Abstract
Salmonella is among the most common foodborne pathogens worldwide, and can lead to acute gastroenteritis. Along with poultry, cattle production is recognized as an important source of human infection. Salmonella transmission from cattle to humans can occur through the environment, or through close contact with sick animals or their derived products. This study aimed to investigate the intestinal carriage of Salmonella spp. within French cattle production. A total of 959 cattle intestinal samples, from one of the largest French slaughterhouses, were analyzed. Isolated strains were genotyped by pulsed field gel electrophoresis (PFGE), and a sub-selection was taken by whole genome sequencing (WGS). Twenty-nine samples were positive for Salmonella spp., yielding an estimated prevalence of 3% in cattle production. Eight different Salmonella serotypes were found: Montevideo was the most prevalent (34%), followed by Mbandaka (24%) and Anatum (14%). PFGE genotyping allowed the clustering of Salmonella isolates according to their serotype. Within the clusters, some isolates presented 100% similarity. To investigate potential epidemiological links between them, WGS and core genome multilocus sequence typing (cgMLST) were used, revealing identical profiles between isolates originating from different areas and/or different animal breeds. This investigation provides new insights on Salmonella serotype epidemiology in cattle production in France.
Keywords: Salmonella, cattle production, whole genome sequencing, France
1. Introduction
Salmonellosis is an important public health problem among the common bacterial foodborne zoonoses, which contributes to economic losses in both developed and developing countries. Worldwide, it remains the second most commonly reported gastrointestinal infection in humans after campylobacteriosis, and is a significant cause of foodborne outbreaks [1]. Nontyphoidal Salmonella spp. are responsible for the highest annual burden and the largest number of deaths both globally and in the European Region [2]. Worldwide, Salmonella spp. were estimated to be responsible for 78 million cases of illness annually, 230,000 deaths and 4 million disability-adjusted life years (DALYs) per year [3,4]. In 2019, 87,923 confirmed cases of salmonellosis in humans were reported in Europe. Nontyphoidal Salmonella are bacterial enteric pathogens associated with animal food reservoirs, predominantly transmitted to humans by contaminated food and water. Among these, Salmonella enterica subspecies enterica is the most important subspecies affecting humans and domestic animals [5].
Salmonella have been widely reported in cattle [6], and dairy cows are known to be reservoirs of Salmonella enterica. Usually, human salmonellosis is attributed to the consumption of contaminated poultry meat and eggs, as well as dairy and beef products [7]. This presence of Salmonella in cattle may result in the contamination of milk or meat in the farm environment (animals, rodents, wild animals, insects, water, etc.), which could lead to direct and indirect infection of people and animals [8,9]. Infected animals may develop symptoms or shed Salmonella in their feces without showing any clinical signs of disease. Clinical manifestations of bovine salmonellosis include mainly apathy, hyperthermia, difficulty breathing, panting, pallor of the mucous membranes, and forthe most commonly reported manifestation, abortion [10,11]. The risk associated with asymptomatic carriage is that cattle may introduce this organism into an abattoir, which can represent a significant food safety hazard of cross-contamination during food processing [8]. Asymptomatic carriage may not impact dairy production, but herds with high S. enterica burden may pose an increased public health risk through contamination of the production environment and possibly milk and meat [7]. Importantly, Salmonella can persist in the environment for several months and be associated with wildlife, which is a reservoir for Salmonella and may serve as a source of contamination.
The overall prevalence in cattle has been reported by the EFSA and ECDC to be 3.34% in the European Union (the prevalence of positive samples at the slaughterhouse was 7.76%) [1]. Moreover, prevalence estimates vary from 2% in Europe to 16% in North America [12,13,14]. Studies have reported 5% in Africa, 4% in Turkey and 2% in Iran [15,16,17]. Fossler et al. showed the presence of Salmonella spp. in more than 90% of the environment on dairy farms in the United States [9].
Salmonella Dublin, Salmonella Newport, and Salmonella Typhimurium are generally associated with salmonellosis in calves and adult cows, causing mild to severe illness. Moreover, asymptomatic carriage and fecal shedding of Salmonella serotypes such as Salmonella Cerro, Salmonella Kentucky, Salmonella Mbandaka, and Salmonella Montevideo have been well documented in dairy animals [10,18].
The objectives of this study were to investigate the occurrence of Salmonella in cattle production in France, serotype distribution, genetic diversity of Salmonella isolates and possible epidemiological links among the isolates, in order to better understand Salmonella epidemiology, and potentially to achieve a better control of Salmonella in cattle production.
2. Materials and Methods
2.1. Sampling Plan
Bovine intestinal content samples were taken from one of the largest slaughterhouses in France. A total of 959 intestinal samples were randomly collected from the slaughter line at the evisceration step, over a period of 6 months with a frequency of 50 samples per week. The 959 samples originated from 282 farms distributed across 32 French areas, representative of cattle production in the country. Among the samples, 476 were from calves (less than 8 months old) and 483 were from adult cattle (Table 1). Samples were transported to the laboratory in isotherm bags and stored at +4 °C until analysis.
Table 1.
