Abstract
Long noncoding RNAs (lncRNA) have been associated with various types of cancer; however, the precise role of many lncRNAs in tumorigenesis remains elusive. Here we demonstrate that the cytosolic lncRNA P53RRA is downregulated in cancers and functions as a tumor suppressor by inhibiting cancer progression. Chromatin remodeling proteins LSH and Cfp1 silenced or increased P53RRA expression, respectively. P53RRA bound Ras GTPase-activating protein-binding protein 1 (G3BP1) using nucleotides 1 and 871 of P53RRA and the RRM interaction domain of G3BP1 (aa 177–466). The cytosolic P53RRA–G3BP1 interaction displaced p53 from a G3BP1 complex, resulting in greater p53 retention in the nucleus, which led to cell-cycle arrest, apoptosis, and ferroptosis. P53RRA promoted ferroptosis and apoptosis by affecting transcription of several metabolic genes. Low P53RRA expression significantly correlated with poor survival in patients with breast and lung cancers harboring wild-type p53. These data show that lncRNAs can directly interact with the functional domain of signaling proteins in the cytoplasm, thus regulating p53 modulators to suppress cancer progression.
Introduction
Long noncoding RNAs (lncRNA) have attracted significant attention because of their emerging role in cancer (1, 2). Tens of thousands of lncRNAs may be encoded in the human genome (3, 4), but the precise role of many of them remains elusive. LncRNAs display enormous variations in expression level and show diversity in subcellular localization (4). Nuclear lncRNAs are involved in transcriptional regulation in cis and in trans, modulation of chromosomal interactions, transcription factor trapping, chromatin looping, gene methylation, recruitment of transcription factor, and chromatin modification (5, 6). Cytoplasmic lncRNAs can influence activity or abundance of interacting proteins, mRNAs, or micro-RNAs (2, 6–8). Nevertheless, research into the functions of lncRNAs, especially those that reside in the cytoplasm, is still at an early stage.
The number of p53-regulated lncRNAs is growing rapidly, pointing to involvement of lncRNAs in p53 signaling (9–11). It has been demonstrated that some lncRNAs play an extensive role in modulating tumor-suppressor pathways has been demonstrated for some lncRNAs (12). The maternally imprinted RNA MEG3 has been found to bind to p53 and activate transcription of a subset of p53-regulated genes (13). The DNA-damage-inducible lncRNA PANDA controls a p53-dependent pathway, by binding to the transcription factor NF-YA and blocking its recruitment to pro-apoptotic genes (14). Although downstream activities of the p53 pathway have been explored, the potential role of lncRNAs in the regulation of p53 activity has not been extensively studied.
Ras-GTPase-activating protein-binding protein 1 (G3BP1) modulates the transduction of signaling stimulated by the oncoprotein Ras (15). The G3BP family of proteins participate in several signaling pathways involved in carcinogenesis, including p53 and Ras signaling as well as the ubiquitin proteasome system (16, 17). Furthermore, G3BP1 is essential for the normal interaction between stress granules and processing bodies and can preserve polyadenylated mRNA during carcinogenesis (18, 19). However, the precise role and mechanism of action of G3BP1 in tumorigenesis is not understood. Here, we report a lncRNA-dependent mechanism that regulates a highly dose-dependent family of RNA binding proteins in the cytoplasm, uncovering a transcriptional regulatory axis that promotes cell death in mammalian cells.
Materials and Methods
Cell culture, chemicals, plasmids, and siRNAs
The normal lung cell lines, HBE (ATCC:CRL-2741) and MRC-5 (ATCC: CCL-171) and the lung cancer cell lines A549 (ATCC: CCL-185), H358 (ATCC: CRL-5807), and H522 (ATCC: CRL-5810) were obtained from the ATCC. The lung cancer cell lines SPCA-1, PC9, 95C, and 95D were obtained from the Cancer Research Institute of Central South University. A549 cells were cultured in DME/F12 1:1(HyClone), 293T cells were cultured in DMEM (Gibco), and the other cell lines were cultured in RPMI1640 (Gibco). All medium was supplemented with 10% (v/v) FBS. All cell lines were maintained at 37C with 5% CO2. All cell lines tested negative for mycoplasma contamination and were passaged less than 10 times after the initial revival from frozen stocks. All cell lines were authenticated prior to use by short tandem repeat profiling.
The chemicals erastin and ferrostatin-1 were purchased from Selleck. The P53RRA (linc00472) lentiviral overexpression construct was built by cloning LINC00472 cDNA that was kindly provided by Hebert Yu (20) into the pCDH-EF1-MCS-BGH-PGK-GFP-T2A-Puro vector (SBI; catalog no. CD550A-1). The FLAG-G3BP1 truncation overexpression constructs were built by the truncated G3BP1 cDNAs into a pLVX-EF1α-IRES-Puro vector (catalog no. 631988; Clontech). Primers used are listed into Supplementary Table S1. The F380L/F382L RNA recognition motif (RRM) mutant of G3BP1 was introduced into the G3BP1 gene using the QuikChange II Site-Directed Mutagenesis Kit (Agilent). The GFP-p53 overexpression constructs were built by cloning p53 cDNA into a pLJM1-EGFP vector (catalog no. 19319; Addgene). Wild-type p53 and mutant p53 plasmids were provided by Drs. Mian Wu (University of Science & Technology of China, Hefei, China) and Yu Zhang (Central South University, Changsha, China). The lentiviral shRNA clones targeting human P53RRA (#1 ATTTCATCTTCATTGGATA; #2 AGATCATTAGCTTCTTAAA), G3BP1 (#1 CATTAACAGTGGTGGGAAA; #2 AGGCTTTGAGGAGATTCAT), p53 (#1 ACTCCAGTGGTAATCTACT; #2 GTCCAGATGAAGCTCCCAG), and the nontargeting control construct were purchased from Genechem (www.genechem.com.cn).
Bisulfite sequencing
Genomic DNA (1 μg) was subjected to bisulfite treatment using the MethylDetector Bisulphite Modification Kit (Active Motif). The amplified fragments were subcloned into the pCR2.1-TOPO vector (Invitrogen). The primers used were as follows: F: GGGGAAAAGTTTGGATTGTTT and R: CTTAACTTAATCTAAACTCCAAAAT. Independent clones were sequenced using the M13 F primer, and only sequences with individual fingerprints were selected for analysis.
RNA pull-down and mass spectrometry analysis
RNA pull-down assays were performed using a Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo Scientific). Fifty pmol of biotin-labeled full-length P53RRA RNA or the antisense P53RRA fragment was heated to 60°C for 10 minutes, slow cooled to 4°C, and then incubated with streptavidin beads at 37°C for 2 hours. The RNA was mixed with 200 μg of cellular protein extracts from lung adenocarcinoma or 293T cells. Proteins bound to P53RRA or antisense P53RRA were subjected to mass spectrometry analysis on a MALDI–TOF instrument (Bruker Daltonics) or examined by Western blotting. The in vitro transcription of P53RRA and its deletion fragments was assessed with primers containing the T7 promoter sequence (Supplementary Table S2) using the TranscriptAid T7 High Yield Transcription Kit (Thermo Scientific).
RNA immunoprecipitation assays
A total of 107 cells were harvested by trypsinization and resuspended in 2 mL of PBS. The cell lysate was pelleted by centrifugation at 4°C and 500 × g for 15 minutes. The cell lysate was resuspended in 1 mL of RIP buffer, split into three fractions (for Input, Mock, and IP), and then centrifuged at 4°C and 13,000 rpm for 10 minutes. Antibodies against normal mouse IgG (Merck Millipore, catalog no. 12–371), normal rabbit IgG (Cell Signaling Technology, catalog no. 2729), G3BP1 (Santa Cruz Biotechnology, catalog no.sc-365338), andG3BP2 (Abcam, catalog no. ab86135) as well as Anti-FLAG M2 Magnetic Beads (Sigma Aldrich, catalog no. M8823) or GFP (Santa Cruz Biotechnology, catalog no. sc-9996) were added to the supernatant and incubated overnight at 4°C with gentle rotation. Next, 40 μL of protein A/G beads were added and the mixture was incubated at 4°C for an additional hour. The beads were pelleted at 2,500 rpm for 30 seconds, washed three times with 500 μL of RIP buffer and one time with PBS, and then resuspended in 1 mL of RNAiso Plus. The total RNA (input control) and RNA precipitated with the isotype control (IgG) for each antibody were assayed simultaneously with all test samples. The coprecipitated RNAs were detected by qRT-PCR for P53RRA (forward, GGTGACTTTCTCGACTCGTCGT and reverse, GATGATGCCAACATGTCTGGTGC).
IHC analysis and lung cancer biopsies
IHC analyses were essentially performed as previously described (21–24) and are also described in the Supplementary Materials and Methods section. Lung cancer biopsies, validated by pathologist Dr. Desheng Xiao (Xiangya Hospital, Central South University, Hunan, China), were obtained from the Department of Pathology at Xiangya Hospital. Written informed consent was obtained from all patients or their relatives, and the institutional ethical committee of our hospital approved the study in accordance with the ethical guidelines from the Declaration of Helsinki. The lung cancer tissue array was purchased from Pantomics.
