Significance
Understanding the basis for exceptional responders represents a key pillar in the framework of precision medicine. In this study, we utilized distinct informatics platforms to analyze the expression profiles of clinically annotated tumor specimens derived from patients afflicted with glioblastoma, the most common form of primary brain cancer. These analyses converged on prognostic contributions from glioblastoma-associated microglia/macrophages. Glioblastoma-associated microglia-secreted interleukin 11 (IL11) to activate a STAT3-MYC signaling axis in glioblastoma cells, facilitating resistance to the standard-of-care chemotherapy, temozolomide. Microglia recruitment and IL11 secretion were dependent on the myeloid-specific phosphoinositide-3-kinase gamma isoform (PI3Kγ). Inhibition or genetic inactivation of PI3Kγ in murine glioblastoma models recapitulated expression profiles observed in specimens derived from exceptional responders, suggesting a potential for clinical translation.
Keywords: glioblastoma, exceptional responders, IL11, PI3Kγ, microglia/macrophages
Abstract
Precision medicine in oncology leverages clinical observations of exceptional response. Toward an understanding of the molecular features that define this response, we applied an integrated, multiplatform analysis of RNA profiles derived from clinically annotated glioblastoma samples. This analysis suggested that specimens from exceptional responders are characterized by decreased accumulation of microglia/macrophages in the glioblastoma microenvironment. Glioblastoma-associated microglia/macrophages secreted interleukin 11 (IL11) to activate STAT3-MYC signaling in glioblastoma cells. This signaling induced stem cell states that confer enhanced tumorigenicity and resistance to the standard-of-care chemotherapy, temozolomide (TMZ). Targeting a myeloid cell restricted an isoform of phosphoinositide-3-kinase, phosphoinositide-3-kinase gamma isoform (PI3Kγ), by pharmacologic inhibition or genetic inactivation disrupted this signaling axis by reducing microglia/macrophage-associated IL11 secretion in the tumor microenvironment. Mirroring the clinical outcomes of exceptional responders, PI3Kγ inhibition synergistically enhanced the anti-neoplastic effects of TMZ in orthotopic murine glioblastoma models. Moreover, inhibition or genetic inactivation of PI3Kγ in murine glioblastoma models recapitulated expression profiles observed in clinical specimens isolated from exceptional responders. Our results suggest key contributions from tumor-associated microglia/macrophages in exceptional responses and highlight the translational potential for PI3Kγ inhibition as a glioblastoma therapy.
Key frameworks in cancer biology have been built on insights gained through genetic analysis of rare hypomorphic mutations/epigenetic events that render hypersensitivity to specific experimental conditions (1). The study of exceptional responders can be viewed in this light, as the tumors in these patients are often characterized by genetic or epigenetic events that confer exquisite sensitivity to the treatment rendered. Most studies define exceptional responders as the <10% of patients who markedly respond to a therapy that confers little benefit to the remaining population (2). Here, we adopt this definition and study the <10% of patients with glioblastoma (World Health Organization grade IV glioma), the most common form of primary brain cancer in adults, who survive beyond 2 y after the standard-of-care treatment with concurrent radiation and temozolomide (TMZ).
Previous studies of exceptional glioblastoma responders have yielded two key observations. First, while glioblastomas harboring isocitrate dehydrogenase (IDH) mutations are generally associated with improved survival (3), such mutations are not required for long-term survivorship (4, 5). Second, in independent studies, clinical specimens from long-term survivors are more likely to harbor promoter methylation attenuating the expression of O6-methylguanine-DNA-methyltransferase (MGMT), a DNA repair protein that confers TMZ resistance (6–9). These results suggest the critical contribution of TMZ efficacy to long-term survivorship. Notably, Food and Drug Administration approval of TMZ for glioblastoma factored into consideration the small “tail” of patients who survived beyond 2 y, since the treatment-associated gain in median survival was minimal (10–12). Beyond these observations, there is little commonality in the various studies of long-term glioblastoma survivorship (12). A central limitation in this literature is that key clinical factors that dominate clinical survival prognostication for glioblastoma patients, such as patient age, neurologic function, and extent of surgical resection (13, 14), are rarely considered in efforts to develop survival gene signatures.
Studies of long-term glioblastoma survivorship have largely focused on interrogating biological processes intrinsic to the tumor cell (12). However, 30 to 50% of the cells in clinical glioblastoma specimens are nonneoplastic, consisting predominantly of microglia or Gr1+ macrophages (15). Importantly, glioblastoma cells are indelibly influenced by these nonneoplastic cells. Microglia are progenies of primitive yolk sac myeloid precursors that reside in the central nervous system (CNS) (16). Macrophages are derived from bone marrow myeloid precursors that circulate in the peripheral blood and enter the brain during tumor development; some differentiate into immune suppressive, Gr1+ tumor-associated macrophages (TAM) (17). While microglia and TAM can be studied as distinct entities in murine glioblastoma models (18), dissociating the contributions of these distinct myeloid progenies is difficult in studies of human clinical glioblastoma specimens (19). As such, microglia/TAM are often grouped into a single entity in human studies.
Many non-CNS tumors secrete chemokines to attract peripheral myeloid cells. These chemokines activate phosphoinositide-3-kinase gamma isoform (PI3Kγ) in myeloid cells to trigger chemotaxis (20) and recruitment into the tumor microenvironment (21, 22). PI3Kγ also regulates myeloid cell transcriptional programming by suppressing proinflammatory transcription and activating immune suppressive and wound healing myeloid cell transcription (23). In mammalian cells, there are three classes of PI3K: Class I PI3K generates secondary messengers to regulate cell growth, movement, and differentiation (24). Classes II and III PI3K play major roles in endocytosis/endosome trafficking (25–27). There are four class I PI3K isoforms (PI3Kα, β, δ, and γ). The current models suggest that PI3Kα, β, and δ (classified as class IA) are primarily activated downstream of receptor tyrosine kinases, while PI3Kγ (classified as class IB) is primarily activated in response to G-protein coupled receptors (GPCR) (28, 29) but can also be activated by receptor tyrosine kinases such as VEGFR (21). While PI3Kγ activation is critical for recruitment and reprogramming of TAM in non-CNS tumors (21–23, 30–33), the relevance of these findings in glioblastoma remain unclear.
Dynamic and complex interactions occur between glioblastoma and microglia/TAM in the tumor microenvironment. Non-transformed microglia isolated from surgical specimens derived from epilepsy patients as well as macrophages cultured from peripheral human blood suppressed glioblastoma tumorigenicity (34), suggesting that native microglia/macrophage serve key roles in immune surveillance against cancer. This surveillance function is lost in microglia/TAM isolated from the murine and human glioblastomas (35). Instead, these microglia/TAM secrete a multitude of growth factors (36) and cytokines, including interleukin 6 (IL6) to promote glioblastoma growth (37). RNA sequencing (RNA-seq) profiling of glioblastoma-associated microglia/TAM revealed distinct subpopulations exhibiting gene expression patterns distinct from native microglia/TAM (38). Notably, single-gene biomarkers intended to proxy cell states in microglia/macrophage are often altered for reasons unrelated to the proxied cell state (39, 40). Reflecting this complexity, there is a notable incongruence in studies correlating clinical survival to the expression of single genes thought to reflect microglia/TAM abundance or cell state, such as the expression of CD204 or ionized calcium-binding adaptor molecule 1 (IBA1) (41). These observations underscore the need for an integrated informatics approach (42, 43).
Here, we used an integrated informatics approach to understand the molecular basis underlying exceptional response after standard-of-care glioblastoma treatment and uncovered contribution from glioblastoma-associated microglia/TAM. Glioblastoma-associated microglia/TAM secrete high levels of the proinflammatory cytokine interleukin 11 (IL11), which in turn, triggers the STAT3-MYC signaling axis in glioblastoma cells to induce stem cell states that confer therapeutic resistance. In murine models, pharmacologic inhibition or genetic inactivation of PI3Kγ inhibited microglial IL11 expression to recapitulate expression signatures observed in clinical specimens derived from exceptional responders. These findings suggest PI3Kγ inhibition as a promising strategy for glioblastoma therapy.
Results
Gene Signatures Implicate Contribution of Glioblastoma-Associated Microglia/Macrophages to Clinical Response.
