Significance
Dogs and humans have coexisted together for thousands of years, but it was not until the Victorian Era that humans practiced selective breeding to produce the modern standards we see today. Strong artificial selection during the breed formation period has simplified the genetic architecture of complex traits and caused an enrichment of identity-by-descent (IBD) segments in the dog genome. This study demonstrates the value of IBD segments and utilizes them to infer the recent demography of canids, predict case-control status for complex traits, locate regions of the genome potentially linked to inbreeding depression, and to identify understudied breeds where there is potential to discover new disease-associated variants.
Keywords: inbreeding depression, fitness, deleterious mutations, complex traits
Abstract
Domestic dogs have experienced population bottlenecks, recent inbreeding, and strong artificial selection. These processes have simplified the genetic architecture of complex traits, allowed deleterious variation to persist, and increased both identity-by-descent (IBD) segments and runs of homozygosity (ROH). As such, dogs provide an excellent model for examining how these evolutionary processes influence disease. We assembled a dataset containing 4,414 breed dogs, 327 village dogs, and 380 wolves genotyped at 117,288 markers and data for clinical and morphological phenotypes. Breed dogs have an enrichment of IBD and ROH, relative to both village dogs and wolves, and we use these patterns to show that breed dogs have experienced differing severities of bottlenecks in their recent past. We then found that ROH burden is associated with phenotypes in breed dogs, such as lymphoma. We next test the prediction that breeds with greater ROH have more disease alleles reported in the Online Mendelian Inheritance in Animals (OMIA). Surprisingly, the number of causal variants identified correlates with the popularity of that breed rather than the ROH or IBD burden, suggesting an ascertainment bias in OMIA. Lastly, we use the distribution of ROH across the genome to identify genes with depletions of ROH as potential hotspots for inbreeding depression and find multiple exons where ROH are never observed. Our results suggest that inbreeding has played a large role in shaping genetic and phenotypic variation in dogs and that future work on understudied breeds may reveal new disease-causing variation.
The unique demographic and selective history of dogs has enabled the persistence of deleterious variation, simplified genetic architecture of complex traits, and caused an increase in both runs of homozygosity (ROH) and identity-by-descent (IBD) segments within breeds (1–6). Specifically, the average FROH was ∼0.3 in dogs (7), compared to 0.005 in humans, computed from the 1000 Genomes populations (8). The large amount of the genome in ROH in dogs, combined with a wealth of genetic variation and phenotypic data (2, 5, 7, 9–11), allow us to test how ROH and IBD influence complex traits and fitness (Fig. 1). Furthermore, many of the deleterious alleles within dogs likely arose relatively recently within a breed, and dogs tend to share similar disease pathways and genes with humans (4, 12, 13), increasing their relevance for complex traits in humans.
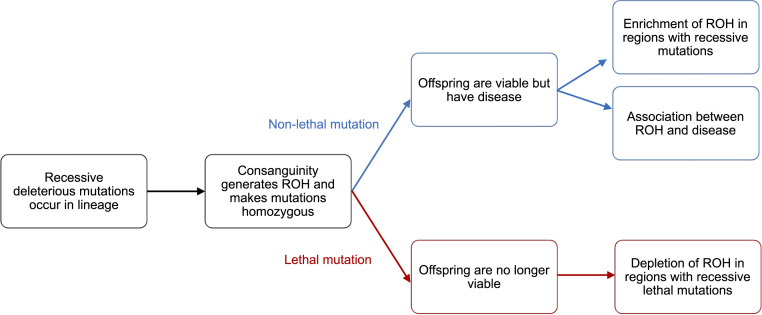
Fig. 1.
Potential mechanisms for associations between ROH and phenotypes that depend on recessive mutations. If a recessive deleterious mutation is nonlethal (blue), it may lead to ROH correlating with disease, while lethal (red) recessive mutations will cause a depletion of ROH.
Despite IBD segments and ROH being ubiquitous in genomes, the extent to which they affect the architecture of complex traits as well as reproductive fitness has remained elusive. Given that ROH are formed by inheritance of the same ancestral chromosome from both parents, there is a much higher probability of the individual to become homozygous for a deleterious recessive variant (8, 14), leading to a reduction in fitness. This prediction was verified in recent work in nonhuman mammals that has shown that populations suffering from inbreeding depression tend to have an increase in ROH (15, 16). ROH in human populations are enriched for deleterious variants (8, 14, 17). However, the extent to which ROH impact phenotypes remains unclear. For example, several studies have associated an increase in ROH with complex traits in humans (18–23), though some associations remain controversial (24–28). Determining how ROH and IBD influence complex traits and fitness could provide a mechanism for differences in complex-trait architecture across populations that vary in their burden of IBD and ROH.
Here, we use IBD segments and ROH from 4,741 breed dogs and village dogs, and 380 wolves to determine the recent demographic history of dogs and wolves and establish a connection between recent inbreeding and deleterious variation associated with both disease and inbreeding depression. This comprehensive dataset contains genotype data from 172 breeds of dog, village dogs from 30 countries, and gray wolves from British Colombia, North America, and Europe. We test for an association with the burden of ROH and case-control status for a variety of complex traits. Remarkably, we also find that the number of disease-associated causal variants identified in a breed is positively correlated with breed popularity rather than burden of IBD or ROH in the genome, suggesting ascertainment biases also exist in databases of dog disease mutations and that many breeds of dog are understudied. Lastly, we identify multiple loci that may be associated with inbreeding depression by examining localized depletions of ROH across dog genomes.
