Abstract
Cancer, a disease of inappropriate cell proliferation, is strongly interconnected with the cell cycle. All cancers consist of an abnormal accumulation of neoplastic cells, which are propagated toward uncontrolled cell division and proliferation in response to mitogenic signals. Mitogenic stimuli include genetic and epigenetic changes in cell cycle regulatory genes and other genes which regulate the cell cycle. This suggests that multiple, distinct pathways of genetic alterations lead to cancer development. Products of both oncogenes (including cyclin-dependent kinase (CDKs) and cyclins) and tumor suppressor genes (including cyclin-dependent kinase inhibitors) regulate cell cycle machinery and promote or suppress cell cycle progression, respectively. The identification of cyclins and CDKs help to explain and understand the molecular mechanisms of cell cycle machinery. During breast cancer tumorigenesis, cyclins A, B, C, D1, and E; cyclin-dependent kinase (CDKs); and CDK-inhibitor proteins p16, p21, p27, and p53 are known to play significant roles in cell cycle control and are tightly regulated in normal breast epithelial cells. Following mitogenic stimuli, these components are deregulated, which promotes neoplastic transformation of breast epithelial cells. Multiple studies implicate the roles of both types of components—oncogenic CDKs and cyclins, along with tumor-suppressing cyclin-dependent inhibitors—in breast cancer initiation and progression. Numerous clinical studies have confirmed that there is a prognostic significance for screening for these described components, regarding patient outcomes and their responses to therapy. The aim of this review article is to summarize the roles of oncogenic and tumor-suppressive components of the cell cycle in breast cancer progression and prognosis.
Keywords: cell cycle, cyclin-dependent kinase, p16, p21, p27, breast cancer, prognosis
1. Introduction
Cancer, a disease of uncontrolled cell division, is known to exhibit a series of changes in the activity of cell cycle regulators [1]. All cancer types arise from a single cell that has transformed due to genetic or regulatory alterations, resulting in uncontrolled cell division in response to mitogenic signals [2]. Mitogenic signals include genetic and epigenetic aberrations in cell cycle regulatory genes. These mitogenic stimuli make oncogenic changes, resulting in cell transformation [3]. The gain-of-function mutations cause the activation of proto-oncogenes, which are normally present in the suppressed state in differentiated cells under epigenetic control [4]. Oncogenic stimuli have the potential to induce transformation of the differentiated cells, causing alterations in genetic material and therefore stimulating the development of certain cancers [5,6,7]. Loss-of-function mutations lead to a decreased expression of tumor suppressor genes, resulting in the diminishment of tumor-protective functions [8,9,10,11]. The collective data obtained suggest that distinct pathways of genetic alteration lead to cancer [11]. Products of both oncogenes and tumor suppressor genes regulate cell cycle machinery [8,12]. There are different phases of the cell cycle, and progression through these phases requires many regulatory components, which include oncogenic genes (CDKs (cyclin-dependent kinases) and cyclins) and tumor suppressor genes (cyclin-dependent kinase inhibitors) [13]. The identification and subsequent functional analysis of cyclins and CDKs enable us to understand the molecular mechanism of cell-cycle machinery [14]. In the pathogenesis of breast cancer, cyclins A, B, C, D1, and E; CDKs; and CDK-inhibitors, such as p21 (Waf1/Cip1), p27 (Kip1), p16 and 53, are known to play important roles in cell cycle control [15] (Figure 1). Each cell cycle phase is tightly regulated in normal cells [15]. After exposure to mitogenic stimuli, however, these regulatory components become deregulated, which predisposes the cellular transformation of breast epithelial cells. Numerous studies implicate the roles of oncogenic and tumor-suppressive components in various human cancer types, including the initiation and development of breast cancer, more specifically [9,16,17]. In addition, several research bodies have confirmed the prognostic significance of oncogenic and tumor suppressor components in regard to therapy or clinical outcomes [18]. Significant information exists on the regulation and roles of the cell cycle components in breast cancer cells, and previous studies may be utilized for therapeutic purposes. Findings from experimental studies also support that alterations in these components are clinically significant [19]. The aim of this review article is to summarize the role of various oncogenic and tumor suppressor components of the cell cycle that are involved in breast cancer progression and prognosis.
Figure 1.
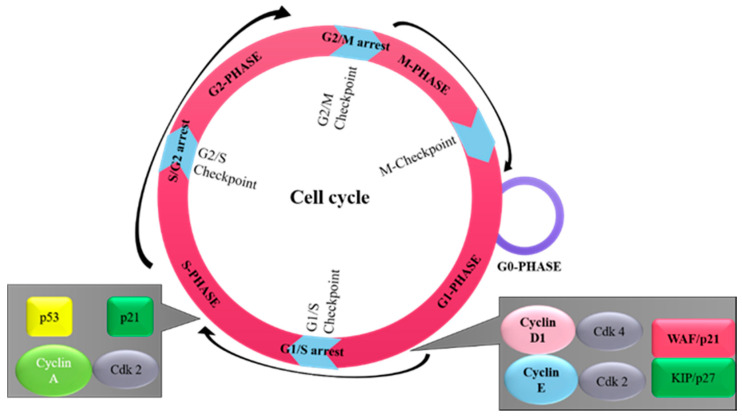
The sequential order of cell cycle events. The cell cycle progresses through four sequential phases: G1-phase (cell increases in size), S-phase (DNA synthesis), G2-phase (prepares to divide), and M-phase (cell division). The phases G1, S, and G2 make up the interphase stage, and span between cell division. There are special proteins and checkpoint systems for the proper progression of the cell cycle. First: G1 checkpoint (at G1/S transition) is the main irreversible decision point for cell division, which assesses for adequate cell size, availability of nutrients, positive molecular signals, and DNA integrity. Second: G2 checkpoint (at G2/M transition) ensures smooth cell division and assesses DNA integrity and successful DNA replication before division. In the case of error, cellular progression will become paused at the G2 checkpoint for repair. Third: the spindle checkpoint (metaphase to anaphase transition), ensures correct attachment of sister chromatids to the spindle microtubules.
2. Overview of Cell Cycle
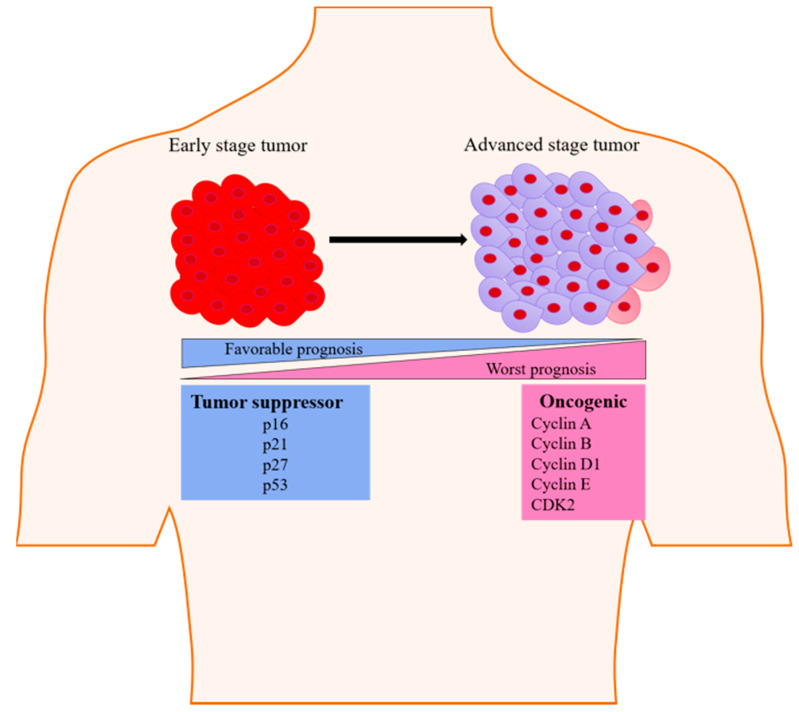
The cell cycle is composed of several phases (Figure 1), including a phase for the preparation of DNA synthesis—G1 phase; a phase for DNA synthesis—S phase; a second preparation phase—G2 phase; and mitosis—M phase. These phases are tightly controlled under physiological conditions [20]. Quiescence (G0) is another phase of the cell cycle found in some differentiated cells, in which the cell undergoes its own distinct biochemical or molecular changes [21]. Under certain pathological stimuli, differentiated cells can leave the G0 phase and re-enter the cell cycle [21]. The transitions between these cell cycles phases are controlled by the function of specific CDKs. This includes CDK1/CDK2, which causes the transition from G2 to mitosis, and CDK2/CDK4/CDK6, which causes the G1 to S phase transition [22]. During cellular division, another group of proteins called cyclins form complexes with specific CDK molecules in their respective phases [22]. The G1 cell cycle phase transition is identified by the activity of CDK4/6-cyclin D and CDK2-cyclin E complexes, S cell cycle phase transition by cyclin A-CDK1/2 complex, and G2-mitosis phase transition by cyclin A-CDK and cyclin B-CDK1 complexes [23]. Many genetic alterations can affect the functional activities of oncogenes or tumor suppressors, including alterations in cyclin E, cyclin D1, and p27. These alterations have been shown to induce a transition from the quiescent state into the active state in breast epithelial cells, subsequently leading to breast epithelial cell transformation (Figure 2) [24].
Figure 2.
Illustration showing the change in the expression status of genes serving as tumor-suppressive and oncogenic markers during tumorigenesis. These changes in expression impact progression into advanced-staged cancer and overall breast cancer prognosis.
3. Oncogenic Components of Cell Cycle
3.1. Cyclin D
The proto-oncogene cyclin D is a crucial regulator for the transition from the G1 to S phase during the cell cycle. It binds with CDK4 and CDK6 and forms an active cyclin D-CDK4/6 complex, which then phosphorylates the retinoblastoma protein (Rb) to promote cell cycle progression [25,26]. Cyclin D may also modulate the activity of various transcription factor proteins and histone deacetylase enzyme [27]. Having a half-life of ~24 min, cyclin D is degraded inside the cell mainly via the activity of 26S proteasome in a ubiquitin-dependent and Skp2 F-box protein-dependent manner [28,29]. In addition, D1-CDK4/6 complex can also impair the functions of mitochondria through the phosphorylation and repression of nuclear respiratory factor 1 (NRF1) and mitochondrial transcription factor A (mtTFA). An earlier report established a molecular link between cyclin D1 and control of mitochondrial function through the inhibition of nuclear respiratory factor 1 [30]. Previously accumulated data underscore the role of cyclin D1 in the tumorigenesis of mammary cancer [31,32]. Overexpression and gene amplification of cyclin D has also been linked to a worsened prognosis and the development of resistance against endocrine therapy in breast cancer (Table 1 and Table 2) [33,34]. A study documented cyclin D1 gene overexpression and copy number amplification in 20% and 50% of human breast cancer cases, respectively [35,36,37]. Furthermore, an enhanced expression of cyclin D1 was also observed in 67.5% of invasive ductal carcinoma cases, where it was strongly correlated with estrogen receptor (ER) and progesterone receptor (PR) expression [38]. Similarly, a study analyzed the immunohistochemical (IHC) positivity of cyclin D1 in invasive ductal and moderately differentiated breast cancer cases, which was associated with significantly poorer prognoses in these patients [39]. Additionally, research data based on in vitro and clinical studies implicated an increased cyclin D1 gene expression and amplification in ~45–50% of breast cancer cases [40]. In another in vitro study, genetic alterations in the cyclin D1 gene and mRNA expression were found in the ER-negative MDA-MB-453 cell line (Table 3), which may be related to malignant transformation [41]. Similarly, cyclin D1 protein expression was examined in infiltrating mammary carcinoma with ER/PR positivity [42]. An abnormal expression of cyclin D1 was displayed in 66% of mammary infiltrating duct carcinomas, suggesting its role in breast tumor metastasis [43]. Zhang et al. [44] determined that enhanced expression of the cyclin D gene was found in ~82% of human breast tumors, and gene amplification was present in ~17% of cases.
Table 1.
