Significance
Transcription–translation coupling is critical for fast and efficient gene expression in many bacteria, yet how coupling between RNA polymerase (RNAP) and ribosome is established and maintained is poorly understood. Combining biochemical and single-molecule fluorescence approaches, we here uncover the roles of nascent mRNA structure and transcription factors in coupling. We find that the inherently paused RNAP facilitates 30S ribosomal subunit recruitment, whereas ligand-induced translational riboswitch folding in the 5′ untranslated region (5′ UTR) impedes it. Transcription factors NusG and RfaH distinctly enhance 30S recruitment and retention, respectively, and actively translating 70S ribosome accelerates the leading RNAP. Taken together, our data yield a unifying dynamic model of transcription–translation coupling, promising to inspire new approaches for the design of antibiotics.
Keywords: transcription–translation coupling, nascent mRNA, translational riboswitch, RNA polymerase, ribosome
Abstract
Bacterial messenger RNA (mRNA) synthesis by RNA polymerase (RNAP) and first-round translation by the ribosome are often coupled to regulate gene expression, yet how coupling is established and maintained is ill understood. Here, we develop biochemical and single-molecule fluorescence approaches to probe the dynamics of RNAP–ribosome interactions on an mRNA with a translational preQ1-sensing riboswitch in its 5′ untranslated region. Binding of preQ1 leads to the occlusion of the ribosome binding site (RBS), inhibiting translation initiation. We demonstrate that RNAP poised within the mRNA leader region promotes ribosomal 30S subunit binding, antagonizing preQ1-induced RBS occlusion, and that the RNAP–30S bridging transcription factors NusG and RfaH distinctly enhance 30S recruitment and retention, respectively. We further find that, while 30S–mRNA interaction significantly impedes RNAP in the absence of translation, an actively translating ribosome promotes productive transcription. A model emerges wherein mRNA structure and transcription factors coordinate to dynamically modulate the efficiency of transcription–translation coupling.
Bacteria and archaea lack the physical barrier separating the transcription and translation machineries in eukaryotes so that the ribosome can initiate translation on a nascent mRNA still being transcribed by RNA polymerase (RNAP). The trailing ribosome is known to then prevent transcription arrest and premature termination and protects the mRNA from degradation by RNases (1, 2). In Escherichia coli (E. coli), the rates of RNA and protein synthesis are closely matching under many conditions in vivo (3, 4), giving rise to a model in which transcription is coupled to the pioneer round of translation. The anti-arrest and antitermination effects of the ribosome depend on this coupling between the transcription and translation complexes being intimate, a feature that has attracted renewed interest in biochemical (4–6) and structural studies (7–10). The actively transcribing RNAP and the lead ribosome have been proposed to be coupled by direct physical interaction (8, 11, 12) or by transcription factors (9, 13, 14). Chemical cross-linking and cryogenic-electron microscopy (cryo-EM) have revealed that the region encompassing the RNAP RNA exit channel can directly interact with the solvent-accessible surface of the 30S subunit, allowing the ribosome to engage the ribosome binding site (RBS) and Shine–Dalgarno (SD) sequence of the nascent mRNA (11, 12). Yet, another recent cryo-EM structure shows a different relative orientation of RNAP and 70S ribosome in this “expressome” complex (8). While these studies support direct physical contacts between RNAP and ribosome, a growing body of work suggests that the interactions between the two machines connected by the flexible mRNA are dynamic (5, 15–17). A recent cryo-EM study even holds that a specific interaction surface is absent in the expressome complex (9).
In addition to being linked via the mRNA, the RNAP and leading ribosome can be bridged by proteins, such as a general transcription factor NusG and its paralog RfaH (13, 18). Their N-terminal domains (NTDs) interact with the RNAP to inhibit pausing by altering the enzyme’s clamp dynamics (19) while their C-terminal domains (CTDs) interact with the 30S subunit of the ribosome (13, 20). While the CTD interactions of RfaH and NusG with the ribosome appear to be very similar (13, 20), their NTDs make distinct contacts to the RNAP and the nucleic acids (19), suggesting functional diversity.
While structural and functional evidence in support of direct and protein-mediated transcription–translation coupling is growing, many key questions remain. How are RNAP progress, initial 30S recruitment, and translation by the 70S ribosome coordinated on a connecting nascent mRNA? How is the initial 30S recruitment affected by a change in SD sequence accessibility of the nascent mRNA? How do NusG and RfaH modulate coordination? How does the ribosome catch up with an RNAP, given that the formation of a translation initiation complex requires multiple steps? While RNAP pausing helps ribosome recruitment, the underlying mechanisms could be distinct, including pausing during RNA synthesis (6) and/or promoting structural rearrangements that expose the RBS (21).
Translational riboswitches are excellent model systems to investigate coordination of transcription and translation and the role of RNAP pausing therein. Riboswitches are found in the 5′ untranslated regions (UTRs) of many mRNAs and contain a ligand-binding aptamer whose conformation rearranges upon ligand binding, thereby modulating downstream gene expression by controlling transcription termination or translation initiation (22). Cotranscriptional folding and ligand-dependent structural rearrangements of both transcriptional and translational riboswitches are often assisted by judicious downstream pausing of RNAP (23–26). In addition, ligand binding typically induces structural rearrangements of translational riboswitches that sequester the SD sequence, thereby inhibiting translation initiation (22, 27–29). Yet, little is known about the interplay between ligand binding and ongoing transcription during ribosome loading and transcription–translation coupling.
Transcription–translation coupling is a fundamental feature of gene expression in E. coli. Moreover, a slim majority (54%) of E. coli genes contain SD sequences expected to affect transcription–translation coupling by mediating initial 30S recruitment (30). Here, to investigate how the dynamics of initial 30S recruitment to a nascent mRNA is affected by the presence of the E. coli transcriptional machinery and SD sequence availability, we used an mRNA from the mesophilic pathogen Bacillus anthracis (Bas) bearing a member of the well-characterized preQ1-sensing translational riboswitch family as a model system (Fig. 1). Previous studies demonstrated that upon preQ1 binding, helix P2 is formed, sequestering the first two nucleotides of the SD sequence (Fig. 1 A and B) and decreasing its accessibility (31, 32). This ligand-induced riboswitching provides us with a convenient platform to modulate SD accessibility without altering the RNA sequence, which could change the nature of the interaction under study. Using single-molecule fluorescence colocalization, we monitor binding of the 30S subunit to a downstream paused elongation complex (PEC). Our results reveal that the proximal RNAP promotes 30S recruitment to the nascent mRNA, whereas preQ1-induced structural sequestration of the SD inhibits 30S binding. The addition of NusG and RfaH unveils distinct mechanisms by which these transcription factors modulate the early steps of translation initiation. Using real-time transcription assays, we observe, conversely, the impact of the translational machinery on transcription. While nonproductive initial 30S recruitment to the RBS is found to delay transcription because of its stochastic interactions with the transcribing RNAP, an actively translating 70S ribosome accelerates the leading RNAP, thus restoring the rate of RNA synthesis. Our results uncover the dynamic coordination between the transcription and translation machineries and the role of the nascent mRNA and transcription factors NusG and RfaH therein. We anticipate that our findings will stimulate the development of new antibiotics that can specifically target transcription–translation coupling.
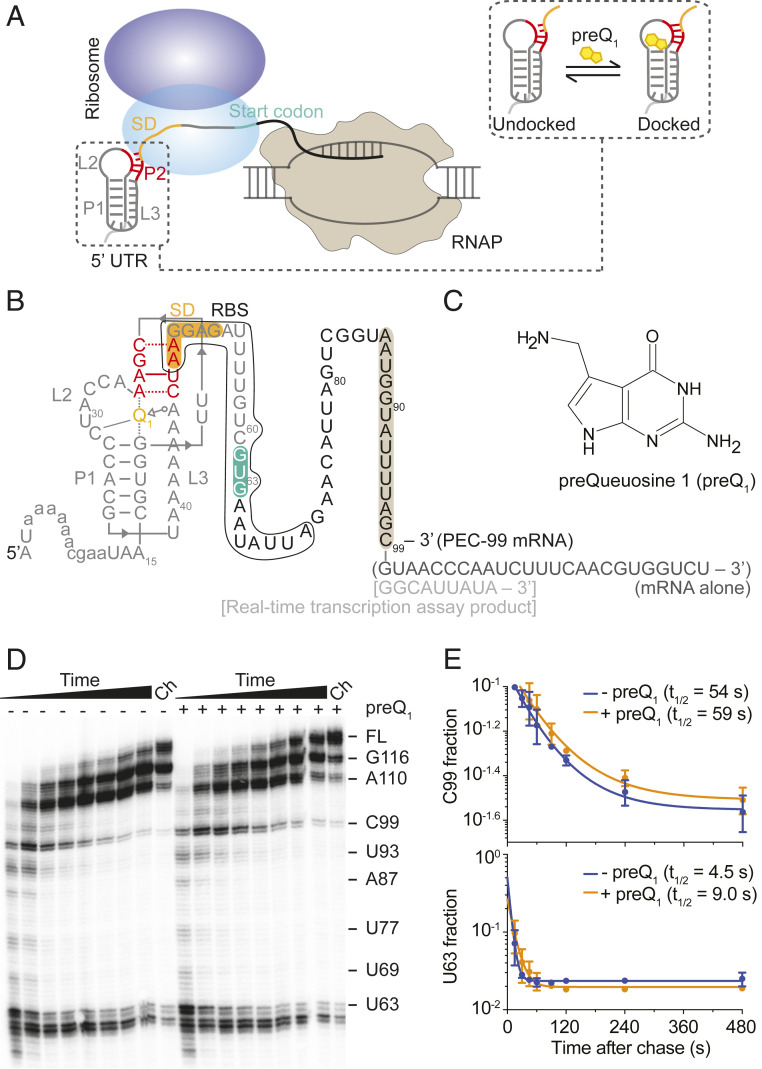
Fig. 1.
