Significance
Following an acute viral infection, the majority of activated virus-specific CD8 T cells die off, while a smaller subset survives and develops into long-lived memory CD8 T cells. The underlying process regulation of this cell fate decision remains incompletely understood. In this study, we identify two main patterns of gene regulatory networks underlying effector and memory CD8 T cell differentiation. Furthermore, enhancer activity correlates with these regulon patterns. Finally, cells destined to become memory CD8 T cells maintain accessibility at enhancers regulating key memory-related genes, and the transcription factor E2A regulates the accessibility of many memory-related enhancers to promote memory cell formation.
Keywords: CD8 T cell, scRNA-seq, epigenetics, LCMV, T cell memory
Abstract
During an acute viral infection, CD8 T cells encounter a myriad of antigenic and inflammatory signals of variable strength, which sets off individual T cells on their own differentiation trajectories. However, the developmental path for each of these cells will ultimately lead to one of only two potential outcomes after clearance of the infection—death or survival and development into memory CD8 T cells. How this cell fate decision is made remains incompletely understood. In this study, we explore the transcriptional changes during effector and memory CD8 T cell differentiation at the single-cell level. Using single-cell, transcriptome-derived gene regulatory network analysis, we identified two main groups of regulons that govern this differentiation process. These regulons function in concert with changes in the enhancer landscape to confer the establishment of the regulatory modules underlying the cell fate decision of CD8 T cells. Furthermore, we found that memory precursor effector cells maintain chromatin accessibility at enhancers for key memory-related genes and that these enhancers are highly enriched for E2A binding sites. Finally, we show that E2A directly regulates accessibility of enhancers of many memory-related genes and that its overexpression increases the frequency of memory precursor effector cells and accelerates memory cell formation while decreasing the frequency of short-lived effector cells. Overall, our results suggest that effector and memory CD8 T cell differentiation is largely regulated by two transcriptional circuits, with E2A serving as an important epigenetic regulator of the memory circuit.
CD8 T cells have a critical role in the defense against viruses, intracellular bacteria, and cancerous cells. Once activated, naïve CD8 T cells rapidly proliferate, while also differentiating into effectors capable of killing infected cells. After clearance of an infection, the vast majority of these effector cells undergo apoptosis, leaving behind a smaller pool of long-lived memory CD8 T cells that provide enhanced protection against reinfection with the same pathogen. Effector CD8 T cells destined to die off are called short-lived effector cells (SLECs) and can be identified by the high expression of killer cell lectin-like receptor G1 (KLRG1) (1). Memory precursor effector cells (MPECs), on the other hand, have high potential to become memory cells and express high levels of interleukin 7 receptor (IL-7R) (2–4). Dynamic changes in gene expression occur during the differentiation of activated CD8 T cells, and it has been proposed that the diverging developmental paths of MPECs and SLECs are controlled in part by the graded expression of various transcription factors (TFs) (5).
The study of individual TFs has greatly improved our understanding of effector and memory CD8 T cell differentiation. However, cell identities tend to be determined by multiple TFs coordinately regulating each other and cell-specific effector genes, forming gene regulatory networks (GRNs) (6). Previous work has been done to construct a genome-wide regulatory network of CD8 T cell differentiation from publicly available expression data (7). However, this study relied on RNA-seq performed on large populations of cells averaged together. Single-cell RNA sequencing (scRNA-seq) has recently emerged as a powerful tool for interrogating the transcriptome in heterogeneous populations of cells. Initial studies of single-cell gene expression in CD8 T cells revealed extensive transcriptional changes as early as the first cell division and identified regulators of CD8 T cell differentiation (8, 9). Recent advances in scRNA-seq technology allow for the analysis of a much greater number of cells than previously possible (10). Furthermore, new computational approaches that combine identification of coexpression modules with cis-regulatory motif analysis can reconstruct complex GRNs from scRNA-seq data (11). Applying this method to effector and memory CD8 T cell differentiation has the potential to more accurately predict the regulatory modules underlying the cell fate decision of CD8 T cells.
While transcriptional differences clearly influence effector and memory CD8 T cell differentiation, the critical role of the epigenetic landscape in regulating this cell fate decision is only beginning to be understood. TFs can have varied effects on different cell types depending on the local chromatin architecture, particularly at enhancer regions, which tend to have greater cell type–specific accessibility (12–14). Therefore, exclusively comparing TF expression levels likely yields an incomplete picture of this differentiation process. Recent evidence supports the idea that the epigenetic landscape influences CD8 T cell differentiation during a range of infection settings and cancer, leading to the identification of new regulators and critical cell-specific enhancers (15–20).
In this study, we used scRNA-seq to explore the transcriptional changes of effector and memory CD8 T cell differentiation at the single-cell level and unveil the GRNs that govern the cell fate decision of CD8 T cells during acute viral infection. We further demonstrated how changes in the enhancer landscape parallel the activity patterns of these transcriptional circuits. Lastly, we found that the chromatin accessibility at these enhancer regions, regulated in part through the direct action of the TF E2A, is a potential underlying mechanism by which the divergent cell fates of MPECs and SLECs are determined.
Results
Single Cell Regulatory Network Analysis Reveals Two Transcriptional Circuits during CD8 T Cell Differentiation.
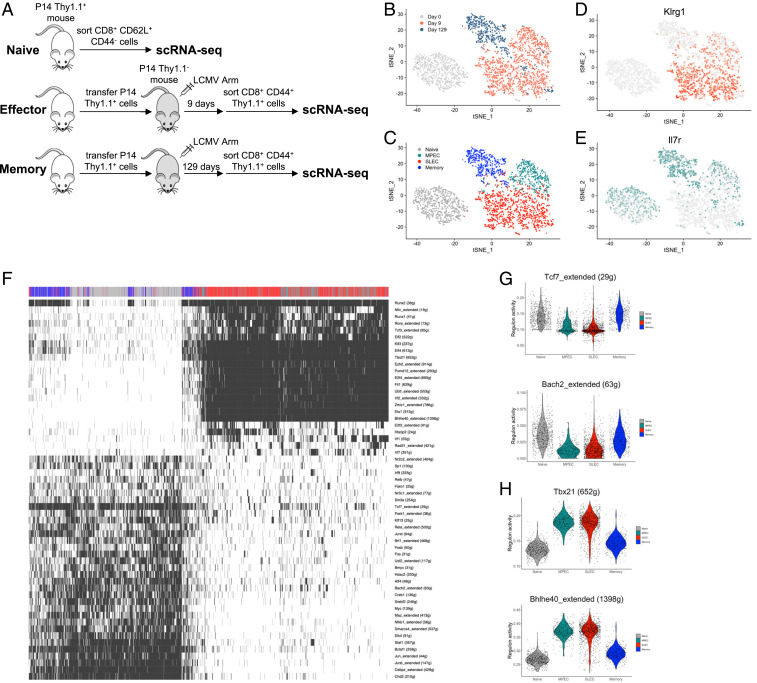
To better understand the cell fate decision process of CD8 T cells responding to an acute viral infection, in particular the underlying transcriptional networks that govern effector and memory T cell formation, we performed scRNA-seq on antigen-specific CD8 T cells during an acute infection with lymphocytic choriomeningitis virus (LCMV). Congenically marked P14 cells, possessing a transgenic H-2Db–restricted T cell receptor (TCR) specific for the gp33–41 peptide of LCMV, were transferred to C57BL/6 mice, followed by infection with the Armstrong strain of LCMV. Naïve, effector, and memory virus-specific CD8 T cells were sorted by flow cytometry on day 0, day 9, and day 129 postinfection (p.i.), respectively, and then single-cell transcriptome libraries were prepared using the 10× Genomics Chromium platform (Fig. 1A). After initial filtering, 762 naïve cells, 1,473 effector cells, and 554 memory cells were used for analysis. The dimensionality reduction technique t-distributed stochastic neighbor embedding (t-SNE) was used to visualize the data. This showed that while cells from each of the three samples largely grouped together with other cells from the same time point, naïve cells were highly distinct from effector and memory cells, suggesting more similar overall gene expression profiles between all antigen-experienced cells (Fig. 1B). Two distinct clusters arose within the day 9 effector cell sample (Fig. 1C). These clusters differed in their expression of surface markers known to distinguish effector CD8 T cells with different memory potential: KLRG1, marking SLECs, and IL-7R, marking MPECs (1) (Fig. 1 D and E).