Clinical relevance of oncogenic and tumor suppressive cell cycle components in breast cancer patients with different molecular subtypes.
| Marker | Expression | Consequences | Receptor Status | Ref |
|---|---|---|---|---|
| Cyclin D | Overexpression | High risk of replace, Local reoccurrence, Metastasis |
ER+/ER- | [45] |
| Overexpression | High tumor grade | ER+/ER-/PR+/PR-/HER2+ | [35] | |
| Overexpression | High proliferation | ER+/ER-/PR+/PR-/TNBC | [38] | |
| Overexpression | High proliferation | ER+/ER-/PR+/PR- | [39] | |
| Overexpression | High proliferation | ER+/ER- | [40] | |
| Overexpression | High proliferation | ER+/ER-/PR+/PR- | [42] | |
| Overexpression | Metastasis | [43] | ||
| Gene amplification Overexpression |
High proliferation | [44] | ||
| Overexpression | High proliferation | ER+ | [46] | |
| Gene amplification | High risk for recurrence | ER+/ER- | [47] | |
| Overexpression, Gene amplification | High proliferation | ER+/ER-/PR+/PR- | [48] | |
| Overexpression | High proliferation, Short overall survival, Large tumor size, Lymph node metastasis |
ER+/ER-/PR+/PR- | [49] | |
| Overexpression, Gene amplification | High proliferation | ER+/ER-/Basal like | [50] | |
| Overexpression | Reduce relapse-free survival | ER+/PR+ | [51] | |
| Overexpression | High proliferation | ER+/PR+/ER-/PR-/HER2+ |
[52] | |
| Overexpression | High proliferation | ER+/PR+/ER-/PR-/HER2+ | [53] | |
| Overexpression | High proliferation | ER+/ER- | [54] | |
| Overexpression | High risk of recurrence | ER+ER-/HER2+/HER2- | [55] | |
| Gene amplification | High proliferation | ER+/ER- | [56] | |
| Gene amplification | Reduce patient survival time, therapy resistance |
ER+ | [57] | |
| Overexpression | Poor prognosis | ER+/PR+/ER-/PR-/HER2+/Basal like | [58] | |
| Overexpression | Invasiveness, metastasis | TNBC | [59] | |
| Overexpression | High proliferation | ER+ | [60] | |
| Reduce expression | High tumor grade, Nodal positive status, Invasion |
ER+/PR+/ER-/PR-/HER2+/HER2- | [61] | |
| Cyclin A | Overexpression, Gene amplification | Poor prognosis | [62] | |
| Overexpression | Relapse, Shorter disease-free survival |
ER+/ER- | [63] | |
| Overexpression | Worst prognosis | ER+/ER- | [64] | |
| Overexpression | Shorter relapse time | ER+/ER- | [65] | |
| Overexpression | Less survival rate, High relapse rate |
ER+/PR+/ER-/PR- | [66] | |
| Overexpression | Short distant metastasis-free survival | ER+/PR+/ER-/PR- | [67] | |
| Overexpression | Poor prognosis | ER+/ER- | [68] | |
| Overexpression | High tumor grade, High proliferation index |
HER2+/HER2- | [69] | |
| Overexpression | Poor survival | ER+/ER- | [70] | |
| Overexpression | Poor prognosis, Decrease survival rate | ER+/PR+/ER-/PR-/HER2+ | [71] | |
| Cyclin B1 | Overexpression | Decrease survival | ER+/PR+/ER-/PR-/HER2+/HER2- | [72] |
| Overexpression | Reduce overall survival, Disease free survival, Lymphatic invasion |
ER+/PR+/ER-/PR-/HER2+/HER2- | [73] | |
| p21(WAF1/Cip1) | Overexpression | High tumor grade, Large tumor size, Positive lymph node, High Ki-67 expression |
ER+/PR+/ER-/PR-/HER2+/HER2- | [74] |
| Overexpression | Favorable prognosis | ER+/PR+/ER-/PR- | [75] | |
| Overexpression | Better survival | ER+/PR+/ER-/PR-/HER2+ | [76] | |
| Overexpression | Better survival | ER+/PR+/ER-/PR-/HER2+/HER2- | [77] | |
| Overexpression | Large tumor size, High tumor grade, Lymph node metastasis |
ER+/PR+/ER-/PR-/HER2+/HER2- | [78] | |
| p27 (Kip1) | Reduced expression | High tumor grade, Lack of tumor differentiation, Poor prognosis | ER+/PR+/ER-/PR-/HER2+/HER2- | [79] |
| Overexpression | Better prognosis | ER+/ER- | [80] | |
| Overexpression | Favorable prognosis | ER+/PR+/HER2+ | [81] | |
| Reduced expression | Poor prognosis | ER+/PR+/ER-/PR-/HER2+/HER2- | [82] | |
| Overexpression | Long disease-free survival, overall survival | ER+/PR+/ER-/PR- | [83] | |
| Reduced expression |
Poor prognosis | ER+/PR+/ER-/PR- | [84] | |
| Reduced expression | Large tumor size, high tumor grade, lymph node metastasis | ER+/ER- | [85] | |
| Overexpression |
Long relapse-free survival,
Overall survival |
ER+/PR+ | [86] | |
| Reduced expression | Poor prognosis | ER+/PR+ | [87] | |
| Reduced expression | Increase proliferation | ER+/PR+/ER-/PR-/HER2+/HER2- | [88] | |
| Overexpression | Favorable prognosis | ER+/PR+/ER-/PR-/HER2+/HER2- | [89] | |
| Lower expression | Worst overall survival, Worst disease-free survival |
ER+/PR+/ER-/PR- | [90] | |
| Lower expression | Worst overall survival | ER+/PR+/ER-/PR- | [91] | |
| p16 (ink4a) | Overexpression | High proliferation index |
ER+/PR+/ER-/PR-/HER2+/HER2- | [92] |
| Overexpression | Favorable prognosis | ER+/PR+/ER-/PR- | [93] | |
| Overexpression | Favorable prognosis | ER-/PR-/HER2- | [94] | |
| Overexpression | Favorable prognosis | ER+/PR+/ER-/PR- | [95] | |
| Overexpression | Disease progression | ER+/PR+/ER-/PR- | [96] | |
| Overexpression | Lung and brain metastasis | ER+/PR+/ER-/PR-/HER2+/HER2- | [97] | |
| Reduced expression | Metastasis | ER+/PR+/ER-/PR-/HER2+/HER2- | [98] | |
| p53 (wild type) | Overexpression | Favorable prognosis | ER+/PR+/ER-/PR-/HER2+/HER2- |
[99] |
| Overexpression | Better disease-free survival | ER+/PR+/ER-/PR- | [100] | |
| Overexpression | Favorable prognosis | ER+/PR+/ER-/PR-/HER2+/HER2-/Basal like |
[101] | |
| Overexpression | Favorable prognosis | ER+/PR+/ER-/PR- | [102] | |
| Overexpression | Worst prognosis | ER-/PR-/HER2- | [103] | |
| Overexpression | Worst prognosis | ER+/PR+/ER-/PR-/HER2+/HER2- | [104] | |
| Overexpression | Worst prognosis associated with ER expression |
ER+/PR+/ER-/PR- | [105] | |
| Overexpression | Worst prognosis associated with HER2+/TNBC subtypes |
ER+/PR+/ER-/PR-/HER2+/HER2- | [106] | |
| Overexpression | Worst prognosis, High tumor grade, Lymph vascular invasion, Lymphocyte infiltration |
ER+/PR+/ER-/PR- | [107] | |
| Overexpression | Better overall survival in TNBC | ER-/PR-/HER2- | [108] | |
| Overexpression | Correlated with HER2 overexpression, High tumor grade |
ER+/PR+/ER-/PR-/HER2+/HER2- | [109] | |
| Overexpression | Worst overall survival, Reoccurrence free survival |
ER+/PR+/ER-/PR- | [110] | |
| p53 (Mutant) | Overexpression | Early relapse | ER+/PR+/ER-/PR-/HER2+/HER2- | [111] |
| Overexpression | Poor prognosis | ER+/PR+/ER-/PR- | [112] | |
| Overexpression | Less 5-years reoccurrence free survival | ER+/PR+/ER-/PR- | [113] |
ER: estrogen receptor; PR: progesterone receptor; Her2+: human epidermal growth factor receptor-2 positive.
Table 2.
Clinical relevance of oncogenic and tumor suppressive cell cycle components in breast cancer cell lines with different molecular subtypes.
| Marker | Expression | Consequences | Model | Histology | Ref |
|---|---|---|---|---|---|
| Cyclin D | Gene amplification mRNA Overexpression |
Proliferation | MCF-7 | ER+ | [33] |
| Overexpression | Malignant transformation | MDA-MB-453 | HER2+ | [41] | |
| Overexpression | Increase proliferation | T-47D, MCF-7 |
ER+/HER2+ | [34] | |
| Overexpression | Increase proliferation | MCF-7 | ER+ | [114] | |
| Overexpression | Increase proliferation | MCF-7, T-47D, MDA-MB-468, BT-549 |
ER+/ER-/PR-/HER2- | [115] | |
| Overexpression | Increase proliferation | HBL-100, MDA-MB-23l, T-47D, MCF-7, MDA-MB-134, HMEC-184 |
ER+/PR+/ER-/PR-/HER2-/HER2+ | [52] | |
| Overexpression | Increase proliferation | MCF-7T, | Tamoxifen-resistant | [116] | |
| Downregulation | Cell death, Growth arrest |
MCF-7, MDA-MB-231, MDA-MB-435, HCC-1937, CAL-148 |
ER+/ER-/PR-/HER2- | [117] | |
| Overexpression | Invasiveness, metastasis | MDA-MB-231 | ER-/PR-/HER2- | [59] | |
| Overexpression | Increase proliferation | MCF-7 | ER+ | [118] | |
| Overexpression | Increase proliferation | MCF-7 | ER+ | [60] | |
| Overexpression | Increase proliferation | MCF-7 | ER+ | [119] | |
| Overexpression | Increase proliferation | ZR75-1-2, ZR-75-1, MDAMB-157, MDA-MB-231, MDA-MB-436, T-47D, BT-20, HBL-100, Hs578T, SK-BR3 |
ER+/ER-/PR+/PR-/HER2-/HER2+ | [120] | |
| Cyclin E | Overexpression, Gene amplification | Increase proliferation | BT-474, BT-474R |
HER2+ | [121] |
| Downregulation | Suppression of cell cycle progression | MCF-7 | ER+ | [122] | |
| Cyclin B | Overexpression | Associated with ER+ status | MCF-7, MDA-MB-231, MDA-MB-436, Hs578 T |
ER+/PR+/ER-/PR-/HER2-/HER2+ | [123,124] |
| Downregulation | Apoptosis, Anti-proliferation |
MCF-7, BT-474, SK-BR-3, MDA-MB-231 |
ER+/ER-/PR+/PR-/HER2-/HER2+ | [125] | |
| p16(WAF1/Cip1) | Reduced expression | Therapy resistance | HCC-1428, T-47D, MCF-7, MDA-MB-436, BT-549, MDA-MB-157, MDA-MB-231, MDA-MB-435S, Hs578T, HCC-1937, BT-20, SK-BR-3 |
ER+/PR+/ER-/PR_/HER2-/HER2+ | [126] |
| Overexpression | Good survival | MCF-7, BT-549, MDA-MB-134, MDA-MB-157, MDA-MB-231, MDA-MB-453, MDA-MB-468, ZR-75-1, BT-20, SK-BR-3, T-47D |
ER+/PR+/ER-/PR-/HER2-/HER2+ | [76] | |
| p21(WAF1/Cip1) | Overexpression | High proliferation rate |
ZR75-1,
ZR75-30, MCF-7, MDA-MB-453, T-47D, Cal51, SK-BR-5, SK-BR-7, CAMA-1, BT-20 |
ER+/PR+/ER-/PR-/HER2+ |
[127] |
| Reduced expression | Trastuzumab resistance | SK-BR-3 | HER2+ | [128] | |
| Overexpression | Cell cycle arrest | MCF-7 | ER+ | [129] | |
| Reduced expression |
Acquired resistance to docetaxel | MCF-7, MDA-MB-231 |
ER+/ER-/PR-/HER2- | [130] | |
| Overexpression | Associated with ER+ status | MCF-7, MDA-MB-231, MDA-MB-436, Hs578T |
ER+/PR+/ER-/PR-/HER2-/HER2+ | [123,124] |
Table 3.
Characteristics of breast cancer cell lines with different molecular subtypes (adopted from Dai, X et al. 2017) [131].
| Cell Line | ER | PR | HER2/neu+ | Subtype | BRAC1 Mutation | p53 Mutation | Tumor |
|---|---|---|---|---|---|---|---|
| CAMA-1 | + | +/- | - | LA | WT | MU | AC |
| HCC1428 | + | + | - | LA | ND | ND | AC |
| MCF-7 | + | - | - | LA | ND | WT | IDC |
| MDA-MB-134 | + | - | - | LA | ND | MU | IDC |
| T-47D | + | + | - | LA | WT | MU | IDC |
| ZR75-1 | + | +/- | - | LA | WT | WT | IDC |
| BT-474 | + | + | + | LB | WT | MU | IDC |
| ZR75-30 | + | - | + | LB | WT | WT | IDC |
| MDA-MB-453 | - | - | + | Her2+ | WT | MU | AC |
| SK-BR-3 | - | - | + | Her2+ | WT | MU | AC |
| SK-BR-5 | - | - | + | Her2+ | WT | MU | AC |
| BT-20 | - | - | - | TNBC | WT | MU | IDC |
| BT-549 | - | - | - | TNBC | WT | MU | IDC |
| CAL-51 | - | - | - | TNBC | WT | MU | AC |
| CAL-148 | - | - | - | TNBC | WT | MU | AC |
| HCC1937 | - | - | - | TNBC | MU | MU | DC |
| Hs578T | - | - | - | TNBC | WT | MU | IDC |
| MDA-MB-157 | - | - | - | TNBC | WT | MU | MC |
| MDA-MB-231 | - | - | - | TNBC | WT | MU | AC |
| MDA-MB-435 | - | - | - | TNBC | WT | MU | AC |
| MDA-MB-436 | - | - | - | TNBC | MU | MU | AC |
| MDA-MB-468 | - | - | - | TNBC | WT | MU | AC |
| SK-BR-7 | - | - | - | TNBC | WT | WT | AC |
WT: wild type; ND: not decided; MU: BRCA1 mutation; AC: adenocarcinoma; DC: ductal carcinoma; IDC: invasive ductal carcinoma; LA: luminal A; LB: luminal B; HER2+: human epidermal growth factor receptor 2-positive; TNBC: triple-negative breast cancer.
A study using a mouse mammary tumor virus model of breast cancer identified CCND1 gene amplification with positive IHC staining in 40% of breast cancer samples [132]. Further, the study identified ectopic overexpression of cyclin Dl and a reversed growth-inhibitory outcome after anti-hormonal therapy in ER-positive breast cancer cases, which provided a potential antitumor mechanism [46]. Kenny et al. [43] showed that ER-positive breast cancer patients had cyclin D1 high expression, and at the same time, also displayed more risk of relapse, metastasis, and early death [34]. Moreover, the data also showed that CCND1 gene amplification alone is a strong predictor of anti-hormonal therapy response in young-age breast cancer patients [47]. Moreover, data from another study indicated amplification of the cyclin D1 gene and noted its correlation with ER-positive invasive lobular breast carcinoma with lymph node metastasis, suggesting a sign of poorer prognosis [48]. An additional study suggested overexpression of cyclin D1 gene in the ER-positive MCF-7 breast tumor cell line, which was responsible for hyperproliferation undergrowth factor-deprived conditions [114]. Another study identified overexpression of cyclin D1 in ER-positive and ER-negative breast cancer samples; however, both shorter overall survival and relapse-free survival were associated only with the ER-negative subgroup [49].
