Significance
Tuberculosis, caused by Mycobacterium tuberculosis, remains a major global public health problem. Deciphering the complex biology of M. tuberculosis will aid in the development of new therapeutics to combat this pathogen. Encapsulins, naturally encapsulating specific cargo proteins, are a recently discovered class of smaller proteinaceous compartments found in bacteria and archaea and needed for essential physiological processes. Here, we report the structural and mechanistic characterization of a native DyP-packaging encapsulin from the Mycobacterium smegmatis, a model system for M. tuberculosis. Our results contribute to the understanding of the assembly and physiological role of the encapsulin systems and provide a rational framework for antituberculosis drug design.
Keywords: encapsulin nanocompartment, dye-decolorizing peroxidase, cryo-electron microscopy, Mycobacterium smegmatis
Abstract
Encapsulins containing dye-decolorizing peroxidase (DyP)-type peroxidases are ubiquitous among prokaryotes, protecting cells against oxidative stress. However, little is known about how they interact and function. Here, we have isolated a native cargo-packaging encapsulin from Mycobacterium smegmatis and determined its complete high-resolution structure by cryogenic electron microscopy (cryo-EM). This encapsulin comprises an icosahedral shell and a dodecameric DyP cargo. The dodecameric DyP consists of two hexamers with a twofold axis of symmetry and stretches across the interior of the encapsulin. Our results reveal that the encapsulin shell plays a role in stabilizing the dodecameric DyP. Furthermore, we have proposed a potential mechanism for removing the hydrogen peroxide based on the structural features. Our study also suggests that the DyP is the primary cargo protein of mycobacterial encapsulins and is a potential target for antituberculosis drug discovery.
Compartmentalization is used by cells to overcome many difficult metabolic and physiological challenges (1). Eukaryotes employ membrane-bound organelles such as the mitochondrion (2); however, most prokaryotes rely on alternative proteinaceous compartments to achieve spatial control (3), one of which is the encapsulin nanocompartment.
Encapsulins are newly identified nanocompartments but have already been applied in various scientific fields due to the unique structures (4, 5). It has been reported that more than 900 putative encapsulin systems in bacteria and archaea exist and are distributed across 15 bacterial and two archaeal phyla (6, 7), suggesting they are functionally diverse. Encapsulins are made of one type of shell protein, as opposed to several as is observed in many bacterial microcompartments (8, 9). The key feature of encapsulin systems is that cargo proteins can be specifically encapsulated and targeted to the encapsulin capsid interior, using a selective C-terminal sequence referred to as targeting peptides (TPs) (10). The functions of the nanocompartment are associated with the functions of its protein cargo. Many functionally diverse cargo proteins are associated with encapsulins, including dye-decolorizing peroxidases (DyPs) (11), ferritin-like proteins (FLP) (12), hydroxylamine oxidoreductase (HAO) (13), and cysteine desulfurases (14). Moreover, it has been shown that some encapsulin systems may possess multiple cargo proteins, which are made up of one core cargo protein and up to three secondary cargo proteins according to the TPs (6). Notably, a large proportion of native cargo proteins are DyP-type peroxidases, conferring the resistance of the cell to oxidative stress (6, 7, 11, 15–18). However, to date, the structural information on the cargo-encapsulated encapsulins is not yet available (SI Appendix, Table S1), and thus, little is known about the structural arrangement and mechanistic features of the cargo proteins loaded in the encapsulins.
Actinobacteria harbors the largest number of encapsulin or encapsulin-like systems (6). DyP-containing encapsulins have already been reported from mycobacteria, including Mycobacterium smegmatis (15) and Mycobacterium tuberculosis (19). These have been considered as potential biomarkers to detect active tuberculosis (TB) (20). In the present study, we have isolated and characterized a DyP-loaded encapsulin system from M. smegmatis, which is commonly used as a model organism in studying the biology of the M. tuberculosis (21). We have determined its complete high-resolution structure by cryogenic electron microscopy (cryo-EM). Our results have revealed the interactions between the CFP-29 (a 29 kDa culture filtrate protein) shell and DyP cargo and a potential antioxidation mechanism. Our study also lays the foundation for the discovery of new diagnosis protocols and treatments of TB.
Results and Discussion
Isolation and Characterization of M. smegmatis DyP-Loaded Encapsulin System.
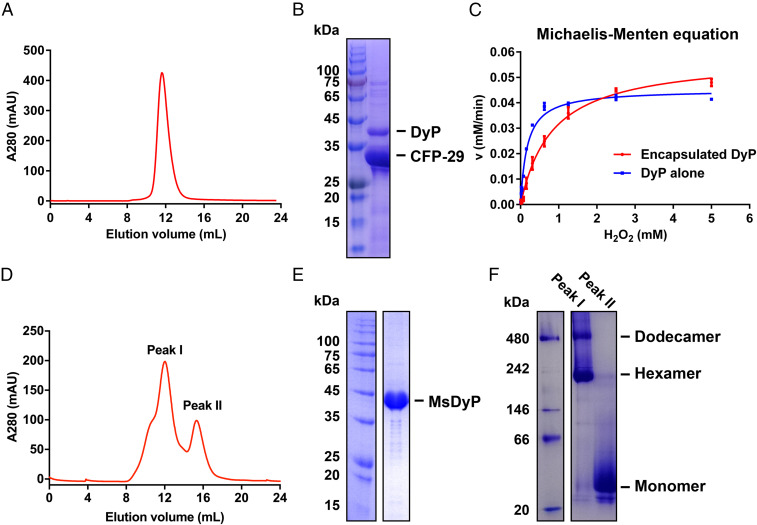
The native encapsulin protein was successfully purified by nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatography and gel filtration (Fig. 1A). SDS-PAGE (sodium dodecyl sulfate–polyacrylamide gel electrophoresis) combined with the mass spectrometry (MS) analysis showed that CFP-29 and DyP proteins are constituents (Fig. 1B and SI Appendix, Table S2). In terms of the peroxidase activity, DyP, both encapsulated and unencapsulated, showed equivalent levels of free radical scavenging capacity with kcat values of 119.54 ± 3.15 s−1 and 92.90 ± 1.61 s−1, respectively, suggesting that the encapsulin shell provides structural protection for DyP without affecting its free radical scavenging capacity (Fig. 1C).
