Significance
Glyphosate is the world’s dominantly used herbicide to control weedy plant species in a wide range of situations, especially in global field crops of soybean, maize, canola, and cotton with genetically engineered glyphosate resistance. Persistent glyphosate selection has led to worldwide evolution of glyphosate-resistant weeds. Several biochemical and physiological mechanisms have been identified that endow glyphosate resistance. To be toxic to plants, glyphosate must be present in the cytoplasm, and thus mechanisms reducing the cytoplasmic glyphosate to a sublethal level could confer resistance. Here, we provide evidence of a plant ABC transporter (ABCC8) that likely serves as a plasma membrane glyphosate exporter, lowering the cytoplasmic glyphosate level and thereby endowing glyphosate resistance.
Keywords: Echinochloa colona, glyphosate resistance, ABC transporter, glyphosate exporter, plasma membrane
Abstract
Glyphosate is the most widely used herbicide in world agriculture and for general vegetation control in a wide range of situations. Global and often intensive glyphosate selection of very large weedy plant populations has resulted in widespread glyphosate resistance evolution in populations of many weed species. Here, working with a glyphosate-resistant (GR) Echinochloa colona population that evolved in a Western Australia agricultural field, we identified an ATP-binding cassette (ABC) transporter (EcABCC8) that is consistently up-regulated in GR plants. When expressed in transgenic rice, this EcABCC8 transporter endowed glyphosate resistance. Equally, rice, maize, and soybean overexpressing the EcABCC8 ortholog genes were made resistant to glyphosate. Conversely, CRISPR/Cas9-mediated knockout of the EcABCC8 ortholog gene OsABCC8 increased rice susceptibility to glyphosate. Subcellular localization analysis and quantification of glyphosate cellular levels in treated ABCC8 transgenic rice plants and isolated leaf protoplasts as well as structural modeling support that EcABCC8 is likely a plasma membrane–localized transporter extruding cytoplasmic glyphosate to the apoplast, lowering the cellular glyphosate level. This is a report of a membrane transporter effluxing glyphosate in a GR plant species, and its function is likely conserved in crop plant species.
Glyphosate is the world’s most widely used herbicide (1, 2), lethal to a wide range of plant species, both annual and perennial. Around one million tons of glyphosate is being used annually for weed control across the world, especially because of high adoption of glyphosate-resistant (GR) transgenic crops. Consequently, glyphosate selection on huge numbers of genetically diverse weedy plant species has resulted in glyphosate resistance evolution, now known in 48 GR weed species worldwide (3).
Glyphosate specifically inhibits the plastidic 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS), disrupting the biosynthesis of aromatic amino acids (4). In evolved GR weeds, resistance can be due to specific EPSPS resistance mutations, EPSPS amplification (5, 6), or due to enhanced glyphosate metabolism (7). Glyphosate is foliar absorbed, readily translocated throughout the plant to meristematic regions, and must move across the plasma membrane (PM) and enter plastids to inhibit EPSPS. Importantly, glyphosate resistance can be unrelated to EPSPS but rather due to reduced glyphosate translocation within the plant (5, 8–10). First observed in GR Lolium rigidum (11), this reduced glyphosate translocation has often been observed in GR Lolium spp (12–14), Conyza spp (15–17), Sorghum halepense (18, 19), Amaranthus palmeri (20, 21), Digitaria insularis (22), and Ambrosia trifida (23). In some GR weed species, there is biochemical evidence that glyphosate is sequestered in vacuoles, or its cellular uptake is reduced (24–26).
There is speculation that if tonoplast or PM ATP-binding cassette (ABC) transporters could move glyphosate out of the cytoplasm then this could endow glyphosate resistance (8). Transcriptome studies revealed a few ABC transporter genes were constitutively higher expressed or induced by glyphosate treatment in GR versus susceptible (S) plants (27, 28). However, there has been no specific evidence of ABC transporter genes endowing glyphosate resistance. In contrast, in human cancer, anticancer drug resistance is well known to be mediated by ABC transporters, including the multidrug resistance proteins (MRPs) (29). For example, HsMRP1 (also known as HsABCC1) can extrude many anticancer drugs from cancer cells, thereby enabling ongoing cell function and division (30, 31).
Plant genomes encode between 120 to 140 ABC transporters, divided into the ABCA, ABCB, ABCC, ABCD, ABCE, ABCF, ABCG, and ABCI subfamilies (32, 33). Plant ABC transporters function in multiple physiological processes such as transport of hormones, lipids, metals, and secondary metabolites, detoxification of xenobiotics, plant–microbe interactions, etc. (32, 33). Laboratory-generated Arabidopsis transporter variants in the ABCB or ABCG family can be resistant to the antibiotic kanamycin (34) or some auxin and/or dinitroaniline herbicides (35, 36) and paraquat (37). The ABCC (also known as MRP) transporter family typically transports organic acids alone, or as glutathione or glucose conjugates, from the cytoplasm into vacuoles. Only some of these transporters have been functionally characterized (32, 33, 38). Multispecificity has been shown for several plant ABCCs (e.g., AtABCC2 transports phytochelatins, phytochelatin conjugates, glutathione conjugates, glucuronate conjugates, chlorophyll catabolites, and auxin conjugates) (32, 39). AtABCC1, AtABCC2, and OsABCC1 can sequester phytochelatin-conjugated arsenic to vacuoles for detoxification (40, 41). AtABCC5, an ortholog of maize ZmMRP4, can transport phytate to vacuoles for Pi storage and stomatal aperture regulation (42).
Echinochloa colona (awnless barnyardgrass, also known as jungle rice) is a global agricultural C4 weed infesting many warm season crops. E. colona is a genetically diverse, resistance-prone weed species, including there being many GR biotypes in various parts of the world (3). Here, we report on our GR E. colona population that we have long studied (43). Using GR versus S plants, our leaf disk study found a trend of increased glyphosate efflux and therefore reduced glyphosate cellular content (44). We speculated whether glyphosate resistance in this GR E. colona could be due to increased ability of plant ABC transporters to extrude glyphosate from the cytoplasm, additionally to the recently discovered glyphosate metabolism (7). Accordingly, and following transcriptomics indications, we cloned and characterized a plant ABC transporter (ABCC8) from GR E. colona and here confirm that heterologous expression of this ABC transporter and overexpression of its orthologs in plants confer glyphosate resistance by reducing the cytoplasmic glyphosate level. This is evidence of a plant ABC transporter conferring field-evolved herbicide resistance.
Results
RNA Sequencing Analysis and Multiple Step Validations Linked Two ABC Transporter Gene Contigs with Glyphosate Resistance in GR E. colona.
According to the selection criterion of twofold change and P < 0.05, initially nine out of 18 differentially expressed candidate contigs with membrane transporter gene annotation in RNA sequencing (RNA-seq) analysis were selected from the GR versus S E. colona samples and validated using RT-qPCR (SI Appendix, Tables S1 and S2). Five of these (SI Appendix, Table S3) were further confirmed using an additional six GR and six S spare samples for RNA-seq and were subjected to several rounds of validation using a series of prephenotyped samples from between and within multiple GR and S populations/lines. This revealed that two ABC transporter contigs, EC_v4.g098055 and EC_v4.g102032 (SI Appendix, Table S3), showed consistently and significantly higher expression in all GR versus S comparisons and lower expression in additional S populations (QBG1 and Grossy). Furthermore, as we have shown that the level of glyphosate resistance in the GR line is influenced by temperature (7), expression of the two ABC transporter contigs were tested for response to temperature. Significantly higher expression was recorded under 35/30 °C than 25/20 °C growth temperatures (SI Appendix, Table S3). These multiple test results indicated that higher expression of the two ABC transporter contigs, EC_v4.g098055 and EC_v4.g102032, might be associated with glyphosate resistance in this GR E. colona population.
Sequence Analysis of the Two Candidate ABC Transporter Genes in GR and S E. colona Revealed No Amino Acid Substitutions.
Full coding sequences of the two ABC transporter contigs were cloned from GR and S E. colona lines. The coding sequence of the contig EC_v4.g098055 was 4,299 base pairs (bp), sharing 90% amino acid sequence identity to Panicum hallii ABCC8 (XP_025810778.1), 87% to Zea mays MRP1 (NP_001105942.2), 77% to Oryza sativa ABCC8 (KAF0901840.1), and 71% to Glycine max ABCC8 (XP_014633115.1). The coding sequence of the contig EC_v4.g102032 was 4,527 bp, sharing 88% sequence identity to P. hallii ABCC10 (XP_025806285.1) and 87% to Dichanthelium oligosanthes ABCC10 (OEL24435.1). Amino acid sequences of contigs EC_v4.g098055 and EC_v4.g102032 only share 39% identity. The phylogenetic tree analysis indicates that contig EC_v4.g098055 has a close evolutionary relationship with ABCC8 and contig EC_v4.g102032 with ABCC10, from various plant species (SI Appendix, Fig. S1 A and B). Based on this, the two ABC transporter genes are named as EcABCC8 and EcABCC10 (National Center for Biotechnology Information [NCBI] accession nos.: MT249005, MT249006), respectively.