Bas mRNA containing a preQ1 riboswitch in the 5′ UTR as a model to study transcription–translation coordination. (A) Schematic representation of transcription–translation coupling on a nascent mRNA containing a structured riboswitch in the 5′ UTR. Binding of the cognate ligand to the riboswitch stabilizes a folded conformation where the SD sequence becomes less accessible to ribosome binding (shown in a dashed box). (B) Secondary structure of the Bas mRNA in our system showing the riboswitch pseudoknot at the 5′ end, the entire RBS, SD sequence (orange), and start codon (green) as indicated. Annotations in parentheses and square brackets indicate modifications introduced for experiments with mRNA alone and during real-time transcription assays, respectively. Sequence protected by the RNAP is highlighted in brown. (C) Chemical structure of the preQ1 ligand. (D) In vitro transcription of Bas mRNA. U63, C99, and full-length RNAs are shown on the right. The Ch lane corresponds to the final chase where 500 µM rNTPs were added at the end and the reaction was incubated for 5 min. (E) Quantification of the half-lives of the C99 and U63 pause sites in the absence or presence of preQ1.
Results
RNAP Pauses Downstream of the preQ1-Sensing Translational Riboswitch.
We previously showed that the ligand-dependent accessibility of the SD region of a translational riboswitch can be monitored directly by single-molecule fluorescence colocalization using a Cy5-labeled anti-SD probe that mimics the 3′ end of the corresponding 16S ribosomal RNA (rRNA) and specifically binds to the SD sequence of surface-attached Cy3-labeled riboswitches (32). We employed this Single-Molecule Kinetic Analysis of RNA Transient Structure (SiM-KARTS) assay to the full-length Bas mRNA (SI Appendix, Note S1) as a function of preQ1 concentration and observed that the probe binding rate constant (kon) gradually decreases as the preQ1 concentration increased, with a half-saturation point K1/2 of ∼160 nM preQ1 (SI Appendix, Fig. S1). This finding is consistent with the previously observed occlusion of the SD region due to preQ1-induced P2 helix formation in the preQ1 riboswitch from Thermoanaerobacter tengcongensis (32), confirming the expectation that preQ1-mediated structural changes decrease the SD/RBS accessibility also in the Bas riboswitch and supporting its use as a mesophilic model system.
RNAP pausing in the vicinity of the start codon was suggested to influence translation initiation in bacteria by reducing structure formation around the RBS and preserving its accessibility for 30S recruitment (21). To investigate the RNAP pausing pattern within Bas mRNA, we used single-round in vitro transcription assays. We observed strong pauses at and near the translation start codon (U60, C61, G62, and U63) and at position C99 located in the open reading frame (ORF) (Fig. 1D). The C99 pause sequence closely matches the previously identified “consensus” pause sequence (G-11G-10Y-1G+1) (21). While pausing at U60 to U63 may play a role in folding of the nascent mRNA to control SD accessibility, pausing at C99 has the potential to impact transcription–translation coupling: when RNAP reaches the C99 pause site, ∼45 nucleotides between the SD sequence and the paused RNAP are available for 30S binding (Fig. 1B); recent cryo-EM studies demonstrated RNAP–ribosome coupling on an mRNA of similar length (9, 10). Transcriptional pausing at C99 was not modulated by preQ1, as indicated by the similar pause half-life in the absence (54 s) and presence of the ligand (59 s; Fig. 1E), consistent with the longer distance of the pause site from the translational riboswitch than previously found for a transcriptional preQ1 riboswitch (23). In contrast to the C99 pause, the duration of the riboswitch-proximal U63 pause instead was extended twofold upon addition of preQ1 (from 4.5 s to 9.0 s, Fig. 1E), a significant effect that strikingly is the opposite of that observed in the transcriptional preQ1 riboswitch (23).
Taken together, these results demonstrate that the PEC at C99 (henceforth, PEC-99) can be used as a model system to study transcription–translation coupling, including the effects of RNAP pausing, contributions of transcription factors, and the influence of preQ1-induced structural changes on ribosome recruitment.
preQ1-Induced Structural Changes Impede 30S Recruitment to the Nascent mRNA in PEC-99.
To investigate the kinetics of 30S binding to PEC-99 and its modulation by mRNA structure, we expanded our SiM-KARTS assay (32) to a fluorescently labeled 30S subunit (33) binding to individual surface-attached, fully assembled, Cy3-labeled PEC-99 in the absence or presence of saturating (5 µM) preQ1. To this end, transcription was initiated using E. coli RNAP on a DNA template that encodes the Bas mRNA with an added upstream DNA sequence without A. A 5′ Cy3-labeled ApU RNA dinucleotide was used to prime transcription and label the mRNA at its 5′ end. To form the halted complex (HC), the RNAP was halted prior to the first templated U13 position by leaving out rUTP (34, 35) (Fig. 2A). The transcript was then extended by adding all four rNTPs and stalled at C99 by using a biotin–streptavidin roadblock (Fig. 2A). A control gel analysis of the transcription product displayed a single band for the 99-nucleotide long nascent mRNA in PEC-99 (SI Appendix, Fig. S2), confirming that, once resuming from the HC, RNAP continues to the pause site without forming any other short transcripts. Single PECs were subsequently immobilized, exploiting the multivalency of streptavidin, on a biotinylated quartz slide and visualized via total internal reflection fluorescence (TIRF) microscopy (Fig. 2B). With this strategy, an observed Cy3 signal indicates the presence of the PEC-99 containing the DNA template, the RNAP stalled at the C99 position, and the 5′ Cy3-labeled nascent mRNA. We note that a small number of unextended HCs might be present on the microscope slide surface along with the PEC-99.
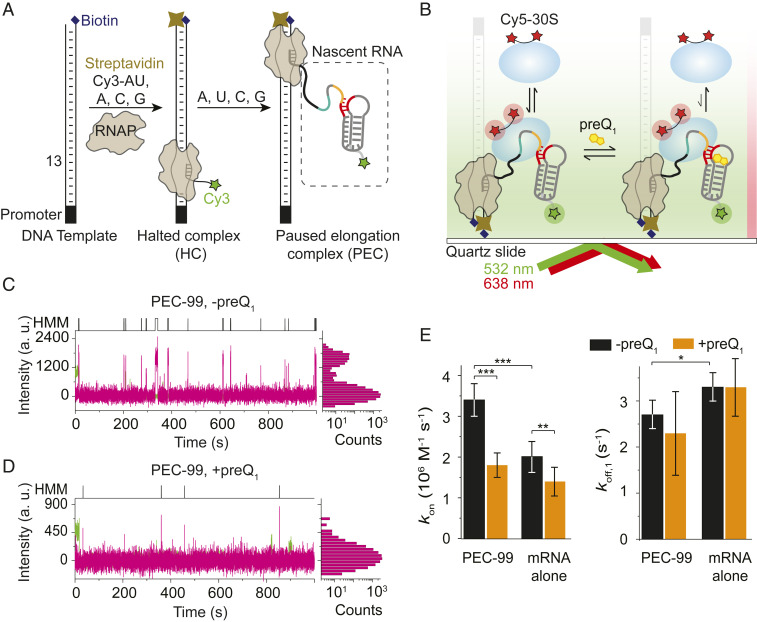
Fig. 2.
30S binding to the nascent mRNA within PEC-99 is dynamic and mediated by the ligand-dependent mRNA structure and the RNAP. (A) Preparation of the paused elongation complex (PEC-99) containing the nascent Cy3-labeled mRNA for monitoring 30S binding using single-molecule colocalization. (B) Single PEC-99s were immobilized on the PEG-coated slide surface via a biotin–streptavidin linkage and imaged by TIRF microscopy to monitor 30S binding in the absence or presence of preQ1. (C and D) Fluorescence intensity versus time traces showing repeated 30S binding events (magenta spikes) in the absence (C) or presence (D) of saturating 5 µM preQ1. Cy5 fluorescence intensity histograms corresponding to individual traces are shown on the right-hand side of the traces. (E) Association (kon) and dissociation rate constants (koff) were calculated from hidden Markov models (shown on top of each trace) for 30S binding to PEC-99 and the corresponding mRNA alone, in the absence or presence of preQ1. Values of binding (kon) and dissociation (koff) rates are reported in SI Appendix, Table S1. Total number of molecules analyzed were N(PEC) = 265, N(PEC + preQ1) = 140, N(mRNA alone) = 64, and N(mRNA alone + preQ1) = 31. (***P < 0.01, **P < 0.025, *P < 0.05).
To detect 30S binding to single PEC-99 complexes, Cy5-labeled 30S was injected onto the slide with immobilized PECs, and binding events were identified by colocalization of Cy3 and Cy5 signals within a diffraction-limited spot. Analysis of individual fluorescence time traces exhibited repeated transient Cy5 colocalizations with Cy3, indicating dynamic binding of 30S to PEC-99 (Fig. 2C). From the distributions of dwell times of 30S in the unbound and bound states (Fig. 2 C, Upper), we derived a binding rate constant of kon = 3.4 ± 0.4 × 106 M−1 ⋅ s−1 and two dissociation rate constants, koff,1 = 2.7 ± 0.3 s−1 (92%) and koff,2 = 0.2 ± 0.05 s−1 (8%) (Fig. 2E and SI Appendix, Table S1), respectively. To test for any nonspecific interaction of 30S with other components of the PEC, we first monitored 30S binding to the surface-immobilized HCs, where the nascent mRNA is only 12 nucleotides long. No traces with characteristic multiple binding events were observed (SI Appendix, Fig. S3), suggesting that the presence of the nascent mRNA containing an SD sequence is necessary for efficient 30S binding. To further test for specificity, we measured 30S binding in the presence of a blocking antisense oligonucleotide (ASO) that occludes the RBS. As expected, we observed significantly fewer 30S binding events (SI Appendix, Fig. S4), resulting in a significantly lower kon of 2.0 ± 0.4 × 106 M−1 ⋅ s−1 (SI Appendix, Table S1). These data demonstrate that the 30S binding events are primarily, albeit not exclusively, dependent on the RBS region in the PEC-99 nascent RNA.
