Significance
Comprehensive assessment of matrisome genes identified collagen IV as one of the many extracellular matrix (ECM) proteins regulating the stem cell pool in planarian tissue homeostasis and regeneration. While collagen IV has been shown to be involved in stem cell biology, our finding links it to pluripotent stem cells in vivo, including self-renewal and differentiation into tissue-specific progenitors. We show a link between the ECM niches in the parenchyma/gut region and EGF/neuregulin-secreting neurons, thus providing mechanistic insight into interactions between cell niches. The conservation of basement membranes between planarian and mammalian gut niches suggests a similar interplay may exist in the mammalian systems, worthy of further investigation.
Keywords: planarian, extracellular matrix, stem cell niche, pluripotency, regeneration
Abstract
The extracellular matrix (ECM) provides a precise physical and molecular environment for cell maintenance, self-renewal, and differentiation in the stem cell niche. However, the nature and organization of the ECM niche is not well understood. The adult freshwater planarian Schmidtea mediterranea maintains a large population of multipotent stem cells (neoblasts), presenting an ideal model to study the role of the ECM niche in stem cell regulation. Here we tested the function of 165 planarian homologs of ECM and ECM-related genes in neoblast regulation. We identified the collagen gene family as one with differential effects in promoting or suppressing proliferation of neoblasts. col4-1, encoding a type IV collagen α-chain, had the strongest effect. RNA interference (RNAi) of col4-1 impaired tissue maintenance and regeneration, causing tissue regression. Finally, we provide evidence for an interaction between type IV collagen, the discoidin domain receptor, and neuregulin-7 (NRG-7), which constitutes a mechanism to regulate the balance of symmetric and asymmetric division of neoblasts via the NRG-7/EGFR pathway.
Across the animal kingdom, stem cell function is regulated by the microenvironment in the surrounding niche (1), where the concentration of molecular signals for self-renewal and differentiation can be precisely regulated (2). The niche affects stem cell biology in many processes, such as aging and tissue regeneration, as well as pathological conditions such as cancer (3). Most studies have been done in tissues with large stem cell populations, such as the intestinal crypt (4) and the hair follicle (5) in mice. Elucidation of the role of the stem cell niche in tissue regeneration requires the study of animals with high regenerative potential, such as freshwater planarians (flatworms) (6). Dugesia japonica and Schmidtea mediterranea are two well-studied species that possess the ability to regenerate any missing body part (6, 7).
Adult S. mediterranea maintain a high number of stem cells (neoblasts)—∼10 to 30% of all somatic cells in the adult worm—with varying potency, including pluripotent cells (8–14). Neoblasts are the only proliferating somatic cells: they are molecularly heterogeneous, but all express piwi-1 (15–18). Lineage-committed neoblasts are “progenitors” that transiently express both piwi-1 and tissue-specific genes (15, 19). Examples include early intestinal progenitors (γ neoblast, piwi-1+/hnf4+) (8, 10, 15, 19–21) and early epidermal progenitors (ζ neoblast, piwi-1+/zfp-1+) (8, 15). Other progenitor markers include collagen for muscles (22), ChAT for neurons (23), and cavII for protonephridia (24, 25). During tissue regeneration, neoblasts are recruited to the wound site, where they proliferate then differentiate to replace the missing cell types (16, 26). Some neoblasts express the pluripotency marker tgs-1, and are designated as clonogenic neoblasts (cNeoblasts) (10, 11). cNeoblasts are located in the parenchymal space adjacent to the gut (11).
Neoblasts are sensitive to γ-irradiation and can be preferentially depleted in the adult planarian (27). After sublethal γ-irradiation, remaining cNeoblasts can repopulate the stem cell pool within their niche (10, 11). The close proximity of neoblasts to the gut suggests gut may be a part of neoblast niche (28, 29). When gut integrity was impaired by silencing gata4/5/6, the egfr-1/nrg-1 ligand-receptor pair, or wwp1, maintenance of non–γ-neoblasts were also disrupted (20, 30, 31), but whether that indicates the gut directly regulates neoblast remains unclear. There is evidence indicating the dorsal-ventral (D/V) transverse muscles surrounding the gut may promote neoblast proliferation and migration, with the involvement of matrix metalloproteinase mt-mmpB (32, 33). The central nervous system has also been implicated in influencing neoblast maintenance through the expression of EGF homolog neuregulin-7 (nrg-7), a ligand for EGFR-3, affecting the balance of neoblast self-renewal (symmetric or asymmetric division) (34).
In other model systems, an important component of the stem-cell niche is the extracellular matrix (ECM) (35). Germline stem cells in Drosophila are anchored to niche supporting cells with ECM on one side, while the opposite side is exposed to differentiation signals, allowing asymmetric cell fate outcomes for self-renewal or differentiation following division (36–38). Few studies have addressed the ECM in planarians, largely due to the lack of genetic tools to manipulate the genome, the absence of antibodies to specific planarian ECM homologs, or the tools required to study cell fate changes. However, the genomes of D. japonica (39–41) and S. mediterranea (41–45), and single-cell RNA-sequencing (scRNA-seq) datasets for S. mediterranea are now available (11, 46–50). A recent study of the planarian matrisome demonstrated that muscle cells are the primary source of many ECM proteins (51), which, together with those produced by neoblasts and supporting parenchymal cells, may constitute components of the neoblast niche. For example, megf6 and hemicentin restrict neoblast’s localization within the parenchyma (51, 52). Functional studies also implicate ECM-modifiers, such as matrix metalloproteases (MMPs) in neoblast migration and regeneration. For example, reducing the activity of the ECM-degrading enzymes mt-mmpA (26, 33), mt-mmpB (53), or mmp-1 (33) impaired neoblast migration, proliferation, or overall tissue growth, respectively. Neoblasts are also likely to interact with ECM components of the niche via cell surface receptors, such as β1 integrin, inactivation of which impairs brain regeneration (54, 55).
Here, we identified planarian ECM homologs in silico, followed by systematic functional assessment of 165 ECM and ECM-related genes by RNA interference (RNAi), to determine the effect on neoblast repopulation in planarians challenged by a sublethal dose of γ-irradiation (10). Surprisingly, multiple classes of collagens were shown to have the strongest effects. In particular, we show that the type IV collagens (COLIV) of basement membranes (BMs), were required to regulate the repopulation of neoblasts as well as lineage progression to progenitor cells. Furthermore, our data support an interaction between COLIV and the discoidin domain receptor (DDR) in neurons that activates signaling of NRG-7 in the neoblasts to regulate neoblast self-renewal versus differentiation. Together, these data demonstrate multifaceted regulation of planarian stem cells by ECM components.
Results
Identification and Characterization of Planarian ECM Genes.
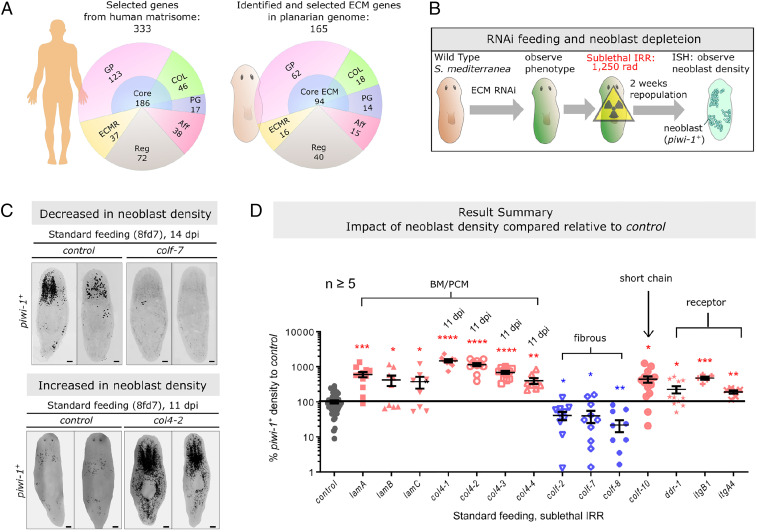
To identify conserved ECM components in the planarian genome, we searched for homologs to sequences in the human/mouse matrisome database that contains all known core ECM and ECM-related proteins (56) (SI Appendix, Fig. S1A). We identified 165 candidates and verified the sequence identities by reverse-BLAST to human and Caenorhabditis elegans counterparts. This represented our primary set of planarian ECM genes for functional assessment (Fig. 1A and SI Appendix, Fig. S1A). An additional 68 genes containing some ECM domains formed a secondary set that was not investigated in this study (Materials and Methods and SI Appendix, Supplementary Methods). Of the primary set, 94 encode core ECM proteins, 15 ECM-affiliated proteins, 40 ECM regulators, and 16 ECM receptors (Fig. 1A, SI Appendix, Fig. S1A, and Dataset S1). The genes in our primary set contained 94 of the 133 core matrisome proteins and 71 of the 167 matrisome-associated proteins identified in a recent in silico assessment of the planarian matrisome (51). In situ hybridization (ISH) and analysis of scRNA-seq data showed these ECM genes were expressed in diverse tissue types, including intestine, muscles, pharynx, parenchymal cells, neurons, pigment cells, and putative immune cells (SI Appendix, Fig. S1C and Dataset S1).
Fig. 1.