Fig. 1.
Purification and characterization of the native encapsulin and MsDyP from M. smegmatis. (A) Size exclusion chromatography. The elution curve of Superose 6 Increase is shown in red, and the encapsulin elution volume is 12 mL. (B) SDS-PAGE analysis of the encapsulin. (Left) The protein ladder. (Right) The elution fraction from A. The bands corresponding to DyP and CFP-29 are labeled with black lines, which were then identified by MS. (C) Analysis of peroxidase activity of DyP alone and DyP-loaded encapsulin at the same heme b concentration. The data represents the mean ± SD (bars) of triplicates for each concentration. (D) Representation of two distinct elution peaks performed on a Superdex 200 10/300 GL column, yielding a dodecamer peak I (dodecamer/hexamer) and peak II (monomer). (E) SDS-PAGE analysis of the mixed fractions of the peak I/II. (F) Blue native polyacrylamide gel electrophoresis analysis of the fractions of the peak I and II.
Spherical Shell Forms Largely Positively Charged Fivefold Pores.
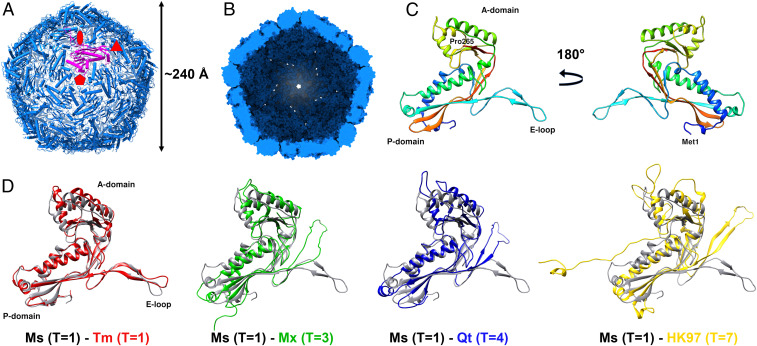
We have determined a 2.5 Å cryo-EM structure of the M. smegmatis encapsulin nanocompartment (Fig. 2 and SI Appendix, Figs. S1 and S2 and Table S3). As the shell-forming proteins of the encapsulin, 60 CFP-29 monomers assemble into a spherical architecture. It is a superstructure with a triangulation number of 1 (T = 1), and its diameter and thickness are ∼240 Å and 25 Å, respectively (Fig. 2 A and B).
Fig. 2.
The overall architecture of the encapsulin shell from M. smegmatis. (A) View down the fivefold symmetry axis. Symmetry axes are marked with red symbols. A single pentamer monomer is shown in purple. (B) View to the inside of the shell from the cross-section. (C) The structure of the shell monomer is organized into three conversed folds, P-domain, A-domain, and E-loop. (D) Structural alignment of shell monomers from M. smegmatis (Ms) T = 1, T. maritima (Tm) T = 1 (PDB 3DKT), M. xanthus (Mx) T = 3 (PDB 4PT2), Q. thermotolerans (Qt) T = 4 (PDB 6NJ8), and bacteriophage HK97 T = 7 (PDB 2FT1).
The CFP-29 monomer consists of three conserved folds, namely the A-domain, the P-domain, and the E-loop (Fig. 2C). The A-domain, including the helical segments, β-sheets, and C terminus, rarely connects to the other monomers in the assembly but mediates interactions at the fivefold symmetry axis. The main body of CFP-29 is formed by the P-domain, which contains the N-terminal helix, spine helix, and a four-stranded β-sheet. It is involved in mediating contacts at the threefold symmetry interface. The E-loops are located at the capsomer interfaces and play an essential role in forming the interface of the twofold symmetry axis by providing a four-stranded β-sheet structure formed by adjacent related subunits. Comparing it to the recently resolved encapsulin shells from Thermotoga maritima (T = 1) (11), Myxococcus xanthus (T = 3) (22), Quasibacillus thermotolerans (T = 4) (12), and the major capsid protein gp5 of the HK97 virus (T = 7) (23), most structural elements are conserved and have the Johnson fold (23), but the orientations of domains are different (Fig. 2D). By comparing with the two different kinds of Johnson folds represented by HK97 gp5 (24) and BPP-1 Bbp17 (25), which have similar three-dimensional (3D) structure arrangement but different topology, CFP-29 adopts the fold represented by HK97 gp5 but not BPP-1 Bbp17 (SI Appendix, Fig. S3). It has been proposed that prokaryotic encapsulins and certain phage capsids have a common evolutionary origin (11). According to the hypothesis of a common viral ancestor that was present before the host organisms diverged (26), CFP-29 in Mycobacterium seems to have evolved from an HK97-like virus ancestor which was proposed to have emerged before the separation of the bacterial and archaeal kingdom (26). However, given the herein-described cellular function of CFP-29 and the diversity in terms of the amino acid sequences within and between all the known encapsulins and virus coat proteins, the structural similarities may also just reflect convergence to one of a limited number of possible solutions to the problem of making a viable capsid (26). In this case, viruses such as HK97 could have originated from a similar CFP-29–like cellular nanocompartment assembly by a switch of the specificity from encapsulating proteins to encapsulation of nucleic acids, perhaps by simply assembling around its own messenger RNA during translation (27).
