Abstract
Enhancers are important noncoding elements, but they have been traditionally hard to characterize experimentally. The development of massively parallel assays allows the characterization of large numbers of enhancers for the first time. Here, we developed a framework using Drosophila STARR-seq to create shape-matching filters based on meta-profiles of epigenetic features. We integrated these features with supervised machine-learning algorithms to predict enhancers. We further demonstrated our model could be transferred to predict enhancers in mammals. We comprehensively validated the predictions using a combination of in vivo and in vitro approaches, involving transgenic assays in mouse and transduction-based reporter assays in human cell lines (153 enhancers in total). The results confirmed our model can accurately predict enhancers in different species without re-parameterization. Finally, we examined the transcription-factor binding patterns at predicted enhancers versus promoters. We demonstrated that these patterns enable the construction of a secondary model effectively discriminating between enhancers and promoters.
Introduction
Enhancers are gene regulatory elements that activate expression of target genes from a distance [1]. The vast majority of enhancers and their spatiotemporal activities remain unknown [2, 3]. Understanding enhancer function and evolution is currently an area of great interest because many variants within distal regulatory elements also have been associated with various traits and diseases during genome-wide association studies [4–6]. Traditionally, regulatory activities of enhancers were experimentally validated using heterologous reporter constructs, leading to a relatively small number of enhancers that are functionally validated in several selected mammalian cell types [7, 8]. These validated enhancers were typically in conserved noncoding regions [9, 10] with particular patterns of chromatin [11], transcription factor (TF) binding [12], or noncoding transcription [13]. When complex computational methods for predicting tissue/cell line-specific enhancers were trained on these validated enhancers, they could be susceptible to potential biases and were difficult to generalize to other tissues or species as the training data were usually not large enough. Some published methods also trained their model based on TF binding sites [12, 14–16]. The TF binding sites provide a larger dataset for training. However, most enhancers do not bind to one or a small group of TFs. In addition, it has remained challenging to assess the performance of different methods for enhancer prediction with a limited number of putative enhancers being validated.
The development of the self-transcribing active regulatory region sequencing (STARR-seq) makes it possible to quantitatively assess the activity of millions of candidate enhancers across entire genomes [17]. In these experiments, plasmids each containing a potential enhancer element downstream of a green fluorescent protein (GFP) gene are transfected into target cells. The differences in the activities of the tested regions are reflected by quantifying the levels of the resulting reporter transcripts through sequencing. STARR-seq confirmed previous findings that active enhancers and promoters are usually located at open chromatin regions where various TFs and cofactors bind [18–20]. In addition, it confirmed that the regulatory regions are often flanked by nucleosomes that contain histone proteins with certain characteristic post-translational modifications, like acetylation on H3K27 (H3K27ac) [21]. These attributes lead to an enriched peak-trough-peak (“double peak”) signal, which has been observed in previous studies [22].
We developed a method to take into account the specific enhancer-associated pattern within different epigenetic signals. Previous ENCODE and modENCODE efforts showed that the chromatin modifications on active promoters and enhancers are conserved across higher eukaryotes [23–29]. We further explored this conservation of epigenetic signal shapes for constructing simple-to-use transferrable statistical models using six epigenetic marks to predict enhancers and promoters in diverse eukaryotic species including fly, mouse, and human.
Working across organisms also allowed us to take advantage of different assays to validate our predictions in a robust fashion using multiple experimental approaches. In the first stage, we predicted enhancers in six different embryonic mouse tissues and tested the activity of these predictions in vivo with transgenic mouse assays. We then proceeded to test the activity of these elements in human cell lines in vitro, e.g. H1 human embryonic stem cells (H1-hESCs), an extensively studied and well-characterized cell line. We showed that the enhancer predictions from our transferrable model are comparable to the prediction accuracy of species-specific models.
Results
Aggregation of epigenetic signals in Drosophila to create metaprofiles
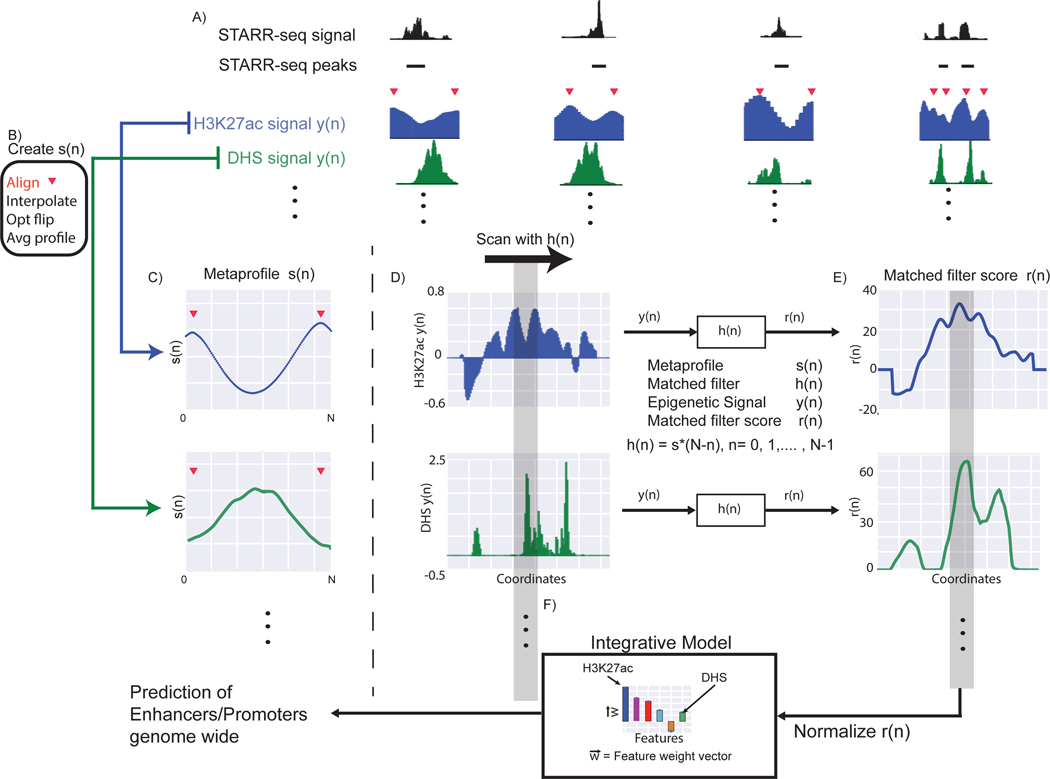
We developed a framework to predict active regulatory elements using the epigenetic signal patterns associated with experimentally validated promoters and enhancers (Fig. 1). The STARR-seq studies on Drosophila cell lines provide the most comprehensive datasets as they were performed genome-wide and performed with multiple core promoters [17, 30]. These peaks typically consist of a mixture of enhancers and promoters. At this stage we did not differentiate between the two sets of regulatory elements. As STARR-seq quantifies enhancer activity in an episomal fashion, not all peaks would be active in the native chromatin environment. Arnold and colleagues showed that the STARR-seq peaks that occur with enriched DNase hypersensitivity or H3K27ac modifications tend to be associated with active genes, whereas other STARR-seq peaks tend to be associated with enrichment of repressive marks such as H3K27me3 [17]. Hence, we took the overlap of the STARR-seq enhancers with H3K27ac and/or DHS peaks to get a high confidence set of enhancers that are active in vivo, based on which we create the representative metaprofiles for each histone modification and DNase signal respectively. During aggregation, we first aligned the two maxima in the H3K27ac signal across active STARR-seq peaks, followed by interpolation of the signal before calculating the average to generate the metaprofile. Then we calculated the dependent metaprofiles for other histone marks following the same procedure (Fig. 1).