Results and Discussion
Global Patterns of Genetic Diversity across Dogs and Wolves.
Dog breeds were initially formed through domestication of one or more ancestral wild populations, in a process involving population bottlenecks. Then, over the last 200 y or so, modern dog breeds were formed (1, 3, 5, 10, 29–32). To examine genetic diversity in dogs and wolves, we merged three previously published genotype array-based datasets (9–11). As an initial quality check, we used principal component analysis (PCA) to examine the relationship between domesticated dogs, village dogs, and wolves (SI Appendix, Fig. S1). We observed a split between dogs and wolves on the first PC. The dogs that fall closest to wolves trace their origins back to Australasia (SI Appendix, Fig. S1), which has been previously shown to be the origin of some of the more ancient dog breeds (5, 29). When we performed a PCA with only breed dogs, they clustered by clade (SI Appendix, Fig. S2). Clades are composed of multiple breeds and were defined in previous work (7, 30). We also observed separation based on the geographic location of wolf populations. Wolves showed clear clustering by the location of where samples originated from, which includes the United States, Mexico, or Europe (SI Appendix, Fig. S3). Lastly, we inferred the recent demographic history of 10 standard breeds of dog, village dogs, and gray wolves from the United States and Europe (SI Appendix, Fig. S4) using patterns of IBD sharing between individuals (33). This analysis shows that all breed dogs experienced a domestication bottleneck followed by another severe bottleneck ∼200 y ago, corresponding with modern breed formation during the Victorian Era (1800s) (SI Appendix, Fig. S4). Though the strength of breed-formation bottleneck varied across breeds, and was less pronounced in mixed breeds, all bottlenecks were followed by a subsequent increase in population size. This breed-formation bottleneck was absent in both the village dogs and gray wolves (SI Appendix, Fig. S4) and has resulted in the majority of breed dogs carrying a large proportion of their genome in ROH (SI Appendix, Fig. S5).
Disease Traits Are Associated with ROH Burden.
We hypothesize that the prevalence of ROH and IBD segments could be associated with recessive genetic disease in each breed (Fig. 1 and SI Appendix, Fig. S2). ROH form when an individual inherits the same segment of their genome identically by descent from both parents (34), and the formation of ROH results in an increased probability of the individual to be homozygous for a deleterious recessive variant (14, 35). Thus, we predict that breeds with large amounts of ROH and IBD segments will have an increased incidence of recessive disease. We tested this hypothesis using data from 4,342 dogs in which we had case-control status for subsets of the data across eight clinical and morphological phenotypes (Fig. 2). Most traits did not have a significant association with ROH burden, even when stratified by breed. However, we observed an excess of associations at a nominal significance level (P < 0.05) compared to what was expected under the null hypothesis of no trait associations (6 observed associations versus 1.45 associations expected under the null, P = 0.0027, binomial test; Fig. 2).
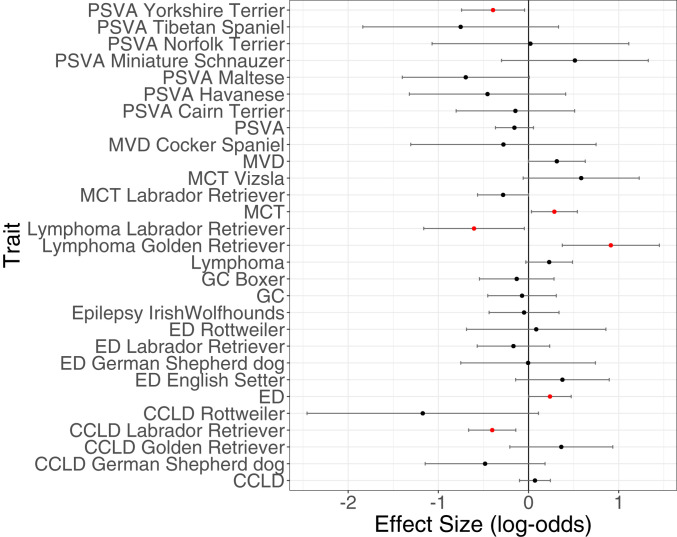
Fig. 2.
Association of ROH burden with eight quantitative traits. Results are presented both stratified by breed and across all breeds. A significant effect of ROH burden on a trait (nominal P < 0.05) is indicated with a red point. An effect size greater than 0 indicates an increase in ROH with the trait or disease status, and less than 0 represents the converse. Phenotype abbreviations are as follows: portosystemic vascular anomalies (PSVA); mitral valve degeneration (MVD); mast cell tumor (MCT); granulomatous colitis (GC); elbow dysplasia (ED); and cranial cruciate ligament disease (CCLD). These results use the SRBOUND correction for populations stratification (see Materials and Methods). Reference SI Appendix for additional information on P values. SI Appendix, Table S1 contains sample sizes, effect sizes, odds ratio, CI, and nominal P values. SI Appendix, Fig. S13 shows the uncorrected results as well as results using the genotype-relatedness matrix.