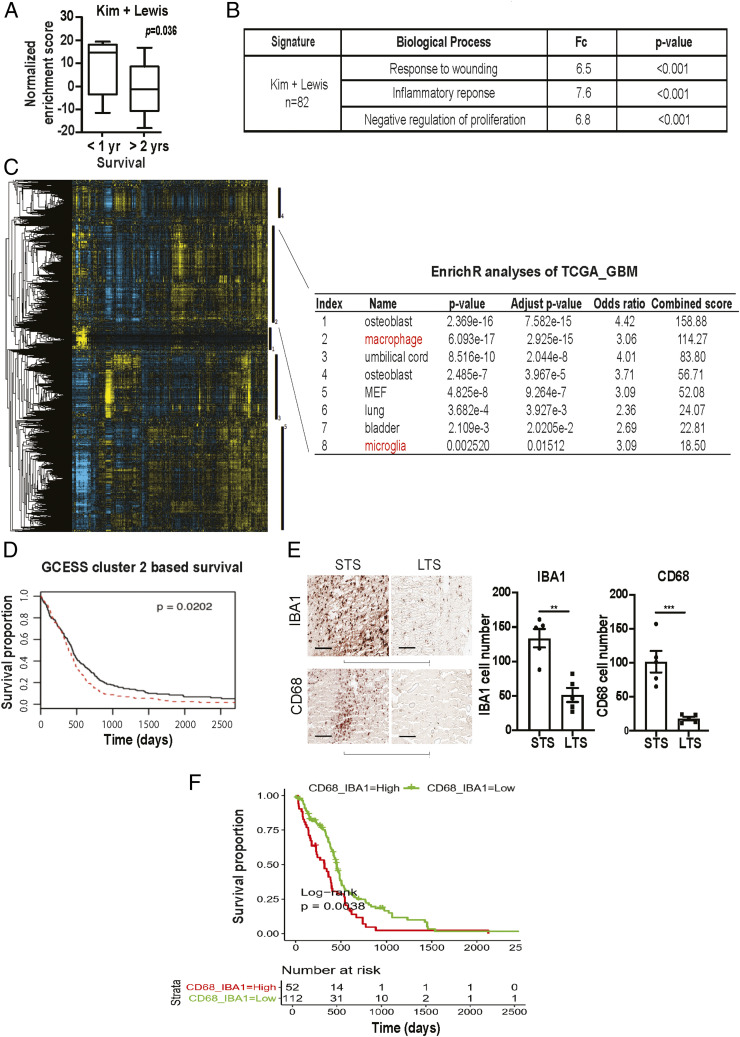
Realizing the hazard of “chance” associations between random gene signatures and clinical survival (44), we began our analysis by interrogating published glioblastoma survival signatures to determine whether these reported associations of gene expression signatures and survival can be recapitulated in independent patient cohorts. For this analysis, we focused on glioblastoma with wild-type IDH (wtIDH), given the paucity of information on survival prognostication in this patient population (45). From the literature, we identified 18 published glioblastoma survival gene signatures (SI Appendix, Table S1) and tested them using the clinically annotated expression profiles derived from wtIDH patients in The Cancer Genome Atlas (TCGA, https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) and the Repository of Molecular Brain Neoplasia Data (REMBRANDT) glioblastoma databases (46). Of the 18 published signatures, only two, Kim et al. (47) and Lewis et al. (48), consistently showed survival association in both patient cohorts (SI Appendix, Table S1). Lowered expression levels of these gene signatures were associated with improved survival. Clinical variables such as age, Karnofsky Performance Score (KPS, a clinical scale commonly used to assess a patient’s functional status), and extent of resection as well as TMZ treatment potently influence patient survival (49). To assess whether survival association of the two gene signatures persists after controlling for these clinical variables, we combined the two signatures into an 82 gene panel (SI Appendix, Table S2) and tested whether the expression of these 82 genes exhibited survival association using a matched cohort design. Of note, the expression of these 82 genes was not enriched in any particular TCGA-defined glioblastoma subtypes (50) (SI Appendix, Fig. S1). In the matched cohort analysis, we identified a total of 10 patients in the Chinese Glioma Genome Atlas (CGGA) database who survived >2 y after gross total resection and TMZ treatment (∼10% of the cohort). Using single sample gene set enrichment analysis (ssGSEA), we then compared the cumulative expression of these 82 genes in the >2 y survivors relative to a cohort of 10 age- and KPS-matched wild-type glioblastoma patients who survived <1 y after gross total resection and TMZ treatment. The expression score of the combined 82 gene signature was lower in the >2 y survivors relative to the <1 y survivors (P = 0.036, Fig. 1A), further confirming the survival associations that we previously observed in the TCGA and the REMBRANDT. Functional annotation (51) of the combined signature revealed enrichment for genes involved in wound healing and inflammatory processes (Fig. 1B and SI Appendix, Table S2).
Fig. 1.
Survival analysis implicates contribution of glioblastoma-associated macrophages/microglia to clinical survival. (A) ssGSEA analysis demonstrating that the ssGSEA expression scores of the 82 survival-associated genes [combining the Kim (47) and Lewis (48) signatures] were significantly lower in the >2 y survivors relative to the <1 y survivors. These cohorts were matched for age and KPS. All patients underwent gross total resection and TMZ treatment. (B) Shown are the pathways enriched for the 82 survival-associated genes. (C and D) GCESS analysis of TCGA glioblastomas. Cluster of genes that are 1) highly correlated to one another and 2) associated with overall survival are shown as a heatmap; each column represents a gene, and each row represents a patient. The five survival-associated gene clusters are indicated by the vertical black bars (to the right of the heatmap). Macrophage/microglia gene signatures are enriched in cluster 2 (C). The higher expression of genes in cluster 2 was associated with poor clinical survival (D). High (in red) and low (in black) expression was defined based on median expression value. n = 539; P = 0.0202. (E) Immunohistochemical staining of macrophage and microglia markers, IBA1 and CD68, in CGGA patient cohort of long-term survival (LTS) and matched short-term survivals (STS). Each row represents an LST with a matched STS. Two representative matched pairs are shown. The statistics were derived from five matched pairs. **P < 0.01; ***P < 0.001. (F) Correlation between the expression of CD68 and IBA1 with the survival of TCGA glioblastoma patients. P = 0.038. n = 226.
Since microglia/bone marrow–derived macrophages are key cell populations that mediate healing and inflammatory processes in the CNS, our functional annotation suggests contributions of these cells in glioblastoma survival prognostication. To further interrogate this hypothesis, we tested whether a distinct survival analysis method, the Gene Cluster Expression Summary Score (GCESS) (52), would yield corroborative evidence. GCESS identifies gene clusters whose expression are 1) highly correlated to one another and 2) associated with overall survival in glioblastoma patients. We further wish to perform this analysis using two independent glioblastoma cohorts, with the rationale that robust findings should be recapitulated in independent datasets. Supporting the hypothesis derived from our functional annotation of the 82 gene panel, GCESS analysis supports the survival contribution from microglia/TAM in both the TCGA (Fig. 1 C and D) and the REMBRANDT datasets (SI Appendix, Fig. S2). Higher expression levels of genes associated with microglia/TAM functions were associated with poor survival.
Together, these analyses suggest that microglia/TAM contributed to poor glioblastoma patient survival. To further validate this hypothesis, we stained glioblastoma specimens prepared from the matched CGGA patients for two microglia/macrophage markers, IBA1 and CD68. Supporting our hypothesis, lower levels of IBA1 and CD68 staining were consistently observed in the exceptional responders relative to the matched control (Fig. 1E). As an independent validation of these CGGA IHC findings, we analyzed the TCGA glioblastoma cohort to test whether CD68 and IBA1 expression correlated with overall survival. Expectedly, the expressions of CD68 and IBA1, both generally accepted microglia markers, were highly correlated (R = 0.76, P < 2.2 × 10−16, SI Appendix, Fig. S2D) in TCGA glioblastomas. The TCGA cohort was divided into those with tumors exhibiting higher or lower than the median value of the sum of CD68 and IBA1 expression. We observed that patients bearing tumors with higher than median CD68 + IBA1 expression exhibited poorer survival relative to those with lower than median CD68 + IBA1 expression (P = 0.0038, Fig. 1F).
Microglia-Secreted IL11 Enhances Glioblastoma Tumorigenicity and TMZ Resistance.
The functional annotation shown in Fig. 1B implicates cell states resembling those of stem cells since they 1) play critical roles in wound healing (53), 2) generally exhibit slower growth kinetics (54), and 3) participate in the inflammatory response (55). Since glioblastoma cells with stem cell properties exhibit features associated with poor clinical outcomes, including increased tumorigenicity (56) and therapeutic resistance (57), our analyses suggest that microglia/macrophage in the tumor microenvironment may induce glioblastoma stem cell states that contribute to shortened survival. Supporting this hypothesis, coculturing of glioblastoma cells with a transformed human microglia cell line (hMG) enhanced the in vitro and in vivo tumorigenicity (SI Appendix, Fig. S3 A and B) of glioblastoma cells as well as their TMZ resistance (SI Appendix, Fig. S3C).
