Abstract
During the previous Ebola and Zika outbreaks, researchers shared their data, allowing many published epidemiological studies to be produced only from open research data, to speed up investigations and control of these infections. This study aims to evaluate the dissemination of the COVID-19 research data underlying scientific publications. Analysis of COVID-19 publications from December 1, 2019, to April 30, 2020, was conducted through the PubMed Central repository to evaluate the research data available through its publication as supplementary material or deposited in repositories. The PubMed Central search generated 5,905 records, of which 804 papers included complementary research data, especially as supplementary material (77.4%). The most productive journals were The New England Journal of Medicine, The Lancet and The Lancet Infectious Diseases, the most frequent keyword was pneumonia, and the most used repositories were GitHub and GenBank. An expected growth in the number of published articles following the course of the pandemics is confirmed in this work, while the underlying research data are only 13.6%. It can be deduced that data sharing is not a common practice, even in health emergencies, such as the present one. High-impact generalist journals have accounted for a large share of global publishing. The topics most often covered are related to epidemiological and public health concepts, genetics, virology and respiratory diseases, such as pneumonia. However, it is essential to interpret these data with caution following the evolution of publications and their funding in the coming months.
Keywords: COVID-19, Data sharing, Supplementary material, Repository, PubMed central
Introduction
Research data are a resource with great value, and their strengths and the benefits of sharing such data are firmly established (Krumholz, 2012; Molloy, 2011, p. krum; Sayogo & Pardo, 2013). Sharing data allows formulating new hypotheses, promoting new discoveries and confirming previous results (Alsheikh-Ali et al., 2011; Piwowar & Chapman, 2010). It also avoids the repetition of many experiments based on existing data, allowing resources to be allocated to other lines of research (Bertagnolli et al., 2017; Zhu, 2019).
There are currently a number of possibilities for sharing the data resulting from investigations. The most common and appreciated procedure used by researchers is the storage of research data as supplementary material together with the article in the publishers' platform (Tenopir et al., 2015). In parallel, some journals more frequently opt to deposit the data underlying the investigations in a recommended repository as part of the manuscript submission process (Federer et al., 2018; Springer Nature, 2020). These repositories must meet a series of requirements relating to access, preservation of data and endurance over time (Wilkinson et al., 2016). Currently, the best known repositories for biomedical researchers are the disciplinary repositories in biological sciences (GenBank, Protein Data Bank) or the health sciences (The Cancer Imaging Archive, Project Data Sphere, ClinicalTrials.gov) that refer to clinical data sets and preserve the anonymity of study. When no discipline-specific data repository is available, generalist repositories, such as the Dryad Digital Repository, Figshare, Harvard Dataverse, Open Science Framework, or Zenodo, are often recommended.
The health emergencies due to epidemic outbreaks caused by the Ebola and Zika viruses showed that to speed up investigations and control of these infections, it was essential that research data be shared quickly and widely. During the Ebola outbreak, researchers shared their epidemiological data and the genetic sequences of the virus in public repositories (Yozwiak et al., 2015), allowing many published epidemiological studies to be produced only from open research data. On the other hand, the fact that in some studies data were published before or at the time of publication strengthens the importance of investigating data-sharing practices during covid-19 (Chretien et al., 2015). With the Zika epidemic, major journals agreed that all Zika-related content should be openly accessible (Wellcome Trust, 2016). For both Ebola and Zika, scientists created public repositories to share data (cdcepi/zika, 2020; Rivers, 2020). Along these same lines of action, the dissemination of research data on COVID-19 cannot be diminished by classic impediments to data sharing, such as restrictions due to intellectual property, confidentiality problems and limitations of technical resources and humans (Chretien et al., 2016; van Panhuis et al., 2014; Whitty et al., 2015). Until now, some initiatives have been undertaken, such as the COVID-19 repository created by the Johns Hopkins University Center for Systems Science and Engineering (JHU CSSE), focused on epidemiological data (CSSEGISandData, 2020).
The objective of this study is based on the evaluation of COVID-19 research data published using the gold open access model through its dissemination as supplementary material or using the green open access model through deposition in repositories during the first five months of the pandemic.
Methods
Analysis of COVID-19 scientific publications through the PubMed Central (PMC) repository
The following search strategy includes the terms used by the World Health Organization (WHO) related to COVID-19 (WHO, 2020) to identify papers through PMC, the most used free full-text repository in biomedicine owned by the US National Institutes of Health, which serves both as an electronic journal platform for Gold Open Access articles provided by journal publishers and as a repository for Green Open Access articles as part of the Public Access Policy.
("2019-nCoV" OR "novel coronavirus" OR "Coronavirus disease 2019" OR "Coronavirus disease 19" OR "COVID-19" OR "COVID19" OR "SARS-CoV-2" OR "Severe acute respiratory syndrome coronavirus 2" OR "Wuhan coronavirus" OR "Wuhan virus" OR "Wuhan pneumonia").
Evaluation of published COVID-19 research data made available through dissemination as supplementary material and in repositories
To recover research data disseminated as supplementary material through scientific publications, the previous COVID-19 search equation was executed in PMC using the filter “Associated Data”. Publications with available research data through disciplinary and generalist repositories were retrieved combining the main COVID-19 search equation with AND ("accession number" OR "accession No" OR "repositor*" OR "data deposition" OR "data available" OR "gse" OR "GenBank" OR "Data bank" OR “Gene Expression Omnibus” OR “GitHub” OR “ArrayExpress” OR “Sequence Read Archive” OR BioProject OR “European Nucleotide Archive” OR “European Molecular Biology Laboratory” OR Zenodo OR Figshare OR Dryad).