We observed a significant association between the burden of ROH and case-control status for five traits: portosystemic vascular anomalies (PSVA) in Yorkshire Terriers (β = −0.394 and P < 0.027), lymphoma within both Labrador (β = −0.604 and P < 0.0340) and Golden Retrievers (β = 0.913 and P < 0.001), cranial cruciate ligament disease (CCLD) in Labrador Retrievers (β = −0.403 and P < 0.003), elbow dysplasia across all breeds (β = 0.238 and P < 0.047), and mast cell tumors across all breeds (β = 0.286 and P < 0.027). For lymphoma in Golden Retrievers, case status is positively associated with the amount of the genome within an ROH [odds-ratio (OR) = 2.491, SI Appendix, Table S1], and on average cases carried more ROH than controls (SI Appendix, Fig. S6). Conversely, ROH appeared to show a protective effect against developing PSVA in Yorkshire Terriers as well as CCLD and lymphoma in Labrador Retrievers. If we restrict our analyses to a single breed, to mitigate the possibility of confounding due to population stratification, four of the six associations remain. The four associations are significantly more than expected under the null of no associations (∼1 expected by chance, P = 0.0222, binomial test) and argue that the association signal between ROH and phenotype is not driven by population stratification at the breed level.
To better understand the potential connection between ROH burden and phenotype, we first intersected the locations of the 15 significant genome-wide association study (GWAS) variants from Hayward et al. with ROH (10). None of these variants fell within an ROH. Next, we took a closer look at the relationship between ROH burden and lymphoma since it was one of our strongest associations. Using a list of human-associated lymphoma-associated genes (36), we observed an enrichment of lymphoma-associated genes overlapping ROH in cases, 760 kB on average, relative to controls, 564 kB on average (SI Appendix, Fig. S7; P = 0.0549). Taken together, these results suggest that the associations between ROH burden and phenotypes are not driven by the previously identified common GWAS variants. Instead, this signal may be driven by individually rare recessive variants potentially clustered in biologically relevant pathways.
To better understand whether the association between ROH and phenotype is driven by common variants shared IBD or recurrent recessive mutations becoming homozygous, we examined within- and between-breed shared ROH (SR) haplotypes within IBD segments (SI Appendix) as well as the amount of the genome IBD. We found that there is more haplotype sharing within breeds than between breeds across all traits (SI Appendix, Fig. S8), for both comparisons. However, there is no difference in either the amount of the genome shared IBD or the amount of SR haplotypes within IBD segments (SI Appendix, Table S2) between cases and controls. This result holds when stratifying across breeds or traits, as well as when aggregating across traits and breeds (SI Appendix, Fig. S8). The lack of an increase of IBD and IBD in ROH in cases suggests that shared ancestry in the past may not explain disease status, providing further evidence that the formation of ROH making deleterious mutations homozygous may be responsible for the associations between ROH and phenotype (Fig. 1).
Breed Popularity, Rather than ROH, Correlates with Mendelian Disease Incidence.
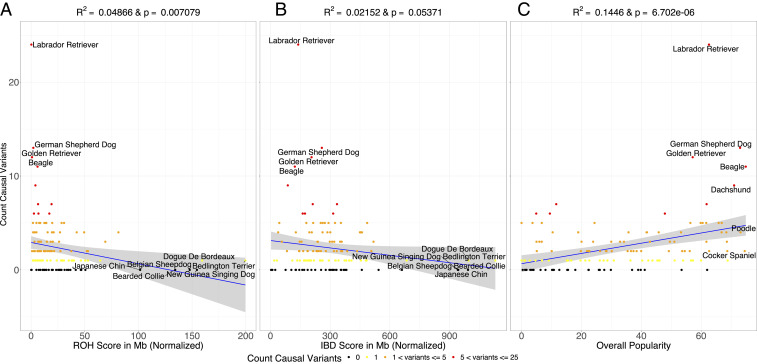
If ROH and IBD increase the homozygosity of disease alleles, we would expect that dogs with the largest amount of ROH and/or IBD would carry the most disease-associated variants because of the increased probability of revealing fully or partially recessive mutations due to excess homozygosity (nonlethal path, Fig. 1). We tested this hypothesis using data from Online Mendelian Inheritance in Animals (OMIA), which included a count of causal variants identified in each breed. To try to mitigate the number of false-positive variants in our analysis, we only included variants that were listed as likely causal (see Materials and Methods) in OMIA. We observed that breeds with the lowest amounts of ROH/IBD have the most identified causal variants (Fig. 3 A and B). Furthermore, those breeds with the most ROH/IBD have no causal variants identified, apart from the Kerry Blue Terrier that had a single reported mutation (Fig. 3 A and B). As this finding was unexpected, we sought additional factors that might explain the variation in the number of OMIA variants per breed. We chose to examine the popularity of different dog breeds over time using data compiled by the American Kennel Club (AKC). We discovered a strong positive correlation between the overall breed popularity and the number of causal variants identified in each breed (R2 = 0.168 and P = 1.145 × 10−06) (Fig. 3C). The most popular breeds, such as the Retrievers, have the most causal variants identified in genomic studies. These results also hold for nonfitness-related traits; however, there are very few of these traits within the OMIA database (SI Appendix, Fig. S9).