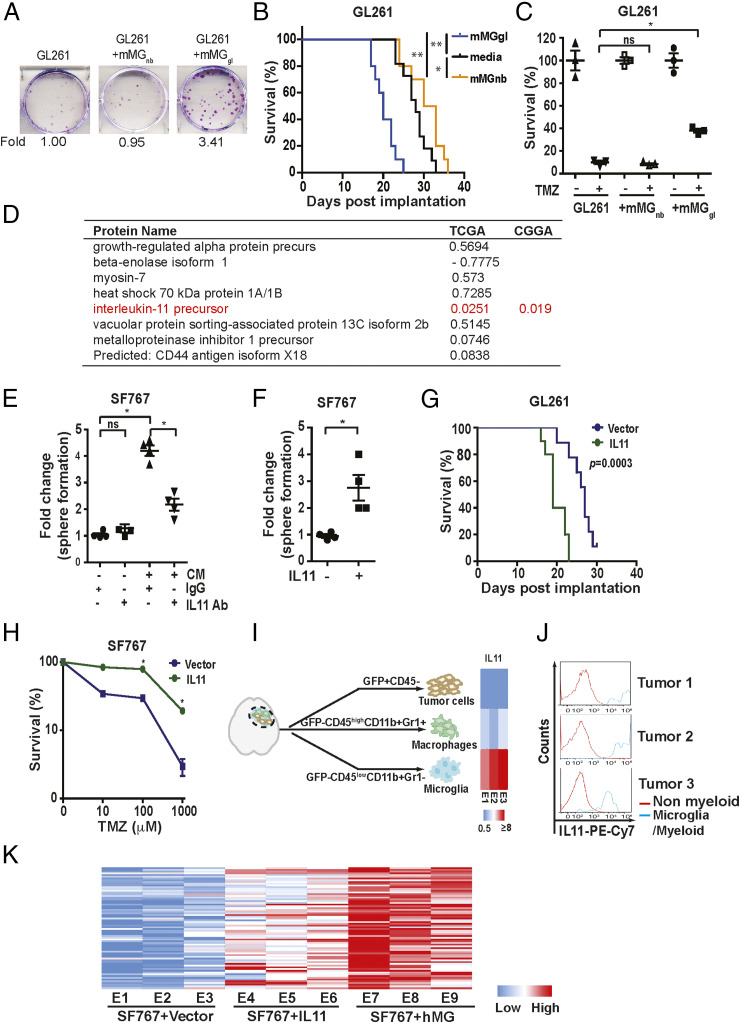
Realizing that the physiology of in vitro cultured microglia may differ from their in vivo roles (58), we tested whether our observations could be recapitulated using murine microglia freshly isolated from a syngeneic glioblastoma model. To this end, murine glioblastoma-associated microglia (mMGgl, GFP-CD45lowCD11b+Gr1− cells, see gating strategy, SI Appendix, Fig. S4) were isolated from murine glioblastomas formed after orthotopic implant of GFP-labeled GL261 cells. Supporting results derived from cultured hMG, coculture of GL261 with freshly isolated mMGgl enhanced in vitro colony formation potency by approximately threefold (Fig. 2A). Further, intracranial coimplantation of GL261 with freshly isolated mMGgl enhanced in vivo tumor growth (Fig. 2B) as demonstrated by the shortened survival. Tumor-promoting effects were not observed when GL261 cells were cocultured or coimplanted with GFP−CD45lowCD11b+Gr1− microglia isolated from normal murine brains (mMGnb, Fig. 2 A and 2B). Moreover, coculturing with mMGgl but not mMGnb enhanced GL261 resistance to TMZ (Fig. 2C). Of note, similar effects were observed when glioblastoma-associated macrophages (defined by GFP−CD45highCD11b+Gr1+) were tested in vivo (SI Appendix, Fig. S3D).
Fig. 2.
Glioblastoma-associated microglia release IL11 to enhance glioblastoma tumorigenicity and TMZ resistance. (A, Upper) Representative images of in vitro colony formation after coculturing of freshly isolated mMGgl or mMGnb with murine GL261 glioblastoma cells. (A, Bottom) Fold change in colony-forming units with or without mMGgl or mMGnb coculturing. (B) Effect of orthotopic coimplantation of mMGgl or mMGnb and GL261 on mice survival. *P < 0.05; **P < 0.0001. n = 10. (C) The effects of coculturing of GL261 with mMGgl or mMGnb on TMZ resistance, assessed by colony formation assays. *P < 0.05. (D) The 10 most abundant proteins identified through unbiased proteomic profiling of CM derived from immortalized hMG. P value of association between mRNA expression of genes encoding these CM proteins and survival in the TCGA are shown under the TCGA column. To avoid multiple comparisons, only IL11 was tested for survival association using the CGGA. P value for this association is shown under the CGGA column. (E) Effect of IL11 neutralizing antibodies on hMG CM-induced neurosphere formation in SF767. *P < 0.05. ns: not significant. (F) The effect of recombinant human IL11 (20 ng/mL) on the neurosphere-forming capacity of SF767 cells. *P < 0.05. (G) Survival of mice bearing GL261 cells ectopically expressing IL11 compared to mice bearing GL261 cells. n = 10. P = 0.0003. (H) Effect of ectopic IL11 expression on TMZ resistance of SF767 cells. In vitro cell viability was determined. *P < 0.05. n = 6. Error bars, SD. (I) GFP-labeled GL261intracranial tumors were sorted into tumor cells (GFP+CD45−), TAM (GFP-CD45highCD11b+Gr1+), and microglia (GFP-CD45lowCD11b+Gr1−). IL11 levels were examined by qRT-PCR, and the results were displayed as a heatmap. (J) IL11 levels in nonmyeloid cells (IBA1−CD68−CD11b−) and tumor-associated microglia/myeloid cells (IBA1+CD68+CD11b+) sorted from three glioblastoma specimens resected from human subjects. (K) The effect of microglia coimplantation and ectopic IL11 expression on the expression of the 82 survival-associated genes. RNA was extracted from subcutaneous tumors formed by SF767-vector, SF767-IL11, and SF767 cells coimplanted with microglia, and gene expression was analyzed by qRT-PCR. ΔCt between gene and actin in each sample was plotted as a heatmap. Three independent tumors were analyzed for each cohort.
To determine the potential role of secreted factors in microglia-mediated tumor growth, glioblastoma cells were cocultured with conditioned medium (CM) derived from hMG (SI Appendix, Fig. S5A) or mMGgl. These experiments demonstrate that CM recapitulated the microglia-mediated tumor growth (SI Appendix, Fig. S5 B and C). To identify the soluble factor responsible for this phenotype, we performed an unbiased proteomic profiling of CM derived from hMG (58) (Fig. 2D). The list of CM proteins was crossed referenced to their messenger RNA (mRNA) expression in clinical specimens and survival outcomes. We reasoned that increased mRNA expression for the CM factor underlying the increased tumorigenicity/resistance to DNA damage would be associated with poor clinical survival. With this rationale, we tested the top 10 most abundant proteins in the hMG media for clinical survival association. Of those tested, only IL11 exhibited the anticipated survival association. In the TCGA dataset, high IL11 mRNA expression was associated with poor survival (Fig. 2D). We then confirmed this association using the CGGA dataset (Fig. 2D). To avoid multiple comparisons, we tested only IL11 in this confirmatory analysis (SI Appendix, Fig. S5D). Importantly, IL11 mRNA and protein expression were elevated in clinical glioblastoma specimens (SI Appendix, Fig. S5E).
Next, we tested whether IL11 is necessary and sufficient for the induction of glioblastoma tumorigenicity and TMZ resistance. Neutralizing antibodies against IL11 suppressed the protumorigenic effect of microglia CM on the SF767, as determined by both neurosphere (Fig. 2E) and colony-forming assays (SI Appendix, Fig. S6A). These results were recapitulated using an independent glioblastoma line, LN229 (SI Appendix, Fig. S6B), a patient-derived glioblastoma line cultured as tumorsphere, MGG4 (SI Appendix, Fig. S6C), as well as a murine glioblastoma line, CT2A (SI Appendix, Fig. S6D). In gain-of-function studies, direct addition of IL11 ligand at concentrations found in clinical glioblastoma (SI Appendix, Fig. S5E) specimens enhanced the neurosphere- and colony-forming potency of SF767 (Fig. 2F and SI Appendix, Fig. S6E) and LN229 (SI Appendix, Fig. S6F) by approximately threefold. Finally, ectopic expression of human IL11 (hence noted as IL11) in the SF767 glioblastoma line, thereby creating autocrine IL11 signaling, enhanced in vivo xenograft formation in a subcutaneous model (SI Appendix, Fig. S6 G and H). In an orthotopic murine glioblastoma model, the median survival of mice bearing GL261 cells ectopically expressing IL11 (designated GL261-IL11) was shortened by ∼8 d relative to mice implanted with GL261-vector cells (Fig. 2G and SI Appendix, Fig. S6G). Additionally, ectopic IL11 expression decreased the minimal number of implanted cells required for in vivo xenograft formation, further supporting the hypothesis that IL11 enhances glioblastoma tumorigenicity (SI Appendix, Fig. S6I). Finally, ectopic IL11 expressing SF767 glioblastoma cells significantly augmented TMZ resistance (Fig. 2H).
To determine the major cell source for IL11 in the glioblastoma microenvironment, we sorted tumor and myeloid cells from GL261 tumors. Glioblastomas formed after orthotopic implant of GFP-labeled GL261 cells were sorted into GFP+CD45− tumor cells, GFP−CD45lowCD11b+Gr1− microglia fraction, and GFP-CD45highCD11b+Grl+ macrophage cells and then subjected to IL11 qRT-PCR. IL11 mRNA expression was approximately an order of magnitude higher in the microglia fraction relative to the tumor and the macrophage fractions (Fig. 2I). To connect these observations to human glioblastomas, we sorted cell populations from freshly resected clinical glioblastoma samples using IBA1+CD68+CD11b+ as markers for microglia/TAM. Analysis of samples secured from three unrelated patients revealed that IL11 expression was largely found in the IBA1+CD68+CD11b+ fraction (Fig. 2J and SI Appendix, Fig. S7), suggesting that the myeloid cell populations were the major cell source of IL11 in the glioblastoma microenvironment.