The search was performed on May 4, 2020, and the selected period covered the five months from December 1, 2019, to April 30, 2020. The documents obtained were imported into Microsoft Access to create a database, and the following information was collected: publication data, document typology, journal title, keywords, number of authors, country of origin and financial support.
Statistical analysis
Quantitative analysis of the research data deposited as supplementary material was performed with SPSS 23.0 according to the different types of formats presented: image files, Excel or CSV tables, Word, pdf, ppt and multimedia files. Compressed files (.zip or.rar) were open to verify the content of the file types. All the associated files were assessed manually, and those files containing information not related to research, such as data availability statements, reference lists, etc., were discarded. In a complementary way, the main repositories used by researchers to deposit COVID-19 research data and named in the papers were counted and classified.
Qualitative variables were presented as absolute values and percentages, whereas quantitative data as means and standard deviations (SD). Differences of bimonthly means of articles per journal and per country were analyzed by the related-samples Wilcoxon signed rank test, giving that, once assessed by the Kolmogorov–Smirnov test, these samples did not follow a normal distribution. A p value of ≤ 0.05 was considered statistically significant.
Additionally, a qualitative study of the content of the research data was performed carrying out an analysis of the keywords present in the articles.
Results
The PMC search related to COVID-19 generated a total of 5,905 records, of which 1,132 papers had associated data and, after discarding those articles containing files with no relevant research information, a final sample of 804 papers (13.6% of the total) included underlying research data. From these 804 works, a 77.4% contained supplementary material. The analysis of publications by fortnights from December 1 to April 31 showed that no articles were published until the 2nd fortnight of January (n = 27), with the first document a letter published by the New England Journal of Medicine on January 18, and this value increased to 237 documents during the 2nd fortnight of April. The percentage of funded articles out of the total showed an oscillating evolution, with values of 63%, 49% and 55% during January, February and March, but in April, this percentage descended to percentages between 38 and 41% by fortnight (Fig. 1).
Fig. 1.
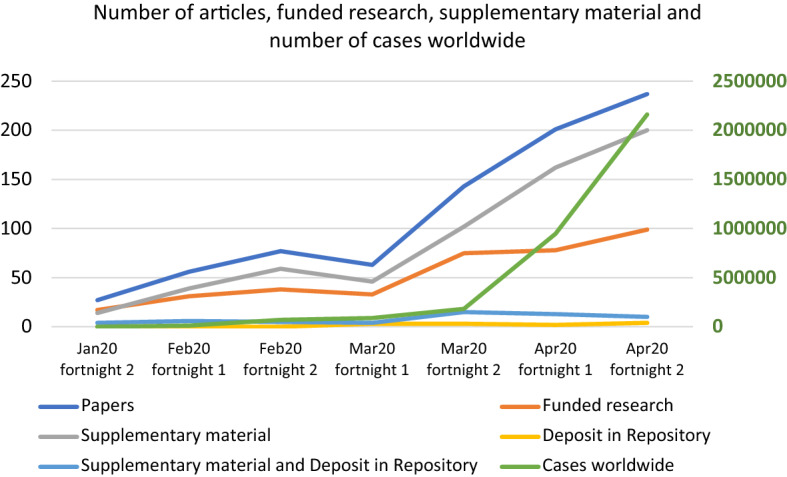
Number of articles, funded research, supplementary material and number of cases worldwide
Table 1 shows the evolution of the publication on COVID-19 pandemics. The most productive journals were the New England Journal of Medicine, Lancet and Lancet Infectious Diseases, publishing 39, 32 and 25 articles, respectively, and the most used repositories were GitHub (21 appearances) and GenBank (20 appearances). A total of 68.8% of the retrieved documents were journal articles, and 20.4% corresponded to letters, while the rest of the typologies were less representative.
Table 1.
Evolution from January to April 2020 of the research data publication on COVID-19 pandemics in the form of supplementary material or deposited in repositories
| Fortnight | Total papers | Journal name | Total papers | Key words | Total papers | Repository | Total papers |
|---|---|---|---|---|---|---|---|
| Jan20 fortnight 2 | 27 | N Engl J Med | 4 | Genome | 4 | GenBank | 3 |
| The Lancet | 4 | MERS–CoV; Wuhan | 3 | ||||
| Emerg Microbes Infect | 4 | Bats; Diagnosis; Emerging; Emerging infectious disease; Pneumonia; Polymerase chain reaction (PCR); Virus | 2 | ||||
| Feb20 fortnight 1 | 56 | J Med Virol | 4 | MERS–CoV | 6 | GenBank | 3 |
| N Engl J Med; Lancet; Euro Surveill; J Virol | 3 | Zoonosis | 5 | BioProject; Github; Sequence Read Archive (SRA) | 2 | ||
| Epidemiology; Pneumonia; SARS-CoV; Wuhan; Antiviral Therapy | 4 | ||||||
| Feb20 fortnight 2 | 77 | The Lancet | 7 | Pneumonia | 7 | Github | 4 |
| The Lancet Infect Dis | 6 | SARS-CoV | 6 | ||||
| J Clin Med | 6 | Epidemic | 5 | ||||
| Mar20 fortnight 1 | 63 | J Infect | 4 | SARS-CoV | 5 | Protein Data Bank (PDB); Electron Microscopy Data Bank (EMDB); GenBank; GISAID | 2 |
| The Lancet | 3 | Spike protein | 4 | ||||
| The Lancet Infect Dis | 3 | Angiotensin converting enzyme 2 (ACE2); MERS–CoV; Pneumonia | 3 | ||||
| Mar20 fortnight 2 | 143 | N Engl J Med | 8 | Pneumonia | 7 | Github | 6 |
| The Lancet Infect Dis | 7 | spike protein | 6 | Gene Expression Omnibus (GEO) | 3 | ||
| Emerg Microbes Infect | 6 | Angiotensin converting enzyme 2 (ACE2); Molecular docking; Pandemics | 5 | ||||
| Apr20 fortnight 1 | 201 | N Engl J Med | 14 | Pandemics | 9 | Github | 4 |
| Eur Heart J | 7 | Outbreak; Public health | 7 | GenBank | 3 | ||
| Int J Nurs Sci | 7 | Polymerase chain reaction (PCR) | 6 | Protein Data Bank (PDB); GISAID | 2 | ||
| Apr20 fortnight 2 | 237 | J Arthroplasty | 12 | Pandemics | 13 | Github | 4 |
| The Lancet | 11 | Antiviral Therapy; Arthroplasty; Clinical characteristics; Diagnosis; E-health; Infectious disease; Statistic model; Therapy | 5 | GenBank | 4 | ||
| The Lancet Infect Dis | 7 | Molecular dynamics; Mortality; Orthopaedic; Pneumonia; Psychological disorders; Public health; SARS; Spike protein; Surveillance | 4 | GISAID; Mendeley Data | 2 |
The retrieved works were published in a total of 335 journals, with a mean number of articles per journal of 2.40 ± 3.80 (range 1–38). The production increased significantly with a statistical difference between the articles/journal mean of the January–February period (0.48 ± 1.41) and the mean of March–April period (1.92 ± 2.83) (Related-samples Wilcoxon signed rank test, p = 0.0001) (Fig. 2).