Fig. 1.
scRNA-seq of CD8 T cells during acute viral infection. (A) Outline of scRNA-seq experiment. (B) t-SNE plot displaying relationship between naïve (day 0), effector (day 9), and memory (day 129) CD8 T cells during acute LCMV infection. (C) t-SNE plot displaying four clusters of CD8 T cells arising during acute LCMW infection. (D and E) t-SNE plots showing expression of Klrg1 (D) and Il7r (E), two classic markers of effector CD8 T cells with low and high potential to become memory CD8 T cells, respectively. (F) Cells were clustered based on binary regulon activity of the 56 regulons that are active in at least 1% of the cells and that correlated (absolute Pearson correlation > 0.30) with at least one other regulon. Cells are colored to indicate that a regulon is active (black) or inactive (white). (G and H) Violin plots showing distribution of continuous regulon activity in naïve and memory TF regulons (Tcf7 and Bach2) (G) or effector TF regulons (Tbx21 and Bhlhe40) (H).
We next used our scRNA-seq data to more thoroughly investigate the GRNs governing effector and memory CD8 T cell differentiation. We took advantage of the recently developed computational method single-cell regulatory network inference and clustering (SCENIC) (11) to infer a regulatory network for each TF based on coexpression in individual cells, identify potential direct targets based on cis-regulatory motif analysis, and determine the activity of these TF regulons in each cell. In this way, cells can be clustered based on overall similarities in TF regulatory network activity, taking into account not just TF expression but also expression of their target genes.
Because we were interested in the global regulatory networks controlling CD8 T cell differentiation, we focused our subsequent analyses on the 56 regulons that had activity patterns that were strongly correlated with other regulons (Fig. 1F). Cells clustered into two groups based on the binary regulon activity of these 56 regulons: MPECs and SLECs clustered together, while naïve and memory cells clustered together separately. Therefore, when classifying each TF regulon as either on or off, MPECs were remarkably similar to SLECs despite their dramatically different eventual fates.
The cluster comprised of naïve and memory cells had higher regulon activity for several TFs involved in T cell quiescence and memory T cell differentiation, including Tcf7 (21–24), Foxo1 (25–30), and Bach2 (31, 32) (Fig. 1 F and G). Additionally, the regulon for the recently identified regulator of memory cell formation, Nr3c1, had higher activity in naïve and memory CD8 T cells (16) (Fig. 1F). Importantly, MPECs and SLECs had surprisingly similar regulon activity for many TFs involved in T cell quiescence, such as Tcf7 and Bach2 (Fig. 1G). The regulons of TFs known to promote effector function and terminal differentiation of CD8 T cells were found to be more active in both MPECs and SLECs (Fig. 1 F and H). For example, the regulon for Tbx21, a key gene promoting the differentiation of SLECs (1, 33–35), had high activity in these cells. This group also included the regulons of Bhlhe40 and Runx2, which were identified in a previous regulatory network study utilizing published bulk RNA-seq, as TFs inhibiting the recall proliferation of CD8 T cells, potentially by driving their terminal differentiation (7). Overall, two main patterns of TF regulon activity arise during effector and memory CD8 T cell differentiation: a naïve/memory GRN that has high activity in naïve cells that is lost in both MPECs and SLECs then reacquired in memory cells and an effector GRN that becomes active in effector CD8 T cells and is dampened, but still increased relative to naïve cells, in long-lived memory CD8 T cells. The unexpected similarity between MPECs and SLECs with respect to their regulon activity suggests additional mechanisms must exist to explain their dramatically distinct cell fate decisions.
Two Major Patterns of Enhancer Activity during Memory CD8 T Cell Differentiation.
Since relative expression of TFs and activity of their regulons appeared to be an incomplete explanation of memory CD8 T cell development, this prompted us to further examine the epigenetic landscape during this T cell differentiation process. In particular, we were interested in the regulation of enhancers throughout this process, as enhancers are critical regulatory elements shown to control the cellular differentiation. Therefore, we performed chromatin immunoprecipitation (ChIP)-seq for the histone modification H3K27Ac, a mark associated with active regulatory regions (36, 37), to measure enhancer activity; assay for transposase-accessible chromatin (ATAC)-seq to identify regions of open chromatin (38); and RNA-seq on naïve CD8 T cells, MPECs, SLECs, and memory CD8 T cells (SI Appendix, Fig. S1A).
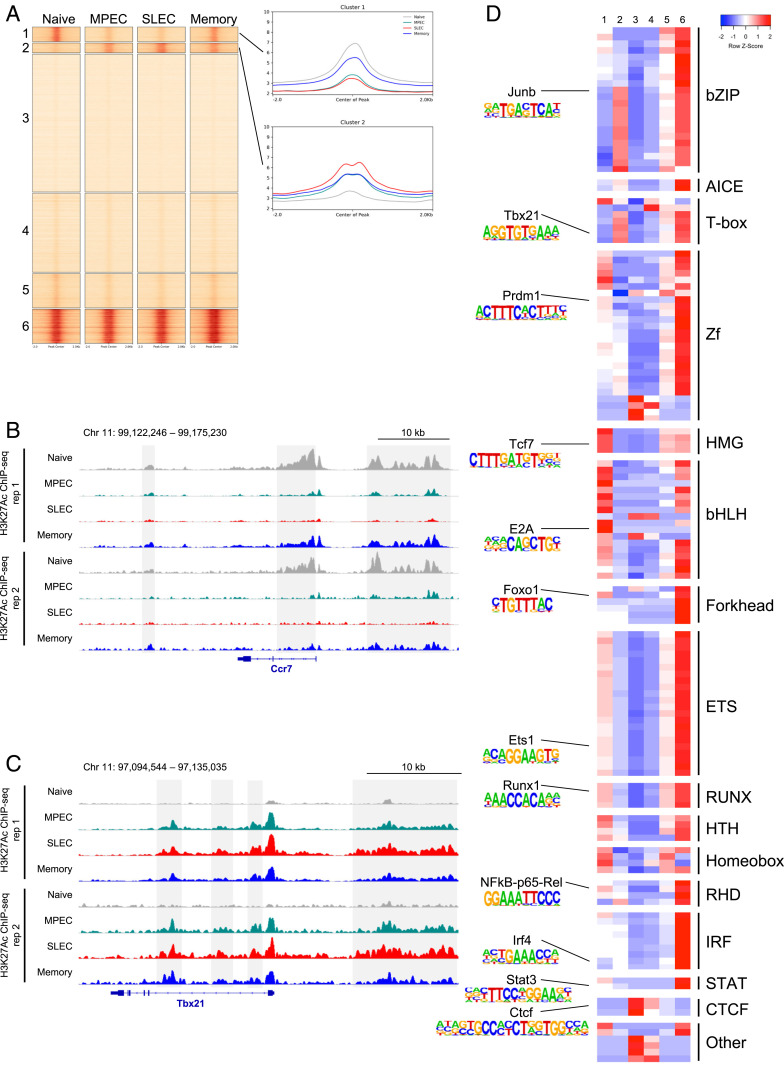
We first investigated changes in enhancer activity during memory CD8 T cell differentiation. To do this, we performed k-means clustering based on H3K27Ac ChIP-seq signal intensity on our pooled replicates at 120,019 putative enhancers that were identified by subtracting known promoter regions from ATAC-seq peaks in our four cell subsets (39) (Fig. 2A). We found that the majority of these sites (110,482/120,019 or 92.1%) had relatively stable H3K27Ac in all four cell populations. Cluster 3 (53,364/120,019 or 44.5%) contains sites that had no enrichment of H3K27Ac ChIP-seq signal, indicating that these sites are likely not actually functional enhancers in CD8 T cells. Cluster 6 (13,235/120,019 or 11.0%) contains sites with very high H3K27Ac enrichment in all four samples and are associated with genes required for general CD8 T cell function and identity, including genes encoding the CD8 glycoprotein (Cd8a and Cd8b1), TFs required for T cell development (Ets1), T cell signaling molecules such as protein kinase c isoenzymes (Prkca, Prkcb, Prkce, Prkch, Prkcq, and Prkcz), and the leukocyte common antigen CD45 (Ptprc).
Fig. 2.