Number of cattle intestinal samples studied in this work, distributed by the month of sampling, age of cattle (cows and calves) and productive aptitude (dairy and beef).
| Month of Sampling | Number of Samples Collected | ||||||
|---|---|---|---|---|---|---|---|
| Cows | Calves | ||||||
| Dairy | Beef | % vs. Total Cows’ Samples | Dairy | Beef | Mixed/ Unknown * |
% vs. Total Calves’ Samples | |
| June | 30 | 11 | 8.5% | 46 | 2 | 10.1% | |
| July | 46 | 54 | 20.7% | 43 | 56 | 1 | 21.0% |
| August | 51 | 49 | 20.7% | 84 | 16 | 21.0% | |
| September | 33 | 67 | 20.7% | 41 | 44 | 17.9% | |
| October | 21 | 69 | 18.6% | 39 | 11 | 10.5% | |
| November | 23 | 14 | 7.7% | 55 | 3 | 12.2% | |
| December | 9 | 6 | 3.1% | 20 | 11 | 1/3 | 7.4% |
| Total number of samples | 213 | 270 | 483 | 328 | 143 | 5 | 476 |
* The mention “mixed” indicates a mixed breed, the mention “unknown” indicates the absence of information about the breed of the animal.
2.2. Salmonella Isolation and Quantification
Samples of 25 g were analyzed for Salmonella detection according to the NF EN ISO 6579 and NF U 47-100 standards [19,20]. Samples were homogenized in a 1:10 sample:broth ratio in a Pulsifier® (Microgen Bioproducts, Surrey, United-Kingdom) with 225 mL of buffered peptone water (Biomérieux, Craponne, France) for pre-enrichment. After incubation, for 18 ± 2 h at 37 °C, 0.1 mL was transferred to modified semi-solid Rappaport-Vassiliadis) (MSRV) medium (Biokar, Beauvais, France) and incubated for 24 h, then 48 h at 41.5 °C. One milliliter of pre-enrichment broth was also inoculated to 10 mL of Muller Kauffmann tetrathionate broth (MKTTn) (Biokar, France) and incubated for 24 h at 37 °C. Cultures obtained from MSRV were inoculated on xylose lysine deoxycholate (XLD) agar (Biokar, France) and Rapid’Salmonella (R’S) agar (BioRad, Paris, France), and cultures obtained from MKTTn on Xylose Lysine Tergitol 4 (XLT4) agar (Biokar, France). Colonies of presumptive Salmonella were subcultured and their identity was confirmed by biochemical assays for glucose fermentation, lactose oxidation, gas, H2S production (triple sugar iron (TSI) agar, Biokar, France), lack of galactosidase (ONPG), and presence of decarboxylase (L-Lysine). All Salmonella isolates were confirmed by serotyping according to the Kauffmann–White scheme using slide agglutination tests [21].
In parallel, Salmonella enumerations were carried out with the miniaturized most probable number (mMPN) technique, according to the XP CEN ISO/TS 6579-2 standard [22].
2.3. Genotyping and Genotypes Comparisons
2.3.1. Pulsed-Field Gel Electrophoresis
Pulsed-field gel electrophoresis (PFGE) of isolates of Salmonella was performed using the XbaI and BlnI restriction enzymes following the procedure developed by the US Center for Disease Control (CDC) [23] and as previously described by Kerouanton et al. [24]. PFGE fingerprints were analyzed (fragment size estimation) using BioNumerics v.7.5 software (Applied Maths, Belgium). Similarities between isolate fingerprints were calculated using the Dice index, with a maximum tolerance of 1%, and a dendrogram was built using the unweighted pair group method with arithmetic mean (UPGMA algorithm) [24].
2.3.2. Whole Genome Sequencing
Whole genome sequencing (WGS) was performed on isolates of Salmonella serotypes collected in at least 2 animals to investigate potential epidemiological links between them. The WGS was performed following DNA extraction from the strains. DNA was extracted from one-day single colony cultures with a QIAamp DNA Mini Kit (QIAGEN, Villebon-sur-Yvette, France), according to the manufacturer’s instructions, and quantified using a Qubit 2.0 fluorometer and a Qubit dsDNA (double-stranded DNA) HS (high-sensitivity) assay kit (Thermo Fisher Scientific, Villebon-sur-Yvette, France). WGS was performed using Illumina technology at the technical center “Institut du Cerveau et de la Moelle épinière”, Pitié-Salpêtrière Hospital, Paris (www.icm-institute.org, accessed on 16 April 2021). Libraries were prepared using a Nextera XT DNA Library Preparation Kit and Nextera XT Index Kit (Illumina, Paris, France). Samples were then sequenced with a NextSeq 500 machine using a NextSeq 500 Mid Output Kit v2 (300 cycles), (Illumina, France). Paired-end raw reads were deposited on the EnteroBase database platform for Salmonella (http://enterobase.warwick.ac.uk/, accessed on 16 April 2021), and were automatically de novo assembled using SPAdes [25] once the sequences were uploaded.
Genomic comparison of the isolated strains was carried out using the cgMLST scheme with 3002 genes included, available on EnteroBase [26]. Similar but nonidentical strains (strains showing different core genome Sequence Type (cgST)) were identified in EnteroBase by using the hierarchical clustering method (HierCC) that allows for grouping of strains into hierarchical clusters (HCs) that can differ up to a specified and fixed number of cgMLST alleles. This number is indicated by the suffix following “HC” (e.g., HC5 for 5 cgMLST allelic differences). To assess the genetic relationship between strains of the same serotype and the population structure of Salmonella isolates, a neighbor-joining tree was created from cgMLST allelic differences in EnteroBase using GrapeTree [27] and the RapidNJ algorithm [28]. The assembly sequences are publicly available from EnteroBase; their accession numbers (barcodes) are listed in Table 3.
2.4. Statistical Analysis
The statistical relationship between the prevalence of Salmonella in calves or adult cattle was analyzed by Chi-Square test. Meanwhile, the statistical relationship between positive dairy and beef cows was analyzed by Fisher’s exact test. p-Value ≤ 0.05 was considered as statistically significant.