Fig. 3.
The correlation between the number of causal variants identified in each breed reported in OMIA and breed demographic characteristics. (A) Within-breed ROH and the total number of causal variants in OMIA. (B) Within-breed IBD and the total number of causal variants in OMIA. (C) Breed popularity over time and the total number of causal variants in OMIA. The shaded regions in each plot represent the CI on the regression line.
The bias toward more popular breeds having more disease-causing variants could be caused by the following: 1) increased numbers of popular breed dogs seen in veterinary offices, 2) increased funding and genomic studies of disease in popular breeds (through clubs or direct-to-consumer genomics), or 3) a combination of both. Notably, many of the breeds with the most IBD and ROH have had no causal variants identified through 2020. Thus, even if there are false-positive associations in OMIA, these false positives cannot explain the lack of OMIA variants in breeds that are less popular, have more ROH, and regions of the genome in IBD segments. Researchers should consider shifting their focus to some of these understudied breeds, as there may be more potential to discover new disease-associated variants. Such breeds include the Bearded Collie, Belgian Sheepdog, Bedlington Terrier, or Dogue de Bordeaux, all of which are prone to serious health conditions according to the AKC (akc.org/dog-breeds).
Ascertainment bias is not unique to OMIA and has been observed in human databases like Online Mendelian Inheritance in Man (OMIM) (37). In the case of human data, authors found that OMIM contains an enrichment of diseases caused by high-frequency recessive alleles because of the method through which these variants have been identified. Many variants were identified in isolated human populations, where there may be elevated levels of relatedness, which increases the probability of mapping higher-frequency deleterious variants (37). This ascertainment bias was not, and could not be discovered, in previous work on ROH in dogs (7), which focused on the more limited, but important, question of whether already identified disease variants were located in ROH.
ROH Reveal Genes with Recessive Lethal Mutations.
Given the relatively high values of FROH observed for breed dogs, much of the genome should be in ROH in at least one of the 4,342 individuals in our study. We hypothesized the genes not contained within an ROH in any individual or showing a deficit of ROH compared to the rest of the genome contain recessive lethal variants because individuals homozygous for these mutations are not viable (lethal path, Fig. 1). Across 4,342 dogs, we observed 27 genes (coordinates available on GitHub) where at least one exon does not overlap an ROH in any individual. To test whether this is unusual, we permuted the locations of the ROH within each individual and recounted the number of genes with an exon not containing an ROH. We found that if ROH were randomly distributed across the genome, we would expect to see ROH in all exons across genes (SI Appendix, Fig. S10). Thus, there are more genes not overlapping ROH than expected by chance (P < 0.0001), suggesting the presence of segregating recessive lethal mutations across breed dogs. This result is similar to what has been shown in inbred Scandinavian wolves (16), where ROH distribution fluctuates nonrandomly across the genome. Thus, these genomic regions could be lacking ROH because strongly deleterious recessive mutations lurk as heterozygotes in the founders of the breed. If offspring become homozygous for these regions, they are no longer viable and are not sampled in our study (Fig. 1).
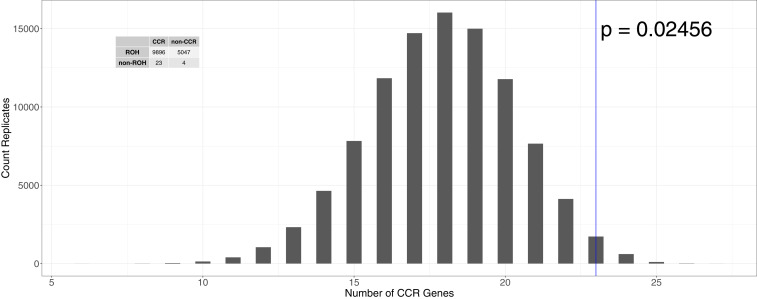
To test whether these 27 genes may have a functional effect, we intersected them with the 90th percentile constrained coding regions (CCRs) identified in human populations (38). CCRs were found to be enriched for disease-causing variants, especially in dominant Mendelian disorders (38). We expected that genes containing recessive lethal mutations would be conserved across species and asked whether the genes not overlapping ROH were enriched for CCRs. After resampling sets of 27 genes and intersecting them with the 90th percentile of CCRs 100,000 times, we would expect to see 18 of 27 genes falling above the 90th percentile of CCRs if exons lacking ROH in dogs were distributed randomly with respect to CCRs. In contrast, 23 out of 27 genes not overlapping ROH in dogs fall above the 90th percentile of the CCR distribution (P = 0.025) (Fig. 4). Additionally, we observed a 2.94-fold enrichment of non-ROH genes relative to ROH genes in CCRs (P = 0.041, Fisher’s exact test) (Fig. 4). Taken together, these results suggest that the genes with an exon not overlapping an ROH in dogs are enriched for exons devoid of variation in humans. Thus, these genes may be targets of strongly deleterious mutations affecting viability. Interestingly, there are some CCRs that do not have any known pathogenic or likely pathogenic variants, suggesting mutations in these exons could cause extreme developmental disorders or potentially be embryonically lethal (38). Studying mutations in these exons without ROH that overlap the CCR data could be fruitful both for identifying new disease phenotypes and for identifying variants with large fitness effects that could be linked to inbreeding depression or embryonic lethality.