Identification of stem cell-regulatory ECM genes in the planarian. (A) Summary of the planarian matrisome identified in reference to the human matrisome. Major ECM groups and the respective number of genes are listed. Aff, ECM specific-affiliated protein; COL, collagen; Core, core ECM; ECMR, ECM-receptor; GP, glycoprotein; PG, proteoglycan; Reg, ECM-regulator. (B) Schematic diagram outlining the RNAi screening (primary), from RNAi feeding, sublethal γ-irradiation, to neoblast repopulation. (C) Representative FISH-confocal (tiled z-projection) images of ECM(RNAi) resulted in reduced neoblast density for colf-7(RNAi) or increased for col4-2(RNAi). Background outside the margin of the worm is removed for presentation consistency. (Scale bars, 100 μm.) (D) Summary of ECM candidate genes with positive or negative effects on neoblast repopulation from RNAi screening, expressed as percentage change relative to control worms. Unless specified, all groups are inspected at 14 dpi. Each dot represents the result of a single animal in each group, and the mean ± SEM is presented. Statistical analysis was assessed using two-tailed unpaired Student t tests. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Planarian Collagen Genes Can Be Classified as Fibrillar, FACIT, or COLIV Types.
Similar to the study of the planarian matrisome (51), we identified 18 collagen genes in the S. mediterranea genome with predicted protein structure and domains with characteristics for fibrillar collagen and COLIV α-chains (SI Appendix, Fig. S1B). We used the same gene nomenclature of the collagen genes as previously reported (51).
From phylogenetic analysis, 11 planaria collagen genes were clustered and predicted to be fibrillar collagens, colf-1 to -11 (SI Appendix, Fig. S2, Upper). The predicted α-chains for colf-8 and colf-11 were most consistent with fibril-forming collagens with no interruptions within the predicted triple helical domain, while others contained multiple interruptions (SI Appendix, Fig. S2, Lower) that were more consistent with vertebrate fibril-associated collagens with interrupted triple helix (FACIT) collagen type. The predicted α-chain for colf-10 had a short helical domain with four interruptions and two predicted TSPN-like domains in the N-terminal region (SI Appendix, Fig. S2, Lower), which resembled the short-chain collagens found in vertebrates and the nonvertebrate chordate Ciona intestinalis (57).
Seven planarian collagen genes, col4-1 to -7, had predicted protein sequences characteristic of COLIV chains. Phylogenetic analysis showed that these genes did not fall into the conventional COLIV α1-like and α2-like clusters (58), but formed a separate cluster with other platyhelminth sequences, within which col4-1 and col4-7 were in one subcluster, and col4-2 to -6 in another (SI Appendix, Fig. S3, Upper). The predicted protein domains showed the characteristic 7S, interrupted triple helix, and NC1 domains of COLIV (SI Appendix, Fig. S3, Lower). With the exception of col4-3, all COLIV chains contained the hydroxylysine and methionine residues used for the formation of a sulfilimine bond in the NC1 domain (59). The presence of these conserved domains suggested conservation of function across species.
Assessment of ECM Function during Neoblast Repopulation.
We performed RNAi of all genes in the primary set (SI Appendix, Fig. S4A) and, following sublethal γ-irradiation (1,250 rad), measured the effect on neoblast repopulation at 11 or 14 d (Fig. 1B and SI Appendix, Fig. S4B). All genes were screened using the standard eight-feed schedule (Materials and Methods). In brief, worms in this schedule were γ-irradiated on the 7th day after the last feed, designated as eight feeds-day 7 (8fd7), corresponding to day 28 of RNAi. Genes with no observable phenotype were rescreened using an alternative feeding schedule (SI Appendix, Fig. S4B).
In control(RNAi) worms on the standard eight-feed schedule, neoblasts repopulated from an average of 13.4 ± 2.50 cells/mm2 at 3 d post–γ-irradiation (dpi) to 169.4 ± 37.81 cells/mm2 at 14 dpi (SI Appendix, Fig. S5). Key changes in neoblast repopulation behavior are summarized in Dataset S1. RNAi of COLIV (col4-1, -2, -3, or -4) and laminin (lamA, -B, or -C) genes, encoding BM and pericellular matrix (PCM) components, resulted in significantly higher neoblast densities than in control(RNAi) worms at 11 or 14 dpi, suggesting these ECM molecules hinder neoblast repopulation (Figs. 1 C and D and 2 A and B and SI Appendix, Fig. S6A). RNAi of two classes of ECM receptor genes, ddr-1, and integrins (itgB1, itgA1, or itgA4), also resulted in higher neoblast densities during repopulation (Fig. 1D and SI Appendix, Fig. S6B), suggesting the BM/PCM proteins interact with neoblast ECM receptors. Conversely, RNAi of fibrillar or fibrillar-related collagen genes, colf-2, colf-7, or colf-8, reduced neoblast densities, suggesting they facilitate neoblast repopulation (Figs. 1 C and D and 2 C and D). An exception was that RNAi of colf-10 resulted in a higher density of neoblasts during repopulation (Fig. 2 C and D). Under the alternative feeding schedule, RNAi of netrin-2 resulted in more condensed local neoblast clusters during repopulation, but the overall neoblast density remains similar to the control group (SI Appendix, Fig. S6C). megf6(RNAi) or hemicentin-1(RNAi) resulted in ectopic neoblast cluster formation at the epidermis (SI Appendix, Fig. S6D), consistent with previous studies (51, 52).
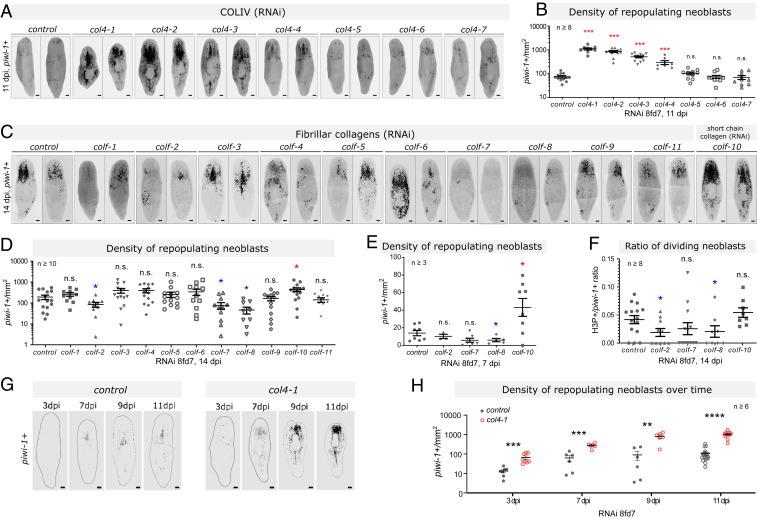
Fig. 2.
Effect of collagen types on the dynamics of neoblast repopulation. (A and B) Representative FISH-confocal images (A) and density quantifications (B) of repopulating neoblast (piwi-1+) in COLIV(RNAi) at 8fd7, 11 dpi. (C and D) Representative FISH-confocal images (C) and density quantifications (D) of repopulating neoblast (piwi-1+) in fibrillar/short chain collagen(RNAi) at 14 dpi. (E) Density of repopulating neoblasts in fibrillar/short chain collagen(RNAi), quantified by number of piwi-1+ cells at 8fd7, 3 dpi. (F) Ratio between H3P+/piwi-1+ cells vs. all piwi-1+ cells. (G and H) Neoblast densities compared between control(RNAi) and col4-1(RNAi) (8fd7) from 3 to 11 dpi. Background outside the margin of the worm is removed for presentation consistency. (Scale bars, 100 μm.) For B–F and H number of animals tested is stated by the number of dots in each group. The mean ± SEM of each group is presented. Statistical significance was assessed using two-tailed unpaired Student t tests. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; n.s., not significant. All FISH images are tiled z-projections.
To investigate the differential effects of collagen types on neoblast behavior, we assessed recovery/repopulation of neoblasts over time in collagen RNAi worms. At 7 dpi, the densities of repopulating neoblasts for colf-2(RNAi) or colf-7(RNAi) worms were similar to control(RNAi) worms (Fig. 2E), indicating the observed change at 14 dpi was the result of a later-stage decline in neoblast proliferation (Fig. 2D). Using histone-H3-phosphate on serine 10 (H3P) as an active cell cycle marker, we confirmed a significant reduction of proliferating neoblasts in colf-2(RNAi) or colf-8(RNAi) worms at 14 dpi (Fig. 2F). For colf-10(RNAi) worms, a higher density of repopulating neoblasts was observed at 3 dpi, but the opposite was seen in colf-8(RNAi) worms (Fig. 2E), suggesting colf-8 or -10 may both regulate neoblast proliferation after the early repopulation phase, but with opposite roles.
Of the COLIV genes, col4-1(RNAi) had the greatest effect at 11 dpi. A time series showed that RNAi of col4-1 increased neoblast density throughout the repopulation process (Fig. 2 G and H), from the earliest time point of repopulation at 3 dpi. This indicated that col4-1 regulates the early repopulation process. It is also possible that in col4-1(RNAi) worms, more neoblasts might have survived after γ-irradiation, meaning there may be a higher density of radiation-resistant neoblasts before γ-irradiation.
Fibrillar and Type IV Collagens Provide Distinct Microenvironments for Neoblast Function.
Fibrillar and BM collagens are normally found in distinct tissue and cell compartments (60). Our finding of opposing effects of the fibrillar and BM collagens on repopulation was intriguing. We therefore investigated the cellular expression and physical location of these proteins with respect to the neoblast microenvironment.