3. Results
3.1. Estimation of Salmonella Prevalence and Quantification
In all, 29 out of 959 samples were positive for Salmonella spp. (Table 2), suggesting a 3% prevalence in cattle production in France. Intestinal contents were taken from animals originating from 32 different breeding areas (Figure 1). Salmonella strains were isolated from animals from 14 of these areas (Table 2).
Table 2.
List of the 29 Salmonella positive samples in this study.
| Sample Name | Cattle Age Category | Breed Type | Cattle Birth Area |
Cattle Breeding Area |
Salmonella Serotype Isolated |
Salmonella Enumeration CFU/g |
|---|---|---|---|---|---|---|
| S16LNR-FLG405 | Calf | Dairy | E | E | S. 1,4,[5],12: i: − | ND |
| S16LNR-FLG412 | Calf | Dairy | C | E | S. 1,4,[5],12: i: − | 22 |
| S16LNR-FLG55 | Calf | Dairy | O | H | S. Anatum | 8 |
| S16LNR-FLG62 | Calf | Dairy | C | C | S. Anatum | 380 |
| S16LNR-FLG66 | Calf | Dairy | F | E | S. Anatum | 2 |
| S16LNR-FLG70 | Calf | Dairy | C | C | S. Anatum | 220 |
| S16LNR-FLG210 | Adult | Dairy | B | B | S. Indiana | 6 |
| S16LNR-FLG43 | Adult | Beef | K | K | S. Mbandaka | >710 |
| S16LNR-FLG250 | Adult | Beef | L | H | S. Mbandaka | ND |
| S16LNR-FLG364 | Calf | Dairy | N | F | S. Mbandaka | 2 |
| S16LNR-FLG372 | Calf | Dairy | S | F | S. Mbandaka | ND |
| S16LNR-FLG377 | Adult | Beef | M | M | S. Mbandaka | ND |
| S16LNR-FLG387 | Calf | Dairy | O | D | S. Mbandaka | ND |
| S16LNR-FLG392 | Calf | Dairy | T | D | S. Mbandaka | ND |
| S16LNR-FLG104 | Adult | Dairy | C | C | S. Montevideo | >710 |
| S16LNR-FLG107 | Adult | Beef | G | G | S. Montevideo | >710 |
| S16LNR-FLG112 | Adult | Dairy | M | M | S. Montevideo | 220 |
| S16LNR-FLG114 | Adult | Dairy | N | N | S. Montevideo | 170 |
| S16LNR-FLG214 | Calf | Dairy | P | A | S. Montevideo | ND |
| S16LNR-FLG222 | Calf | Dairy | Q | A | S. Montevideo | 19 |
| S16LNR-FLG256 | Adult | Beef | N | N | S. Montevideo | ND |
| S16LNR-FLG261 | Adult | Beef | I | I | S. Montevideo | 8 |
| S16LNR-FLG374 | Calf | Dairy | N | F | S. Montevideo | ND |
| S16LNR-FLG401 | Adult | Dairy | F | F | S. Montevideo | >710 |
| S16LNR-FLG52 | Calf | Beef | E | E | S. Ohio | ND |
| S16LNR-FLG244 | Adult | Beef | L | L | S. Ohio | 8 |
| S16LNR-FLG05 | Adult | Dairy | J | J | S. Stourbridge | ND |
| S16LNR-FLG219 | Calf | Beef | R | A | S. Virchow | ND |
| S16LNR-FLG228 | Calf | Dairy | P | A | S. Virchow | ND |
ND: Non detectable (under the detection limit of the method). Cattle birth and breading areas were anonymized; each letter was randomly attributed to a specific area. If animals were born and bred in the same French area, letters of birth and breeding areas were identical. In the same way, if several animals harbored the same letter as birth and/or breeding area, it means that the animals originated from the same area of birth and/or breeding.
Figure 1.
Geographical distribution of the sampled cattle from breeding areas throughout the country.
Among the positive samples, 55% (16/29) and 45% (13/29) of isolates were from intestinal contents of calves and adults, respectively, which represents a prevalence of 3.4% (16/476) for calves and 2.7% (13/483) for adult cattle. No statistical difference was found for Salmonella carriage according to animal age (p > 0.05). Salmonella was detected among several cattle breeds (Table 2) but there was no statistical difference between Salmonella positive dairy cows (3.7%; 20/541) and beef cows (2.2%; 9/413) (p > 0.05).
Among all the positive samples, 55% (16/29) were quantified using the mMPN method. The range of Salmonella concentrations found in all samples varied from 2 CFU/g to >710 CFU/g (Table 2). More precisely, 27% of samples (8/29) had a load range between 2 and 100 CFU/g, 14% (4/29) between 100 and 700 CFU/g, and 14% (4/29) greater than 710 CFU/g.