Alignment of the EcABCC8 coding sequences between GR and S E. colona samples showed only one single nucleotide polymorphism (SNP), and this did not cause any amino acid change. No SNPs were found in EcABCC10 in GR and S E. colona sequences. In addition, EcABCC8 and EcABCC10 sequences from the two supplementary S populations (QBG1 and Crossy) were also cloned and compared with the S. Three silent SNPs in EcABCC10 and four in EcABCC8 were found. These results are indicative that glyphosate resistance could be due to overexpression but not mutation of the two ABC transporter genes in this GR E. colona population.
Heterologous Expression of EcABCC8 and Overexpression of Its Orthologs in Planta Confer Glyphosate Resistance.
Heterologous expression of EcABCC8 in rice.
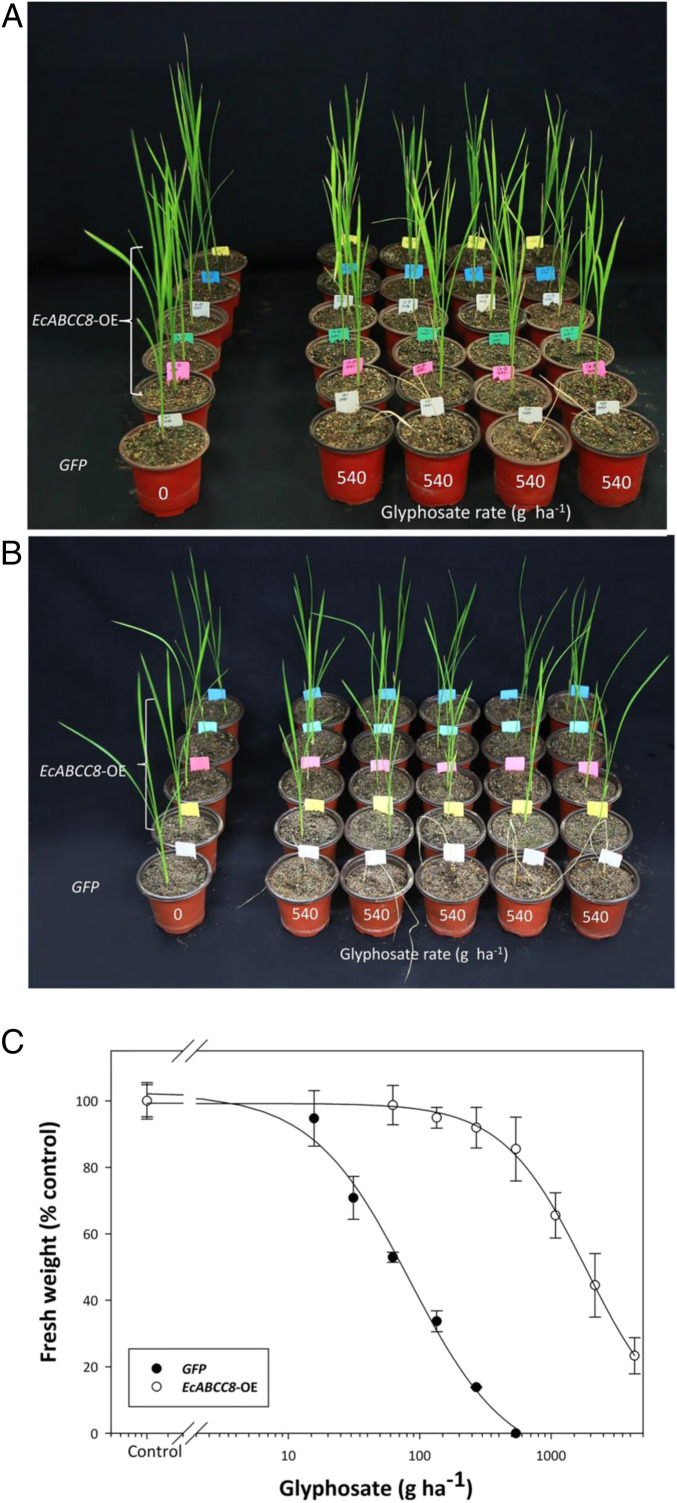
The two ABC transporter genes (EcABCC8 and EcABCC10) were used to transform rice for functional characterization in comparison to rice expressing the GFP gene. Treatment of GFP and EcABCC10-OE seedlings (24 from four T1 lines) with glyphosate at the field rate of 540 g ⋅ ha−1 caused 100% mortality (SI Appendix, Fig. S2), and therefore, no further analysis on EcABCC10-OE lines was conducted. Conversely, 30 T1 rice seedlings, from five EcABCC8-OE lines, survived this glyphosate treatment. Segregation revealed a resistance versus susceptibility ratio of 22:8, indicating single gene 3:1 inheritance (Fig. 1A). There was no resistance to the primary glyphosate metabolite aminomethyl phosphonic acid (AMPA) (50 mM) or to a widely used alternative herbicide glufosinate (250 g ⋅ ha−1), relative to the GFP control (SI Appendix, Fig. S3). Subsequently, 20 T2 seedlings from four EcABCC8-OE lines were treated at the same glyphosate field rate (540 g ⋅ ha−1), and all survived (Fig. 1B). Quantification of glyphosate dose response in one homozygous T2 EcABCC8-OE line (Fig. 1C) revealed a GR50 value (herbicide dose causing 50% growth reduction) of 1,847 ± 282 in comparison to 84 ± 12 g ⋅ ha−1 for the GFP control line, giving 22-fold (P = 0.005) glyphosate resistance. These results clearly established that heterologous expression of the ABC transporter gene EcABCC8 in rice conferred glyphosate resistance.
Fig. 1.
Heterologous expression of EcABCC8 in rice endows glyphosate resistance. Growth of T1 (A) (five lines) and T2 (B) (four lines) transgenic rice seedlings expressing EcABCC8 (EcABCC8-OE) or GFP control, 3 wk after glyphosate treatment. Only glyphosate surviving T1 seedlings from EcABCC8-OE lines were shown in A, and the ratio of surviving to dead plants was 22:8. (C) Glyphosate dose response of EcABCC8-OE and the GFP control. The three- to four-leaf stage seedlings were foliar sprayed with glyphosate, and results were assessed 3 wk after treatment. Data points are means ± SE (n = 3).
Homologous expression of EcABCC8 orthologs in rice, maize, and soybean.
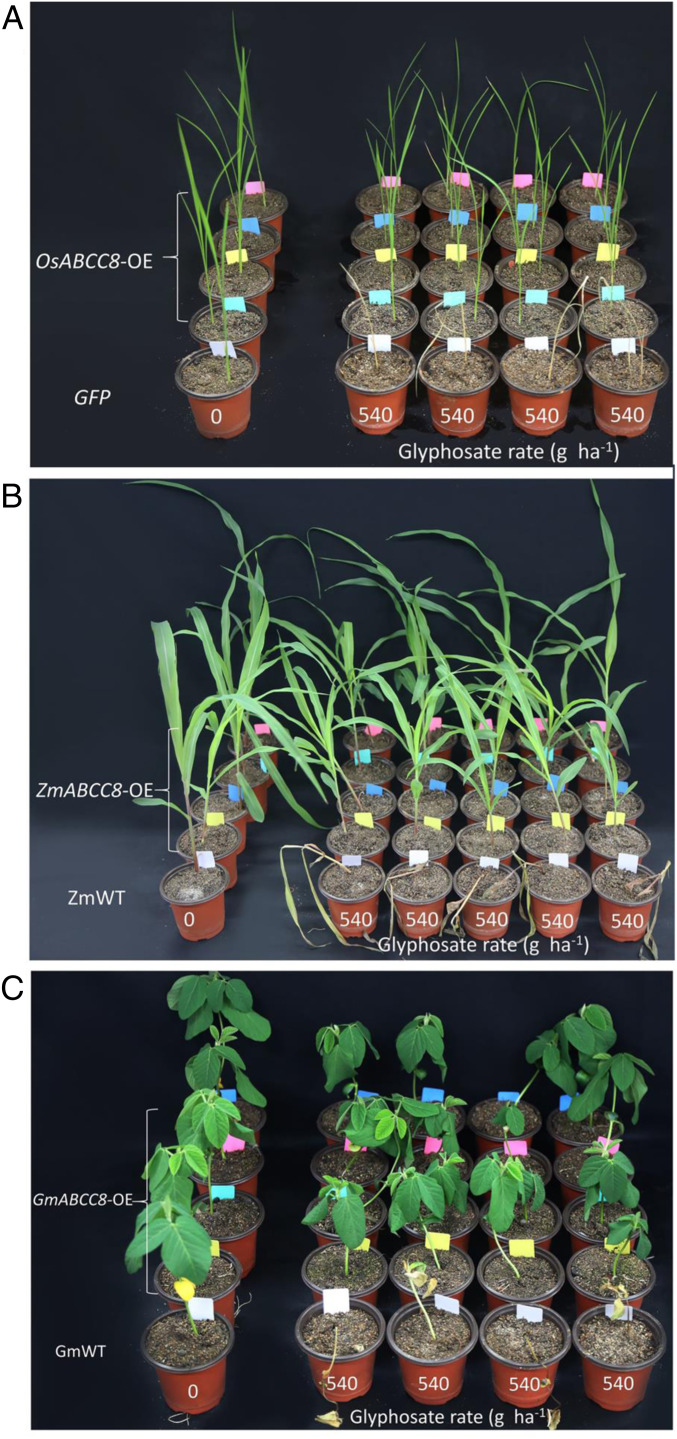
OsABCC8 (LOC_Os06g36650), ZmABCC8 (Zm00001d046226), and GmABCC8 (Glyma.07G011600.1) are orthologous genes of EcABCC8 in rice, maize, and soybean, sharing 77, 87, and 71% identity in protein sequence to EcABCC8, respectively. Four T1 lines each from rice, maize, and soybean (with six to seven seedlings per line) overexpressing the respective ortholog gene were tested for glyphosate resistance. In each of these three crop species, overexpression of the ABCC8 orthologs endowed resistance at the glyphosate field rate (540 g ⋅ ha−1), relative to their respective GFP or wild-type (WT) controls (Fig. 2). The ratio of surviving to dead plants was 22:6, 19:5, and 21:7 in T1 rice, maize, and soybean, respectively, close to single gene 3:1 inheritance. One T1 line each from OsABCC8-OE rice, GmABCC8-OE maize, and ZmABCC8-OE soybean was used for quantifying glyphosate resistance levels. GR50 values from the glyphosate dose responses (SI Appendix, Fig. S4) are 1,330 ± 222 versus 93 ± 18 g ⋅ ha−1 for OsABCC8-OE versus GFP plants, 568 ± 83 versus 41 ± 1.5 g ⋅ ha−1 for GmABCC8-OE versus GmWT, and 5,860 ± 140 versus 337 ± 42 g ⋅ ha−1 for ZmABCC8-OE versus ZmWT. Based on the GR50 ratio, a level of glyphosate resistance of 14-, 13.8- and 17-fold was obtained for OsABCC8-OE (P = 0.005), GmABCC8-OE (P = 0.006), and ZmABCC8-OE (P = 0.003) line, respectively. These results suggest that the ABCC8 transporter EcABCC8 and its orthologs have conserved function in plant species.