Correlation of high cyclin D1-related elevation with Rb phosphorylation was also observed in >100 high-grade breast carcinomas [115]. Furthermore, a separate study also demonstrated a strong positive correlation between cyclin D gene amplification and higher expression in basal-like and ER-positive breast cancer subtypes, and suggested that cyclin D1 was an independent predictor for prognosis in ER-positive breast cancers [50]. The ABCSG Trial 05 and 06 documented an increased expression of cyclin D1, which was associated with the poorer clinical outcome and shorter overall survival of breast cancer patients [51]. A separate investigation determined the cyclin D1 positivity in proliferative disease without atypia, atypical ductal hyperplasia, low-grade ductal carcinoma in situ (DCIS), high-grade DCIS, and invasive carcinoma. The results showed that cyclin D1 was significantly higher in proliferative disease than normal breast epithelium, and even higher in DCIS than proliferative disease [52]. Additionally, another research group demonstrated an association between high cyclin D1 gene expression and high-grade tumor development, increased Ki-67 expression, and poorer survival in the ER-positive breast cancer group [53].
The majority of invasive lobular carcinomas showed cyclin D1 overexpression at the protein levels, suggesting its role in the progression of invasive lobular carcinoma [54]. Another study showed that ER-positive patients with moderate cyclin D1 expression had benefited from anti-hormonal therapy (tamoxifen), whereas those with high cyclin D1 expression had not benefited from tamoxifen, suggesting its role as a predictive marker for tamoxifen resistance [55]. Further results suggest that the silencing of cyclin D1 expression may reduce the development and progression of tamoxifen-resistant tumors [116]. Cisplatin drug targets cyclin D1, and treatment of ER-positive MCF-7 breast cancer cells with cisplatin increased cell death or growth arrest by decreasing the cyclin D in MCF-7 cells [117]. Using techniques fluorescent in situ hybridization (FISH) and IHC, researchers observed that CCND1 had increased amplification in high-grade infiltrating ductal carcinoma in comparison to low-grade infiltrate ductal carcinoma [56]. Cyclin D1 overexpression has been found to have a strong correlation with receptor status, suggesting that cyclin D1 expression could be a biomarker for good prognoses [57,58].
Additionally, expression of cyclin D2 was found to be very rare in breast cancer cases, in comparison to normal human mammary epithelial cells [47,59]; its role in cancer is yet to be elucidated [133]. Cyclin D3 has also been reported to be overexpressed in breast cancer samples, but there are limited research data on its relationship to disease outcomes [118,133,134]. Furthermore, experimental evidence has also shown elevated cyclin D1 protein levels and deposition of cyclin D3 in breast cancer samples [60]. Another study identified that 64 breast cancer cases out of 82 had cyclin D1 gene amplification, and 36 out of 86 cases had cyclin D3 gene amplification [62]. Expression of cyclin D1 was evaluated in different molecular breast cancer subtypes, and results showed a stronger intensity of positive cyclin D1 staining in the ER positive/PR positive subtype than in triple-negative breast cancer (TNBC) cases, and negative cyclin D1 staining was seen in human epidermal growth factor receptor 2-positive (HER2-positive) molecular subtypes. Further, TNBC cases with a low amount of cyclin D1 expression had higher tumor grade, tumor stage, and more positive lymph nodes with lymphovascular invasion, proposing that cyclin D1 expression may be a key factor to consider for aid in breast cancer management [61]. Lundberg et al. [135] determined CCND1 amplification and its association with worst 15-year survival with ER+/LN−/HER2−(1.66; 1.14–2.41), luminal A (HR = 1.68; 95% CI, 1.15–2.46), and luminal B (1.37; 1.01–1.86) breast cancer subtypes [135]. Overexpressed cyclin D1 induced Dicer expression in luminal A and basal-like breast cancer subtypes [136]. In another study, lower levels of cyclin D led to a decrease in MDA-MB-231 cells’ motility which resulted due to the decrease in phosphorylation of filamin A protein [137]. These studies found that cyclin D1 can also contribute to cellular proliferation and migration through non-canonical functions.
3.2. Cyclin A
Cyclin A protein forms complexes with both CDK1 and CDK2, which functions in both the S to G2 phase transition and the G2 to M phase transition of the cell cycle [51]. In the S phase, the cyclin-A-CDK complexes phosphorylate the components of the DNA replication machinery, subsequently initiating replication [51]. While in the mitosis phase, cyclin A/CDK2 coordinates centrosomal and nuclear mitotic events. However, it is thought to contribute to the stability of other cyclin molecules [51]. The increased expression of cyclin A gene has been found in different types of human tumors, including breast cancer, which suggests that cyclin A may potentially serve as a prognosis marker for the disease (Table 1 and Table 2). Studies have shown that microinjection of cyclin A into Xenopus oocytes and mammalian cells stimulates the breast tumor epithelial cells and induce the transition into M phase of the cell cycle [46,58,119,138,139]. A great number of tumors have shown a strong statistical correlation between cyclin A gene amplification and cyclin A protein levels [62]. Findings suggested that assessment of cyclin A and/or E2-promoter binding factor 1 (E2F1) expression levels associated with Ki-67 might be a useful tool for improved prognostic evaluation in negative lymph node breast cancer patients [63]. Another study showed that cyclin A is an independent prognostic factor and predictor of both breast cancer recurrence and response to tamoxifen therapy [64]. Lastly, overexpressed cyclin A was observed to be significantly correlated with breast cancer patients with earlier relapse, higher risk, and shorter overall survival rate, when compared to the breast cancer patients with better prognoses. Therefore, cyclin A may potentially be an accurate marker for tumor proliferation and prognosis in breast cancer [65].
3.3. Cyclin E
Cyclin E protein, a regulatory subunit for CDK-2, is thought to be a rate-limiting factor for the G1 to S phase cell cycle transition [140]. Cyclin E protein and its associated kinase (CDK2) experience well-regulated activation in normal cells. In actively-dividing tumor epithelial cells, however, the cyclin E and CDK complex remains activated throughout the cell cycle [141]. The deregulation in the expression of the cyclin E gene was found responsible for breast cancer tumorigenesis [46,60,114]. Previous data have demonstrated that higher levels of cyclin E gene amplification have been found in breast cancer tissues (Table 1) [62]. Another study observed an 8-fold amplification of the cyclin E gene and a 64-fold overexpression of its mRNA in human breast cancer cells, which provides evidence for aberrant cyclin E expression during tumorigenesis [120]. Further, a multivariate analysis correlated an elevated cyclin E level with poor patient outcome and showed that patients with elevated levels of cyclin E had a greater hazard ratio, as compared to those with low levels of cyclin E [142]. In addition, a relation between cyclin E gene expression and an ER-positive status was also observed in patients with breast cancer. In additional studies [89,90,91], cyclin E expression was greater in the ER-negative group and correlated with increased risks of death and relapse, suggesting that cyclin E may be responsible for ER-independent tumor growth. Similarly, cyclin E overexpression in breast cancer cells was associated with ER-negative tumors, HER2-positive tumors, and high-grade tumors with increased proliferation indexes [68,69]. A cohort study performed on 34 HER2-positive patients subjected to trastuzumab (Herceptin)-based therapy observed that the cyclin E gene copy number or mRNA overexpression was associated with diminished therapeutic benefits and lower rates of progression-free survival, as compared to non-overexpressing cyclin E patients [121]. Moreover, cyclin E expression was associated with a poor prognosis and closely related with cyclin D1 and p27Kip1 expression [70]. Similarly, high expression of cyclin E measured by IHC was a significant factor of poor prognosis and associated with a higher risk of death in the node-positive breast cancer group, as illustrated in a separate multivariate analysis [71].
3.4. Cyclin B
Two types of mammalian cyclin B regulate the G2-to-mitosis phase progression in the cell cycle, which do so by forming complexes with CDK1 kinase [143]. The available data suggest that breast cancer patients experience cyclin B gene amplification and overexpression at both the mRNA level and protein level (Table 1 and Table 2) [62]. Its increased expression has been correlated with a large tumor size, a high tumor grade, lymph node involvement, an ER-negative/PR-negative status, and a HER2-positive status [74]. Its overexpression has also been linked with younger age at diagnosis and higher expression levels of cyclin A, cyclin E, and Ki-67 [144]. Both univariate and multivariate analyses significantly identified an increased breast cancer death rate correlated with cyclin B1 overexpression, suggesting that it serves as a remarkable prognostic factor [72].
A meta-analysis investigated the significance between cyclin B protein and clinicopathological characteristics in breast cancer patients. Observations showed that overexpressed cyclin B was associated with poorer rates in disease-free survival (DFS), disease-specific survival (DSS), and overall survival (OS), along with a positive association with lymphatic invasion [73]. Androic et al. [100] observed apoptosis induction and growth reduction in different breast cancer cell lines, namely MCF-7, MDA-MB-231, BT-474, and SK-BR-3 (Table 3), in the absence of cyclin B. The suppression of cyclin B via small interfering RNA (siRNA) caused G2/M cell cycle phase arrest in breast cancer cell lines [73,125]. The HER2-positive invasive breast cancer samples used for the determination of cyclin B1 expression showed a direct correlation between positive cyclin B1 staining and higher tumor grade, large tumor size, positive lymph node counts, younger age, and higher Ki-67 expression. Thus, due to its relation with an aggressive phenotype, cyclin B1 might be considered a strong independent prognostic factor in breast cancer [74].
3.5. CDK2
Cyclin-dependent kinase 2 binds and forms complexes with cyclin E or cyclin A proteins and exclusively promotes the G1 to S and G2 to M phase transition within the cell cycle [145,146]. It has been observed that fulvestrant inhibited cyclin E-CDK2 activity, which in turn promoted the arrest of MCF-7 cells in quiescence (G0) [147]. Similarly, the findings suggest that the suppression of the cell-cycle progression through the G1 cell cycle phase by pentagalloylglucose (5GG) treatment in MCF-7 cells was mediated by blocking cyclin E/CDK2 activity [122].
4. Tumor Suppressive Components of Cell Cycle
4.1. p16(INK4A/MTS-1/CDKN2A)
The tumor suppressor p16, also known as INK4A/MTS-1/CDKN2A, has widespread importance in oncology due to its CDK-inhibitory function [148]. The frequently occurring SNP (single nucleotides polymorphism) mutations and deletions of the p16 gene in breast cancer cells suggest an important role in tumorigenesis [149]. The p16 protein molecule binds to and inactivates the cyclin D-CDK4/6 complexes, leads to subsequent Rb protein inactivation, and consequently results in cell cycle arrest [149]. Archived breast tumors of different histological subtypes provided evidence that aberrant p16 gene expression is the most common abnormality in human breast cancer (Table 1) [150]. Furthermore, an abnormal expression of p16 was found in ER-negative, pre-menopause breast cancer patients, in comparison to ER-positive patients. The abnormal p16 expression these researchers observed was closely associated with a high proliferative index [108]. An earlier study suggests that abnormal p16 expression may act as a predictor of poor response to hormonal therapy [92]. Another study found p16 protein-positive expression in the luminal A subtype of breast cancer patients, and higher expression was associated with breast cancer progression from DCIS to invasive ductal carcinoma (IDC) [96]. Abou-Bakr et al. [110] investigated the p16 expression in basal-like breast carcinoma grade III with histopathological findings in line with IDC. Results suggested that the p16 protein demonstrated high IHC intensity in basal-like carcinoma, which subsequently was associated with brain and lung metastasis [97]. A study by Arima et al. [126] found low p16 expression in resistant TNBC carcinoma [126]. Both p16-positive and p16-negative cells in the stromal cells of invasive lobular carcinoma reflected high nodal involvement, early recurrence, and metastatic propensity. Additionally, restoration of p16 expression in stromal fibroblasts suppressed cancer cell migration and invasion. Thus, these findings proposed positive stromal p16 expression as a treatment strategy to prevent nodal or distant metastasis [98].
4.2. p21 (WAF1/CIP1/SDI1/MDA-6)
The CDK-inhibitor p21 (also known as WAF1/CIP1/SDI1/MDA-6) activates the CDK4 and proliferating cell nuclear antigen, which results in G1 phase arrest [151]. Both in vivo and in vitro experimental models demonstrated that overexpression of p21WAF1/CIP1 resulted in G1 cell cycle phase arrest and effectively suppressed tumor growth (Table 1 and Table 2) [151]. Data on lymph node-negative breast cancer patients suggested that detection of p21 indicates the presence of a parameter that may act as a tumor suppressor and benefit patient survival [75]. Another study identified p21-positive tumor cell nuclei in more than 30% of the breast carcinomas, which was remarkably associated with a low histological grade and node-negative status [76]. The findings strongly suggested that p21WAF1/CIP1 gene expression might be used as a key prognostic biomarker for breast cancer, allowing therapy options to be adjusted more appropriately for individual cancer patients [77]. Breast cancer mastectomy used for measuring p21WAF1/CIP1 expression showed its upregulation in larger tumors in patients who presented with higher tumor dedifferentiation grades, more lymph node metastases, and shorter disease-free survival rates [78]. Moreover, an in vitro study where ER-positive or ER-negative breast cancer cell lines were immunostained for evaluation of p21 found a direct correlation between p21WAF1/CIP1 and ER expression [123,124].