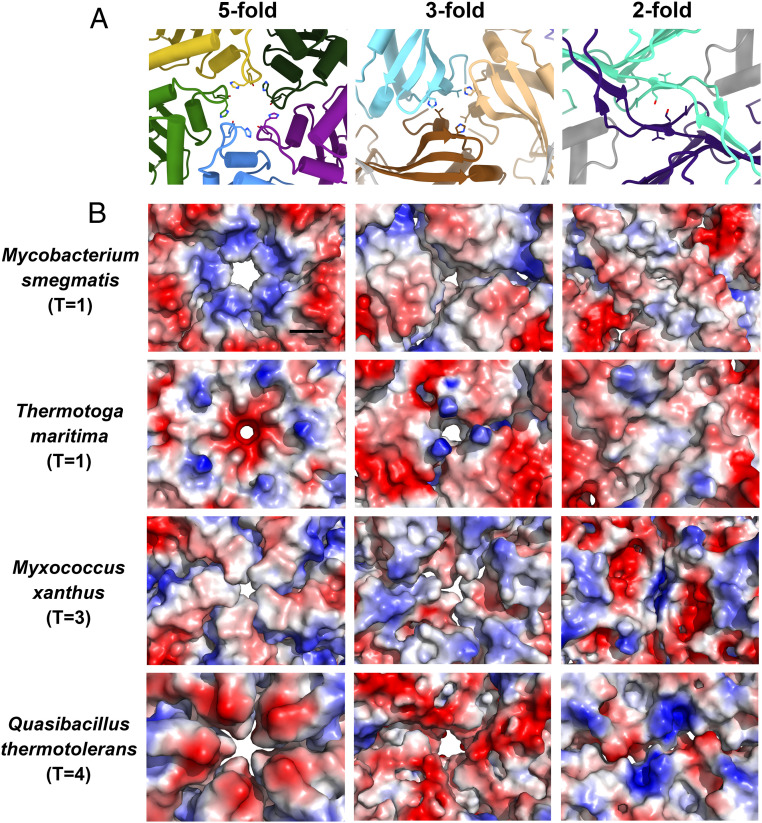
Similar to previous studies (11, 12, 22), the M. smegmatis encapsulin shell also forms several pore openings (Fig. 3A). Two holes are located at the points of the threefold and fivefold symmetry axes. There is also a hole at the interface formed by related two E-loop regions. However, a major difference is that the diameter of the fivefold pore in the M. smegmatis encapsulin is the largest among its known counterparts (Fig. 3B). After the subtraction of the Van der Waals radii of the surface atoms, the diameter of the mycobacterial pentameric pore is about 6.8 Å. Around this pore, several histidine residues exist as a ring, resulting in an array of a large number of positive charges.
Fig. 3.
The pores on the encapsulin capsids. (A) Cartoon representation of the pores down the fivefold, threefold, and twofold symmetry axes on the encapsulin shell of M. smegmatis. (B) Electrostatic surface representation of the pores down the fivefold, threefold, and twofold symmetry axes on the encapsulin capsids from M. smegmatis, T. maritima (PDB 3DKT), M. xanthus (PDB 4PT2), and Q. thermotolerans (PDB 6NJ8). (The black scale bar is 10 Å.)
Charged pores of the encapsulin may mediate substrate specificity, regulating the flux of substrates in and out of encapsulin that are available to the protein cargo (28). Previous studies suggest that the encapsulins from T. maritima (11), M. xanthus (22), and Q. thermotolerans (12) tend to package a ferritin-like protein to regulate the metabolism of cationic iron. Coincidentally, their pentameric pores have negative charges distributed on the surface. Besides, the DyP-loaded encapsulin of M. tuberculosis only has minimal peroxidase activity with the neutral guaiacol as a substrate compared with the negatively charged ABTS as a substrate (19). It is worth noting that the pentameric pore of the encapsulin shell of the M. smegmatis, which is closely related to M. tuberculosis, also has many positive charges distributed on the surface (Fig. 3B). Similar observations that the charges aid ionized substrate transport have also been presented in the carboxysome shell proteins and bicarbonate transport systems (29, 30).
Dodecameric DyP Cargo in the Encapsulin Shell.
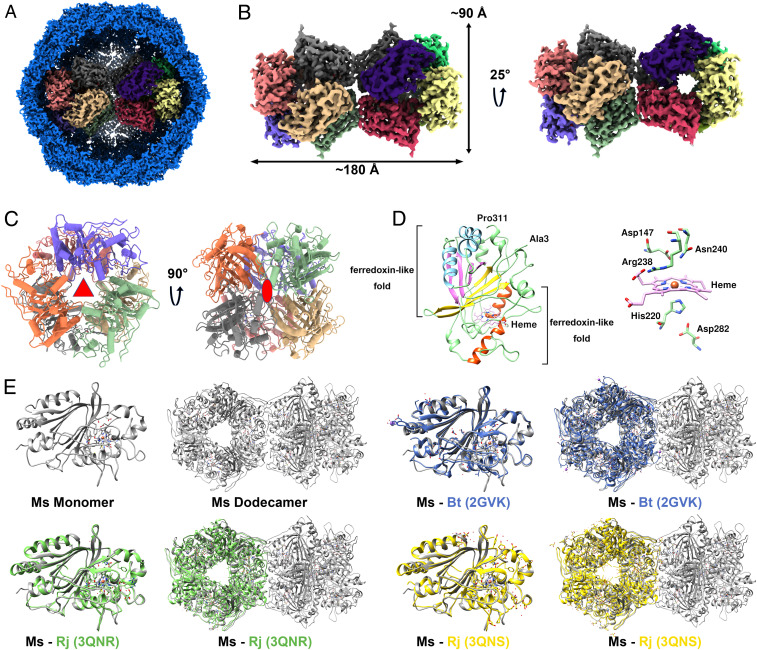
DyP proteins are heme-containing superfamily enzymes and have been identified in the genomes of fungi, bacteria, and archaea (31). DyP protein has been suggested to be the cargo of the encapsulin (11, 17), but this hypothesis lacks direct experimental evidence because only the low-resolution structure of the DyP-packaging encapsulin has been solved. Here, we determined the structure of dodecameric DyP as the cargo of encapsulin at 3.7 Å by cryo-EM (Fig. 4 and SI Appendix, Figs. S1 and S2 and Table S3). Because of the flexible interactions between the DyP and the encapsulin shell (Movie S1), we performed local refinement for the encapsulin shell and the DyP cargo. Hence, a composited map consisting of the individual encapsulin shell and dodecameric DyP was obtained. However, ∼26% of encapsulin particles are present as a single hexamer (SI Appendix, Fig. S1), which are likely to be assembly intermediates of the dodecameric DyP encapsulin. Considering the low ratio of the particles containing a single DyP hexamer, we focused on resolving the structure of the dodecameric DyP.