Figure 1: Flowchart of the Matched-filter model.
A) We identified the “double peak” pattern in the H3K27ac signal close to STARR-seq peaks. The red triangles denote the position of the two maxima in the double peak. B) We aggregated the H3K27ac signal around these regions after aligning the flanking maxima, using interpolation and smoothing on the H3K27ac signal, and averaged the signal across different STARR-seq peaks to create the metaprofile in C). The same operations were performed on other histone signals and DHS to create metaprofiles in other dependent epigenetic signals. D) Matched filters were used to scan the histone and/or DHS datasets to identify the occurrence of the corresponding pattern in the genome. E) The matched filter scores are high in regions where the profile occurs (grey region shows an example) but low when only noise is present in the data. The individual matched filter scores from different epigenetic datasets were combined using integrated model in F) to predict active promoters and enhancers in a genome-wide fashion.
Match of a metaprofile is predictive of regulatory activity
To calculate the matched filter scores, we first smoothened the input signal track for each epigenetic mark. Then we scanned the H3K27ac signal track to find each pair of local maximum points that are between 300 and 1,100 basepairs. Due to the variability of the distance between the double peaks, we interpolate each double peak region before convolving it with the filter to get an initial score (Supplementary Fig. 1). If there are multiple overlapping double peak regions, we used the highest score within a 1,500 bp region as the prediction for the regulatory potential. We then calculate the matched filter scores for other epigenetic marks based on those same double peak regions (See Online methods).
We calculated the matched filter score for all 30 epigenetic modification signals available in the Drosophila cell lines on STARR-seq peaks and a negative control set (Supplementary Fig. 2). The negatives are randomly chosen non-STARR-seq-peak regions in the genome that had the same length distribution as the enhancers from the STARR-seq (see Model assessment in Online methods). Interestingly, the distribution of matched filter scores for STARR-seq peaks are unimodal for each histone mark except for H3K4me1, H3K4me3, and H2Av, which are bimodal. We looked at the degree to which the matched filter scores for promoters and enhancers are higher than the matched filter scores for the rest of the genome (Supplementary Fig. 2), as this is a measure of the signal-to-noise ratio for regulatory region prediction. We observed that the H3K27ac matched filter score is the most accurate feature for predicting active regulatory regions identified using STARR-seq (Supplementary Table 1), consistent with previous studies [21, 31, 32]. In addition, several histone acetylations marks, as well as H1, H3K4 methylations, and DHS were the most accurate prediction features, whereas other histone marks like H3K79m1 and H4K20me1 were not well suited as their matched filter scores for positive regions and negative regions were not distinguishable.
To quantitatively evaluate whether the occurrence of the epigenetic metaprofiles could be used to predict active enhancers and promoters, we did a ten-fold cross validation assessing the average areas under the receiver operating characteristic (ROC) (AUROC) and the areas under the precision-recall (PR) (AUPR) curves. Comparing the matched filter result with the peak calling result, we found that the AUROC and AUPR of the matched filter scores for different histone modifications were higher than those of the peaks of corresponding histone marks (Fig. 2), suggesting that the matched filter score is more accurate for predicting active STARR-seq peaks than the simple enrichment of the signals.
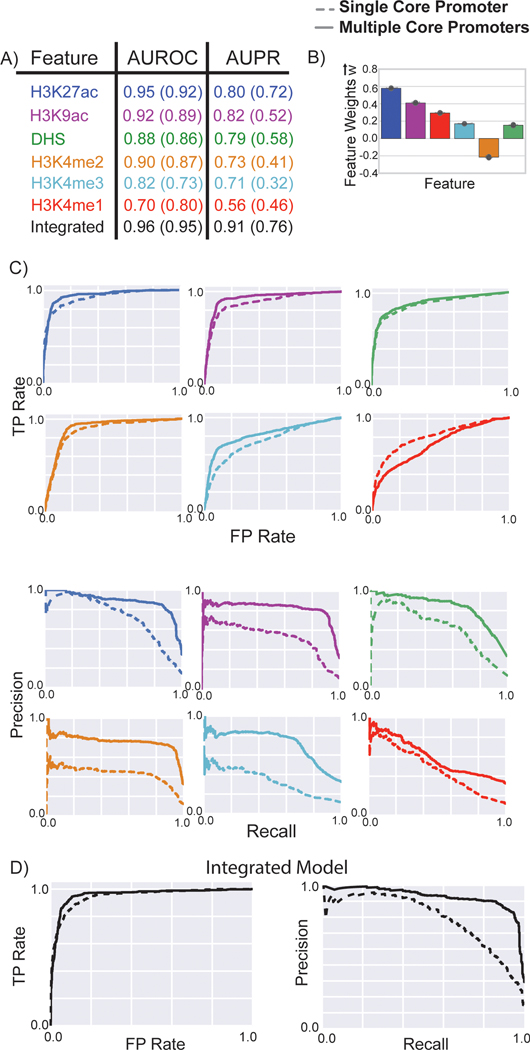
Figure 2: Performance of matched filters and integrated models for predicting STARR-seq peaks, comparing to peak-based models.