We next determined if IL11 expression or presence of microglia contributed to increased expression of the 82 survival-associated genes (SI Appendix, Table S2) that we collated through our informatics analysis (Fig. 1A). To this end, intracranial glioblastoma xenografts derived from SF767-vector, SF767-IL11, or SF767 cells coimplanted with hMG were isolated and analyzed by qRT-PCR. Supporting our hypothesis, >50% of the 82 genes showed a greater than twofold increase in expression when comparing glioblastomas formed by SF767-IL11 with those formed by SF767-vector cells (Fig. 2K, gene list shown in SI Appendix, Table S2). Impressively, >90% of the 82 genes showed a greater than twofold increase when comparing glioblastoma formed after SF767/microglia coimplantation with those formed by SF767-vector cells. As controls, we showed that ectopic IL11 expression or microglia coimplantation did not systemically affect the expression of a set of randomly selected genes (SI Appendix, Fig. S6J).
IL11 Induces Glioblastoma STAT3-MYC Signaling.
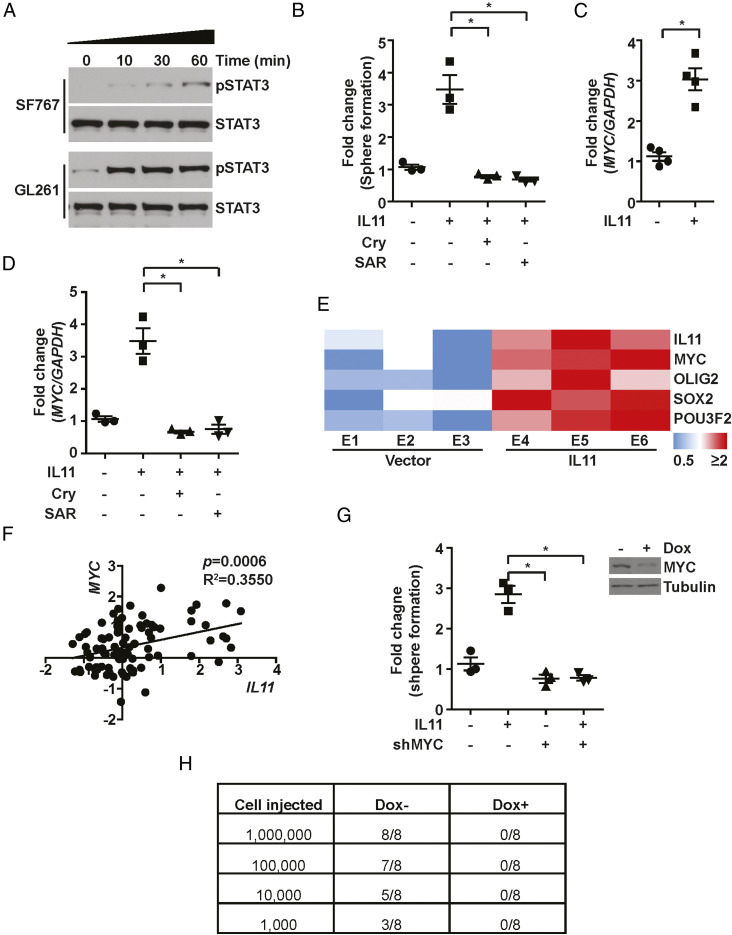
In several cancer types, IL11 facilitates tumorigenesis through the activation of the STAT3 pathway (53, 59–62). However, the role of STAT3 in IL11-mediated glioblastoma tumorigenesis remains an open question. IL11 treatment of independent glioblastoma lines induced a time-dependent increase in the accumulation of the active, tyrosine-705 phosphorylated form of STAT3 (p-STAT3), suggesting that IL11 induces STAT3 activation in glioblastoma cells (Fig. 3A). Moreover, inhibitors of STAT3 activation, including cryptotanshinone (54) and the JAK2 inhibitor SAR317461 (55), abolished the effect of IL11 on tumorsphere formation (Fig. 3B), suggesting STAT3 activation is required for the protumorigenic effects of IL11.
Fig. 3.
IL11 promotes glioblastoma tumorigenicity via STAT3-MYC signaling. (A) The effects of IL11 stimulation on phospho-STAT3 (Y705) levels in SF767 and GL261 cells. Cells were treated with IL11 (20 ng/mL) for the indicated time prior to immunoblot assay. (B) The effect of IL11 treatment (20 ng/mL) on neurosphere formation in SF767 in the presence or absence of STAT3 inhibitor Cryptotanshinone (Cry) and JAK2 inhibitor SAR317461 (SAR). *P < 0.05. (C) The effect of IL11 treatment (20 ng/mL) on MYC expression, assessed by qRT-PCR. *P < 0.05. (D) The effect of cryptotanshinone (Cry) and SAR317461 (SAR) treatment on IL11-induced MYC mRNA expression, assessed by qRT-PCR. *P < 0.05. (E) qRT-PCR analysis of MYC, OLIG2, SOX2, and POU3F2 expression in tumors that formed after orthotopic implantation of SF767 and SF767 cells that ectopically expressed IL11. ΔCt between gene and actin in each sample was plotted as a heatmap. Three independent tumors were analyzed for each cohort. (F) Correlation between IL11 (x-axis) and MYC (y-axis) mRNA expression in CGGA glioblastoma patients. R2 = 0.3550, P = 0.0006. n = 226. (G) The effect of MYC silencing on IL11-induced in vitro neurosphere formation. A dox-inducible shMYC lentivirus was used for stable knockdown of IL11 in SF767 cells. (H) The effect of MYC silencing on IL11-enhanced tumorigenicity in vivo. Serial dilutions of SF767-IL11 cells with or without MYC silencing were injected subcutaneously into Nu/Nu mice, and the tumor formation was examined 1 mo after injection. Tumors that grew in size over 50 mm3 were scored.
A key downstream effector of STAT3 is MYC (57), a master transcriptional regulator that governs a network of transcriptional factors essential for glioblastoma tumorigenicity, including OLIG2, SOX2, and POU3F2 (63, 64). IL11 treatment of the SF767 glioblastoma line in vitro increased MYC expression by approximately threefold (Fig. 3C). This increase was abolished by inhibitors that prevent STAT3 activation (Fig. 3D), suggesting an IL11-STAT3-MYC signaling axis. Confirming this observation, the SF767-IL11 xenografts showed increased expression of MYC as well as its downstream effectors, OLIG2, SOX2, and POU3F2 (63) (Fig. 3E). Supporting a clinical connection, IL11 mRNA expression positively correlated with MYC mRNA expression in two independent clinical glioblastomas cohorts, including the CGGA (R2 = 0.3550, P = 0.0006; Fig. 3F) and the TCGA (R = 0.37, P = 6.3 × 10−06, SI Appendix, Fig. S5F). Finally, doxycycline-induced silencing of MYC expression suppressed the effect of IL11 on glioblastoma tumorigenicity in vitro (Fig. 3G). For in vivo experiments, mice were implanted with serially diluted SF767-IL11 cells harboring a doxycycline-inducible shMYC construct (dox-shMYC) and randomized to doxycycline or vehicle treatment. By day 30, the vehicle-treated group formed glioblastoma xenografts in a dilution-dependent manner (Fig. 3H), while no xenografts were observed in the doxycycline-treated group. Of note, doxycycline treatment did not affect the growth kinetics of SF767-IL11-dox-shScramble (SI Appendix, Fig. S8). Collectively, these results suggest that IL11 in the tumor microenvironment induces activation of a STAT3-MYC axis in glioblastoma cells.
PI3Kγ Inhibition Suppresses Glioblastoma Tumorigenicity and TMZ Resistance.