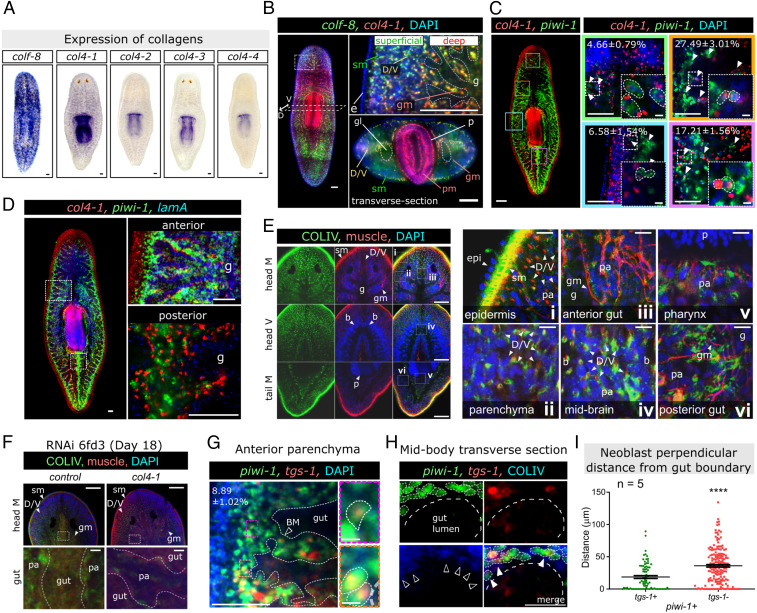
Most of the collagen genes were expressed in muscle cells (51) (SI Appendix, Figs. S7 and S8A). The expression patterns of fibrillar collagen genes, colf-1 to -6, -8, and -9 were similar in all mature muscles and subclusters of the subepidermal, gut, pharyngeal, and intestinal types, while colf-7 and colf-10 were detected predominantly in the pharynx muscles and subepidermal layer, respectively (SI Appendix, Fig. S7). COLIV genes, col4-1, col4-2, and col4-3, were expressed across all subclusters of muscle cells (SI Appendix, Fig. S8A, circled) and in the muscle-specialized neoblast subcluster (SI Appendix, Fig. S8B), with col4-1 as the most highly expressed COLIV gene in neoblasts (SI Appendix, Fig. S8A). col4-4 and col4-7 showed a similar, but weaker, pattern of expression, and col4-6 was not detected in any of the muscle subgroups (SI Appendix, Fig. S8A). col4-1, but not other collagen genes, was expressed in pluripotent (tgs-1+) and other tissue-committed neoblast subclusters, including γ- (intestinal, pharyngeal; cathepsin+ and hnf4+), ζ- (epidermal; cathepsin+ and zfp-1+), and protonephridia (pou2/3+ and cavII+) neoblasts (Fig. 3B and SI Appendix, Fig. S8 A and B).
Fig. 3.
Fibrillar and type IV collagens have different environmental distributions. (A) Wild-type colorimetric-ISH of colf-8 and col4-1 to -4. Background outside the margin of the worm is removed for presentation consistency. (B–H) FISH/immunofluoresent (IF) confocal-images of single z-plane. (B) Coexpression of colf-8 and col4-1 by FISH in whole animal (Left), head coronal section (Upper Right), and midbody transverse section (Lower Right). Dotted line: gut-parenchymal boundary. The “superficial” and “deep” parenchymal regions are indicated. (C) Coexpression of col4-1 with piwi-1 by FISH. Inner head (green frame), anterior deep parenchyma (orange frame), lateral epidermis, and superficial (blue frame), and posterior deep parenchyma (purple frame). White arrowheads and small panels: col4-1+/piwi-1+ neoblasts. The percentage of col4-1+/piwi-1+ cells within all col4-1+ cells is indicated in each panel (value ± SEM n = 5). (D) Neoblasts and muscles produce col4-1 and gut enterocytes produce lamA. (E) αCOLIV and muscle (α6G10) stains BM and PCM-like structures surrounding subepidermal (i), D/V (ii), anterior gut (iii), brain (iv), pharynx (v), and posterior gut (vi) muscles. (F) αCOLIV and α6G10 after six RNA feeds (18 d post-RNAi, 6fd3: 6-feeds-day3). (Lower) Gut-parenchyma. (G) Dorsal view of parenchymal region showing tgs-1+/piwi-1+ (outlined in Inset images) and other (piwi-1+/tgs-1−) neoblasts relative to the position of the gut BM (white dotted lines). (H) BM and PCM-like structures (αCOLIV, hollow arrowheads) costained with piwi-1+/tgs-1+ and other neoblasts (piwi-1+/tgs-1−). White outlines: neoblasts; arrowheads: COLIV-contacting neoblasts. (I) Quantifications of tgs-1+/piwi-1+ and piwi-1+/tgs-1− cells’ closest perpendicular distance to gut boundary (254 neoblasts are measured in the anterior parenchyma (region shown in G) from five animals, each dot represents one neoblast). Mean ± SEM is presented, and statistical significance was assessed using two-tailed unpaired Student t tests. ****P < 0.0001. For B–F: b, brain; D/V, D/V muscles; g, gut; gl, gut lumen; gm, gut muscles; head M, middle head section; head V, ventral head section; p, pharynx; pa, parenchyma; pm, pharyngeal muscles; sm, subepidermal muscles; tail M, middle tail section. (Scale bars: C small panels and G small panels, 10 μm; others, 100 μm.)
To validate the scRNA-seq collagen expression findings, we performed ISH, which showed cells in the subepidermal/superficial parenchyma expressed colf-8, and cells in the deep parenchyma expressed col4-1, -2, and -3 (Fig. 3 A and B). From cell quantification using double-fluorescent ISH (dFISH) of col4-1 and piwi-1, we estimated 27.5 ± 3.01% and 17.2 ± 1.56% of col4-1+ cells within the anterior and posterior deep parenchyma, respectively, were neoblasts (Fig. 3C). Reducing the number of neoblasts by sublethal γ-irradiation decreased col4-1 expression at 3 dpi (SI Appendix, Fig. S8 C and D). These findings suggest neoblasts may contribute to the COLIV matrix in the parenchyma.
Different Cell Types Contribute to the Neoblast Microenvironment.
Laminin genes encode another class of the ECM component that negatively affects neoblast proliferation, as shown using RNAi (SI Appendix, Fig. S6A). LamA is expressed in adjacent intestinal cells, but not in piwi-1+ neoblasts (Fig. 3D). Using an antibody known to stain planarian COLIV proteins (61) and costained with an antibody for muscle (α6G10) (62), we detected BM-like structures within the deep parenchyma (Fig. 3E), enclosing the subepidermal muscles (Fig. 3 E, i), D/V muscles (Fig. 3 E, ii), gut branches (Fig. 3 E, iii and vi), D/V muscles through the brain (Fig. 3 E, iv), and surrounding the pharynx (Fig. 3 E, v). Suppression of COLIV production reduced COLIV secretion (SI Appendix, Fig. S8E), specifically in subepidermal, gut, and D/V muscles in the parenchyma (Fig. 3F). Dense COLIV patches were observed in some deep gut-parenchymal regions (SI Appendix, Fig. S8F), indicating altered ECM structure. tgs-1+ pluripotent neoblasts in the deep parenchyma contributed 8.89 ± 1.02% of the neoblast population in the anterior parenchyma (Fig. 3G) and were located close to the gut BM (Fig. 3H). At midcoronal sections of the animal, the majority of tgs-1+ neoblasts are located closer to the gut boundary compared to tgs-1− neoblasts (Fig. 3 G and I). Thus, a COLIV/laminin-containing BM environment may provide a niche for tgs-1+ neoblasts, with laminins contributed by intestinal cells.
col4-1 Regulates the Proportion of Pluripotent and Progenitor Cell Pools.
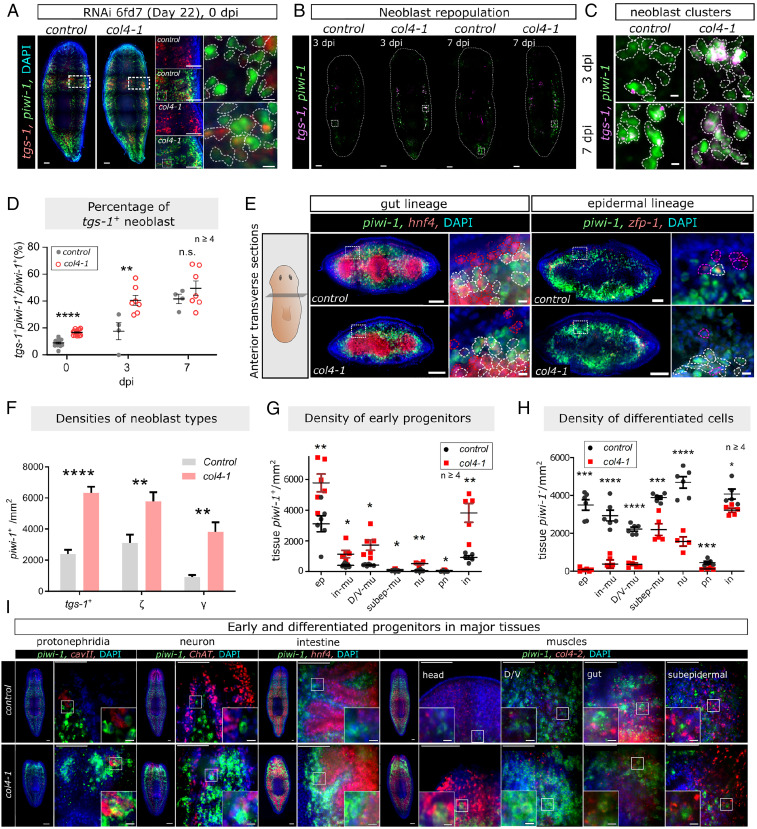
Under homeostatic (non–γ-irradiated) conditions, eight-feed RNAi of colf-2, -7, or -8 resulted in no change in neoblast proliferation (SI Appendix, Fig. S9 A and B) and muscle fiber density (SI Appendix, Fig. S9 C and D), whereas itgB1(RNAi) disrupted muscle fiber density (SI Appendix, Fig. S9C). In contrast, col4-1(RNAi) resulted in an increase in the neoblast population that was noticeable after six RNAi feeds (SI Appendix, Fig. S9 E and F). The proportion of tgs-1+ neoblasts increased from 8.66 ± 0.87% in control(RNAi) worms to 15.31 ± 0.51% in col4-1(RNAi) worms, following the standard RNAi feeding protocol (Fig. 4A). After sublethal γ-irradiation, the proportion of tgs-1+ neoblasts in control(RNAi) worms increased to 17.82 ± 3.98% at 3 dpi, and 46.94 ± 6.81% by 7 dpi (Fig. 4 B–D). In col4-1(RNAi) worms, there were more tgs-1+ neoblasts than in control(RNAi) at 0 (before γ-irradiation) and 3 dpi, but not at 7 dpi (Fig. 4D). This suggests that in a COLIV-deficient niche, there is a higher proportion of tgs-1+ neoblasts prior to γ-irradiation and more neoblasts survive the γ-irradiation.