Fig. 2.
Overexpression of EcABCC8 ortholog genes in crop plants confers glyphosate resistance. Growth response to glyphosate of T1 plants overexpressing OsABCC8 (OsABCC8-OE) in rice (A), ZmABCC8 (ZmABCC8-OE) in maize (B), and GmABCC8 (GmABCC8-OE) in soybean (C), relative to GFP or untransformed WT controls. Plants at the four- to six-leaf stage were foliar sprayed with a field-relevant glyphosate rate (540 g ⋅ ha−1), and photos were taken 3 wk after treatment. Note only glyphosate surviving T1 seedlings from ABCC8 overexpressing lines are shown. The ratio of surviving to dead plants was 22:6, 19:5, and 21:7 for OsABCC8-OE rice, ZmABCC8-OE maize, and GmABCC8-OE soybean, respectively, close to single gene 3:1 inheritance.
The apparently weaker effect of GmABCC8 overexpression on soybean growth at the glyphosate field rate (540 g ⋅ ha−1) (Fig. 2C) is likely due to intrinsically higher sensitivity of soybean to glyphosate and relatively lower sequence homology of GmABCC8 (71%) to EcABCC8 compared to OsABCC8 (77%) and ZmABCC8 (87%).
CRISPR/Cas9 Knockout of OsABCC8 Increases Glyphosate Susceptibility in Rice.
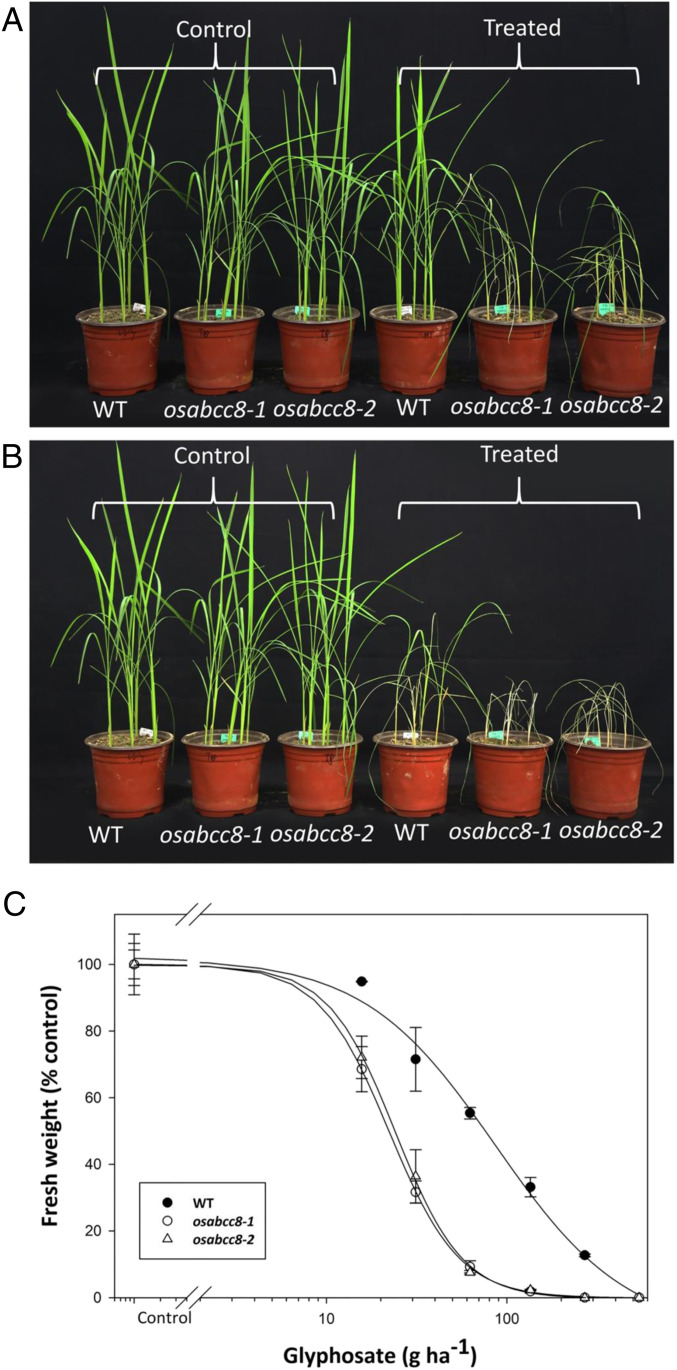
To further confirm the function of the ABC transporter ABCC8, we generated OsABCC8 nonfunctional, knockout (KO) rice mutants. Among seven T1 nonfunctional KO mutants, four of them had a nucleotide insertion (allele1, SI Appendix, Fig. S5) and three had a nucleotide deletion in the OsABCC8 gene (allele2, SI Appendix, Fig. S5), leading to a frame shift with premature transcription termination. The OsABCC8 gene was sequenced in 12 to 18 randomly chosen T2 plants derived from each of the seven T1 nonfunctional KO lines, and as expected, these T2 lines all had identical deletion (osabcc8-1) or insertion (osabcc8-2) variants in OsABCC8.
Glyphosate susceptibility of these two T2 nonfunctional KO variants was tested in comparison to the WT line. As expected, WT seedlings were S to glyphosate, with growth reduction (in shoot fresh weight) by 17 ± 3.3% at glyphosate as low as 26 g ⋅ ha−1 (Fig. 3A) and marked reduction (48 ± 5.3% in shoot fresh weight) at 105 g ⋅ ha−1 (Fig. 3B). However, the two OsABCC8-KO variants were even more glyphosate S than the WT, suffering 39 ± 4.2% and 42 ± 7.2% growth reduction, respectively, at 26 g ⋅ ha−1 and 100% mortality at 105 g ⋅ ha−1 (Fig. 3 A and B).
Fig. 3.
KO of the EcABCC8 ortholog gene in rice increases susceptibility to glyphosate. Growth response of the osabcc8-1 and osabcc8-2 KO mutants versus WT rice seedlings to glyphosate treatment at (A) 26 g ⋅ ha−1 and (B) 105 g ⋅ ha−1. (C) Glyphosate dose response of the two KO versus WT lines. Plants at the three- to four-leaf stage were foliar treated with glyphosate, and results were assessed 3 wk after treatment. Data points are means ± SE (n = 3).
The GR50 value estimated from glyphosate dose response was 86 ± 9.6 g ⋅ ha−1 for WT and 24 ± 1.7 and 22 ± 2.1 g ⋅ ha−1 for the two nonfunctional OsABCC8-KO variants, respectively, giving up to 3.9-fold (P < 0.033) increased glyphosate susceptibility (Fig. 3C). From these results, it is clear that KO of the OsABCC8 gene increases glyphosate susceptibility. In contrast, as shown in Fig. 2A, overexpression of the OsABCC8 gene in rice confers glyphosate resistance.