In addition, p21WAF1/CIP1 also plays multifaceted roles in breast cancer. For instance, p21WAF1/CIP1 expression induced cell invasion and had correlation with OS and distant metastasis-free survival in breast cancer patients mediated via controlling TGFβ/Smad signaling [152]. A study measured high p21WAF1/CIP1 levels in the cytoplasm of metastatic breast cancer cells where it was associated with elevated p53 levels and poor prognoses [153]. Multiple studies identified that phosphorylation of p21WAF1/CIP1 by AKT1 disrupted its binding with proliferating cell nuclear antigen (PCNA) and induced its cytoplasmic accumulation. Accumulated p21WAF1/CIP1 regulates the ERBB2-mediated proliferation of breast cancer cells and breast carcinogenesis [154,155]. Further, downregulation of p21WAF1/CIP1 promoted EMT, enhanced the cell viability and migration potential in response to long non-coding RNA plasmacytoma variant translocation 1 (PVT1) in distinct MDA-MB-231, MDA-BA-468 breast cancer cell lines [156]. Similarly, another study using breast cancer mouse models has shown that invasion is accompanied by an upregulation of p21WAF1/CIP1, indicating its oncogenic role [157]. The overexpression of p21WAF1/CIP1 has also been found to be associated with a poor response to tamoxifen treatment in MCF-7 cells [158]. Similarly, Akt-dependent phosphorylated p21WAF1/CIP1 enhanced doxorubicin resistance in SUM159 TNBC cells [159]. Another study demonstrated that p21WAF1/CIP1 inhibited apoptosis in breast cancer. The overexpression of p21WAF1/CIP1 in breast cancer decreased cell sensitivity to infrared-induced apoptosis through inhibition of CDKs [160].
4.3. p27 (Kip1)
Tumor suppressor p27, an important regulator for the G1 to S transition in the cell cycle, is known to coordinate the activation of the cyclin E-CDK2 complex with the accumulation of cyclin D-CDK4, which initiates the exit of cells from the cell cycle in response to anti-mitogenic signals [161]. The downregulation of p27 gene expression is strongly correlated with higher tumor grade and phenotypes with lower tumor differentiation (Table 1) [79]. Reduced levels of p27 protein is also an indicator of poor clinical outcomes in a majority of lymph node-negative breast cancer patients [79].
Multiple sources of evidence suggest that p27 induced G1 cell cycle phase arrest, mediated by transforming growth factor-β (TGF-β), rapamycin, and cyclic adenosine monophosphate (cAMP) [48,60,114,127]. Previous studies also demonstrated that high expression levels of p27 in human breast cancer cells inversely correlated with the degree of malignancy in the human breast [127]. Moreover, a high expression of p27 was noticed in breast cancer patients, which was significantly correlated with an ER-positive status and inversely associated with shorter survival [80]. A univariate Kaplan–Meier analysis indicated that the decreased expression of p27 was significantly correlated with a worse clinical course [81]. A flow cytometry study using resistant breast tumor cells demonstrated a higher S-phase fraction and increased CDK2 activity in low p27-expressed cells, which was reversed after an exogenous addition of p27 [128].
Immunostaining of breast tumor indicated that downregulation of p27 correlated with HER2 gene overexpression in primary breast carcinomas, which may be significant in selecting patients for HER2-positive/neu antibody therapy in the future [82]. A separate study found that tamoxifen treatment caused MCF-7 cell cycle arrest due to an upregulation of p27 levels [129]. Another evaluation of p27 expression observed that it was a significant predictor for 5-year breast cancer survival, and that reduced p27 expression correlated with a high histologic grade, an advanced TNM stage (tumor size, lymph node status, metastatic status), and negative hormone receptor status [83,84]. A reduced expression of p27 was also observed in docetaxel-resistant breast cancer cells (MCF-7 and MDA-MB-231 cell lines) [130]. Another univariate analysis showed a remarkable relationship between low p27 expression and increased tumor grade, nuclear pleomorphism, and mitosis, along with decreased tubule formation in ER-negative and ductal/no special type tumor status [85].
High p27 expression independently predicted superior relapse-free survival and overall survival, and subsequently suggested its use as an independent predictor in hormonal therapy response [86]. An immunohistochemically retrospective investigation of 216 breast carcinomas found that p27-negative patients had a poorer prognosis than those in other categories, highlighting that the examination of p27 expression may identify breast carcinoma patients who would benefit from adjuvant therapy [87]. Further, in the lymph node-negative population, decreased p27 immunoreactivity was associated with higher tumor grade, more HER2-positive overexpression, greater lymph node positive populations, lower expression of thymidylate synthase, higher Ki-67 expression, and poorer disease-free survival [88]. In hormonal receptor- positive carcinoma, lower p27Kip1 was correlated with decreased overall survival [hazard ratio (HR) = 1.42; 95% confidence intervals (CI) = 1.05 to 1.94; disease-free survival HR = 1.27; and 95% CI = 0.99 to 1.63], as compared to carcinoma with higher p27Kip1 expression treated with adjuvant therapy (doxorubicin and cyclophosphamide) [90]. An inverse correlation was also observed between p27Kip1 expression and the degree of breast tumor malignancy [162]. Breast cancer patients in Taiwan were evaluated for the expression of p27Kip1, and both univariate and multivariate analyses showed that lower p27Kip1 expression correlated with OS in ER/PR positive tumors. Therefore, p27Kip1 may be considered an independent prognosis marker for breast cancer in Taiwan [91].
Another meta-analysis study showed a significant association between high p27 expression and OS, DFS, and RFS in lymph node-negative and lymph node-positive breast cancer patients [163]. In addition, Austrian Breast and Colorectal Cancer Study Group Trial 06 enrolled early-stage breast cancer patients with an ER/PR hormonal-positive status for evaluation of p27Kip1 expression and observed its impact on the clinicopathological features of women receiving adjuvant tamoxifen for 5 years. Observations confirmed that high p27Kip1 expression was significantly associated with longer disease-free survival (0.22; 95% CI, 0.11–0.42; p < 0.001) and overall survival (0.39; 95% CI, 0.21–0.72; p = 0.002) as compared to women with low p27 expression [164].
4.4. p53 (Wild Type)
Tumor suppressor p53 protein plays a key role in coordinating the response of cells to several stress conditions, including oncogenic activation, hypoxia, and DNA damage [165]. In response to mitogenic stress, p53 activates apoptosis in normal cells. This same activation of apoptosis by p53 has also been observed in anticancer therapy response. A mutated version of p53 protein that does not respond appropriately during oncogenic stress allows cell transformation, resulting in tumor initiation [165]. After immunohistochemical evaluation of p53 expression in primary breast cancer specimens, it was assessed that p53 overexpression was associated with an advanced-stage tumor, metastatic spread, and lower concentrations of progesterone receptors (Table 1) [89]. An increased cytoplasmic accumulation of p53 was observed in breast cancer patients as well. These patient samples demonstrated high proliferative activity with median Ki-67 fractions increased by up to 75%, along with a 74% increase in median S-phase fraction compared to the control group [93].
Utilizing invasive ductal carcinoma samples, Yang et al. (2013) [143] calculated DFS and its correlation with p53 expression. The Cox regression and multivariate analysis showed that p53 expression acted as a predictive factor of DFS [100]. Additionally, several studies also associated positive p53 expression with worsened prognoses. For instance, a Kaplan–Meier analysis of TNBC invasive ductal carcinoma samples showed that a positive p53 expression was correlated with worse overall survival (79.6% vs. 89.6%, log-rank test p = 0.025) and the patients had a 2.2 times higher mortality risk than that of p53-negative patients (HR: 2.222; 95% CI: 1.147–4.308) [102]. Similarly, p53 overexpression tested by IHC on modified radical mastectomy samples obtained from TNBC patients also showed lower overall survival rates (p = 0.021, log-rank test) compared to the patient group with low p53 expression. Moreover, the multivariate analysis proposed p53 overexpression as having the strongest prognostic significance in TNBC patients (<50 years) [103].
In a retrospective study of a large number of luminal/HER2-negative breast cancer patients, the data demonstrated that a p53 expression of ≥50% (present in 9% patients) was associated with shorter disease-free survival, in comparison to patients with p53 expression of <50%. Therefore, p53 overexpression was classified as a prognostic marker for unfavorable characteristics [104]. Another study performed on ER-positive and ER-negative invasive breast cancer determined an association of p53 overexpression with ER status. Results showed that in ER-negative breast tumors, a higher p53 expression was associated with DFS and OS than in ER-positive breast tumors [105].
Expression of the p53 gene was also evaluated in all breast cancer subtypes, i.e., luminal A, luminal B, HER2-positive, TNBC, and basal-like, and the findings suggested that p53 had a higher expression within HER2-positive and TNBC subtypes than in luminal A and luminal B subtypes. The overexpression of p53 in HER2-positive and TNBC subtypes also had significance in early-onset, high-grade tumors, and an increased proliferative index [106]. In invasive breast carcinoma grade II and III samples, positive p53 expression was significantly related with increased tumor grade (p < 0.006), lymphovascular invasion (p < 0.003), and lymphocytic infiltration (p < 0.004). These results indicate that p53 overexpression is a marker for a poor prognosis and a compromised immune response in more aggressive breast cancer types [107].
Better overall survival was observed in p53-overexpressed TNBC cases than in p53-negative TNBC patients who underwent neoadjuvant chemotherapy [108]. Another study concluded that p53 overexpression was inversely correlated with ER/PR expression and positively correlated with HER2-positive overexpression in high-grade tumors with nodal metastasis [109]. In a randomized stage II clinical trial on lymph node-positive patients who received four cycles of cyclophosphamide and one dose of doxorubicin adjuvant therapy, epithelial p53 expression was evaluated (using monoclonal antibodies DO7 and 1801). After univariate analysis, this study stated that positive p53 IHC was associated with worse OS and RFS in lymph node-positive patients [110].
4.5. p53 (Mutant)
A study performed by Marchetti et al. [154] found an “Arg72Pro” p53 variant in 23% of primary breast cancer patients. The patients positive for the Arg72Pro variant had relapsed within 10 months of the median DFS, compared to those that showed a wild-type p53 status [111]. Lenora W.M. et al. [155] also found a higher nuclear expression of mutant p53 using PAb1801 monoclonal antibody in young breast cancer patients. Kaplan–Meier curves and a log-rank test analysis correlated mutant p53 expression with a poor prognosis among distinct ethnic populations. Similarly, TNBC patients with abnormal mRNA expression of mutant p53 in a separate study were more likely to experience less 5-year reoccurrence-free survival. Mutant p53, therefore, may be considered a potential prognostic marker in TNBC patients [113].
5. Future Perspectives
Aside from great improvements in diagnostic tools and the increased availability of multiple therapeutic options, breast cancer cure rates remain poor. In the GLOBOCAN-2018 report, 2.1 million new breast cancer cases (11% of all total cancer types) were diagnosed in 185 countries [166]. In India, 144,000 breast cancer cases with a 5-year prevalence of 396,000 and 70,000 deaths were reported in 2012 [166,167]. As per the GLOBOCAN-2018 report, 162,000 breast cancer cases (27.7% of all new cancers), a 5-year prevalence of 405,000 breast cancer cases, and 87,000 deaths were observed in the Indian population [166]. Knowledge of reliable biomarkers related to disease prognosis and therapy decisions can improve cancer management. In this regard, the oncogenic and tumor suppressor components of the cell cycle may serve as such markers. Data from the multiple studies provided above support this notation.
Although these markers are detectable by expression-profiling experiments, the lack of reproductivity of the described results from various studies delays their use in the clinical setting. In addition to technique sensitivity issues, the non-reproducibility of the results might be due to the variation in sample selection methods or variations in study designs. Studies having a smaller number of patients, different-aged patients, varying tumor grades, varying tumor sizes or metastatic potential, and different patient ethnicities can also lead to non-reproducible results. Therefore, larger-scale validation studies involving greater demographic, ethnic, and clinicopathological variabilities are required in the future, before we can apply their suggestive use in patient management. High throughput technologies, such as next-generation RNA sequencing [168,169,170] and single-cell RNA sequencing [171,172], give results with high coverage and depth, and cover potential sensitivity issues. The use of these technologies may help to identify reliable biomarkers for cancer management. Thus, the data presented in the review article propose the use of cell cycle components as biomarkers in breast cancer management.
6. Conclusions
Previous experimental studies have described several oncogenic and tumor-suppressive genes involved in cell cycle regulation and progression among various subtypes of human breast cancer. It is well-established that multiple genetic alterations are required for tumorigenesis, yet continued research regarding the specific and sequential mechanisms involved—and how they affect clinical outcomes—may continue to guide new therapeutic strategies for more effective cancer treatments. Current research supports the notion that these regulatory cell cycle genes are useful prognostic biomarkers in breast cancer tumorigenesis, and the clinical relevance of these suggestive biomarkers has been established by several studies, as described above. The accurate measurement of cell cycle component expression and their correlation with clinical symptoms and prognoses may provide valuable insight for the future of both breast cancer management and anti-cancer therapeutics.
Acknowledgments
The authors (D.K., N.G., and V.K.G.) would like to gratefully acknowledge the Department of Histopathology, Post Graduate Institute of Medical Education and Research and University Institute of Engineering and Technology, Panjab University, Chandigarh, Punjab for providing platform support to compile this manuscript.