Fig. 4.
COLIV controls tgs-1+ neoblast population size and lineage commitment. (A and B) Confocal images of tgs-1+/piwi-1+ and tgs-1−/piwi-1+ neoblasts at non–γ-irradiated (0 dpi), 3 and 7 d dpi after receiving sublethal γ-irradiation, repopulating neoblast clones shown in C, quantified in D. (E) FISH of prepharyngeal transverse sections showing γ (hnf4+/piwi-1+) and ζ-neoblasts (zfp-1+/piwi-1+) (white outlines). Nonneoblast progenitors of gut (hnf4+/piwi-1−) and epidermis (zfp-1+/piwi-1−): red and magenta outlines, respectively. (F) Densities of tgs-1+, ζ- and γ-neoblasts. (G, H) Densities of piwi-1+ (G) and piwi-1− (H) cells within early epidermal progenitors (ep) (zfp-1+), protonephridia (pn) (cavII+), neuron (nu) (ChAT+), intestine (in) (hnf4+), D/V muscles (D/V-mu), gut muscles (in-mu), and subepidermal muscles (subep-mu) (col4-2+). (I) Lineage-committed neoblasts within tissue compartments. White box and magnified Insets highlight piwi-1 and tissue marker in control(RNAi) and col4-1(RNAi) worms. For C, D, G, and H, the number of animals tested is indicated by the number of data points in each group. Mean ± SEM is presented for each assessment, and statistical significance was assessed using two-tailed unpaired Student t tests. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Confocal images are shown as tiled z-projection (A–C) or single z-plane (E and I). (Scale bars: A Right, C and E small panels, and I small panels, 10 μm; others, 100 μm.)
Following col4-1(RNAi), quantitative analyses showed increased densities for ζ- (piwi-1+/zfp-1+) and γ- (piwi-1+/hnf4+) neoblasts (Fig. 4 E and F). Higher percentages of tgs-1+ and γ-types were observed compared to control(RNAi). The ζ-neoblast proportion remained unchanged, while the proportion of the “other” lineages (piwi-1+/tgs-1−/hnf4−/zfp-1−) decreased (SI Appendix, Fig. S9G). The proportion (SI Appendix, Fig. S9H) and density (Fig. 4G) of piwi-1+ lineage-specific progenitor cells expressing cavII (protonephridia), ChAT (nervous system), or col4-2 (D/V, gut, subepidermal, and head tip muscles) generally increased. Corresponding decreases of density were observed in the respective differentiated (piwi-1−) cell types (Fig. 4H). The changes in the progenitor/differentiated cell proportions were quantified using dFISH with piwi-1, together with markers for the protonephridia (cavII), neurons (ChAT), intestine (hnf4), or muscle (col4-2) (Fig. 4I), as well as epidermal progenitors (zfp-1) (Fig. 4E). Overall, these findings suggested that the COLIV environment regulates the proportions between stem cell and postmitotic tissues.
COLIV Deficiency Impairs Differentiation of Tissue Progenitor Cells.
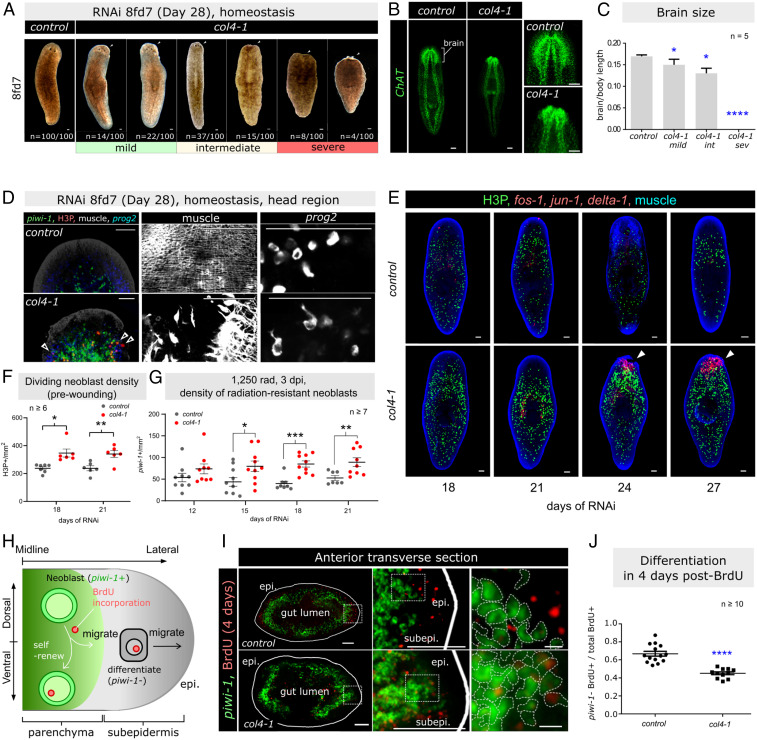
RNAi of col4-1 led to a spectrum of morphological phenotypes, classified as: 1) mild with no obvious tissue loss, 2) intermediate with reduction of head size anterior to the eyespots, and 3) severe with complete loss of the head (Fig. 5A). Assessment of ChAT expression in worms with intermediate phenotype showed smaller brains (Fig. 5 B and C). Associated with this phenotype was ectopic proliferation of neoblasts (piwi-1+/H3P+) in the epidermal progenitor (prog2+) compartment, where neoblasts are absent in control(RNAi) worms, and fewer epidermal progenitor cells (prog2+) and muscle fibers (6G10+) (Fig. 5D and SI Appendix, Fig. S10 A–E). The reduction in progenitor cells correlated with severity of anterior tissue regression (Fig. 5C). Similarly, in col4-1(RNAi) worms with intermediate phenotypes, all three muscle types (circular, longitudinal, and diagonal) were reduced in the regressing regions, although muscles appeared to be intact in the region posterior to the eyespots (SI Appendix, Fig. S10E)
Fig. 5.
Impaired neoblast differentiation disrupted tissue homeostasis in col4-1(RNAi) worms. (A) col4-1(RNAi) head regression phenotypes, ranked as “mild,” “intermediate,” to “severe.” White arrowhead: site of tissue regression (n = 100 worms per group). Background outside the margin of the worm is removed for presentation consistency. (B) Regression of the brain in col4-1(RNAi) animals with the intermediate phenotype. (C) Quantification of brain-to-body length ratio (n = 5 worms per group. int, intermediate; sev, severe). (D, Left) Head region of control(RNAi) and intermediate col4-1(RNAi) worms stained for neoblasts (piwi-1+), proliferating neoblasts (H3P+), epidermal progenies (prog2+), and muscle fibers (6G10+). White arrowheads: actively proliferating neoblasts in epidermal region (Center) Muscle fiber network. (Right) Epidermal progeny. (E) FISH/IF of wounding genes (fos-1, jun-1, delta-1), proliferating neoblasts (H3P+), and muscle (6G10+) from 18 to 27 d of RNAi (5fd6 to 8fd6). (F) Quantification of dividing neoblast density (H3P+) in RNAi animals when no detectable wounding was observed (18 of 21 d of RNAi). (G) Density of neoblasts (piwi-1+) for RNAi worms fed until 3fd3/4fd3/5fd3/6fd3, respectively, and then sublethally γ-irradiated and harvested at 3 dpi (in total 12 to 21 d of RNAi). (H) Schematic diagram of BrdU pulse-chase labeling of neoblast in mild phenotype col4-1(RNAi) worms. (I) BrdU and piwi-1. White line: epidermal edge. (Right) Lateral edge of neoblast compartment. Epi, epidermis; subpei, subepidermal region. (J) Quantification of the ratios of piwi-1− BrdU+ cells to total BrdU+ cells, 4 d after pulse labeling. For F, G, and J, the number of animals tested is indicated by the number of data points in each group. Mean ± SEM is presented, and statistical significance was assessed using two-tailed unpaired Student t tests. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. (Scale bars in I, Right, 10 μm; all others, 100 μm.) B, D, and E are z-projection images; I is single-plane image.
While tissue regression and muscle defects occurred primarily in the head tip in col4-1(RNAi) worms, epidermal progenitors were decreased throughout the body (SI Appendix, Fig. S10A). Detailed analyses of progenitor cell density in regions posterior to the regressing site showed fewer progenitor cells in the head region posterior to the eyespots and the midbody prepharyngeal region (SI Appendix, Fig. S10 B and C). Neoblast numbers were not affected, leading to an overall change in the progenitor/neoblast ratio (SI Appendix, Fig. S10D).