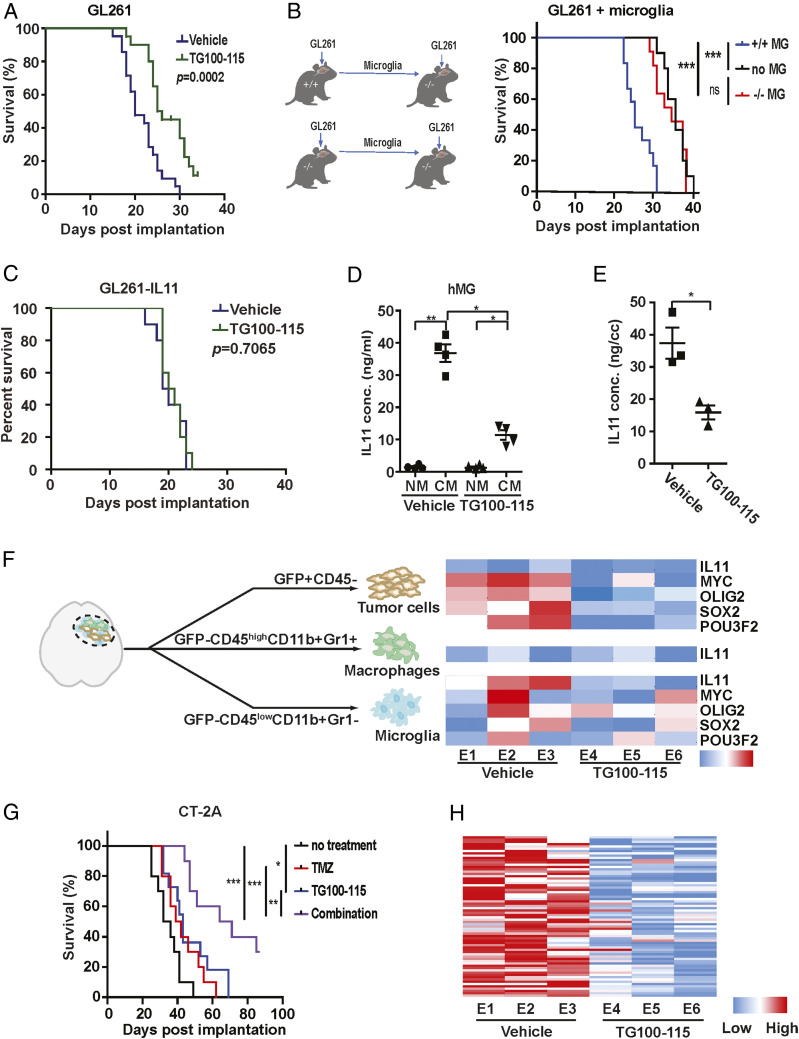
PI3Kγ activation promotes accumulation of protumor myeloid cells (20–22) and immunosuppressive reprogramming of these cells in the tumor microenvironment (23). Our results suggest these myeloid-specific effects mediate key aspects of glioblastoma tumorigenicity. In this context, we hypothesized that PI3Kγ inhibition may impede these functions to suppress glioblastoma growth. Confirming this hypothesis, while treatment with the PI3Kγ inhibitor, TG100-115, did not affect glioblastoma growth in vitro (SI Appendix, Fig. S9A), TG100-115 treatment prolonged the median survival of GL261 bearing mice by ∼5 d relative to mice treated with vehicle (Fig. 4A). Furthermore, PI3Kγ −/− C57BL/6 implanted with GL261 glioblastomas exhibited a ∼5 d increase in survival relative to GL261-implanted PI3Kγ +/+ (SI Appendix, Fig. S9B), confirming the survival benefit associated with PI3Kγ inhibition. Importantly, TG100-115 treatment of GL261 implanted in PI3Kγ −/− mice did not prolong survival (SI Appendix, Fig. S9C), suggesting that the TG100-115 associated survival benefit was attributable to PI3Kγ inhibition. To confirm that PI3Kγ in the microglia population drove this survival difference, microglia (GFP-CD45lowCD11b+Gr1−) were isolated from glioblastomas formed after orthotopic implantation of GL261 cells into PI3Kγ +/+ and PI3Kγ −/− mice. The isolated microglia were then coimplanted with a new batch of GL261 into PI3Kγ −/− mice. Mice implanted with GL261 and microglia isolated from PI3Kγ +/+ mice exhibited shortened survival by ∼6 d relative to mice implanted with GL261 and microglia isolated from PI3Kγ −/− mice (Fig. 4B).
Fig. 4.
PI3Kγ inhibition suppresses microglia density and glioblastoma tumorigenicity. (A) The effect of TG100-115 treatment on the survival of mice orthotopically implanted with GL261. n = 10, P = 0.0002. (B) The survival of PI3Kγ−/− mice after orthotopic coimplant of GL261 cells with the microglia isolated from PI3Kγ+/+ or PI3Kγ−/− mice. n = 10. ***P < 0.0001; ns: not significant. (C) The effect of TG100-115 treatment on the survival of mice orthotopically implanted with GL26-IL11. n = 10, P = 0.7065. (D) The enzyme-linked immunosorbent assay (ELISA) analysis of IL levels in CM of TG100-115–treated immortalized hMG. (E) The ELISA analysis of IL11 protein level in orthotopically implanted GL261 tumors with or without TG100-115 treatment. *P < 0.05. (F) qRT-PCR analysis of IL11, MYC, OLIG2, SOX2, and POU3F2 in tumor cells, TAM, and microglia isolated from GL261 tumors that underwent TG100-115 treatment (Right). Orthotopic tumors were resected after treatment and sorted as shown in Left. Three independent tumors were analyzed for the treated and untreated cohorts. (G) The survival of mice orthotopically implanted with CT-2A tumor after treatment with TG100-115, TMZ, or a combination of these two drugs (combo). n = 10. *P < 0.05; **P < 0.01; ***P < 0.0001. (H) The effect of TG100-115 treatment on the expression of the 82 survival-associated genes. RNA was extracted from orthotopic GL261 tumors with or without TG100-115 treatment and analyzed by qRT-PCR. ΔCt between gene and actin in each sample was plotted as a heatmap. Three independent tumors were analyzed for the treated and untreated cohorts.
Given the importance of IL11 in microglia-mediated protumorigenic signaling to glioblastoma in our murine model, we next determined whether ectopic IL11 overexpression in glioblastoma cells induces an autocrine loop that circumvent the tumor-suppressive effects of TG100-115 treatment. Supporting IL11 as a key mediator of myeloid-induced protumorigenic effect, TG100-115 did not significantly alter the survival of C57BL/6 mice implanted with GL261 that ectopically expressed IL11 (Fig. 4C),
Since our murine studies suggest microglia as the predominant myeloid population in the glioblastoma microenvironment (SI Appendix, Figs. S10 and S11) and a major source of IL11 (Fig. 2I), we determined the effects of TG100-115 on microglia IL11 production. TG100-115 treatment of cultured hMG in vitro suppressed the level of IL11 secreted by approximately threefold without affecting cell viability (Fig. 4D and SI Appendix, Fig. S9D). To characterize the effects of TG100-115 on IL11 production in vivo, murine GL261 glioblastomas were isolated after vehicle or TG100-115 treatment. The TG100-115–treated glioblastomas showed an approximately threefold decrease in IL11 level relative to the vehicle-treated samples (Fig. 4E). Similarly, IL11 levels in GL261 glioblastomas formed after orthotopic implant into PI3Kγ −/− mice were approximately threefold lower relative to those implanted into PI3Kγ +/+ mice (SI Appendix, Fig. S9E).
To further demonstrate that TG100-115 suppressed microglial IL11-associated protumorigenic signaling, GFP-labeled GL261 glioblastoma formed with or without TG100-115 treatment were sorted into tumor (GFP+CD45−), microglia (GFP−CD45lowCD11b+Gr1−), and bone marrow–derived macrophage (GFP−CD45highCD11b+Gr1+) fractions and then analyzed for IL11, MYC, SOX2, OLIG2, and POU3F2 mRNA expression. TG100-115 treatment did not significantly alter the levels of IL11 expression in the tumor or TAM (Fig. 4F). However, TG100-115 suppressed microglia IL11 expression by approximately threefold (Fig. 4F). While TG100-115 treatment did not significantly affect the expression of MYC, SOX2, OLIG2, and POU3F2 in microglia, the expression of these genes in the tumor cells was suppressed by approximately twofold after TG100-115 treatment (Fig. 4F). As these experiments were performed using the same number of sorted cells, the decreased IL11 expression in TG100-115–treated microglia further supported PI3Kγ inhibition-suppressed microglia IL11 secretion in vivo.
We next examined the effects of PI3Kγ inhibition on microglia density in the glioblastoma tumor microenvironment. TG100-115 did not significantly alter the proportion of the CD45highCD11b+Gr1− population (SI Appendix, Fig. S9F), suggesting that microglia density was unaffected by TG100-115 treatment. Consistent with our previous results (23), the percentage of CD45highCD11b+Gr1lo bone marrow–derived macrophages were reduced by two- to fourfold. Confirming these results, the microglia densities were comparable to GL261 glioblastomas formed after orthotopic implant into PI3Kγ −/− and PI3Kγ +/+ mice (SI Appendix, Fig. S9G).
Since the aggregate of our results suggests that microglia IL11 signaling modulate glioblastoma tumorigenicity and therapeutic resistance, one implication of the above results is that PI3Kγ inhibition transformed the glioblastoma microenvironment from one associated with poor TMZ response to one mimicking that of “exceptional” response. We tested this hypothesis. Although TG100-115 treatment failed to affect the TMZ sensitivity of GL261 cells in vitro (SI Appendix, Fig. S9H), substantial TMZ sensitization was detected in vivo (SI Appendix, Fig. S9I). The survival of mice bearing intracranial CT-2A glioblastomas was prolonged by either TG100115 (∼7 d) or TMZ treatment (∼6 d), but the combination of these treatments extended survival by 31 d (P < 0.0001, Fig. 4G). This result was recapitulated using the GL261 model (SI Appendix, Fig. S9I).
GL261 tumors treated with TG100-115 (Fig. 4H) or implanted into PI3Kγ −/− mice (SI Appendix, Fig. S12A) consistently showed decreased expression of the 82 survival-associated gene panel (SI Appendix, Table S2), further supporting our hypothesis. TG100-115 treatment or GL261 engraftment in PI3Kγ −/− mice did not systemically affect the expression of a set of randomly selected genes (SI Appendix, Fig. S12B). In aggregate, these results suggest that PI3Kγ inhibition converts the glioblastoma tumor microenvironment into one that more closely resembles those observed in “exceptional responders.”