To examine the effect of ligand-induced riboswitching, we measured 30S binding to PEC-99 prepared in the presence of a saturating preQ1 concentration. We observed sparser 30S binding events (Fig. 2D) compared to the absence of the ligand (Fig. 2C), leading to an 48% decrease in the value of kon to 1.8 ± 0.3 × 106 M−1 ⋅ s−1 (Fig. 2E and SI Appendix, Table S1), very similar to the rate constant when blocking the SD via ASO (SI Appendix, Table S1). This result is consistent with our data indicating that preQ1 occludes the RBS due to the formation of helix P2 (SI Appendix, Fig. S1). A plausible explanation for the remaining 30S binding events upon blocking the SD/RBS via preQ1 or ASO is the known ability of the 30S to interact with the mRNA at loosely defined, nonspecific “standby sites” (30, 36). In particular, the A-rich L3 region upstream of the SD sequence (Fig. 1B) may be recognized by the ribosomal protein S1 present in our preparation (SI Appendix, Fig. S5), which is known to facilitate 30S binding to structured mRNAs during translation initiation (30, 37). Due to the structural sequestration in the presence of either preQ1 or ASO, unfolding and accommodation of the mRNA RBS into the standby site of the 30S subunit is disfavored (36, 37), leading to the observed reduction in binding events. In contrast, the 30S dissociation rate constants are similar in the absence and presence of preQ1 (Fig. 2E and SI Appendix, Table S1), indicating that ligand-induced nascent mRNA structural rearrangement does not destabilize the mRNA–30S interaction once formed.
Together, these results demonstrate that the 30S subunit interacts with PEC-99 primarily through the SD sequence of the nascent mRNA in a way that preQ1-induced riboswitching decreases accessibility and therefore impedes 30S recruitment but not 30S retention.
The Proximally Paused RNAP Facilitates 30S Recruitment to the Nascent mRNA.
Next, to investigate the role of RNAP in 30S recruitment to the nascent mRNA, we compared 30S binding to PEC-99 with that to an RNA alone transcript containing the same RNA sequence as PEC-99, with an additional 25 nucleotides for immobilization to the slide surface through hybridization with a biotinylated capture ASO (Fig. 1B). Strikingly, we observed significantly fewer 30S binding events, interrupted by long unbound times (SI Appendix, Fig. S6), leading to a 41% decrease in kon value to 2.0 ± 0.3 × 106 M−1 ⋅ s−1 (Fig. 2E), indicating that the RBS is as little accessible in the RNA-only transcript as it is in PEC-99 with a blocked SD sequence. These data support the notion that RNAP is promoting 30S binding to the RBS in PEC-99. One reason might be the presence of RNAP at the C99 position, which may prevent structure formation in the RBS region of the nascent mRNA, thereby making it more accessible to the 30S (21). A direct contact between the 30S subunit and the RNAP paused at C99 is possible because the 30S mRNA channel can contact up to 20 nucleotides downstream of the SD region (38), thereby placing the interacting surfaces of RNAP and 30S in PEC-99 sufficiently close for 30S recruitment (Fig. 1B). This notion is also consistent with a recent study showing that the RNAP–ribosome interaction is dynamic and mediated by the intervening mRNA (9). Notably, the 30S dissociation rate constant koff,1 from the RNA-only control (3.3 ± 0.3 s−1, 95%) is ∼20% higher than that of the PEC-99 nascent mRNA (2.7 ± 0.3 s−1, 92%) (Fig. 2E and SI Appendix, Table S1), further supporting direct physical interaction between the RNAP and the bound 30S on the nascent mRNA in PEC-99. By contrast, while preQ1-induced structural changes of the RNA-only transcript impede 30S binding as they do in PEC-99, the 30S dissociation rate constant again remains unchanged (Fig. 2E and SI Appendix, Table S1).
Taken together, these results suggest that RNAP specifically facilitates dynamic 30S recruitment to the nascent mRNA in PEC-99 but does not impact the kinetic stability of the complex.
Transcription Factors NusG and RfaH Serve Distinct Roles in 30S Recruitment and Retention.
To investigate how transcription factors NusG and RfaH, thought to bridge RNAP and ribosome (13, 18), influence 30S recruitment, we tested 30S binding to PEC-99 in the presence of equimolar concentrations of NusG or RfaH. Upon addition of NusG, we observed a significant increase (∼25%) in the 30S binding rate constant (kon) value to 4.3 ± 0.1 × 106 M−1 ⋅ s−1, while both dissociation rate constants (koff,1 and koff,2) only slightly decreased to 2.4 ± 0.03 s−1 (88%) and 0.2 ± 0.04 s−1 (12%) (Fig. 3A and SI Appendix, Fig. S7 and Table S1). This result indicates that NusG assists in initial 30S recruitment but does not significantly stabilize the bound 30S subunit. We note that a slight decrease (12%) in the 30S dissociation rate constant in the presence of NusG may be due to the stabilization of the bound 30S by simultaneous interaction of NusG with the RNAP in PEC-99 and the bound 30S but is not statistically significant (P > 0.05). In contrast, upon addition of RfaH, the kon remained unchanged, while the dissociation significantly slowed (koff,1 = 1.1 ± 0.1 s−1 (86%) and koff,2 = 0.04 ± 0.01 s−1 (14%), Fig. 3A and SI Appendix, Table S1). These data suggest that RfaH, in contrast to NusG, plays no role in initial 30S binding to RBS but instead stabilizes the bound 30S.
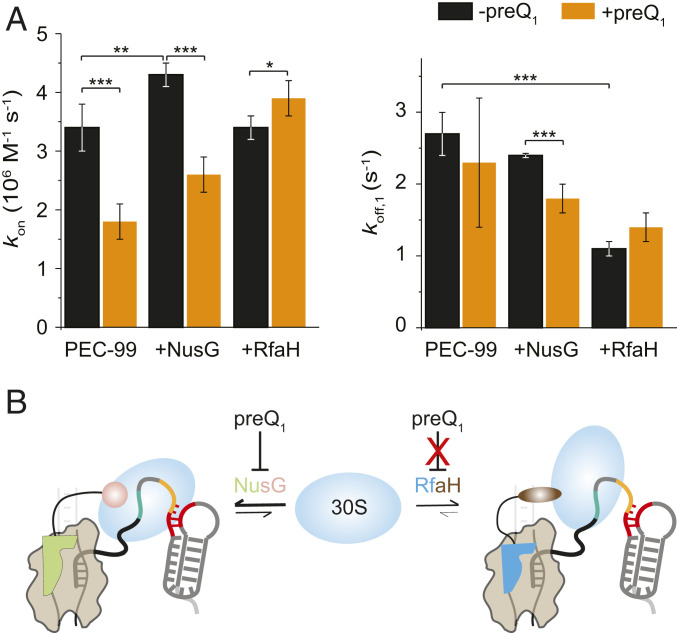
Fig. 3.
NusG and RfaH assist initial 30S binding and stabilize the bound 30S on the nascent mRNA, respectively. (A) Association (kon) and dissociation rate constants (koff) calculated from hidden Markov models for 30S binding to the mRNA in PEC-99 in the presence of equimolar concentrations of NusG or RfaH in the absence or presence of preQ1. Values of binding (kon) and dissociation (koff) rates are reported in SI Appendix, Table S1. N(PEC) = 265, N(PEC + preQ1) = 140, N(PEC + NusG) = 253, N(PEC + NusG + preQ1) = 252, N(PEC + RfaH) = 253, and N(PEC + RfaH + preQ1) = 332. (***P < 0.01, **P < 0.025, *P < 0.05.) (B) A model highlighting the differential roles of NusG and RfaH in the initial 30S binding and stabilization of the 30S subunit on the nascent mRNA.
Experiments performed in the presence of preQ1 revealed further differences between NusG and RfaH. In particular, preQ1-induced riboswitching has significant impact on the ability of NusG both to recruit 30S to the mRNA and then to retain it, as revealed by a 40% decrease in kon and a 25% decrease in koff,1 upon addition of the ligand (Fig. 3A and SI Appendix, Fig. S7 and Table S1). We note that RNAP-bound NusG can still promote 30S binding by interaction of its flexibly tethered CTD with the S10 protein of 30S. However, the effect of preQ1 on decreasing the kon is more prominent than the increase induced by NusG. By contrast, experiments performed with RfaH did not reveal significant differences in the 30S binding or dissociation rate constants in the absence and presence of preQ1 (Fig. 3A and SI Appendix, Table S1). Thus, while preQ1 significantly interferes with NusG’s ability to recruit and retain the 30S subunit, it has no such effect on RfaH (Fig. 3A). These differences could be due in part to much higher affinity of RfaH to RNAP as compared to NusG (19).
Taken together, these results reveal that NusG facilitates initial 30S binding, while RfaH helps to stabilize the bound 30S on the mRNA. Furthermore, the effect of NusG is modulated by preQ1 addition, which specifically reduces RBS accessibility, suggesting that NusG promotes specific 30S–RBS interactions. By contrast, the effect of RfaH is not influenced by preQ1-mediated SD occlusion, consistent with the cellular role of RfaH in mediating 30S recruitment to mRNAs lacking SD elements (13).
Ribosome Binding and Translation of the Nascent mRNA Regulate Transcription Speed.
Our results so far show that RNAP facilitates 30S recruitment to the nascent mRNA. The next question we addressed is whether, conversely, a 30S transiently bound to, or a 70S ribosome translating, the nascent mRNA has an impact on the speed of transcription. To detect changes in transcription rate during ribosome binding and translation, we employed a real-time transcription assay where individual halted transcription elongation complexes were attached to a microscope slide via a 5′ biotin moiety on the nascent mRNA and visualized using a double-stranded DNA template labeled with Cy3 at the downstream 5′ end of the template strand (Fig. 4A). Transcription was resumed by flowing a mixture of all four rNTPs during movie collection. In this experiment, the completion of transcription can be visualized by a protein-induced fluorescence enhancement (PIFE) signal when RNAP reaches the end of the DNA template (35, 39), synthesizing an additional 96-nucleotide stretch of mRNA after the halt position (Fig. 1B). The transcription time is therefore defined by the delay between rNTP injection and onset of PIFE signal (Fig. 4B).