To assess whether anterior regression may be due to impaired tissue patterning, we analyzed the expression of anterior (notum-1) and midline (slit-1) patterning genes, which showed no observable changes (SI Appendix, Fig. S10F). Apoptotic cell density was similar to control(RNAi) in mild col4-1(RNAi) worms, but significantly increased at the anterior regions in the intermediate and severe phenotypes (SI Appendix, Fig. S10 G and H). Thus, cell death was correlated with severity of tissue regression. Tissue dysmorphology was not restricted to the anterior region, as we observed slight dorsal and ventral bloating at the prepharyngeal region (Fig. 5A). Unlike the previously described edemas caused by protonephridial defects, increased tissue differentiation and parenchymal edema were not observed (63, 64). Instead, FISH of the gut marker mat showed fusion and outgrowth of gut branches in these worms (SI Appendix, Fig. S10I), accompanied by invasion of neoblasts (piwi-1+) into the gut (hnf4+) compartment (SI Appendix, Fig. S10J). These worms also showed thinner and disrupted BM as assessed by electron microscopy in the lateral subepidermis (SI Appendix, Fig. S10K), and by immunostaining with anti-COLIV antibody in the neoblast-gut compartment (SI Appendix, Fig. S10L).
Tissue wounding induces temporary neoblast hyperproliferation and differentiation as part of the normal regenerative mechanism. Thus, the col4-1(RNAi) phenotypes described above may induce wounding signals. Serial assessment (SI Appendix, Fig. S11A) of the early wounding signals (fos-1, jun-1, and delta-1) (65) showed induction after col4-1(RNAi) treatment from day 21 (6fd6) (Fig. 5E and SI Appendix, Fig. S11B), that was concentrated at the head region at day 24 (7fd6), concomitant with anterior muscle regression (Fig. 5E). Ablation of neoblasts by lethal γ-irradiation suppressed activation of wound signals at day 21 of RNAi (SI Appendix, Fig. S11C), suggesting the wounding signals either came primarily from neoblasts, or from postmitotic tissues but downstream of neoblast activities. Next, we assessed neoblasts prior to tissue wounding and observed that there was an increase in radiation-resistant neoblast density and neoblast proliferation rate as early as days 15 (4fd6) and 18 (5fd6), respectively (Fig. 5 E–G). Early increase of radiation-resistant neoblast, reduction of epidermal progenitor, but no change in apoptosis on day 21 of RNAi, suggests the observed COLIV–neoblast relationship is independent of known tissue wounding pathways.
To further assess the relationship between COLIV and neoblasts, we employed an in vitro neoblast culturing system on decellularized planarian ECM (SI Appendix, Fig. S12A) (66, 67). col4-1(RNAi) and control(RNAi) worms from the standard feeding schedule (8fd7) were decellularized, leaving an intact ECM scaffold devoid of cells, as indicated by absence of piwi-1 or DAPI signals (SI Appendix, Fig. S12 B and C). Seeding of neoblast-enriched cell populations from normal donor worms onto these scaffolds showed no significant differences between the two scaffold types in cell attachment following seeding or proliferation after 2 d of culture (SI Appendix, Fig. S12D). However, the relative proportion of piwi-1+ neoblasts was significantly higher in the col4-1(RNAi) matrix than the control one (SI Appendix, Fig. S12 E and F). This was consistent with the notion that a COLIV-deficient ECM scaffold promotes proliferation and maintains “stemness” of the neoblasts, while suppressing differentiation to tissue progenitor cells.
Results thus far pointed to an impairment of differentiation of neoblasts as the primary cause of tissue regression. Thus, we used BrdU pulse-labeling with a time chase to trace the lineage of proliferating neoblasts and their progeny in the parenchyma and subepidermal regions (Fig. 5H). After 4 d postpulse in control(RNAi) worms, most of the labeled cells had migrated away from the parenchyma and entered the subepidermal region (Fig. 5I), while in col4-1(RNAi) worms many labeled cells remained within the parenchyma, and the proportion of differentiated cells, represented by BrdU+ cells that were negative for piwi-1, was reduced (Fig. 5J). Together, these data supported a general impairment of progenitor cell differentiation and survival. As proper lineage commitment and differentiation of pluripotent neoblasts are important for both normal tissue turnover and regeneration in the planarian, we next investigated the effect of col4-1(RNAi) on the formation of a blastema in tissue regeneration.
COLIV Is Required for Blastema Growth and Regeneration.
Amputation activates a switch from tissue maintenance to regeneration. This leads to hyperproliferation of neoblasts, in particular the tgs-1+ neoblasts (10, 11), followed by rapid differentiation to tissue progenitors and formation of the blastema, with complete tissue restoration by 7 d postamputation (dpa), when the neoblast population reestablishes a normal state (6, 49, 65).
We studied the expression of col4-1 during regeneration in wild-type worms amputated at prepharyngeal and postpharyngeal regions, which produces head, trunk, and tail fragments at key stages of the regenerative process (SI Appendix, Fig. S13). There was a gradual increase of col4-1 in the growing blastema but not in old tissues, while the number of H3P+ proliferating neoblasts decreased as the blastema grew in size (SI Appendix, Fig. S14A). While the FISH signal intensity may not directly correlate to the actual gene expression level, it showed a general trend of up-regulation in the blastema.
In col4-1(RNAi) worms following the standard 8fd7 protocol, blastema formation appeared normal until 3 dpa, but was unable to progress to regeneration of the missing tissues, such as the eye spots, which were reestablished by 9 dpa in control(RNAi) worms (SI Appendix, Fig. S14B). When blastema progression was delayed from 3 dpa, the head blastema regressed from 9 dpa, as measured by the head blastema/body size ratio (SI Appendix, Fig. S14C). Consistent with col4-1 FISH, we observed enrichment of COLIV deposition in the blastema of control(RNAi) worms (SI Appendix, Fig. S14D), which was reduced in col4-1(RNAi) worms (SI Appendix, Fig. S14D), accompanied with fewer prog2+ epidermal progeny and 6G10+ muscle fibers (SI Appendix, Fig. S14 E and F). Regenerating ChAT+ brain lobes were substantially smaller and fused at the midline (SI Appendix, Fig. S14 E and G). The proportion of proliferative cells (H3P+) was higher in col4-1(RNAi) than control(RNAi) worms (SI Appendix, Fig. S14 H and I). Neoblasts, which were scarce in control(RNAi) head blastemas, were found in the regenerating head blastemas of col4-1(RNAi) worms at 2 dpa (SI Appendix, Fig. S14 J and K). Similarly, higher densities of cycling neoblasts were observed at 2 and 6 dpa (SI Appendix, Fig. S14L), suggesting more cells retained stem cell characteristics in a COLIV-deficient blastema.
Collectively, our findings indicated that col4-1 was required spatially and temporally to regulate neoblast number and differentiation, fine-tuning tissue homeostasis and regeneration. Next, we addressed whether a physical interaction between neoblasts and a COLIV-containing ECM niche might regulate neoblast behavior.
Integrin or DDR RNAi Worms Phenocopy COLIV Deficiency.
Integrins function as heterodimers of α- and β-chains. S. mediterranea has four α (itgA1-4) and one β (itgB1) integrin genes (54, 55). One would predict that the β1 chain could dimerize with each of the α-chains. S. mediterranea has two ddr genes, ddr-1 and ddr-2. DDR are collagen receptors (68). In the neoblast repopulation assay, ddr-1, itgA1, itgA4, or itgB1(RNAi) showed a similar hyperproliferation phenotype to col4-1(RNAi) (SI Appendix, Fig. S6B).
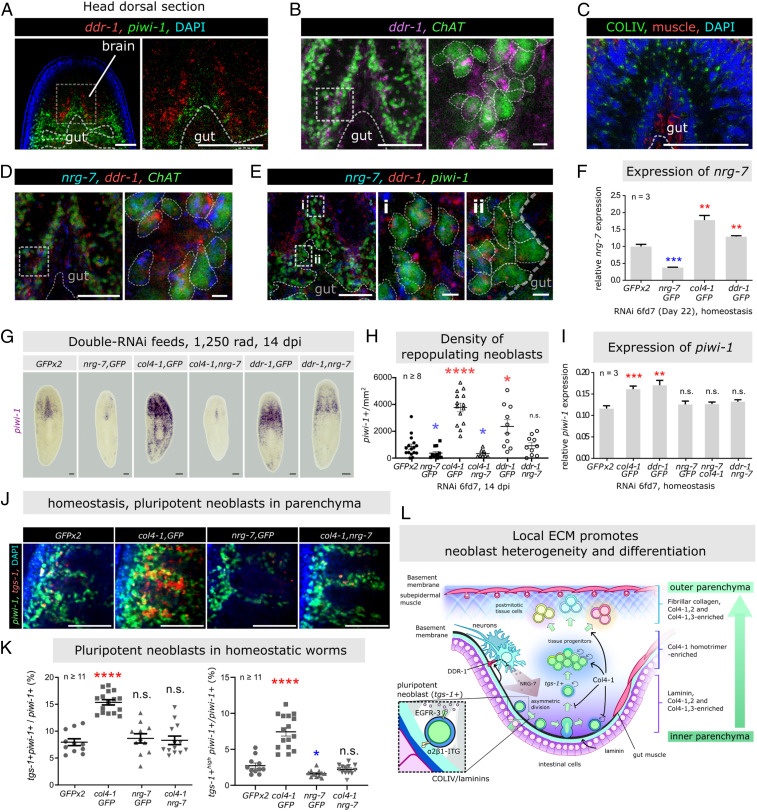
Analysis of the scRNA-seq dataset showed integrin genes were expressed by diverse tissue types, including neoblasts, whereas ddr-1 was expressed primarily by neurons (SI Appendix, Fig. S15A), particularly PC2+ neurons (SI Appendix, Fig. S15B). These expression patterns were confirmed by ISH (SI Appendix, Fig. S15C). In the parenchyma, piwi-1+/itgA4+ neoblasts were located within a COLIV-containing ECM environment (SI Appendix, Fig. S15D), supporting the notion that COLIV and neoblast integrins can interact directly. The parenchyma is dorsal to the neuronal system (brain, ventral nerve cord, and multiple transverse neurons) and enclosing the intestinal system (SI Appendix, Fig. S15E), suggesting both neurons and intestinal cells could serve as niche cells for neoblasts. We confirmed ddr-1 was expressed in the neuronal system (ChAT+) immediately adjacent to neoblasts (piwi-1+) and the gut (structure indicated by DAPI) microenvironment (Fig. 6 A and B). Neurons in the brain were also surrounded by COLIV from the gut and D/V muscles, thus likely to be in contact with COLIV (Fig. 6C).