Fig. 4.
Histogram of the expected number of genes that fall into the top 10% CCRs over 100,000 randomly drawn sets of 27 genes. The empirical data are demarcated by the blue line (P = 0.025). The contingency table shows the count of genes classified as to whether all exons overlap an ROH (“ROH”) and whether any exons overlap a CCR (“CCR”). There is a 2.94-fold enrichment of genes with at least one exon without an ROH (“non-ROH” genes) in CCRs (P = 0.041) relative to genes where all exons overlap an ROH.
We also tested whether our ROH analyses could be affected by low single nucleotide polymorphism (SNP) density, since the analyses thus far used only SNP genotype data. Because whole-genome sequence data would have an increased density of SNPs, we repeated our analyses using two sets of sequence data. The first dataset represents samples from four different breeds of dog: Pug (n = 15) with ∼47× coverage (39), Labrador Retriever (n = 10) with ∼30× coverage (40), Tibetan Mastiff (n = 9) with ∼15× coverage (41), and Border Collie (n = 7) with ∼24× coverage (40). The second dataset was previously published (see ref. 16) and contains 220 samples from human populations. We find that of the 27 genes with at least one exon not overlapping an ROH in any dog, 3 genes ANKH, FYTTD1, and PRMT2 have exons not overlapping an ROH in any of the three data sets (SI Appendix, Table S3). One of these genes, ANKH, has known Mendelian phenotypes that have been reported in OMIM and is also a 95th percentile CCR (42). It should also be noted that these three genes ANKH, FYTTD1, and PRMT2 all reside toward the end of the chromosome in dogs (SI Appendix, Fig. S11). Nevertheless, the relative distribution of ROH and these three genes not containing ROH were concordant across both VCFTools and PLINK (SI Appendix, Fig. S11). We further examined the locations of exons devoid of ROH and observed that ROH tend to not occur at all within the genes, or ROH occur in the exons toward the end of the gene (SI Appendix, Fig. S12).
Conclusions
Here, we have shown how the population history of dogs has increased the number of regions of the genome carried in ROH and IBD segments, affecting phenotypes and fitness. Our work contributes to a burgeoning number of studies associating ROH burden with complex traits and directly shows this association in dogs (7, 43, 44). Our findings have implications for understanding the architecture of complex traits in other species, such as humans. Specifically, the fact that we find a relationship between ROH and certain phenotypes (Fig. 2), suggests that recessive mutations play a role in some traits. Much of the existing GWAS in humans has largely suggested that complex traits are highly polygenic with many additive effects (45–48). These differences across species likely reflect differences in genetic architecture driven by the demographic history of the populations combined with natural selection. Nevertheless, searching for recessive variants underlying complex traits in humans may be a fruitful avenue of research. Furthermore, variation in the amount of the genomes in ROH across human populations (8, 14, 17, 35) could lead to population-specific architectures for complex traits. For example, causal variants in populations with a higher burden of ROH may be more recessive and less polygenic than in populations with fewer ROH. Future research will be needed to fully elucidate which mutations are directly responsible for severe inbreeding depression and the functional impact of these deleterious mutations. Additional work could examine which models of trait architecture (e.g., the degree of dominance and mutational target size) and demography could generate the association with ROH burden that we detected. In conclusion, the joint analysis of IBD and ROH provides considerable information about demography and selection in the genome and how they influence phenotype.
Materials and Methods
Genomic Data.
Autosomal genotype data were aggregated from two published studies (9, 10), and all original data files are publicly available through Dryad. The Fitak et al. data (9) were lifted over to CanFam3.1, then merged with Hayward et al. data (10) using PLINK (49). There were 210 sites that failed the lift over and 24,739 sites identified by PLINK as allele flips between Hayward and Fitak data. We retained sites that were present in 90% of individuals across the two data sets, which resulted in 117,288 sites of the 118,077 sites in the Fitak (∼99%) data and ∼73% of the 160,725 sites in the Hayward data being retained. SNPRelate (50) was used to perform PCA (SI Appendix, Fig. S1), identify duplicate individuals, and compute relatedness between individuals. Duplicate individuals and potential hybrid individuals were removed from the data set. The final data set contained 4,414 breed dogs, 327 village dogs, and 380 wolves. Code for merging data and final files are available on GitHub.
AKC Data.
We used AKC registration data from 1926 to 2005 to compute breed popularity. These data were curated from previous work (51) and contain information for ∼150 recognized breeds. To compute popularity through time, we dropped the first entry for each breed, as this number potentially reflects older dogs and new litters, then used the remaining data as the total number of new registrants per year. The popularity score is the integral from the second entry through 2005 (available on GitHub).