Fig. 2.
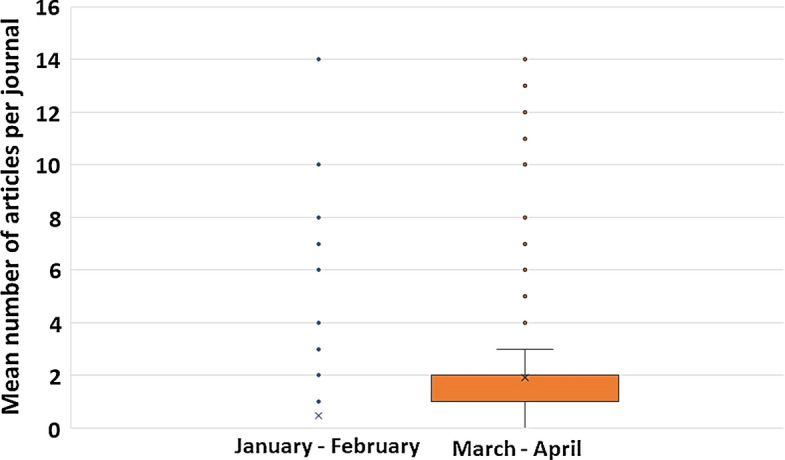
Bimonthly mean production of articles per journal. Related-samples Wilcoxon signed rank test: p = 0.0001
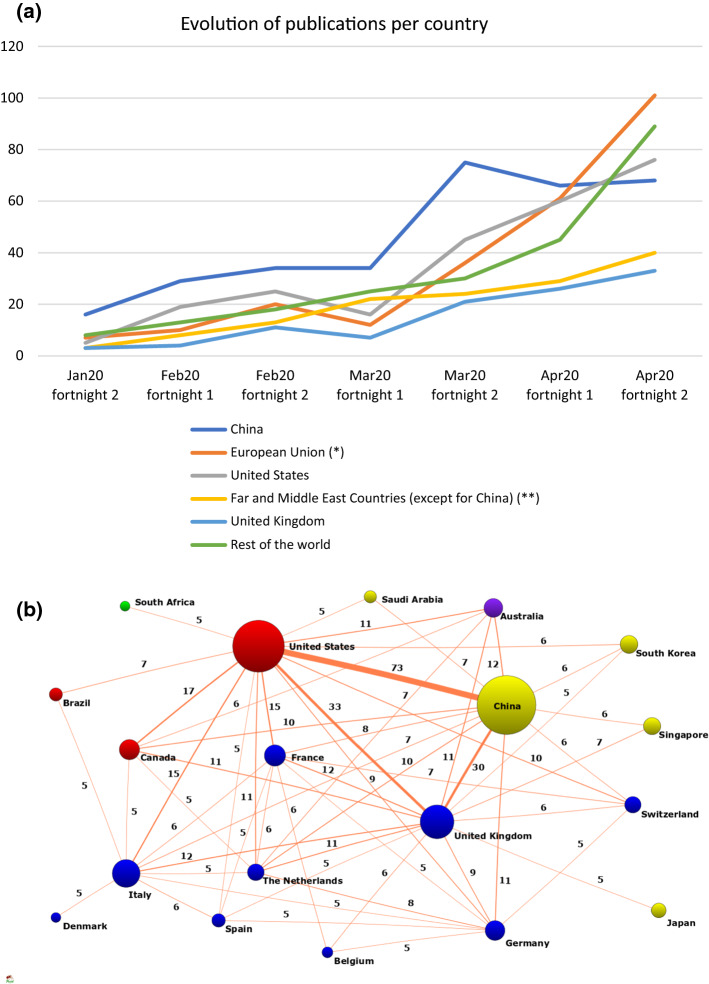
The evolution of the publication of articles during the pandemics per country showed that of the 27 papers published in January, China participated in 16, and its production grew exponentially until the second fortnight of March, while in the same period, the United States of America (USA) took part in 5 papers and the United Kingdom (UK) and France in 3 papers. Nevertheless, the production of the European Union (EU) countries started to increase since the first fortnight of March, and at the end of April, the EU countries, especially Italy (71), France (40) and Germany (36), became the most productive. The USA followed a similar pace, and the Far and Middle East countries experienced milder growth, as did the UK (Fig. 3a).