Fig. 4.
The structure of DyP cargo and structural alignment with other DyPs. (A) The composite map of the complete DyP-loaded encapsulin. (B) Overall structure of the dodecamer DyP. (C) The symmetric structure of one DyP hexamer. The threefold and twofold axis of the hexamer are labeled with red symbols. (D) Cartoon representation of DyP monomer structure. Heme and five important residues surrounding the prosthetic group in a stick model are shown. (E) Structural comparison of monomeric and oligomeric DyPs from M. smegmatis (MsDyP), R. jostii RHA1 (RjDyP), and B. thetaiotaomicron (BtDyP). The DyPs are displayed as follows: MsDyP (gray), BtDyP (PDB 2GVK, blue), RjDyP (PDB 3QNR, green and PDB 3QNS, yellow).
In the present study, the dodecameric DyP stretches across the interior of the encapsulin, with a length of 180 Å and a height of 90 Å (Fig. 4B). This is a dodecameric structure of a DyP-type peroxidase, which also has a twofold axis between two hexamers. In the hexamer, it can be divided into two layers, and each layer is formed by three monomers with a threefold axis. There is a large hole running through the center of the two layers. This hole contains a large number of positive charges, attracting and channeling negatively charged ions (described below). These two layers are also arranged in the form of a twofold axis of symmetry within the hexamer (Fig. 4C).
The protomer in this dodecameric DyP has also a core structure, which consists of N-terminal and C-terminal ferredoxin-like fold domains, each including two helices and a four-stranded β-sheet. Near the ferredoxin-like fold domain, there is a heme prosthetic group. His220 functions as the ligand for the Fe atom of the heme (Fig. 4D). The heme-binding pocket is essential to confer the catalytic activities of DyP (32). This active site is also involved in other residues, including Asp147, Arg238, Asn240, and Asp282, which are well conserved among the DyPs (SI Appendix, Fig. S4). Additionally, the secondary structural elements and overall topology of M. smegmatis DyP are remarkably conserved compared to the DyP structures of Bacteroides thetaiotaomicron (33) and Rhodococcus jostii RHA1 (34), with a homology of 50.49 and 53.35% at the amino acid levels, respectively. However, the former mainly exists as a dodecamer in the encapsulin, but the latter forms a hexameric state. Even so, the assembly of the hexamers from R. jostii RHA1 and B. thetaiotaomicron is also highly similar to that of the hexamer in the M. smegmatis dodecameric DyP (Fig. 4E). Similarly, the purified nonencapsulated DyP, which was overexpressed alone in M. smegmatis, is also mainly in the hexameric state (Fig. 1 D–F).
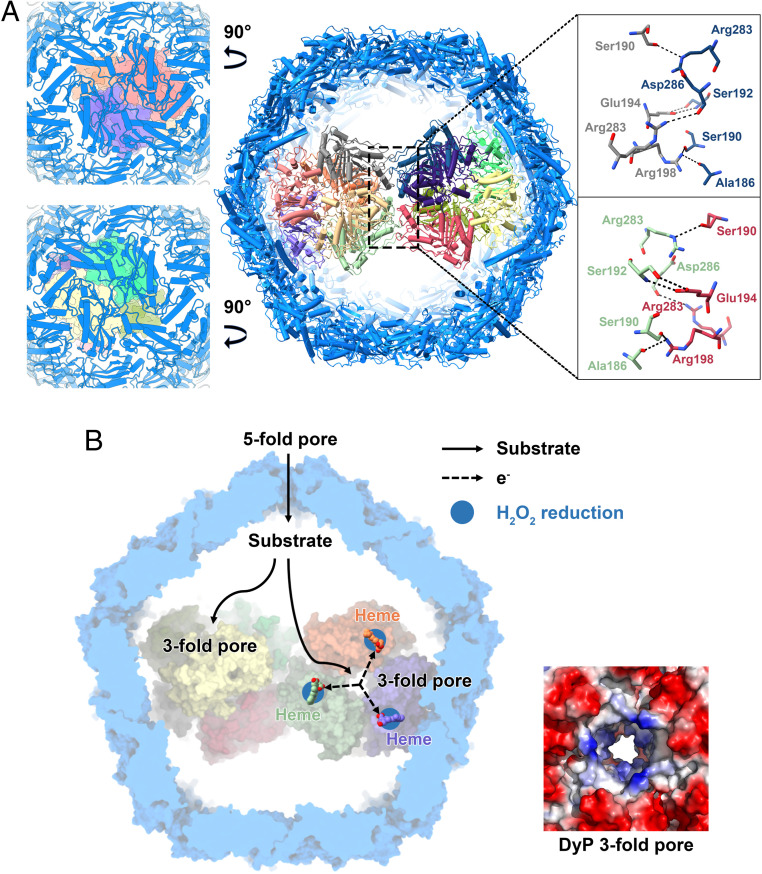
Although the native or foreign cargo proteins can be encapsulated into the encapsulin via the TP binding (10), the interactions between them have not thoroughly been investigated. In the present study, two copies of the DyP hexamer are stacked together, and several hydrogen bonds are observed between the two hexamers (Fig. 5A). Here, the threefold symmetry of the individual hexamers is lost, but new symmetry is gained. The dodecameric arrangement of DyP enzymes appears to obey D2 symmetry, which becomes accommodated in an icosahedral symmetry shell and also makes equivalent interactions of both ends with the interior of the icosahedral shell (Fig. 5A). Nevertheless, according to the observation from our cryo-EM analysis (Movie S1), the dodecamer is not rigid, so it is difficult to observe the exact interactions between the DyP and the encapsulin shell. In addition, due to the flexibility of TP, we also have not observed it in our structure, which is in line with the previous study (12).
Fig. 5.
DyP encapsulin coassembly and the model for the potential pathway of the endogenous substrate. (A) The interactions between the two hexamers of DyP and the views through the shell into the DyP. The residues involved in interactions are denoted by the dotted boxes, and hydrogen bonds are indicated by dashed lines. (B) The potential substrate transport pathway (Left) and the electrostatic surface representations of the threefold channel of the DyP hexamer (Right).