Patterns of enhancer activity during memory CD8 T cell differentiation. (A) k-means clustering of H3K27Ac ChIP-seq signal at 120,019 nonpromoter chromatin accessible regions in naïve CD8 T cells, MPECs, SLECs, and memory CD8 T cells. On the right, the average H3K27Ac profile is shown for each of the four samples in clusters 1 and 2. (B and C) H3K27Ac ChIP-seq data are shown at the Ccr7 (B) and Tbx21 (C) loci. Enhancers showing representative patterns of activity are highlighted in gray. (D) Motif analysis was performed on the six clusters from A using the database of known TF motifs in HOMER. Each row displays the row-normalized −log P value, with red denoting high enrichment of a given motif in that cluster and blue denoting low enrichment.
Interestingly, two clusters were found to have enhancer activity that changes over the course of the viral infection: cluster 1 (5,699/120,019 or 4.7%), which has high enhancer activity in naïve and memory CD8 T cells but low activity in both MPECs and SLECs and cluster 2 (3,838/120,019 or 3.2%), which has low activity in naïve cells and increased activity in all antigen-experienced cells, with the highest activity in SLECs. Cluster 1 contains enhancers associated with genes related to CD8 T cell quiescence, homeostasis, and lymph node homing, such as the TFs Bach2, Foxo1, Foxp1, Id3, Lef1, and Tcf7 and the cytokine and homing receptors Ccr7 (Fig. 2B), Il7r, and Sell. Cluster 2 contains enhancers associated with genes related to CD8 T cell activation and effector function. Examples include the TFs Batf, Bhlhe40, Prdm1, and Tbx21 (Fig. 2C); the chemokines Ccl3, Ccl5, and Ccl9; the effector molecules Gzmb, Gzmk, Ifng, and Prf1; and other genes such as Cd44 and Klrg1. Furthermore, by comparing relative gene expression levels, we found that genes undergoing significant changes during memory CD8 T cell differentiation largely exhibited one of these two patterns of expression (SI Appendix, Fig. S1 B and C). These results suggest that there are two dominant transcriptional circuits regulating gene expression in CD8 T cells as they undergo differentiation into memory CD8 T cells.
Super Enhancers Exhibit Two Patterns of Activity during Memory CD8 T Cell Differentiation.
A small proportion of regulatory regions, termed super enhancers (SEs), possess exceedingly high binding levels of Mediator, master regulator TFs, and histone modifications associated with enhancer activity such as H3K27Ac (40–42). These SEs have been proposed to play an important role in determining cell identity and are found near cell type–specific master regulator TFs (40, 42). They can be identified by stitching together enhancers within 12.5 kb of each other and then ranking these enhancer regions by enrichment of Mediator, H3K27Ac, or master regulator TF ChIP-seq signal (40–42).
Given the particular importance of SEs in regulating expression of genes that determine cell fate, we explored the changes in their activity during memory CD8 T cell differentiation. First, we identified 1,149, 399, 358, and 723 SEs in naïve, MPECs, SLECs, and memory CD8 T cells, respectively, using the rank ordering of super enhancers algorithm (40, 42) (SI Appendix, Fig. S2A). Genes nearest to SEs for each subset tended to have slightly higher average expression in that subset (SI Appendix, Fig. S2B). However, the majority of SEs for each subset were shared with at least one other subset, likely contributing to the relatively similar average expression levels (SI Appendix, Fig. S2C). Naïve CD8 T cells had the most unique SEs (518/1,289 or 40.2%), while MPECs, SLECs, and memory CD8 T cells only had 17, 24, and 63 unique SEs, respectively.
As much of the critical regulation may occur because of the relative activity of each SE, rather than its presence or absence, we next investigated the changes in enhancer activity, as measured by H3K27Ac signal intensity in our four subsets. Using hierarchical clustering, we identified two distinct patterns of SE activity occurring during memory CD8 T cell differentiation (SI Appendix, Fig. S2 D and E). Similar to the patterns we observed of RNA expression (SI Appendix, Fig. S1 B and C) and overall enhancer activity (Fig. 2A), cluster 1 SEs started with high activity in naïve cells that was lost in both effector cell subsets then regained in memory cells, while cluster 2 SEs gained activity after naïve cells were activated. These results demonstrate that, at both typical enhancers and SEs, there are two dominant regulatory patterns that coordinate effector and memory CD8 T cell differentiation.
Regulation of CD8 T Cell Enhancer Activity.
After identifying the patterns of enhancer and SE activity arising during memory CD8 T cell differentiation, we sought to determine the TFs responsible for controlling these regulatory regions. To do this, we performed motif analysis on the six clusters of enhancers using the hypergeometric optimization of motif enrichment (HOMER) motif database (43). The region at the center of each enhancer, corresponding to the overlapping ATAC-seq peak, was searched for enrichment of known TF binding motifs. TF motif enrichment was compared in each cluster for motifs that were significantly enriched in at least one of the six clusters (−log P value ≥ 9) (Fig. 2D).
Several known TF motifs were found to be more enriched in the cluster 1 enhancers (high activity in naïve and memory CD8 T cells but low activity in MPECs and SLECs) than the cluster 2 enhancers (low activity in naïve CD8 T cells and high activity in activated cells). High mobility group; basic helix–loop–helix; and some forkhead motifs, which are bound by the memory T cell–promoting TFs TCF7 (21, 22, 24), E2A (44, 45), and Foxo1 (25–29), respectively, were all more enriched in cluster 1 than cluster 2. Additionally, GATA motifs were more highly enriched in cluster 1. Though less is known about the role of these TFs in the CD8 T cell response to viral infection, GATA3 (46) has been shown to regulate expression of Il7r, which is required for maintenance of both naïve and memory CD8 T cells. E26 transformed-specific motifs were also slightly enriched in cluster 1 relative to cluster 2; however, these were the most highly enriched motifs in all clusters except clusters 3 and 4, suggesting that TFs from this family likely play important roles in all CD8 T cell enhancers. The binding motifs of some repressive TFs were also found to be enriched in cluster 1 enhancers. The transcriptional repressor Blimp-1 (encoded by Prdm1) motif was enriched in cluster 1, indicating that expression of this protein might inhibit activity of enhancers critical for memory T cell formation. This is consistent with the observation that deficiency in Blimp-1 results in higher expression of several memory-related genes, such as Id3, Ccr7, and Sell (47), all of which have at least one nearby enhancer in cluster 1. The RUNX motif was also enriched in this cluster. Recent work demonstrated a role for Runx3 in acting on promoters and enhancers of Tcf7 and Bcl6 to promote deposition of the repressive histone modification H3K27Me3 (48).
Cluster 2 enhancers, with low activity in naïve cells and high activity in activated cells, had higher enrichment of many other known TF motifs. Basic leucine zipper domain TFs from the AP-1 family that bind to the consensus AP-1 motif (5′-TGA G/C TCA-3′) were highly enriched in cluster 2. AP-1 factors are induced by TCR signaling, and recent work has shown that the memory-promoting TF BACH2 competes with AP-1 factors for binding to these sites in order to attenuate TCR-induced activation (31, 49). Additionally, the AP-1-IRF composite element motif was enriched in cluster 2, consistent with the known role of the AP-1 factor BATF cooperating with IRF4 to regulate CD8 T cell effector function (50–52). Interferon regulatory factors (IRFs) alone, on the other hand, have similar enrichment in the two clusters of dynamically utilized enhancers. Cluster 2 also had high enrichment of the binding motif for T-bet, a TF that drives formation of terminally differentiated effector cells (1, 33, 34, 47, 53). Together, AP-1 factors, including those cooperating with IRF proteins, and T-bet likely play a major role in the regulation of these cluster 2 effector enhancers.
Finally, cluster 3 and 4 regions, which mostly lack H3K27Ac enrichment, have high enrichment of the CTCF binding motif, indicating that these sites might structurally regulate the three-dimensional chromatin architecture (54). Overall, these results suggest that cluster 1 naïve/memory enhancers are positively regulated by TCF7, E2A, Foxo1, and GATA-3 and negatively regulated by Blimp-1 and Runx3, whereas cluster 2 effector enhancers are regulated by AP-1, AP-1-IRF complexes, and T-bet.
Accessibility at E2A- and TCF7-Occupied Enhancers Is Maintained in Memory Precursor Effector Cell Differentiation.