The performance of the matched filters of different epigenetic marks and the integrated model for predicting all STARR-seq peaks was compared using ten-fold cross validation. A) The area under the receiver-operating characteristic (AUROC) and the precision-recall (AUPR) curves were used to measure the accuracy of different matched filters and the integrated model. B) Weights of the different features in the integrated model are plotted; the mean value is displayed in the bar plot while the error bars show the standard deviation of feature weights measured by ten-fold cross validation. These weights may be used as a proxy for the importance of each feature in the integrated model. C-D) The individual ROC and PR curves for each matched filter and the integrated model are shown. The performance of these features and the integrated model for predicting the STARR-seq peaks using multiple core promoters and a single core promoter were compared to the performance of peak-based models. The colored numbers within the parentheses in A) refer to the AUROC and AUPR for predicting the peaks using a single STARR-seq core promoter; the colored numbers outside the parentheses refer to the performance of the model for predicting peaks from multiple core promoters; the gray numbers in the parentheses refer to the performance of the peak-based models.
Integration of matched filter scores of multiple epigenetic features
We first combined the matched filter scores from all 30 measured histone marks along with the DHS in statistical models such as random forest and SVM (Supplementary Fig. 3). We evaluated the performance of the integrated model using ten-fold cross validation. For each fold of validation, 90% of the positives and negatives were used to build metaprofiles for each epigenetic marks, generate matched filter scores, and train the integrative model. The remaining 10% data were used to test model accuracy. The integrated models with 30 epigenetic features displayed high accuracy (Average AUROC=0.97 and AUPR=0.93 for SVM model with multiple core promoters). We obtained the feature coefficients or GINI score of each epigenetic mark from the integrated models.
We then built an integrated model with combined matched filter scores of six commonly available and discriminative epigenetic marks (H3K27ac, H3K4me1, H3K4me2, H3K4me3, H3K9ac, and DHS) associated with active regulatory regions using a linear support vector machine (SVM) [33]. The selection of these six features was based on their matched filter score performance, their importance in the integrated model, and data availability (See Feature Selection in Online methods). We then assessed the performances of different statistical approaches including random forest, ridge regression and Naïve Bayes and SVM to combine the features. While all these approaches performed similarly (Supplementary Fig. 4), we used a linear SVM in our framework because its performances are most stable in cross validations.
We found that the simplified SVM model had a high performance similar to that of the full SVM model using all 30 epigenetic marks, with an AUROC of 0.96 (0.97 in the full model) and an AUPR of 0.91 (0.93 in the full model). We also trained an SVM model using all STARR-seq peaks (including those with no DHS and H3K27ac signals) with the same six features. We found that H3K27ac still had the highest GINI score in random forest, albeit a slightly smaller coefficient in SVM (See supplementary material). In general, the integrated model trained on the six features achieved good performance upon cross-validation, and this set of input features allowed the integrated model to be applied to a variety of cell lines and tissues, as many relevant ChIP-seq and DNase experiments have been performed by the Roadmap Epigenomics Mapping [34] and the ENCODE [35] Consortia in a wide variety of samples.
Distinct epigenetic signals associated with promoters and enhancers
We proceeded to create individual metaprofiles and machine learning models for the two classes of regulatory activators – promoters (or proximal) and enhancers (or distal). We assessed the performance of the matched filters for predicting active regulatory regions within each category (Fig. 3). We also combined the peaks identified from multiple STARR-seq experiments of S2 cells and reassessed the performance of the matched filters at predicting promoters and enhancers, respectively. Merging the STARR-seq peaks from multiple core promoters led to higher AUROC and AUPR for the matched filters of most histone marks (Supplementary Table 2). The highest matched filter scores were typically observed on promoters, and the matched filters for each of the six features tended to perform better for promoter prediction. Similar to previous studies [36, 37], we observed that the H3K4me1 metaprofile was very predictive for enhancers but was close to random for predicting promoters. In contrast, the H3K4me3 metaprofile could be utilized to predict promoters and not enhancers. The histogram for matched filter scores showed that the H3K4me1 matched filter score was higher near enhancers while the H3K4me3 matched filter score tended to be higher near promoters. The mixture of these two populations led to bimodal distributions for H3K4me1 and H3K4me3 matched filter scores when calculated over all regulatory regions (Supplementary Fig. 2).
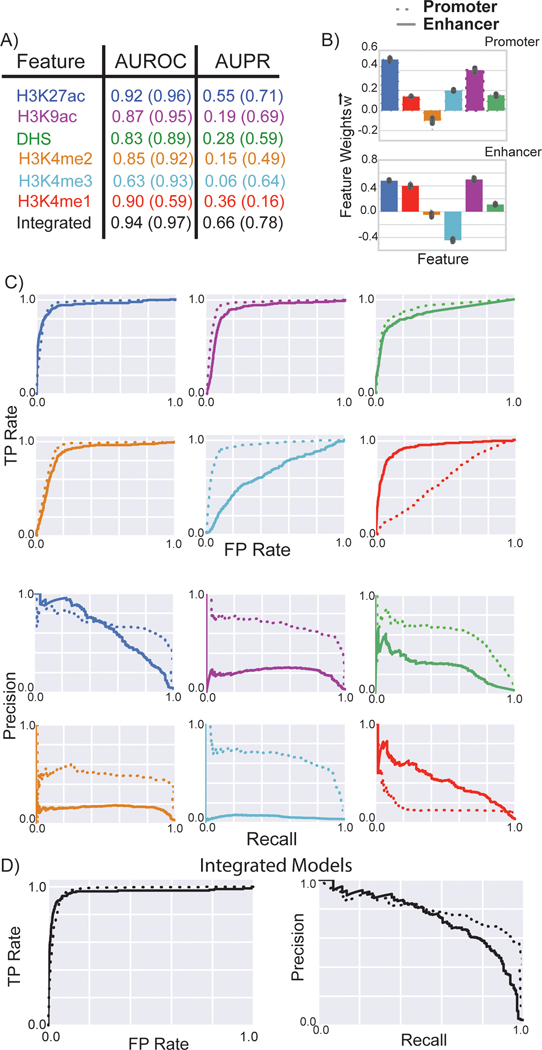
Figure 3: Performance of matched filters and integrated models for predicting promoters and enhancers.