Fig. 3.
a Evolution of publications per authors’ country of affiliation. b International collaboration network. (The size of the node is proportional to the number of documents generated by each country. The color of the nodes is the same in all countries belonging to the same continent. The width of the lines represents the numbers papers in collaboration between the countries.)
Figure 3b shows the international collaboration, where the size of the spheres represents the number of works with the participation of one country. China, with 323 authorships (39.68%), followed by the USA with 249 (30.58%) and the UK (n = 106, 13.02%), are the most productive countries in this study. The colour of the spheres identifies the continent where a country is located, and the thickness of the lines shows the degree of collaboration. China and the USA have the most intense collaboration (73 works in common).
Regarding the production per country, 88 different countries published COVID-19 related scientific data, with a mean number of articles per country of 14.60 ± 44.46 (range 1–322). The production per country also increased, with a significant difference between the articles/country mean of the January–February period (2.78 ± 10.05) and the March–April mean (11.82 ± 34.64) (Related-samples Wilcoxon signed rank test, p = 0.0001) (Fig. 4).
Fig. 4.
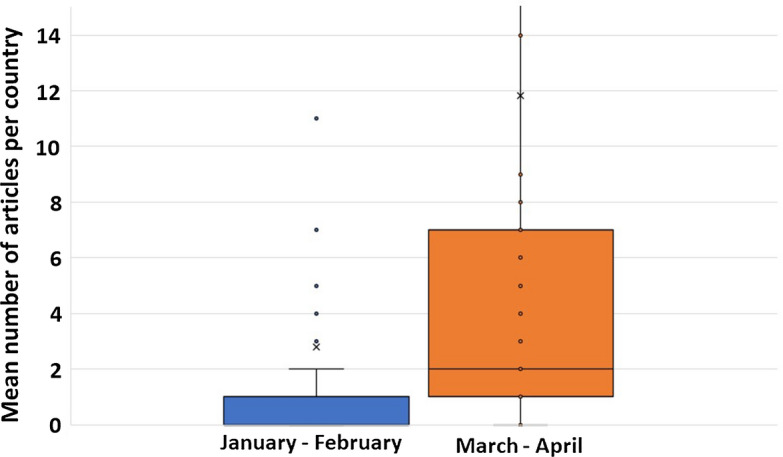
Bimonthly mean production of articles per country. Related-samples Wilcoxon signed rank test: p = 0.0001
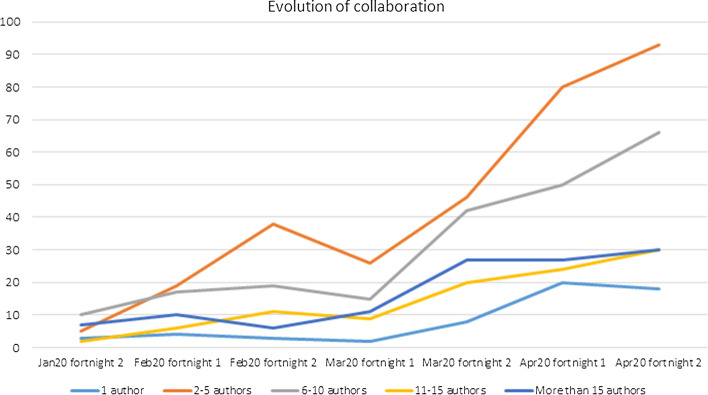
The study of the degree of collaboration between authors shows that most of the articles were signed by 2 to 5 authors, followed by 6 to 10, and Fig. 5 shows that this difference is growing. Nevertheless, papers signed by a single author are scarce (Fig. 5). Regarding the journal articles, there is one document signed by 127 authors, 54 documents signed by 4 authors, 55 documents signed by 5 authors and 31 articles signed by a single author. However, the letters are also signed by quite a few authors; there are 21 letters signed by 3 authors, 22 signed by 5 authors and 8 letters signed by 12 authors (Lucas-Dominguez et al., 2020).
Fig. 5.
Evolution of collaboration according to the number of authors signing the papers
The analysis of supplementary material showed that the most frequent types of files were mainly PDF and DOC, which were found in 42.75% and 32.56% of the articles, respectively (Table 2). These files were mostly figures and tables related to DNA, RNA, protein sequence analysis and molecular signalling pathways; other frequently observed documents are checklists, clinical protocols and informed consents.
Table 2.