OMIA Data.
We downloaded all “Likely Causal” variants listed on OMIA. The “Likely Causal” criteria is met if there is at least one publication to be listed where the variant is associated with a disorder. If a variant had been identified in multiple breeds, it was counted in each breed. We were able to use all the genic coordinates from the “Likely Causal” category of variants from OMIA because we do not require that the causal variant is present in our data. The total number of causal variants per breed was downloaded from OMIA and is available here: https://github.com/jaam92/DogProject_Jaz/tree/master/LocalRscripts/OMIA.
Detecting IBD Segments.
To identify IBD segments, we used the software IBDSeq. (52) on its default settings. We set the minimum IBD segment length to 4cM, as that is the suggested length to reliably detect IBD segments in genotype data when using IBDSeq. We restricted to unrelated individuals, defined as being no closer than third-degree relatives, because IBDSeq only considers that individuals have only zero or one haplotype IBD.
Detecting ROH.
VCFTools, which implements the procedure from Auton et al. (53), was used to discover ROH in all individuals. We only kept ROH that contained at least 50 SNPs, were at least 100 kb long, and where SNP coverage was within one SD of mean SNP coverage across all remaining ROH. A file that contains the final ROH and scripts for running quality control can be found here: https://github.com/jaam92/DogProject_Jaz/tree/master/LocalRscripts/ROH. We also detected ROH using PLINK (reference SI Appendix) to assess the concordance of our results.
Computing IBD and ROH Scores.
We computed each population’s IBD and ROH scores using an approach similar to that of Nataksuka et al. (54). A population’s IBD score was calculated by computing the total length of all IBD segments between 4 and 20 cM and normalizing by the sample size. A population’s ROH score was computed using all ROH that passed quality control and normalizing by the sample size.
Association Test and Effect Size Estimates.
For these analyses, we only used the subset of breed-dog data from Hayward et al. where we had phenotype information (10), allowing us to follow the same strict requirements for each clinical phenotype as originally done. For breed-specific analyses, we required there to be at least 10 cases and 10 controls per breed. For the “all breed” analyses, we include all individuals combined in the same analysis, rather than conducting a meta-analysis of all the results from the individual breed analyses. If there were more cases than controls, in either the breed-specific or all-breed analyses, the cases were down sampled to match the sample size of controls (reference SI Appendix, Table S1 for sample sizes).
We computed the association between FROH and each trait using a general linear mixed model, specifically a logistic mixed model, which is implemented in the R package GMMAT (55). Following the protocol from Hayward et al. (10), we did not include covariates in the association test and included the SNP-based or ROH-based kinship matrix as a random effect in the model to control for population stratification due to covariation of the amount of ROH per breed with the incidence of the phenotype in the breed. P values were determined using a Wald test with a significance threshold of P = 0.05. Note, we use nominal P values in the manuscript and SI Appendix, these are not corrected for multiple testing (reference SI Appendix, SI Text for rationale). For more details on clinical-trait ascertainment see ref. 10. We generated the kinship matrix two different ways: 1) using the R package PC-Relate (56) on the SNP genotype matrix, and 2) by computing the total amount of the genome within an ROH that is shared between two individuals (SR),
Here, ∈ {0, 1} and equals 1 if the genotype (G) at the jth site falls within an ROH shared by both individuals and equals 0 otherwise. SR was computed for each pair of individuals and bounded between 0 (no sharing) and 1 (complete sharing) as follows:
where the max amount of sharing in the equation above is the total length of the autosome and minimum is smallest SR value in base pairs. Results reported in the main text are for SRBOUND. We also compared these results to those when not using a kinship or ROH matrix (SI Appendix, Fig. S13).
Identifying Depletions of ROH.
To find the number of genes expected to contain at least one exon without an ROH, the ROH in each individual were permuted to a new location on the same chromosome using BEDTools shuffle (57). Next, we created a bed file containing the permuted ROH locations, intersected this file with the exon locations from CanFam3.1, and counted the number of genes with at least one exon where we did not observe any overlap with an ROH. To control for edge effects along the chromosomes, we concatenated all 38 chromosomes into a single chromosome with a total length equivalent to the sum of all the autosome lengths. As such, the shuffled locations of ROH could occur at the ends of chromosomes. We repeated our permutation test 10,000 times to create a null distribution. The P value was computed as the proportion of permuted datasets with as many or more genes with an exon not overlapping an ROH relative to what was seen empirically (27 genes).
To examine the overlap between genes lacking ROH and CCRs from Havrilla et al. (38), we used BEDTools (57) to intersect non-ROH with the top 10% (90th percentile) of CCRs and exon ranges for CanFam3.1 (31), which came from Ensembl (58). Then, we tabulated the total number of genes where there was at least one exon where we never observed any overlap (including partial overlap) with an ROH (non-ROH genes) and the converse (ROH genes), as well as the count of whether these non-ROH and ROH genes fell within a CCR. Significance of the ratio of non-ROH genes relative to ROH genes within a CCR was assessed using Fisher’s exact test. We computed the expected number of non-ROH genes within the 90th percentile CCRs by randomly sampling an equal number of genes from the entire gene set and intersecting the randomly sampled genes with CCRs. We repeated this random sampling of genes 100,000 times to build the null distribution and computed a P value as the proportion of sets of the 27 genes that had at least 23 genes containing an exon in the 90th percentile of CCR genes.