The performance of the matched filters of different epigenetic marks and the integrated model for predicting active promoters and enhancers were compared using ten-fold cross validation. A) The numbers within parentheses refer to the AUROC and AUPR for predicting promoters; the numbers outside the parentheses refer the performance of the models for predicting enhancers. B) Weights of the different features in the integrated models for promoter and enhancer prediction are plotted; the mean value is displayed in the bar plot while the error bars show the standard deviation of feature weights measured by ten-fold cross validation. C-D) The ROC and PR curves for each matched filter and the integrated model are shown. The performance of these features and the integrated model for predicting the active promoters and enhancers using multiple core promoters were compared.
We again trained different statistical models to learn the combination of features associated with promoters and enhancers respectively. These integrated models outperformed the individual matched filters at predicting active enhancers and promoters (Fig. 3 and Supplementary Fig. 5). In addition, the weights of the individual features identified the difference in the roles of H3K4me1 and H3K4me3 matched filter scores at discriminating active promoters and enhancers from inactive regions in the genome. The trained promoter-specific model has a high weight for H3K4me3, which is considered as a marker for promoters from previous studies [31], but a lower weight for H3K4me1, which is considered as a marker for enhancers [31]. This is reversed in the enhancer-specific model, indicating the unique features captured for different identification task. (See supplementary material). We also created two integrated models utilizing matched filter scores of all 30 histone marks as features for predicting enhancers and promoters. The additional histone marks provided independent information regarding the activity of promoters and enhancers as these features increased the accuracy of these models (Supplementary Fig. 6).
Application of the STARR-seq model to predict enhancers in mammalian species
One of the important findings of previous ENCODE and model organism ENCODE efforts was the conserved patterns of chromatin marks close to regulatory elements across hundreds of millions of years of evolution [23–29]. The relationship of chromatin marks to gene expression was very similar, for instance, in worms, flies, mice and humans. Therefore, one could build a statistical model relating chromatin modification to gene expression that would work without re-parameterization across different organisms. This motivated us to transfer our well-parameterized model based on the STARR-seq data from flies to mammalian systems, eg. mouse and human, and test our model performance.
We started by making genome-wide predictions of regulatory regions in mouse. Predictions were made in six different tissues (forebrain, midbrain, hindbrain, limb, heart and neural tube) at the embryonic day 11.5 (e11.5) stage (Predictions are available through our website at http://matchedfilter.gersteinlab.org). Using our model, we predicted 31K to 39K regulatory regions in individual tissues in mouse, with each region ranging from 300bp to 1,100bp. Similarly, we performed a genome-wide prediction of regulatory regions in the ENCODE top-tier human cell lines, including H1-hESC, GM12878, K562, HepG2, A549 and MCF-7. In H1-hESC, for example, we predicted 43,463 active regulatory regions, of which 22,828 (52.5%) were within 2kb of the TSS and were labeled as promoters. Most of the predicted regulatory regions were also present near active genes (Figure S7).
Validation in vivo in mouse
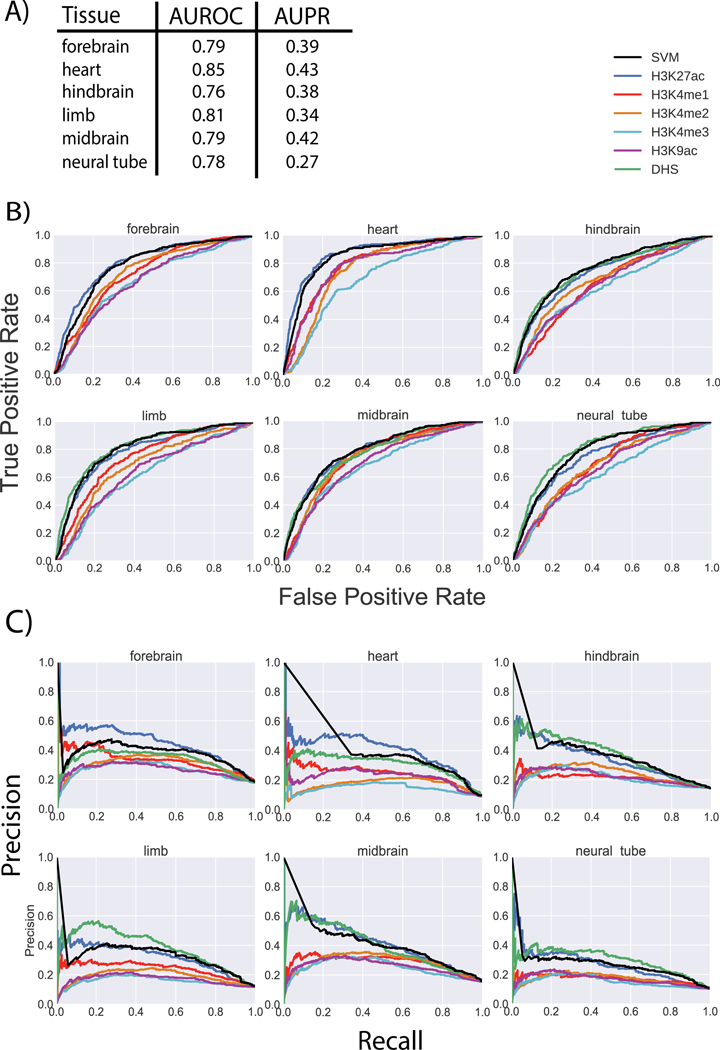
To test the activity of predicted mouse enhancers in vivo, we performed transgenic mouse enhancer assays (Supplementary Fig. 8) in e11.5 mice for 133 regions, including 102 regions selected based on the H3K27ac signals rank of the corresponding mouse tissues and another 31 regions selected by an ensemble approach from human homolog sequences (Supplementary Table 3–8). In addition, we included other published transgenic mouse experiments from the VISTA database for validation. In total, we had 1,253 positive regions and 8,631 negative regions pulling together from different tissues. This large set of validated enhancers allowed us to comprehensively evaluate the predictability of the matched filter scores of each epigenetic mark, as well as the integrated SVM model (Fig. 4). On average, the integrated model trained with Drosophila STARR-seq data achieved an AUROC of 0.80. We also did a similar evaluation with publicly available FIREWACh assay data [38] in mouse, and the result was consistent (Supplementary Fig. 9). For comparison, we trained an integrated model based directly on the validated mouse enhancers. We observed a similar prediction accuracy upon cross validation (see Supplementary Fig. 10 and supplemental materials).
Figure 4: Performance of matched filters and integrated model for predicting active enhancers in mice.