Fig. 6.
COLIV regulates neoblast potency in tissue maintenance via EGF signaling. (A and B) Animal’s anterior dorsal view showing expression of ddr-1 in relation to neoblasts (piwi-1+) (A), brain neurons (ChAT+) (B), and gut (visualized by DAPI, structure highlighted by dotted lines). Box highlighted some ddr-1+/ChAT+ neurons. (C) Brain structure stained with αCOLIV and α6G10 (muscle). (D) nrg-7+/ddr-1+ neurons, enlarged Right. (E) Gut-neoblast niche enclosed by nrg-7–producing ddr-1+ neurons as niche cells. Boxes: neural-committed neoblasts (i) and other neoblasts (ii). (F) Expression of nrg-7 in the indicated RNAi groups (six feeds–day 7, non–γ-irradiated) by qRT-PCR. (G and H) Neoblast repopulation in col4-1/ddr-1 and nrg-7 double RNAi worms (6 feeds, 1,250 rad γ-irradiated, 14 dpi). (G) FISH piwi-1+ neoblasts, density quantified in H. (I) qRT-PCR quantification of piwi-1 expression in non–γ-irradiated double RNAi worms. (J) Location of pluripotent (tgs-1+/piwi-1+) and other (tgs-1−/piwi-1+) neoblasts in anterior parenchyma (dorsal view). (K) Proportions of tgs-1+ (Left) and tgs-1+high (Right) pluripotent subtypes. For all qRT-PCR assays, gene-expression levels are normalized by housekeeping gene gapdh. Each group is done with biological triplicate (each contained cDNA pooled from three worms) and experimental triplicate. For all qRT-PCR and cell density quantifications with the mean ± SEM presented. Statistical significance was assessed using two-tailed unpaired Student t tests. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. For H and K, the number of data points in each group represents number of animals tested. All images shown are tiled single z-plane confocal. Background outside the margin of the worm is removed for presentation consistency. (Scale bars: B, Right and E small panels, 10 μm; all others, 100 μm.) (L) A model of ECM-neoblast niche in homeostasis. Pluripotent neoblasts and their progenitors surrounded by Col4-1 homotrimers as their territorial ECM niches. Territorial and interterritorial ECM niches act together to restrict population sizes of pluripotent neoblasts in the inner parenchyma, and promotes neoblast differentiation in outer parenchyma. Overall, a broad heterogenicity of neoblasts of different types and potencies can be maintained in close proximity.
PC2+ neurons and some epidermal progenitors express nrg-7 and binding of NRG-7 to its receptor (EGFR-3) on neoblasts regulates asymmetric cell division and self-renewal in neoblast-repopulating worms (34). We hypothesized that there was a relationship between DDR and nrg-7 expression in neurons. ddr-1+/ChAT+ neurons in the brain and ventral nerve cord also expressed nrg-7 (Fig. 6D), demonstrating the presence of ddr-1+/nrg-7+ neurons surrounding the parenchyma, including the gut-neoblast microenvironment (Fig. 6E). Additionally, we detected a small population of nrg-7+ neoblasts adjacent to the brain compartment, consistent with neural lineage-committed neoblasts (Fig. 6 E, i), but we did not detect nrg-7 in other neoblasts (Fig. 6 E, ii). Thus, an interrelationship between COLIV, neurons, and neoblasts may exist in the niche; we therefore investigated this with respect to col4-1 expression and function.
COLIV Regulates Neoblast Pluripotency in Tissue Maintenance via EGF Signaling.
We hypothesized that NRG-7/EGF signaling was downstream of the COLIV/DDR-1 interaction in neurons. RNAi of col4-1 or ddr-1 increased nrg-7 expression as detected by qRT-PCR and FISH (Fig. 6F and SI Appendix, S15F), including in ddr-1+ neurons (SI Appendix, Fig. S15F). Double RNAi of col4-1 and nrg-7 resulted in neoblast hyperproliferation similar to single nrg-7(RNAi) worms in the repopulation assay (Fig. 6 G and H), whereas double RNAi of ddr-1 and nrg-7 restored neoblast proliferation to control(RNAi) levels (Fig. 6 G and H). These data support a functional relationship between these three genes, where nrg-7 was suppressed by COLIV/DDR interaction in the niche surrounding neurons, consistent with our hypothesis.
RNAi of col4-1 and ddr-1 resulted in higher piwi-1 expression in homeostatic worms. While nrg-7(RNAi) caused no significant changes, double RNAi of nrg-7 with col4-1 or ddr-1 restored piwi-1 expression to the levels in control(RNAi) worms (Fig. 6I), supporting a role for NGR-7 downstream of DDR or Col4-1. We propose that this occurs via a direct ECM/receptor interaction at the surface of neurons.
Finally, as RNAi of col4-1 increased the proportion of tgs-1+ neoblasts, we assessed tgs-1+ neoblast proportions in nrg-7(RNAi) worms and found no effect compared with control(RNAi) worms (Fig. 6 J and K). Interestingly, the tgs-1+ neoblasts appeared to be lower in signal intensity compared to all other groups (Fig. 6J). Using a gray-value cutoff of 100 to define a high tgs-1 intensity (tgs-1+high), we found significantly fewer of these cells in nrg-7(RNAi) worms. Thus, nrg-7(RNAi) alters the pluripotent neoblast characteristics that may contribute to poor neoblast repopulation. While col4-1(RNAi) resulted in higher proportions of both total tgs-1+ and tgs-1+high neoblasts, double RNAi of col4-1 and nrg-7 restored the proportions to those of control(RNAi) worms (Fig. 6 J and K). Together, these data support a model where COLIV/DDR1 interactions at a neuronal niche limit self-renewal of pluripotent cells at the neoblast niche via the NRG-7/EGF pathway.
Discussion
In this study, we assessed the role of ECM and ECM-related genes in neoblast function during planarian stem cell maintenance, repopulation, and differentiation. Collagens emerged as the most influential class, with BM and fibrillar collagens showing contrasting outcomes. BM is known to play a role in stem cell regulation, involving its major components, COLIV, and laminins (58). For example, COLIV induces trophoectoderm differentiation of mouse embryonic stem cells (69) and germline stem cell maintenance in Drosophila (70), and laminins facilitate clonal expansion of a single human blastocyst inner cell mass or blastomere cell in culture (71). Here, we describe the role of COLIV in the regulation of stem cell pluripotency in the planarian. COLIV maintains a constant ratio of neoblast subtypes with high and low pluripotency. It also promotes differentiation of pluripotent cells, as the density of tissue progenitor cells is reduced in col4-1(RNAi) worms. While we have not studied all tissue progenitor types, COLIV is likely to have general effect.
The planarian COLIV α-chains contain the typical features of mammalian COLIV α-chains, including the 7S domain, Gly-X-Y repeats, NC1 (C4) domain, and a sulfilimine bond (72). Although the chains do not cluster with the typical α1 and α2 classes in a hierarchical analysis with mammalian COLIV chains, they fall into a separate cluster of two classes, suggesting a similar requirement for the assembly of heterotrimers, comprising combinations of the α1 and α2 classes.
The similarity in the neoblast repopulation outcomes of col4-1, -2, and -3(RNAi), with col4-1(RNAi) being the most severe, suggests col4-1 α-chains form heterotrimers with col4-2 and/or col4-3 α-chains. In support of this notion, the combined RNAi of the genes for all three α-chains had a similar effect to RNAi of col4-1 alone. scRNA-seq showed col4-1, -2, and -3 were all expressed by muscle progenitors. As chain trimerization occurs before secretion, Col4-1/2 and Col4-1/3 heterotrimers can be formed. Nonmuscle cells, including neoblasts, express col4-1 in much higher levels than col4-2 and -3, and thus are likely to produce Col4-1 homotrimers. The redundancy of multiple COLIV types could explain why col4-1(RNAi) alone led to only partial reduction of differentiation, together with incomplete removal of COLIV by knockdown. Other non-COLIV extrinsic pathways that influence differentiation may also act in parallel. For example, RNAi of cystic kidney diseases gene NPHP8 or inhibition of tubule flow increased the number of protonephridial lineage-committed neoblasts and differentiated progenitors (63). col4-1(RNAi) resulted in fewer protonephridia (Fig. 4 H and I), which may impair tubule flow, triggering NPHP8-related phenotypes that partially compensate for the cell loss.
COLIV has been shown in vitro to direct the differentiation of embryonic stem cells into mesoderm lineages, including hematopoietic, endothelial, and smooth muscle cells, in the mouse (73–75) and human (76). How COLIV contributes to the mammalian stem cell niche is not clear. In the adult Drosophila germline stem cell niche, COLIV (Viking) produced by niche-surrounding adipose cells interacted directly with the germline stem cell via β-integrin and focal adhesion kinase, bringing COLIV close to the cell surface (70). During Drosophila development, VIKING binds BMP (Dpp) and promotes the presentation of Dpp receptors at the cell surface for signaling, which is an important process in the control of the early germline stem cell population (77, 78).
In the planarian, while neoblasts are the only proliferating somatic cells, how their cell fates are regulated is poorly understood. γ-Irradiation eliminates the proliferating neoblasts and varying the dosage controls lethal and nonlethal outcomes, suggesting that neoblasts are heterogeneous in terms of cycling state (10). Pluripotent neoblasts are more resistant to γ-irradiation, which suggests COLIV might regulate the cell cycle to maintain a small pool of slow-cycling piwi-1+/tgs-1+ neoblasts. However, a reduction of COLIV in the niche led to amplification/enrichment of this piwi-1+/tgs-1+ neoblast pool, resulting in a hyperproliferative phenotype after sublethal γ-irradiation.