Quantitative real-time PCR and chromatin immunoprecipitation
The details of these procedures have been described previously (21,22,25). Primers are listed in Supplementary Tables S3 and S4. The antibodies used are as follows: H3K4me3 (Active Motif, catalog no. 39915), Histone H3K27me3 (Active Motif, catalog no. 39155), LSH (Santa Cruz Biotechnology, catalog no. sc-46665), and Cfp1 (Santa Cruz Biotechnology, catalog no. 25391).
Nude mice and study approval
The xenograft tumor formation assay was essentially performed as previously described (22). All procedures for animal study were approved by the Institutional Animal Care and Use Committee of the Central South University of Xiangya School of Medicine and confirm to the legal mandates and federal guidelines for the care and maintenance of laboratory animals. SCID mice (Hunan SJA Laboratory Animal Co. Ltd.) were injected with the indicated cells in the mammary fat pad (10 mice/group). Injected mice were imaged from both the dorsal and ventral sides every three days. Data were analyzed using Student t test; a P value < 0.05 was considered significant. Additional methods can be found in the supplemental section.
Results
The lncRNA P53RRA is a cytosolic lncRNA that is silenced in multiple cancers
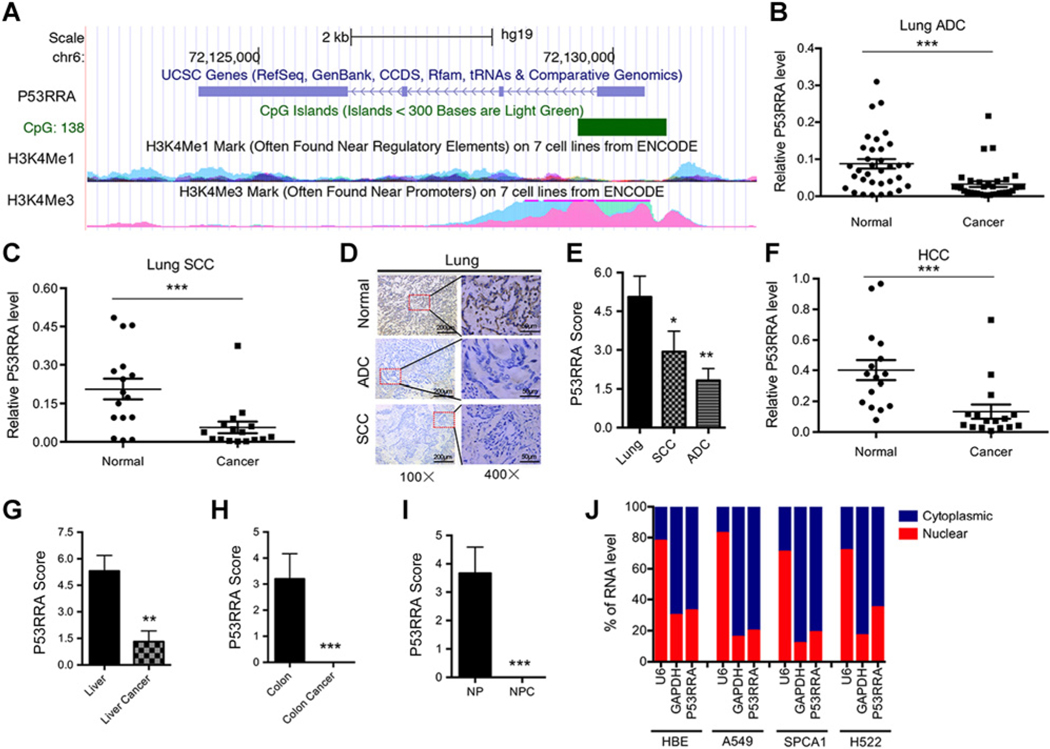
To investigate the role of lncRNAs in lung cancer, we first examined lncRNA expression profiles in two non–small cell lung cancer tissue samples and adjacent normal tissues using the Arraystar LncRNA Microarray (V2.0). Thirteen lncRNAs were upregulated and 18 were downregulated by more than five-fold, including p53-related lncRNA (P53RRA; Supplementary Table S5). Using 5′- and 3′-rapid amplification of the cDNA ends, P53RRA was found to be 2,933-nucleotide (nt) long. The gene is identical to the sequence C6orf155 in GeneBank and uc003pfz.1 in the UCSC database (hg19 chr6: 72,124,149–72,130,448) and shows enrichment of H3K4me3-modified histones at the transcription start site and H3K4me1-modified histones at the promoter region (Fig. 1A).
Figure 1.
P53RRA is a cytosolic lncRNA that is downregulated in multiple cancers. A, A schematic representation of P53RRA (annotated in RefSeq as LINC00472) with associated UCSC Genome Browser tracks as well as H3K4Me1 and H3K4me3 ChIP-seq coverage. B and C, qRT-PCR showed the decreased expression of P53RRA in 47 paired lung cancer and corresponding normal lung tissue samples (B, ADC; C, SCC). D and E, In situ hybridization (D) and the relative scores (E) used to analyze the decrease of P53RRA expression in lung cancer. F, qRT-PCR revealed that the expression of P53RRA was decreased in hepatocellular carcinoma (HCC). G–I, In situ hybridization showed that the relative expression of P53RRA was reduced in liver cancer (G), colon cancer (H), and nasopharyngeal carcinoma (NPC; I). J, The P53RRA expression levels in the nuclear and cytosolic fractions derived from HBE, A549, SPCA1, and H522 lung cancer cells. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Using an independent panel of 47 primary lung tumors and normal lung tissues, we found that P53RRA was downregulated in both lung adenoma (ADC) and squamous carcinoma (SCC; Fig. 1B and C). In situ hybridization showed that P53RRA was located primarily in the cytoplasm in normal lung tissues and the level of P53RRA was significantly decreased in both lung ADC and SCC (Fig. 1D and E). The level of P53RRA was also decreased in liver cancers compared with normal liver tissues (Fig. 1F). In situ hybridization analysis also indicated that P53RRA localized to the cytoplasm of normal liver cells and that its level was decreased in liver cancer (Fig. 1G; Supplementary Fig. S1A). Similar findings were obtained in colon cancer and nasopharyngeal carcinoma (Fig. 1H and I; Supplementary Fig. S1B and S1C). Quantitative RT-PCR (qRT-PCR) detected significantly higher levels of P53RRA expression in HBE and H1299 cells, and reduced levels in A549 and SPCA1 cells (Supplementary Fig. S1D). Furthermore, the localization of P53RRA in the cytoplasm was confirmed by the assessment of the nuclear and cytoplasmic fractions in HBE, A549, SPCA1, and H522 cells using RT-PCR (Fig. 1J). Our data suggest that P53RRA might exert its biological function in the cytoplasm and that its expression is suppressed in human cancers.
Finally, nine GEO datasets representing different cancer types were used to compare expression levels between normal breast tissues and tumors. Higher levels of P53RRA were detected in normal tissues compared with tumor tissues (Supplementary Fig. S1E), and higher P53RRA expression was associated with low grade and less aggressive disease (Supplementary Fig. S1F). Overall, these results were in agreement with our own findings.
Epigenetic silencing of P53RRA by DNA methylation is mediated by LSH, whereas Cfp1 is involved in the activation of P53RRA
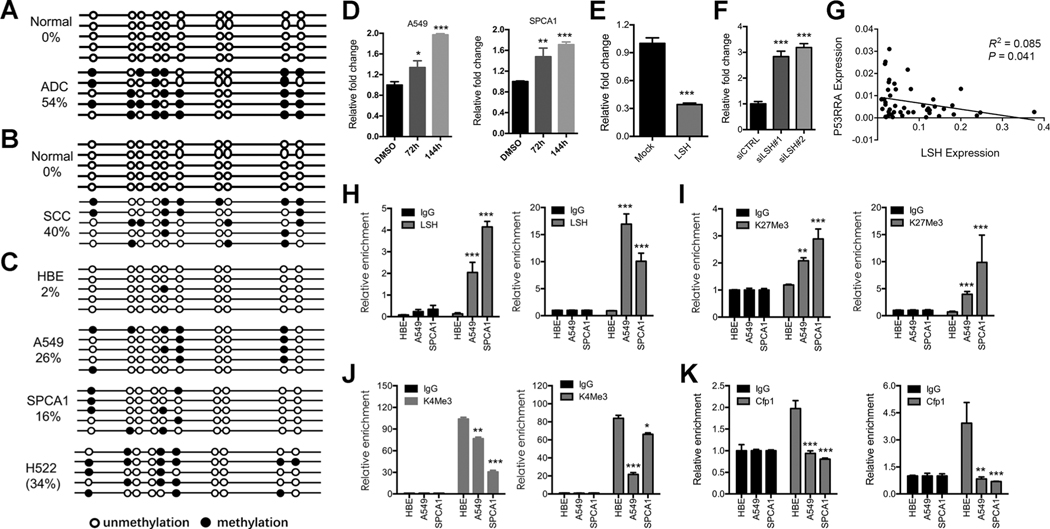
Abnormal DNA methylation has been found in cancer cells, especially at silenced tumor suppressor genes (26, 27). We detected increased level of DNA methylation around the transcription start site of the P53RRA gene in lung cancer tissues compared with control lung tissues (Fig. 2A and B; Supplementary Fig. S2A). This increase was found in A549, SPCA1, and H522, but not in HBE cells (Fig. 2C). After treatment of A549 and SPCA1 cells with the DNA demethylating drug azacytidine, P53RRA expression increased, indicating that DNA methylation represses P53RRA expression (Fig. 2D).