Discussion
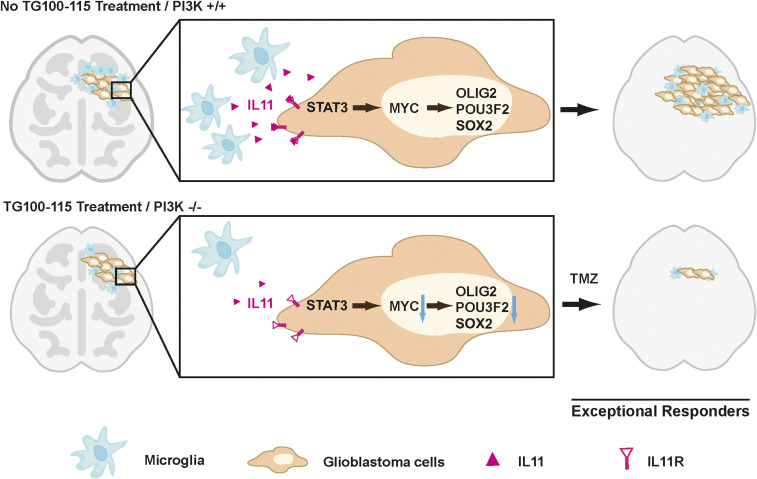
Glioblastomas are diagnosed and graded by pathological landmarks, including two canonical microenvironmental features, microvascular proliferation, and necrosis. Necrotic regions portend a poor prognosis and contain substantial inflammatory components, including microglia and TAM. No consensus exists in terms of the prognostic importance of these cell types (23). The available studies presented conflicting results (23, 65–67) and are largely limited by study designs without considerations for clinical variables that potently influence survival (such as age, KPS, and extent of resection) or validation in independent cohorts. We designed our study to address these limitations. Orthogonal informatics interrogation and cross-validation using three independent patient cohorts unveiled an inverse association between the density of glioblastoma-associated microglia/macrophages and survival. Our results suggest that clinical response to TMZ is, at least in part, modulated by soluble factors released by these glioblastoma-associated microglia/macrophages, including contributions by IL11 (68). To date, IL11’s role in glioblastoma pathogenesis and contribution to the exceptional response remains poorly described. We now show that the microglia/macrophage-secreted IL11 activates STAT3-MYC signaling in glioblastoma cells to promote transition into a cancer stem cell state, resulting in tumorigenicity and TMZ resistance (Fig. 5).
Fig. 5.
Mechanisms underlying “exceptional response” to TMZ therapy in glioblastoma patients. Glioblastoma-associated microglia secrete high levels of the inflammatory cytokine IL11. The secreted IL11 triggers activation of a STAT3-MYC signaling axis in glioblastoma cells. This signaling induces a change in glioblastoma cell state into a stem cell–like state, causing enhanced tumorigenicity and TMZ resistance. Such signaling is absent in “exceptional responders” and can be suppressed by the PI3Kγ inhibitor, TG100-115, or by PI3Kγ knockout.
Microglia and macrophage in their native state exhibit anti-glioblastoma activities (69). In this context, our findings would suggest that by the time of the clinical presentation, when glioblastoma patients bear significant tumor burden, most microglia/macrophages in the tumor microenvironment have been reprogrammed to promote tumor growth (41). The emerging literature on glioblastoma-associated microglia/TAM is largely supportive of this hypothesis (38–40). Further supporting this notion, microglia derived from the normal mice were not protumorigenic (Fig. 2B) but became protumorigenic after in vivo coimplantation with GL261 glioblastoma cells (Fig. 4B). This reprogramming results in the altered secretion of a multitude of cytokines, including IL11. Importantly, these cytokines play key roles in inducing peritumoral edema that often contributes to clinical deterioration (70). As such, PI3Kγ inhibition has the potential benefit of mitigating deleterious effects resulting from peritumoral edema.
We previously demonstrated that stochastic fluctuation in MYC expression influenced glioblastoma cell fate in terms of the equilibrium between entry and exit from the cancer stem cell states (63). Similar findings have been reported for other master regulatory transcription factors in glioblastomas (64, 71) and for other cancers (30, 72). Our current study suggests that this equilibrium is influenced by glioblastoma microenvironmental factors, including microglia secretion of IL11. Specifically, IL11 activates STAT3 signaling in glioblastoma cells, resulting in increased MYC expression. In turn, increased MYC expression shifts the equilibrium toward the cancer stem cell states. This dependence on microenvironment signaling is reminiscent of postnatal stem cells, which require signaling from surrounding cells to maintain the capacity of self-renewal and multipotency (73). In this context, reprogramming of microglia and TAM may serve as a mechanism by which glioblastoma cells reconstitute the signaling required for induction/maintenance of cancer stem cell states.
It remains unclear whether the tumor microenvironment of the exceptional responders is dictated by the intrinsic genetic/epigenetic landscape of the tumor, patient-specific genetics/physiology, or a combination of the two. Recent findings that distinct PIK3CA mutations induce tumor microenvironment with differential capacity for synapse formation suggest the primacy of tumor genetics in shaping its microenvironment (74). Applied to our findings, this model would suggest that microglia/macrophage recruitment is driven by mutations within the cancer genome through cell-autonomous mechanisms. On the other hand, natural polymorphisms in the somatic genome and patient-to-patient variation in neuroendocrine regulation modulate macrophage migration (21) and function (70). As such, aspects of patient physiology independent from tumor genetic/epigenetic composition may contribute to the recruitment and reprogramming of microglia/macrophage. Thus, noncell-autonomous factors may contribute to microglia/macrophage functions in the glioblastoma tumor microenvironment.
Our finding that glioblastoma cells leveraged cellular constituents in their microenvironment to protect themselves from the deleterious effects of cancer therapeutics is reminiscent of findings reported in breast and ovarian cancer, where the percentage of stromal cells in the clinical specimen prognosticated survival after systemic therapy (75). These findings bear implications in cancer evolution as it relates to therapeutic resistance (76, 77). Cancer evolution models built on temporal ordering of clonal or subclonal mutations typically do not directly account for contributions from the microenvironment (78). While these analyses are of great value in understanding selective pressures imposed on cell-autonomously driven processes, noncell-autonomous phenotypes may be invisible to most such analyses. Thus, the trajectory of cancer evolution in response to therapy cannot be fully modeled without understanding the cooperativity of constituents within the complex cancer ecosystem.
In summary, our study suggests clinical glioblastoma specimens derived from exceptional responders to TMZ are characterized by a microenvironment with decreased accumulation of microglia/TAM. This state is recapitulated by PI3Kγ inhibition, which suppresses microglial IL11-mediated protumorigenic signaling. As such, PI3Kγ inhibition warrants consideration in clinical translation as a potential glioblastoma therapeutic.
Materials and Methods
Cell Lines, Reagents, and Molecular/Cell Assays.
Cell lines and reagents are described in SI Appendix. Tumorigenicity was assessed by either crystal violet (63) or limiting dilution assays (79). qPCR, immunoblotting, and immunohistochemistry were performed as previously described (63) and in details in SI Appendix. Clinical specimen collection/analysis was approved by the Institutional Review Board (IRB) at the University of California San Diego Human Research Protections Program (IRB121046) and described in detail under SI Appendix, Clinical Glioblastoma Specimen Analysis. Patient capacity for informed consent was determined by the treating surgeon during consent for the surgical procedure. Once the patient is determined capable of consent, a dedicated study coordinator reviews the IRB-approved consent form with the patient, answers questions from the patient, and obtains written consent in the presence of a witness. Animal studies were performed in accordance with the Animal Care and Use Rules at the University of California San Diego under protocol S11253 or University of Minnesota under protocol 1707–349371A and described under SI Appendix, Xenograft and Allograft Models.
Literature Search and Statistical Analysis.
The MEDLINE database was queried for studies reporting an mRNA expression signature associated with survival in adult glioblastoma patients. The initial search performed using the following Medical Subject Heading terms yielded 102 articles: (“prognostic” OR “survival”) AND (“gene signature”) OR (“signature” and “gene expression”) OR (“gene signatures”) AND (“glioma” OR “glioblastoma” OR “GBM”). Two independent reviewers evaluated these articles to identify mRNA signatures associated with glioblastoma survival. A total of 18 signatures were identified (SI Appendix, Table S2) and analyzed as described below.
Cohorts of patients with wtIDH were identified in the following manner: IDH status in the TCGA dataset was based on DNA sequencing information, and IDH status in the REMBRANDT was assigned based on a published mRNA-based gene classifier (46, 80). The ssGSEA (81) score was calculated for each sample for each signature. This enrichment score was normalized to the mean of 10,000 dataset permutations. Cox proportional hazard regression was carried out on these normalized enrichment scores to determine survival association.
An mRNA expression dataset of age- and KPS-matched wtIDH glioblastoma patients was collaged using the CGGA with <1 y survival (n = 10) and >2 y survival (n = 10). Age was matched to ± 5 y, and KPS was precisely matched. The ssGSEA enrichment score was calculated for the combined Kim (47) and Lewis (48) signatures for the <1 y and >2 y survivors and compared using t test.