It is likely that neoblasts interact both directly and indirectly with ECM components, as RNAi of neoblast integrins and neural DDR both resulted in neoblast hyperproliferation, the latter through NRG-7 up-regulation. These interactions might be related to cell proliferation and stem cell quiescence, as in the slow-cycling quiescent adult stem cells in mammalian tissues that are mobilized to proliferate for self-renewal and differentiation for tissue repair (79). Because NRG-7 is involved, these interactions are also likely to regulate the self-renewal/differentiation choice (34), but the precise mechanism requires further investigation.
Laminin normally coexists with COLIV in BM structures (58). RNAi of lamA, -B, and -C caused hyperproliferation of neoblasts in the repopulation assay, consistent with cooperation among these ECM molecules. The composition of the BM appears to be finely tuned in the planarian, with COLIV and laminin produced by different cell types, which might provide different and specific cell niches, even for cells with close proximity (Fig. 6L). Long-lived, quiescent adult stem cells can give rise to transitory, short-lived amplifying stem cells, and they both can coexist in close proximity within the niche (80). How such cell type heterogeneity is achieved is not well understood.
In the mammalian intestine, adult stem cells reside at the bottom of crypts and divide continuously. Supporting cells, such as the Paneth and the surrounding mesenchyme, create a specialized metabolic environment that, together with a range of molecular signals, regulates stem cell renewal and differentiation (81). The role of the ECM in this intestinal crypt system is not well understood. The mammalian intestinal crypt contains stem cells with diverse potency and lineage-committed progenitors (82), and Lgr5high intestinal stem cell clones compete against each other (4). In the planarian parenchyma, pluripotent and various subclasses of neoblasts coexist, with possible feedback mechanisms as exemplified in the intestinal crypt (83–85). A COLIV-containing ECM niche restricts the population of pluripotent neoblasts, while promoting lineage progression and the differentiation of functional tissue cells. Supporting neurons, via interaction with a COLIV environment, could regulate the supply of NRG-7 for neoblast self-renewal (Fig. 6L). It is possible that this role for COLIV applies beyond the parenchyma.
The need for COLIV in the neoblast lineage progression from pluripotent to tissue-specific progenitors is best illustrated in the regression of the anterior tissue (head) in col4-1–deficient worms. The same interpretation can be applied to the formation of the blastema after amputation. COLIV is up-regulated in blastema, which may promote local neoblast differentiation to form new tissues.
Understanding how the ECM niche signals to the neoblast is of great interest from many aspects, such as providing clues to mimic planarian regeneration in mammalian systems, in vitro generation of specific progenitor cells, and applications in tissue engineering. This study showed that the neoblast fate-controlling mechanism involves COLIV/integrins and neuronal DDR/NRG-7 in the niche. While the precise mechanism and context of the response transduction need further investigation, our findings support COLIV being a key mechanism in the regulation of symmetric and asymmetric division of neoblasts via the Col4-1/DDR interaction and NRG-7/EGF pathway in self-renewal and differentiation.
Materials and Methods
The authors declare that all data supporting the findings of this study are available within the article and its SI Appendix files, or from the corresponding author upon reasonable request. The detailed materials and methods including identification and selection of planarian ECM genes, phylogenetic analysis of planarian collagens, planarian maintenance, γ-irradiation and RNAi feedings, quantitative real-time PCR, single-cell transcriptomic data analysis, whole-mount ISH and antibody staining, TUNEL and BrdU pulse-chase experiment, neoblast culturing on planarian decellularized matrix scaffold, transmission electron microcopy, and statistical quantifications, are all available in SI Appendix, Supplementary Methods.
Supplementary Material
Acknowledgments
We thank members of the B.J.P. and D.C. laboratories for insightful discussions and comments for this study. This work was supported by the S. Y. and H. Y. Cheng Professor endowment in Stem Cell Biology and Regenerative Medicine (to D.C.). B.J.P. was supported by Ontario Institute for Cancer Research Grant IA-026.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2021251118/-/DCSupplemental.
Data Availability
All study data are included in the article and supporting information. Original images for figure panels where the background was removed can be viewed in the SI Appendix.
References
- 1.Scadden D. T., The stem-cell niche as an entity of action. Nature 441, 1075–1079 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Spradling A., Drummond-Barbosa D., Kai T., Stem cells find their niche. Nature 414, 98–104 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Fane M., Weeraratna A. T., How the ageing microenvironment influences tumour progression. Nat. Rev. Cancer 20, 89–106 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snippert H. J., et al., Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Tanimura S., et al., Hair follicle stem cells provide a functional niche for melanocyte stem cells. Cell Stem Cell 8, 177–187 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Reddien P. W., Sánchez Alvarado A., Fundamentals of planarian regeneration. Annu. Rev. Cell Dev. Biol. 20, 725–757 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Umesono Y., Tasaki J., Nishimura K., Inoue T., Agata K., Regeneration in an evolutionarily primitive brain—The planarian Dugesia japonica model. Eur. J. Neurosci. 34, 863–869 (2011). [DOI] [PubMed] [Google Scholar]
- 8.van Wolfswinkel J. C., Wagner D. E., Reddien P. W., Single-cell analysis reveals functionally distinct classes within the planarian stem cell compartment. Cell Stem Cell 15, 326–339 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karami A., Tebyanian H., Goodarzi V., Shiri S., Planarians: An in vivo model for regenerative medicine. Int. J. Stem Cells 8, 128–133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wagner D. E., Wang I. E., Reddien P. W., Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science 332, 811–816 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeng A., et al., Prospectively isolated tetraspanin(+) neoblasts are adult pluripotent stem cells underlying Planaria regeneration. Cell 173, 1593–1608.e20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayashi T., Asami M., Higuchi S., Shibata N., Agata K., Isolation of planarian X-ray-sensitive stem cells by fluorescence-activated cell sorting. Dev. Growth Differ. 48, 371–380 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Reddien P. W., Bermange A. L., Murfitt K. J., Jennings J. R., Sánchez Alvarado A., Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev. Cell 8, 635–649 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu S. J., Hallows S. E., Currie K. W., Xu C., Pearson B. J., A mex3 homolog is required for differentiation during planarian stem cell lineage development. eLife 4, e07025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu S. J., Pearson B. J., (Neo)blast from the past: New insights into planarian stem cell lineages. Curr. Opin. Genet. Dev. 40, 74–80 (2016). [DOI] [PubMed] [Google Scholar]
- 16.O. C. Guedelhoefer, 4th, Sánchez Alvarado A., Amputation induces stem cell mobilization to sites of injury during planarian regeneration. Development 139, 3510–3520 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shibata N., et al., Inheritance of a nuclear PIWI from pluripotent stem cells by somatic descendants ensures differentiation by silencing transposons in planarian. Dev. Cell 37, 226–237 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Reddien P. W., Oviedo N. J., Jennings J. R., Jenkin J. C., Sánchez Alvarado A., SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science 310, 1327–1330 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Scimone M. L., Kravarik K. M., Lapan S. W., Reddien P. W., Neoblast specialization in regeneration of the planarian Schmidtea mediterranea. Stem Cell Reports 3, 339–352 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barberán S., Fraguas S., Cebrià F., The EGFR signaling pathway controls gut progenitor differentiation during planarian regeneration and homeostasis. Development 143, 2089–2102 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Wenemoser D., Reddien P. W., Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Dev. Biol. 344, 979–991 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Witchley J. N., Mayer M., Wagner D. E., Owen J. H., Reddien P. W., Muscle cells provide instructions for planarian regeneration. Cell Rep. 4, 633–641 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishimura K., Kitamura Y., Taniguchi T., Agata K., Analysis of motor function modulated by cholinergic neurons in planarian Dugesia japonica. Neuroscience 168, 18–30 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Sánchez Alvarado A., Newmark P. A., Robb S. M., Juste R., The Schmidtea mediterranea database as a molecular resource for studying platyhelminthes, stem cells and regeneration. Development 129, 5659–5665 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Rink J. C., Vu H. T., Sánchez Alvarado A., The maintenance and regeneration of the planarian excretory system are regulated by EGFR signaling. Development 138, 3769–3780 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abnave P., et al., Epithelial-mesenchymal transition transcription factors control pluripotent adult stem cell migration in vivo in planarians. Development 144, 3440–3453 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dubois F., Wolff E., [On a localized irradiation method allowing to highlight the migration of regenerative cells in planarians] [in French]. C. R. Seances Soc. Biol. Fil. 141, 903–906 (1947). [PubMed] [Google Scholar]