Potential Antioxidation Mechanism of the Mycobacterial DyP-Loaded Encapsulin.
The pathogenesis of M. tuberculosis is contingent upon an ability to evade the host immune response and the bactericidal reactive oxygen species generated by the host (35, 36). A connection to oxidative stress is seen in the encapsulin found in M. tuberculosis. It has been reported that the M. tuberculosis encapsulin could potentially encapsulate the DyP, bacterioferritin B (BrfB), or 7,8-dihydroneopterin aldolase (FolB) via a unique C-terminal extension (19). Each of these three cargo proteins has independent antioxidant activity (37–40). However, it is unknown which cargo protein(s) that mycobacterial encapsulin encapsulates in vivo. In the present study, we successfully isolated a native encapsulin of M. smegmatis after exposure to stationary phase stress, which can be thought of as simulating the existing condition of M. tuberculosis inside the host (41). According to the structural information here, it has been identified as a native DyP cargo-loaded encapsulin system. This structural information suggests that the DyP protein is the primary cargo and protects M. tuberculosis from host oxidative assault including hydrogen peroxide (H2O2). It has been reported that the free DyPs and even in the encapsulated form can successfully catalyze the H2O2-mediated oxidation of various substrates (18). Therefore, these data suggest that mycobacterial DyP-containing encapsulins play a key role in combating oxidative stress.
Dye-decolorizing (DyP-type) peroxidases as a relatively new class of peroxidases were found to have broad substrate specificity, as also artificial electron donors (e.g., ABTS, a classical peroxidase substrate) (34), nonphenolic lignin compounds (42), azo-dyes (43), manganese (44), aromatic sulphides (45), and β-carotene (46) can be oxidized. In the present structure, the fivefold pore of the CFP-29 encapsulin is the largest channel on the encapsulin surface and contains a large number of positive charges (Fig. 3B). Therefore, besides the H2O2, the endogenous substrates of mycobacterial DyP-loaded encapsulins are likely the negatively charged aromatic compounds, and the origin of them transported through the encapsulin is the fivefold pore here (Fig. 5B). Current studies on DyPs showed that high-turnover oxidation of dyes and other bulky substrates occurs via long-range electron transfer from the enzyme surface (47). In line with the fivefold pore of the encapsulin, the threefold central channels of both DyP hexamers are highly positively charged. It might guide the negatively charged substrates flowing into the hexamer interior and accomplish the electron transfer to the heme group somewhere on the surface of the enzyme (Fig. 5B).
Conclusions.
We have determined a complete high-resolution structure of the native DyP-packaging encapsulin isolated from M. smegmatis and depicted its assembly mechanism and structural features. Our results reveal that the shell plays a role in stabilizing the dodecameric DyP. The structural information suggests that the DyP is the primary cargo protein of mycobacterial encapsulins and is a potential target for antituberculosis drug discovery. We have proposed a potential mechanism for removing H2O2. Further, our study provides insight into the assembly and physiological role of the cargo-loaded encapsulin systems.
Materials and Methods
Bacteria Cell Culture and Membrane Preparation.
Bacteria cell culture and membrane preparation were performed with some modifications to that described in previously published protocols (48, 49). M. smegmatis strain mc2 155 was grown in Luria-Bertani (LB) culture medium with 25 μg/mL carbenicillin and 1% (vol/vol) Tween 80 (Aladdin) at 220 rpm 37 °C. The growth curve was detected by OD600. Cells were harvested in the late stationary phase by centrifugation at 4,000 × g for 10 min (yielding about 40 g wet weight per 6 L). Cell pellets were washed three times with Buffer A (20 mM 3-(N-morpholino)propanesulfonic acid [MOPS], pH 7.4, 100 mM NaCl, 1 mM ethylenediaminetetraacetic acid, and 1 mM phenylmethylsulfonyl fluoride). For membrane preparation, cell pellets were then resuspended in Buffer A and lysed through an ATS AH-basic high-pressure homogenizer (1,000 to 1,100 bar). The remaining nucleic acids in retentate were digested by DNase (1 U/μL) and RNase A (10 mg/mL) for 1 h at 4 °C. The cell lysate was centrifuged at 23,700 × g for 10 min to remove unbroken cells and cell debris. The clarified supernatant was collected and ultracentrifuged at 150,000 × g in a Type 45 Ti Rotor (Beckman) for 2 h. The membrane pellets were collected for encapsulin purification.
Encapsulin Purification.
It was reported that there is a ring of histidines along the cytosolic face of the encapsulin shell of Thermotoga maritima (11). Based on this, Ni-NTA affinity chromatography and gel filtration were used to isolate the native encapsulin protein in the present study. Protein purification was always performed at 4 °C unless otherwise noted. Membranes were ground in Buffer A containing 1% (wt/vol) Brij 35. Encapsulin were extracted from the membrane with slow stirring for 2 h. Insoluble components were pelleted by centrifugation at 39,200 × g for 45 min. The cleared supernatant was loaded onto a 5 mL Ni-NTA column pre-equilibrated in Buffer B (20 mM MOPS, pH 7.4, 100 mM NaCl, and 0.05% [wt/vol] Brij 35) for affinity chromatography. The resin was washed after protein binding using Buffer B containing 50 mM imidazole. Finally, encapsulin was eluted with Buffer B containing 500 mM imidazole and concentrated to a final volume of 0.5 mL by ultrafiltration using the 100 kDa cutoff filter (Millipore). Additional purification was performed by gel filtration chromatography (Superose 6 Increase 10/300 GL, GE Healthcare) pre-equilibrated in Buffer A. The fractions (corresponding to elution volume between 11 and 13 mL) were pooled for SDS-PAGE and peroxidase activity analysis. Protein was concentrated and quantified by a NanoDrop 2000 instrument (Thermo Fisher Scientific) and then stored at −80 °C.
Characterization of the Components of Encapsulin.
SDS-PAGE was used to investigate the components of encapsulin. The electrophoretic bands were analyzed by MS at the National Center for Protein Science. All experiments were performed in triplicate.