Regulation of the EcABCC8 Expression in E. colona May Involve DNA Methylation.
In order to investigate possible mechanisms regulating EcABCC8 expression in E. colona, the 1, 990 bp promoter sequences of the EcABCC8 were obtained from five plants of each GR and S populations. Sequence alignment showed only three SNPs in the promoter region comparing the GR and S samples (SI Appendix, Fig. S6). Global DNA methylation analysis identified three differentially methylated regions in the EcABCC8 gene between the GR and S samples (SI Appendix, Fig. S7). Compared to S, the level of methylation in the GR EcABCC8 gene was lower in two promoter regions at the CHH context (0.25 versus 0.39 and 0.54) and higher in one exon region under the CG contexts (0.55 versus 0.36) (SI Appendix, Fig. S7). These results indicate that epigenetic mechanisms may be involved in EcABCC8 expression regulation, that is, a lower level of methylation in the promoter region is likely related to a higher level of EcABCC8 expression in GR versus S plants.
Tissue Expression, Subcellular Location, and Function of ABCC8.
ABCC8 is expressed in both above- and below-ground plant tissue.
The RT-qPCR results demonstrated that the EcABCC8 gene expresses in leaf, stem, and root tissues of GR and S E. colona plants, with up to 10-fold higher expression in GR than in S (SI Appendix, Fig. S8). EcABCC8 expression in leaf and stem of both GR and S plants was up to threefold higher than in the root (SI Appendix, Fig. S8).
EcABCC8 is likely localized to the PM.
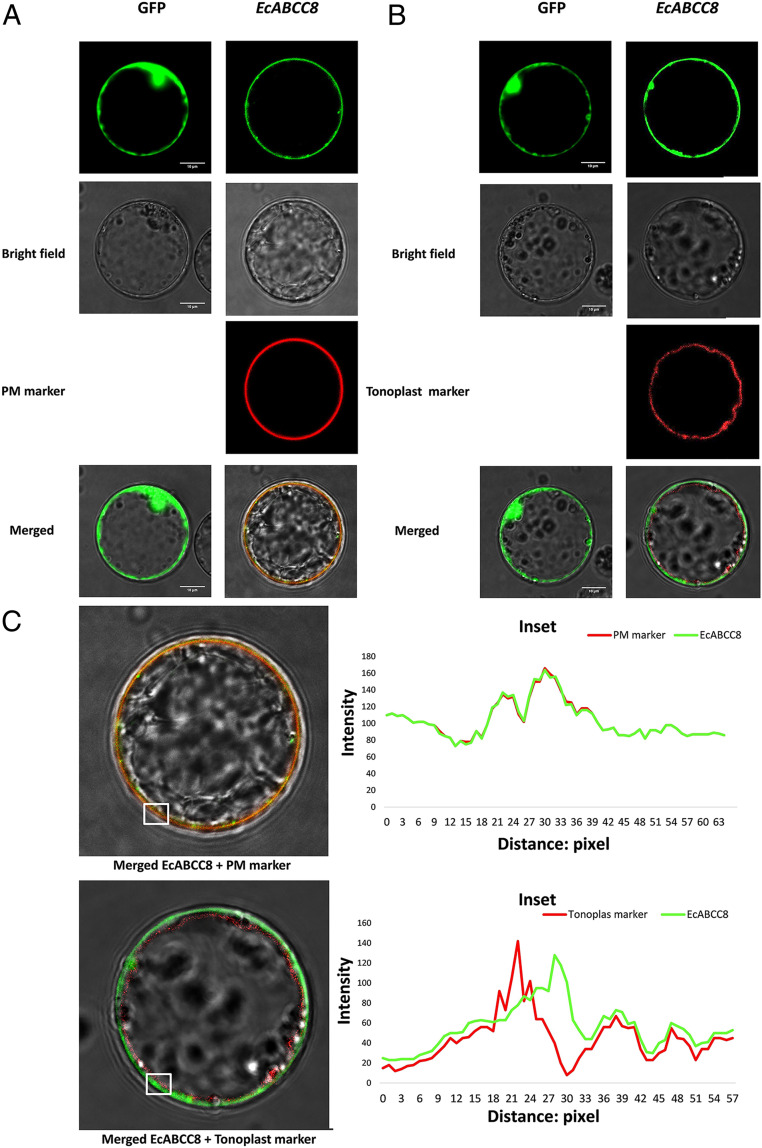
Rice and Arabidopsis protoplast transit expression systems were used to determine the subcellular localization of EcABCC8. Multiphoton confocal laser scanning microscopy examination of transformed rice protoplasts showed that protoplasts expressing 35S:GFP alone had strong fluorescent signals in cytoplasm around the periphery of the large vacuole in the cytoplasm (Fig. 4A). When expressed together, green and red fluorescent signals respectively from the 35S:EcABCC8-GFP and the PM marker 35S:SCAMP1-mRFP completely merged to the PM (Fig. 4A). However, this was not the case for the coexpressed tonoplast marker 35S:AtTPK3-mRFP, which had red fluorescence outlining the large central vacuole, close to but clearly distinguishable from the green fluorescence of the 35S:EcABCC8-GFP (Fig. 4B). In addition, line intensity scan analysis also revealed complete overlapping of fluorescence distribution of EcABCC8 with the PM marker (Fig. 4 C, Upper) and separation from the tonoplast marker (Fig. 4 C, Lower), supporting that EcABCC8 is more likely a PM- than a tonoplast-located transporter. Similar results were also obtained with the Arabidopsis protoplasts (SI Appendix, Fig. S9).
Fig. 4.
Subcellular location of EcABCC8. (A) Colocalization of the EcABCC8 and the PM marker and (B) lack of colocalization of the EcABCC8 and the tonoplast marker in rice protoplasts. (C) Line scan analysis showing overlapping of fluorescence distribution of EcABCC8 (green) and the PM maker (red) (Upper) and separation of EcABCC8 (green) and the tonoplast marker (red) (Lower) in areas of interest (boxed). (Scale bars, 10 μm.)
In addition, subcellular location of the EcABCC8 ortholog GmABCC8 was also probed using the same markers and is also likely localized to the PM (SI Appendix, Fig. S10).
ABCC8 enhances glyphosate efflux, reducing glyphosate cellular level in rice leaf discs.
To avoid complication by any other glyphosate resistance mechanisms in the GR E. colona populations (e.g., AKR-mediated glyphosate metabolism, see ref. 7), glyphosate efflux and net content were examined in 1 mm leaf discs of EcABCC8-OE versus GFP transgenic rice seedlings, in contrast to the ortholog gene KO (osabcc8-1 versus WT) rice seedlings.
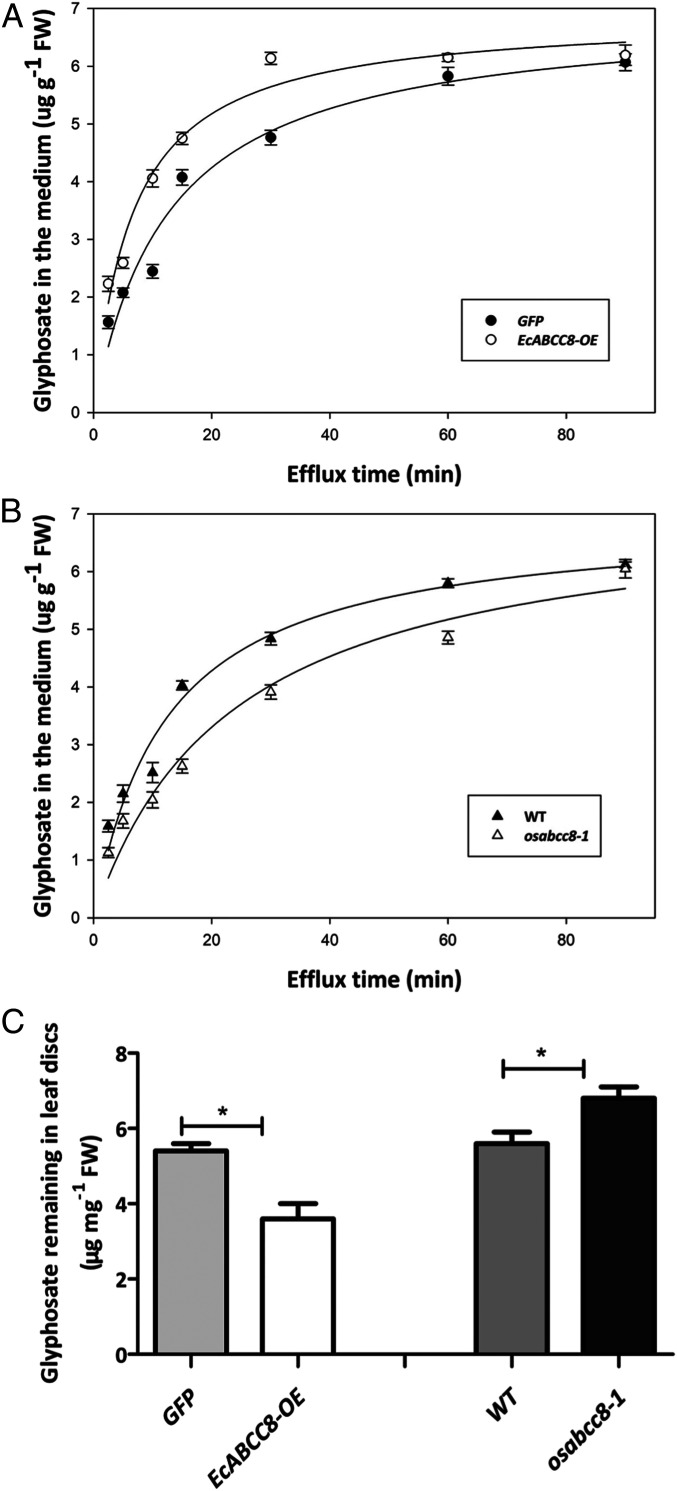
Glyphosate efflux from leaf discs of rice seedlings was rapid over the first 30 min and then slowed (Fig. 5A). The rate of glyphosate efflux to the apoplast (external solution) was nearly twofold faster in EcABCC8-OE leaf discs (b = 0.15 ± 0.02 [μg ⋅ g−1 fresh weight (FW) min−1]) than in GFP control (b = 0.08 ± 0.02), leading to significantly (P = 0.016) lower glyphosate content in EcABCC8-OE than in the GFP samples (Fig. 5A). In contrast, glyphosate efflux rate was twofold slower in leaf discs of the nonfunctional osabcc8-1 (b = 0.04 ± 0.01) than in the WT control (b = 0.08 ± 0.01) (Fig. 5B), resulting in significantly (P = 0.026) higher glyphosate content in osabcc8-1 than in the WT samples (Fig. 5C). These results are consistent with the hypothesis that the EcABCC8 codes for a PM transporter that can move glyphosate out of the cytoplasm into the apoplast, therefore reducing the glyphosate level in the cytoplasm.