Supplementary Material
Acknowledgments
We thank Eduardo Amorim, Arun Durvasula, Nelson Freimer, Malika Kumar-Freund, Jesse Garcia, Jacqueline Robinson, and Janet Sinsheimer for helpful discussions about data analysis and curation as well as Bob Wayne and the reviewers for comments on the manuscript. This material is based upon work supported by the NSF Graduate Research Fellowship under Grant DGE-1650604 awarded to J.A.M. as well as NIH Grant R35GM119856 awarded to K.E.L.
Footnotes
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2019116118/-/DCSupplemental.
Data Availability
SNP genotype data from the original projects (9, 10) are available on Dryad (https://datadryad.org/stash/dataset/doi:10.5061/dryad.g68k008 and https://datadryad.org/stash/dataset/doi:10.5061/dryad.266k4). Code used to process original data and generate intermidiate files is available on GitHub (https://github.com/jaam92/DogProject_Jaz) (59).
References
- 1.Marsden C. D., et al., Bottlenecks and selective sweeps during domestication have increased deleterious genetic variation in dogs. Proc. Natl. Acad. Sci. U.S.A. 113, 152–157 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyko A. R., et al., A simple genetic architecture underlies morphological variation in dogs. PLoS Biol. 8, e1000451 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freedman A. H., Lohmueller K. E., Wayne R. K., Evolutionary history, selective sweeps, and deleterious variation in the dog. Annu. Rev. Ecol. Evol. Syst. 47, 73–96 (2016). [Google Scholar]
- 4.Boyko A. R., The domestic dog: Man’s best friend in the genomic era. Genome Biol. 12, 216 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vonholdt B. M., et al., Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature 464, 898–902 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parker H. G., et al., Genomic analyses reveal the influence of geographic origin, migration, and hybridization on modern dog breed development. Cell Rep. 19, 697–708 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sams A. J., Boyko A. R., Fine-scale resolution of runs of homozygosity reveal patterns of inbreeding and substantial overlap with recessive disease genotypes in domestic dogs. G3 (Bethesda) 9, 117–123 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mooney J. A.et al.; Costa Rica/Colombia Consortium for Genetic Investigation of Bipolar Endophenotypes , Understanding the hidden complexity of Latin American population isolates. Am. J. Hum. Genet. 103, 707–726 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitak R. R., Rinkevich S. E., Culver M., Genome-wide analysis of SNPs is consistent with no domestic dog ancestry in the endangered Mexican wolf (Canis lupus baileyi). J. Hered. 109, 372–383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayward J. J., et al., Complex disease and phenotype mapping in the domestic dog. Nat. Commun. 7, 10460 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stronen A. V., et al., North-South differentiation and a region of high diversity in European wolves (Canis lupus). PLoS One 8, e76454 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Awano T., et al., Genome-wide association analysis reveals a SOD1 mutation in canine degenerative myelopathy that resembles amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. U.S.A. 106, 2794–2799 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shearin A. L., Ostrander E. A., Leading the way: Canine models of genomics and disease. Dis. Model. Mech. 3, 27–34 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szpiech Z. A., et al., Long runs of homozygosity are enriched for deleterious variation. Am. J. Hum. Genet. 93, 90–102 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson J. A., et al., Genomic signatures of extensive inbreeding in Isle Royale wolves, a population on the threshold of extinction. Sci. Adv. 5, eaau0757 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kardos M., et al., Genomic consequences of intensive inbreeding in an isolated wolf population. Nat. Ecol. Evol. 2, 124–131 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Szpiech Z. A., et al., Ancestry-dependent enrichment of deleterious homozygotes in runs of homozygosity. Am. J. Hum. Genet. 105, 747–762 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keller M. C.et al.; Schizophrenia Psychiatric Genome-Wide Association Study Consortium , Runs of homozygosity implicate autozygosity as a schizophrenia risk factor. PLoS Genet. 8, e1002656 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Assié G., LaFramboise T., Platzer P., Eng C., Frequency of germline genomic homozygosity associated with cancer cases. JAMA 299, 1437–1445 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Bacolod M. D., et al., The signatures of autozygosity among patients with colorectal cancer. Cancer Res. 68, 2610–2621 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lencz T., et al., Runs of homozygosity reveal highly penetrant recessive loci in schizophrenia. Proc. Natl. Acad. Sci. U.S.A. 104, 19942–19947 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghani M.et al.; Alzheimer’s Disease Genetics Consortium , Association of long runs of homozygosity with Alzheimer disease among African American individuals. JAMA Neurol. 72, 1313–1323 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McQuillan R.et al.; ROHgen Consortium , Evidence of inbreeding depression on human height. PLoS Genet. 8, e1002655 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson E. C.et al.; Schizophrenia Working Group of the Psychiatric Genomics Consortium , No reliable association between runs of homozygosity and schizophrenia in a well-powered replication study. PLoS Genet. 12, e1006343 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spain S. L., Cazier J.-B., Houlston R., Carvajal-Carmona L., Tomlinson I.; CORGI Consortium , Colorectal cancer risk is not associated with increased levels of homozygosity in a population from the United Kingdom. Cancer Res. 69, 7422–7429 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Enciso-Mora V., Hosking F. J., Houlston R. S., Risk of breast and prostate cancer is not associated with increased homozygosity in outbred populations. Eur. J. Hum. Genet. 