MsDyP Expression and Purification.
The coding sequence of MsDyP (Dyp-type peroxidase, MSMEG_5829) was amplified from the genomic DNA of M. smegmatis strain mc2 155 and inserted into the modified shuttle vector pMV-261 with a C-terminal 10 × histidine tag. The recombinant plasmid was sequenced and transformed into mc2 155 competent cells by electroporation. The colony was inoculated into the LB culture, with 50 μg/mL kanamycin and 1% (vol/vol) Tween 80, that was induced with 0.2% acetamide and 25 μg/mL hemin at 16 °C for 4 d. Cells were harvested by centrifugation at 4,000 × g for 10 min. For his-tagged DyP purification, the cell pellets were resuspended in Buffer A and disrupted through the high-pressure homogenizer at 800 bar. After centrifugation at 39,200 × g for 40 min, the clarified lysate was then loaded onto a 2 mL Ni-NTA column pre-equilibrated with Buffer A. After resin washing, MsDyP was eluted with Buffer A containing 500 mM imidazole, concentrated using concentrators with the appropriate 30 kDa cutoff (Millipore), and finally purified by size exclusion chromatography (Superdex 200 10/300 GL, GE Healthcare). Fractions were collected and analyzed by SDS-PAGE and blue native polyacrylamide gel electrophoresis.
Peroxidase Activity of the Sample.
The peroxidase activity was carried out utilizing H2O2 as the electron acceptor and pyrogallol (Sigma P0381-25g) as an electron donor, with minor modifications as described previously (18, 19). Oxidation of pyrogallol was carried out in the presence of 2 mM H2O2, monitoring at 430 nm (ε 2,470 M−1 cm−1) in 50 mM Hepes buffer pH 5.5 at 25 °C, using a BioTek Gen5 Spectrophotometer. Steady-state kinetic assays were conducted in triplicate in the presence of 0.005 to 5 mM H2O2, with pyrogallol kept at 320 mM. Kinetic parameters were determined by nonlinear curve fitting using GraphPad Prism 6.0 software.
Cryo-EM Sample Preparation.
The purified encapsulin sample was concentrated to 10 mg/mL before sample preparation. After concentration, 4 μL of the sample solutions was applied to a glow-discharged Quantifoil R 1.2/1.3 200 mesh Holey Carbon gold grid, blotted for 3.0 to 4.0 s, and immediately plunge frozen in liquid ethane using a Thermo Fisher Scientific Vitrobot Mark IV.
Cryo-EM Data Acquisition.
The encapsulin dataset was collected on a Thermo Fisher Scientific 300 keV Titan Krios microscope equipped with a Gatan GIF K2 direct electron detector. The images were acquired using the automated data collection program SerialEM (50), which were recorded in super-resolution mode with a super-resolution pixel size of 0.52 Å (a physical pixel size of 1.04 Å) at a nominal magnification of 130 k× and a slit width of 20 eV. Each movie was recorded in 40 frames with a total exposure time of 6.4 s and a total exposure dose of 60 e−/Å2. The nominal defocus range was set to 1.5 to 2.5 μm.
Image Processing.
The dose-fractionated movies were aligned, summed, dose weighted, distortion corrected, and binned by twofold in Fourier space (giving a pixel size of 1.04 Å) using MotionCor2 (51) to generate unweighted and weighted micrographs for each movie. Next, the unweighted aligned micrographs were used for contrast transfer function (CTF) estimation using Gctf 1.06 (52). According to the Gctf results, micrographs with Thon rings fit values worse than 5.5 Å, or the difference between two defocus values larger than 500 Å were eliminated from further processing, resulting in the retention of 2,003 micrographs. Gautomatch 0.53 (https://www2.mrc-lmb.cam.ac.uk/download/gautomatch-053/) was used to automatically pick particles from parts of micrographs without a template, and the resulting particles stack was subjected to reference-free two-dimensional (2D) classification using RELION (REgularized LIkelihood OptimizatioN) 3.0.5 (53–55). Five representative classes were then used for template-based particle picking in Gautomatch, yielding a stack of 110,017 particles. The following image processing steps were all performed in RELION 3.0.5 unless otherwise stated. The stack of 110,017 particles was extracted using 360-pixel box size from dose-weighted distortion-corrected micrographs and binned by fourfold (giving a pixel size of 4.16 Å), which was then subjected to several rounds of 2D classification. Poorly populated classes with low angular accuracy were removed, resulting in 76,820 particles remaining. The good 2D class averages with representative multiple views were selected for ab initio reconstruction and generated an initial model using cryoSPARC 2.4.0 (56). Then, the remaining 76,820 particles after 2D classification were subjected to 3D classification using the initial model low-pass filtered to 60 Å. Visual inspection of the resulted 3D maps showed that three classes had good encapsulin shell features, and only one class had good DyP features inside the encapsulin shell. For the encapsulin shell, 68,701 particles corresponding to the three classes were recentered, re-extracted, unbinned (giving a pixel size of 1.04 Å), and subjected to a 3D auto-refinement, followed by postprocessing, CTF refinement, and 3D auto-refinement again with icosahedral symmetry imposed. This procedure yielded a postprocessed 2.5 Å map based on the gold-standard Fourier shell correlation (FSC) 0.143 criterion. For the DyP cargo protein, 25,020 particles corresponding to the only one class were recentered, re-extracted, unbinned (giving a pixel size of 1.04 Å), and subjected to 3D auto-refine. After refinement, focused 3D classification was performed for DyP, and two classes corresponding to 13,937 particles had good DyP features. These particles were used for signal subtraction to keep only the DyP signal. Local 3D auto-refinement was performed for these subtracted particles with C2 symmetry. The resulting map was then used for generating 12 masks covering 12 DyP monomers using the Segment Map tool in University of California, San Francisco (UCSF) Chimera (57) and mask creation in RELION to search for local symmetry operators. The subtracted particles were subjected to final local 3D auto-refinement with the converged local symmetry operators and regularization parameter T = 12, yielding a postprocessed map with a resolution of 3.7 Å based on the gold-standard FSC = 0.143 criterion.