Type of files of supplementary materials analysed
| Type of files | N° papers | N° files | Average of files/articles | % articles /total | % files /total |
|---|---|---|---|---|---|
| 348 | 530 | 1.52 | 42.75% | 41.31% | |
| DOC/DOCX | 265 | 394 | 1.49 | 32.56% | 30.71% |
| JPEG/JPG/TIF/TIFF/PNG/GIF | 31 | 79 | 2.55 | 3.81% | 6.16% |
| XLS/XLSX | 65 | 110 | 1.69 | 7.99% | 8.57% |
| MOV/MP4/WMV/MPG/AVI | 25 | 45 | 1.80 | 3.07% | 3.51% |
| PPT/PPTX | 10 | 12 | 1.20 | 1.23% | 0.94% |
| EPS | 1 | 2 | 2.00 | 0.12% | 0.16% |
| HTML | 2 | 3 | 1.50 | 0.25% | 0.23% |
| CSV | 8 | 45 | 5.63 | 0.98% | 3.51% |
| XML/GZ | 29 | 32 | 1.10 | 3.56% | 2.49% |
| TXT/PL | 1 | 1 | 1.00 | 0.12% | 0.08% |
| M | 1 | 3 | 3.00 | 0.12% | 0.23% |
| MAT | 1 | 4 | 4.00 | 0.12% | 0.31% |
| SuppDOI | 10 | 10 | 1.00 | 1.23% | 0.78% |
| 7z | 1 | 1 | 1.00 | 0.12% | 0.08% |
| FLV | 3 | 3 | 1.00 | 0.37% | 0.23% |
Discussion
This study has revealed the availability of research data published as supplementary material or deposited in repositories from the articles on COVID-19 indexed in PMC, as well as the evolution, documentary typology, subject matter and financing during the first months of the pandemic. The visualization by fortnights showed an increase in the scientific production of COVID-19 related studies. To statistically demonstrate this increase, we perform a bimonthly comparison since, as expected after the declaration of this crisis as an official pandemics, March 2020 supposed a critical moment, which demonstrated empirically the high speed of response of research worldwide. Nevertheless, despite the extraordinary number of articles published and their continuous increase, only 13.6% contained supplementary material or data deposited in repositories. Research data sharing has evolved equally in case of the attachment as supplementary material of the article, while the deposit of material in a repository was invariable over time. While the number of articles has increased exponentially since the 2nd fortnight of March 2020, coinciding with the increase in the number of cases confirmed by COVID-19 as the pandemic progresses, the response of funding has grown modestly, although we need to keep in mind that the results of these works may not yet have been published as their funding began in early 2020. In addition to the low percentage of deposited data, it has been reported that patient-level COVID-19 data is not publicly available. In the current era of global interaction via the Internet, it would be desirable that electronic patient records, conveniently anonymized, were also available to researchers (Rios et al., 2020).
In parallel to the growth in the number of articles, numerous journals have published related articles (n = 335). High-impact generalist journals (such as N Engl J Med, Lancet and Science) have published more articles than journals on infectious diseases, public health, critical care medicine and respiratory systems. In total, 20.40% of published articles are letters, reviews (6.47%) or editorials (2.99%), without appreciating changes in the percentages of letters regarding journal articles (68,78%) throughout the fortnights (ranging between 20 and 30%). In a previous study, only 46% of papers were found to be articles or reviews (Aleixandre-Benavent et al., 2020), and in another study that analyzed the open data in 140 articles from five high impact journals, most of the published papers were opinion papers, case reports, and reviews (Gkiouras et al., 2020). It seems that all journals have something to convey to their readers regarding COVID-19. However, the information disseminated through peer-reviewed journals and the data sets published as supplementary material or deposited in online repositories are both vital for researchers and decision-makers (Dye et al., 2016; Modjarrad et al., 2016; Whitty et al., 2015).
The frequency and evolution of the keywords collected by fortnights shows the chronology of the main events that have marked this health crisis. On December 31, the WHO was alerted of a cluster of pneumonia cases of unknown aetiology in Wuhan. The 1st fortnight of January, MERS, SARS, and influenza viruses were ruled out as causative pathogens of this emerging outbreak, and the origin of the zoonosis in a bat was investigated as a reservoir. China publicly shared the gene sequence of the novel coronavirus SARS-CoV-2, establishing polymerase chain reaction diagnostic testing. During the 2nd half of January, the National Genomics Data Center of China launched the 2019 novel coronavirus database to release the genome of SARS-CoV-2, and the NIH started working on vaccines. On January 30, the WHO declared the COVID-19 outbreak a Public Health Emergency of International Concern. The 1st fortnight of February, the disease was officially called COVID-19, and scientific researchers discovered the SARS-CoV-2 spike protein binding to its human cell receptor protein called ACE2 (Scudellari, 2020). The 1st fortnight of March, the WHO declared a pandemic and announced that no pharmaceutical therapies had yet been shown to be safe and effective for the treatment of COVID-19 (Li et al., 2020; WHO, 2020). Following these events, some words are related to the specific topic of the journals, but more general terms stand out in almost all journals: epidemiological concepts (such as pandemics and outbreak), respiratory diseases (such as pneumonia and SARS-CoV, since the lung is a target organ in this infection), biological markers (such as spike proteins, ACE2, polymerase chain reaction), virology and genetics (genome of the virus). Since the articles analysed were published at the beginning of the outbreak, it is possible that a large part of them are notifications and reports of the place where the outbreak occurred, laboratory data, information obtained from previous outbreaks with similar organisms, mechanisms of transmission, natural history of infection, populations at risk, treatments being used to control the disease, diagnostic tests and genetic sequence information of the virus. Although early case studies of COVID-19 usually contain few patients, these are very important because they contain critical information about the contacts that transmitted the infection and those that the patient had subsequently, allow estimation of incubation periods, describe clinical manifestations, provide key laboratory and radiological information and facilitates decision making in concomitant diseases (Heymann, 2020).
A significant feature of the modern response to epidemics is the ability to efficiently exploit all available data, which can facilitate evidence-based research and decision-making (Campos et al., 2015; Cori, 2017; WHO Ebola Response Team, 2014). For example, analysis of data on epidemics can be used to predict outbreaks in other regions and the most significant factors associated with the disease, such as biological, environmental and climatic factors (humidity, temperature and rainfall), quality housing, transport conditions and population density, among others (Wu et al., 2018). However, experiences with data storage and use in epidemics have not always been conclusive. Thus, it has been reported that data sharing was important during the influenza virus A subtype H1N1 epidemic of 2009; however, it was not as significant with the Middle East Respiratory Syndrome (MERS) epidemic, first reported in Saudi Arabia in 2012, and the Ebola epidemics in 2014–2016, which revealed gaps in the open availability of the genetic sequences of the virus (Yozwiak et al., 2015). The Ebola, dengue and Zika epidemics have evidenced the need to develop infrastructures for the proper management of data of interest to public health (D’Agostino et al., 2018), as well as the establishment of codes of conduct that should govern the exchange of data on new biological threats (Capua, 2016).