3.2. Serotype
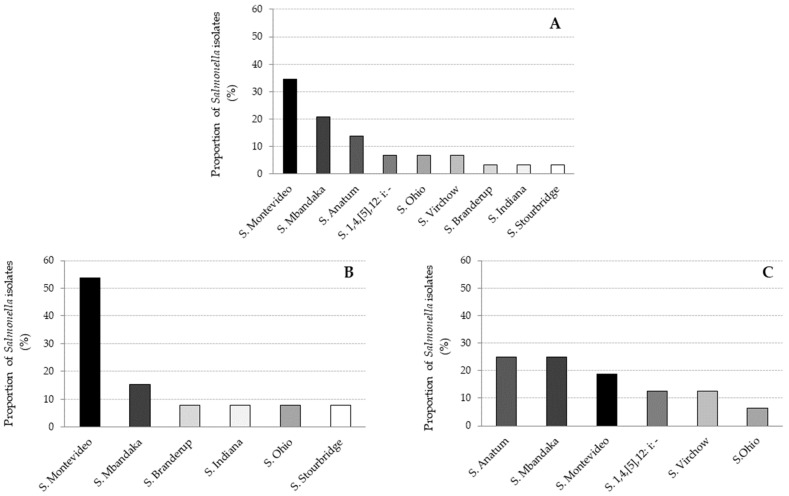
Eight serotypes of Salmonella were identified among the 29 positive samples. Salmonella Montevideo was the most prevalent serotype (34%; 10/29), followed by Salmonella Mbandaka (24%; 7/29) and Salmonella Anatum (14%; 4/29).
The other serotypes, Salmonella Stourbridge, Salmonella Ohio, Salmonella Indiana, Salmonella Virchow and Salmonella Typhimurium monophasic variant (1,4,[5],12: i: −), were more rarely isolated (<10%) (Figure 2A). The distribution of Salmonella serotypes was slightly different between adult cattle (Figure 2B) and calves (Figure 2C). In adults, S. Montevideo was the most prevalent serotype, while in calves, S. Anatum and S. Mbandaka predominated. Moreover, S. Anatum was only found in calves.
Figure 2.
Main Salmonella serotypes found in the intestinal contents from all sampled animals (A), adult cattle (B) and calves (C). Each color represents one serotype of Salmonella.
3.3. Genotyping
As expected, pulsed-field genotyping clustered Salmonella isolates according to their serotypes (Figure 3A). The S. Stourbridge isolate was not genotyped by PFGE since no growth was observed during subculture. Within the clusters, some isolates, such as S. Anatum or S. Virchow, presented 100% similarity using two restriction enzymes (XbaI and BlnI), while S. Montevideo and S. Mbandaka showed higher diversity between the isolates, since they showed from 67% to 83% similarity (Figure 3A). Interestingly, clustered isolates of S. Anatum and S. Ohio originated from different birth areas and/or from different animal breeds, independently of the serotype.
Figure 3.
Dendrogram of pulsed-field gel electrophoresis (PFGE) cluster analysis using the XbaI and BlnI restriction enzymes (A) and cgMLST Grape Tree (B) of selected isolates of Salmonella.
Among the 29 collected isolates of Salmonella, 27 were sequenced; S. Indiana and S. Stourbridge isolates were excluded for the WGS, since these serotypes were isolated from only one animal and no link can be drawn with other animals. The genomes of Salmonella strains were compared using the core-genome MLST approach (cgMLST scheme available on EnteroBase, https://enterobase.warwick.ac.uk, accessed on 16 April 2021). The 27 sequenced strains harbored a unique cgST profile (Table 3), indicating that they are all different and showed at least one cgMLST allelic variation between one another. As previously described in this study using PFGE (Figure 3A), with WGS (prediction serotype, SISTR1 and using the Sequence Type MLST), the strains belonging to the same serotype clustered together (Table 3 and Figure 3B). This clustering of the strains according to their serotype was also illustrated at the HC200 level, since each strain of the same serotype harbored the same HC200|cluster (Table 3). Here, we considered that the isolates clustered in the same node on the neighbor-joining tree (Figure 3B) and belonging to the same HC5|cluster (up to five allelic variations between strains of the same HC5|cluster) are highly probably epidemiologically linked.
Table 3.
Twenty-seven Salmonella strains sequenced in this study, serotypes, sequence type (ST) and core genome MLST (cg MLST) profiles.
| Strain Name | Genome Accession Number (BarCode) in EnteroBase |
SISTR1 Serovar | Sequence Type (ST) MLST |
Sequence Type (ST) cgMLST |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Serotype | Serogroup | H1 | H2 | Achtman Scheme | Core Genome Sequence Type (cgST) |
HC0 | HC2 | HC5 | HC10 | HC200 | ||
| S16LNR-FLG405 | SAL_BB2134AA | 4: i: − | B | i | − | 34 | 215775 | 215775 | 215775 | 215775 | 2 | 2 |
| S16LNR-FLG412 | SAL_BB2133AA | 4: i: − | B | i | − | 34 | 215772 | 215772 | 215772 | 215772 | 215772 | 2 |
| S16LNR-FLG55 | SAL_BB2119AA | Anatum | E1 | e,h | 1,6 | 64 | 215763 | 215763 | 215763 | 215760 | 215760 | 5 |
| S16LNR-FLG62 | SAL_BB2117AA | Anatum | E1 | e,h | 1,6 | 64 | 215760 | 215760 | 215760 | 215760 | 215760 | 5 |
| S16LNR-FLG66 | SAL_BB2118AA | Anatum | E1 | e,h | 1,6 | 64 | 215761 | 215761 | 215761 | 215761 | 215761 | 5 |