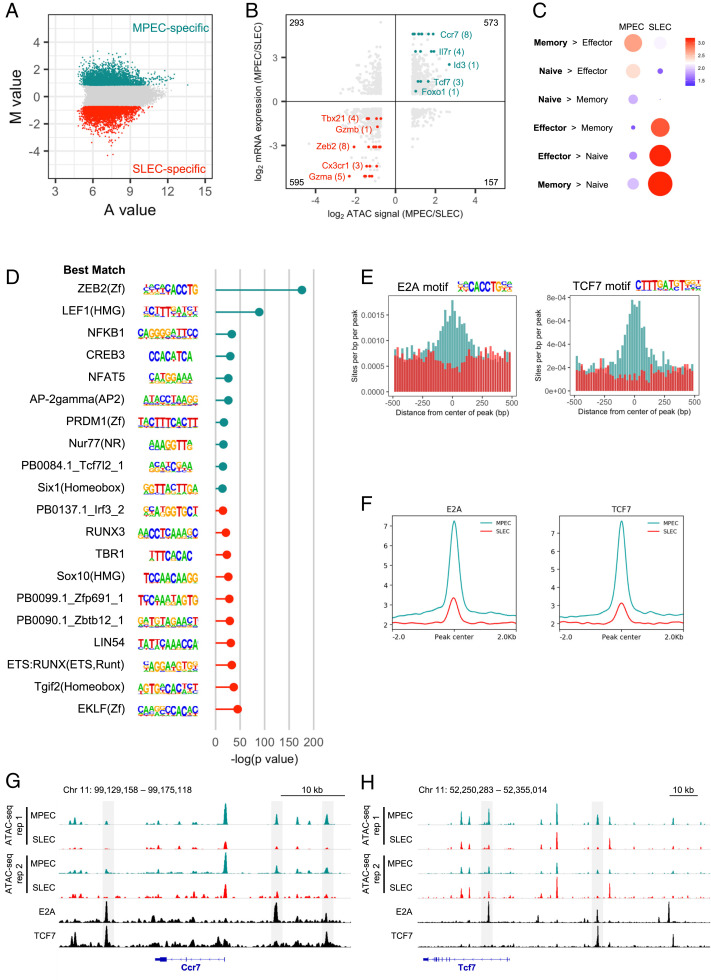
Earlier, we identified a group of regular enhancers (Fig. 2A, cluster 1) and SEs (SI Appendix, Fig. S2C, cluster 1) with high activity in naïve and memory CD8 T cells but low activity in both MPECs and SLECs. A natural question that arises from this observation is how are MPECs able to regain activity of these enhancers on their way to becoming memory cells, while SLECs cannot? We hypothesized that differences in chromatin accessibility might explain this phenomenon. To investigate these differences, we first normalized the ATAC-seq signal between MPECs and SLECs and then identified the top 5,000 MPEC- and SLEC-specific enhancers based on M value, a metric proportional to the log2 ratio of ATAC-seq signal in MPECs divided by ATAC-seq signal in SLECs (Fig. 3A). After pairing each enhancer with its nearest gene, we found that MPEC-specific enhancers were enriched for genes with higher expression in MPECs than SLECs (Ccr7, Foxo1, Il7r, Id3, and Tcf7), and SLEC-specific enhancers were enriched for genes with higher expression in SLECs than MPECs (Cx3cr1, Gzma, Gzmb, Tbx21, and Zeb2), suggesting that these enhancers do regulate gene expression in each subset (Fig. 3B). Furthermore, gene ontology analysis using Genomic Regions Enrichment of Annotations Tool (GREAT) (55) showed that MPEC-specific enhancers were located near genes in memory > effector, naïve > effector, and naïve > memory gene sets; white SLEC-specific enhancers were located near genes in effector > memory, effector > naïve, and memory > naïve gene sets (56) (Fig. 3C). Together, these results suggest that the MPEC- and SLEC-specific ATAC-seq peaks we identified are enriched for enhancers controlling the expression of genes critical to each subset’s function and cell fate.
Fig. 3.
E2A and TCF7 binding sites are enriched in enhancers maintained in an open state by MPECs but not SLECs. (A) MA plot showing M value (log2 read density in MPEC ÷ read density in SLEC) versus A value (0.5 × log2 read density in MPEC × read density in SLEC) of the merged set of MPEC and SLEC nonpromoter ATAC-seq peaks after normalization. The top 5,000 peaks are highlighted for MPECs (cyan) and SLECs (red). (B) The log2 ratio of normalized ATAC-seq signal is compared to the log2 ratio of RNA expression for MPECs and SLECs. Only the top 5,000 MPEC- and SLEC-specific nonpromoter ATAC-seq peaks that are associated with genes that are significantly differently expressed between MPECs and SLECs are included, with the total number in each quadrant noted in each corner. Some notable genes with higher ATAC-seq signal and RNA expression in MPECs (cyan) or SLECs (red) are highlighted, with the number of enhancers in parentheses. (C) Gene ontology analysis for the MPEC- and SLEC-specific enhancers was performed using GREAT (55) and compared to published microarray gene sets of naïve, day 8 effector, and memory P14 CD8 T cells (56). Circle size denotes significance of gene set enrichment. Color represents fold enrichment. (D) The top 10 significantly enriched de novo motifs for each subset are shown, along with their significance (−log P value), ranked from most MPEC-specific (Top) to most SLEC-specific (Bottom). On the left is the best match known motif according to HOMER. (E) The distributions of the E2A and TCF7 motifs in the top 5,000 MPEC- and SLEC-specific enhancers are shown for the regions ± 500 bp from the center of each enhancer. (F) E2A (SRR3984693) (57) and TCF7 (SRR1024054) (58) ChIP-seq enrichment was assessed at the top 5,000 MPEC- and SLEC-specific enhancers (cyan and red, respectively). (G and H) MPEC and SLEC ATAC-seq data and E2A and TCF7 ChIP-seq data are shown at the Ccr7 (G) and Tcf7 (H) loci. Highlighted in gray are enhancers with higher ATAC-seq signal in MPECs than SLECs as well as E2A and/or TCF7 peaks.
We then compared the DNA sequences of these two groups of enhancers by de novo motif analysis to identify short regions of DNA enriched in one of the groups that correspond to TF binding sites (Fig. 3D). Among the best matches of known TF motifs for the top de novo motifs were ZEB2, E2A, and ZEB1, and for the second-most significant, de novo motifs were LEF1, TCF7L2, and TCF7. Given the previously reported roles for E2A (44, 45) and TCF7 (21, 22, 24) in memory CD8 T cell formation, we predicted that these two TFs were binding to and regulating MPEC-specific enhancers. By searching specifically for the E2A and TCF7 binding motifs, we found that both were highly enriched in MPEC-specific enhancers compared to SLEC-specific enhancers (Fig. 3E). Using published E2A (57) and TCF7 (58) ChIP-seq data, we confirmed that both of these TFs also have much greater binding to MPEC-specific enhancers (Fig. 3F).
Several genes critical for memory CD8 T cell fate and function were found to have MPEC-specific enhancers bound by E2A and/or TCF7 (Fig. 3 G and H). Ccr7, a chemokine receptor responsible for T cell homing to lymph nodes, possesses three enhancers with higher accessibility in MPECs than SLECs, as well as E2A and TCF7 peaks (Fig. 3G). Of particular interest, Tcf7 itself had two nearby enhancers sharing this pattern, a +7.5 kb enhancer bound by E2A and a −30 kb enhancer bound by both E2A and TCF7 (Fig. 3H). E2A is likely an important regulator of Tcf7 expression and has previously been shown to have greater binding to these enhancers in naïve CD8 T cells than in vitro–activated CD8 T cells (45). The higher level of ID2, an inhibitor of E proteins like E2A, in SLECs than MPECs has been proposed to repress memory formation through decreased E2A activity (45, 59, 60). This potentially leads to lower levels of TCF7 and loss of chromatin accessibility at E2A- and TCF7-regulated enhancers, explaining why SLECs cannot regain activity at these enhancers, reexpress critical memory-related genes, and survive long term. Overall, these data demonstrate that enhancers that lose accessibility in SLECs compared to MPECs are bound by E2A and TCF7 and suggest a mechanism explaining the opposing cell fate decisions of these two effector CD8 T cell subsets.