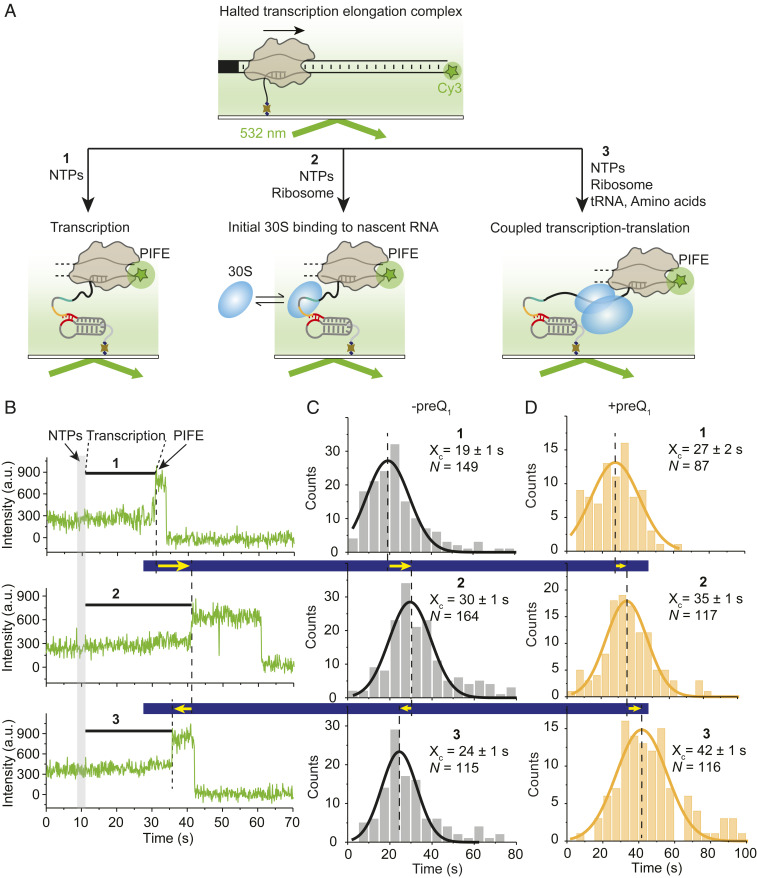
Fig. 4.
30S binding and translation of the nascent mRNA modulate the transcription rate. (A) An experimental approach to analyze the transcription speed in the absence (condition one) or the presence of nontranslating (condition two; −tRNA, −amino acids) and translating ribosome (condition three; +tRNA, +amino acids). (B) Cy3 intensity versus time traces showing real-time transcription progress in the absence (condition one) and presence of nontranslating (condition two) and translating (condition three) ribosomes. The end of the transcription is indicated by the occurrence of a PIFE signal. (C and D) Histograms of transcription times constructed from N number of molecules for conditions one, two, and three in the absence (C) or presence (D) of preQ1. Xc represents the mean transcription time ± SD from Gaussian fitting.
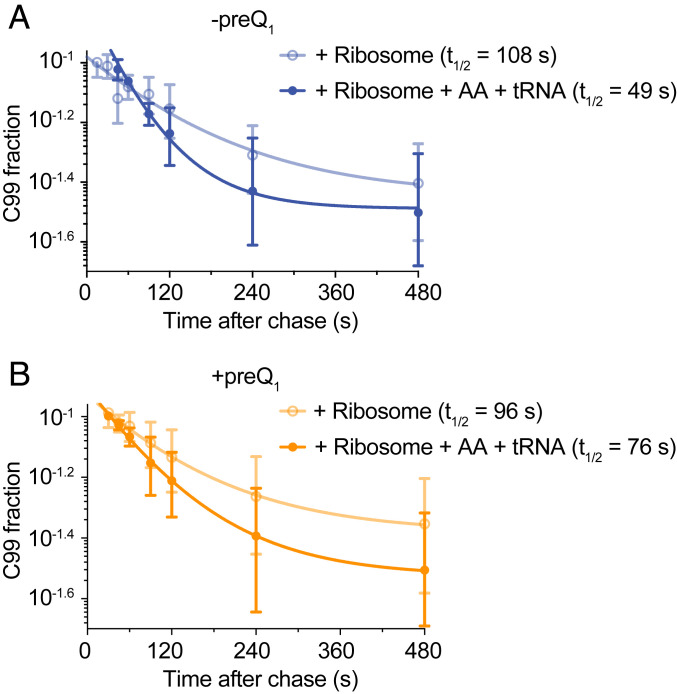
First, we determined the transcription time of RNAP alone (condition one in Fig. 4B). Analysis of individual complexes revealed an approximately twofold enhancement in Cy3 intensity, indicating the end of transcription ∼20 s after rNTP injection (condition one in Fig. 4B). A total of 149 such individual transcription times were fit with a Gaussian distribution, revealing a mean of 19 ± 1 s (Fig. 4C) or an average transcription rate of 5.1 nt/s. This rate is comparable to rNTP concentration-dependent transcription rates previously measured by single-molecule analysis (35). When the two ribosome subunits were added along with translation initiation factors (TIFs) but without transfer RNAs (tRNAs) or amino acids, where the 30S subunit will only transiently bind without the ability to initiate mRNA translation (condition two in Fig. 4A), the average transcription time increased to 30 ± 1 s (Fig. 4 B and C, Middle). This slower transcription rate of 3.2 nt/s suggests that a 30S subunit dynamically binding to the nascent mRNA, and therefore only transiently interacting with the RNAP, slows down transcription. A control experiment to verify the binding kinetics of 30S to the nascent mRNA in PEC-99 (Fig. 2) under the same buffer conditions as condition two (Fig. 4) indicated no significant change in kon, whereas the bound 30S was slightly stabilized as indicated by a 26% decrease in koff (SI Appendix, Table S1). This may be due to the presence of the TIFs that stabilize 30S binding relative to that of 30S subunit alone. By contrast to condition two, when adding the two ribosomal subunits with both TIFs and tRNAs/amino acids (condition three in Fig. 4A), allowing the ribosome to initiate translation on the nascent mRNA, the transcription time significantly decreased to 24 ± 1 s, reflecting a recovery in the transcription rate to 4 nt/s. A similar trend was observed during single-round in vitro transcription assays (SI Appendix, Fig. S8 and Fig. 5); notably, RNAP escapes from the C99 pause significantly more slowly in the presence of nontranslating 30S subunit (pause half-life was 108 s) than in its absence (54 s; compare Fig. 5 and Fig. 1D). By contrast, and consistent with the recovery of transcription speed observed under translation conditions (condition three), addition of the full ribosome, TIFs, and tRNAs/amino acids facilitated RNAP pause escape, reducing the C99 pause half-life to 49 s (SI Appendix, Fig. S8 and Fig. 5). To further test whether the observed recovery of the transcription speed was due to transcription–translation coupling, we measured the transcription rate in the presence of translation inhibitor chloramphenicol, known to stall the ribosome at early steps of elongation (40). Upon addition of chloramphenicol, RNAP arrival at the end of the DNA template was dramatically delayed (transcription time 41 ± 2 s; SI Appendix, Fig. S9); that is, the average transcription rate was reduced to 2.3 nt/s (SI Appendix, Fig. S10), more than by transiently binding 30S subunit (3.2 nt/s). This result supports the notion that stalling a translating ribosome dramatically slows transcription through coupling with RNAP (4) but is opposite to findings from a recent in vivo study (41). Chloramphenicol binds tightly to the translating ribosome and arrests translation (42), so we also used another antibiotic, fusidic acid, that binds in a reversible manner and more gradually delays ribosome translocation (43). Measurement of the transcription speed in the presence of fusidic acid under coupling conditions (condition three) revealed two populations of RNAP with different transcription speeds (SI Appendix, Figs. S9 and S10). A fraction (∼38%) of RNAPs move fast with an average transcription time of 21 ± 2 s (4.6 nt/s), while the remainder moves at a significantly slower speed with an average transcription time of 50 ± 2 s (1.9 nt/s). We suggest that the former fraction of RNAPs finish the translation-coupled transcription before fusidic acid binds, whereas the fraction of RNAPs move slower because of coupling with the slow-moving ribosome in the presence of reversibly binding fusidic acid.
Fig. 5.
Impact of the ribosome under nontranslating and translating conditions on RNAP progression. Quantification of the half-life of the C99 pause observed in SI Appendix, Fig. S8 in the absence (A) or presence of preQ1 (B) under nontranslating (condition two) and translating (condition three) ribosome conditions.
Taken together, these data demonstrate that cotranscriptional 30S binding slows transcription because of transient RNAP–30S interactions mediated by the nascent mRNA, while transcription–translation coupling reinstates faster transcription as the pioneering ribosome follows the leading RNAP, unless an antibiotic blocks its movement.
preQ1-Induced Riboswitching Slows Transcription Both Directly and via the Ribosome.
The observed strong effects of the ribosome on the speed of transcription prompted us to use our real-time transcription assays to ask whether preQ1-induced riboswitching, which impedes 30S binding to the nascent mRNA (Fig. 2), further modulates RNAP progress. Notably, the inclusion of preQ1 slows down transcription under all three conditions tested (Fig. 4D and SI Appendix, Fig. S10)—in the absence of the ribosome (condition one) from a transcription time of 19 ± 1 s (transcription rate of 5.1 nt/s) to 27 ± 1 s (3.6 nt/s), in the presence of the ribosome and TIFs but without tRNAs/amino acids (condition two) from a transcription time of 30 ± 1 s (transcription rate of 3.2 nt/s) to 35 ± 1 s (2.7 nt/s), and in the presence of a translating ribosome with all cofactors (condition three) from a transcription time of 24 ± 1 s (transcription rate of 4.0 nt/s) to a remarkably slow 42 ± 1 s (2.3 nt/s). This general decrease in transcription speed upon preQ1 addition is consistent with the ligand effects observed during the earlier stages of transcription, such as at the pause U63 right downstream of the preQ1 riboswitch where the ligand slows RNAP progress approximately twofold (Fig. 1 B and E). Most notably, however, while translation condition three significantly accelerates transcription in the absence of preQ1 relative to condition two, where the 30S subunit is expected to bind only transiently, in the presence of preQ1, this recovery was no longer observed (Fig. 4D). In fact, transcription with preQ1 added was as slow (2.3 nt/s) as when the ribosome was inhibited with chloramphenicol (2.3 nt/s, SI Appendix, Fig. S10).
Taken together, these results support the notion that, upon binding of its cognate ligand, the translational preQ1 riboswitch slows transcription, first, immediately downstream in direct fashion and, second, farther downstream by instead disrupting transcription–translation coupling, most likely by occluding the SD sequence and thereby suppressing ribosome loading and translation initiation.
Discussion
In this work, combining biochemical and single-molecule fluorescence assays, we have studied the mechanism of the coordination between the transcription and translation machineries on a nascent mRNA containing a preQ1-sensing riboswitch in its 5′ UTR (Fig. 1A). Our results provide dynamic information missing from a recent flurry of structural studies (7–10, 12) regarding the initial 30S recruitment to the cotranscriptionally folded nascent transcript and how it is modulated by the ligand-dependent structure of the mRNA as well as the presence of RNAP and transcription factors (Fig. 6). Furthermore, by tracking the real-time progress of RNAP along the DNA (Fig. 4), we reveal the importance of transcription–translation coupling for rapid RNA synthesis.