| S16LNR-FLG70 | SAL_BB2121AA | Anatum | E1 | e,h | 1,6 | 64 | 215770 | 215770 | 215763 | 215760 | 215760 | 5 |
| S16LNR-FLG43 | SAL_BB2111AA | Mbandaka | C1 | z10 | e,n,z15 | 413 | 215755 | 215755 | 215755 | 215755 | 215755 | 4 |
| S16LNR-FLG250 | SAL_ZA0098AA | Mbandaka | C1 | z10 | e,n,z15 | 413 | 199394 | 199394 | 199394 | 199394 | 199394 | 4 |
| S16LNR-FLG364 | SAL_ZA0099AA | Mbandaka | C1 | z10 | e,n,z15 | 413 | 199395 | 199395 | 199395 | 199395 | 199395 | 4 |
| S16LNR-FLG372 | SAL_ZA0100AA | Mbandaka | C1 | z10 | e,n,z15 | 413 | 199398 | 199398 | 199398 | 199398 | 199398 | 4 |
| S16LNR-FLG377 | SAL_ZA0101AA | Mbandaka | C1 | z10 | e,n,z15 | 413 | 199396 | 199396 | 199396 | 199396 | 199396 | 4 |
| S16LNR-FLG387 | SAL_ZA0105AA | Mbandaka | C1 | z10 | e,n,z15 | 413 | 199401 | 199401 | 199401 | 199401 | 199401 | 4 |
| S16LNR-FLG392 | SAL_ZA0102AA | Mbandaka | C1 | z10 | e,n,z15 | 413 | 199399 | 199399 | 199399 | 199399 | 199399 | 4 |
| S16LNR-FLG104 | SAL_BB2120AA | Montevideo | C1 | g,m,[p],s | − | 39 | 215759 | 215759 | 215759 | 215759 | 215759 | 16 |
| S16LNR-FLG107 | SAL_BB2122AA | Montevideo | C1 | g,m,[p],s | − | 39 | 215766 | 215766 | 215766 | 215766 | 215766 | 16 |
| S16LNR-FLG112 | SAL_BB2124AA | Montevideo | C1 | g,m,[p],s | − | 39 | 239489 | 239489 | 239489 | 239489 | 215766 | 16 |
| S16LNR-FLG114 | SAL_BB2125AA | Montevideo | C1 | g,m,[p],s | − | 39 | 239490 | 239490 | 239489 | 239489 | 215766 | 16 |
| S16LNR-FLG214 | SAL_BB2123AA | Montevideo | C1 | g,m,[p],s | − | 39 | 215764 | 215764 | 215764 | 215764 | 215764 | 16 |
| S16LNR-FLG222 | SAL_BB2131AA | Montevideo | C1 | g,m,[p],s | − | 39 | 215771 | 215771 | 215771 | 215771 | 215771 | 16 |
| S16LNR-FLG256 | SAL_BB2130AA | Montevideo | C1 | g,m,[p],s | − | 39 | 215769 | 215769 | 215769 | 215769 | 215769 | 16 |
| S16LNR-FLG261 | SAL_BB2129AA | Montevideo | C1 | g,m,[p],s | − | 39 | 239491 | 215773 | 215773 | 215773 | 215773 | 16 |
| S16LNR-FLG374 | SAL_BB2135AA | Montevideo | C1 | g,m,[p],s | − | 39 | 215773 | 215773 | 215773 | 215773 | 215773 | 16 |
| S16LNR-FLG401 | SAL_BB2132AA | Montevideo | C1 | g,m,[p],s | − | 39 | 215774 | 215774 | 215774 | 215774 | 215774 | 16 |
| S16LNR-FLG244 | SAL_BB2128AA | Ohio | C1 | b | l,w | 72 | 215767 | 215767 | 215767 | 215767 | 215767 | 621 |
| S16LNR-FLG052 | SAL_BB2115AA | Ohio | C1 | b | l,w | 72 | 239488 | 239488 | 239488 | 239488 | 239488 | 621 |
| S16LNR-FLG219 | SAL_BB2127AA | Virchow | C1 | r | 1,2 | 9 | 215768 | 215768 | 215768 | 215768 | 215765 | 715 |
| S16LNR-FLG228 | SAL_BB2126AA | Virchow | C1 | r | 1,2 | 9 | 215765 | 215765 | 215765 | 215765 | 215765 | 715 |
HC: Hierarchical clustering.
Within S. Anatum, three isolates (S16LNR-FLG55, S16LNR-FLG62 and S16LNR-FLG70) were grouped in the same HC5|cluster, HC5|215760. Among these isolates, two (S16LNR-FLG55 and S16LNR-FLG70) harbored the same HC2|cluster, indicating a maximum of two allelic variations. Interestingly, within the three clustered isolates, two were originated from animals bred in the same geographic area, indicating a high probability of an epidemiological link between these isolates.
For S. Typhimurium monophasic variant isolates (S16LNR-FLG405 and S16LNR-FLG412), S. Mbandaka isolates (S16LNR-FLG372 and S16LNR-FLG364), and S. Montevideo isolates (S16LNR-FLG214 and S16LNR-FLG222), although the PFGE genotypes presented 100% similarity (Figure 3), WGS and cgMLST results differentiated the strains up to the HC200 level (a maximum of 200 cgMLST allelic variations). Therefore, it did not allow for inferring any epidemiological links between these isolates, despite a common breeding area for S. Typhimurium monophasic variant and S. Mbandaka isolates.
Regarding S. Virchow isolates, WGS and cgMLST confirmed the genotyping results found using PFGE. The two strains clearly grouped in the same HC10|cluster (Table 3), suggesting a possible epidemiological link between the strains isolated from animals bred in the same geographic area (Table 2).