Figure 2.
DNA methylation is involved in the epigenetic silencing of P53RRA. A–C, Genomic DNA derived from human lung ADC (A) and SCC tissues (B), corresponding normal lung tissues, and HBE, A549, SPCA1, and H522 cells (C) was examined by bisulfite sequencing of the P53RRA promoter regions. D, Real-time RT-PCR analyses detecting the P53RRA levels in lung cancer cells after the treatment with 5-azacytidine for 72 hours. E and F, The P53RRA levels were detected in xenografts derived from H522 cells (E) and A549 cells either stably expressing LSH or with LSH depleted (F). G, The correlation between LSH and P53RRA in lung cancer tissues was analyzed. H, ChIP analysis was used to detect the presence of LSH in the promoter region of P53RRA using two primer sites. I and J, ChIP analysis for the detection of the presence H3K27Me3 (I) and H3K4Me3 (J) at the transcription start site of P53RRA using two separate primer sites. K, ChIP analysis for the detection of Cfp1 binding to the promoter region of P53RRA using two primer sites. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Because LSH is crucial for transcriptional repression at DNA methylated genes and plays an important role in cancer progression (23, 25, 28–31), we hypothesized that LSH is involved in the regulation of P53RRA transcription. Overexpression of LSH further decreased P53RRA levels by increasing DNA methylation (Fig. 2E), which specifically at the P53RRA promoter region (Supplementary Fig. S2B and S2C). In contrast, depletion of LSH increased P53RRA expression levels, and DNA methylation in the P5RRA promoter region decreased (Fig. 2F; Supplementary Fig. S2D and S2E), suggesting that LSH represses P53RRA. Furthermore, the expression of LSH and P53RRA were inversely correlated in lung cancers (Fig. 2G). Chromatin immunoprecipitation (ChIP) assays revealed that LSH is bound to the P53RRA promoter in A549 and SPCA1 lung cancer cells, but not in HBE cells, suggesting a direct role for LSH (Fig. 2H).
A high percentage of DNA-methylated cancer genes are pre-marked with H3K27me3 modifications, whereas hypo-methylated genes show H3K4me3 modifications (28, 32). We found that H3K27me3 was significantly increased in both A549 and SPCA1 cells compared with HBE cells (Fig. 2I), whereas H3K4me3 was significantly reduced in both A549 and SPCA1 cells compared with HBE cells (Fig. 2J). The conserved subunit CxxC finger protein 1 (Cfp1) binds to unmethylated CpGs and is a component of the Set1 complex, which mediates H3K4me3 modifications (33, 34). The binding of Cfp1 was reduced in A549 and SPCA1 cells compared with HBE cells, consistent with the potential role of Cfp1 in depositing H3K4me3 modifications (Fig. 2K). Thus, DNA hypermethylation is associated with the silencing of P53RRA and a dynamic epigenetic switch accompanied by DNA hypo-demethylation is linked to increased transcript levels.
Overexpression of Cfp1 in A549 cells increased the level of P53RRA RNA (Supplementary Fig. S3A–S3E), whereas the depletion of Cfp1 in SPCA1 cells decreased P53RRA RNA levels independent of changes in DNA methylation (Supplementary Fig. S3F–S3I), indicating that Cfp1 is a key activator of P53RRA expression.
P53RRA functions as a tumor suppressor in vitro and in vivo
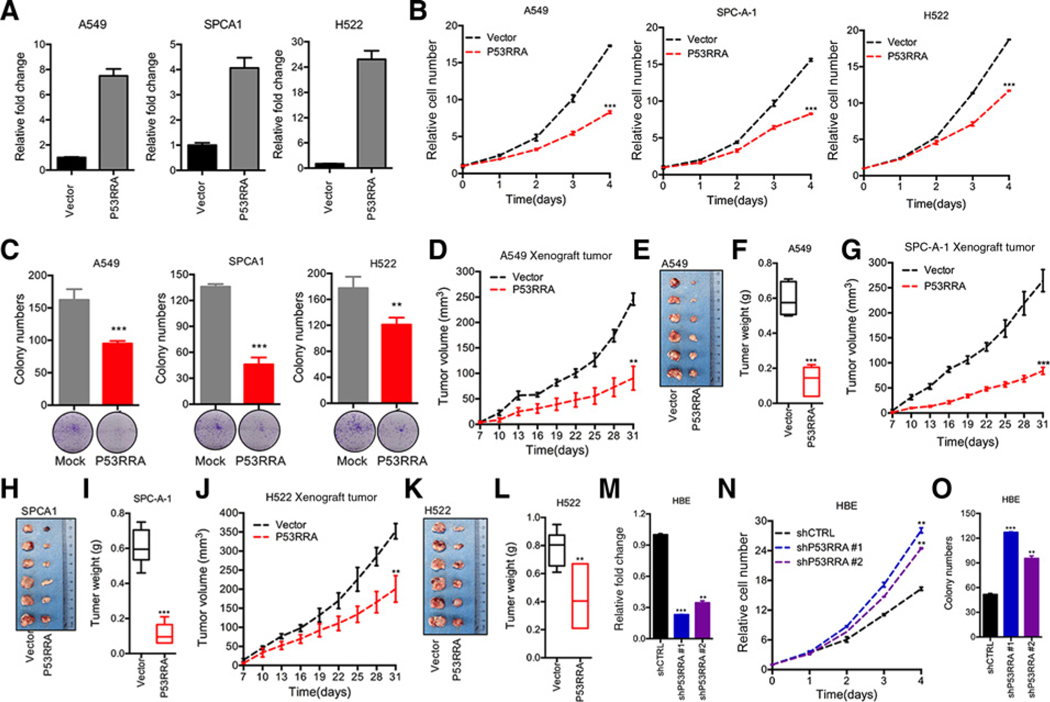
To uncover the physiological role of P53RRA in cancer, we stably overexpressed P53RRA in A549, SPCA1, and H522 lung cancer cell lines (Fig. 3A). P53RRA overexpression significantly decreased the growth of all cell lines in vitro (Fig. 3B). The stable expression of P53RRA in A549, SPCA1, and H522 cell lines significantly reduced colony formation (Fig. 3C).
Figure 3.
P53RRA functions as a tumor suppressor. A, qRT-PCR analysis was conducted to detect P53RRA levels in A549, SPCA1, and H359 cells that were stably transfected with the P53RRA expression plasmid. B, P53RRA inhibited cell growth in A549, SPCA1, and H522 cells, as demonstrated by MTT assay. C, P53RRA inhibited colony formation in cells as indicated. D–L, Nude mice are shown after injection of A549, SPCA1, and H522 cells stably expressing the control vector or P53RRA expression plasmids. Tumor formation (D, G, and J) was monitored at the indicated time points, tumor formation (E, H, and K) was shown, and tumor weight (F, I, and L) was recorded (n = 6). M, qRT-PCR analysis was conducted to detect the P53RRA levels in HBE cells that were stably transfected with two distinct target gene shRNA expression vectors or a control vector (shCTRL). N and O, Depletion of P53RRA in HBE cells promoted cell proliferation (N) and colony formation (O). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To address whether P53RRA can also play a role in cancer in vivo, we used a xenograft model. We found that that A549-P53RRA cells produced significantly smaller tumors after 1 month of growth compared with A549 cells (Fig. 3D–F). The injection of SPCA1-P53RRA and H522-P53RRA cells (2 × 106) demonstrated that P53RRA overexpression significantly decreased both tumor size and tumor weight (Fig. 3G–L).
To substantiate our findings, we stably knocked down P53RRA in HBE cells. The knockdown approach successfully reduced P53RRA mRNA using two separate sequences (Fig. 3M). The depletion of P53RRA significantly increased the growth and colony formation of HBE cells in vitro (Fig. 3N and O). Thus, our data suggest a role of P53RRA as a tumor suppressor.
P53RRA represses metabolic-related genes at the transcriptional level
To gain insight into the function of P53RRA, we used RNA sequencing following the stable expression of P53RRA in H522 cells. Using two independent biological replicates, we obtained an average of 8.4 million unique mapped reads and 17,442 unique transcripts [with fragments per kilobase of exon per million reads (FPKM) >1 in both replicates] per condition. We identified 921 mRNAs with two-fold greater counts (upregulated) and 738 mRNAs with two-fold fewer counts (down-regulated) in cells stably expressing P53RRA compared with cells expressing the control vector (Supplementary Fig. S4A). Further, a volcano plot further showed 227 upregulated and 126 downregulated genes (Supplementary Fig. S4B). Gene ontology analysis identified that these genes were significantly enriched for pathways related to metabolic processes, cell components and molecular function, cell cycle, and cell death (Supplementary Fig. S4C–S4E).