Abbreviations
| 5GG | Pentagalloylglucose |
| ABCSG | Austrian breast and colorectal cancer study group |
| AC | Adenocarcinoma |
| c-AMP | Cyclic adenosine monophosphate |
| CCND1 | Cyclin D1 |
| CDC2 kinase | Cell division control 2 kinase |
| CDKs | Cyclin-dependent kinases |
| CI | Confidence interval |
| DC | Ductal carcinoma |
| DCIS | Ductal carcinoma in situ |
| DFS | Disease-free survival |
| DSS | Disease-specific survival |
| E2F1 | E2 promoter factor 1 |
| ER | Estrogen receptor |
| FISH | Fluorescent in situ hybridization |
| GLOBOCAN | Global cancer observatory |
| HDAC3 | Histone deacetylase 3 |
| HER2+ | Human epidermal growth factor receptor2+ |
| HR | Hazard ratio |
| IDC | Invasive ductal carcinoma |
| IHC | Immunohistochemistry |
| ILC | Invasive lobular carcinoma |
| LA | Luminal A |
| MU | Mutation |
| NBE | Normal breast epithelium |
| ND | Not decided |
| OS | Overall survival |
| PD | Proliferative disease |
| PR | Progesterone receptor |
| Rb | Retinoblastoma |
| RFS | Recurrence free survival |
| siRNA | Small interfering RNA |
| TGF-β | Transforming growth factor-β |
| TNBC | Triple negative breast cancer |
| WT | Wild type |
Author Contributions
N.G. conceived the study; D.K. designed the study design and wrote the first draft; V.K.G. performed literature search and collection. E.N.S. performed language editing and suggested for quality improvement. A.B. supervised the project and performed the final editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
There exists no conflict of interest amongst authors regarding the publication of this manuscript.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zhivotovsky B., Orrenius S. Cell cycle and cell death in disease: Past, present and future. J. Intern. Med. 2010;268:395–409. doi: 10.1111/j.1365-2796.2010.02282.x. [DOI] [PubMed] [Google Scholar]
- 2.Witsch E., Sela M., Yarden Y. Roles for Growth Factors in Cancer Progression. Physiology. 2010;25:85–101. doi: 10.1152/physiol.00045.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.You J.S., Jones P.A. Cancer Genetics and Epigenetics: Two Sides of the Same Coin? Cancer Cell. 2012;22:9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baylin S.B., Jones P.A. Epigenetic Determinants of Cancer. Cold Spring Harb. Perspect. Biol. 2016;8:a019505. doi: 10.1101/cshperspect.a019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh A., Goel N. Yogita Integrative Analysis of Multi-Genomic Data for Kidney Renal Cell Carcinoma. Interdiscip. Sci. Comput. Life Sci. 2020;12:12–23. doi: 10.1007/s12539-019-00345-8. [DOI] [PubMed] [Google Scholar]
- 6.Goel N., Khandnor P. Materials Today: Proceedings. Elsevier; Amsterdam, The Netherlands: 2020. TCGA: A multi-genomics material repository for cancer research; pp. 1492–1495. [Google Scholar]
- 7.Lee S.Y., Ju M.K., Jeon H.M., Lee Y.J., Kim C.H., Park H.G., Han S.I., Kang H.S. Oncogenic Metabolism Acts as a Prerequisite Step for Induction of Cancer Metastasis and Cancer Stem Cell Phenotype. Oxidative Med. Cell. Longev. 2018;2018:1–28. doi: 10.1155/2018/1027453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kashyap D., Tuli H.S., Garg V.K., Goel N., Bishayee A. Oncogenic and Tumor-Suppressive Roles of MicroRNAs with Special Reference to Apoptosis: Molecular Mechanisms and Therapeutic Potential. Mol. Diagn. Ther. 2018;22:179–201. doi: 10.1007/s40291-018-0316-1. [DOI] [PubMed] [Google Scholar]
- 9.Kashyap D., Kaur H. Cell-free miRNAs as non-invasive biomarkers in breast cancer: Significance in early diagnosis and metastasis prediction. Life Sci. 2020;246:117417. doi: 10.1016/j.lfs.2020.117417. [DOI] [PubMed] [Google Scholar]
- 10.Mitra S., Bal A., Kashyap D., Kumar S., Shrivastav S., Das A., Singh G. Tumour angiogenesis and c-Met pathway activation –implications in breast cancer. APMIS. 2020;128:316–325. doi: 10.1111/apm.13031. [DOI] [PubMed] [Google Scholar]
- 11.Wang L.-H., Wu C.-F., Rajasekaran N., Shin Y.K. Loss of Tumor Suppressor Gene Function in Human Cancer: An Overview. Cell. Physiol. Biochem. 2018;51:2647–2693. doi: 10.1159/000495956. [DOI] [PubMed] [Google Scholar]
- 12.Lee E.Y.H.P., Muller W.J. Oncogenes and Tumor Suppressor Genes. Cold Spring Harb. Perspect. Biol. 2010;2:a003236. doi: 10.1101/cshperspect.a003236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Velez A.M.A., Howard M.S. Tumor-suppressor genes, cell cycle regulatory checkpoints, and the skin. N. Am. J. Med Sci. 2015;7:176–188. doi: 10.4103/1947-2714.157476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deshpande A., Sicinski P., Hinds P.W. Cyclins and cdks in development and cancer: A perspective. Oncogene. 2005;24:2909–2915. doi: 10.1038/sj.onc.1208618. [DOI] [PubMed] [Google Scholar]
- 15.Law M.E., Corsino P.E., Narayan S., Law B.K. Cyclin-Dependent Kinase Inhibitors as Anticancer Therapeutics. Mol. Pharmacol. 2015;88:846–852. doi: 10.1124/mol.115.099325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yixiao F., Mia S., Shifeng H., Chengfu Y., Zongyue Z., Linghuan Z., Xiaojuan J., Wei L., Bo H., Wenping L., et al. Breast cancer development and progression: Risk factors, cancer stem cells, signaling pathways, genomics, and molecular pathogenesis. Genes Dis. 2018;5:77–106. doi: 10.1016/j.gendis.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aggarwal V., Kashyap D., Sak K., Tuli H.S., Jain A., Chaudhary A., Garg V.K., Sethi G., Yerer M.B. Molecular Mechanisms of Action of Tocotrienols in Cancer: Recent Trends and Advancements. Int. J. Mol. Sci. 2019;20:656. doi: 10.3390/ijms20030656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mehta S., Shelling A., Muthukaruppan A., Lasham A., Blenkiron C., Laking G., Print C. Predictive and prognostic molecular markers for cancer medicine. Ther. Adv. Med Oncol. 2010;2:125–148. doi: 10.1177/1758834009360519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caldon C.E., Daly R.J., Sutherland R.L., Musgrove E.A. Cell cycle control in breast cancer cells. J. Cell. Biochem. 2005;97:261–274. doi: 10.1002/jcb.20690. [DOI] [PubMed] [Google Scholar]
- 20.Haase S.B., Wittenberg C. Topology and Control of the Cell-Cycle-Regulated Transcriptional Circuitry. Genetics. 2014;196:65–90. doi: 10.1534/genetics.113.152595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheung T.H.T., Rando T.A. Molecular regulation of stem cell quiescence. Nat. Rev. Mol. Cell Biol. 2013;14:329–340. doi: 10.1038/nrm3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Enders G.H. Mammalian Interphase Cdks: Dispensable Master Regulators of the Cell Cycle. Genes Cancer. 2012;3:614–618. doi: 10.1177/1947601913479799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertoli C., Skotheim J.M., De Bruin R.A.M. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 2013;14:518–528. doi: 10.1038/nrm3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cynthia O., Paschal W., Debu T. Oncogenes and tumor suppressor genes in breast cancer: Potential diagnostic and therapeutic applications. Oncologist. 2004;9:361–377. doi: 10.1634/theoncologist.9-4-361. [DOI] [PubMed] [Google Scholar]
- 25.Kato J., Matsushime H., Hiebert S.W., Ewen E.M., Sherr C.J. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev. 1993;7:331–342. doi: 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- 26.Wiman K.G. The retinoblastoma gene: Role in cell cycle control and cell differentiation. FASEB J. 1993;7:841–845. doi: 10.1096/fasebj.7.10.8393817. [DOI] [PubMed] [Google Scholar]
- 27.Coqueret O. Linking cyclins to transcriptional control. Gene. 2002;299:35–55. doi: 10.1016/S0378-1119(02)01055-7. [DOI] [PubMed] [Google Scholar]
- 28.Diehl A.J., Zindy F., Sherr C.J. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 1997;11:957–972. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- 29.Yu Z.-K., Gervais J.L.M., Zhang H. Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21CIP1/WAF1 and cyclin D proteins. Proc. Natl. Acad. Sci. USA. 1998;95:11324–11329. doi: 10.1073/pnas.95.19.11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang C., Li Z., Lu Y., Du R., Katiyar S., Yang J., Fu M., Leader J.E., Quong A., Novikoff P.M., et al. Cyclin D1 repression of nuclear respiratory factor 1 integrates nuclear DNA synthesis and mitochondrial function. Proc. Natl. Acad. Sci. USA. 2006;103:11567–11572. doi: 10.1073/pnas.0603363103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall M., Peters G. Advances in Cancer Research. Elsevier; Amsterdam, The Netherlands: 1996. Genetic Alterations of Cyclins, Cyclin-Dependent Kinases, and Cdk Inhibitors in Human Cancer; pp. 67–108. [DOI] [PubMed] [Google Scholar]
- 32.Yamamoto M., Tamakawa S., Yoshie M., Yaginuma Y., Ogawa K. Neoplastic hepatocyte growth associated with cyclin D1 redistribution from the cytoplasm to the nucleus in mouse hepatocarcinogenesis. Mol. Carcinog. 2006;45:901–913. doi: 10.1002/mc.20204. [DOI] [PubMed] [Google Scholar]
- 33.Hodges L.C., Cook J.D., Lobenhofer E.K., Li L., Bennett L., Bushel P.R., Aldaz C.M., Afshari A.C., Walker C.L. Tamoxifen functions as a molecular agonist inducing cell cycle-associated genes in breast cancer cells. Mol. Cancer Res. 2003;1:300–311. [PubMed] [Google Scholar]
- 34.Kenny F.S., Hui R., Musgrove E.A., Gee J.M., Blamey R.W., Nicholson R.I., Sutherland R.L., Robertson J.F. Overexpression of cyclin D1 messenger RNA predicts for poor prognosis in estrogen receptor-positive breast cancer. Clin Cancer Res. 1999;5:2069–2076. [PubMed] [Google Scholar]
- 35.Mohammadizadeh F., Hani M., Ranaee M., Bagheri M. Role of cyclin D1 in breast carcinoma. J. Res. Med Sci. 2013;18:1021–1025. [PMC free article] [PubMed] [Google Scholar]
- 36.Barnes D.M., Gillett C.E. Cyclin D1 in Breast Cancer. Breast Cancer Res. Treat. 1998;52:1–15. doi: 10.1023/A:1006103831990. [DOI] [PubMed] [Google Scholar]
- 37.A Velasco-Velázquez M., Li Z., Casimiro M., Loro E., Homsi N., Pestell R.G. Examining the role of cyclin D1 in breast cancer. Futur. Oncol. 2011;7:753–765. doi: 10.2217/fon.11.56. [DOI] [PubMed] [Google Scholar]
- 38.Ravikumar G., Ananthamurthy A. Cyclin D1 expression in ductal carcinoma of the breast and its correlation with other prognostic parameters. J. Cancer Res. Ther. 2014;10:671–675. doi: 10.4103/0973-1482.138135. [DOI] [PubMed] [Google Scholar]
- 39.Assem M., Youssef E.A., Rashad R.M., Yahia M.A.-H. Immunohistochemical Expression of Cyclin D1 in Invasive Ductal Carcinoma of Human Breast. Oncomedicine. 2017;2:80–87. doi: 10.7150/oncm.18416. [DOI] [Google Scholar]
- 40.Buckley M.F., Sweeney K.J., Hamilton A.J., Sini R.L., Manning D.L., Nicholson I.R., DeFazio A., Watts C.K., Musgrove A.E., Sutherland R.L. Expression and amplification of cyclin genes in human breast cancer. Oncogene. 1993;8:2127–2133. [PubMed] [Google Scholar]
- 41.Lebwohl E.D., Muise-Helmericks R., Sepp-Lorenzino L., Serve S., Timaul M., Bol R., Borgen P., Rosen N. A truncated cyclin D1 gene encodes a stable mRNA in a human breast cancer cell line. Oncogene. 1994;9:1925–1929. [PubMed] [Google Scholar]
- 42.Zukerberg L.R., Yang I.W., Gadd M., Thor A.D., Koerner F.C., Schmidt E.V., Arnold A. Cyclin D1 (PRAD1) protein expression in breast cancer: Approximately one-third of infiltrating mammary carcinomas show overexpression of the cyclin D1 oncogene. Mod. Pathol. 1995;8:560–567. [PubMed] [Google Scholar]
- 43.Khan N., Syed D.N., Ahmad N., Mukhtar H. Fisetin: A Dietary Antioxidant for Health Promotion. Antioxid. Redox Signal. 2013;19:151–162. doi: 10.1089/ars.2012.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosendahl A.H., Perks C.M., Zeng L., Markkula A., Simonsson M., Rose C., Ingvar C., Holly J.M., Jernström H. Caffeine and Caffeic Acid Inhibit Growth and Modify Estrogen Receptor and Insulin-like Growth Factor I Receptor Levels in Human Breast Cancer. Clin. Cancer Res. 2015;21:1877–1887. doi: 10.1158/1078-0432.CCR-14-1748. [DOI] [PubMed] [Google Scholar]
- 45.Zhang S.-Y., Caamano J., Cooper F., Guo X., Klein-Szanto A.J. Immunohistochemistry of Cyclin D1 in Human Breast Cancer. Am. J. Clin. Pathol. 1994;102:695–698. doi: 10.1093/ajcp/102.5.695. [DOI] [PubMed] [Google Scholar]