The performance of the Drosophila STARR-seq-based matched filters and the integrated model for predicting active enhancers identified by transgenic mouse enhancer assays in six different tissues of e11.5 mice. A) The AUROC and AUPR are shown for the integrated SVM model in six tissues. The weights of the different features in the integrated model are the same as the weights shown in Figure 3 for enhancers. B) The individual ROC curves of each feature and the integrated SVM model for each tissue are shown. C) The individual PR curves of each feature and the integrated SVM model for each tissue are shown.
Validation in human cell lines
We proceeded to validate our STARR-seq based model for predicting human enhancers using a cell line-based transduction assay. We randomly selected 20 predicted intergenic enhancers for validation. Insertion of 11 of the putative enhancers into the HIV vector resulted in a significant increase in eGFP expression (P-value < 0.05 for both directions) in H1-hESCs (Supplementary Table 9, extended data table). The positive enhancers displayed a significant increase in gene expressions in both orientation. In contrast, the negatives displayed much lower levels of gene expression (Supplementary Fig. 11). The activity of these tested enhancers also showed cell type specificity. More than half of the predicted enhancers show activity in H1-hESC (Supplementary Fig. 12), but less in A549 and TZM-bl, which are derived from tumor cells (See Supplementary table 9 and Supplementary material). Overall, 16 of the 20 tested predictions displayed a statistically significant increase in gene expression of the reporter gene in at least one of the cell lines. Given the promoter specificity of enhancers in such assays, we anticipate that some of the elements that could not be validated in this particular vector would function as enhancers in a more natural biological context (e.g., with the cognate promoter or in the absence of surrounding HIV vector sequences).
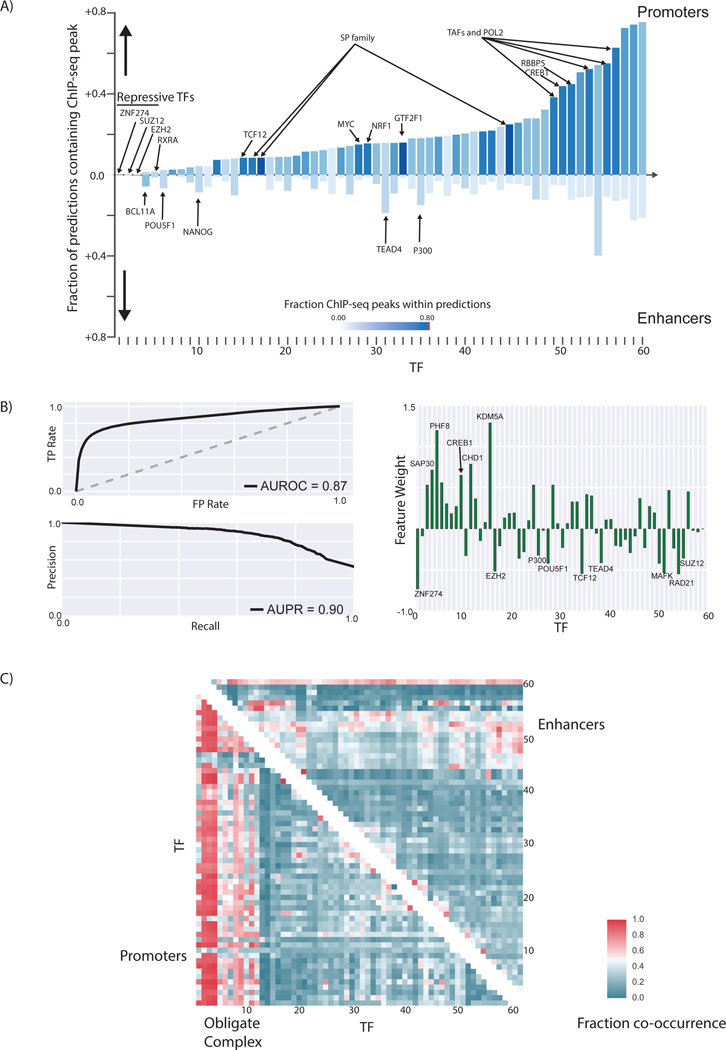
TFs exhibit different binding patterns at enhancers and promoters
We further studied the differences in TF binding at promoters and enhancers (Fig. 5) We focused on the human H1-hESC cell line as there is large amount of functional genomic assays from the ENCODE [35] and Roadmap Epigenomics Mapping Consortium [34] of this cell line. Together, the consortia have generated ChIP-Seq data for 60 transcription-related factors in the H1-hESC cell line, including a few chromatin remodelers and histone modification enzymes. Collectively, we call these transcription-related factors “TFs” for simplicity.
Figure 5: Differences in TF binding patterns at enhancers and promoters.
A) The fraction of predicted promoters and enhancers that overlap with ENCODE ChIP-seq peaks for different TFs in H1-hESC are shown. The names of all TFs in the figure can be viewed in Figure S35. B) The AUROC and AUPR for a logistic regression model created using the pattern of TF binding at each regulatory region to distinguish enhancers from promoters are shown. The weight of each feature in the logistic regression model could be used to identify the most important TFs that distinguish enhancers from promoters. C) The patterns of TF co-binding at active promoters and enhancers are shown. The TFs co-occur at promoters regions tend to form obligate complexes. The names of all the TFs in this graph can be viewed in Figure S36.
We showed that the patterns of TFs binding within regulatory regions could be utilized in a logistic regression model to distinguish active enhancers from promoters with high accuracy (AUPR = 0.90, AUROC = 0.87) (Fig. 5). We were also able to identify the most important features that distinguish promoters from enhancers. In addition to TATA box-associated factors such as TAF1, TAF7, and TBP, the RNA polymerase-II binding patterns as well as chromatin remodelers such as KDM5A and PHF8 are some of the most important factors that distinguish promoters from enhancers in H1-hESCs. This provides a framework that can be utilized to identify the most important TFs associated with active enhancers and promoters in each cell type.
We found that although most promoters and enhancers contain multiple TF binding sites, the pattern of TF binding at promoters was different from that at enhancers and that TF binding at enhancers displayed more heterogeneity: more than 70% of the promoters bound to the same set of 2–3 sequence-specific TFs, which was not observed for enhancers (Fig. 5c). The majority of the promoters contained peaks for several TATA-associated factors (TAF1, TAF7, and TBP). These TF co-associations could lead to mechanistic insights of cooperativity between TFs. Similarly, CTCF and ZNF143 may function cooperatively as they are observed to co-occur frequently at distal regulatory regions, consistent with the previous report [39].