T-Bet– and AP-1–Bound Enhancers Become Accessible in Memory CD8 T Cells.
Memory CD8 T cells possess the ability to more rapidly express effector molecules upon reinfection than naïve CD8 T cells. We hypothesized that this could be due to a set of enhancers existing in a poised state: accessible in memory cells but inactive or having low activity until the cells are restimulated. We therefore compared the ATAC-seq signal in naïve and memory CD8 T cells to identify the memory-specific enhancers that potentially endow these cells with their enhanced protective abilities. After normalizing the ATAC-seq signal across these two samples, we identified the top 5,000 naïve- and memory-specific enhancers (SI Appendix, Fig. S3A). Naïve-specific enhancers were enriched for genes with higher expression in naïve cells than memory cells (Ccr7, Ccr9, Id3, and Tcf7), while the memory-specific enhancers were enriched for genes with higher expression in memory cells than naïve cells (Ccl4, Ccl5, Gzma, Ifng, Prdm1, Prf1, Stat4, Tbx21, and Xcl1) (SI Appendix, Fig. S3B). Gene ontology analysis using GREAT (55) showed that naïve- and memory-specific enhancers are enriched for genes highly expressed in the corresponding subset (SI Appendix, Fig. S3C).
Next, we performed de novo motif analysis to determine which TFs regulate these enhancers (SI Appendix, Fig. S3D). Two memory-specific motifs stood out as being highly significant. The top de novo motif was consistent with the known binding motif of T-bet, and the second-most significant de novo motif was consistent with the AP-1 family binding motif. Searching specifically for the motifs of T-bet and JunB, an AP-1 family member, we confirmed that both were highly enriched in memory-specific enhancers compared to naïve-specific enhancers (SI Appendix, Fig. S3E). When comparing T-bet (53) and JunD (31) ChIP-seq enrichment from published datasets at the top 5,000 naïve- and memory-specific enhancers, we found that memory-specific enhancers had much more T-bet and JunD binding (SI Appendix, Fig. S3F).
Several genes critical for the cytotoxic function of CD8 T cells were found to have memory-specific enhancers bound by T-bet and/or JunD (SI Appendix, Fig. S3 G and H). We identified the chemokines Ccl3 and Ccl4 and the cytokine Ifng as genes with multiple enhancers possessing higher accessibility in memory cells than naïve cells as well as a JunD (a member of the AP-1 TF family) and/or T-bet binding peak (SI Appendix, Fig. S3 G and H). Additionally, Stat4 and Tbx21 have several nearby enhancers that share this pattern, indicating that this transcriptional circuit may be self-reinforcing, as STAT4 signaling induces T-bet expression (1, 61), and T-bet binds enhancers to regulate the expression of both Stat4 and itself. Overall, these data suggest that the enhancers acquired by memory CD8 T cells that contribute to their enhanced function relative to naïve cells are regulated by AP-1 factors and T-bet.
E2A Promotes Chromatin Accessibility at Naïve- and Memory-Specific Enhancers.
Thus far, we have found that the E2A binding motif is highly enriched in MPEC-specific enhancers, and E2A has greater binding to these enhancers based on published E2A ChIP-seq data (SRR3984693) (57). Additionally, E2A is a potential regulator of the next most significant transcription factor TCF7 identified in our motif analysis of MPEC-specific enhancers. Therefore, E2A might serve as an important TF in the prevention of terminal differentiation in MPECs by maintaining accessibility at memory enhancers.
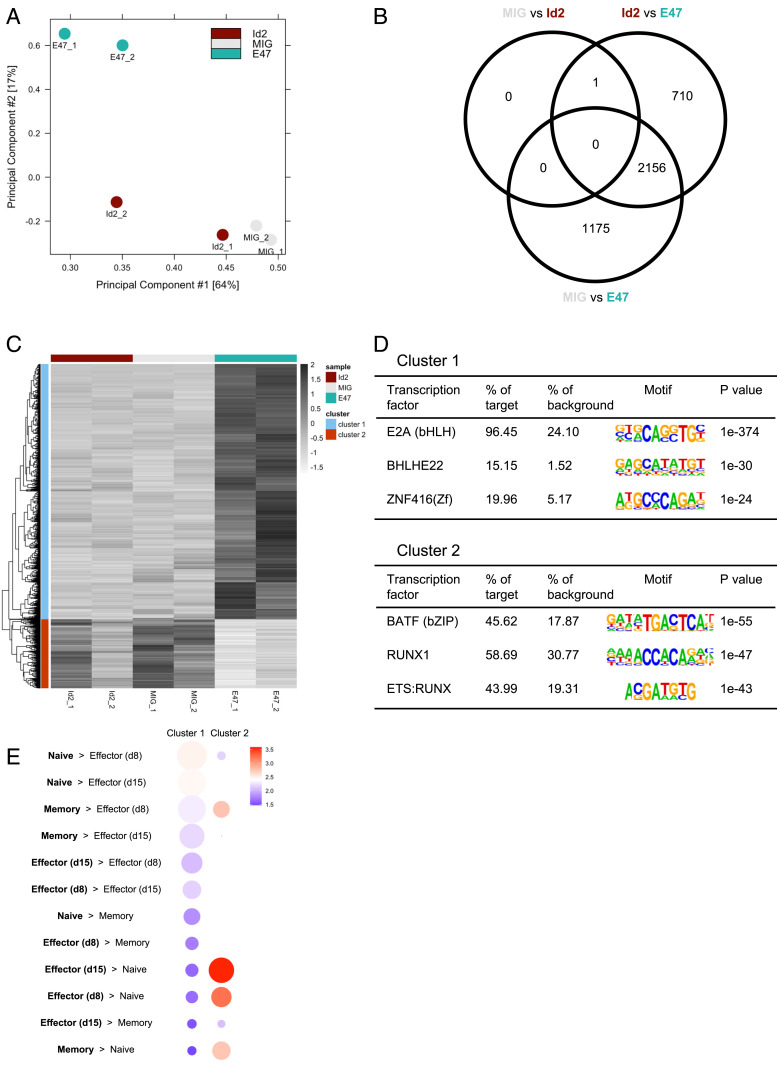
To directly determine the effect of E2A on the enhancer landscape of CD8 T cells, we performed RNA-seq and ATAC-seq on activated CD8 T cells overexpressing E47, one of the two alternative splice variants of E2A that also has higher endogenous expression than the E proteins E12 and HEB, or ID2, a well-known inhibitor of E2A activity (62). We first confirmed overexpression of E47 or ID2 by RNA-seq (SI Appendix, Fig. S4A). Interestingly, the cells with E47 overexpression also expressed more Id2 than the empty vector control, suggesting a feedback mechanism by which Id2 expression is enhanced to counteract the high E47 activity. As E protein activity is heavily influenced by the levels of their inhibitors, the ID proteins, we looked at the ratio of expression levels for these molecules in our three conditions. We found a Tcf3:Id2 ratio of 0.0925, 0.896, and 2.63 in the MIG-Id2, empty MIG, and MIG-E47 samples, respectively, demonstrating a broad range of relative levels for comparison (SI Appendix, Fig. S4B). When comparing our three conditions, we found 2,022 genes with significantly different expression between MIG and MIG-E47 (588 higher in MIG and 1,434 higher in MIG-E47), 2,192 genes with significantly different expression between MIG-Id2 and MIG-E47 (849 higher in MIG-Id2 and 1,343 higher in MIG-E47), and 105 genes with significantly different expression between MIG and MIG-Id2 (32 higher in MIG and 73 higher in MIG-Id2) (SI Appendix, Fig. S4C). Overall, 2,541 genes showed significant changes in expression between at least two of the conditions. Hierarchical clustering of these differentially expressed genes separated them into two distinct groups: 1,653 genes had higher expression in MIG-E47 than the other two groups (cluster 1), while 888 genes had lower expression in MIG-E47 than the other two groups (cluster 2) (SI Appendix, Fig. S4C). Cluster 1 contains several genes expressed in memory CD8 T cells, such as Cxcr5, Id3, Il6ra, Nr4a3, Pou6f1, and Zeb1 (SI Appendix, Fig. S4D). Cluster 2 contains more effector-related genes, including Bhlhe40, Ccl5, Cxcr6, Gzmk, Junb, and Tbx21 (SI Appendix, Fig. S4E). Overall, forced expression of E47 increases expression of more genes than it decreases, while forced expression of Id2 does not have a substantial effect on gene expression in this setting.