- 46.Wilcken N.R., Prall O.W., Musgrove A.E., Sutherland R.L. Inducible overexpression of cyclin D1 in breast cancer cells reverses the growth-inhibitory effects of antiestrogens. Clin. Cancer Res. 1997;3:849–854. [PubMed] [Google Scholar]
- 47.Jirström K., Stendahl M., Rydén L., Kronblad A., Bendahl P.-O., Stal O., Landberg G. Adverse Effect of Adjuvant Tamoxifen in Premenopausal Breast Cancer with Cyclin D1 Gene Amplification. Cancer Res. 2005;65:8009–8016. doi: 10.1158/0008-5472.CAN-05-0746. [DOI] [PubMed] [Google Scholar]
- 48.Courjal F., Louason G., Speiser P., Katsaros D., Zeillinger R., Theillet C. Cyclin gene amplification and overexpression in breast and ovarian cancers: Evidence for the selection of cyclin D1 in breast and cyclin E in ovarian tumors. Int. J. Cancer. 1996;69:247–253. doi: 10.1002/(SICI)1097-0215(19960822)69:4<247::AID-IJC1>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 49.Umekita Y., Ohi Y., Sagara Y., Yoshida H. Overexpression of cyclinD1 predicts for poor prognosis in estrogen receptor-negative breast cancer patients. Int. J. Cancer. 2002;98:415–418. doi: 10.1002/ijc.10151. [DOI] [PubMed] [Google Scholar]
- 50.Elsheikh S., Green A.R., Aleskandarany M.A., Grainge M., Paish C.E., Lambros M.B.K., Reis-Filho J.S., Ellis I.O. CCND1 amplification and cyclin D1 expression in breast cancer and their relation with proteomic subgroups and patient outcome. Breast Cancer Res. Treat. 2007;109:325–335. doi: 10.1007/s10549-007-9659-8. [DOI] [PubMed] [Google Scholar]
- 51.Rudas M., Lehnert M., Huynh A., Jakesz R., Singer C., Lax S., Schippinger W., Dietze O., Greil R., Stiglbauer W., et al. Cyclin D1 Expression in Breast Cancer Patients Receiving Adjuvant Tamoxifen-Based Therapy. Clin. Cancer Res. 2008;14:1767–1774. doi: 10.1158/1078-0432.CCR-07-4122. [DOI] [PubMed] [Google Scholar]
- 52.Alle K.M., Henshall S.M., Field A.S., Sutherland R.L. Cyclin D1 protein is overexpressed in hyperplasia and intraductal carcinoma of the breast. Clin. Cancer Res. 1998;4:847–854. [PubMed] [Google Scholar]
- 53.Heiss C., Schanz A., Amabile N., Jahn S., Chen Q., Wong M.L., Rassaf T., Heinen Y., Cortese-Krott M., Grossman W., et al. Nitric Oxide Synthase Expression and Functional Response to Nitric Oxide Are Both Important Modulators of Circulating Angiogenic Cell Response to Angiogenic Stimuli. Arter. Thromb. Vasc. Biol. 2010;30:2212–2218. doi: 10.1161/ATVBAHA.110.211581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oyama T., Kashiwabara K., Yoshimoto K., Arnold A., Koerner F. Frequent overexpression of the cyclin D1 oncogene in invasive lobular carcinoma of the breast. Cancer Res. 1998;58:2876–2880. [PubMed] [Google Scholar]
- 55.Ahnström M., Nordenskjöld B., Rutqvist L.E., Skoog L., Stål O. Role of cyclin D1 in ErbB2-positive breast cancer and tamoxifen resistance. Breast Cancer Res. Treat. 2005;91:145–151. doi: 10.1007/s10549-004-6457-4. [DOI] [PubMed] [Google Scholar]
- 56.Simpson J.F., Quan D.E., O’Malley F., Odom-Maryon T., Clarke P.E. Amplification of CCND1 and expression of its protein product, cyclin D1, in ductal carcinoma in situ of the breast. Am. J. Pathol. 1997;151:161–168. [PMC free article] [PubMed] [Google Scholar]
- 57.Tobin N.P., Bergh J. Analysis of Cyclin D1 in Breast Cancer: A Call to Arms. Curr. Breast Cancer Rep. 2012;4:171–173. doi: 10.1007/s12609-012-0083-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boström P., Söderström M., Palokangas T., Vahlberg T., Collan Y., Carpén O., Hirsimäki P. Analysis of cyclins A, B1, D1 and E in breast cancer in relation to tumour grade and other prognostic factors. BMC Res. Notes. 2009;2:140. doi: 10.1186/1756-0500-2-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dai M., Al-Odaini A.A., Fils-Aimé N., Villatoro A.M., Guo J., Arakelian A., Rabbani A.S., Ali S., Lebrun J.J. Cyclin D1 cooperates with p21 to regulate TGFβ-mediated breast cancer cell migration and tumor local invasion. Breast Cancer Res. 2013;15:R49. doi: 10.1186/bcr3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Russell A.F., Thompson A.M., Hendley J., Trute L., Armes E.J., Germain D.S. Cyclin D1 and D3 associate with the SCF complex and are coordinately elevated in breast cancer. Oncogene. 1999;18:1983–1991. doi: 10.1038/sj.onc.1202511. [DOI] [PubMed] [Google Scholar]
- 61.Khandeparkar S.G.S., Lengare P.V., Joshi A.R., Gogate B.P., Solanke S.G., Gore S.H. Immunohistochemical expression of cyclin D1 in invasive breast carcinoma and its correlation with clinicopathological parameters. Indian J. Pathol. Microbiol. 2020;63:376. doi: 10.4103/IJPM.IJPM_106_20. [DOI] [PubMed] [Google Scholar]
- 62.Husdal A., Bukholm G., Bukholm I.R.K. The Prognostic Value and Overexpression of Cyclin A Is Correlated with Gene Amplification of both Cyclin A and Cyclin E in Breast Cancer Patient. Cell. Oncol. 2006;28:107–116. doi: 10.1155/2006/721919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baldini E., Camerini A., Sgambato A., Prochilo T., Capodanno A., Pasqualetti F., Orlandini C., Resta L., Bevilacqua G., Collecchi P. Cyclin A and E2F1 overexpression correlate with reduced disease-free survival in node-negative breast cancer patients. Anticancer. Res. 2007;26:4415–4421. [PubMed] [Google Scholar]
- 64.Michalides R., Van Tinteren H., Balkenende A., Vermorken J.B., Benraadt J., Huldij J., Van Diest P. Cyclin A is a prognostic indicator in early stage breast cancer with and without tamoxifen treatment. Br. J. Cancer. 2002;86:402–408. doi: 10.1038/sj.bjc.6600072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Poikonen P., Sjöström J., Amini R.-M., Villman K., Ahlgren J., Blomqvist C. Cyclin A as a marker for prognosis and chemotherapy response in advanced breast cancer. Br. J. Cancer. 2005;93:515–519. doi: 10.1038/sj.bjc.6602735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nielsen N., Arnerlöv C., Emdin S., Landberg G. Cyclin E overexpression, a negative prognostic factor in breast cancer with strong correlation to oestrogen receptor status. Br. J. Cancer. 1996;74:874–880. doi: 10.1038/bjc.1996.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sieuwerts A.M., Look M.P., Gelder M.E.M.-V., Timmermans M., Trapman A.M., Garcia R.R., Arnold M., Goedheer A.J., De Weerd V., Portengen H., et al. Which Cyclin E Prevails as Prognostic Marker for Breast Cancer? Results from a Retrospective Study Involving 635 Lymph Node–Negative Breast Cancer Patients. Clin. Cancer Res. 2006;12:3319–3328. doi: 10.1158/1078-0432.CCR-06-0225. [DOI] [PubMed] [Google Scholar]
- 68.Donnellan R., Kleinschmidt I., Chetty R. Cyclin E immunoexpression in breast ductal carcinoma: Pathologic correlations and prognostic implications. Hum. Pathol. 2001;32:89–94. doi: 10.1053/hupa.2001.21141. [DOI] [PubMed] [Google Scholar]
- 69.Potemski P., Kusinska R., Watala C., Pluciennik E., Bednarek A.K., Kordek R. Cyclin E expression in breast cancer correlates with negative steroid receptor status, HER2 expression, tumor grade and proliferation. J. Exp. Clin. Cancer Res. 2006;25:59–64. [PubMed] [Google Scholar]
- 70.Han S., Park K., Bae B.-N., Kim K.H., Kim H.-J., Kim Y.-D. Prognostic implication of cyclin E expression and its relationship with cyclin D1 and p27Kip1 expression on tissue microarrays of node negative breast cancer. J. Surg. Oncol. 2003;83:241–247. doi: 10.1002/jso.10268. [DOI] [PubMed] [Google Scholar]
- 71.Potemski P., Kusińska R., Pasz-Walczak G., Piekarski J.H., Watała C., Płuciennik E., Bednarek A.K., Kordek R. Prognostic relevance of cyclin E expression in operable breast cancer. Med Sci. Monit. 2009;15:34–40. [PubMed] [Google Scholar]
- 72.Niméus-Malmström E., Koliadi A., Ahlin C., Holmqvist M., Holmberg L., Amini R.-M., Jirström K., Wärnberg F., Blomqvist C., Fernö M., et al. Cyclin B1 is a prognostic proliferation marker with a high reproducibility in a population-based lymph node negative breast cancer cohort. Int. J. Cancer. 2009;127:961–967. doi: 10.1002/ijc.25091. [DOI] [PubMed] [Google Scholar]
- 73.Sun X., Zhangyuan G., Shi L., Wang Y., Sun B., Ding Q. Prognostic and clinicopathological significance of cyclin B expression in patients with breast cancer. Medicine. 2017;96:e6860. doi: 10.1097/MD.0000000000006860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Aaltonen K., Amini R.-M., Heikkilä P., Aittomäki K., Tamminen A., Nevanlinna H., Blomqvist C. High cyclin B1 expression is associated with poor survival in breast cancer. Br. J. Cancer. 2009;100:1055–1060. doi: 10.1038/sj.bjc.6604874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Göhring U.-J., Schöndorf T., Kiecker V.R., Becker M., Kurbacher C., Scharl A. Immunohistochemical detection of H-ras protooncoprotein p21 indicates favorable prognosis in node-negative breast cancer patients. Tumor Biol. 1999;20:173–183. doi: 10.1159/000030061. [DOI] [PubMed] [Google Scholar]
- 76.Domagala W., Welcker M., Chosia M., Karbowniczek M., Harezga B., Bartkova J., Bartek J., Osborn M. p21/WAF1/Cip1 expression in invasive ductal breast carcinoma: Relationship to p53, proliferation rate, and survival at 5 years. Virchows Arch. 2001;439:132–140. doi: 10.1007/s004280100410. [DOI] [PubMed] [Google Scholar]
- 77.Fayed Y.M., El-Sheikh S.A.-M., Abulkheir I.L.H., Khorshed I.N.A.-H. Immunohistochemical expression of p27 in ductal carcinoma of breast and its correlation with HER2/neu expression and hormonal status. Egypt. J. Pathol. 2012;32:33–41. doi: 10.1097/01.XEJ.0000415799.59629.63. [DOI] [Google Scholar]
- 78.Wei C.-Y., Tan Q.-X., Zhu X., Qin Q.-H., Zhu F.-B., Mo Q.-G., Yang W.-P. Expression of CDKN1A/p21 and TGFBR2 in breast cancer and their prognostic significance. Int. J. Clin. Exp. Pathol. 2015;8:14619–14629. [PMC free article] [PubMed] [Google Scholar]
- 79.Alkarain A., Jordan R., Slingerland J. p27 Deregulation in Breast Cancer: Prognostic Significance and Implications for Therapy. J. Mammary Gland. Biol. Neoplasia. 2004;9:67–80. doi: 10.1023/B:JOMG.0000023589.00994.5e. [DOI] [PubMed] [Google Scholar]
- 80.Tsuchiya A., Zhang G.J., Kanno M. Prognostic impact of cyclin-dependent kinase inhibitor p27kip1 in node-positive breast cancer. J. Surg. Oncol. 1999;70:230–234. doi: 10.1002/(SICI)1096-9098(199904)70:4<230::AID-JSO6>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 81.Traub F., Mengel M., Lück H.J., Kreipe H.H., Von Wasielewski R. Prognostic impact of Skp2 and p27 in human breast cancer. Breast Cancer Res. Treat. 2006;99:185–191. doi: 10.1007/s10549-006-9202-3. [DOI] [PubMed] [Google Scholar]
- 82.Newman L., Xia W., Yang H.-Y., Sahin A., Bondy M., Lukmanji F., Hung M.-C., Lee M.-H. Correlation of p27 protein expression with HER-2/neu expression in breast cancer. Mol. Carcinog. 2001;30:169–175. doi: 10.1002/mc.1025. [DOI] [PubMed] [Google Scholar]
- 83.Deepu M., Daniel F.R., Giorgio I., Anne Z.J., Joan C., Amber G., Rachael A.S., Silvia C.F., Michele P., Franco M. Loss of p27KIP1 expression in fully-staged node-negative breast cancer: Association with lack of hormone receptors in T1a/b, but not T1c infiltrative ductal carcinoma. Anticancer Res. 2011;12:4401–4405. [PMC free article] [PubMed] [Google Scholar]
- 84.Leivonen M., Nordling S., Lundin J., Von Boguslawski K., Haglund C. p27 expression correlates with short-term, but not with long-term prognosis in breast cancer. Breast Cancer Res. Treat. 2001;67:15–22. doi: 10.1023/A:1010623326118. [DOI] [PubMed] [Google Scholar]
- 85.Barnes A., Pinder S., Bell J., Paish E., Wencyk P., Robertson J., Elston C., Ellis I. Expression of p27kip1 in breast cancer and its prognostic significance. J. Pathol. 2003;201:451–459. doi: 10.1002/path.1464. [DOI] [PubMed] [Google Scholar]
- 86.Pohl G., Rudas M., Dietze O., Lax S., Markis E., Pirker R., Zielinski C.C., Hausmaninger H., Kubista E., Samonigg H., et al. High p27Kip1 Expression Predicts Superior Relapse-Free and Overall Survival for Premenopausal Women With Early-Stage Breast Cancer Receiving Adjuvant Treatment With Tamoxifen Plus Goserelin. J. Clin. Oncol. 2003;21:3594–3600. doi: 10.1200/JCO.2003.02.021. [DOI] [PubMed] [Google Scholar]