Given that P53RRA is potentially linked to metabolic and cell death genes, we validated the expression of downstream target genes. Overexpression of P53RRA significantly increased the mRNA expression of seven genes and decreased the expression level of 14 genes in A549, SPCA1, and H522 cells (Supplementary Fig. S5A–S5C) in agreement with our RNA sequencing data. Moreover, depletion of P53RRA in HBE cells inversely changed the mRNA levels of metabolic genes that were regulated by P53RRA (Supplementary Fig. S5D).
We further explored the correlation between P53RRA levels and the expression of nine selected genes downstream target genes in 56 clinical lung cancer samples. We found a positive correlation between P53RRA and RIMBP3C, TIGAR, GPR162, and TIAI, and a negative correlations between P53RRA and SLC1A5, SLC7A11, HNRNPC, and BATF3 in normal, tumor-adjacent lung tissues and in lung cancer tissues (Supplementary Fig. S6A). A Kaplan–Meier analysis of a cohort of these patients with lung cancer showed that lower expression of the metabolic genes SLC7A11, CS, SLC1A5, SLC2A4, and SLC2A14 and higher expression of TIA1 was linked to overall survival in all lung cancers (Supplementary Fig. S6B).
P53RRA promotes cell-cycle arrest, apoptosis, and ferroptosis
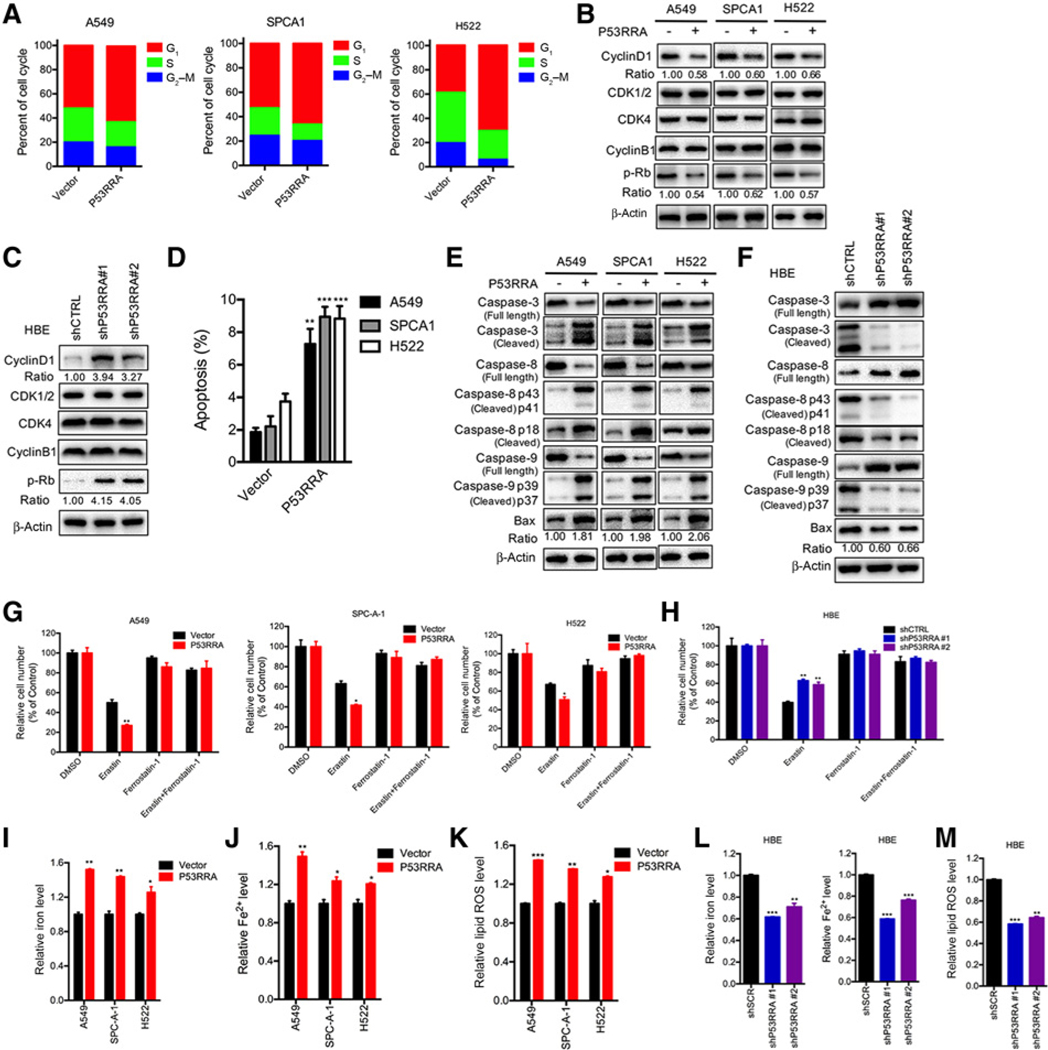
Because GO analysis indicated that P53RRA affects cell cycle and apoptosis progression, we next tested whether P53RRA plays a role in this biological process. Overexpression of P53RRA reproducibly resulted in cell-cycle redistribution with a significant increase in the number of cells in the G1 phase and a significant decrease in other stages of the cell cycle in synchronized A549, SPCA1, and H522 cells (Fig. 4A; Supplementary Fig. S7A–S7C). Depletion of P53RRA promoted cell-cycle progression (Supplementary Fig. S7D and S7E). In addition, depletion of P53RRA increased the levels of the cyclin D1 protein and phosphorylated Rb (Fig. 4B and C), whereas overexpression of P53RRA decreased these levels, further supporting a role for P53RRA in cell-cycle regulation.
Figure 4.
P53RRA induces cell-cycle arrest, apoptosis, and ferroptosis. A, FACS showed that P53RRA affected cell-cycle progression in A549, SPCA1, and H522 cells. B, Overexpression of P53RRA inhibited cell-cycle regulators in A549, SPCA1, and H522 cells. C, Depletion of P53RRA in HBE cells promoted regulators of cell-cycle progression. D, P53RRA promoted apoptosis in A549, SPCA1, and H522 cells, as demonstrated using FACS. E, P53RRA affected the activities of caspase-3, caspase-8, and caspase-9 in A549, SPCA1, and H522 cells stably overexpressing P53RRA. F, Depletion of P53RRA in HBE cells affected the activities of caspase-3, caspase-8, and caspase-9 in HBE cells. G and H, A549, SPCA1, and H522 (H) cells stably overexpressing P53RRA and HBE cells with depletion of P53RRA (I) in response to erastin (5 μmol/L) ± ferrostatin (1 μmol/L). J and K, Total iron levels were analyzed in the presence of P53RRA in A549, SPCA1, and H522 cells. The levels of total iron (I) and ferrous iron (J) were analyzed in the presence of P53RRA. K, The level of lipid ROS in A549, SPCA1, and H522 cells was measured by C11-BODIPY staining coupled with flow cytometry. L and M, The levels of total iron and ferrous iron (L) and lipid ROS (M) were analyzed in HBE cells after the depletion of P53RRA. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Moreover, we found that P53RRA induced a greater level of both early apoptosis and late apoptosis as measured by Annexin V and PI staining (Fig. 4D; Supplementary Fig. S7F), whereas depletion of P53RRA decreased apoptosis in HBE cells (Supplementary Fig. S7G). Expression of P53RRA increased the levels of cleaved caspase-3, cleaved caspase-9, and Bax and decreased the levels of cleaved caspase-8 in A549, SPCA1, and H522 cells (Fig. 4E), whereas depletion of P53RRA in HBE cells decreased the levels of cleaved caspase-3, cleaved caspase-9, and Bax and increased the level of cleaved caspase-8 (Fig. 4F).
To characterize the role of P53RRA in ferroptosis, a novel mode of non-apoptotic cell death, we assessed the role of several metabolic genes. We found that erastin induced growth inhibition. We observed that P53RRA increased erastin-induced growth inhibition in A549, SPCA1, and H522 cells (Fig. 4G), whereas depletion of P53RRA decreased erastin-induced growth inhibition (Fig. 4H). The intracellular concentrations of iron and lipid reactive oxygen species (ROS) are two surrogate markers for ferroptosis (35, 36). We found that P53RRA increased the intracellular concentrations of iron (Fig. 4I and J) and lipid ROS in A549, SPCA1, and H522 cells (Fig. 4K), whereas the depletion of P53RRA in HBE cells decreased the intracellular concentrations of iron and lipid ROS (Fig. 4L and M). Taken together, these findings imply that P53RRA promotes cell-cycle arrest, apoptosis, and ferroptosis.