Discussion
In this study, we developed a framework using transferable supervised machine learning models trained on regulatory regions identified by STARR-seq to accurately predict active enhancers in a cell-type-specific manner. The rich amount of whole-genome STARR-seq experiments established the characteristic pattern flanking active regulatory regions within certain histone modifications [17]. This motivated us to train a shape-matching and filtering model that could be used to identify these patterns in the ChIP-seq signals. As the chromatin marks and epigenetic profiles associated with active regulatory regions are highly conserved among organisms [23–29], we showed that a well-parameterized model in one model organism can be transferred to another with high prediction accuracy.
While STARR-seq provides a genome-wide unbiased test of the enhancer activity of putative sequences, it is intrinsically episomal and thus not completely revealing the enhancer activity in the native chromatin environment. Selecting for chromosomally active enhancers using H3K27ac and DHS could introduce subtle biases in model training. To address this issue, we employed very different experiment techniques and provided orthogonal validations. This included in vivo transgenic assays and in vitro transduction assays, in which the predicted regions were tested for regulatory activity in the native chromatin environment. With these orthogonal validations, we were able to comprehensively assess our tissue-specific predictions in six different tissues in mouse. With multiple comparisons to other published methods trained directly on mouse data, we showed that the matched filter model is transferable with high accuracy in predicting active enhancers in mouse tissues. The in vitro transduction assays were performed in H1-hESCs and three other human cell lines to validate the human regulatory elements predictions. The majority of the predicted elements displayed a significant increase in expression of the reporter gene, further confirming the predictability of our model in mammalian organisms.
Recently, genome-wide STARR-seq has been applied to mammalian systems like HeLa-S3 cells [40]. In the future, we expect that more extensive whole-genome STARR-seq dataset will become available on mammalian systems. It could be advantageous to re-train the matched filter model on state-of-art datasets. With the setup of our framework, re-training the model with newly generated datasets should be straightforward. We envision that our framework would benefit from these datasets and generate more comprehensive regulatory element annotations across eukaryotic species.
Online methods:
Creation of metaprofile
A metaprofile is a template used to estimate the signal distribution on active enhancers for one epigenetic signal. We evaluated whether we could utilize the metaprofiles to predict active promoters and enhancers using matched filters (Fig.1). Matched filter is a well-established pattern recognition algorithm that uses a shape-matching filter to recognize the occurrence of a template in the presence of stochastic noise [41]. We started with creating the metaprofiles, which we generally denote as , based on experimentally validated active enhancers. The STARR-seq studies on Drosophila cell lines provide the most comprehensive datasets as they were performed genome wide and with multiple core promoters [17, 30]. These peaks typically consist of a mixture of enhancers and promoters. At this stage, we did not differentiate between the two sets of regulatory elements. As STARR-seq quantifies enhancer activity in an episomal fashion, not all peaks would be active in the native chromatin environment. Arnold and colleagues showed that the STARR-seq peaks that occur with enriched DNase hypersensitivity or H3K27ac modifications tend to be associated with active genes, whereas other STARR-seq peaks tend to be associated with an enrichment of repressive marks such as H3K27me3 [17]. Hence, we took the overlap of the STARR-seq enhancers with H3K27ac and/or DHS peaks to get a high-confidence set of enhancers that are active in vivo, based on which we will create representative metaprofiles for each histone modifications and DNase signals, respectively.
We utilized the smoothed histone signal tracks for the Drosophila S2 cell line provided by the modENCODE consortium [42] to create metaprofiles for ChIP-seq signals. The genome-wide profile for open chromatin (DNase-seq or DHS) for the S2 cell line was calculated based on experiments by the Stark lab [17]. To create the metaprofiles, we aligned active STARR-seq peaks with identifiable “double peak” patterns of the H3K27ac signal and aggregated the signals in the S2 cell line (Fig.1B). Aggregation of signals over a large number of enhancers reduced the noise in the metaprofiles. To identify double peak regions, we initially identified the minimum in the H3K27ac signal track closest to the middle of the STARR-seq peaks. A minimum was accepted if it had the lowest signal within a 100 base pair region in the H3K27ac signal track. We then proceeded to identify the flanking maxima (both sides of the minimum) within a total of 2-kilo base pair region of the STARR-seq peak (1kb on each direction from the center of the STARR-seq peak). These maxima were accepted only if they had the highest signal within a 100 base pair region in the H3K27ac signal track.
Approximately 70% of the active STARR-seq peaks contained an identifiable double peak within the H3K27ac signal, although there was variability in the distance between the two maxima of the double peak in the ChIP-chip signal (Supplementary Fig. 1a). While the minimum tended to occur in the center of these two maxima on average, the distance between the two maxima in the double peaks varied between 300 and 1,100 base pairs. During aggregation, we first aligned the two maxima in the H3K27ac signal across active STARR-seq peaks. We then interpolated the signal with a cubic spline fit so that the signal track contained an equal number of points for each double peak region. All interpolation and smoothing steps were performed using the scipy module in Python. The aggregated signal tracks were averaged to create the metaprofile for the double peak regions. While the signal tracks were aggregated based on identifying the double peak regions in the H3K27ac signal track, the same set of operations could be performed with any epigenetic mark expected to have the double peak pattern flanking regulatory regions.
We calculated the metaprofiles of ~30 other epigenomic datasets (histone marks and DHS signal). These metaprofiles were calculated by aggregating the corresponding ChIP-seq or DHS signals based on the same regions where H3K27ac double peak were identified, so the matched filter scores of each epigenetic mark were calculated on the same regions in the integrated model. We observe that the metaprofiles for some epigenetic marks also show a double peak pattern, and the maxima across different histone modification signals tended to align with each other on average, likely because these epigenetic marks flank enhancers in a similar pattern as H3K27ac (Supplementary Fig. 1). This indicates that a large number of histone modifications would simultaneously co-occur on the nucleosomes flanking an active enhancer or promoter. In contrast, the repressive histone marks did not contain a double peak pattern, so they did not have the same epigenetic template associated with enhancers. The DHS signal, as expected, displayed a single peak at the center of the H3K27ac double peak.