Fig. 5.
Glyphosate efflux from leaf discs to the external solution. Glyphosate efflux from leaf discs of rice seedlings (A) expressing EcABCC8 (EcABCC8-OE) versus GFP control and (B) osabcc8-1 KO mutant versus WT. (C) Glyphosate content in leaf discs after efflux. Data points are means ± SE (n = 3). Significance of difference by the Student’s t test is indicated by *P < 0.05. The experiments were repeated with similar results.
ABCC8 reduces glyphosate accumulation in rice leaf protoplasts.
To mimic the in vivo situation, rice seedlings of each EcABCC8-OE versus GFP and OsABCC8-KO versus WT were glyphosate treated at a low rate (68 g ⋅ ha−1), and then leaf protoplasts were isolated, and glyphosate content was quantified. By 2 and 6 h after foliar glyphosate treatment, the glyphosate content in leaf protoplasts was up to 4.2-fold less (P ≤ 0.016) in EcABCC8-OE than in GFP lines (SI Appendix, Fig. S11A) and up to 1.9-fold higher (P ≤ 0.024) in OsABCC8-KO than in WT lines (SI Appendix, Fig. S11A).
To avoid complication by the in vivo approach, leaf protoplasts were also isolated from each line and then treated in vitro with 60 μM glyphosate and followed by glyphosate quantification. Although the absolute glyphosate content is understandably different, relative glyphosate levels in ECABCC8 versus GFP and the nonfunctional KO versus WT was found to be similar, between the two treatment approaches. By 1 and 2 h after glyphosate treatment, the protoplast glyphosate level was up to 3.1-fold less (P ≤ 0.032) in EcABCC8-OE than in GFP lines and up to 2.2-fold higher (P ≤ 0.028) in OsABCC8-KO than in WT lines (SI Appendix, Fig. S11B).
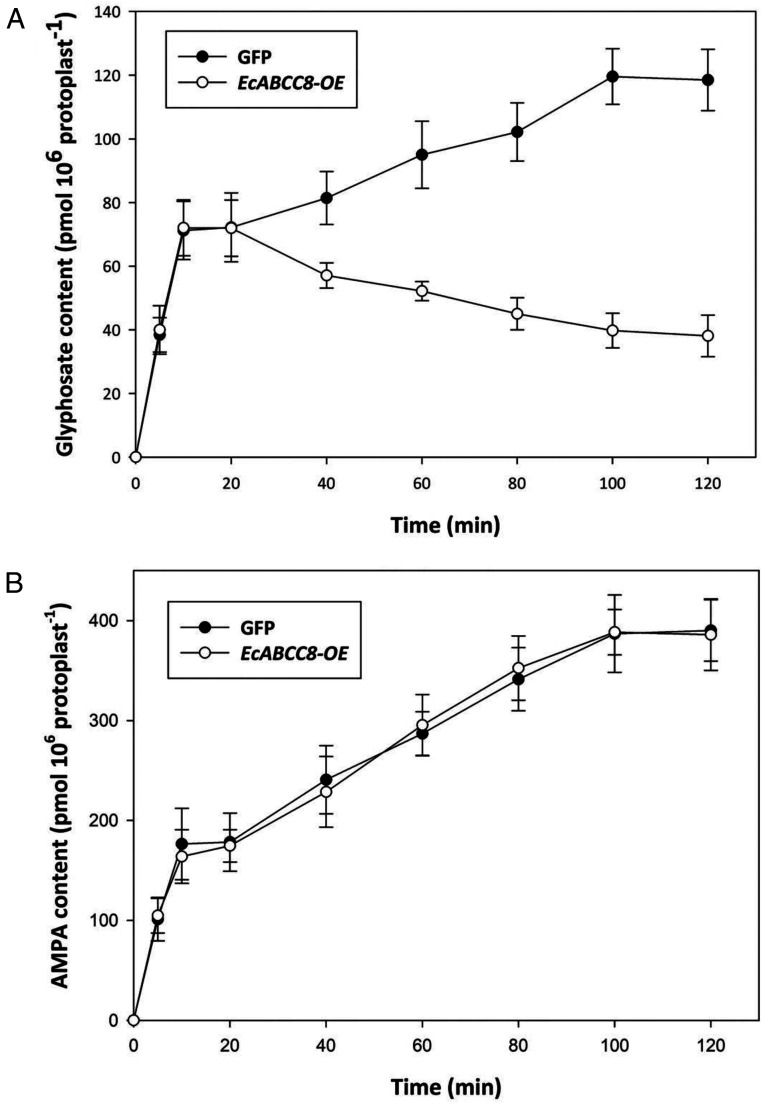
Measurement of time-dependent glyphosate accumulation in isolated rice protoplasts of GFP revealed that the glyphosate level rapidly increased in the first 10 min (initial uptake) following treatment and remained unchanged for 10 min and then slowly increased until 100 min (Fig. 6A). This is similar to glyphosate uptake in broad bean protoplasts when measured for a shorter timespan at glyphosate concentration of 0.1 mM (45). However, glyphosate protoplast accumulation in EcABCC8-OE displayed a distinct pattern with clear departure from that of GFP 20 min after treatment because of ongoing decrease in glyphosate levels over time (Fig. 6A). This result also implies that after rapid glyphosate uptake in the early phase, there then was glyphosate extrusion by the EcABCC8 transporter evident in the later slow uptake phase (after 20 min in our experimental conditions).
Fig. 6.
Glyphosate and AMPA levels in rice protoplasts. Time-dependent glyphosate (A) and AMPA (B) accumulation in rice protoplasts of EcABCC8-OE versus GFP. Glyphosate or AMPA was present at 60 µM. Data points are means ± SE (n = 3). The experiment was repeated with similar results.
As a control, time-dependent accumulation of the glyphosate metabolite AMPA was also measured in isolated rice protoplasts of EcABCC8-OE and GFP lines. However, no difference in AMPA levels between the two lines was observed throughout the experiment period (Fig. 6B). This is expected, as EcABCC8-OE plants are not resistant to AMPA (SI Appendix, Fig. S3).
Thus, consistent results from the in vivo and in vitro treatment situations in contrasting EcABCC8-OE and OsABCC8-KO lines confirmed that the ABC transporter ABCC8 reduces glyphosate accumulation in the cytoplasm.
3D Reconstruction Reveals Structural Interactions of the EcABCC8 and Glyphosate.