By performing ATAC-seq on cells from these three conditions, we found that increased E47 leads to a substantial change in the overall enhancer landscape when compared to the MIG and MIG-Id2 groups (Fig. 4A). MIG-E47 had 3,331 enhancers with significantly different ATAC-seq signal compared to MIG and 2,867 enhancers with significantly different ATAC-seq signal compared to MIG-Id2, with 2,156 enhancers shared between the two groups (Fig. 4B). Only one enhancer, on the other hand, showed significantly different accessibility when comparing MIG and MIG-Id2. Similar to our expression data, hierarchical clustering demonstrated two primary patterns of changes in enhancer accessibility in our groups: 3,182 enhancers that open upon overexpression of E47 (cluster 1) and 860 enhancers that close (cluster 2) (Fig. 4C). To determine which of these changes is because of a direct effect of E47, we performed motif analysis on these two clusters of enhancers (Fig. 4D). Significant enrichment of the E2A binding site was found in cluster 1, with 96% of these enhancers containing this motif. Surprisingly, the AP-1 binding site was most significantly enriched in cluster 2 enhancers, suggesting that the effect of E2A on these enhancers may be indirect. We then used GREAT to determine if genes near these two groups of enhancers are important for different aspects of CD8 T cell function (Fig. 4E). Cluster 1 enhancers were significantly enriched for genes with higher expression in naïve CD8 T cell than effector CD8 T cells, such as Ccr7, Id3, Lef1, and Tcf7. Cluster 2 enhancers had more significant enrichment of genes with higher expression in effector and memory CD8 T cells than naïve cells, such as Bhlhe40, Cd44, Gzmk, and Prdm1. These results support a model in which E47 maintains accessibility of naïve CD8 T cell enhancers, while also indirectly preventing the opening of effector CD8 T cell enhancers, thereby allowing a fraction of effector CD8 T cells to return to quiescence and develop into memory cells after clearance of an infection.
Fig. 4.
E2A regulates the accessibility of enhancers regulating memory T cell genes. (A) PCA plot comparing ATAC-seq signal in CD8 T cells transduced with empty MIG, MIG-Id2, and MIG-E47. (B) Venn diagram showing the number of enhancers with significantly different ATAC-seq signal in MIG versus MIG-E47, MIG versus MIG-Id2, and MIG-Id2 versus MIG-E47. (C) Heatmap showing hierarchical clustering of enhancers with significantly different ATAC-seq signal in the three groups. (D) Motif analysis with top three significantly enriched TF binding motifs in enhancers more open (Top) or more closed (Bottom) in E47 overexpressing cells. (E) Gene ontology analysis for cluster 1 and cluster 2 enhancers was performed using GREAT (55) and compared to published microarray gene sets of naïve, day 8 effector, day 15 effector, and memory P14 CD8 T cells (56). Circle size denotes significance of gene set enrichment. Color represents fold enrichment.
E2A Overexpression Promotes Memory CD8 T Cell Formation.
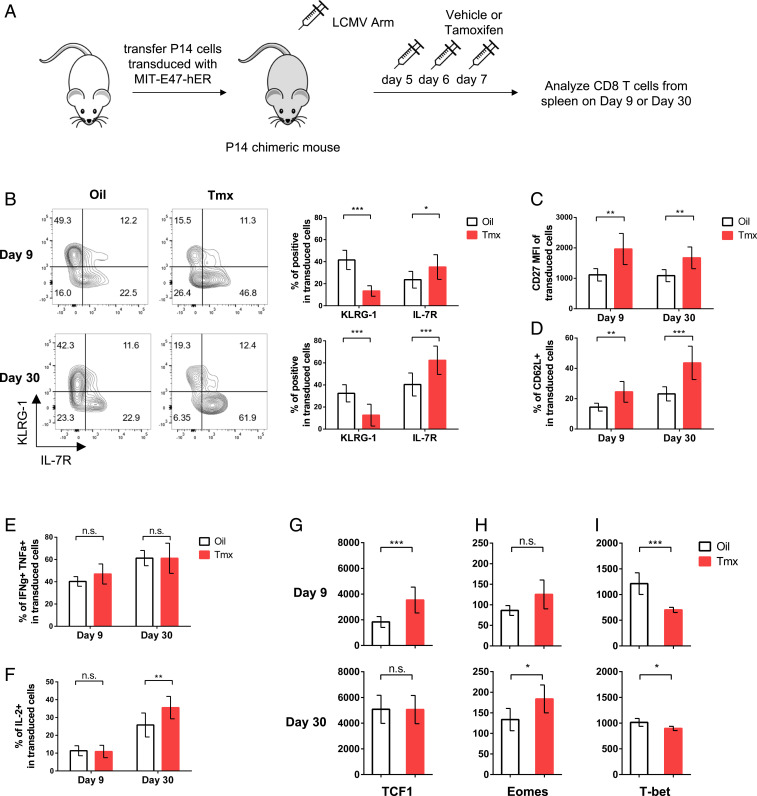
To test whether E2A activity could enhance the formation of virus-specific memory CD8 T cells in vivo, we transferred transduced P14 cells to mice and infected them with LCMV Armstrong. However, preliminary experiments showed that enforced E47 expression from the beginning of an immune response led to low numbers of transferred cells, possibly due to decreased expansion and/or increased apoptosis. To mitigate this effect, we constructed a vector to express E47 fused to a modified human estrogen receptor (hER), as has been done previously (63). This allows us to selectively turn on E47 activity at a specific time by injecting tamoxifen. We first treated two separate groups of mice that received empty retroviral vector-transduced P14 T cells with either corn oil or tamoxifen intraperitoneally (i.p.) on days 5, 6, and 7 p.i., which resulted in no detectable phenotypic changes (SI Appendix, Fig. S5 A and B). Next, we administered the same treatments to two groups of mice that received E47-hER retroviral vector-transduced P14 T cells and analyzed cells from spleens on days 9 and 30 p.i. (Fig. 5A). Increased E47 activity on days 5 to 7 p.i. led to significantly increased frequencies of MPECs and subsequent decreased frequencies of SLECs (Fig. 5B). In the same vein, enforced E47 activity accelerated memory development with increased formation of IL-7R+KLRG1− central memory CD8 T cells (Fig. 5B). Consistently, we also observed increased expression of CD27 and CD62L (Fig. 5 C and D and SI Appendix, Fig. S5 C and D). Although the frequencies of IFN-γ- and TNF-α-producing cells were similar between the two groups, IL-2 production, another key memory signature, was significantly increased in the tamoxifen-treated group at day 30 p.i. (Fig. 5 E and F and SI Appendix, Fig. S5 E and F). Lastly, increased E47 activity drove significantly increased TCF1 expression, whereas it suppressed T-bet expression in effector CD8 T cells (Fig. 5 G–I). While transitioning toward memory phase, tamoxifen-treated T cells maintained a heightened expression of Eomes and reduced expression of T-bet, but the expression of TCF1 was indistinguishable from the control group (Fig. 5 G–I). Taken together, these results demonstrate that enforced E2A activity before the peak of the CD8 T cell response diminishes terminal differentiation and promotes memory cell formation.
Fig. 5.
E2A activity promotes memory CD8 T cell formation. (A) Diagram showing experimental outline. (B) Representative flow cytometry plots and corresponding bar graphs showing frequencies of KLRG-1 and IL-7R expression in P14 cells transduced with MIT-E47-hER in mice that received vehicle control (empty box) or tamoxifen (Tmx) (red box) at day 9 (Top) or day 30 (Bottom) post-LCMV Armstrong infection. (C and D) Bar graphs show the mean fluorescence intensity (MFI) of CD27 and frequencies of CD62L expression in transduced P14 cells. (E and F) Bar graphs show the frequencies of IFN-γ/TNF-α double positive and IL-2-producing cells in transduced P14 cells. (G–I) Bar graphs show the expression of TCF1, Eomes, and T-bet in transduced CD8 T cells at day 9 (Top) or day 30 (Bottom). Data (mean ± SD) are pooled from three independent experiments and analyzed using multiple t test (*P < 0.05; **P < 0.01; ***P < 0.001; n.s., not significant).
