8304 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang Y., et al., Nonlegumes respond to rhizobial Nod factors by suppressing the innate immune response. Science 341, 1384–1387 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Lakshmanan V., et al., Microbe-associated molecular patterns-triggered root responses mediate beneficial rhizobacterial recruitment in Arabidopsis. Plant Physiol. 160, 1642–1661 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Millet Y. A., et al., Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell 22, 973–990 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rallapalli G., et al., EXPRSS: An Illumina based high-throughput expression-profiling method to reveal transcriptional dynamics. BMC Genomics 15, 341 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denoux C., et al., Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol. Plant 1, 423–445 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castrillo G., et al., Root microbiota drive direct integration of phosphate stress and immunity. Nature 543, 513–518 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rushton P. J., Somssich I. E., Ringler P., Shen Q. J., WRKY transcription factors. Trends Plant Sci. 15, 247–258 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Couto D., Zipfel C., Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16, 537–552 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Gigolashvili T., et al., The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 50, 886–901 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Pascale A., Proietti S., Pantelides I. S., Stringlis I. A., Modulation of the root microbiome by plant molecules: The basis for targeted disease suppression and plant growth promotion. Front. Plant Sci. 10, 1741 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma K.-W., et al., Coordination of microbe-host homeostasis by crosstalk with plant innate immunity. Nature Plants, 10.1038/s41477-021-00920-2 (2021) in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winkelmüller T. M., et al., Gene expression evolution in pattern-triggered immunity within Arabidopsis thaliana and across Brassicaceae species. bioRxiv (2020). [DOI] [PMC free article] [PubMed]
- 29.Gómez-Gómez L., Felix G., Boller T., A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J. 18, 277–284 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Finkel O. M., et al., A single bacterial genus maintains root growth in a complex microbiome. Nature 587, 103–108 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karasov T. L., et al., Arabidopsis thaliana and Pseudomonas pathogens exhibit stable associations over evolutionary timescales. Cell Host Microbe 24, 168–179.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng W., et al., Assembly, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 15, 323–337 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Price M. N., et al., Mutant phenotypes for thousands of bacterial genes of unknown function. Nature 557, 503–509 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Lakshmanan V., Castaneda R., Rudrappa T., Bais H. P., Root transcriptome analysis of Arabidopsis thaliana exposed to beneficial Bacillus subtilis FB17 rhizobacteria revealed genes for bacterial recruitment and plant defense independent of malate efflux. Planta 238, 657–668 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Vogel C., Bodenhausen N., Gruissem W., Vorholt J. A., The Arabidopsis leaf transcriptome reveals distinct but also overlapping responses to colonization by phyllosphere commensals and pathogen infection with impact on plant health. New Phytol. 212, 192–207 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Gourion B., Berrabah F., Ratet P., Stacey G., Rhizobium-legume symbioses: The crucial role of plant immunity. Trends Plant Sci. 20, 186–194 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Cianciotto N. P., White R. C., Expanding role of type II secretion in bacterial pathogenesis and beyond. Infect. Immun. 85, e00014-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE156426).