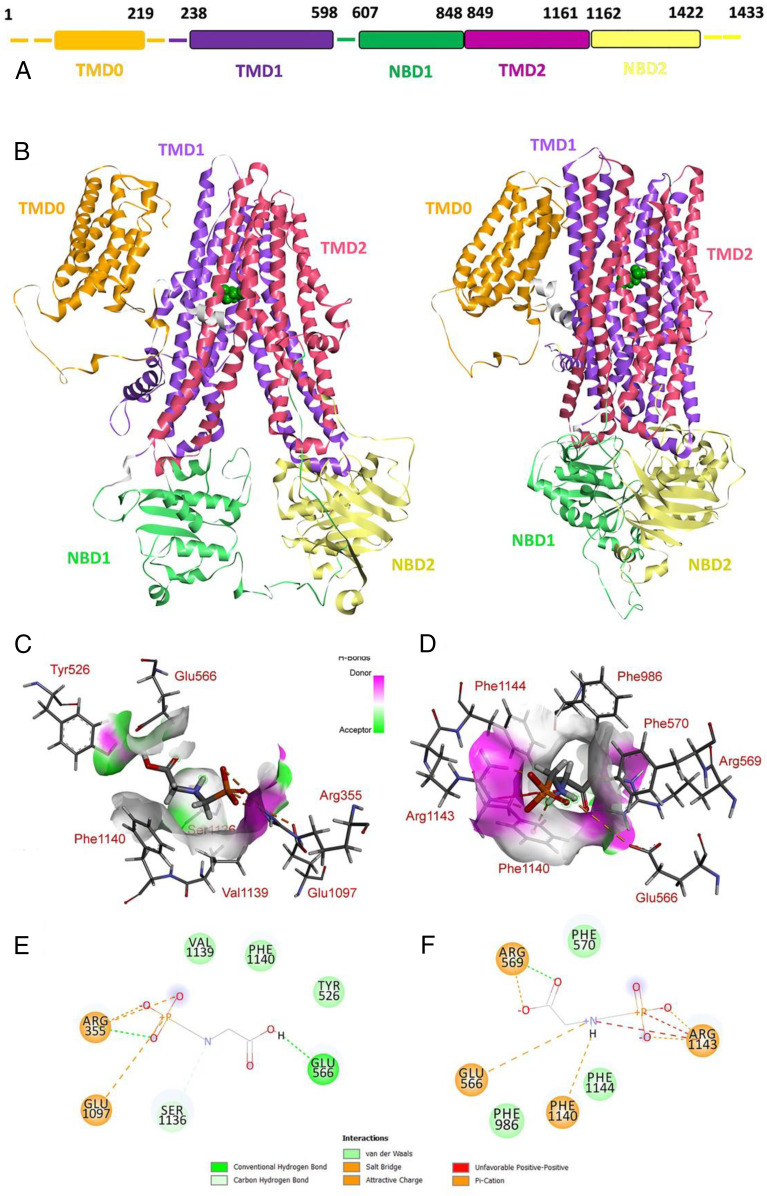
The plant ABC transporter EcABCC8 is a monosubunit protein containing three transmembrane domains (TMDs) and two intracellular nucleotide-binding domains (NBDs) (Fig. 7A). The overall spatial organization of EcABCC8 is very similar to its mammalian homolog MRP1 (46, 47). The characteristic TMD0 domain is located in space closely to TMD1 (Fig. 7B) in both “open” and close” (inward facing and outward facing) conformations.
Fig. 7.
3D reconstruction showing structural interactions of the EcABCC8 and glyphosate. (A) Schematic view of the domain structure of EcABCC8. (B) General view of EcABCC8 in the inward-facing (Left) and outward-facing (Right) conformations with glyphosate (green Van der Waals representation) bound into the cargo-binding site. Structural domains in EcABCC8 composition are highlighted by different colors. Spatial structure of the contact interface between the glyphosate and EcABCC8 in the cargo-binding site in (C) the inward-facing and (D) the outward-facing conformations. Protein contact surface is colored by H-bond donor/acceptor distribution, binding site amino acids represented by sticks, and intermolecular contacts indicated by dotted lines. 2D diagram of molecular interactions between the glyphosate and EcABCC8 in the cargo-binding site in (E) in the inward-facing and (F) the outward-facing conformations.
Results of blind docking of the glyphosate molecule into EcABCC8 and the molecular dynamics (MD) investigations reveal the binding mechanism which is typical for ABCC8/MRP1 proteins and consists of ligand interactions in the substrate interaction area (cargo site) of these exporters. The glyphosate molecule primarily interacts with EcABCC8 in an inward-facing conformation. The correspondent binding site consists of residues Arg355, Tyr526, Glu566, Glu1097, Ser1136, Val1139, and Phe1140 (Fig. 7C). A transfer of glyphosate from the intracellular space between the NBD into the primary binding cargo site results in a reduction of total energy by 2,100 kJ ⋅ mol−1, establishing a favorable condition for this process to occur. In the predicted binding site, glyphosate directly forms several favorable interactions, particularly the salt bridge with Arg355, two attractive charges with Arg355 and Glu1097, two conventional hydrogen bonds with Arg355 and Glu566, and one carbon hydrogen bond with Ser1136 (Fig. 7D). The free interaction energy between the glyphosate and EcABCC8 calculated from the MD ensemble is −97 kJ ⋅ mol−1.
As a result of EcABCC8 transition from the inward-facing (intracellular space) to outward-facing (extracellular space) conformation, the rearrangement of glyphosate and amino acid microenvironment occurs, and glyphosate appears to interact with several other residues located closely to the primary binding site. These include residues Glu566, Arg569, Phe570, Phe986, Phe1140, and Phe1144 (Fig. 7E), involving five electrostatic interactions and one conventional H-bond (Fig. 7F). Some of these residues correspond to residues in the bovine MRP1 substrate binding site (e.g., residues Phe1140 and Arg1143 of EcABCC8 correspond to Trp1245 and Arg1248 of MRP1). One of the results of the conformational change is the appearance of the two unfavorable contacts between the positively charged nitrogen and the phosphorus of glyphosate and the amino group of Arg1143. The presence of such a contact may cause the subsequent release of the glyphosate molecule from EcABCC8 into the extracellular space. The free energy of glyphosate/EcABCC8 interaction after EcABCC8 switching to the outward-facing conformation decreased by 138 kJ ⋅ mol−1, indicating this stage as a rate-limiting step for low molecular weight compound transport.
Release of the glyphosate molecule from the intracellular space of the EcABCC8 transporter into the extracellular space is accompanied by a decrease of total energy of the investigated system (including the ligand, protein, membrane, and water–salt solution) by 3,500 kJ ⋅ mol−1. This proves the high probability of glyphosate release despite strong binding of glyphosate to EcABCC8.
Discussion
Here, in our well-characterized GR E. colona population (43), we present several lines of evidence demonstrating that GR plants overexpress a PM-located ABC transporter (EcABCC8) that functions as a cytoplasmic glyphosate exporter. EcABCC8 is able to move glyphosate from the cytoplasm to the extracellular space (apoplast). In this manner, the cellular glyphosate level is lowered, therefore conferring glyphosate resistance. This is evidence of an ABC transporter endowing plants with herbicide resistance.
Using RNA-seq and RT-qPCR validation, we show that higher EcABCC8 transcript levels are consistently associated with glyphosate resistance in multiple GR versus multiple S E. colona lines/populations and under different temperature scenarios (SI Appendix, Table S3). Among others (e.g., transcription activation), the higher expression of EcABCC8 may involve epigenetic regulation, as a lower CHH methylation level in the promoter regions and a higher CG methylation level in the EcABCC8 gene body was observed in GR versus S sequences (SI Appendix, Fig. S7). Generally, the lower the methylation level, the higher the expression level (48, 49). However, in the gene body (exon) region, higher methylation (CG sites) levels could result in higher gene expression (50–52).
In human medicine, it is known that human ABCC-type transporters (e.g., HsABCC1 or HsMRP1) are PM located and are able to pump certain chemotherapeutic drugs and other xenobiotics out of cancer cells, thereby conferring resistance to anticancer drugs (30). We propose that in plants, EcABCC8 serves (serendipitously) as a PM-embedded glyphosate exporter. Indeed, colocalization of the EcABCC8 (and the ortholog GmABCC8) with the PM marker (Fig. 4A and SI Appendix, Fig. S10) and lack of colocalization with the known tonoplast marker in rice and Arabidopsis leaf protoplasts (Fig. 4 B and C and SI Appendix, Fig. S9) supports this.
Although glyphosate uptake into plant cells can be via both active (at low concentrations) and passive (at high concentrations) mechanisms (8), once glyphosate enters the cytoplasm (pH around seven) it disassociates as anions and cannot freely efflux (25, 26). Therefore, cellular glyphosate efflux must be a transporter-mediated active process. Given that EcABCC8 is a PM-embedded glyphosate exporter, the EcABCC8-OE plants will extrude a higher amount of glyphosate to the apoplast, lowering the glyphosate level in the cytoplasm. This was confirmed by quantification of glyphosate efflux in leaf discs and glyphosate content in protoplasts of EcABCC8-OE in contrast with the ortholog KO rice plants (Figs. 5 and 6 and SI Appendix, Fig. S11). Especially, time-dependent glyphosate accumulation in protoplasts of EcABCC8-OE showed a clear opposite trend to that of GFP (decrease versus increase) after the initial uptake of glyphosate (Fig. 6A). We are aware that overexpression of transporters may lead to a mislocalization of membrane proteins (42). However, this functional analysis data on glyphosate cellular distribution, together with ABCC8 subcellular location, strongly suggest a PM localization of EcABCC8 and therefore provide evidence that EcABCC8 and its orthologs confer glyphosate resistance by serving to lower the intracellular glyphosate level.