The geographical production of papers has followed a pace very much in line with the expansion of the disease, with China leading the publication in the first fortnight of January and growing until March and being overtaken by the EU and USA in April, coinciding with the virus expansion in these areas.
The analysis of supplementary material showed that three-quarters of the documents were PDF and DOC, containing mostly textual or graphic materials complementary to the research, and a percentage that barely reached 10% (73 papers) were files with reusable data formats (xls and csv), which is equivalent to 1.2% of the 5,905 records published and analysed in this work. Furthermore, 68.78% of the documents retrieved in this work were originals or revisions (6.47%), which could include underlying research data. It is not unreasonable to say that this is a very low percentage that does not respond to recommendations and calls for sharing research data. The previous study by Gkiouras et al., (2020) also showed a low percentage, since only one out of the 140 articles on Covid-19 published in high-impact journals (0.7%), provided complete open data. This percentage may be diminished by classic impediments to data sharing, such as restrictions due to intellectual property, confidentiality problems, limitations of technical resources and humans, and the lack of incentives for researchers who deposit their data. Co-authoring an article and being cited frequently are often the only rewards for sharing information that can take years to collect and months of hard work to select. In short, we are in an era in which the scientific community is still debating the pros and cons of data sharing (Aleixandre-Benavent et al., 2018; Chretien et al., 2016; Sixto-Costoya et al., 2020; Vidal-Infer et al., 2019; Walport & Brest, 2011). This low percentage does not extend to gene sequence repositories, where scientists often share the sequences at the same time as they are discovered (Chretien et al., 2016; Pham-Kanter et al., 2014).
In global public health emergencies, it should be mandatory to disseminate any information that may be of value in fighting the crisis. For this to be done efficiently, there is a need to develop agreed global standards for sharing data and results for scientists, institutions and governments (Capua, 2016; McNutt, 2016; Modjarrad et al., 2016). Establishing data sharing as the gold standard of any published work may be crucial to contain the current and future possible health emergencies that may come from emerging biological threats. To overcome existing misgivings about data sharing, embargo periods could be established, but these should not delay data use, and codes of conduct for data on epidemics should be established (Capua, 2016; Research Data Alliance, 2020). The International Committee of Medical Journal Editors confirmed that the pre-publication dissemination of critical public health information in the context of WHO-declared health emergencies will not prejudice the publication of works (ICMJE 2020). An example of the importance of the data repository for some government institutions is provided by the NIH strategic plan for the decade 2017–2027, which involves improvements in data infrastructures, integration of individual data sets, new tools and resources for data analysis, and incorporation of FAIR principles (findability, accessibility, interoperability and reusability) (Wilkinson et al., 2016; National Institutes of Health Office of Strategic Coordination, 2020).
Accelerated reporting and data repository should include both clinical trials and epidemiological surveillance studies, observational studies, information on the virus and its genetic sequences, and disease control programmes (Moorthy et al., 2020). These emerging data, properly integrated, allow for the refinement of risk assessment and the provision of recommendations to countries for the management of the epidemic (Heymann, 2020; Yozwiak et al., 2015). To facilitate research on the disease, journals and publishers around the world issued a joint statement promising full cooperation in data exchange. These measures include, in addition to providing open access to all peer-reviewed research, sharing research findings prior to peer review, including protocols, results, and data (“Calling all coronavirus researchers”, 2020). However, the emergency initiative to share research findings prior to peer review by means of preprints implies a reduction in quality control that can lead to the dissemination of erroneous information. Some data series may have errors, which could generate misleading conclusions, with the consequences that this may have for the health of the population (Rinott et al., 2020). Therefore, researchers who reuse Covid-19 data should be cautious because the fact that they are findable does not guarantee their quality. It has been reported that some of the first published articles on COVID-19 had to be withdrawn because of quality issues, and the Retraction Watch blog has created a special list for COVID-19 publications (Retraction Watch, 2020).
Limitations and future work
We have analysed only papers on COVID-19 indexed in PMC, so additional documents included in other repositories may have been omitted. Future work should look at other possible literature sources and explore whether funding had a positive effect on the publication and storage of free reusable data.
Conclusion
During the current pandemic, there has been a massive publication of articles, and many journals have released them for open access. However, the deposit of supplementary material and data in repositories amounts to only 13.6%, and reusable data reaches just 1.2%. From these percentages, it can be deduced that data sharing is not a common practice, even in health emergencies, such as the present one. Therefore, greater awareness and more efficient infrastructures are necessary. High-impact generalist journals have accounted for a large share of global publishing. The topics most often covered are related to epidemiological and public health concepts, genetics, virology and respiratory diseases, such as pneumonia. However, it is essential to interpret these data with caution and to follow the evolution of publications and their funding in the coming months.
Acknowledgements
Authors would like to thank Dr. Daniel López-Padilla for his valuable statistical assistance to this work.
Author contributions
All listed authors meet ICMJE requirements: (1) Substantial contributions to the conception or design of the work, or the acquisition, analysis, or interpretation of data for the work; AND (2) Drafting the work or revising it critically for important intellectual content; AND (3) Final approval of the version to be published; AND (4) Agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Funding
This work benefited from assistance by Spanish Ministry of Science and Innovation (PID2019-105708RB-C22, PID2019-108579RB-I00 and BES-2016–079394) and the CIBERONC (CB16/12/00350).
Data availability
The data generated and used during this research are openly available from Zenodo.org public repository at https://doi.org/10.5281/zenodo.3967025.