P53RRA interacts with Ras GTPase-activating protein-binding protein 1
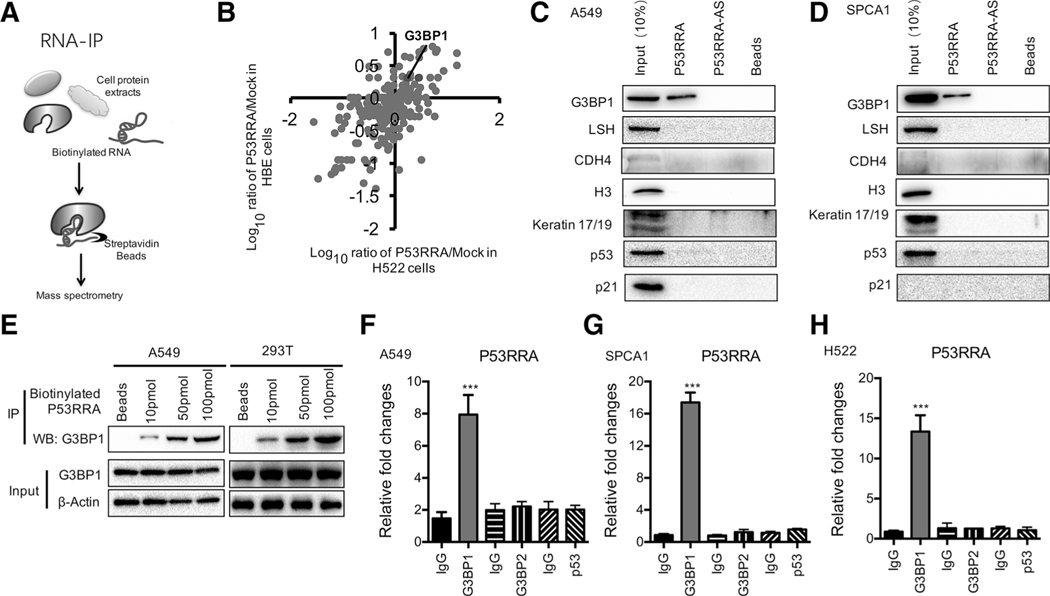
Because lncRNAs may function through their interactions with other proteins (37, 38), we hypothesized that P53RRA might interact with certain cellular proteins to regulate biological functions. Using RNA pulldown assays (Fig. 5A) and mass spectrometry analysis, we identified a P53RRA-protein complex in cell lysates generated from HBE-P53RRA and H522-P53RRA cells, antisense of P53RRA served as a negative control in these experiments. Among the many proteins identified G3BP1 and G3BP2 were most interesting (Fig. 5B; supplementary Table S6). We validated the presence of G3BP1 in an intact complex from independent RNA pulldown assays in A549 cells (Fig. 5C) and SPCA1 cells (Fig. 5D), whereas other proteins, such as LSH, CDH4, Histone H3, keratin17/19, and p53, did not show enrichment in the biotion-P53RRA pulldown. The interaction of G3BP1 and P53RRA was demonstrated to be dose-dependent using biotin-P53RRA in A549 and 293T cells (Fig. 5E). RNA immune-precipitation (RIP) assays confirmed an enrichment of P53RRA in the complexes precipitated with the antibody against G3BP1, but not G3BP2 or p53 in A549, SPCA1, and H522 cells (Fig. 5F–H). Taken together, these findings suggest that G3BP1 is a proven and novel binding partner of P53RRA.
Figure 5.
P53RRA interacts with G3BP1. A, A schematic of the RNA pull-down experiment for the identification of proteins associated with P53RRA. B, Differentially expressed genes were annotated with their average fold change in the H522-P53RRA and HBE-P53RRA groups. C and D, The relative mRNA represents the RNA levels associated with G3BP1 relative to an input control from three experiments. Immunoblot analysis of five proteins from the proteomics screen after pull-down shows the specific association of G3BP1 but not LSH, CDH4, H3, keratin 17/19, or p53 with P53RRA in A549 (C) and SPCA1 (D) cells. E, RNA pull-down assays using biotinylated P53RRA indicated that P53RRA interacts with G3BP1 in a dose-dependent manner in A549 and 293T cells. F–H, RIP assays show the association of G3BP1, but not G3BP2 or p53, with P53RRA in A549 (F), SPCA1 (G), and H522 (H) cells. ***, P < 0.001.
The P53RRA–G3BP1 interaction in the cytoplasm is critical for activation of the p53 signaling pathway
Based on the interaction of P53RRA and G3BP1, we examined whether P53RRA affects G3BP1 expression and cellular localization. However, overexpression or depletion of P53RRA did not affect G3BP1 expression or localization (Supplementary Fig. S8A–S8C). Further, overexpression of G3BP1 did not change P53RRA expression or cellular localization (Supplementary Fig. S8D–S8F). However, G3BP1 expression did promote cell growth and colony formation (Supplementary Fig. S8G and S8H). Consistent with these findings, depletion of G3BP1 in HBE cells did not affect P53RRA expression and localization (Supplementary Fig. S8I–S8K) but attenuated cell growth and colony formation (Supplementary Fig. S8L and S8M). Moreover, overexpression and knockdown experiments demonstrated that G3BP1 regulates several P53RRA target genes (Supplementary Fig. S8N and S8O), suggesting a functional role of the P53RRA–G3BP1 complex in the regulation of these metabolic genes.
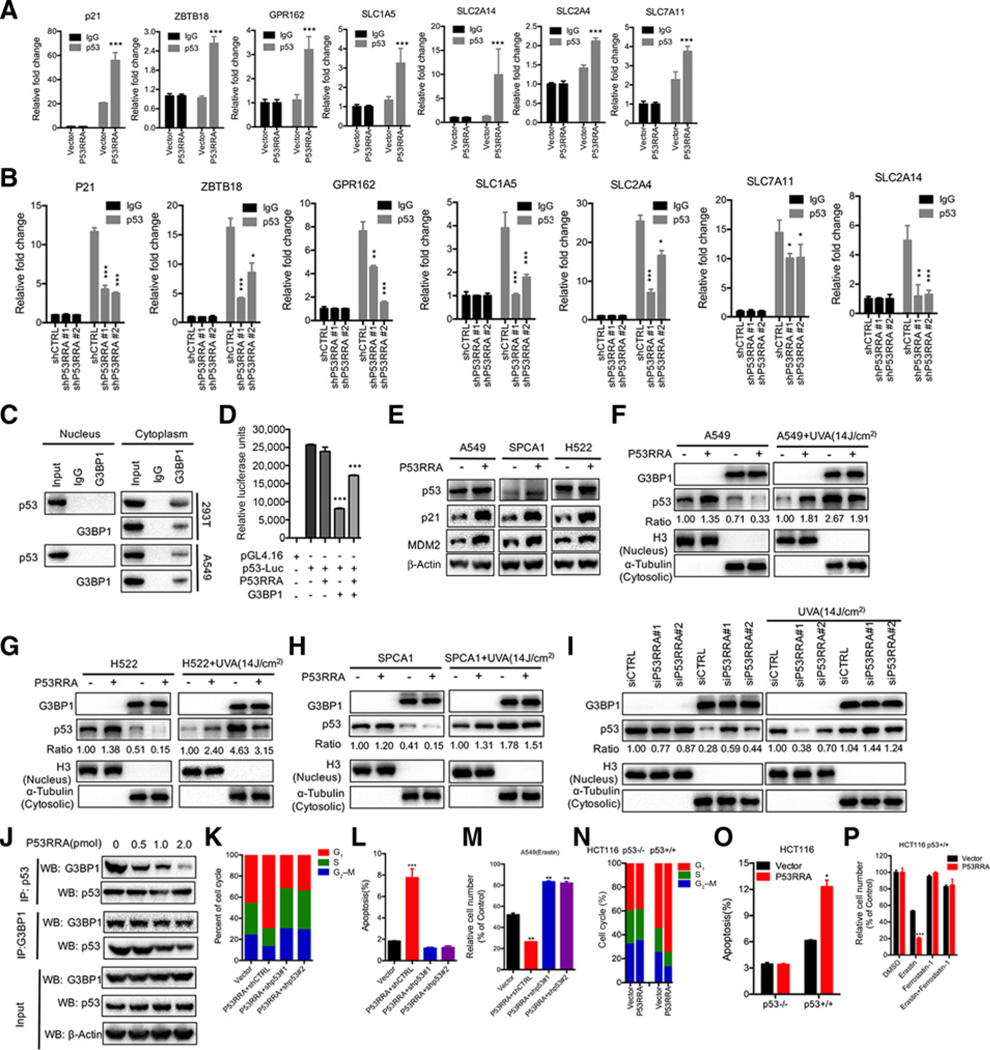
Most metabolic genes, including SLC7A11 and TIGAR, are targets of p53 (39). We first confirmed that p53 could regulate these genes using by overexpressing p53 in A549 cells and assessing the cells by RT-PCR and ChIP (Supplementary Fig. S9A and S9B). We found that G3BP1 also regulated the recruitment of p53 to these metabolic genes (Supplementary Fig. S10A and S10B). Using a ChIP assay for p53 in A549, H522, and SPCA1 cells that highly expressed P53RRA, we observed recruitment of p53 to the promoter regions in all three cell lines (Fig. 6A; Supplementary Fig. S11A and S11B). Conversely, depletion of P53RRA in HBE cells reduced the recruitment of p53 to promoters (Fig. 6B). These findings suggest a potential role for p53 in the downstream function of the P53RRA–G3BP1 interaction.
Figure 6.