Model Building and Refinement.
For the encapsulin shell, the T. maritima encapsulin structure (Protein Data Bank [PDB] 3DKT) was docked into the final 2.5 Å map. For the DyP cargo protein, Streptomyces coelicolor DyP-type peroxidase structure (PDB 4GU7) was docked into the final 3.7 Å map. Both structures were docked as rigid bodies using UCSF Chimera. Then Ms-Enc and Ms-DyP models were built manually in Coot (58) by mutating amino acid residues and further refined using real-space refinement in Phenix (59). The symmetry matrices were generated using UCSF Chimera. MolProbity (60) was used for structure validation. All the figures were prepared using the UCSF Chimera (57), UCSF ChimeraX (61), and PyMOL (62).
Supplementary Material
Acknowledgments
We thank Dr. Chao Peng of the Mass Spectrometry System at the National Facility for Protein Science in Shanghai, Zhangjiang Lab, Shanghai Advanced Research Institute for data collection and analysis and Prof. Kaixia Mi (Chinese Academy of Sciences [CAS] Key Laboratory of Pathogenic Microbiology and Immunology, Institute of Microbiology, CAS) for sharing the strain M. smegmatis mc2 155. We are also grateful to Boling Zhu, Xiaojun Huang, and other staff members from the Center for Biological Imaging, Institute of Biophysics, CAS for their technical support on cryo-EM. This work was supported by grants from the National Key Research and Development Program of China (grants 2017YFC0840300 and 2020YFA0707500 to Z.R.), the Strategic Priority Research Program of the CAS (grants XDB08020200 to Z.R. and XDB37020203 to Q.W.), the National Natural Science Foundation of China (grants 81520108019 and 813300237 to Z.R. and 31971118 to Q.W.), the Tianjin Natural Science Foundation (grant 20JCQNJC01430 to H.G.), and the Science and Technology Commission of Shanghai Municipality (grant YDZX20203100001571).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2025658118/-/DCSupplemental.
Data Availability
The 3D cryo-EM density map and coordinate data have been deposited in the PDB database. The accession numbers for the 3D cryo-EM density map of encapsulin shell and the cargo protein DyP in the present study are EMD-30130 and EMD-30131, respectively. The accession numbers for the 3D cryo-EM density map of the complete DyP-loaded encapsulin is EMD-30132. The accession numbers for the coordinates for the encapsulin shell and cargo protein DyP in this study are PDB 7BOJ and PDB 7BOK respectively. All other study data are included in the article and/or supporting information.
References
- 1.Diekmann Y., Pereira-Leal J. B., Evolution of intracellular compartmentalization. Biochem. J. 449, 319–331 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Martin W., Evolutionary origins of metabolic compartmentalization in eukaryotes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 847–855 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cornejo E., Abreu N., Komeili A., Compartmentalization and organelle formation in bacteria. Curr. Opin. Cell Biol. 26, 132–138 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gabashvili A. N., et al., Encapsulins—bacterial protein nanocompartments: Structure, properties, and application. Biomolecules 10, 966 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stupka I., Heddle J. G., Artificial protein cages–Inspiration, construction, and observation. Curr. Opin. Struct. Biol. 64, 66–73 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Giessen T. W., Silver P. A., Widespread distribution of encapsulin nanocompartments reveals functional diversity. Nat. Microbiol. 2, 17029 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Tracey J. C., et al., The discovery of twenty-eight new encapsulin sequences, including three in anammox bacteria. Sci. Rep. 9, 20122 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kerfeld C. A., Aussignargues C., Zarzycki J., Cai F., Sutter M., Bacterial microcompartments. Nat. Rev. Microbiol. 16, 277–290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sutter M., Greber B., Aussignargues C., Kerfeld C. A., Assembly principles and structure of a 6.5-MDa bacterial microcompartment shell. Science 356, 1293–1297 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rurup W. F., Snijder J., Koay M. S., Heck A. J., Cornelissen J. J., Self-sorting of foreign proteins in a bacterial nanocompartment. J. Am. Chem. Soc. 136, 3828–3832 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Sutter M., et al., Structural basis of enzyme encapsulation into a bacterial nanocompartment. Nat. Struct. Mol. Biol. 15, 939–947 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Giessen T. W., et al., Large protein organelles form a new iron sequestration system with high storage capacity. eLife 8, e46070 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xing C. Y., et al., A self-assembled nanocompartment in anammox bacteria for resisting intracelluar hydroxylamine stress. Sci. Total Environ. 717, 137030 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Nichols R. J., et al., Discovery and characterization of a novel family of prokaryotic nanocompartments involved in sulfur metabolism. bioRxiv [Preprint] (2020). 10.1101/2020.05.24.113720. [DOI] [PMC free article] [PubMed]
- 15.Kirykowicz A. M., Woodward J. D., Shotgun EM of mycobacterial protein complexes during stationary phase stress. Current Res. Struct. Biol. 2, 204–212 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leite F. L., Reinhardt T. A., Bannantine J. P., Stabel J. R., Envelope protein complexes of Mycobacterium avium subsp. paratuberculosis and their antigenicity. Vet. Microbiol. 175, 275–285 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Putri R. M., et al., Structural characterization of native and modified encapsulins as nanoplatforms for in vitro catalysis and cellular uptake. ACS Nano 11, 12796–12804 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rahmanpour R., Bugg T. D., Assembly in vitro of Rhodococcus jostii RHA1 encapsulin and peroxidase DypB to form a nanocompartment. FEBS J. 280, 2097–2104 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Contreras H., et al., Characterization of a Mycobacterium tuberculosis nanocompartment and its potential cargo proteins. J. Biol. Chem. 289, 18279–18289 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacLean E., et al., A systematic review of biomarkers to detect active tuberculosis. Nat. Microbiol. 4, 748–758 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Gong H., et al., An electron transfer path connects subunits of a mycobacterial respiratory supercomplex. Science 362, eaat8923 (2018). [DOI] [PubMed] [Google Scholar]