Genotyping by PFGE showed two other clusters of S. Montevideo with about 100% similarity, the first was composed of four strains (S16LNR-FLG104, S16LNR-FLG107, S16LNR-FLG112 and S16LNR-FLG114) and the second of two strains of S. Montevideo (S16LNR-FLG261 and S16LNR-FLG374). Within the first PFGE cluster, two strains belonged to the same HC2 cluster (S16LNR-FLG112 and S16LNR-FLG114), and three (S16LNR-FLG107, S16LNR-FLG112 and S16LNR-FLG114) grouped in the same HC10|cluster (Table 3). The fourth isolate, S16LNR-FLG104, belonged to different HC10|cluster, indicating that it differs from the others three strains by at least 10 allelic variations. Regarding the second PFGE cluster within S. Montevideo isolates, two (S16LNR-FLG261 and S16LNR-FLG374) harbored different cgST profiles; however, they belonged to the same HC0|cluster, HC0|215773. These cgMLST results indicate that all alleles of the core genes present in the strains are identical (same HC0|cluster), but the strains differed in the presence/absence of some core genes (cgST different). Here, an epidemiological link could be probable, despite different birth and breeding areas.
4. Discussion
Foodborne illnesses are a major public health concern and result in considerable economic burden. Salmonella has the ability to adapt to a variety of animal hosts, and to humans, and can be transmitted through contaminated food such as eggs, meat, raw vegetables, or through water [29]. More specifically, bovine salmonellosis is responsible for public and animal health problems and serious economic losses due to high mortality, and is often caused by Salmonella Dublin [11,30].
The prevalence of Salmonella in cattle found in this study (3%) is consistent with the data found in the literature for Europe, where asymptomatic carriage of Salmonella in cattle is generally less than 5% based on fecal samples [8,31]. It is possible that the incidence of salmonellosis was underestimated, since fecal samples are not necessarily the most sensitive source to detect the presence of Salmonella, and some positive cattle may have been missed by the detection method used [8,17,32]. In addition, prevalence may be underestimated because the animals sampled at the slaughterhouse are healthy, which does not take into account symptomatic cattle carrying Salmonella. The prevalence of Salmonella in apparently healthy cattle is of significant concern to public health [17], especially that they are able to carry more than 700 CFU per gram of Salmonella, as shown in this study, and thus may cross-contaminate the meat products during processing.
In this investigation, neither the age of animals (calf: 3.4%; adult: 2.7%) nor the type of cattle (dairy cow: 3.7%; beef cow: 2.2%) were discriminating factors for Salmonella carriage, despite some studies showing the opposite [32]. It was also reported that the prevalence of Salmonella in animal and environmental samples varied among seasons, with an increase in the presence of Salmonella on dairy farms when the seasonal temperature increased [33]. This intensification was even observed during the period from August to October in an Irish slaughterhouse [8].
Within the 29 of 959 samples collected that were positive for Salmonella enterica, eight different serotypes were isolated. Salmonella Montevideo was identified as the most prevalent, followed by S. Mbandaka and S. Anatum. Several studies have highlighted the presence of theses serotypes, and particularly S. Montevideo as the predominant serotype in cattle [12,13,14,34]. Although S. Montevideo and S. Dublin are the most frequently reported serotypes in North America and Europe, no S. Dublin was found in this investigation [8,31,35,36]. These results can be explained by the possible presence of symptoms in animals infected with S. Dublin, which would have resulted in the eviction of these animals from slaughter. Nevertheless, other serotypes such as S. Virchow and S. Typhimurium monophasic variant, commonly found in cattle, were also isolated [31]. These results also show variability in the serotypes of Salmonella in cattle production between calves and adult cattle.
The geographic distribution of breeding areas did not influence Salmonella carriage. However, PFGE and WGS genotyping highlighted genetic similarities, suggesting potential epidemiological links or cross-contaminations between animals. The available metadata were not sufficiently detailed to identify the pathways of contamination of the cattle by Salmonella. Nevertheless, this investigation made it possible to assess whether the contamination occurred at the place of birth of the animals, or at the breeding farm, and to put forward the hypothesis of possible contamination during transport or at the slaughterhouse (when no links between the strains were found). As an example, certain strains of S. Anatum presented strong evidence of an epidemiological link between isolates originating from different areas of birth and breeding. Proximity between the areas might suggest possible circulation of the same S. Anatum strain throughout these areas of France. However, in the case of S. Virchow, contamination appeared to have taken place on the farm, at the breeding step, since the areas of birth of the calves were different. Linking exhaustive epidemiological data and WGS genotyping would allow the establishment of reliable epidemiological links, which are needed to understand Salmonella contamination in cattle production.
5. Conclusions
This investigation made it possible, for the first time, to evaluate the intestinal carriage of Salmonella by cattle in France and to identify S. Montevideo as the most common serotype. The use of WGS to genotype strains enabled the identification of possible epidemiological links among the Salmonella strains in cattle. The knowledge gained in this investigation about the prevalence and diversity of Salmonella serotypes will help to improve understanding about the dissemination of Salmonella in French cattle production, to adapt the current control measures, and to prevent public health problems.
Acknowledgments
The authors would like to thank the slaughterhouse in which the survey was conducted for having accepted to host this study and all the people who helped during sampling.