Cellular elimination of glyphosate may not be easily detectable at the whole plant level. Indeed, no significant difference in 14C-glyphosate uptake and translocation was observed in the GR versus S plants in our previous study (53). This is likely because 1) the 14C-glyphosate foliar uptake assay measures the total amount of glyphosate within the leaf tissue (e.g., in apoplast and symplast), and the herbicide in the apoplast portion may not be easily removed with gentle leaf wash, and 2) glyphosate translocation in the GR population was confounded by the recently discovered glyphosate metabolism (7).
Theoretically, the PM-based extruding mechanism increases glyphosate sequestration to the apoplast, resulting in more glyphosate available for upward movement with the transpiration stream and hence accumulation in leaf tips and edges. Indeed, in glyphosate-treated EcABCC8-OE rice seedlings, localized leaf tip damage is evident as compared to extended damage in the whole leaf in GFP control seedlings (SI Appendix, Fig. S12). Therefore, it is anticipated that GR plants with more glyphosate translocation to the leaf apical area relative to the S counterpart (e.g., in Lolium spp) (12, 14) may likely use the transporter mechanism similar to EcABCC8.
The plant ABCC (MRP) transporters studied so far are largely tonoplast located, sequestering organic anions and xenobiotics mostly as conjugates into vacuoles (33, 38). It is interesting to know whether the exported glyphosate by EcABCC8 is conjugated or not. Our data measure glyphosate that remained in the protoplast, not exported to the apoplast (except for the leaf disc efflux data), and hence cannot address this question. However, glyphosate is water soluble and is unlikely to form glutathione (GSH) and other conjugates. Our structural modeling also supports the view that glyphosate can directly bind to the inward-facing EcABCC8 from the intracellular space (inside the PM, cytoplasm) and is released from the outward-facing transporter to the extracellular space (outside the PM, apoplast) with favorable interaction energy changes (Fig. 7).
As glyphosate is obviously a serendipitous (nonplant) substrate of the ABCC8, the endogenous functions of ABCC8 in planta (e.g., its physiological substrates and whether it can transport GSH or others) remain to be revealed. In addition, ABC transporters usually rely on energization by ATP. However, our experimental system does not answer the question of whether EcABCC8-mediated glyphosate efflux is indeed ATP driven or occurs by facilitated diffusion through the pore of EcABCC8, or if, alternatively, EcABCC8 is a regulator of a so far unknown glyphosate exporter. Indeed, the mammalian sulfonylurea receptor is a non-ATP–dependent ABCC/MRP type transporter regulating K channel (54). Further experiments using PM vesicles or electron cryomicroscopy structures of free EcABCC8 and in complex with glyphosate will better address these above questions.
As ABC transporters are known to transport diverse substrates (55), we examined whether EcABCC8 could export the glyphosate metabolite AMPA. However, EcABCC8-OE plants were found not to be resistant to AMPA (SI Appendix, Fig. S3) and did not reduce the cellular AMPA levels (Fig. 6B). Our modeling shows that by structural analogy to glyphosate, AMPA can reach the cargo site of ECABCC8. However, AMPA being smaller than glyphosate has fewer stabilizing contacts with amino acids in the binding site. The AMPA-ЕсАBСС8 complex is therefore functionally unstable (as evidenced by the small value of free interaction energy of −29 kJ ⋅ mol−1 as compared to –97 kJ ⋅ mol−1 for glyphosate). In addition, we also found that EcABCC8-OE plants remain S to the herbicide glufosinate (SI Appendix, Fig. S3). Likewise, the GR E. colona is glufosinate S (43).
Glyphosate resistance in this studied population of E. colona involves multiple resistance mechanisms. These include target site EPSPS Pro106Thr mutation (56), non-target site AKR-catalyzed glyphosate metabolism to AMPA (7), and ABC transporter-mediated glyphosate extrusion to the extracellular space (this study). Given that temperature had a positive effect on glyphosate resistance in this population (7), and EcAKR/ABCC8 expression responds to temperature, non-target site resistance via AKR and EcABCC8 gene overexpression likely plays a more important role than the EPSPS mutation whose effect may be diluted by multiple S alleles in polypoid species (57) such as E. colona (58).
Identification and characterization of the ABC transporter EcABCC8 will facilitate discovery of the transporters involved in glyphosate resistance in other plant species. In addition, plant species with varying levels of glyphosate tolerance (or uptake) may be also in part ascribable to the abundance of ABCC8 orthologs, as was demonstrated in the current study that up-regulation of ABCC8 provides glyphosate resistance in different plant species (Fig. 2).
Identification of ABCC8-associated cell types will help understand its roles in specific cellular processes, as demonstrated for AtABCC5 in guard cell signaling and phytate storage (42). Given that the ABCC8 expresses in both above- and below-ground tissues (SI Appendix, Fig. S8), it is tempting to speculate implication of this ABC transporter in other plant cellular processes beyond xenobiotic detoxification, such as root exudation of plant metabolites for soil nutrient acquisition and plant–soil microbe interactions (59) and allelopathy (60).
In summary, here we show in a GR E. colona population that overexpression of the ABCC8 transporter endows resistance to glyphosate. This finding that an ABC transporter can endow herbicide resistance will catalyze studies to investigate ABCC8-like and other membrane bound transporters for their capacity to endow plant species with resistance to glyphosate or other herbicides/xenobiotics by moving toxic compounds out of the cytosol.
Materials and Methods
Detailed information on plant material, RNA-seq data analysis and selection of candidate transporter contigs, rice genetic transformation with the two ABC transporter genes EcABCC8 and EcABCC10, homologous overexpression of EcABCC8 orthologs in other crop plants, rice OsABCC8 gene KO by CRISPR/Cas9 gene editing, global DNA methylation analysis for E. colona, subcellular localization of ABCC8, glyphosate efflux and content in leaf discs of transgenic rice seedlings, glyphosate quantification in leaf protoplasts of transgenic rice plants, and structural reconstruction of EcABCC8 variant are described in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (31901905), the Australian Grains Research and Development Corporation, the Natural Science Foundation of Hunan Province, China (2020JJ5238), China Agriculture Research System (CARS-16-E19), and the Scientific Research Fund of Hunan Provincial Education Department (19B254).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2100136118/-/DCSupplemental.
Data Availability
The EcABCC8 and EcABCC10 complementary DNA sequences data have been deposited in the GenBank database (access nos. MT249005 and MT249006). All data used in the study are included in the paper and SI Appendix. All protocols are described in SI Appendix, Materials and Methods or in the references therein. Plant materials are available upon request by qualified researchers to the corresponding author.
References
- 1.Duke S. O., Powles S. B., Sammons R. D., Glyphosate-How it became a once in a hundred year herbicide and its future. Outlooks Pest Manag. 29, 247–251 (2018). [Google Scholar]
- 2.Duke S. O., Powles S. B., Glyphosate: A once-in-a-century herbicide. Pest Manag. Sci. 64, 319–325 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Heap I., International Survey of Herbicide Resistant Weeds. http://www.weedscience.org. Accessed 17 March 2021.