- 22.McHugh C. A., et al., A virus capsid-like nanocompartment that stores iron and protects bacteria from oxidative stress. EMBO J. 33, 1896–1911 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wikoff W. R., et al., Topologically linked protein rings in the bacteriophage HK97 capsid. Science 289, 2129–2133 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Zhou Z. H., Chiou J., Protein chainmail variants in dsDNA viruses. AIMS Biophys. 2, 200–218 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X., et al., A new topology of the HK97-like fold revealed in Bordetella bacteriophage by cryoEM at 3.5 A resolution. eLife 2, e01299 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bamford D. H., Grimes J. M., Stuart D. I., What does structure tell us about virus evolution? Curr. Opin. Struct. Biol. 15, 655–663 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Heinemann J., et al., Fossil record of an archaeal HK97-like provirus. Virology 417, 362–368 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Nichols R. J., Cassidy-Amstutz C., Chaijarasphong T., Savage D. F., Encapsulins: Molecular biology of the shell. Crit. Rev. Biochem. Mol. Biol. 52, 583–594 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Kerfeld C. A., et al., Protein structures forming the shell of primitive bacterial organelles. Science 309, 936–938 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Tsai Y., et al., Structural analysis of CsoS1A and the protein shell of the Halothiobacillus neapolitanus carboxysome. PLoS Biol. 5, e144 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colpa D. I., Fraaije M. W., van Bloois E., DyP-type peroxidases: A promising and versatile class of enzymes. J. Ind. Microbiol. Biotechnol. 41, 1–7 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Yoshida T., et al., Anabaena sp. DyP-type peroxidase is a tetramer consisting of two asymmetric dimers. Proteins 84, 31–42 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Zubieta C., et al., Crystal structures of two novel dye-decolorizing peroxidases reveal a beta-barrel fold with a conserved heme-binding motif. Proteins 69, 223–233 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Roberts J. N., et al., Characterization of dye-decolorizing peroxidases from Rhodococcus jostii RHA1. Biochemistry 50, 5108–5119 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Manca C., Paul S., Barry C. E. III, Freedman V. H., Kaplan G., Mycobacterium tuberculosis catalase and peroxidase activities and resistance to oxidative killing in human monocytes in vitro. Infect. Immun. 67, 74–79 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slauch J. M., How does the oxidative burst of macrophages kill bacteria? Still an open question. Mol. Microbiol. 80, 580–583 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goulding C. W., et al., Regulation by oligomerization in a mycobacterial folate biosynthetic enzyme. J. Mol. Biol. 349, 61–72 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Sugano Y., Muramatsu R., Ichiyanagi A., Sato T., Shoda M., DyP, a unique dye-decolorizing peroxidase, represents a novel heme peroxidase family: ASP171 replaces the distal histidine of classical peroxidases. J. Biol. Chem. 282, 36652–36658 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Pandey R., Rodriguez G. M., A ferritin mutant of Mycobacterium tuberculosis is highly susceptible to killing by antibiotics and is unable to establish a chronic infection in mice. Infect. Immun. 80, 3650–3659 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaur A., et al., Coordination of frontline defense mechanisms under severe oxidative stress. Mol. Syst. Biol. 6, 393 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smeulders M. J., Keer J., Speight R. A., Williams H. D., Adaptation of Mycobacterium smegmatis to stationary phase. J. Bacteriol. 181, 270–283 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liers C., et al., Substrate oxidation by dye-decolorizing peroxidases (DyPs) from wood- and litter-degrading agaricomycetes compared to other fungal and plant heme-peroxidases. Appl. Microbiol. Biotechnol. 97, 5839–5849 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Sezer M., et al., Distinct structural and redox properties of the heme active site in bacterial dye decolorizing peroxidase-type peroxidases from two subfamilies: Resonance Raman and electrochemical study. Biochemistry 52, 3074–3084 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Brown M. E., Barros T., Chang M. C., Identification and characterization of a multifunctional dye peroxidase from a lignin-reactive bacterium. ACS Chem. Biol. 7, 2074–2081 (2012). [DOI] [PubMed] [Google Scholar]
- 45.van Bloois E., Torres Pazmiño D. E., Winter R. T., Fraaije M. W., A robust and extracellular heme-containing peroxidase from Thermobifida fusca as prototype of a bacterial peroxidase superfamily. Appl. Microbiol. Biotechnol. 86, 1419–1430 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheibner M., et al., Novel peroxidases of Marasmius scorodonius degrade beta-carotene. Appl. Microbiol. Biotechnol. 77, 1241–1250 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Linde D., et al., Basidiomycete DyPs: Genomic diversity, structural-functional aspects, reaction mechanism and environmental significance. Arch. Biochem. Biophys. 574, 66–74 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Winter N., et al., Characterization of the gene encoding the immunodominant 35 kDa protein of Mycobacterium leprae. Mol. Microbiol. 16, 865–876 (1995). [DOI] [PubMed] [Google Scholar]
- 49.Rosenkrands I., et al., Identification and characterization of a 29-kilodalton protein from Mycobacterium tuberculosis culture filtrate recognized by mouse memory effector cells. Infect. Immun. 66, 2728–2735 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Zheng S. Q., et al., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakane T., Kimanius D., Lindahl E., Scheres S. H., Characterisation of molecular motions in cryo-EM single-particle data by multi-body refinement in RELION. eLife 7, e36861 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zivanov J., Nakane T., Scheres S. H. W., A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scheres S. H., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Pettersen E. F., et al., UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Afonine P. V., et al., Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williams C. J., et al., MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goddard T. D., et al., UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schrodinger, The PyMOL Molecular Graphics System (Version 1.3r, Schrodinger LLC, 2010).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 3D cryo-EM density map and coordinate data have been deposited in the PDB database. The accession numbers for the 3D cryo-EM density map of encapsulin shell and the cargo protein DyP in the present study are EMD-30130 and EMD-30131, respectively. The accession numbers for the 3D cryo-EM density map of the complete DyP-loaded encapsulin is EMD-30132. The accession numbers for the coordinates for the encapsulin shell and cargo protein DyP in this study are PDB 7BOJ and PDB 7BOK respectively. All other study data are included in the article and/or supporting information.