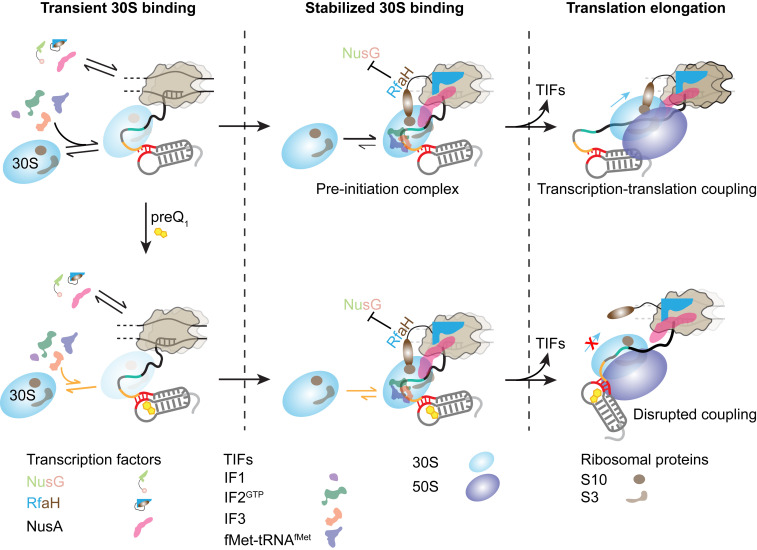
Fig. 6.
Model for the coordination between the transcription and translation machineries on a nascent mRNA as a function of ligand binding to the riboswitch. The 30S subunit can dynamically bind to the nascent mRNA as soon as the RBS emerges from the RNAP exit channel. Transcription factors and TIFs assist in initial binding and retention of the 30S on the mRNA, resulting in the stabilization of a preinitiation complex. Transient 30S binding to the nascent mRNA can slow down the transcription because of the transient RNAP–30S interaction. However, during translation elongation, the ribosome (70S) can follow the leading RNAP, establishing transcription–translation coupling and maintaining optimal transcription speed. preQ1, which binds to the 5′ UTR of the mRNA and occludes the RBS, impedes 30S binding. Consequently, in the presence of preQ1 transcription–translation coupling is disrupted, leading to slow transcription.
In particular, we found that the 30S subunit binds dynamically to the nascent mRNA upon RNAP pausing at residue 99 and that preQ1-induced structural rearrangement of the riboswitch leads to fewer 30S binding events because of decreased RBS accessibility (Fig. 2 D and E), highlighting the importance of the nascent mRNA in the interactions between the transcription machinery and the recruited 30S subunit. Comparison of 30S binding to the mRNA in PEC-99 or the corresponding mRNA alone revealed a role for RNAP in facilitating 30S recruitment during transcription (Fig. 2E), indicating dynamic interactions between RNAP and mRNA-bound 30S, in agreement with recent cryo-EM data showing variable interaction surfaces of RNAP and ribosome on an mRNA of similar length (9). Overall, our data suggest that RNAP and the 30S subunit stochastically interact during nascent mRNA synthesis (5, 11, 16), consistent with recent cryo-EM structural studies (9, 10).
Stochastic coupling could be modulated by accessory factors that bridge RNAP and ribosomal subunits, either during the assembly of translation preinitiation complexes or throughout the first round of protein synthesis. Two paralogous transcription elongation factors, NusG and RfaH, have been proposed to couple transcription and translation (13, 18). However, the cellular functions of RfaH and NusG are entirely different from each other, as these factors compete for binding to RNAP to control expression of nonoverlapping sets of genes via distinct interactions with RNAP, nucleic acids, and Rho transcription termination factor (44). Expanding this current understanding, our results provide direct evidence for different roles of NusG and RfaH in assisting 30S binding to and stabilization on the nascent mRNA, respectively (Fig. 3 and SI Appendix, Table S1). It has been suggested that an RNAP-bound NusG can promote 30S binding by interaction of its flexibly tethered CTD with the S10 protein of 30S. Alternatively, free NusG can first bind to the 30S subunit and then deliver it to the transcribing RNAP, facilitating initial 30S recruitment. Our finding that NusG specifically facilitates 30S recruitment to the mRNA (Fig. 3) supports these models and suggests that the factor can indeed simultaneously interact with RNAP and the mRNA-bound 30S, consistent also with recent observations on a similarly sized mRNA (9, 10).
In stark contrast to NusG, we show that RfaH does not promote 30S recruitment but instead plays another important role by stabilizing the bound 30S on the nascent mRNA. This result suggests that an RNAP-bound RfaH interacts with 30S when it is already bound to the mRNA, a scenario fully consistent with unusual metamorphic properties of RfaH. Unlike NusG, the free RfaH cannot bind to 30S because the RfaH CTD is folded as an α-helical hairpin in which the S10-interacting residues are buried (45). Following recruitment to the transcription elongation complex, the RfaH CTD refolds into a β-barrel that binds to S10 similarly to the NusG CTD (13). Another difference between RfaH and NusG is highlighted by our observation that RfaH appears to make the RNAP–30S interaction insensitive to preQ1. Upon binding to preQ1, the mRNA rearranges to mask the SD element, thus blocking 30S binding. However, in vivo data indicate that RfaH mediates ribosome recruitment to SD-less mRNAs (13), rationalizing our observation that an SD made less accessible by preQ1 addition has only negligible effects on RfaH–ribosome interactions. Finally, stabilization of 30S–mRNA binding by RfaH could be particularly important for activation of gene expression by RfaH. RfaH and other specialized paralogs of NusG activate expression of very long virulence operons (46); RfaH-dependent operons lack SD elements, are enriched in rare codons, and are thought to be translated by a single ribosome that does not dissociate between ORFs (13). Furthermore, RfaH is required for expression of gigantic proteins, such as a 5,559–amino acid long nonfibrial adhesin encoded by the Salmonella pathogenicity island IV (47). Our results suggest that a large part of the dramatic RfaH effects on gene expression, currently thought to be explained by inhibition of Rho-dependent termination through the exclusion of NusG, could instead be due to stabilization of the ribosome on mRNA during RfaH-mediated transcription–translation coupling.
During active transcription, the 30S subunit can start transiently binding to the nascent mRNA as soon as the RBS emerges from the RNAP exit channel; while the length of the nascent mRNA is still short enough, an RBS-bound 30S will therefore directly interact with the RNAP (9). Under this 30S loading condition, repeated 30S binding to the RBS leads to stochastic coupling of the 30S with the transcribing RNAP (12, 15), slowing down the RNAP (Fig. 4). However, a translating 70S ribosome can directionally follow the transcribing RNAP, establishing transcription–translation coupling (9, 10) and therefore recovering the transcription rate (Fig. 4). We observe that disruption of this transcription–translation coupling by the structure-dependent inhibition of translation initiation by a ligand-bound translational riboswitch or the presence of a translation inhibitor such as chloramphenicol leads to decreased transcription speed (SI Appendix, Fig. S10), thereby minimizing synthesis of additional intervening mRNA between the RNAP and the ribosome. We note that such transcription–translation coupling is also important for preventing Rho-dependent premature transcription termination (25), invoking the idea that a riboswitch can influence translation and transcription at multiple levels. Our data are consistent with previous studies demonstrating stochastic coupling between the independently moving RNAP and the trailing ribosome (5, 17). Together, these findings demonstrate the impact of transcription–translation coupling on transcription efficiency and reveal the ability of a riboswitch-harboring, ligand-sensing nascent mRNA to regulate the entire gene expression process.
Considering our discoveries in light of the prior findings above, we propose a general model for cotranscriptional 30S recruitment to a nascent mRNA that results in transcription–translation coupling (Fig. 6). The 30S subunit transiently binds to the nascent mRNA as soon as the RBS emerges from the RNAP RNA exit channel, forming short-lived “standby site” complexes (36, 48). The proximal RNAP facilitates this initial 30S recruitment by, first, influencing mRNA folding wherein the RBS is exposed and, second, stochastically interacting with the transiently binding 30S directly. Transcription factors and TIFs assist and stabilize this initial 30S recruitment. In particular, the highly conserved transcription factor NusG and its paralog RfaH promote 30S binding and stabilize the mRNA-bound 30S, respectively, by their simultaneous bridging interactions with both RNAP and 30S (9, 10). Other transcription factors such as NusA have also been shown recently to mediate such RNAP–ribosome interactions (10); additionally, ribosomal proteins are known to interact with the intervening mRNA and modulate transcription–translation coupling through mRNA structural rearrangement (9). These interactions eventually lead to the development of a stable translation preinitiation complex. The stochastic RNAP–30S interactions established during this initial 30S recruitment to the RBS impact, conversely, mRNA synthesis by decreasing the speed of RNAP and therefore allowing the fully assembled 70S ribosome to start transcription–coupled translation of the nascent mRNA to recover a faster transcription speed. Riboswitch- or translation inhibitor-induced disruption in transcription–translation coupling leads to slowed transcription and suppresses the synthesis of additional intervening mRNA. Together, our data demonstrate the importance of transcription–translation coupling in productive mRNA synthesis and how a ligand-sensing translational riboswitch can help coordinate this coupling to control bacterial gene expression. Accordingly, we anticipate that these insights may inspire new approaches for the design of antibacterials.
Methods
Bacterial Strains, Plasmids, and Growth Conditions.
E. coli JM109 competent cells (Promega, L2001) were used for maintenance of all DNA templates. The strain carrying the pKK3535 (gift from Joseph Puglisi, Stanford University School of Medicine, Stanford, CA) plasmid was used for purifying the mutant ribosome. All strains were grown at 37 °C in Luria-Bertani (LB) media supplemented with 100 µg/mL Ampicillin. Plasmids used in this study are listed in the SI Appendix, Table S2.
Preparation of DNA Templates.
A 191-nucleotide DNA template including the preQ1 riboswitch from Bas under the control of the T7A1 promoter was cloned into a pUC19 plasmid. In addition, 10 nucleotides not found in the wild-type sequence were inserted after the promoter sequence in order to generate a 12-nucleotide stretch in which the RNA transcript lacks any uracil residues (EC-12) except for the +2 position dependent of the ApU dinucleotide used to initiate the transcription. Transcription templates for in vitro transcription were generated by PCR using the « T7A1-Bas_PreQ1-Fwd2 » forward oligonucleotide and the according reverse oligonucleotides (SI Appendix, Table S3). Oligonucleotides used in this study are listed in the SI Appendix, Table S3.