The cytoplasmic P53RRA–G3BP1 interaction is critical for activation of p53 signaling. A, ChIP assays indicate that p53 is recruited to the promoters of metabolic genes after the overexpression of p53RRA in A549 cells. p21 was used as a positive control. B, ChIP assays indicate that p53 is recruited to the promoters of metabolic genes in P53RRA depleted in HBE cells. C, Coimmunoprecipitation assays showed that G3BP1 interacted with p53 in the cytoplasm in 293T and A549 cells. D, P53RRA does not affect p53 transcriptional activity, as indicated by a luciferase assay. E, Western blot analysis indicated that P53RRA promotes p53 signaling. F–H, The nuclear and cytoplasmic extracts from A549 (F), H522 (G), and SPCA1 (H) cells overexpressing P53RRA showed that p53 was sequestered in the nucleus, whereas UVA treatment induced greater accumulation of p53 in the cytoplasm. I, p53 was increased in the cytoplasm after depletion of P53RRA, whereas UVA treatment induced greater accumulation of p53 in the cytoplasm. J, Co-IP assays indicated that P53RRA decreased the interaction of G3BP1 and p53 in a dose-dependent manner in 293T cells. K–M, The effect of P53RRA on cell-cycle progression (K), apoptosis (L), and erastin-induced ferroptosis (M) in A549 cells after the depletion of p53. N–P, The effect of P53RRA on the cell-cycle progression (N), apoptosis (O), and erastin-induced ferroptosis (P) in HCT116 p53+/+ and HCT116 p53−/− cells after overexpression of P53RRA. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To address the hypothesis that p53 activity might be affected by the intact complex of P53RRA-G3BP1, we first confirmed that G3BP1 interacted with p53 in the cytoplasm in A549 and 293T cells (Fig. 6C). Overexpression of G3BP1 sequestered more p53 in the cytoplasm in both the absence and presence of UVA treatment, which itself induces the redistribution of p53 into the cytoplasm (Supplementary Fig. S12A). Depletion of G3BP1 in A549 cells attenuated p53 redistribution in the absence and presence of UVA treatment that induced the redistribution of p53 into the cytoplasm (Supplementary Fig. S12 B). To test whether P53RRA affects the transactivation activity of p53, we used a reporter gene assay. Although P53RRA expression had no further effect on p53 reporter expression, G3BP1 expression decreased p53 transactivation activity (Fig. 6D). Based on these indications, we hypothesized that the intact complex of P53RRA–G3BP1 may affect p53 protein levels. We found that overexpression of P53RRA slightly increased the p53 protein levels in A549, SPCA1, and H522 cells. Interestingly, the levels of p21 protein, a direct target of p53, increased in these cells. Similar findings were also observed for MDM2 protein levels (Fig. 6E), indicating that P53RRA promotes the classic p53 signaling pathway.
Because G3BP1 expression sequestered p53 in the cytoplasm, we hypothesized that in the presence of P53RRA less G3BP1 may complex with p53 and therefore more p53 may be available in the nucleus. Indeed, expression of P53RRA decreased the level of cytosolic p53, but not G3BP1, in A549, H522, and SPCA1 cells (left panels of Fig. 6F–H). UVA treatment induced the redistribution of p53 into the cytoplasm, whereas P53RRA enhanced the level of p53 in the nucleus in both A549 and SPCA1 cells (right panels of Fig. 6F–H). The depletion of P53RRA in HBE cells induced greater retention of p53 in the cytoplasm (Fig. 6I). Finally, we confirmed that P53RRA decreased the intact complex of p53-G3BP1 in a dose-dependent manner after P53RRA was introduced into the cells that were also transfected with G3BP1 and p53 (Fig. 6J).
p53 is known to regulate lncRNA expression (9, 10, 40, 41). However, we did not find evidence that p53 regulates P53RRA RNA levels (Supplementary Fig. S13A and S13B), suggesting that P53RRA acts upstream of the p53 signaling pathway. In addition, we demonstrated that depletion of p53 increased the percentage of cells in S phase, decreased apoptosis, and attenuated erastin-induced ferroptosis through changes in metabolic genes after introduction of P53RRA into A549 cells (Fig. 6K–M; Supplementary Fig. S13C–S13F). We further showed that knockout of p53 increased the percentage of cells in S phase, decreased cell growth, colony formation, apoptosis, and attenuated erastin-induced ferroptosis through changes these metabolic genes after introduction of P53RRA into HCT116 p53+/+, but not HCT116 p53−/− cells (Fig. 6N–P; Supplementary Fig. S13F–S13M), thus suggesting that P53RRA regulates these downstream effects at least partly through p53. Together, these findings suggest that the effects of P53RRA on cell growth, proliferation, cell-cycle progression, and apoptosis are indeed dependent on the presence of p53.
Altogether, our data suggest that P53RRA regulates p53 target genes through its interaction with G3BP1 in the cytoplasm, which leads to less sequestration of p53 in the cytoplasm and more p53 retention in the nucleus.
P53RRA interacts with the RRM domain of G3BP1 via residues critical to RNA binding
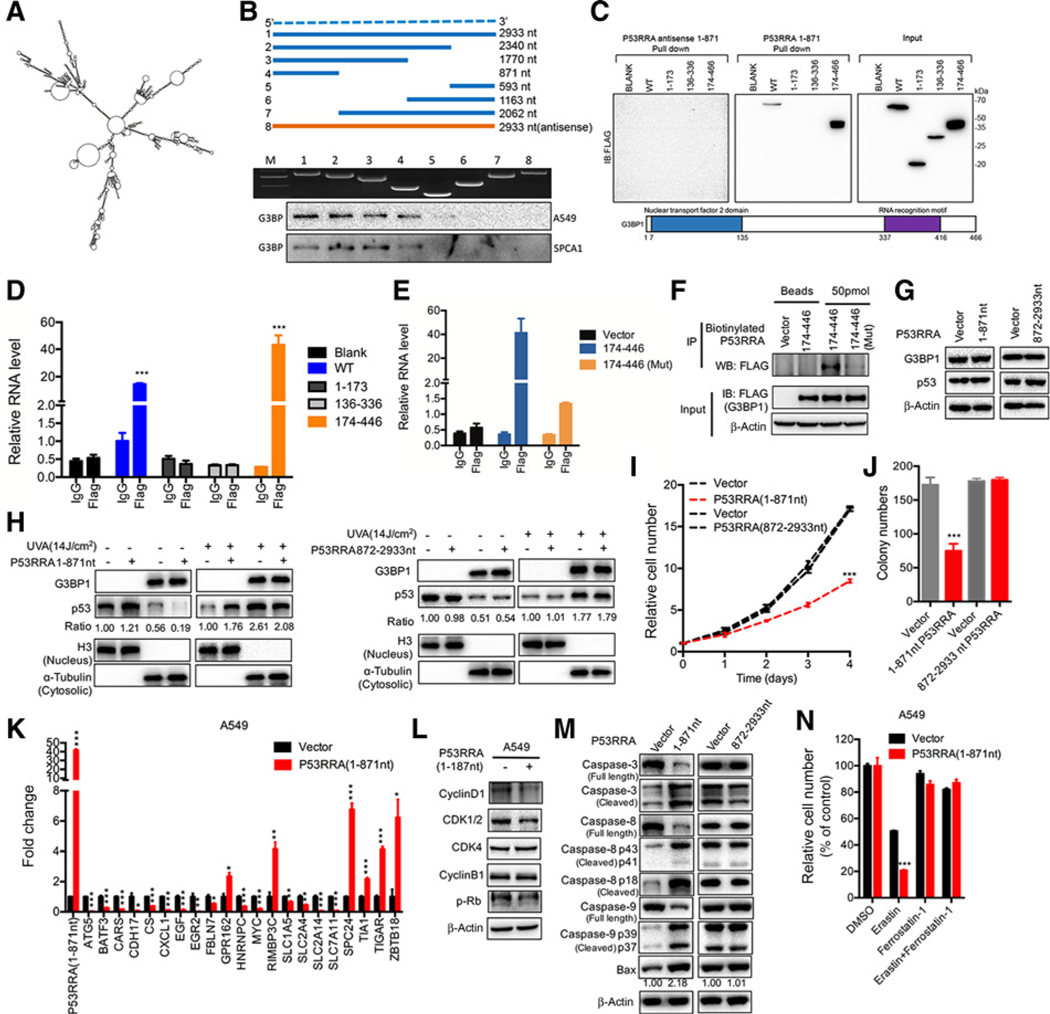
To determine specific domains that are responsible for the interaction between P53RRA and G3BP1, we performed RPA with different fragments of P53RRA using software (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi; Fig. 7A). One region (1–2340 nt) of P53RRA bound to G3BP1 as efficiently as the full-length P53RRA. Further mapping of this region indicated that the 1–871 nt domain was required for the P53RRA–G3BP1 interaction in A549 and SPCA1 cells (Fig. 7B).
Figure 7.