- 28.Rossi L., Salvetti A., Planarian stem cell niche, the challenge for understanding tissue regeneration. Semin. Cell Dev. Biol. 87, 30–36 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Forsthoefel D. J., et al., An RNAi screen reveals intestinal regulators of branching morphogenesis, differentiation, and stem cell proliferation in planarians. Dev. Cell 23, 691–704 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flores N. M., Oviedo N. J., Sage J., Essential role for the planarian intestinal GATA transcription factor in stem cells and regeneration. Dev. Biol. 418, 179–188 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henderson J. M., et al., Identification of HECT E3 ubiquitin ligase family genes involved in stem cell regulation and regeneration in planarians. Dev. Biol. 404, 21–34 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Dingwall C. B., King R. S., Muscle-derived matrix metalloproteinase regulates stem cell proliferation in planarians. Dev. Dyn. 245, 963–970 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Isolani M. E., et al., Planarians as a model to assess in vivo the role of matrix metalloproteinase genes during homeostasis and regeneration. PLoS One 8, e55649 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lei K., et al., Egf signaling directs neoblast repopulation by regulating asymmetric cell division in planarians. Dev. Cell 38, 413–429 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gattazzo F., Urciuolo A., Bonaldo P., Extracellular matrix: A dynamic microenvironment for stem cell niche. Biochim. Biophys. Acta 1840, 2506–2519 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spradling A. C., et al., Stem cells and their niches: Integrated units that maintain Drosophila tissues. Cold Spring Harb. Symp. Quant. Biol. 73, 49–57 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Morrison S. J., Spradling A. C., Stem cells and niches: Mechanisms that promote stem cell maintenance throughout life. Cell 132, 598–611 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamashita Y. M., Jones D. L., Fuller M. T., Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science 301, 1547–1550 (2003). [DOI] [PubMed] [Google Scholar]
- 39.An Y., et al., Draft genome of Dugesia japonica provides insights into conserved regulatory elements of the brain restriction gene nou-darake in planarians. Zoological Lett. 4, 24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishimura O., et al., Unusually large number of mutations in asexually reproducing clonal planarian Dugesia japonica. PLoS One 10, e0143525 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rozanski A., et al., PlanMine 3.0-improvements to a mineable resource of flatworm biology and biodiversity. Nucleic Acids Res. 47, D812–D820 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robb S. M., Gotting K., Ross E., Sánchez Alvarado A., SmedGD 2.0: The Schmidtea mediterranea genome database. Genesis 53, 535–546 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robb S. M., Ross E., Sánchez Alvarado A., SmedGD: The Schmidtea mediterranea genome database. Nucleic Acids Res. 36, D599–D606 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brandl H., et al., PlanMine—A mineable resource of planarian biology and biodiversity. Nucleic Acids Res. 44, D764–D773 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grohme M. A., et al., The genome of Schmidtea mediterranea and the evolution of core cellular mechanisms. Nature 554, 56–61 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fincher C. T., Wurtzel O., de Hoog T., Kravarik K. M., Reddien P. W., Cell type transcriptome atlas for the planarian Schmidtea mediterranea. Science 360, eaaq1736 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Plass M., et al., Cell type atlas and lineage tree of a whole complex animal by single-cell transcriptomics. Science 360, eaaq1723 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Swapna L. S., Molinaro A. M., Lindsay-Mosher N., Pearson B. J., Parkinson J., Comparative transcriptomic analyses and single-cell RNA sequencing of the freshwater planarian Schmidtea mediterranea identify major cell types and pathway conservation. Genome Biol. 19, 124 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wurtzel O., et al., A generic and cell-type-specific wound response precedes regeneration in planarians. Dev. Cell 35, 632–645 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Molinaro A. M., Pearson B. J., In silico lineage tracing through single cell transcriptomics identifies a neural stem cell population in planarians. Genome Biol. 17, 87 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cote L. E., Simental E., Reddien P. W., Muscle functions as a connective tissue and source of extracellular matrix in planarians. Nat. Commun. 10, 1592 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lindsay-Mosher N., Chan A., Pearson B. J., Planarian EGF repeat-containing genes megf6 and hemicentin are required to restrict the stem cell compartment. PLoS Genet. 16, e1008613 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nykvist P., et al., Distinct recognition of collagen subtypes by alpha(1)beta(1) and alpha(2)beta(1) integrins. Alpha(1)beta(1) mediates cell adhesion to type XIII collagen. J. Biol. Chem. 275, 8255–8261 (2000). [DOI] [PubMed] [Google Scholar]
- 54.Seebeck F., et al., Integrins are required for tissue organization and restriction of neurogenesis in regenerating planarians. Development 144, 795–807 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bonar N. A., Petersen C. P., Integrin suppresses neurogenesis and regulates brain tissue assembly in planarian regeneration. Development 144, 784–794 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hynes R. O., Naba A., Overview of the matrisome—An inventory of extracellular matrix constituents and functions. Cold Spring Harb. Perspect. Biol. 4, a004903 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vizzini A., et al., Cloning and expression of a type IX-like collagen in tissues of the ascidian Ciona intestinalis. Biochim. Biophys. Acta 1577, 38–44 (2002). [DOI] [PubMed] [Google Scholar]
- 58.Fidler A. L., et al., Collagen IV and basement membrane at the evolutionary dawn of metazoan tissues. eLife 6, e24176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vanacore R., et al., A sulfilimine bond identified in collagen IV. Science 325, 1230–1234 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Frantz C., Stewart K. M., Weaver V. M., The extracellular matrix at a glance. J. Cell Sci. 123, 4195–4200 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bansal D., et al., Cytoplasmic poly (A)-binding protein critically regulates epidermal maintenance and turnover in the planarian Schmidtea mediterranea. Development 144, 3066–3079 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ross K. G., et al., Novel monoclonal antibodies to study tissue regeneration in planarians. BMC Dev. Biol. 15, 2 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thi-Kim Vu H., et al., Stem cells and fluid flow drive cyst formation in an invertebrate excretory organ. eLife 4, e07405 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin A. Y., Pearson B. J., Planarian yorkie/YAP functions to integrate adult stem cell proliferation, organ homeostasis and maintenance of axial patterning. Development 141, 1197–1208 (2014). [DOI] [PubMed] [Google Scholar]
- 65.Wenemoser D., Lapan S. W., Wilkinson A. W., Bell G. W., Reddien P. W., A molecular wound response program associated with regeneration initiation in planarians. Genes Dev. 26, 988–1002 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lei K., McKinney S. A., Ross E. J., Lee H.-C., Alvarado A. S., Cultured pluripotent planarian stem cells retain potency and express proteins from exogenously introduced mRNAs. bioRxiv [Preprint] (12 March 2019). 10.1101/573725.
- 67.Schürmann W. P., Peter R., Planarian cell culture: A comparative review of methods and an improved protocol for primary cultures of neoblasts. Belg. J. Zool. 131, 123–130 (2001). [Google Scholar]
- 68.Fu H. L., et al., Discoidin domain receptors: Unique receptor tyrosine kinases in collagen-mediated signaling. J. Biol. Chem. 288, 7430–7437 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schenke-Layland K., et al., Collagen IV induces trophoectoderm differentiation of mouse embryonic stem cells. Stem Cells 25, 1529–1538 (2007). [DOI] [PubMed] [Google Scholar]
- 70.Weaver L. N., Drummond-Barbosa D., Maintenance of proper germline stem cell number requires adipocyte collagen in adult Drosophila females. Genetics 209, 1155–1166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rodin S., et al., Clonal culturing of human embryonic stem cells on laminin-521/E-cadherin matrix in defined and xeno-free environment. Nat. Commun. 5, 3195 (2014). [DOI] [PubMed] [Google Scholar]
- 72.Khoshnoodi J., Pedchenko V., Hudson B. G., Mammalian collagen IV. Microsc. Res. Tech. 71, 357–370 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yamashita J., et al., Flk1-positive cells derived from embryonic stem cells serve as vascular progenitors. Nature 408, 92–96 (2000). [DOI] [PubMed] [Google Scholar]
- 74.Nishikawa S. I., Nishikawa S., Hirashima M., Matsuyoshi N., Kodama H., Progressive lineage analysis by cell sorting and culture identifies FLK1+VE-cadherin+ cells at a diverging point of endothelial and hemopoietic lineages. Development 125, 1747–1757 (1998). [DOI] [PubMed] [Google Scholar]
- 75.Xiao Q., Zeng L., Zhang Z., Hu Y., Xu Q., Stem cell-derived Sca-1+ progenitors differentiate into smooth muscle cells, which is mediated by collagen IV-integrin alpha1/beta1/alphav and PDGF receptor pathways. Am. J. Physiol. Cell Physiol. 292, C342–C352 (2007). [DOI] [PubMed] [Google Scholar]
- 76.Gerecht-Nir S., Ziskind A., Cohen S., Itskovitz-Eldor J., Human embryonic stem cells as an in vitro model for human vascular development and the induction of vascular differentiation. Lab. Invest. 83, 1811–1820 (2003). [DOI] [PubMed] [Google Scholar]
- 77.Pastor-Pareja J. C., Xu T., Shaping cells and organs in Drosophila by opposing roles of fat body-secreted collagen IV and perlecan. Dev. Cell 21, 245–256 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Van De Bor V., et al., Companion blood cells control ovarian stem cell niche microenvironment and homeostasis. Cell Rep. 13, 546–560 (2015). [DOI] [PubMed] [Google Scholar]
- 79.Buczacki S. J., et al., Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature 495, 65–69 (2013). [DOI] [PubMed] [Google Scholar]
- 80.Arai F., Suda T., “Quiescent stem cells in the niche” in StemBook, Girard L., Ed. (Harvard Stem Cell Institute, Cambridge, MA, 2008) ID NBK27036. [PubMed] [Google Scholar]
- 81.Gehart H., Clevers H., Tales from the crypt: New insights into intestinal stem cells. Nat. Rev. Gastroenterol. Hepatol. 16, 19–34 (2019). [DOI] [PubMed] [Google Scholar]
- 82.Grün D., et al., Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature 525, 251–255 (2015). [DOI] [PubMed] [Google Scholar]
- 83.Farin H. F., et al., Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature 530, 340–343 (2016). [DOI] [PubMed] [Google Scholar]
- 84.Kay S. K., et al., The role of the Hes1 crosstalk hub in Notch-Wnt interactions of the intestinal crypt. PLOS Comput. Biol. 13, e1005400 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen K. Y., et al., A Notch positive feedback in the intestinal stem cell niche is essential for stem cell self-renewal. Mol. Syst. Biol. 13, 927 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information. Original images for figure panels where the background was removed can be viewed in the SI Appendix.