Ligand Preparation.
The preQ1 ligand used in this study was synthesized as described previously (27) and was generously provided by George Garcia at the University of Michigan. The concentrations of the preQ1 stock solutions were measured by ultraviolet (UV)–visible (vis) spectrophotometry using a NanoDrop 2000 spectrophotometer. Final concentrations were measured by UV–vis spectrophotometry using the molar extinction coefficient of preQ1 at 256 nm (11,200 M−1 ⋅ cm−1).
Single-Round in Vitro Transcription Assays.
HCs were prepared in transcription buffer (20 mM Tris HCl, pH 8.0, 20 mM NaCl, 20 mM MgCl2, 14 mM 2-mercaptoethanol, and 0.1 mM ethylenediaminetetraacetic acid [EDTA]) containing 25 µM adenosine 5′-triphosphate (ATP)/cytidine 5′-triphosphate (CTP) mix, 50 nM α32P-GTP (3,000 Ci/mmol), 10 µM ApU dinucleotide primer (TriLink), and 50 nM DNA template. A total of 100 nM E. coli RNAP holoenzyme (New England Biolabs, M0551S) was added to the reaction mixture and incubated for 10 min at 37 °C. The reaction mixture was then passed through a G-50 (GE Healthcare, 27533001) column to remove any free nucleotides. To complete the transcription reaction, a mixture containing all four rNTPs (25 µM) was added concomitantly with heparin (450 µg/mL) to prevent the reinitiation of transcription. preQ1 (when present) was added to 10 µM. The reaction mixture was incubated at 37 °C, and aliquots were quenched at the desired times into an equal volume of loading buffer (95% formamide, 1 mM EDTA, 0.1% sodium dodecyl sulphate [SDS], 0.2% bromophenol blue, and 0.2% xylene cyanol). Sequencing ladders were prepared by combining the HC with a chase solution containing 250 µM of each rNTP, in addition to one 3′-OMe rNTP (at 25 µM for 3′-OMe GTP and 15 µM for 3′-OMe ATP, uridine 5′-triphosphate, and CTP). Reaction aliquots were denatured before loading 5 µL each onto a denaturing 8 M urea, 6% polyacrylamide sequencing gel. The gel was dried and exposed to a phosphor screen (typically overnight), which was then scanned on an Amersham Typhoon PhosphorImager (GE Lifesciences). Gel images were analyzed with ImageLab (Bio-Rad Laboratories) software.
Time Pausing Analysis.
The half-life of transcriptional pausing was determined by calculating the fraction of the RNA pause species compared with the total amount of RNA for each time point, which was analyzed with pseudofirst-order kinetics to extract the half-life (49). For each determination, we have subtracted the background signal. Error bars in transcription quantification represent the SD of the mean from at least two independent replicates.
Preparation of Bas mRNA for anti-SD Probe Binding Assay.
Bas full-length mRNA (SI Appendix, Note S1) was produced by in vitro transcription using a pUC19 plasmid with an engineered T7 promoter. The Bas pUC19 plasmid was linearized with HindIII (New England Biolabs) digestion for run-off transcription. Transcription reactions were performed in the presence of 120 mM Hepes-KOH (pH 7.5 at 25 C), 25 mM MgCl2, 2 mM spermidine, 40 mM dithiothreitol (DTT), 30 mM total rNTPs, 0.01% (wt/vol) Triton X-100, 200 nM linearized plasmid, 0.01 U/µl pyrophosphatase, and 0.07 mg/mL T7 RNAP in a total volume of 1 mL. Transcription reactions were incubated at 37 °C for 4 h. The enzyme was removed using phenol/chloroform extraction, and the resulting solution was spun in an Amicon 100 MWCO spin column to reduce the volume to 100 µl. mRNA was purified by denaturing polyacrylamide gel containing 7 M urea and detected using brief 254 nm UV radiation and gel-eluted overnight. The mRNA band from PAGE was subjected to crush and soak treatment to extract the mRNA out of the gel using crush and soak buffer (300 mM sodium acetate and 1 mM EDTA, pH 8.0). The resulting RNA solutions were ethanol precipitated and resuspended in Tris EDTA (TE) buffer at pH 7.0. The sequences of the Bas mRNA used in this study are listed in SI Appendix, Note S1.
preQ1-Dependent anti-SD Probe Binding Assay.
The kinetics of preQ1-dependent anti-SD probe binding to the SD sequence of the in vitro transcribed Bas mRNA was studied using the SiM-KARTS technique previously established by Rinaldi et al. (32). The full-length Bas mRNA was heat annealed at 70° C for 2 min and at room temperature for 10 min with the biotinylated capture strand and TYE547-LNA oligo at 1:1:1 ratio in the presence of 1× SiM-KARTS buffer (50 mM Tris HCl, pH 7.5, 0.6 M NaCl, and 20 mM MgCl2). The stock solution for this annealed complex was made to be 100 nM, which was used to refold the riboswitch with different concentrations of preQ1 (0 to 10 µM). The diluted complex with varying concentrations of preQ1 was chilled on ice. The chilled solution was flowed over an assembled microfluidic channel on a quartz slide coated with biotinylated bovine serum albumin and streptavidin. The chilled 40 pM RNA complex solution (100 µL) was flowed over the slide and allowed to equilibrate for 10 min. Excess mRNA was washed off the slide with 1× SiM-KARTS buffer with or without preQ1. An oxygen-scavenging system consisting of 5 mM protocatechuic acid and 50 nM protocatechuate-3,4-dioxygenase with or without preQ1, to slow photobleaching, and 2 mM Trolox, to reduce photoblinking, as well as 50 nM Cy5-labeled anti-SD probe was flowed over the slide and allowed to equilibrate for 5 min. Both Cy5 and TYE563 dyes were directly and simultaneously excited using 638 nm (red) and 532 nm (green) diode lasers, respectively. Emission from both fluorophores was simultaneously recorded using an intensified charge-coupled device camera (I-PentaMAX, Princeton Instruments) at 100 ms time resolution using the Micro-Manager software (https://www.micro-manager.org/). Fluorescence time traces were extracted from the raw movie files using Interactive Data Language (Research Systems) and analyzed using custom made MATLAB (The MathWorks, Inc.) scripts. Traces exhibiting significant binding events were manually selected using the following criteria: a single photobleaching step of the TYE563 signal to localize the mRNA molecule on the slide surface and at least one Cy5 colocalization signal per trajectory corresponding to anti-SD-binding events with a signal to noise ratio of at least 3:1. Suitable traces were compiled and processed with hidden Markov model analysis, and it was performed on the Cy5 intensity using the segmental k-means algorithm in the QuB software. A two-state model was used with an unbound and bound state to idealize the data. Kinetics plots were constructed by extracting the dwell times in the bound and unbound states. The normalized cumulative distributions of bound dwell times were fit with a double exponential, and unbound dwell times were fit with a single exponential association function in OriginLab 8.5 from which on and off rates were calculated. Rate constants for the anti-SD probe as a function of preQ1 concentration were fit with a standard Hill coefficient of 1.
Preparation of the Paused Transcription Elongation Complex.
The paused transcription elongation complexes (PEC-99) containing the nascent Bas mRNA were prepared following a method described previously (34, 50), with minor modifications. The DNA template containing the sequence of the Bas mRNA with the preQ1 riboswitch at its 5′ end and a biotin moiety at the 5′ end of the antisense DNA strand was used to prepare the PEC-99. The RNAP was stalled at the end of the transcription (C99 position) by a biotin–streptavidin roadblock. In vitro transcription reactions were performed in two steps to allow the specific incorporation of Cy3 at the 5′ end of the mRNA. Before transcription, the DNA template was mixed with excess streptavidin and incubated at 37 °C for 5 min. Transcription reactions were performed in the same transcription buffer (20 mM Tris HCl, pH 8.0, 20 mM NaCl, 20 mM MgCl2, 14 mM 2-mercaptoethanol, and 0.1 mM EDTA) as described above. Reactions were initiated by adding 100 µM AU-Cy3 dinucleotide (GE Healthcare) and 25 µM ATP/CTP/GTP nucleotides to a mixture of 50 nM DNA template containing biotin–streptavidin roadblock and 100 nM E. coli RNAP holoenzyme and incubating at 37 °C for 10 min, therefore yielding a Cy3-labeled HC. The reaction mixture was then passed through a G-50 column to remove any free nucleotides. The transcription was resumed upon addition of all four rNTPs at a final concentration of 100 µM and incubated at 37 °C for 5 min. To prevent any reinitiation of transcription, 450 µg/mL heparin was used in the reaction mixture. preQ1 (when present) was added during the transcription of the mRNA at a final concentration of 5 µM, and the same concentration of preQ1 was added during subsequent dilutions. To prepare the Cy3-labeled mRNA alone transcripts, the DNA template was assembled corresponding to the same sequence of the Bas mRNA until the C99 position and fused to a capture sequence. After transcription, the mRNA alone transcripts were hybridized to a 5′ biotinylated capture probe (CP probe) that is complementary to the 3′ end capture sequence of the mRNA. The CP probe was mixed to a ratio of 10:1 with RNA transcript and incubated at 37 °C for 5 min. The PEC-99 as well as the mRNA alone transcripts were immobilized on the biotinylated microscope slide surface by using biotin–streptavidin linkage.
To verify the presence of any PEC complexes other than the PEC-99, we performed a gel analysis. Radiolabeled HCs were prepared as described above and separated into three aliquots. Streptavidin (250 nM) or a mock solution were then added to two separate tubes and incubated for 5 min at 37 °C. To complete the transcription reaction, rNTPs were added to attain a final concentration of 100 µM, concomitantly with heparin (450 µg/mL) to prevent the reinitiation of transcription. The mixtures were incubated for 10 min at 37 °C, then quenched with an equal volume of loading buffer (95% formamide, 1 mM EDTA, 0.1% SDS, 0.2% bromophenol blue, and 0.2% xylene cyanol) and heated for 2 min at 95 °C before loading onto a 6% denaturing polyacrylamide gel.
30S Purification and Fluorescent Labeling.