The 1–187 nt of P53RRA interacts with the G3BP1 RRM domain via residues critical to RNA binding, inhibiting cell growth and affecting target gene expression, cell cycle, and apoptosis. A, The predicted secondary structure of P53RRA. B, Deletion mapping of the G3BP1-binding domain in P53RRA. Top, diagrams of full-length P53RRA and the deletion fragments. Middle, the in vitro–transcribed full-length P53RRA and deletion fragments with the correct sizes are indicated. Bottom, immunoblot analysis for G3BP1 in the protein samples pulled down by different P53RRA constructs. C, The immunoblot analysis of FLAG-tagged G3BP1 [wild-type (WT) vs. domain truncation mutants] retrieved by in vitro–transcribed biotinylated P53RRA. The domain structure of G3BP1 is shown below. D, RIP assays show the association of the G3BP1 RRM domain with P53RRA. E, The isolated G3BP1 RRM domain is sufficient for binding to P53RRA, as demonstrated using the RIP-assay. F, RIP assays using biotinylated P53RRA indicated that P53RRA interacted with the G3BP1 RRM domain via residues critical to RNA binding. G, Overexpression of truncated in A549 cells did not change G3BP1 and p53 protein levels. H, p53 was sequestered in the nucleus in the presence of 1–871 nt of P53RRA, whereas UVA treatment induced greater accumulation of p53 in the cytoplasm. I and J, MTT assays (I) and plate colony formation assays (J) were used to assess A549 cells that were stably transfected with 1–871 nt of P53RRA and 872–2933 nt of P53RRA. K, The RNA expression of genes was measured by qPCR in A549 cells overexpressing 1–187 nt of P53RRA. L, The effect of 1–187 nt P53RRA on the regulators of cell-cycle progression in A549 cells. M, The truncation 1–187 nt of P53RRA in A549 cells affected the activities of caspase-3, caspase-8, and caspase-9 in HBE cells. N, The truncation 1–187 nt of P53RRA in A549 cells in response to erastin (5 μmol/L) ± ferrostatin-1 (1 μmol/L). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To map the region or regions within G3BP1 that interact with P53RRA, we tested the binding of G3BP1 truncation variants to P53RRA, with antisense P53RRA serving as a negative control (left panel of Fig. 7C). The RRM, but not the nuclear transport factor 2 domain, of G3BP1 was required for P53RRA binding. Finally, the interaction motifs of G3BP1 with P53RRA were confirmed by RIP followed by real-time PCR. The G3BP1 anti-body enriched P53RRA by approximately 20-fold compared with the IgG control (Fig. 7D). Moreover, the relative enrichment was further increased for the RRM domain of G3BP1 (174–466 aa; Fig. 7D), indicating that nucleotides 1–871 nt of P53RRA form an intact complex with the RRM of G3BP1. Thus, the RRM domain of G3BP1 is necessary and sufficient for P53RRA binding. The F380L/F382L double mutation in the RRM domain of G3BP1 caused a significant loss of interaction with the 1–871 nt sequence of P53RRA (Fig. 7E). This mutant of G3BP1 did not bind the 1–871 nt form of P53RRA using the RIP-assay with biotinylated-P53RRA, suggesting that these residues are critical to the interaction (Fig. 7F). Therefore, the F380 and F382 residues in the RRM domain of G3BP1 and nucleotides 1–871 of P53RRA are critical for the P53RRA–G3BP1 interaction.
Furthermore, neither the 1–871 nt nor the 872–2933 nt of truncated form of P53RRA affected G3BP1 protein level (Fig. 7G). However, only the 1–871 nt, but not the 872–2933 nt form of P53RRA, resulted in less sequestration of p53 in the cytoplasm (Fig. 7H). In addition, only the 1–871 nt form of P53RRA, but not the 872–2933 nt form, decreased cell growth and colony formation (Fig. 7I–J) and the expression of metabolic genes in A549 cells (Fig. 7K). Moreover, the 1–871 nt of P53RRA induced cell-cycle arrest by decreasing the cyclin D1 protein level (Fig. 7L) and increasing the levels of cleaved caspase-3 and caspase-9 (Fig. 7M) and erastin-induced ferroptosis (Fig. 7N) in A549 cells.
Finally, a Kaplan–Meier analysis for a cohort of these lung cancers showed that higher expression of P53RRA and G3BP1 was associated with longer overall survival in all patients with lung cancer (Supplementary Fig. S14A) and ADC (Supplementary Fig. S14B). However, lower p53 expression was associated with longer survival in all lung cancers and ADCs (Supplementary Fig. S14C). Higher P53RRA expression is closely linked to overall survival in breast cancer (Supplementary Fig. S14D). In 50% or more of human cancer types, p53 is directly inactivated by mutations (42). We found that high P53RRA expression concurrent with wild-type p53 was strongly linked with overall survival in breast cancer, whereas a higher level of P53RRA with mutated p53 was slightly linked with overall survival (Supplementary Fig. S14E and S14F), suggesting that P53RRA may exert its function through wild-type p53, but not through mutant p53. Moreover, we found that cooperation between P53RRA and wild-type p53 significantly decreased cell growth and colony formation compared with P53RRA and mutant p53 (Supplementary Fig. S14G and S14H). Finally, we also show that P53RRA together with wild-type p53 significantly attenuated the percentage of cells in S phase and increased apoptosis compared with P53RRA in p53 mutant cells (Supplementary Fig. S14I and S14J).
In conclusion, we have characterized the lncRNA P53RRA as a tumor suppressor. Based on our findings, we propose a model for P53RRA-mediated interaction with G3BP1 and its inhibition of lung tumorigenesis (Supplementary Fig. S15).
Discussion
The number of known p53-regulated lncRNAs is growing rapidly, indicating the widespread involvement of lncRNAs downstream of p53 activation (9, 10, 40, 43). Distant p53-bound enhancer regions generate enhancer RNAs that are required for the regulation of multiple genes by p53 (44). The p53-induced lncRNA LED binds to and activates enhancers, thus supporting p53 induced cell-cycle arrest (41). Here, we demonstrate that the lncRNA P53RRA induces cell-cycle arrest, apoptosis and ferroptosis and acts as a potential tumor suppressor by displacing p53 from a cytosolic G3BP1 complex. Moreover, we demonstrate that LSH is directly recruited to the P53RRA promoter region to silence P53RRA and that DNA methylation is involved in the repression of P53RRA, while recruitment of Cfp1, a modulator of H3K4Me3 (33, 34, 45), activate P53RRA expression, indicating the delicate balance in chromatin modification involved in the regulation of P53RRA.
The regulation of the p53 tumor-suppressor pathway by lncRNAs has been a topic of intense interest. The lncRNA LED is silenced in a subset of wild-type p53 leukemia cells, suggesting its potential role as tumor suppressor (41). Here, we demonstrate that P53RRA is suppressed in a subset of wild-type p53 breast and lung cancers. The maternally imprinted RNA MEG3 binds to p53 and activates the p53-dependent transcription of a subset of genes (13). We show that P53RRA inhibits ferroptosis-regulated genes in a p53-dependent manner by interacting with G3BP1. G3BP1 sequesters p53 in the cytoplasm in response to IR (17). We show that P53RRA interacts with G3BP1 in the cytoplasm. Moreover, the same domain of G3BP1 responsible for the interaction with p53 is critical for the interaction with P53RRA p53 redistributes to the cytoplasm in response to IR and UVA (46), whereas expression of P53RRA results in its binding to G3BP1, via the RRM motif, leading to increased p53 retention in the nucleus. MDM2 can sometimes activate the p53 pathway (47), and P53RRA increases the level of both MDM2 and p21, the latter being a target of p53 activity. Our data suggest that expression of P53RRA enhances p53-dependent regulation of a subset of metabolic genes.
As a tumor suppressor, p53 regulates cellular biological process and various metabolic pathways. For example, p53 can limit the production of ROS and induce apoptosis or ferroptosis (48, 49). Sequestered cytoplasmic p53 may initiate apoptosis and inhibit autophagy (46, 50, 51). We demonstrate that the lncRNA P53RRA leads to higher retention of p53 in the nucleus, which in turn triggers apoptosis and ferroptosis.
The ferroptotic mode of programmed necrosis was recently discovered as an apoptosis-independent form of cell death (52). Ferroptosis is characterized by the iron-dependent lethal accumulation of lipid ROS (30, 35, 53). Here, we demonstrate that P53RRA increases lipid ROS and iron concentrations, consistent with its role in ferroptosis. Moreover, the expression of several metabolic genes, including SCL7A11, which is linked to ferroptosis through its role in controlling iron concentration, decreased in the presence of P53RRA. The ferroptosis-inducer erastin inhibits ferroptosis by inhibiting the cystine/glutamate antiporter system (54). Here, we demonstrated that P53RRA promotes ferroptosis by retaining p53 in the nucleus.
In summary, we have uncovered a previously unknown function of a specific lncRNA in the p53 signaling pathway that promotes apoptosis and ferroptosis.
Supplementary Material
Significance:
A cytosolic lncRNA functions as a tumor suppressor by activating the p53 pathway.
Acknowledgments
This project has been funded in whole or in part with federal funds from the Frederick National Laboratory for Cancer Research, NIH, under contract HHSN261200800001E. This research was supported in part by the Intramural Research Program of NIH, Frederick National Lab, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the U.S. Government.
This manuscript has been read and approved by all the authors and is not submitted or under consideration for publication elsewhere.