Discussion
In this study, we explored memory CD8 T cell differentiation during acute viral infection through analysis of the transcriptome at the single-cell level and changes in the enhancer landscape. This work revealed dynamic changes in gene expression as well as two major patterns of TF regulon activity, as naïve CD8 T cells differentiate into memory cells. Furthermore, our work supports a model in which the epigenetic landscape is a key factor for the cell fate decision of effector CD8 T cells, as was also proposed in a recent study of CD8 T cells responding to bacterial infection (16). In our model, SLECs lose accessibility at E2A- and TCF7-bound enhancers regulating genes that are critical for memory potential and are therefore unable to regain activity at these enhancers after clearance of the infection, accounting for their inability to form long-lived memory cells.
Our regulon-based scRNA-seq analysis using SCENIC identified several TFs with known roles in CD8 T cell differentiation. However, some other TFs, that we and others (15, 16) identified through investigation of the epigenome, did not appear in this analysis or appeared in the opposite cluster as expected, such as E2A (encoded by Tcf3). One major limitation of this analysis is the reliance on coexpression patterns for initial identification of potential regulons. Several TFs that mediate effector and memory CD8 T cell differentiation are regulated primarily by posttranslational modifications or inhibition by other proteins, causing them to be hidden from an expression-based analysis, as their activity does not directly correlate with their expression. Incorporation of this information will be necessary for future specification of the CD8 T cell regulatory network. Additionally, single-cell ATAC-seq and other technological advances hold great promise for deciphering the heterogeneity of the epigenome in individual effector CD8 T cells (63).
Through our clustering analysis of H3K27Ac enrichment in naïve, MPEC, SLEC, and memory CD8 T cells, we identified two major patterns of enhancer activity, which strongly correlated with our regulon activity results. The first cluster, composed of enhancers near genes important for memory potential, had high activity in naïve and memory cells and low activity in MPECs and SLECs and seems to be positively regulated by TCF7, E2A, Foxo1, and GATA3 and negatively regulated by Blimp-1 and Runx3. The second cluster, made up largely of enhancers located near effector-related genes, had negligible activity in naïve cells, moderate activity in MPECs and memory cells, and high activity in SLECs and appears to be regulated by AP-1, AP-1-IRF complexes, and T-bet. Similar patterns were also observed during infection with Listeria (16) based on H3K4Me1 enrichment—a marker of the presence of an enhancer (64, 65).
Though both MPECs and SLECs lose activity at enhancers for memory-related genes, this loss is only temporary for MPECs. After viral clearance, MPECs differentiate into long-lived memory CD8 T cells, in part by regaining activity of these enhancers and expression of their target genes. A natural hypothesis arising from this phenomenon would be that these enhancers remain in an open, poised state in MPECs, while they become permanently closed and repressed in SLECs. We provided evidence supporting this notion and found that E2A and TCF7 bind to and potentially regulate these enhancers. Recently, other groups utilizing comparable model systems have made similar observations (15, 16). Furthermore, we showed that E2A can regulate the accessibility of enhancers controlling critical memory T cell genes in CD8 T cells, and that its activity near the peak of the CD8 T cell response can skew their differentiation toward memory formation. Additionally, the Tcf7 locus contained two enhancers, +7.5 kb and –30 kb, that are bound by E2A or E2A and TCF7, respectively, in which SLECs had severely diminished accessibility relative to MPECs. This could provide MPECs with a self-reinforcing circuit in which E2A enhances expression of TCF7, TCF7 binds to its own enhancer to stabilize its expression, and they both then act on many other enhancers to orchestrate the memory CD8 T cell transcriptional program.
Our results are consistent with the previously proposed decreasing potential model of memory CD8 T cell differentiation (5, 66). In this model, successive stimulations by antigen and cytokines progressively drive CD8 T cells to terminal differentiation. Fitting our results into this framework, naïve cells are activated and acquire an effector program via AP-1 and T-bet. If limited stimulation occurs, these cells retain their memory potential by maintaining accessibility at E2A- and TCF7-regulated enhancers. Repetitive stimulation eventually leads to inhibition of these core components of the naïve/memory GRN and subsequent permanent loss of these enhancers, preventing the cells from ever regaining their memory potential. It would be interesting to know how early effector CD8 T cells establish the epigenetic structure of terminal differentiation and lose memory formation potential. With advancements in single-cell transcriptomics and epigenomics, future studies are warranted to elucidate the temporal cell fate bifurcation in early effector CD8 T cells.
Overall, this work supports a model in which CD8 T cell differentiation is governed by the parallel action of two transcriptional circuits: one controlling the loss and subsequent recovery of memory gene expression and one responsible for the acquisition of effector-related genes. Though expression of these memory-related genes is largely lost in both MPECs and SLECs, they can be reexpressed as MPECs become memory cells because the accessibility of these enhancers is maintained in MPECs but lost in SLECs, possibly due to the action of TCF7 and E2A. Understanding the regulation of this group of enhancers may be the key to successfully eliciting a robust memory CD8 T cell response in vaccines or reprogramming tumor-infiltrating lymphocytes for optimal efficacy of adoptive cell transfer-based cancer therapies.
Materials and Methods
Generation of P14 Chimeric Mice and Infection.
Mouse handling conformed to the requirements of the Institutional Animal Care and Use Guidelines of Medical College of Wisconsin. P14 mice, possessing a transgenic TCR specific for the gp33–41 peptide of LCMV with the congenic marker Thy1.1, were bred in house. C57BL/6 mice that were used as recipients were purchased from Charles River Laboratories. P14 chimeric mice were made by adoptively transferring 100,000 P14 Thy1.1+ CD8 T cells to C57BL/6 mice (Thy1.2+). These mice were then infected with 2 × 105 plaque-forming unit LCMV Armstrong by i.p. injection.
For experimental procedures, see SI Appendix, Supplemental Methods.
Supplementary Material
Acknowledgments
We thank Kirthi Pulakanti, Sridhar Rao, and members of the Cui laboratory for helpful discussions. This work is supported by NIH Grants AI125741 (W.C.), AI148403 (W.C.), DK108557 (D.M.S.), and DK127526 (M.Y.K.) as well as an American Cancer Society Research Scholar Grant (W.C.) and an Advancing a Healthier Wisconsin Endowment Grant (W.C.). D.M.S. and M.Y.K. are members of the Medical Scientist Training Program at the Medical College of Wisconsin (MCW), which is partially supported by a training grant from the National Institute of General Medical Sciences (T32-GM080202). This research was completed in part with computational resources and technical support provided by the Research Computing Center at MCW.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2013452118/-/DCSupplemental.
Data Availability
scRNA-seq, bulk RNA-seq, H3K27Ac ChIP-seq, and ATAC-seq data have been deposited in the Gene Expression Omnibus database with the accession codes GSE130130, GSE150442, and GSE142344. All other raw data are available from the corresponding author upon request.