Other statistical analyses are found under SI Appendix, Statistical Analysis. All statistical analyses were carried out in MATLAB (Statistics Toolbox Release 2012b, The MathWorks Inc.). Genes in the signature were compared to a list of 100 genes selected at random from the TCGA. Genes were selected without replacement using the random number generator from R version 3.1. None of the selected genes overlapped with the genes in any of the signatures of interest.
Flow Cytometry Staining, Analysis, and Sorting.
Single-cell suspensions (106 cells in 100 µl total volume) were incubated with LIVE/DEAD Fixable Aqua Dead Cell Stain at 4 °C for 15 min (Life Technologies), FcR-blocking antibody at 4 °C for 10 min (BD Biosciences), and various fluorescently labeled antibodies at 4 °C for 1 h. Primary antibodies against CD45-Alexa700 (mouse, 30-F11), CD11b-APC (mouse, M1/70), Gr1-FITC (mouse, RB6-8C5), CD3e-eFluor780 (mouse, 145–2C11), CD68-PE (human, eBioY1/82A), and CD11b-APC (human, ICRF44) were purchased from eBioscience; F4/80-PE-Cy7 (mouse, BM8), CD4-PEdazzle (mouse, GK1.5), and CD8-BV605 (mouse, 53–13) were purchased from BioLegend. IBA1-FITC (human) was from Abcam. For intracellular staining, cells were fixed and permeabilized using Flow Cytometry Permeabilization/Wash Buffer I (R&D) and then incubated with rabbit IL11 antibody (R&D) followed by PE-Cy7 conjugated anti-Rabbit secondary antibody (eBioscience), or cells were permeabilized using Permeabilization/Wash Buffer I (R&D) and then incubated with anti-IBA1-FITC antibody (Abcam). Multicolor fluorescence-activated cell sorting (FACS) analysis was performed on a BD Canto RUO19 Color Analyzer at the Flow Cytometry Core at the University of California San Diego Center for AIDS Research. All data analysis was performed using FlowJo (Tree Star, Inc.).
For cell sorting, single-cell suspensions were stained with LIVE/DEAD Fixable Aqua Dead Cell Stain (Life Technologies) to exclude dead cells and anti-CD11b-APC (mouse, M1/70, eBioscience) and anti-Gr1-PE (RB6-8C5, eBioscience). FACS sorting was performed using FACS Aria 11 color high-speed sorter at the Flow Cytometry Core at the University of California San Diego Center for AIDS Research. GFP-tagged SF767 tumor cells were sorted based on GFP positivity. GFP-tagged GL261 cells were sorted into GFP+CD45− (glioblastoma), GFP−CD45lowCD11b+Gr1− (microglia), and GFP-CD45highCD11b+Gr1+ (macrophages). Microglia were isolated from both GL261 glioblastomas (mMGgl) and normal murine brain (mMGnb). Cells from freshly resected glioblastoma specimens were sorted into IBA1−CD68−CD11b− (nonmyeloid cells) and IBA1+CD68+CD11b+ (microglia/myeloid cells).
Please refer to SI Appendix for other full detailed materials and methods.
Supplementary Material
Acknowledgments
The work is supported by NIH grants NS097649-01 and CA240953-01, the Doris Duke Charitable Foundation Clinical Scientist Development award (https://www.ddcf.org/), the Sontag Foundation Distinguished Scientist Award (http://www.sontagfoundation.org), the Burroughs Wellcome Fund Career Awards for Medical Scientists (https://www.bwfund.org/), the Kimmel Scholar award (http://www.kimmel.org), and a Discovery Grant from the American Brain Tumor Association (https://www.abta.org/). We thank Dr. Bing Ren for his helpful comments. We are grateful to Brian Hirshman, Valya Ramakrishnan, Florina Grigore, Mihir Gupta, and Johnny Akers for assistance in qRT-PCR, informatics analysis, animal care, and graphical design.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. W.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009290118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Kalb R., et al., Hypomorphic mutations in the gene encoding a key Fanconi anemia protein, FANCD2, sustain a significant group of FA-D2 patients with severe phenotype. Am. J. Hum. Genet. 80, 895–910 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Czapski B., Baluszek S., Herold-Mende C., Kaminska B., Clinical and immunological correlates of long term survival in glioblastoma. Contemp. Oncol. (Pozn.) 22, 81–85 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fu Y., et al., Glioma-derived mutations in IDH: From mechanism to potential therapy. Biochem. Biophys. Res. Commun. 397, 127–130 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Amelot A., et al., IDH-mutation is a weak predictor of long-term survival in glioblastoma patients. PLoS One 10, e0130596 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van den Bent M. J., et al., Interlaboratory comparison of IDH mutation detection. J. Neurooncol. 112, 173–178 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Hegi M. E., et al., MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 352, 997–1003 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Smrdel U., et al., Long-term survival in glioblastoma: Methyl guanine methyl transferase (MGMT) promoter methylation as independent favourable prognostic factor. Radiol. Oncol. 50, 394–401 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sonoda Y., [Radiation and temozolomide therapy]. Nihon Rinsho 73 (suppl. 2), 619–622 (2015). [PubMed] [Google Scholar]
- 9.Zhou Z., Howard T. A., Villano J. L., Long-term daily temozolomide with dose-dependent efficacy in MGMT promotor methylation negative recurrent high-grade astrocytoma. Cancer Chemother. Pharmacol. 80, 1043–1046 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Friedman H. S., Kerby T., Calvert H., Temozolomide and treatment of malignant glioma. Clin. Cancer Res. 6, 2585–2597 (2000). [PubMed] [Google Scholar]
- 11.van Genugten J. A., Leffers P., Baumert B. G., Tjon-A-Fat H., Twijnstra A., Effectiveness of temozolomide for primary glioblastoma multiforme in routine clinical practice. J. Neurooncol. 96, 249–257 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gately L., McLachlan S. A., Philip J., Rathi V., Dowling A., Molecular profile of long-term survivors of glioblastoma: A scoping review of the literature. J. Clin. Neurosci. 68, 1–8 (2019). [DOI] [PubMed] [Google Scholar]
- 13.W. J. Curran, Jr, et al., Recursive partitioning analysis of prognostic factors in three Radiation Therapy Oncology Group malignant glioma trials. J. Natl. Cancer Inst. 85, 704–710 (1993). [DOI] [PubMed] [Google Scholar]
- 14.Stummer W.et al.; ALA-Glioma Study Group , Fluorescence-guided surgery with 5-aminolevulinic acid for resection of malignant glioma: A randomised controlled multicentre phase III trial. Lancet Oncol. 7, 392–401 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Simmons G. W., et al., Neurofibromatosis-1 heterozygosity increases microglia in a spatially and temporally restricted pattern relevant to mouse optic glioma formation and growth. J. Neuropathol. Exp. Neurol. 70, 51–62 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ginhoux F., Garel S., The mysterious origins of microglia. Nat. Neurosci. 21, 897–899 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Hoyer F. F., et al., Tissue-specific macrophage responses to remote injury impact the outcome of subsequent local immune challenge. Immunity 51, 899–914.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quarta A., et al., Murine iPSC-derived microglia and macrophage cell culture models recapitulate distinct phenotypical and functional properties of classical and alternative neuro-immune polarisation. Brain Behav. Immun. 82, 406–421 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Hambardzumyan D., Bergers G., Glioblastoma: Defining tumor niches. Trends Cancer 1, 252–265 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stephens L., et al., A novel phosphoinositide 3 kinase activity in myeloid-derived cells is activated by G protein beta gamma subunits. Cell 77, 83–93 (1994). [DOI] [PubMed] [Google Scholar]
- 21.Schmid M. C., et al., Receptor tyrosine kinases and TLR/IL1Rs unexpectedly activate myeloid cell PI3kγ, a single convergent point promoting tumor inflammation and progression. Cancer Cell 19, 715–727 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmid M. C., et al., PI3-kinase γ promotes Rap1a-mediated activation of myeloid cell integrin α4β1, leading to tumor inflammation and growth. PLoS One 8, e60226 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaneda M. M., et al., PI3Kγ is a molecular switch that controls immune suppression. Nature 539, 437–442 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang S., Denley A., Vanhaesebroeck B., Vogt P. K., Oncogenic transformation induced by the p110beta, -gamma, and -delta isoforms of class I phosphoinositide 3-kinase. Proc. Natl. Acad. Sci. U.S.A. 103, 1289–1294 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hawkins P. T., Stephens L. R., PI3K signalling in inflammation. Biochim. Biophys. Acta 1851, 882–897 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Yoshioka K., et al., Endothelial PI3K-C2α, a class II PI3K, has an essential role in angiogenesis and vascular barrier function. Nat. Med. 18, 1560–1569 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Chen P. H., Yao H., Huang L. J., Cytokine receptor endocytosis: New kinase activity-dependent and -independent roles of PI3K. Front. Endocrinol. (Lausanne) 8, 78 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirsch E., Ciraolo E., Ghigo A., Costa C., Taming the PI3K team to hold inflammation and cancer at bay. Pharmacol. Ther. 118, 192–205 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Okkenhaug K., Graupera M., Vanhaesebroeck B., Targeting PI3K in cancer: Impact on tumor cells, their protective stroma, angiogenesis, and immunotherapy. Cancer Discov. 6, 1090–1105 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li M., et al., Remodeling tumor immune microenvironment via targeted blockade of PI3K-γ and CSF-1/CSF-1R pathways in tumor associated macrophages for pancreatic cancer therapy. J. Control. Release 321, 23–35 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Yuan S., et al., Tumor-associated macrophages affect the biological behavior of lung adenocarcinoma A549 cells through the PI3K/AKT signaling pathway. Oncol. Lett. 18, 1840–1846 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gunderson A. J., et al., Bruton tyrosine kinase-dependent immune cell cross-talk drives pancreas cancer. Cancer Discov. 6, 270–285 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Venner K., [AIDS: Its early detection in dental practice]. Schweiz. Monatsschr. Zahnmed. 99, 143 (1989). [PubMed] [Google Scholar]