Author Contributions
L.B. (Laetitia Bonifait), A.T., and M.C. contributed to the conceptualization and methodology of this study. All authors carried out sampling. L.B. (Laetitia Bonifait), A.T., L.B. (Louise Baugé), F.L.G. and S.R. carried out bacteriological analysis, analyzed data, and performed typing experiments. L.B. (Laetitia Bonifait), A.T. and M.C. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the French National Reference Laboratory for Salmonella, Anses, Ploufragan site.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Genome assemblies of Salmonella isolates sequenced in this study are publicly available from the Salmonella database on EnteroBase platform (http://enterobase.warwick.ac.uk/, accessed on 16 April 2021), their accession numbers (barcode) are listed in the paper.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.EFSA-ECDC The European Union One Health 2019 Zoonoses Report. EFSA J. 2021:6406. doi: 10.2903/j.efsa.2021.6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO European Region The burden of foodborne diseases in the WHO european Region. WHO J. 2017:1–36. [Google Scholar]
- 3.Havelaar A.H., Kirk M.D., Torgerson P.R., Gibb H.J., Hald T., Lake R.J., Praet N., Bellinger D.C., de Silva N.R., Gargouri N., et al. World Health Organization Global Estimates and Regional Comparisons of the Burden of Foodborne Disease in 2010. PLoS Med. 2015;12:e1001923. doi: 10.1371/journal.pmed.1001923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO . Estimates of the Global Burden of Foodborne Diseases. WHO; Geneva, Switzerland: 2015. p. 255. [Google Scholar]
- 5.Uzzau S., Brown D.J., Wallis T., Rubino S., Leori G., Bernard S., Casadesus J., Platt D.J., Olsen J.E. Host adapted serotypes of Salmonella enterica. Epidemiol. Infect. 2000;125:229–255. doi: 10.1017/S0950268899004379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hughes L.E., Gibson E.A., Roberts H.E., Davies E.T., Davies G., Sojka W.J. Bovine salmonellosis in England and Wales. Br. Vet. J. 1971;127:225–238. doi: 10.1016/S0007-1935(17)37588-7. [DOI] [PubMed] [Google Scholar]
- 7.Salaheen S., Sonnier J., Kim S.W., Haley B.J., van Kessel J.A.S. Interaction of Salmonella enterica with Bovine Epithelial Cells Demonstrates Serovar-Specific Association and Invasion Patterns. Foodborne Pathog. Dis. 2020 doi: 10.1089/fpd.2019.2765. [DOI] [PubMed] [Google Scholar]
- 8.McEvoy J.M., Doherty A.M., Sheridan J.J., Blair I.S., McDowell D.A. The prevalence of Salmonella spp. in bovine faecal, rumen and carcass samples at a commercial abattoir. J. Appl. Microbiol. 2003;94:693–700. doi: 10.1046/j.1365-2672.2003.01898.x. [DOI] [PubMed] [Google Scholar]
- 9.Fossler C.P., Wells S.J., Kaneene J.B., Ruegg P.L., Warnick L.D., Bender J.B., Godden S.M., Halbert L.W., Campbell A.M., Zwald A.M. Prevalence of Salmonella spp on conventional and organic dairy farms. J. Am. Vet. Med. Assoc. 2004;225:567–573. doi: 10.2460/javma.2004.225.567. [DOI] [PubMed] [Google Scholar]
- 10.Nielsen L.R. Review of pathogenesis and diagnostic methods of immediate relevance for epidemiology and control of Salmonella Dublin in cattle. Vet. Microbiol. 2013;162:1–9. doi: 10.1016/j.vetmic.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 11.Guizelini C.C., Tutija J.F., Morais D.R., Bacha F.B., Ramos C.A.N., Leal C.R.B., Zaquetti M.E., Lemos R.A.A. Outbreak investigation of septicemic salmonellosis in calves. J. Infect. Dev. Ctries. 2020;14:104–108. doi: 10.3855/jidc.12087. [DOI] [PubMed] [Google Scholar]
- 12.Gutema F.D., Agga G.E., Abdi R.D., de Zutter L., Duchateau L., Gabriel S. Prevalence and Serotype Diversity of Salmonella in Apparently Healthy Cattle: Systematic Review and Meta-Analysis of Published Studies, 2000–2017. Front. Vet. Sci. 2019;6:102. doi: 10.3389/fvets.2019.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutema F.D., Agga G.E., Abdi R.D., de Zutter L., Duchateau L., Gabriel S. Corrigendum: Prevalence and Serotype Diversity of Salmonella in Apparently Healthy Cattle: Systematic Review and Meta-Analysis of Published Studies, 2000–2017. Front. Vet. Sci. 2019;6:184. doi: 10.3389/fvets.2019.00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Callaway T.R., Keen J.E., Edrington T.S., Baumgard L.H., Spicer L., Fonda E.S., Griswold K.E., Overton T.R., VanAmburgh M.E., Anderson R.C., et al. Fecal prevalence and diversity of Salmonella species in lactating dairy cattle in four states. J. Dairy Sci. 2005;88:3603–3608. doi: 10.3168/jds.S0022-0302(05)73045-9. [DOI] [PubMed] [Google Scholar]
- 15.Thomas K.M., de Glanville W.A., Barker G.C., Benschop J., Buza J.J., Cleaveland S., Davis M.A., French N.P., Mmbaga B.T., Prinsen G., et al. Prevalence of Campylobacter and Salmonella in African food animals and meat: A systematic review and meta-analysis. Int. J. Food Microbiol. 2020;315:108382. doi: 10.1016/j.ijfoodmicro.2019.108382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halimi H.A., Seifi H.A., Rad M. Bovine salmonellosis in northeast of Iran: Frequency, genetic fingerprinting and antimicrobial resistance patterns of Salmonella spp. Asian Pac. J. Trop. Biomed. 2014;4:1–7. doi: 10.1016/S2221-1691(14)60199-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cetin E., Temelli S., Eyigor A. Salmonella prevalence and serovar distribution in healthy slaughter sheep and cattle determined by ISO 6579 and VIDAS UP Salmonella methods. J. Food Sci. Technol. 2019;56:5317–5325. doi: 10.1007/s13197-019-04002-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Kessel J.A., Karns J.S., Wolfgang D.R., Hovingh E., Schukken Y.H. Dynamics of Salmonella serotype shifts in an endemically infected dairy herd. Foodborne Pathog. Dis. 2012;9:319–324. doi: 10.1089/fpd.2011.1054. [DOI] [PubMed] [Google Scholar]