- 87.Nohara T., Ryo T., Iwamoto S., Gon G., Tanigawa N. Expression of Cell-Cycle Regulator p27 Is Correlated to the Prognosis and ER Expression in Breast Carcinoma Patients. Oncology. 2001;60:94–100. doi: 10.1159/000055303. [DOI] [PubMed] [Google Scholar]
- 88.Spataro V.J., Litman H., Viale G., Maffini F., Masullo M., Golouh R., Martínez-Tello F.J., Grigolato P., Shilkin K.B., Gusterson B.A., et al. Decreased immunoreactivity for p27 protein in patients with early-stage breast carcinoma is correlated with HER-2/neuoverexpression and with benefit from one course of perioperative chemotherapy in patients with negative lymph node status. Cancer. 2003;97:1591–1600. doi: 10.1002/cncr.11224. [DOI] [PubMed] [Google Scholar]
- 89.Davidoff A.M., Herndon E.J., Glover N.S., Kerns B.J., Pence J.C., Iglehart J.D., Marks J.R. Relation between p53 overexpression and established prognostic factors in breast cancer. Surgery. 1991;110:259–264. [PubMed] [Google Scholar]
- 90.Porter P.L., Barlow W.E., Yeh I.-T., Lin M.G., Yuan X.P., Donato E., Sledge G.W., Shapiro C.L., Ingle J.N., Haskell C.M., et al. p27 Kip1 and Cyclin E Expression and Breast Cancer Survival After Treatment With Adjuvant Chemotherapy. J. Natl. Cancer Inst. 2006;98:1723–1731. doi: 10.1093/jnci/djj467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chu J.-S., Huang C.-S., Chang K.-J. p27 expression as a prognostic factor of breast cancer in Taiwan. Cancer Lett. 1999;141:123–130. doi: 10.1016/S0304-3835(99)00084-1. [DOI] [PubMed] [Google Scholar]
- 92.Cui S.-P., Wang H.-L., Peng W., Liu H.-J., Hou L., Zhang B. Aberrant expression and correlative analysis of P16 in breast cancers. Beijing da xue xue bao. Yi xue ban = J. Peking Univ. Heal. Sci. 2012;44:755–759. [PubMed] [Google Scholar]
- 93.Emig R., Magener A., Ehemann V., Meyer A., Stilgenbauer F., Volkmann M., Wallwiener D., Sinn H.P. Aberrant cytoplasmic expression of the p16 protein in breast cancer is associated with accelerated tumour proliferation. Br. J. Cancer. 1998;78:1661–1668. doi: 10.1038/bjc.1998.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pan Y., Yuan Y., Liu G., Wei Y. P53 and Ki-67 as prognostic markers in triple-negative breast cancer patients. PLoS ONE. 2017;12:e0172324. doi: 10.1371/journal.pone.0172324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Al-Joudi F.S., Iskandar A.Z., Rusli J. The expression of p53 in invasive ductal carcinoma of the breast: A study in the North-East States of Malaysia. Med. J. Malays. 2008;63:96–99. [PubMed] [Google Scholar]
- 96.Shan M., Zhang X., Liu X., Qin Y., Liu T., Liu Y., Wang J., Zhong Z., Zhang Y., Geng J., et al. P16 and P53 Play Distinct Roles in Different Subtypes of Breast Cancer. PLoS ONE. 2013;8:e76408. doi: 10.1371/journal.pone.0076408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Abou-Bakr A.A., Eldweny I.H. P16 expression correlates with basal-like triple-negative breast carcinoma. Ecancermedicalscience. 2013;7:317. doi: 10.3332/ecancer.2013.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Harbhajanka A., Lamzabi I., Bitterman P., Reddy V.B., Ghai R., Gattuso P. Correlation of p16 Expression on Cancer and Stromal Cells With Clinicopathologic and Immunohistochemical Features of Lobular Breast Carcinoma. Appl. Immunohistochem. Mol. Morphol. 2019;27:658–662. doi: 10.1097/PAI.0000000000000679. [DOI] [PubMed] [Google Scholar]
- 99.Salmani H., Hosseini A., Azarnezhad A., Ahmad H. PTEN and p53 gene expressions in breast cancer specimens and their clinicopathological significance. Middle East J. Cancer. 2018;9:105–111. doi: 10.30476/mejc.2018.42112. [DOI] [Google Scholar]
- 100.Yang P., Du C.W., Kwan M., Liang S.X., Zhang G.J. The impact of p53 in predicting clinical outcome of breast cancer patients with visceral metastasis. Sci. Rep. 2013;3:srep02246. doi: 10.1038/srep02246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bertheau P., Lehmann-Che J., Varna M., Dumay A., Poirot B., Porcher R., Turpin E., Plassa L.-F., De Roquancourt A., Bourstyn E., et al. p53 in breast cancer subtypes and new insights into response to chemotherapy. Breast. 2013;22:S27–S29. doi: 10.1016/j.breast.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 102.Li J.-P., Zhang X.-M., Zhang Z., Zheng L.-H., Jindal S., Liu Y.-J. Association of p53 expression with poor prognosis in patients with triple-negative breast invasive ductal carcinoma. Medicine. 2019;98:e15449. doi: 10.1097/MD.0000000000015449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lee D.-S., Kim S.H., Suh Y.J., Kim H.K., Shim B.Y. Clinical Implication of p53 Overexpression in Breast Cancer Patients Younger than 50 Years with a Triple-negative Subtype Who Undergo a Modified Radical Mastectomy. Jpn. J. Clin. Oncol. 2011;41:854–866. doi: 10.1093/jjco/hyr066. [DOI] [PubMed] [Google Scholar]
- 104.Kikuchi S., Nishimura R., Osako T., Okumura Y., Nishiyama Y., Toyozumi Y., Arima N. Definition of p53 overexpression and its association with the clinicopathological features in luminal/HER2-negative breast cancer. Anticancer. Res. 2013;33:3891–3898. [PubMed] [Google Scholar]
- 105.Coates A.S., Millar E.K., O’Toole S.A., Molloy T.J., Viale G., Goldhirsch A., Regan M.M., Gelber R.D., Sun Z., Castiglione-Gertsch M., et al. Prognostic interaction between expression of p53 and estrogen receptor in patients with node-negative breast cancer: Results from IBCSG Trials VIII and IX. Breast Cancer Res. 2012;14:R143. doi: 10.1186/bcr3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Abubakar M., Guo C., Koka H., Sung H., Shao N., Guida J., Deng J., Li M., Hu N., Zhou B., et al. Clinicopathological and epidemiological significance of breast cancer subtype reclassification based on p53 immunohistochemical expression. NPJ Breast Cancer. 2019;5 doi: 10.1038/s41523-019-0117-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Muhammad E.M.S., Ahmad A.N., Guirguis M.N., Ali A.-E.M. Immunohistochemical P53 Expression in Breast Carcinoma with Correlation to Clinico-Pathological Parameters. Med. J. Cairo Univ. 2012;80:179–189. [Google Scholar]
- 108.Bae S.Y., Lee J.H., Bae J.W., Jung S.P. Differences in prognosis by p53 expression after neoadjuvant chemotherapy in triple-negative breast cancer. Ann. Surg. Treat. Res. 2020;98:291–298. doi: 10.4174/astr.2020.98.6.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nema S., Mehta P., Narang S. Role of p53 and Ki-67 in prognostication of carcinoma breast. Indian J. Pathol. Oncol. 2019;6:261–265. doi: 10.18231/j.ijpo.2019.051. [DOI] [Google Scholar]
- 110.Lara J.F., Thor A.D., Dressler L.G., Broadwater G., Bleiweiss I.J., Edgerton S., Cowan D., Goldstein L.J., Martino S., Ingle J.N., et al. p53 Expression in Node-Positive Breast Cancer Patients: Results from the Cancer and Leukemia Group B 9344 Trial (159905) Clin. Cancer Res. 2011;17:5170–5178. doi: 10.1158/1078-0432.CCR-11-0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Marchetti P., Cannita K., Ricevuto E., De Galitiis F., Di Rocco Z.C., Tessitore A., Bisegna R., Porzio G., De Rubeis G.P., Ventura T., et al. Prognostic value of p53 molecular status in high-risk primary breast cancer. Ann. Oncol. 2003;14:704–708. doi: 10.1093/annonc/mdg197. [DOI] [PubMed] [Google Scholar]
- 112.Loo L.W.M., Gao C., Shvetsov Y.B., Okoro D.R., Hernandez B.Y., Bargonetti J. MDM2, MDM2-C, and mutant p53 expression influence breast cancer survival in a multiethnic population. Breast Cancer Res. Treat. 2018;174:257–269. doi: 10.1007/s10549-018-5065-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kim J.-Y., Park K., Jung H.H., Lee E., Cho E.Y., Lee K.H., Bae S.Y., Lee S.K., Kim S.W., Lee J.E., et al. Association between Mutation and Expression of TP53 as a Potential Prognostic Marker of Triple-Negative Breast Cancer. Cancer Res. Treat. 2016;48:1338–1350. doi: 10.4143/crt.2015.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zwijsen R.M., Klompmaker R., Wientjens E.B., Kristel P.M., Van Der Burg B., Michalides R.J. Cyclin D1 triggers autonomous growth of breast cancer cells by governing cell cycle exit. Mol. Cell. Biol. 1996;16:2554–2560. doi: 10.1128/MCB.16.6.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lodén M., Stighall M., Nielsen N.H., Roos G., Emdin S.O., Östlund H., Landberg G. The cyclin D1 high and cyclin E high subgroups of breast cancer: Separate pathways in tumorogenesis based on pattern of genetic aberrations and inactivation of the pRb node. Oncogene. 2002;21:4680–4690. doi: 10.1038/sj.onc.1205578. [DOI] [PubMed] [Google Scholar]
- 116.Kilker R.L., Planas-Silva M.D. Cyclin D1 Is Necessary for Tamoxifen-Induced Cell Cycle Progression in Human Breast Cancer Cells. Cancer Res. 2006;66:11478–11484. doi: 10.1158/0008-5472.CAN-06-1755. [DOI] [PubMed] [Google Scholar]
- 117.Yde C.W., Issinger O.-G. Enhancing cisplatin sensitivity in MCF-7 human breast cancer cells by down-regulation of Bcl-2 and cyclin D1. Int. J. Oncol. 2006;29:1397–1404. doi: 10.3892/ijo.29.6.1397. [DOI] [PubMed] [Google Scholar]
- 118.Kilker R.L., Hartl M.W., Rutherford T.M., Planas-Silva M.D. Cyclin D1 expression is dependent on estrogen receptor function in tamoxifen-resistant breast cancer cells. J. Steroid Biochem. Mol. Biol. 2004;92:63–71. doi: 10.1016/j.jsbmb.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 119.Joung Y.H., Lim E.J., Lee M.Y., Park J.-H., Ye S.-K., Park E.U., Kim S.Y., Zhang Z., Lee K.J., Park N.K., et al. Hypoxia activates the cyclin D1 promoter via the Jak2/STAT5b pathway in breast cancer cells. Exp. Mol. Med. 2005;37:353–364. doi: 10.1038/emm.2005.45. [DOI] [PubMed] [Google Scholar]
- 120.Keyomarsi K., Pardee A.B. Redundant cyclin overexpression and gene amplification in breast cancer cells. Proc. Natl. Acad. Sci. USA. 1993;90:1112–1116. doi: 10.1073/pnas.90.3.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Scaltriti M., Eichhorn P.J., Cortés J., Prudkin L., Aura C., Jiménez J., Chandarlapaty S., Serra V., Prat A., Ibrahim Y.H., et al. Cyclin E amplification/overexpression is a mechanism of trastuzumab resistance in HER2+breast cancer patients. Proc. Natl. Acad. Sci. USA. 2011;108:3761–3766. doi: 10.1073/pnas.1014835108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen W.-J., Chang C.-Y., Lin J.-K. Induction of G1 phase arrest in MCF human breast cancer cells by pentagalloylglucose through the down-regulation of CDK4 and CDK2 activities and up-regulation of the CDK inhibitors p27(Kip) and p21(Cip) Biochem. Pharmacol. 2003;65:1777–1785. doi: 10.1016/S0006-2952(03)00156-4. [DOI] [PubMed] [Google Scholar]
- 123.Chen X., Danes C., Lowe M., Herliczek T.W., Keyomarsi K. Activation of the Estrogen-Signaling Pathway by p21WAF1/CIP1 in Estrogen Receptor-Negative Breast Cancer Cells. J. Natl. Cancer Inst. 2000;92:1403–1413. doi: 10.1093/jnci/92.17.1403. [DOI] [PubMed] [Google Scholar]
- 124.Fritah A., Saucier C., Mester J., Redeuilh G., Sabbah M. p21WAF1/CIP1 Selectively Controls the Transcriptional Activity of Estrogen Receptor α. Mol. Cell. Biol. 2005;25:2419–2430. doi: 10.1128/MCB.25.6.2419-2430.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Androic I., Krämer A., Yan R., Rödel F., Gätje R., Kaufmann M., Strebhardt K., Yuan J. Targeting cyclin B1 inhibits proliferation and sensitizes breast cancer cells to taxol. BMC Cancer. 2008;8:391. doi: 10.1186/1471-2407-8-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Arima Y., Hayashi N., Hayashi H., Sasaki M., Kai K., Sugihara E., Abe E., Yoshida A., Mikami S., Nakamura S., et al. Loss of p16 expression is associated with the stem cell characteristics of surface markers and therapeutic resistance in estrogen receptor-negative breast cancer. Int. J. Cancer. 2012;130:2568–2579. doi: 10.1002/ijc.26271. [DOI] [PubMed] [Google Scholar]
- 127.Maria K., Vassilis G.G., George Z.R., Petros L., Christos M., John G., Christos K. High expression levels of p27 correlate with lymph node status in a subset of advanced invasive breast carcinomas: Relation to E-cadherin alterations, proliferative activity, and ploidy of the tumors. Cancer. 2002;94:2454–2465. doi: 10.1002/cncr.10505. [DOI] [PubMed] [Google Scholar]