- 34.Sarkar S., et al., Microglia induces Gas1 expression in human brain tumor-initiating cells to reduce tumorigenecity. Sci. Rep. 8, 15286 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Razavi S. M., et al., Immune evasion strategies of glioblastoma. Front. Surg. 3, 11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nijaguna M. B., et al., Glioblastoma-derived macrophage colony-stimulating factor (MCSF) induces microglial release of insulin-like growth factor-binding protein 1 (IGFBP1) to promote angiogenesis. J. Biol. Chem. 290, 23401–23415 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Couto M., et al., The interplay between glioblastoma and microglia cells leads to endothelial cell monolayer dysfunction via the interleukin-6-induced JAK2/STAT3 pathway. J. Cell. Physiol. 234, 19750–19760 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Szulzewsky F., et al., Human glioblastoma-associated microglia/monocytes express a distinct RNA profile compared to human control and murine samples. Glia 64, 1416–1436 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Bowman R. L., Joyce J. A., Therapeutic targeting of tumor-associated macrophages and microglia in glioblastoma. Immunotherapy 6, 663–666 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Coniglio S. J., Segall J. E., Review: Molecular mechanism of microglia stimulated glioblastoma invasion. Matrix Biol. 32, 372–380 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Roesch S., Rapp C., Dettling S., Herold-Mende C., When immune cells turn bad-tumor-associated microglia/macrophages in glioma. Int. J. Mol. Sci. 19, 436 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Speranza M. C., et al., Preclinical investigation of combined gene-mediated cytotoxic immunotherapy and immune checkpoint blockade in glioblastoma. Neuro-oncol. 20, 225–235 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Müller S., et al., Single-cell profiling of human gliomas reveals macrophage ontogeny as a basis for regional differences in macrophage activation in the tumor microenvironment. Genome Biol. 18, 234 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimoni Y., Association between expression of random gene sets and survival is evident in multiple cancer types and may be explained by sub-classification. PLOS Comput. Biol. 14, e1006026 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burgenske D. M., et al., Molecular profiling of long-term IDH-wildtype glioblastoma survivors. Neuro-oncol. 21, 1458–1469 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J., et al., Epigenetic suppression of EGFR signaling in G-CIMP+ glioblastomas. Oncotarget 5, 7342–7356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim Y. W., et al., Identification of prognostic gene signatures of glioblastoma: A study based on TCGA data analysis. Neuro-oncol. 15, 829–839 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lewis C. A., et al., SREBP maintains lipid biosynthesis and viability of cancer cells under lipid- and oxygen-deprived conditions and defines a gene signature associated with poor survival in glioblastoma multiforme. Oncogene 34, 5128–5140 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Karnofsky D. A., Abelmann W. H., Craver L. F., Burchenal J. H., The use of the nitrogen mustards in the palliative treatment of carcinoma–With particular reference to bronchogenic carcinoma. Cancer 1, 634–656 (1948). [Google Scholar]
- 50.Verhaak R. G.et al.; Cancer Genome Atlas Research Network , Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17, 98–110 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Scott M. C., et al., Comparative transcriptome analysis quantifies immune cell transcript levels, metastatic progression, and survival in osteosarcoma. Cancer Res. 78, 326–337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blando J. M., et al., Cooperation between Stat3 and Akt signaling leads to prostate tumor development in transgenic mice. Neoplasia 13, 254–265 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shin D. S., et al., Cryptotanshinone inhibits constitutive signal transducer and activator of transcription 3 function through blocking the dimerization in DU145 prostate cancer cells. Cancer Res. 69, 193–202 (2009). [DOI] [PubMed] [Google Scholar]
- 55.Mukthavaram R., et al., Effect of the JAK2/STAT3 inhibitor SAR317461 on human glioblastoma tumorspheres. J. Transl. Med. 13, 269 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu Z., Pestell T. G., Lisanti M. P., Pestell R. G., Cancer stem cells. Int. J. Biochem. Cell Biol. 44, 2144–2151 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kiuchi N., et al., STAT3 is required for the gp130-mediated full activation of the c-myc gene. J. Exp. Med. 189, 63–73 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stansley B., Post J., Hensley K., A comparative review of cell culture systems for the study of microglial biology in Alzheimer’s disease. J. Neuroinflammation 9, 115 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ernst M., et al., STAT3 and STAT1 mediate IL-11-dependent and inflammation-associated gastric tumorigenesis in gp130 receptor mutant mice. J. Clin. Invest. 118, 1727–1738 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Putoczki T. L., et al., Interleukin-11 is the dominant IL-6 family cytokine during gastrointestinal tumorigenesis and can be targeted therapeutically. Cancer Cell 24, 257–271 (2013). [DOI] [PubMed] [Google Scholar]
- 61.Dimitriadis E., Sharkey A. M., Tan Y. L., Salamonsen L. A., Sherwin J. R., Immunolocalisation of phosphorylated STAT3, interleukin 11 and leukaemia inhibitory factor in endometrium of women with unexplained infertility during the implantation window. Reprod. Biol. Endocrinol. 5, 44 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lewis V. O., et al., The interleukin-11 receptor alpha as a candidate ligand-directed target in osteosarcoma: Consistent data from cell lines, orthotopic models, and human tumor samples. Cancer Res. 69, 1995–1999 (2009). [DOI] [PubMed] [Google Scholar]
- 63.Kozono D., et al., Dynamic epigenetic regulation of glioblastoma tumorigenicity through LSD1 modulation of MYC expression. Proc. Natl. Acad. Sci. U.S.A. 112, E4055–E4064 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suvà M. L., et al., Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell 157, 580–594 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Le Rhun E., et al., Molecular targeted therapy of glioblastoma. Cancer Treat. Rev. 80, 101896 (2019). [DOI] [PubMed] [Google Scholar]
- 66.Gutmann D. H., Kettenmann H., Microglia/brain macrophages as central drivers of brain tumor pathobiology. Neuron 104, 442–449 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Poon C. C., et al., Differential microglia and macrophage profiles in human IDH-mutant and -wild type glioblastoma. Oncotarget 10, 3129–3143 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kohsaka S., et al., STAT3 inhibition overcomes temozolomide resistance in glioblastoma by downregulating MGMT expression. Mol. Cancer Ther. 11, 1289–1299 (2012). [DOI] [PubMed] [Google Scholar]
- 69.Sarkar S., et al., Therapeutic activation of macrophages and microglia to suppress brain tumor-initiating cells. Nat. Neurosci. 17, 46–55 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Hambardzumyan D., Gutmann D. H., Kettenmann H., The role of microglia and macrophages in glioma maintenance and progression. Nat. Neurosci. 19, 20–27 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lathia J. D., Mack S. C., Mulkearns-Hubert E. E., Valentim C. L., Rich J. N., Cancer stem cells in glioblastoma. Genes Dev. 29, 1203–1217 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Friedmann-Morvinski D., Verma I. M., Dedifferentiation and reprogramming: Origins of cancer stem cells. EMBO Rep. 15, 244–253 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Crane G. M., Jeffery E., Morrison S. J., Adult haematopoietic stem cell niches. Nat. Rev. Immunol. 17, 573–590 (2017). [DOI] [PubMed] [Google Scholar]
- 74.Yu K., et al., PIK3CA variants selectively initiate brain hyperactivity during gliomagenesis. Nature 578, 166–171 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Natrajan R., et al., Microenvironmental heterogeneity parallels breast cancer progression: A histology-genomic integration analysis. PLoS Med. 13, e1001961 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Noch E. K., Ramakrishna R., Magge R., Challenges in the treatment of glioblastoma: Multisystem mechanisms of therapeutic resistance. World Neurosurg. 116, 505–517 (2018). [DOI] [PubMed] [Google Scholar]
- 77.Osuka S., Van Meir E. G., Overcoming therapeutic resistance in glioblastoma: The way forward. J. Clin. Invest. 127, 415–426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim H., et al., Whole-genome and multisector exome sequencing of primary and post-treatment glioblastoma reveals patterns of tumor evolution. Genome Res. 25, 316–327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ramakrishnan V., et al., Radiation-induced extracellular vesicle (EV) release of miR-603 promotes IGF1-mediated stem cell state in glioblastomas. EBioMedicine 55, 102736 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Baysan M., et al., G-cimp status prediction of glioblastoma samples using mRNA expression data. PLoS One 7, e47839 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Subramanian A., et al., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.