References
- 1.Joshi N. S., et al., Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity 27, 281–295 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaech S. M., et al., Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat. Immunol. 4, 1191–1198 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Schluns K. S., Kieper W. C., Jameson S. C., Lefrançois L., Interleukin-7 mediates the homeostasis of naïve and memory CD8 T cells in vivo. Nat. Immunol. 1, 426–432 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Huster K. M., et al., Selective expression of IL-7 receptor on memory T cells identifies early CD40L-dependent generation of distinct CD8+ memory T cell subsets. Proc. Natl. Acad. Sci. U.S.A. 101, 5610–5615 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaech S. M., Cui W., Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 12, 749–761 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emmert-Streib F., Dehmer M., Haibe-Kains B., Gene regulatory networks and their applications: Understanding biological and medical problems in terms of networks. Front. Cell Dev. Biol. 2, 38 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu G., Chen J., A genome-wide regulatory network identifies key transcription factors for memory CD8+ T-cell development. Nat. Commun. 4, 2830 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arsenio J., et al., Early specification of CD8+ T lymphocyte fates during adaptive immunity revealed by single-cell gene-expression analyses. Nat. Immunol. 15, 365–372 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kakaradov B., et al., Early transcriptional and epigenetic regulation of CD8+ T cell differentiation revealed by single-cell RNA sequencing. Nat. Immunol. 18, 422–432 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng G. X. Y., et al., Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aibar S., et al., SCENIC: Single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Love M. I., et al., Role of the chromatin landscape and sequence in determining cell type-specific genomic glucocorticoid receptor binding and gene regulation. Nucleic Acids Res. 45, 1805–1819 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gertz J., et al., Distinct properties of cell-type-specific and shared transcription factor binding sites. Mol. Cell 52, 25–36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arvey A., Agius P., Noble W. S., Leslie C., Sequence and chromatin determinants of cell-type-specific transcription factor binding. Genome Res. 22, 1723–1734 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scott-Browne J. P., et al., Dynamic changes in chromatin accessibility occur in CD8+ T cells responding to viral infection. Immunity 45, 1327–1340 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu B., et al., Epigenetic landscapes reveal transcription factors that regulate CD8+ T cell differentiation. Nat. Immunol. 18, 573–582 (2017). Correction in: Nat. Immunol.18, 705 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sen D. R., et al., The epigenetic landscape of T cell exhaustion. Science 354, 1165–1169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pauken K. E., et al., Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 354, 1160–1165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mognol G. P., et al., Exhaustion-associated regulatory regions in CD8+ tumor-infiltrating T cells. Proc. Natl. Acad. Sci. U.S.A. 114, E2776–E2785 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He B., et al., CD8+ T cells utilize highly dynamic enhancer repertoires and regulatory circuitry in response to infections. Immunity 45, 1341–1354 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou X., et al., Differentiation and persistence of memory CD8(+) T cells depend on T cell factor 1. Immunity 33, 229–240 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou X., Xue H.-H., Cutting edge: Generation of memory precursors and functional memory CD8+ T cells depends on T cell factor-1 and lymphoid enhancer-binding factor-1. J. Immunol. 189, 2722–2726 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao D.-M., et al., Constitutive activation of Wnt signaling favors generation of memory CD8 T cells. J. Immunol. 184, 1191–1199 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeannet G., et al., Essential role of the Wnt pathway effector Tcf-1 for the establishment of functional CD8 T cell memory. Proc. Natl. Acad. Sci. U.S.A. 107, 9777–9782 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerdiles Y. M., et al., Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat. Immunol. 10, 176–184 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ouyang W., Beckett O., Flavell R. A., Li M. O., An essential role of the Forkhead-box transcription factor Foxo1 in control of T cell homeostasis and tolerance. Immunity 30, 358–371 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao R. R., Li Q., Gubbels Bupp M. R., Shrikant P. A., Transcription factor Foxo1 represses T-bet-mediated effector functions and promotes memory CD8(+) T cell differentiation. Immunity 36, 374–387 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim M. V., Ouyang W., Liao W., Zhang M. Q., Li M. O., The transcription factor Foxo1 controls central-memory CD8+ T cell responses to infection. Immunity 39, 286–297 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hess Michelini R., Doedens A. L., Goldrath A. W., Hedrick S. M., Differentiation of CD8 memory T cells depends on Foxo1. J. Exp. Med. 210, 1189–1200 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tejera M. M., Kim E. H., Sullivan J. A., Plisch E. H., Suresh M., FoxO1 controls effector-to-memory transition and maintenance of functional CD8 T cell memory. J. Immunol. 191, 187–199 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roychoudhuri R., et al., BACH2 regulates CD8(+) T cell differentiation by controlling access of AP-1 factors to enhancers. Nat. Immunol. 17, 851–860 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richer M. J., Lang M. L., Butler N. S., T cell fates zipped up: How the Bach2 basic leucine zipper transcriptional repressor directs T cell differentiation and function. J. Immunol. 197, 1009–1015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Intlekofer A. M., et al., Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 6, 1236–1244 (2005). Correction in: Nat. Immunol.7, 113 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Intlekofer A. M., et al., Requirement for T-bet in the aberrant differentiation of unhelped memory CD8+ T cells. J. Exp. Med. 204, 2015–2021 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takemoto N., Intlekofer A. M., Northrup J. T., Wherry E. J., Reiner S. L., Cutting edge: IL-12 inversely regulates T-bet and eomesodermin expression during pathogen-induced CD8+ T cell differentiation. J. Immunol. 177, 7515–7519 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Creyghton M. P., et al., Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 107, 21931–21936 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heintzman N. D., et al., Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., Greenleaf W. J., Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ye T., et al., seqMINER: An integrated ChIP-seq data interpretation platform. Nucleic Acids Res. 39, e35 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lovén J., et al., Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hnisz D., et al., Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whyte W. A., et al., Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heinz S., et al., Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.D’Cruz L. M., Lind K. C., Wu B. B., Fujimoto J. K., Goldrath A. W., Loss of E protein transcription factors E2A and HEB delays memory-precursor formation during the CD8+ T-cell immune response. Eur. J. Immunol. 42, 2031–2041 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Masson F., et al., Id2-mediated inhibition of E2A represses memory CD8+ T cell differentiation. J. Immunol. 190, 4585–4594 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y., et al., GATA-3 controls the maintenance and proliferation of T cells downstream of TCR and cytokine signaling. Nat. Immunol. 14, 714–722 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xin A., et al., A molecular threshold for effector CD8(+) T cell differentiation controlled by transcription factors Blimp-1 and T-bet. Nat. Immunol. 17, 422–432 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shan Q., et al., The transcription factor Runx3 guards cytotoxic CD8+ effector T cells against deviation towards follicular helper T cell lineage. Nat. Immunol. 18, 931–939 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rincón M., Flavell R. A., AP-1 transcriptional activity requires both T-cell receptor-mediated and co-stimulatory signals in primary T lymphocytes. EMBO J. 13, 4370–4381 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kurachi M., et al., The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nat. Immunol. 15, 373–383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grusdat M., et al., IRF4 and BATF are critical for CD8+ T-cell function following infection with LCMV. Cell Death Differ. 21, 1050–1060 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xin G., et al., A critical role of IL-21-induced BATF in sustaining CD8-T-cell-mediated chronic viral control. Cell Rep. 13, 1118–1124 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dominguez C. X., et al., The transcription factors ZEB2 and T-bet cooperate to program cytotoxic T cell terminal differentiation in response to LCMV viral infection. J. Exp. Med. 212, 2041–2056 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Phillips J. E., Corces V. G., CTCF: Master weaver of the genome. Cell 137, 1194–1211 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McLean C. Y., et al., GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaech S. M., Hemby S., Kersh E., Ahmed R., Molecular and functional profiling of memory CD8 T cell differentiation. Cell 111, 837–851 (2002). [DOI] [PubMed] [Google Scholar]
- 57.Leong Y. A., et al., CXCR5(+) follicular cytotoxic T cells control viral infection in B cell follicles. Nat. Immunol. 17, 1187–1196 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Steinke F. C., et al., TCF-1 and LEF-1 act upstream of Th-POK to promote the CD4(+) T cell fate and interact with Runx3 to silence Cd4 in CD8(+) T cells. Nat. Immunol. 15, 646–656 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cannarile M. A., et al., Transcriptional regulator Id2 mediates CD8+ T cell immunity. Nat. Immunol. 7, 1317–1325 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Yang C. Y., et al., The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat. Immunol. 12, 1221–1229 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiao Z., Casey K. A., Jameson S. C., Curtsinger J. M., Mescher M. F., Programming for CD8 T cell memory development requires IL-12 or type I IFN. J. Immunol. 182, 2786–2794 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Omilusik K. D., et al., Sustained Id2 regulation of E proteins is required for terminal differentiation of effector CD8+ T cells. J. Exp. Med. 215, 773–783 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buenrostro J. D., et al., Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heintzman N. D., et al., Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311–318 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Birney E.et al.; ENCODE Project Consortium; NISC Comparative Sequencing Program; Baylor College of Medicine Human Genome Sequencing Center; Washington University Genome Sequencing Center; Broad Institute; Children’s Hospital Oakland Research Institute , Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ahmed R., Gray D., Immunological memory and protective immunity: Understanding their relation. Science 272, 54–60 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
scRNA-seq, bulk RNA-seq, H3K27Ac ChIP-seq, and ATAC-seq data have been deposited in the Gene Expression Omnibus database with the accession codes GSE130130, GSE150442, and GSE142344. All other raw data are available from the corresponding author upon request.