18, 909–914 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siraj A. K., et al., Colorectal cancer risk is not associated with increased levels of homozygosity in Saudi Arabia. Genet. Med. 14, 720–728 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Hosking F. J., et al., Genome-wide homozygosity signatures and childhood acute lymphoblastic leukemia risk. Blood 115, 4472–4477 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Shannon L. M., et al., Genetic structure in village dogs reveals a Central Asian domestication origin. Proc. Natl. Acad. Sci. U.S.A. 112, 13639–13644 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parker H. G., et al., Genetic structure of the purebred domestic dog. Science 304, 1160–1164 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Lindblad-Toh K., et al., Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438, 803–819 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Ostrander E. A., Wayne R. K., Freedman A. H., Davis B. W., Demographic history, selection and functional diversity of the canine genome. Nat. Rev. Genet. 18, 705–720 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Browning S. R., Browning B. L., Accurate non-parametric estimation of recent effective population size from segments of identity by descent. Am. J. Hum. Genet. 97, 404–418 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McQuillan R., et al., Runs of homozygosity in European populations. Am. J. Hum. Genet. 83, 359–372 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pemberton T. J., et al., Genomic patterns of homozygosity in worldwide human populations. Am. J. Hum. Genet. 91, 275–292 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skibola C. F., Curry J. D., Nieters A., Genetic susceptibility to lymphoma. Haematologica 92, 960–969 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amorim C. E. G., et al., The population genetics of human disease: The case of recessive, lethal mutations. PLoS Genet. 13, e1006915 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Havrilla J. M., Pedersen B. S., Layer R. M., Quinlan A. R., A map of constrained coding regions in the human genome. Nat. Genet. 51, 88–95 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marchant T. W., et al., Canine brachycephaly is associated with a retrotransposon-mediated missplicing of SMOC2. Curr. Biol. 27, 1573–1584.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Plassais J., et al., Whole genome sequencing of canids reveals genomic regions under selection and variants influencing morphology. Nat. Commun. 10, 1489 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phung T. N., Wayne R. K., Wilson M. A., Lohmueller K. E., Complex patterns of sex-biased demography in canines. Proc. Biol. Sci. 286, 20181976 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamosh A., Scott A. F., Amberger J. S., Bocchini C. A., McKusick V. A., Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 33, D514–D517 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clark D. W., et al., Associations of autozygosity with a broad range of human phenotypes. Nat. Commun. 10, 4957 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ceballos F. C., Joshi P. K., Clark D. W., Ramsay M., Wilson J. F., Runs of homozygosity: Windows into population history and trait architecture. Nat. Rev. Genet. 19, 220–234 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Visscher P. M., et al., 10 years of GWAS discovery: Biology, function, and translation. Am. J. Hum. Genet. 101, 5–22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manolio T. A., et al., Finding the missing heritability of complex diseases. Nature 461, 747–753 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang J., et al., Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42, 565–569 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boyle E. A., Li Y. I., Pritchard J. K., An expanded view of complex traits: From polygenic to omnigenic. Cell 169, 1177–1186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang C. C., et al., Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng X., et al., A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28, 3326–3328 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghirlanda S., Acerbi A., Herzog H., Serpell J. A., Fashion vs. function in cultural evolution: The case of dog breed popularity. PLoS One 8, e74770 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Browning B. L., Browning S. R., Detecting identity by descent and estimating genotype error rates in sequence data. Am. J. Hum. Genet. 93, 840–851 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Auton A., et al., Global distribution of genomic diversity underscores rich complex history of continental human populations. Genome Res. 19, 795–803 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakatsuka N., et al., The promise of discovering population-specific disease-associated genes in South Asia. Nat. Genet. 49, 1403–1407 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen H., et al., Control for population structure and relatedness for binary traits in genetic association studies via logistic mixed models. Am. J. Hum. Genet. 98, 653–666 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Conomos M. P., Reiner A. P., Weir B. S., Thornton T. A., Model-free estimation of recent genetic relatedness. Am. J. Hum. Genet. 98, 127–148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cunningham F., et al., Ensembl 2019. Nucleic Acids Res. 47, D745–D751 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mooney J., Yohannes A., Lohmueller K., The impact of identity-by-descent on fitness and disease in dogs. GitHub. https://github.com/jaam92/DogProject_Jaz. Deposited 6 January 2021. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
SNP genotype data from the original projects (9, 10) are available on Dryad (https://datadryad.org/stash/dataset/doi:10.5061/dryad.g68k008 and https://datadryad.org/stash/dataset/doi:10.5061/dryad.266k4). Code used to process original data and generate intermidiate files is available on GitHub (https://github.com/jaam92/DogProject_Jaz) (59).