Declarations
Conflict of interest
The authors report no conflicts of interest.
References
- Aleixandre-Benavent R, Castelló-Cogollos L, Valderrama-Zurián J-C. Information and communication during the early months of Covid-19. Infodemics, misinformation and the role of information professionals. El profesional de la información. 2020 doi: 10.3145/epi.2020.jul.08. [DOI] [Google Scholar]
- Aleixandre-Benavent R, Lucas-Domínguez R, Sixto-Costoya A, Vidal-Infer A. The sharing of research data in the cell & tissue engineering area: Is It a common practice? Stem Cells and Development. 2018;27(11):717–722. doi: 10.1089/scd.2018.0036. [DOI] [PubMed] [Google Scholar]
- Alsheikh-Ali AA, Qureshi W, Al-Mallah MH, Ioannidis JPA. Public availability of published research data in high-impact journals. PLoS ONE. 2011;6(9):e24357. doi: 10.1371/journal.pone.0024357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertagnolli MM, Sartor O, Chabner BA, Rothenberg ML, Khozin S, Hugh-Jones C, et al. Advantages of a truly open-access data-sharing model. The New England Journal of Medicine. 2017;376(12):1178–1181. doi: 10.1056/NEJMsb1702054. [DOI] [PubMed] [Google Scholar]
- Calling all coronavirus researchers: keep sharing, stay open. (2020). Nature, 578(7793), 7. https://doi.org/10.1038/d41586-020-00307-x. [DOI] [PubMed]
- Campos GS, Bandeira AC, Sardi SI. Zika virus outbreak, Bahia Brazil. Emerging Infectious DiseasesEmerging Infectious Diseases. 2015;21:1885–1886. doi: 10.3201/eid2110.150847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capua I. A code of conduct for data on epidemics. Nature. 2016;534(7607):326–326. doi: 10.1038/534326c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- cdcepi/zika. (2020). HTML, CDC Epidemic Prediction Initiative. https://github.com/cdcepi/zika. Accessed 19 July 2020.
- Chretien J-P, Riley S, George DB. Mathematical modeling of the West Africa Ebola epidemic. eLife. 2015;4:e09186. doi: 10.7554/eLife.09186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chretien J-P, Rivers CM, Johansson MA. Make Data Sharing Routine to Prepare for Public Health Emergencies. PLoS medicine. 2016;13(8):e1002109. doi: 10.1371/journal.pmed.1002109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cori A, et al. Key data for outbreak evaluation: building on the Ebola experience. Philosophical Transactions of the Royal Society B. 2017;372:20160371. doi: 10.1098/rstb.2016.0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CSSEGISandData. (2020). CSSEGISandData/COVID-19. https://github.com/CSSEGISandData/COVID-19. Accessed 19 July 2020.
- D'Agostino M, Samuel NO, Sarol MJ, de Cosio FG, Marti M, Luo T, Brooks I, Espinal M. Open data and public health. Revista Panamericana de Salud Publica. 2018;42:e66. doi: 10.26633/RPSP.2018.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye C, Bartolomeos K, Moorthy V, Kieny MP. Data sharing in public health emergencies: A call to researchers. Bulletin of the World Health Organization. 2016;94(3):158. doi: 10.2471/BLT.16.170860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federer LM, Belter CW, Joubert DJ, Livinski A, Lu Y-L, Snyders LN, Thompson H. Data sharing in PLOS ONE: An analysis of data availability statements. PLoS ONE. 2018;13(5):e0194768. doi: 10.1371/journal.pone.0194768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkiouras K, Nigdelis MP, Grammatikopoulou MG, Goulis DG. Tracing open data in emergencies: The case of the COVID-19 pandemic. European Journal of Clinical Investigation. 2020;50:e13323. doi: 10.1111/eci.13323Heymann. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann DL. Data sharing and outbreaks: Best practice exemplified. Lancet. 2020;395(10223):469–470. doi: 10.1016/S0140-6736(20)30184-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ICMJE. (2020). Recommendations for the conduct, reporting, editing, and publication of scholarly work in medical journals. ICMJE. http://www.icmje.org/recommendations/browse/publishing-and-editorial-issues/overlapping-publications.html. Accessed 19 July 2020. [PubMed]
- Krumholz HM. Open science and data sharing in clinical research basing informed decisions on the totality of the evidence. Circulation-Cardiovascular Quality and Outcomes. 2012;5(2):141–142. doi: 10.1161/CIRCOUTCOMES.112.965848. [DOI] [PubMed] [Google Scholar]
- Lucas-Dominguez, R., Alonso-Arroyo, A., Vidal-Infer, A., & Aleixandre-Benavent, R. (2020, July 30). Raw data belonged to the study: The sharing of research data facing the COVID-19 pandemic. Zenodo. 10.5281/zenodo.3967025. [DOI] [PMC free article] [PubMed]
- Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, et al. Early transmission dynamics in wuhan, china, of novel coronavirus-infected pneumonia. New England Journal of Medicine. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNutt M. Data sharing. Science. 2016;351(6277):1007–1007. doi: 10.1126/science.aaf4545. [DOI] [PubMed] [Google Scholar]
- Modjarrad K, Moorthy VS, Millett P, Gsell P-S, Roth C, Kieny M-P. Developing global norms for sharing data and results during public health emergencies. Plos Medicine. 2016;13(1):e1001935. doi: 10.1371/journal.pmed.1001935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molloy JC. The open knowledge foundation: Open data means better science. Plos Biology. 2011;9(12):e1001195. doi: 10.1371/journal.pbio.1001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorthy V, Restrepo AMH, Preziosi M-P, Swaminathan S. Data sharing for novel coronavirus (COVID-19) Bulletin of the World Health Organization. 2020;98(3):150–150. doi: 10.2471/BLT.20.251561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institutes of Health Office of Strategic Coordination. NIH strategic plan for data science (2020). Cited 2020 January, 16. Available from: https://datascience.nih.gov/sites/default/files/NIH_Strategic_Plan_for_Data_Science_Final_508.pdf.