Mutant E. coli pKK3535 plasmid strains with an extension at the helix 44 of the 16s rRNA was obtained from Puglisi Laboratory. This extension allows labeling of the 30S using a fluorescently labeled DNA oligonucleotide complementary to the extended portion of the helix 44. Single salt-washed ribosomes were prepared using previously described protocols with minor modifications (32, 33). Briefly, the pKK3535 strain containing mutated ribosomes was grown in LB medium at 37 °C to an OD600 of 0.8 to 1 starting from an overnight culture. The cells were then cooled at 4 °C for 45 min and pelleted at 5,000 rpm for 15 min. All subsequent steps were performed on ice or at 4 °C. The cell pellet was resuspended in buffer A (20 mM Tris HCl [pH 7.05 at 25 °C], 100 mM NH4Cl, 10 mM MgCl2, 0.5 mM EDTA, and 6 mM 2-mercaptoethanol), and the cells were lysed in a single pass using a M-110L Microfluidizer Processor (Microfluidics). The lysate was cleared by centrifugation at 16,000 rpm in a JA-20 Rotor. The clarified lysate was then pelleted over a 35 mL sucrose cushion (1.1 M sucrose, 20 mM Tris HCl [pH 7.0 at 25 °C], 500 mM NH4Cl, 10 mM MgCl2, and 0.5 mM EDTA) in a Beckman Type 45 Ti Rotor overnight at 37,000 rpm. The pellet was washed twice with 1 mL buffer B (20 mM Tris HCl [pH 7.0 at 25 °C], 500 mM NH4Cl, 10 mM MgCl2, and 0.5 mM EDTA) and resuspended in 6 mL buffer B by gentle stirring. The one salt-washed 70S ribosomes were then dialyzed against low magnesium buffer E (50 mM Tris HCl [pH 7.0 at 25 °C], 150 mM NH4Cl, 1 mM MgCl2, and 6 mM 2-mercaptoethanol) three times. The low Mg2+ ions facilitate the two subunits to dissociate from each other and remain as individual subunits in the solution. Next, 100 A260 units of the dissociated ribosome is loaded onto a 36 mL previously prepared sucrose gradient (to prepare the gradient, buffer E + 20% sucrose is frozen at −80 °C and then gently thawed at room temperature without mixing). The gradients were then loaded onto a swinging bucket (SW 28) rotor and centrifuged at 20,000 rpm for 18 h. The gradients were then fractionated using a Brandel Gradient fractionator coupled with a UV signal monitor. Appropriate fractions were pooled together as pure 30S and pure 50S fractions. The 30S and 50S fractions were then palleted separately for 12 h at 66,000 rpm in a Beckman Type 70 Ti Rotor. Pelleted subunits were resuspended in storage buffer (50 mM Tris HCl [pH 7.5 at 25 °C], 70 mM NH4Cl, 30 mM KCl, 7 mM MgCl2, and 6 mM 2-mercaptoethanol) and flash frozen with liquid nitrogen and stored at −80 °C. S1-depleted 30S subunits (30S ΔS1) were prepared by incubation of the purified 30S subunits with polyU resin according to a method described by Lauber et al. (51). S1-reconstituted 30S subunits were prepared by incubating 30S ΔS1 with either half or fully stoichiometric purified protein S1 on ice for 5 min (52). S1 content in our 30S preparations was assessed by a composite nondenaturing 3% polyacrylamide:0.5% agarose gel following a method established by Dahlberg et al. (53). The gel was stained with SYBR-Gold and imaged using an Amersham Typhoon scanner (GE Lifesciences). Gel images were quantified with ImageLab (Bio-Rad Laboratories) software.
To observe direct binding of the 30S to the nascent mRNA, we doubly labeled the E. coli 30S with Cy5 by hybridizing a dual Cy5-labeled DNA oligonucleotide to an engineered extension in the helix 44 of the 16S rRNA (33). The 30S labeling was performed with a 10-fold excess of dual Cy5-labeled DNA oligonucleotide at a final 30S concentration of 1 μM and a buffer composition (50 mM Tris-OAc [pH 7.5 at 25 °C], 100 mM KCl, 5 mM NH4OAc, 0.5 mM Ca(OAc)2, 5 mM Mg(OAc)2, 6 mM 2-mercaptoethanol, 0.5 mM EDTA, 5 mM putrescine, and 1 mM spermidine) which has been optimized for the activity of purified ribosomes (54). The reaction was protected from light and incubated for 10 min at 37 °C and then 60 min at 30 °C and finally cooled gradually to room temperature. Excess fluorescent oligonucleotides were then removed by spin column (Millipore, UFC510024), and the solution containing the labeled 30S was flash frozen in aliquots and stored at −80 °C. The final concentration of the 30S in the recovered solution was determined spectrophotometrically using the extinction coefficient ε260 = 14492753.62 M−1 ⋅ cm−1 for 30S and ε650 = 250000 M−1 ⋅ cm−1 for Cy5.
Single-Molecule Measurements of 30S Binding Kinetics.
For single-molecule experiments, we immobilized the Cy3-labeled PEC-99 on a PEG/biotin-PEG coated quartz microscope slide surface prepared following established protocol (55). The PEC-99 solution was diluted to a final concentration of 50 pM in the transcription buffer (20 mM Tris HCl, pH 8.0, 20 mM NaCl, 20 mM MgCl2, 14 mM 2-mercaptoethanol, and 0.1 mM EDTA) and flowed on the biotinylated microscope slide and incubated for 10 min to allow immobilization. For experiments with the mRNA alone transcripts, a solution containing the mRNA annealed with the biotinylated CP probe at its 3′ end was flowed on the streptavidin coated slide surface and incubated for 10 min to allow immobilization. The excess unbound complex was then washed off the slide with transcription buffer. An oxygen-scavenging system (OSS) consisting 44 mM glucose, 165 U/mL glucose oxidase from Aspergillus niger, 2,170 U/mL catalase from Corynebacterium glutamicum to slow photobleaching (56), and 5 mM Trolox to reduce photoblinking (57) as well as dual Cy5-labeled 30S (2 nM) was flowed onto the slide and allowed to equilibrate for 5 min before imaging with a prism-based TIRF microscope as described previously (31, 37, 58). preQ1 (when present) was added to the imaging solution at a final concentration of 5 µM. For 30S binding experiments in the presence of a blocking strand, it was added to the imaging solution to attain a final concentration of 50 nM. For experiments with NusG and RfaH, a final concentration of 2 nM and 1 nM, respectively, was attained in imaging solution. For experiments in the presence of translation initiation factor mix, 2 µl factor mix (NEB, P0762) from PURExpress Δ Ribosome Kit was added to the imaging solution. Both Cy3 and Cy5 dyes were directly and simultaneously excited using 532 nm green and 638 nm red lasers, respectively. Emission from both fluorophores was simultaneously recorded at 100 ms time resolution using a digital CMOS camera (Hamamatsu, C13440-20CU) using a custom acquisition script written in MATLAB. Locations of molecules and fluorophore over time traces were extracted from raw movie files using MATLAB (MathWorks). Genuine fluorescence time traces for individual molecules were selected manually and analyzed using custom MATLAB (MathWorks) scripts using the following criteria: single-step photobleaching of Cy3 and at least two Cy5 intensity spikes of more than twofold of the background intensity. Dual Cy5 labeling of 30S allowed us to distinguish 30S dissociation events (single-step Cy5 intensity decrease) from photobleaching events (double-step Cy5 intensity decrease). Traces showing binding events were idealized using a two-state hidden Markov model for the unbound and bound states in QuB (59). From the idealized traces, dwell times of 30S in the bound (τbound) and the unbound (τunbound) states were calculated. Cumulative of bound and unbound dwell-time distributions were plotted and fitted in OriginLab with single exponential or double exponential functions to obtain the lifetimes in the bound and unbound states. The dissociation rates (koff) were calculated as the inverse of the τbound, whereas the association rates (kon) were calculated by dividing the inverse of the τunbound by the concentration of 30S used during the data collection. The statistical significance of differences in the rate constants was determined using the two-tailed Student’s t test. P values < 0.05 were considered significant.
Real-Time Transcription Assay.
A DNA template containing the sequence of the Bas mRNA same as above and a Cy3 molecule at its 3′ end was used to monitor the real-time transcription in different experimental conditions. First, the HCs were prepared by adding 100 µM AU-biotin dinucleotide (GE Healthcare) and 25 µM ATP/CTP/GTP nucleotides to a mixture of 50 nM DNA template and 100 nM E. coli RNAP holoenzyme and incubating at 37 °C for 10 min. The reaction mixture was then passed through a G-50 column to remove any free nucleotides. The biotin moiety present in the 5′ end of the nascent mRNA allows immobilization of the HCs on a biotinylated slide surface via biotin–streptavidin linkage. The slide surface was coated with streptavidin (0.2 mg/mL) for 10 to 15 min prior to flowing the HC onto the slide chamber.
For the real-time transcription assay, the inlet consists of a cut pipet tip acting as a 100 µl reservoir, and the outlet is connected to a 3 mL syringe to pull in the solutions from the reservoir. After immobilizing the HC on the slide, the 50 µl OSS solution was flowed and allowed to equilibrate for 5 min before restarting the transcription. The transcription was then restarted at ∼10 s of the movie collection by slowly flowing 50 µl imaging solution containing all four rNTPs and the OSS in a buffer of 40 mM Tris HCl, pH 8.0, 330 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, and 0.1 mM DTT as described previously (35, 39). The end of the transcription was characterized by the occurrence of PIFE signal due to the presence of RNAP in close proximity to the Cy3 molecule at the end of the template. For experimental conditions where the transcription is slow, we occasionally observed traces with a gradual increase in Cy3 intensity before a sharp increase (PIFE) as observed before (39). For experiments in the nontranslating ribosome condition, 2 µl PURExpress Solution B (New England Biolabs, P0760) was added in the imaging solution. For experiments in translating ribosome condition, 2 µl each PURExpress Solution B (New England Biolabs, P0760), E. coli tRNA (New England Biolabs, N6842), and amino acid mix (New England Biolabs, N6843) were added in the imaging solution. preQ1 (when present) was added in the imaging solution at a final concentration of 5 µM. Chloramphenicol and fusidic acid (when present) were added in the imaging solution to attain final concentrations of 20 and 200 µM, respectively. Traces showing a PIFE signature were selected manually and analyzed using custom MATLAB (MathWorks) scripts. Transcription time was calculated from the time difference between the rNTP injection and the occurrence of the PIFE signal. Transcription rate was calculated by dividing the length of the nascent mRNA by the transcription time.