- 128.Nahta R., Takahashi T., Ueno N.T., Hung M.-C., Esteva F.J. P27kip1 Down-Regulation Is Associated with Trastuzumab Resistance in Breast Cancer Cells. Cancer Res. 2004;64:3981–3986. doi: 10.1158/0008-5472.CAN-03-3900. [DOI] [PubMed] [Google Scholar]
- 129.Cariou S., Donovan J.C.H., Flanagan W.M., Milic A., Bhattacharya N., Slingerland J.M. Down-regulation of p21WAF1/CIP1 or p27Kip1 abrogates antiestrogen-mediated cell cycle arrest in human breast cancer cells. Proc. Natl. Acad. Sci. USA. 2000;97:9042–9046. doi: 10.1073/pnas.160016897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Brown I., Shalli K., McDonald S.L., Moir E.S., Hutcheon A.W., Heys S.D., Schofield A.C. Reduced expression of p27 is a novel mechanism of docetaxel resistance in breast cancer cells. Breast Cancer Res. 2004;6:1–7. doi: 10.1186/bcr918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dai X., Cheng H., Bai Z., Li J. Breast Cancer Cell Line Classification and Its Relevance with Breast Tumor Subtyping. J. Cancer. 2017;8:3131–3141. doi: 10.7150/jca.18457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dickson C., Fantl V., Gillett C., Brookes S., Bartek J., Smith R., Fisher C., Barnes D., Peters G. Amplification of chromosome band 11q13 and a role for cyclin D1 in human breast cancer. Cancer Lett. 1995;90:43–50. doi: 10.1016/0304-3835(94)03676-A. [DOI] [PubMed] [Google Scholar]
- 133.Zhang P. The cell cycle and development: Redundant roles of cell cycle regulators. Curr. Opin. Cell Biol. 1999;11:655–662. doi: 10.1016/S0955-0674(99)00032-0. [DOI] [PubMed] [Google Scholar]
- 134.Sutherland R.L., Musgrove E.A. Cyclins and Breast Cancer. J. Mammary Gland. Biol. Neoplasia. 2004;9:95–104. doi: 10.1023/B:JOMG.0000023591.45568.77. [DOI] [PubMed] [Google Scholar]
- 135.Lundberg A., Lindström L.S., Li J., Harrell J.C., Darai-Ramqvist E., Sifakis E., Foukakis T., Perou C.M., Czene K., Bergh J., et al. The long-term prognostic and predictive capacity of cyclin D1 gene amplification in 2305 breast tumours. Breast Cancer Res. 2019;21:34. doi: 10.1186/s13058-019-1121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yu Z., Wang L., Wang C., Ju X., Wang M., Chen K., Loro E., Li Z., Zhang Y., Wu K., et al. Cyclin D1 induction of Dicer governs microRNA processing and expression in breast cancer. Nat. Commun. 2013;4:1–10. doi: 10.1038/ncomms3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhong Z., Yeow W.-S., Zou C., Wassell R., Wang C., Pestell R.G., Quong J.N., Quong A.A. Cyclin D1/Cyclin-Dependent Kinase 4 Interacts with Filamin A and Affects the Migration and Invasion Potential of Breast Cancer Cells. Cancer Res. 2010;70:2105–2114. doi: 10.1158/0008-5472.CAN-08-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ravazoula P., Batistatou A., Aletra C., Ladopoulos J., Kourounis G., Tzigounis B. Immunohistochemical expression of glucose transporter Glut1 and cyclin D1 in breast carcinomas with negatine lymph nodes. Eur. J. Gynaecol. Oncol. 2003;24:544–546. [PubMed] [Google Scholar]
- 139.Ates C., Sevinçli S., Pohl T. Electromagnetically induced transparency in strongly interacting Rydberg gases. Phys. Rev. A. 2011;83:6380–6385. doi: 10.1103/PhysRevA.83.041802. [DOI] [Google Scholar]
- 140.Leng X., Connell-Crowley L., Goodrich D., Harper J. S-Phase entry upon ectopic expression of G1 cyclin-dependent kinases in the absence of retinoblastoma protein phosphorylation. Curr. Biol. 1997;7:709–712. doi: 10.1016/S0960-9822(06)00301-0. [DOI] [PubMed] [Google Scholar]
- 141.Hwang H.C., Clurman E.B. Cyclin E in normal and neoplastic cell cycles. Oncogene. 2005;24:2776–2786. doi: 10.1038/sj.onc.1208613. [DOI] [PubMed] [Google Scholar]
- 142.Keyomarsi K., Tucker S.L., Buchholz T.A., Callister M., Ding Y., Hortobagyi G.N., Bedrosian I., Knickerbocker C., Toyofuku W., Lowe M., et al. Cyclin E and Survival in Patients with Breast Cancer. N. Engl. J. Med. 2002;347:1566–1575. doi: 10.1056/NEJMoa021153. [DOI] [PubMed] [Google Scholar]
- 143.Dorée M., Galas S. The cyclin-dependent protein kinases and the control of cell division. FASEB J. 1994;8:1114–1121. doi: 10.1096/fasebj.8.14.7958616. [DOI] [PubMed] [Google Scholar]
- 144.Singhai R., Patil A.V., Patil V.W., Gurav P.D. Triple-negative (ER, PgR, HER-2/neu) breast cancer in Indian women. Breast Cancer Targets Ther. 2011;3:9–19. doi: 10.2147/BCTT.S17094. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 145.Kaldis P., Aleem E. Cell Cycle Sibling Rivalry: Cdc2 Versus Cdk2. Cell Cycle. 2005;4:1491–1494. doi: 10.4161/cc.4.11.2124. [DOI] [PubMed] [Google Scholar]
- 146.Hakem A., Sasaki T., Kozieradzki I., Penninger J.M. The Cyclin-dependent Kinase Cdk2 Regulates Thymocyte Apoptosis. J. Exp. Med. 1999;189:957–968. doi: 10.1084/jem.189.6.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Carroll J.S., Prall O.W.J., Musgrove E.A., Sutherland R.L. A Pure Estrogen Antagonist Inhibits Cyclin E-Cdk2 Activity in MCF-7 Breast Cancer Cells and Induces Accumulation of p130-E2F4 Complexes Characteristic of Quiescence. J. Biol. Chem. 2000;275:38221–38229. doi: 10.1074/jbc.M004424200. [DOI] [PubMed] [Google Scholar]
- 148.Li J., Poi M.J., Tsai M.-D. Regulatory Mechanisms of Tumor Suppressor P16INK4Aand Their Relevance to Cancer. Biochemistry. 2011;50:5566–5582. doi: 10.1021/bi200642e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Baker S.J., Reddy E.P. CDK4: A Key Player in the Cell Cycle, Development, and Cancer. Genes Cancer. 2012;3:658–669. doi: 10.1177/1947601913478972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Geradts J., Wilson P. High frequency of aberrant p16(INK4A) expression in human breast cancer. Am. J. Pathol. 1996;149:15–20. [PMC free article] [PubMed] [Google Scholar]
- 151.Gartel A.L., Serfas M.S., Tyner A.L. p21--Negative Regulator of the Cell Cycle. Exp. Biol. Med. 1996;213:138–149. doi: 10.3181/00379727-213-44046. [DOI] [PubMed] [Google Scholar]
- 152.Dai M., Al-Odaini A.A., Arakelian A., Rabbani A.S., Ali S., Lebrun J.-J. A novel function for p21Cip1 and acetyltransferase p/CAF as critical transcriptional regulators of TGFβ-mediated breast cancer cell migration and invasion. Breast Cancer Res. 2012;14:R127. doi: 10.1186/bcr3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Winters Z., Hunt N., Bradburn M., Royds J., Turley H., Harris A., Norbury C. Subcellular localisation of cyclin B, Cdc2 and p21WAF1/CIP1 in breast cancer: Association with prognosis. Eur. J. Cancer. 2001;37:2405–2412. doi: 10.1016/S0959-8049(01)00327-6. [DOI] [PubMed] [Google Scholar]
- 154.Li Y., Dowbenko D., Lasky L.A. AKT/PKB Phosphorylation of p21Cip/WAF1 Enhances Protein Stability of p21Cip/WAF1 and Promotes Cell Survival. J. Biol. Chem. 2002;277:11352–11361. doi: 10.1074/jbc.M109062200. [DOI] [PubMed] [Google Scholar]
- 155.Rossig L., Jadidi A.S., Urbich C., Badorff C., Zeiher A.M., Dimmeler S. Akt-Dependent Phosphorylation of p21Cip1 Regulates PCNA Binding and Proliferation of Endothelial Cells. Mol. Cell. Biol. 2001;21:5644–5657. doi: 10.1128/MCB.21.16.5644-5657.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wang L., Wang R., Ye Z., Wang Y., Li X., Chen W., Zhang M., Cai C. PVT1 affects EMT and cell proliferation and migration via regulating p21 in triple-negative breast cancer cells cultured with mature adipogenic medium. Acta Biochim. Biophys. Sin. 2018;50:1211–1218. doi: 10.1093/abbs/gmy129. [DOI] [PubMed] [Google Scholar]
- 157.Qian X., Hulit J., Suyama K., Eugenin A.E., Belbin T.J., Loudig O., Smirnova T., Zhou Z.N., Segall J., Locker J., et al. p21CIP1 mediates reciprocal switching between proliferation and invasion during metastasis. Oncogene. 2012;32:2292–2303. doi: 10.1038/onc.2012.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Pérez-Tenorio G., Berglund F., Merca A.E., Nordenskjöld B., Rutqvist L.E., Skoog L., Stål O. Cytoplasmic p21WAF1/CIP1 correlates with Akt activation and poor response to tamoxifen in breast cancer. Int. J. Oncol. 2006;28:1031–1042. doi: 10.3892/ijo.28.5.1031. [DOI] [PubMed] [Google Scholar]
- 159.Vincent A.J., Ren S., Harris L.G., Devine D.J., Samant R.S., Fodstad O., Shevde L.A. Cytoplasmic translocation of p21 mediates NUPR1-induced chemoresistance. FEBS Lett. 2012;586:3429–3434. doi: 10.1016/j.febslet.2012.07.063. [DOI] [PubMed] [Google Scholar]
- 160.Sohn D., Essmann F., Schulze-Osthoff K., Jänicke R.U. p21 Blocks Irradiation-Induced Apoptosis Downstream of Mitochondria by Inhibition of Cyclin-Dependent Kinase–Mediated Caspase-9 Activation. Cancer Res. 2006;66:11254–11262. doi: 10.1158/0008-5472.CAN-06-1569. [DOI] [PubMed] [Google Scholar]
- 161.Neganova I., Lako M. G1 to S phase cell cycle transition in somatic and embryonic stem cells. J. Anat. 2008;213:30–44. doi: 10.1111/j.1469-7580.2008.00931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Fredersdorf S., Burns J., Milne A.M., Packham G., Fallis L., Gillett C.E., Royds J.A., Peston D., Hall P.A., Hanby A.M., et al. High level expression of p27kip1 and cyclin D1 in some human breast cancer cells: Inverse correlation between the expression of p27kip1 and degree of malignancy in human breast and colorectal cancers. Proc. Natl. Acad. Sci. USA. 1997;94:6380–6385. doi: 10.1073/pnas.94.12.6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Guan X., Wang Y., Xie R., Chen L., Bai J., Lu J., Kuo M.T. p27Kip1 as a prognostic factor in breast cancer: A systematic review and meta-analysis. J. Cell. Mol. Med. 2009;14:944–953. doi: 10.1111/j.1582-4934.2009.00730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Filipits M., Rudas M., Heinzl H., Jakesz R., Kubista E., Lax S., Schippinger W., Dietze O., Greil R., Stiglbauer W., et al. Low p27 Expression Predicts Early Relapse and Death in Postmenopausal Hormone Receptor–Positive Breast Cancer Patients Receiving Adjuvant Tamoxifen Therapy. Clin. Cancer Res. 2009;15:5888–5894. doi: 10.1158/1078-0432.CCR-09-0728. [DOI] [PubMed] [Google Scholar]
- 165.Zilfou J.T., Lowe S.W. Tumor Suppressive Functions of p53. Cold Spring Harb. Perspect. Biol. 2009;1:a001883. doi: 10.1101/cshperspect.a001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 167.Mathur P., Sathishkumar K., Chaturvedi M., Das P., Sudarshan K.L., Santhappan S., Nallasamy V., John A., Narasimhan S., Roselind F.S., et al. Cancer Statistics, 2020: Report From National Cancer Registry Programme, India. JCO Glob. Oncol. 2020;6:1063–1075. doi: 10.1200/GO.20.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chen F., Li Y., Qin N., Wang F., Du J., Wang C., Du F., Jiang T., Jiang Y., Dai J., et al. RNA-seq analysis identified hormone-related genes associated with prognosis of triple negative breast cancer. J. Biomed. Res. 2020;34:129–138. doi: 10.7555/JBR.34.20190111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yu Z., Wang Z., Yu X., Zhang Z. RNA-Seq-Based Breast Cancer Subtypes Classification Using Machine Learning Approaches. Comput. Intell. Neurosci. 2020;2020:1–13. doi: 10.1155/2020/4737969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Pennock N.D., Jindal S., Horton W., Sun D., Narasimhan J., Carbone L., Fei S.S., Searles R., Harrington C.A., Burchard J., et al. RNA-seq from archival FFPE breast cancer samples: Molecular pathway fidelity and novel discovery. BMC Med Genom. 2019;12:1–18. doi: 10.1186/s12920-019-0643-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhou S., Huang Y.-E., Liu H., Zhou X., Yuan M., Hou F., Wang L., Jiang W. Single-cell RNA-seq dissects the intratumoral heterogeneity of triple-negative breast cancer based on gene regulatory networks. Mol. Ther.-Nucleic Acids. 2021;23:682–690. doi: 10.1016/j.omtn.2020.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Karaayvaz M., Cristea S., Gillespie S.M., Patel A.P., Mylvaganam R., Luo C.C., Specht M.C., Bernstein B.E., Michor F., Ellisen L.W. Unravelling subclonal heterogeneity and aggressive disease states in TNBC through single-cell RNA-seq. Nat. Commun. 2018;9:1–10. doi: 10.1038/s41467-018-06052-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.