- Pham-Kanter G, Zinner DE, Campbell EG. Codifying collegiality: Recent developments in data sharing policy in the life sciences. PLoS ONE. 2014;9(9):e108451. doi: 10.1371/journal.pone.0108451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwowar HA, Chapman WW. Public sharing of research datasets: A pilot study of associations. Journal of Informetrics. 2010;4(2):148–156. doi: 10.1016/j.joi.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Research Data Alliance. (2020, June 30). The Value of RDA for COVID-19. RDA. https://www.rd-alliance.org/value-rda-covid-19. Accessed 19 July 2020.
- Retraction Watch. Retracted coronavirus (COVID-19) papers. Cited 2020 January, 16. Available from: https://retractionwatch.com/retracted-coronavirus-covid-19-papers/.
- Rinott E, Kozer E, Shapira Y, Bar-Haim A, Youngster I. Ibuprofen use and clinical outcomes in COVID-19 patients. Clinical Microbiology and Infection: The Official Publication of the European Society of Clinical Microbiology and Infectious Diseases. 2020;26(9):1259.e5–1259.e7. doi: 10.1016/j.cmi.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios RS, Zheng KI, Zheng M-H. Data sharing during COVID-19 pandemic: What to take away. Expert Review of Gastroenterology & Hepatology. 2020;14(12):1125–1130. doi: 10.1080/17474124.2020.1815533. [DOI] [PubMed] [Google Scholar]
- Rivers, C. (2020). cmrivers/ebola. PHP. https://github.com/cmrivers/ebola. Accessed 19 July 2020
- Sayogo DS, Pardo TA. Exploring the determinants of scientific data sharing: Understanding the motivation to publish research data. Government Information Quarterly. 2013;30:S19–S31. doi: 10.1016/j.giq.2012.06.011. [DOI] [Google Scholar]
- Scudellari M. Coronavirus piece by piece. Nature. 2020;581(7808):252–255. doi: 10.1038/d41586-020-01444-z. [DOI] [PubMed] [Google Scholar]
- Sixto-Costoya A, Aleixandre-Benavent R, Lucas-Dominguez R, Vidal-Infer A. Title: The emergency medicine facing the challenge of open science. Data. 2020;5:28. doi: 10.3390/data5020028. [DOI] [Google Scholar]
- Springer Nature. (2020). Recommended Data Repositories. Scientific Data. https://www.nature.com/sdata/policies/repositories. Accessed 19 July 2020.
- Tenopir C, Dalton ED, Allard S, Frame M, Pjesivac I, Birch B, et al. Changes in data sharing and data reuse practices and perceptions among scientists worldwide. PLoS ONE. 2015;10(8):e0134826. doi: 10.1371/journal.pone.0134826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Panhuis WG, Paul P, Emerson C, Grefenstette J, Wilder R, Herbst AJ, et al. A systematic review of barriers to data sharing in public health. BMC Public Health. 2014;14:1144. doi: 10.1186/1471-2458-14-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal-Infer A, Aleixandre-Benavent R, Lucas-Domínguez R, Sixto-Costoya A. The availability of raw data in substance abuse scientific journals. Journal of Substance Use. 2019;24(1):36–40. doi: 10.1080/14659891.2018.1489905. [DOI] [Google Scholar]
- Walport M, Brest P. Sharing research data to improve public health. Lancet. 2011;377(9765):537–539. doi: 10.1016/S0140-6736(10)62234-9. [DOI] [PubMed] [Google Scholar]
- Wellcome Trust. (2016). Sharing data during Zika and other global health emergencies | Wellcome. https://wellcome.ac.uk/news/sharing-data-during-zika-and-other-global-health-emergencies. Accessed 19 July 2020.
- Whitty CJM, Mundel T, Farrar J, Heymann DL, Davies SC, Walport MJ. Providing incentives to share data early in health emergencies: the role of journal editors. Lancet. 2015;386(10006):1797–1798. doi: 10.1016/S0140-6736(15)00758-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. (2020). Coronavirus Disease (COVID-19) - events as they happen. https://www.who.int. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/events-as-they-happen. Accessed 19 July 2020.
- WHO Ebola Response Team Ebola virus disease in West Africa–the first 9 months of the epidemic and forward projections. New England Journal of Medicine. 2014;371:1481–1495. doi: 10.1056/NEJMoa1411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson MD, Dumontier M, Aalbersberg IJJ, Appleton G, Axton M, Baak A, et al. The FAIR Guiding Principles for scientific data management and stewardship. Scientific Data. 2016;3:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Kao SC, Shih CH, Kan MH. Open data mining for Taiwan's dengue epidemic. Acta Tropica. 2018;183:1–7. doi: 10.1016/j.actatropica.2018.03.017. [DOI] [PubMed] [Google Scholar]
- Yozwiak NL, Schaffner SF, Sabeti PC. Make outbreak research open access. Nature. 2015;518(7540):477–479. doi: 10.1038/518477a. [DOI] [PubMed] [Google Scholar]
- Zhu Y. Open-access policy and data-sharing practice in UK academia. Journal of Information Science. 2019 doi: 10.1177/0165551518823174. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data generated and used during this research are openly available from Zenodo.org public repository at https://doi.org/10.5281/zenodo.3967025.