Statistical details.
Statistical details of individual experiments such as number of molecules analyzed and definition of error bars are indicated in the main text, figures, and figure legends.
Supplementary Material
Acknowledgments
We thank Dr. Catherine Scull for critical reading of the manuscript and Dr. Katelyn Green and Jacqueline Kunesh for technical assistance. This work was supported by NIH Grants GM062357, GM118524, and GM131922 to N.G.W. and Grant R01 GM067153 to I.A.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. S.A.W. is a guest editor invited by the Editorial Board.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2023426118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix. Study data have been deposited in the University of Michigan Deep Blue Data repository (DOI: https://doi.org/10.7302/dxhn-0q11).
References
- 1.Conn A. B., Diggs S., Tam T. K., Blaha G. M., Two old dogs, one new trick: A review of RNA polymerase and ribosome interactions during transcription-translation coupling. Int. J. Mol. Sci. 20, 2595 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ray-Soni A., Bellecourt M. J., Landick R., Mechanisms of bacterial transcription termination: All good things must end. Annu. Rev. Biochem. 85, 319–347 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Vogel U., Jensen K. F., The RNA chain elongation rate in Escherichia coli depends on the growth rate. J. Bacteriol. 176, 2807–2813 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Proshkin S., Rahmouni A. R., Mironov A., Nudler E., Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science 328, 504–508 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stevenson-Jones F., Woodgate J., Castro-Roa D., Zenkin N., Ribosome reactivates transcription by physically pushing RNA polymerase out of transcription arrest. Proc. Natl. Acad. Sci. U.S.A. 117, 8462–8467 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Landick R., Carey J., Yanofsky C., Translation activates the paused transcription complex and restores transcription of the trp operon leader region. Proc. Natl. Acad. Sci. U.S.A. 82, 4663–4667 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Reilly F. J., et al., In-cell architecture of an actively transcribing-translating expressome. Science 369, 554–557 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kohler R., Mooney R. A., Mills D. J., Landick R., Cramer P., Architecture of a transcribing-translating expressome. Science 356, 194–197 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Webster M. W., et al., Structural basis of transcription-translation coupling and collision in bacteria. Science 369, 1355–1359 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Wang C., et al., Structural basis of transcription-translation coupling. Science 369, 1359–1365 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan H., et al., Transcription-translation coupling: Direct interactions of RNA polymerase with ribosomes and ribosomal subunits. Nucleic Acids Res. 45, 11043–11055 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demo G., et al., Structure of RNA polymerase bound to ribosomal 30S subunit. eLife 6, e28560 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burmann B. M., et al., An α helix to β barrel domain switch transforms the transcription factor RfaH into a translation factor. Cell 150, 291–303 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Washburn R. S., et al., Escherichia coli NusG links the lead ribosome with the transcription elongation complex. iScience 23, 101352 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen M., Fredrick K., RNA polymerase’s relationship with the ribosome: Not so physical, most of the time. J. Mol. Biol. 432, 3981–3986 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen M., Fredrick K., Measures of single- versus multiple-round translation argue against a mechanism to ensure coupling of transcription and translation. Proc. Natl. Acad. Sci. U.S.A. 115, 10774–10779 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li R., Zhang Q., Li J., Shi H., Effects of cooperation between translating ribosome and RNA polymerase on termination efficiency of the Rho-independent terminator. Nucleic Acids Res. 44, 2554–2563 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saxena S., et al., Escherichia coli transcription factor NusG binds to 70S ribosomes. Mol. Microbiol. 108, 495–504 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang J. Y., et al., Structural basis for transcript elongation control by NusG family universal regulators. Cell 173, 1650–1662.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burmann B. M., et al., A NusE:NusG complex links transcription and translation. Science 328, 501–504 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Larson M. H., et al., A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science 344, 1042–1047 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sherwood A. V., Henkin T. M., Riboswitch-mediated gene regulation: Novel RNA architectures dictate gene expression responses. Annu. Rev. Microbiol. 70, 361–374 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Widom J. R., et al., Ligand modulates cross-coupling between riboswitch folding and transcriptional pausing. Mol. Cell 72, 541–552.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gabizon R., Lee A., Vahedian-Movahed H., Ebright R. H., Bustamante C. J., Pause sequences facilitate entry into long-lived paused states by reducing RNA polymerase transcription rates. Nat. Commun. 9, 2930 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bastet L., et al., Translational control and Rho-dependent transcription termination are intimately linked in riboswitch regulation. Nucleic Acids Res. 45, 7474–7486 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mironov A. S., et al., Sensing small molecules by nascent RNA: A mechanism to control transcription in bacteria. Cell 111, 747–756 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Roth A., et al., A riboswitch selective for the queuosine precursor preQ1 contains an unusually small aptamer domain. Nat. Struct. Mol. Biol. 14, 308–317 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Greenlee E. B., et al., Challenges of ligand identification for the second wave of orphan riboswitch candidates. RNA Biol. 15, 377–390 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winkler W. C., Nahvi A., Roth A., Collins J. A., Breaker R. R., Control of gene expression by a natural metabolite-responsive ribozyme. Nature 428, 281–286 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Saito K., Green R., Buskirk A. R., Translational initiation in E. coli occurs at the correct sites genome-wide in the absence of mRNA-rRNA base-pairing. eLife 9, e55002 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suddala K. C., et al., Single transcriptional and translational preQ1 riboswitches adopt similar pre-folded ensembles that follow distinct folding pathways into the same ligand-bound structure. Nucleic Acids Res. 41, 10462–10475 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rinaldi A. J., Lund P. E., Blanco M. R., Walter N. G., The Shine-Dalgarno sequence of riboswitch-regulated single mRNAs shows ligand-dependent accessibility bursts. Nat. Commun. 7, 8976 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dorywalska M., et al., Site-specific labeling of the ribosome for single-molecule spectroscopy. Nucleic Acids Res. 33, 182–189 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chauvier A., et al., Transcriptional pausing at the translation start site operates as a critical checkpoint for riboswitch regulation. Nat. Commun. 8, 13892 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duss O., et al., Real-time assembly of ribonucleoprotein complexes on nascent RNA transcripts. Nat. Commun. 9, 5087 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mustoe A. M., Corley M., Laederach A., Weeks K. M., Messenger RNA structure regulates translation initiation: A mechanism exploited from bacteria to humans. Biochemistry 57, 3537–3539 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lund P. E., Chatterjee S., Daher M., Walter N. G., Protein unties the pseudoknot: S1-mediated unfolding of RNA higher order structure. Nucleic Acids Res. 48, 2107–2125 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Culver G. M., Meanderings of the mRNA through the ribosome. Structure 9, 751–758 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Rodgers M. L., Woodson S. A., Transcription increases the cooperativity of ribonucleoprotein assembly. Cell 179, 1370–1381.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi J., et al., Dynamics of the context-specific translation arrest by chloramphenicol and linezolid. Nat. Chem. Biol. 16, 310–317 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu M., Mori M., Hwa T., Dai X., Disruption of transcription-translation coordination in Escherichia coli leads to premature transcriptional termination. Nat. Microbiol. 4, 2347–2356 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harvey R. J., Koch A. L., How partially inhibitory concentrations of chloramphenicol affect the growth of Escherichia coli. Antimicrob. Agents Chemother. 18, 323–337 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uemura S., et al., Real-time tRNA transit on single translating ribosomes at codon resolution. Nature 464, 1012–1017 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Artsimovitch I., Knauer S. H., Ancient transcription factors in the news. MBio 10, e01547-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Belogurov G. A., et al., Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol. Cell 26, 117–129 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sevostyanova A., Belogurov G. A., Mooney R. A., Landick R., Artsimovitch I., The β subunit gate loop is required for RNA polymerase modification by RfaH and NusG. Mol. Cell 43, 253–262 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Main-Hester K. L., Colpitts K. M., Thomas G. A., Fang F. C., Libby S. J., Coordinate regulation of Salmonella pathogenicity island 1 (SPI1) and SPI4 in Salmonella enterica serovar Typhimurium. Infect. Immun. 76, 1024–1035 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Smit M. H., van Duin J., Translational standby sites: How ribosomes may deal with the rapid folding kinetics of mRNA. J. Mol. Biol. 331, 737–743 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Landick R., Wang D., Chan C. L., Quantitative analysis of transcriptional pausing by Escherichia coli RNA polymerase: His leader pause site as paradigm. Methods Enzymol. 274, 334–353 (1996). [DOI] [PubMed] [Google Scholar]
- 50.Frieda K. L., Block S. M., Direct observation of cotranscriptional folding in an adenine riboswitch. Science 338, 397–400 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lauber M. A., Rappsilber J., Reilly J. P., Dynamics of ribosomal protein S1 on a bacterial ribosome with cross-linking and mass spectrometry. Mol. Cell. Proteomics 11, 1965–1976 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dahlberg A. E., Dingman C. W., Peacock A. C., Electrophoretic characterization of bacterial polyribosomes in agarose-acrylamide composite gels. J. Mol. Biol. 41, 139–147 (1969). [DOI] [PubMed] [Google Scholar]
- 53.Dahlberg A. E., Two forms of the 30 S ribosomal subunit of Escherichia coli. J. Biol. Chem. 249, 7673–7678 (1974). [PubMed] [Google Scholar]
- 54.Blanchard S. C., Kim H. D., Gonzalez R. L. Jr, Puglisi J. D., Chu S., tRNA dynamics on the ribosome during translation. Proc. Natl. Acad. Sci. U.S.A. 101, 12893–12898 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roy R., Hohng S., Ha T., A practical guide to single-molecule FRET. Nat. Methods 5, 507–516 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aitken C. E., Marshall R. A., Puglisi J. D., An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J. 94, 1826–1835 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rasnik I., McKinney S. A., Ha T., Nonblinking and long-lasting single-molecule fluorescence imaging. Nat. Methods 3, 891–893 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Chauvier A., Cabello-Villegas J., Walter N. G., Probing RNA structure and interaction dynamics at the single molecule level. Methods 162-163, 3–11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blanco M., Walter N. G., Analysis of complex single-molecule FRET time trajectories. Methods Enzymol. 472, 153–178 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix. Study data have been deposited in the University of Michigan Deep Blue Data repository (DOI: https://doi.org/10.7302/dxhn-0q11).