Matched filter algorithm
The epigenetic signal at enhancers and promoters can be approximated as the linear superposition of background noise and the metaprofile s(n) learned in Figure 1. To identify the occurrence of the metaprofile with the presence of noise, we adopted the canonical signaling processing method known as matched-filter. The matched filter process convolves the signal y(n) with the filter h(n). Before calculating the matched filter score, interpolation of signal was used to ensure that the scanned region contained the same number of points as the metaprofile.
where stands for convolution, and r(n) is the resulting matched filter score. The matched filter is defined as the conjugated reverse of the metaprofile template:
where N is the total number of points in the template, and * denotes the complex conjugate.
As shown in Supplementary Figure 1, there was a large amount of variability in the span (distance between the two peaks in the histone signal) of the regulatory region in the epigenetic signal. As a result, we scanned different spans of the genome with the matched filter (distance between the two peaks were allowed to vary between 300 and 1,100 base pairs) and took the highest score as the matched filter score for that region. Matched filter recognizes the given template in a signal in the presence of noise with the highest signal-to-noise ratio [41]. At positive regions, the presence of the metaprofile within the signal leads to high matched filter scores. At background regions where the signal is mostly comprised of noise, the matched filter score is low.
Statistical learning models
We built an integrated model to include matched filter scores from multiple epigenetic signals for more accurate enhancer prediction. The matched filter scores from each epigenetic signal are first normalized. The distribution of matched filter scores in random negative regions for a particular histone mark is approximately Gaussian and it represents the background distribution in the genome. The Z-scores of matched filter scores with respect to the negatives (random regions of genome) were used as input features for training different statistical learning models. The Z-score of the matched filter score is defined as:
where r is the matched filter scores, and are the mean and standard deviation of the Gaussian fit to the matched filter scores for random regions in genome.
We have tested different statistical learning models, including the support vector machine (SVM) [43], ridge regression [44], random forest [45], and Gaussian naïve bayes [46] models. For SVM, we utilized a linear kernel to distinguish between positives and negatives. The linear SVM identifies a decision boundary that maximally separates the regulatory regions and the random regions of the genome from the decision boundary. Ridge regression is a linear regression technique that prevents overfitting by penalizing large weights for each feature. Random forest is an ensemble learning method that operates by constructing a large number of decision trees and outputting the mean prediction of different decision trees. We used thousand trees for creating our enhancer and promoter prediction models. The naïve Bayes classifier is a family of simple probabilistic classifiers that assumes that all the features are independent of one another. We used scikit-learn [47] with default parameters for training and assessing the performance of all the statistical models. In the main text, we discussed the results of the support vector machine (SVM) model, which showed high performance, and low variance in performance upon cross validation.
Feature selection
We selected the features to use in our framework by assessing their individual performance with matched filter, their importance in the integrative model, and their general data availability in mammalian systems. Specifically, the ability to distinguish enhancers from negative regions of each feature is shown in Supplementary Figure 2 and Supplementary table 1. We found that some histone marks like H3K27ac give very different score distributions for the enhancer regions and random regions, while other histone marks like H3K79m1 and H4K20me1 have indistinguishable score distributions on these two categories of regions.
For the importance of each feature in the integrative model, we trained an SVM model, a random forest model, and a ridge regression using all 30 epigenetic marks, and assessed the importance of each feature using their feature coefficient or GINI score. Among these 30 features, H3K27ac, H3K4me1, H3K4me3 and H3K9ac showed high feature coefficients or high GINI score in all three models; DHS and H3K4me2 had high GINI scores and were also widely used in previous literature to identify promoters and enhancers. In contrast, other histone marks like the repressive mark H3K27me3 show little contribution to the integrated model as indicated by the GINI score and the feature coefficients.
Finally, as the 30 histone marks we tested were from Drosophila experimental data, many of them were unavailable in even top-tier tissues and cell lines for mouse and human. For example, H2BKac performed well with matched filter, and had a very high feature coefficient in each model, but the ChIP-seq experiment data is generally unavailable in mammalian cell lines. As our goal was to build a model with broad applicability across organisms, we decided to not include these epigenetic marks (e.g., H2BK5ac, H4ac and H4K12ac) for now, but if more study is done on these histone marks in the future, we can easily include them in our framework. After filtering, we found six features that satisfied all three above criteria, namely, H3K27ac, H3K4me1, H3K4me1, H3K4me3, H3K9ac, and DHS. Integrating these six features in the linear SVM model yielded a high performance (AUROC of 0.96, AUPR of 0.91) similar to that of the complete SVM model using all 30 epigenetic marks (AUROC of 0.97, AUPR of 0.93). We subsequently tested the performance of this simplified model in Drosophila cells, mouse tissue, and human cells.
In the 6-feature model, the DHS signal has lower weight than the other 5 features (Fig. 2). It should be noted that the matched filter on DHS signal performed well on its own. The lower weight is likely due to the fact that the information in DHS is redundant with the information contained within the histone mark (e.g., the DHS peaks usually occur at the trough region between two maxima in the histone signal). Despite the redundancy, the combination of the DHS and histone signals was more predictive of regulatory activity because the reinforcing signals strengthened the prediction as compared to the uncorrelated noise.
Model assessment
In order to assess the accuracy of the matched filter model for predicting enhancers and promoters, we used a ten-fold cross validation. The STARR-seq positives and negatives were randomly divided into ten groups. For each fold of cross validation on a single histone mark, the profiles were created with 90% of the STARR-seq positives, and the remaining 10% of the positives were used for testing the accuracy of the model. Similarly, In the integrative SVM model, the SVM was trained on 90% of the data in each fold of cross validation, whereas the remaining 10% of the positives were used to test accuracy.
We quantified our model performance with area under receiver-operating characteristic (ROC), and area under precision-recall (PR) curves. In the ROC curve, the true positive (TP) rate was plotted against the false positive (FP) rate at different thresholds in the statistical model. The TP rate is defined as the number of true positives identified by the model divided by the total number of positives. The FP rate is defined as the fraction of negatives misclassified as positives by the model, divided by the total number of negatives. When comparing the performance of two different classifiers in the ROC curve, the classifier with a higher TP rate at the same FP rate is considered to be a better classifier. The area under the ROC (AUROC) is a single measure for the accuracy of a model, as models with higher AUROC are generally considered to be better models.