This work was supported by the National Basic Research Program of China (2015CB553903 to Y. Tao); the National Natural Science Foundation of China (81672787 and 81372427 to Y. Tao, 81672307 to X. Wang, 81672991 to S. Liu, 81302354 to Y. Shi, 81422051 and 81472593 to Y. Cheng), and the Fundamental Research Funds for the Central Universities (2015zzts099 to C. Mao).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
References
- 1.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009;458:223–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huarte M. The emerging role of lncRNAs in cancer. Nat Med 2015; 21:1253–61. [DOI] [PubMed] [Google Scholar]
- 3.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 2015;47:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell 2013;154:26–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 2016;539:452–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engreitz JM, Ollikainen N, Guttman M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol 2016;17:756–70. [DOI] [PubMed] [Google Scholar]
- 7.Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 2013;493:231–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du Z, Sun T, Hacisuleyman E, Fei T, Wang X, Brown M, et al. Integrative analyses reveal a long noncoding RNA-mediated sponge regulatory network in prostate cancer. Nat Commun 2016;7:10982. doi: 10.1038/ncomms10982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanchez Y, Segura V, Marin-Bejar O, Athie A, Marchese FP, Gonzalez J, et al. Genome-wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature. Nat Commun 2014;5:5812. doi: 10.1038/ncomms6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marin-Bejar O, Marchese FP, Athie A, Sanchez Y, Gonzalez J, Segura V, et al. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol 2013;14:R104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmitt AM, Garcia JT, Hung T, Flynn RA, Shen Y, Qu K, et al. An inducible long noncoding RNA amplifies DNA damage signaling. Nat Genet 2016;48:1370–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arab K, Park YJ, Lindroth AM, Schafer A, Oakes C, Weichenhan D, et al. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol Cell 2014;55:604–14. [DOI] [PubMed] [Google Scholar]
- 13.Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, et al. Activation of p53 by MEG3 non-coding RNA. J Biol Chem 2007;282:24731–42. [DOI] [PubMed] [Google Scholar]
- 14.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet 2011;43:621–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duchesne M, Schweighoffer F, Parker F, Clerc F, Frobert Y, Thang MN, et al. Identification of the SH3 domain of GAP as an essential sequence for RasGAP-mediated signaling. Science 1993;259:525–8. [DOI] [PubMed] [Google Scholar]
- 16.Tourriere H, Gallouzi IE, Chebli K, Capony JP, Mouaikel J, van der Geer P, et al. RasGAP-associated endoribonuclease G3Bp: selective RNA degradation and phosphorylation-dependent localization. Mol Cell Biol 2001; 21:7747–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim MM, Wiederschain D, Kennedy D, Hansen E, Yuan ZM. Modulation of p53 and MDM2 activity by novel interaction with Ras-GAP binding proteins (G3BP). Oncogene 2007;26:4209–15. [DOI] [PubMed] [Google Scholar]
- 18.Aulas A, Caron G, Gkogkas CG, Mohamed NV, Destroismaisons L, Sonenberg N, et al. G3BP1 promotes stress-induced RNA granule interactions to preserve polyadenylated mRNA. J Cell Biol 2015;209:73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Somasekharan SP, El-Naggar A, Leprivier G, Cheng H, Hajee S, Grunewald TG, et al. YB-1 regulates stress granule formation and tumor progression by translationally activating G3BP1. J Cell Biol 2015;208:913–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen Y, Katsaros D, Loo LW, Hernandez BY, Chong C, Canuto EM, et al. Prognostic and predictive values of long non-coding RNA LINC00472 in breast cancer. Oncotarget 2015;6:8579–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi Y, Tao Y, Jiang Y, Xu Y, Yan B, Chen X, et al. Nuclear epidermal growth factor receptor interacts with transcriptional intermediary factor 2 to activate cyclin D1 gene expression triggered by the oncoprotein latent membrane protein 1. Carcinogenesis 2012;33:1468–78. [DOI] [PubMed] [Google Scholar]
- 22.Jiang Y, Yan B, Lai W, Shi Y, Xiao D, Jia J, et al. Repression of Hox genes by LMP1 in nasopharyngeal carcinoma and modulation of glycolytic pathway genes by HoxC8. Oncogene 2015;34:6079–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao D, Huang J, Pan Y, Li H, Fu C, Mao C, et al. Chromatin remodeling factor LSH is upregulated by the LRP6-GSK3beta-E2F1 axis linking reversely with survival in gliomas. Theranostics 2017;7:132–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jia J, Shi Y, Chen L, Lai W, Yan B, Jiang Y, et al. Decrease in lymphoid specific helicase and 5-hydroxymethylcytosine is associated with metastasis and genome instability. Theranostics 2017;7:3920–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chuang SS, Chen SW, Chang ST, Kuo YT. Lymphoma in Taiwan: Review of 1347 neoplasms from a single institution according to the 2016 Revision of the World Health Organization Classification. J Formos Med Assoc 2017;116:620–5. [DOI] [PubMed] [Google Scholar]
- 26.Klutstein M, Nejman D, Greenfield R, Cedar H. DNA methylation in cancer and aging. Cancer Res 2016;76:3446–50. [DOI] [PubMed] [Google Scholar]
- 27.Baylin SB, Jones PA. A decade of exploring the cancer epigenome—biological and translational implications. Nat Rev Cancer 2011;11: 726–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tao Y, Liu S, Briones V, Geiman TM, Muegge K. Treatment of breast cancer cells with DNA demethylating agents leads to a release of Pol II stalling at genes with DNA-hypermethylated regions upstream of TSS. Nucleic Acids Res 2011;39:9508–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tao Y, Xi S, Shan J, Maunakea A, Che A, Briones V, et al. Lsh, chromatin remodeling family member, modulates genome-wide cytosine methylation patterns at nonrepeat sequences. Proc Natl Acad Sci U S A 2011;108: 5626–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang Y, Mao C, Yang R, Yan B, Shi Y, Liu X, et al. EGLN1/c-Myc induced lymphoid-specific helicase inhibits ferroptosis through lipid metabolic gene expression changes. Theranostics 2017;7:3293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He X, Yan B, Liu S, Jia J, Lai W, Xin X, et al. Chromatin remodeling factor LSH drives cancer progression by suppressing the activity of fumarate hydratase. Cancer Res 2016;76:5743–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet 2007; 39:232–6. [DOI] [PubMed] [Google Scholar]
- 33.Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature 2010;464:1082–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clouaire T, Webb S, Skene P, Illingworth R, Kerr A, Andrews R, et al. Cfp1 integrates both CpG content and gene activity for accurate H3K4me3 deposition in embryonic stem cells. Genes Dev 2012;26:1714–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dixon SJ, Stockwell BR. The role of iron and reactive oxygen species in cell death. Nat Chem Biol 2014;10:9–17. [DOI] [PubMed] [Google Scholar]
- 36.Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 2012;149:1060–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011;477:295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010;142:409–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang L, Kon N, Li T, Wang SJ, Su T, Hibshoosh H, et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature 2015;520:57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Younger ST, Kenzelmann-Broz D, Jung H, Attardi LD, Rinn JL. Integrative genomic analysis reveals widespread enhancer regulation by p53 in response to DNA damage. Nucleic Acids Res 2015;43:4447–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leveille N, Melo CA, Rooijers K, Diaz-Lagares A, Melo SA, Korkmaz G, et al. Genome-wide profiling of p53-regulated enhancer RNAs uncovers a subset of enhancers controlled by a lncRNA. Nat Commun 2015; 6:6520. doi: 10.1038/ncomms7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joerger AC, Fersht AR. The p53 pathway: origins, inactivation in cancer, and emerging therapeutic approaches. Annu Rev Biochem 2016;85: 375–404. [DOI] [PubMed] [Google Scholar]
- 43.Evans JR, Feng FY, Chinnaiyan AM. The bright side of dark matter: lncRNAs in cancer. J Clin Invest 2016;126:2775–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit E, Oude Vrielink JA, et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell 2013;49:524–35. [DOI] [PubMed] [Google Scholar]
- 45.Clouaire T, Webb S, Bird A. Cfp1 is required for gene expression-dependent H3K4 trimethylation and H3K9 acetylation in embryonic stem cells. Genome Biol 2014;15:451. doi: 10.1186/s13059-0140451-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chipuk JE, Bouchier-Hayes L, Kuwana T, Newmeyer DD, Green DR. PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science 2005;309:1732–5. [DOI] [PubMed] [Google Scholar]
- 47.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004;303:844–8. [DOI] [PubMed] [Google Scholar]
- 48.Kruiswijk F, Labuschagne CF, Vousden KH. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat Rev Mol Cell Biol 2015;16:393–405. [DOI] [PubMed] [Google Scholar]
- 49.Maddocks OD, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, et al. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature 2013;493:542–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature 2009;458:1127–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D’Amelio M, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol 2008;10:676–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shaw AT, Winslow MM, Magendantz M, Ouyang C, Dowdle J, Subramanian A, et al. Selective killing of K-ras mutant cancer cells by small molecule inducers of oxidative stress. Proc Natl Acad Sci U S A 2011;108:8773–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 2014;156:317–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dixon SJ, Patel DN, Welsch M, Skouta R, Lee ED, Hayano M, et al. Pharmacological inhibition of cystine-glutamate exchange induces endoplasmic reticulum stress and ferroptosis. Elife 2014;3:e02523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.