- 4.Holländer H., Amrhein N., The site of the inhibition of the shikimate pathway by glyphosate: I. Inhibition by glyphosate of phenylpropanoid synthesis in buckwheat (Fagopyrum esculentum moench). Plant Physiol. 66, 823–829 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sammons R. D., Gaines T. A., Glyphosate resistance: State of knowledge. Pest Manag. Sci. 70, 1367–1377 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaines T. A., et al., Mechanisms of evolved herbicide resistance. J. Biol. Chem. 295, 10307–10330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan L., et al., Aldo-keto reductase metabolizes glyphosate and confers glyphosate resistance in Echinochloa colona. Plant Physiol. 181, 1519–1534 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaner D. L., Role of translocation as a mechanism of resistance to glyphosate. Weed Sci. 57, 118–123 (2009). [Google Scholar]
- 9.Powles S. B., Yu Q., Evolution in action: Plants resistant to herbicides. Annu. Rev. Plant Biol. 61, 317–347 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Jugulam M., Shyam C., Non-target-site resistance to herbicides: Recent developments. Plants 8, 417 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lorraine-Colwill D., et al., Investigations into the mechanism of glyphosate resistance in Lolium rigidum. Pestic. Biochem. Physiol. 74, 62–72 (2002). [Google Scholar]
- 12.Wakelin A. M., Lorraine-Colwill D. F., Preston C., Glyphosate resistance in four different populations of Lolium rigidum is associated with reduced translocation of glyphosate to meristematic zones. Weed Res. 44, 453–459 (2004). [Google Scholar]
- 13.Yu Q., Cairns A., Powles S., Glyphosate, paraquat and ACCase multiple herbicide resistance evolved in a Lolium rigidum biotype. Planta 225, 499–513 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Perez-Jones A., Park K. W., Polge N., Colquhoun J., Mallory-Smith C. A., Investigating the mechanisms of glyphosate resistance in Lolium multiflorum. Planta 226, 395–404 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Feng P. C. C., et al., Investigations into glyphosate-resistant horseweed (Conyza canadensis): Retention, uptake, translocation, and metabolism. Weed Sci. 52, 498–505 (2004). [Google Scholar]
- 16.Koger C. H., Reddy K. N., Role of absorption and translocation in the mechanism of glyphosate resistance in horseweed (Conyza canadensis). Weed Sci. 53, 84–89 (2005). [Google Scholar]
- 17.Dinelli G., et al., Physiological and molecular bases of glyphosate resistance in Conyza bonariensis biotypes from Spain. Weed Res. 48, 257–265 (2008). [Google Scholar]
- 18.Vila-Aiub M. M., et al., Glyphosate resistance in perennial Sorghum halepense (Johnsongrass), endowed by reduced glyphosate translocation and leaf uptake. Pest Manag. Sci. 68, 430–436 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Riar D. S., Norsworthy J. K., Johnson D. B., Scott R. C., Bagavathiannan M., Glyphosate resistance in a Johnsongrass (Sorghum halepense) biotype from Arkansas. Weed Sci. 59, 299–304 (2011). [Google Scholar]
- 20.Nandula V. K., et al., Multiple resistance to glyphosate and pyrithiobac in Palmer Amaranth (Amaranthus palmeri) from Mississippi and response to flumiclorac. Weed Sci. 60, 179–188 (2012). [Google Scholar]
- 21.Palma-Bautista C., et al., Reduced absorption and impaired translocation endows glyphosate resistance in Amaranthus palmeri harvested in glyphosate-resistant soybean from Argentina. J. Agric. Food Chem. 67, 1052–1060 (2019). [DOI] [PubMed] [Google Scholar]
- 22.de Carvalho L. B., et al., Pool of resistance mechanisms to glyphosate in Digitaria insularis. J. Agric. Food Chem. 60, 615–622 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Nandula V., et al., Glyphosate resistance in giant ragweed (Ambrosia trifida L.) from Mississippi is partly due to reduced translocation. Am. J. Plant Sci. 06, 2104–2113 (2015). [Google Scholar]
- 24.Ge X., d’Avignon D. A., Ackerman J. J. H., Sammons R. D., Rapid vacuolar sequestration: The horseweed glyphosate resistance mechanism. Pest Manag. Sci. 66, 345–348 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ge X., et al., Vacuolar glyphosate-sequestration correlates with glyphosate resistance in ryegrass (Lolium spp.) from Australia, South America, and Europe: A 31P NMR investigation. J. Agric. Food Chem. 60, 1243–1250 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Ge X., D’Avignon A., Ackerman J. J. H., Ostrander E., Sammons R. D., “Application of 31P-NMR spectroscopy to glyphosate studies in plants: Insights into cellular uptake and vacuole sequestration correlated to herbicide resistance” in Handbook on Herbicides: Biological Activity, Classification and Health and Environmental Implications, Kobayashi D., Watanabe E. (Nova Science Publishers, Inc., Hauppauge, NY, 2013), pp. 55–84. [Google Scholar]
- 27.Peng Y., et al., Characterization of the horseweed (Conyza canadensis) transcriptome using GS-FLX 454 pyrosequencing and its application for expression analysis of candidate non-target herbicide resistance genes. Pest Manag. Sci. 66, 1053–1062 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Piasecki C., et al., Transcriptomic analysis identifies new non-target site glyphosate-resistance genes in Conyza bonariensis. Plants 8, 26 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leslie E. M., Deeley R. G., Cole S. P. C., Multidrug resistance proteins: Role of P-glycoprotein, MRP1, MRP2, and BCRP (ABCG2) in tissue defense. Toxicol. Appl. Pharmacol. 204, 216–237 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Cole S. P., et al., Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science 258, 1650–1654 (1992). [DOI] [PubMed] [Google Scholar]
- 31.Cole S. P. C., Targeting multidrug resistance protein 1 (MRP1, ABCC1): Past, present, and future. Annu. Rev. Pharmacol. 54, 95 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Kang J., et al., Plant ABC transporters. Arabidopsis Book 9, e0153 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hwang J. U., et al., Plant ABC transporters enable many unique aspects of a terrestrial plant’s lifestyle. Mol. Plant 9, 338–355 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Mentewab A., Stewart C. N. Jr., Overexpression of an Arabidopsis thaliana ABC transporter confers kanamycin resistance to transgenic plants. Nat. Biotechnol. 23, 1177–1180 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Ito H., Gray W. M., A gain-of-function mutation in the Arabidopsis pleiotropic drug resistance transporter PDR9 confers resistance to auxinic herbicides. Plant Physiol. 142, 63–74 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Windsor B., Roux S. J., Lloyd A., Multiherbicide tolerance conferred by AtPgp1 and apyrase overexpression in Arabidopsis thaliana. Nat. Biotechnol. 21, 428–433 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Xi J., Xu P., Xiang C. B., Loss of AtPDR11, a plasma membrane-localized ABC transporter, confers paraquat tolerance in Arabidopsis thaliana. Plant J. 69, 782–791 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Klein M., Burla B., Martinoia E., The multidrug resistance-associated protein (MRP/ABCC) subfamily of ATP-binding cassette transporters in plants. FEBS Lett. 580, 1112–1122 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Lu Y. P., et al., AtMRP2, an Arabidopsis ATP binding cassette transporter able to transport glutathione S-conjugates and chlorophyll catabolites: Functional comparisons with Atmrp1. Plant Cell 10, 267–282 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song W. Y., et al., Arsenic tolerance in Arabidopsis is mediated by two ABCC-type phytochelatin transporters. Proc. Natl. Acad. Sci. U.S.A. 107, 21187–21192 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song W. Y., et al., A rice ABC transporter, OsABCC1, reduces arsenic accumulation in the grain. Proc. Natl. Acad. Sci. U.S.A. 111, 15699–15704 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagy R., et al., The Arabidopsis ATP-binding cassette protein AtMRP5/AtABCC5 is a high affinity inositol hexakisphosphate transporter involved in guard cell signaling and phytate storage. J. Biol. Chem. 284, 33614–33622 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gaines T. A., Cripps A., Powles S. B., Evolved resistance to glyphosate in Junglerice (Echinochloa colona) from the tropical Ord Riverr-region in Australia. Weed Technol. 26, 480–484 (2012). [Google Scholar]
- 44.Goh S. S., “Quantitative estimation of fitness cost associated with glyphosate resistance in Echinochloa colona,” PhD thesis, The University of Western Australia, Perth WA, Australia (2016).
- 45.Denis M. H., Delrot S., Carrier-mediated uptake of glyphosate in broad bean (Vicia faba) via a phosphate transporter. Physiol. Plant. 87, 569–575 (1993). [Google Scholar]
- 46.Johnson Z. L., Chen J., Structural basis of substrate recognition by the multidrug resistance protein MRP1. Cell 168, 1075–1085.e9 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Johnson Z. L., Chen J., ATP binding enables substrate release from multidrug resistance protein 1. Cell 172, 81–89.e10 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Smith Z. D., Meissner A., DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 14, 204–220 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Henderson I. R., Jacobsen S. E., Epigenetic inheritance in plants. Nature 447, 418–424 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Wang H., et al., CG gene body DNA methylation changes and evolution of duplicated genes in cassava. Proc. Natl. Acad. Sci. U.S.A. 112, 13729–13734 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jones P. A., Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13, 484–492 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Cokus S. J., et al., Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452, 215–219 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goh S. S., et al., Non-target-site glyphosate resistance in Echinochloa colona from Western Australia. Crop Prot. 112, 257–263 (2018). [Google Scholar]
- 54.Moreau C., Prost A. L., Dérand R., Vivaudou M., SUR, ABC proteins targeted by KATP channel openers. J. Mol. Cell. Cardiol. 38, 951–963 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Remy E., et al., A major facilitator superfamily transporter plays a dual role in polar auxin transport and drought stress tolerance in Arabidopsis. Plant Cell 25, 901–926 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McElroy J. S., Hall N. D., Echinochloa colona with reported resistance to glyphosate conferred by aldo-keto reductase also contains a Pro-106-Thr EPSPS target site mutation. Plant Physiol. 183, 447–450 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu Q., Ahmad-Hamdani M. S., Han H., Christoffers M. J., Powles S. B., Herbicide resistance-endowing ACCase gene mutations in hexaploid wild oat (Avena fatua): Insights into resistance evolution in a hexaploid species. Heredity 110, 220–231 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Han H., Yu Q., Widderick M. J., Powles S. B., Target-site EPSPS Pro-106 mutations: Sufficient to endow glyphosate resistance in polyploid Echinochloa colona? Pest Manag. Sci. 72, 264–271 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Canarini A., Kaiser C., Merchant A., Richter A., Wanek W., Root exudation of primary metabolites: Mechanisms and their roles in plant responses to environmental stimuli. Front. Plant. Sci. 10, 157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weston L. A., Ryan P. R., Watt M., Mechanisms for cellular transport and release of allelochemicals from plant roots into the rhizosphere. J. Exp. Bot. 63, 3445–3454 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The EcABCC8 and EcABCC10 complementary DNA sequences data have been deposited in the GenBank database (access nos. MT249005 and MT249006). All data used in the study are included in the paper and SI Appendix. All protocols are described in SI Appendix, Materials and Methods or in the references therein. Plant materials are available upon request by qualified researchers to the corresponding author.