- 19.Afnor NF EN ISO 6579, Horizontal Method for the Detection of Salmonella spp. [(accessed on 16 April 2021)];2002 Available online: https://www.boutique.afnor.org/
- 20.Afnor N.F. U 47-100, Méthodes D’analyse en Santé Animale—Recherche par L’isolement et Identification de tout Sérovar ou de Sérovar(s) Spécifié(s) de Salmonelles dans L’environnement des Productions Animales. Decitre; Lyon, France: 2007. [Google Scholar]
- 21.Grimont P.A., Weill F.X. Antigenic Formulae of the Salmonella Serovars. [(accessed on 16 April 2021)];WHO Collab. Center Ref. Res. Salmonella. 2007 9:1–167. Available online: https://www.pasteur.fr/sites/default/files/veng_0.pdf. [Google Scholar]
- 22.Afnor N.F. EN ISO 6579-2, Microbiology of Food Animal Feed—Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella—Part 2: Enumeration by a Miniaturized Most Probable Number Technique, XP CEN ISO/TS 6579-2. [(accessed on 16 April 2021)];2013 Available online: https://www.boutique.afnor.org/
- 23.Ribot E.M., Fair M.A., Gautom R., Cameron D.N., Hunter S.B., Swaminathan B., Barrett T.J. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 2006;3:59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- 24.Kerouanton A., Marault M., Lailler R., Weill F.X., Feurer C., Espie E., Brisabois A. Pulsed-field gel electrophoresis subtyping database for foodborne Salmonella enterica serotype discrimination. Foodborne Pathog. Dis. 2007;4:293–303. doi: 10.1089/fpd.2007.0090. [DOI] [PubMed] [Google Scholar]
- 25.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alikhan N.F., Zhou Z., Sergeant M.J., Achtman M. A genomic overview of the population structure of Salmonella. PLoS Genet. 2018;14:e1007261. doi: 10.1371/journal.pgen.1007261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Z., Alikhan N.F., Sergeant M.J., Luhmann N., Vaz C., Francisco A.P., Carrico J.A., Achtman M. GrapeTree: Visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018;28:1395–1404. doi: 10.1101/gr.232397.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simonsen M., Mailund T., Pedersen C.N.S. Rapid Neighbour-Joining; Proceedings of the 8th International Workshop on Algorithms in Bioinformatics (WABI 2008); Karlsruhe, Germany. 15–19 September 2008; pp. 113–122. [Google Scholar]
- 29.Majowicz S.E., Musto J., Scallan E., Angulo F.J., Kirk M., O’Brien S.J., Jones T.F., Fazil A., Hoekstra R.M. The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 2010;50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 30.Costa L.F., Paixao T.A., Tsolis R.M., Baumler A.J., Santos R.L. Salmonellosis in cattle: Advantages of being an experimental model. Res. Vet. Sci. 2012;93:1–6. doi: 10.1016/j.rvsc.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 31.Ketema L., Ketema Z., Kiflu B., Alemayehu H., Terefe Y., Ibrahim M., Eguale T. Prevalence and antimicrobial susceptibility profile of Salmonella serovars isolated from slaughtered cattle in Addis Ababa, Ethiopia. BioMed Res. Int. 2018;2018:9794869. doi: 10.1155/2018/9794869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cummings K.J., Warnick L.D., Alexander K.A., Cripps C.J., Grohn Y.T., McDonough P.L., Nydam D.V., Reed K.E. The incidence of salmonellosis among dairy herds in the Northeastern United States. J. Dairy Sci. 2009;92:3766–3774. doi: 10.3168/jds.2009-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pangloli P., Dje Y., Ahmed O., Doane C.A., Oliver S.P., Draughon F.A. Seasonal incidence and molecular characterization of Salmonella from dairy cows, calves, and farm environment. Foodborne Pathog. Dis. 2008;5:87–96. doi: 10.1089/fpd.2007.0048. [DOI] [PubMed] [Google Scholar]
- 34.Blau D.M., McCluskey B.J., Ladely S.R., Dargatz D.A., Fedorka-Cray P.J., Ferris K.E., Headrick M.L. Salmonella in dairy operations in the United States: Prevalence and antimicrobial drug susceptibility. J. Food Prot. 2005;68:696–702. doi: 10.4315/0362-028X-68.4.696. [DOI] [PubMed] [Google Scholar]
- 35.Harvey R.R., Friedman C.R., Crim S.M., Judd M., Barrett K.A., Tolar B., Folster J.P., Griffin P.M., Brown A.C. Epidemiology of Salmonella enterica Serotype Dublin Infections among Humans, United States, 1968–2013. Emerg. Infect. Dis. 2017;23:9. doi: 10.3201/eid2309.170136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ung A., Baidjoe A.Y., Van Cauteren D., Fawal N., Fabre L., Guerrisi C., Danis K., Morand A., Donguy M.P., Lucas E., et al. Disentangling a complex nationwide Salmonella Dublin outbreak associated with raw-milk cheese consumption, France, 2015 to 2016. Eur. Surveill. 2019;24:3. doi: 10.2807/1560-7917.ES.2019.24.3.1700703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Genome assemblies of Salmonella isolates sequenced in this study are publicly available from the Salmonella database on EnteroBase platform (http://enterobase.warwick.ac.uk/, accessed on 16 April 2021), their accession numbers (barcode) are listed in the paper.