In the PR curve, the precision was plotted against recall at different thresholds in the statistical model. The recall is the same as the TP rate of the model (i.e., the number of true positives identified by the model divided by the total number of positives). The precision is the fraction of positives predicted by the model that are correct (i.e., the number of true positives identified by the model divided by the total number of positives predicted by the model). The area under the PR curve (AUPR) is another measure of performance of a model. If the AUPR is high, the corresponding model has a low false discovery rate and can better distinguish the positives from the negatives. PR curves are particularly useful to assess the performance of classifiers in skewed or imbalanced data sets in which one of the classes is observed much more frequently compared to the other class [9]. For such skewed datasets, the AUROC for two different models may be very similar even though they actually differ in performance with respect to their precision. Hence, the area under the PR (AUPR) curve is a better reflection of the performance difference between two models with a similar AUROC in skewed datasets.
In Figure 2, the positives are defined as the active peaks (intersecting with DHS or H3K27ac peaks) from a single STARR-seq experiment (single core promoter) or the union of active peaks from multiple STARR-seq experiments (multiple core promoters). The negatives are randomly chosen non-STARR-seq-peak regions in the genome that had the same lengths distribution as the enhancers from the STARR-seq. We required most of the regions to contain some H3K27ac signals, as negatives with no H3K27ac signal at all wouldn’t provide enough information for training. We typically chose 5 to 10x the number of negatives as compared to the number of positives in Figures 2, 3, and 4, as the number of enhancers and promoters in the genome (positives) is far less than the number of negatives; moreover, the area under the PR curve is dependent on the ratio of negatives to positives during the ten-fold cross validation.
To evaluate the impact of the training sample size on model performance, we did a saturation analysis where we down-sampled the training data to different levels of fractions and evaluated the model performance on the remaining data. For each down-sampling fraction from 10% to 90% with 10% as the step, we performed the ten-fold cross-validations. In each fold, the whole model including the aggregation of signals was based on the training data set. The performance was tested on the remaining data and was independent of the training data. We found that the average AUPR increased with an increasing size of training data. The AUPR of the SVM model started to saturate with 80%−90% of the experiment data for training (Supplementary Fig. 4). The average AUROC remained comparable, although the variances decreased with increasing training data size. This might suggest that a five-fold cross-validation would be sufficient.
Promoters and Enhancers
In the STARR-seq experiment, each peak functions as an enhancer within the plasmid environment in the S2 cell line. However, to delineate the native role of the region, we classified them as promoters and enhancers based on their distance to the transcription start sites in the genome. In Figure 3, the active promoters were defined as active STARR-seq peaks (multiple core promoter) within 1 kb of TSS (Ensembl release 78); enhancers were defined as active STARR-seq peaks more than 1kb from any TSS in Drosophila. However, a few of the promoters may also regulate distal genes in addition to their promoter activity [48].
Validating enhancers in mammalian species
We downloaded tissue specific epigenetics data from the ENCODE portal (https://www.encodeproject.org). The histone signals were converted to log-fold enrichment (with respect to control signal). We ran the integrated matched-filter to get the enhancer and promoter predictions for six different mouse tissues (forebrain, midbrain, hindbrain, limb, heart and neural tube) at the embryonic day 11.5 (e11.5) stage (Genome-wide predictions are available through our website at https://goo.gl/E8fLNN). These tissues were selected as their epigenetic signals have been highly studied in mouse ENCODE, providing us with a rich source of raw data that could be utilized for making enhancer and promoter predictions. In addition, the VISTA database contains close to 100 validated enhancers that could be used to test predictions in each of these tissues. Using our model, we predicted 31K to 39K regulatory regions in individual tissues in mouse, with each region ranging from 300bp to 1,100bp. Notably, a consistent proportion of two-thirds (66–70%) of these predicted regulatory regions were distal regulatory elements for all six tissues, with the other one-third (30–34%) being proximal regulators (Supplementary Table 10). These numbers agree with a previous enhancer evolution study [49], and suggest that the amount of enhancers and promoters are likely comparable in different tissues.
Similarly, we performed a genome-wide prediction of regulatory regions in the ENCODE top-tier human cell lines, including H1-hESC, GM12878, K562, HepG2, A549 and MCF-7. Predicted active regions within 2kb of any TSS were annotated as promoters, and regions that were more than 2kb from any TSS were annotated as enhancers. The distribution of the expression of the closest gene (GENCODE v19 TSS [50]) from the ENCODE RNA-seq dataset for H1-hESCs was compared to the expression of all genes from H1-hESCs. The Wilcoxon test was used to measure the significance of changes in gene expression.
To assess the predictions, we ranked all the tested candidate elements by either the matched-filter scores of individual features, or the final prediction (probability of being an enhancer) from the integrated SVM model. We then took the labels of the candidate elements from the experiment readout to assess the predictions using ROC and PR curves.
Validation in mouse embryos
In Figure 4, the enhancers were tested by transgenic mouse reporter assays [9, 51]. Predicted enhancers were PCR amplified and cloned into a plasmid upstream of a minimal hsp68 promoter and a lacZ reporter gene. Resulting plasmids were linearized and injected into single-cell FVB/NCrl strain Mus musculus embryos. After reimplantation into surrogate mothers, resulting embryos were collected at embryonic day 11.5 (e11.5), stained for b-galactosidase activity, and imaged. Elements were scored positive for enhancer activity if at least three resulting transgenic embryos had reporter gene expression in the same tissue and pattern. Elements were scored negative if at least five transgenic embryos were recovered and no reproducible staining patterns was observed.
Validation in human cell lines
We used a third-generation, self-inactivating (SIN) HIV-1 based vector system in which the enhanced GFP (eGFP) reporter was driven by the DNA element of interest to test putative enhancers after stable transduction of four cell lines, including H1-hESCs (Supplementary Fig. 11). The predicted enhancers were PCR amplified from human genomic DNA and separately inserted immediately upstream of a basal Oct-4 promoter of 142 bp within the SIN HIV vector. Each putative enhancer was tested in triplicate for both forward and reverse orientation in H1-hESCs. We used empty SIN HIV vector and FG12 as the negative and the positive controls, respectively. Note that the empty vector had the basal Oct-4 promoter, along with the IRES-eGFP reporter cassette. We assessed putative enhancer activity by flow cytometric readout of eGFP expression 48–72 h post-transduction, normalized to the negative control.
We selected a total of 23 predicted intergenic enhancers for validation. These predictions were chosen at random to ensure that they truly represented the whole spectrum of predicted enhancers and not just the top tier of predicted enhancers. Of these 23 putative enhancers, 20 were successfully PCR-amplified and cloned into the SIN HIV vector in both directions. To measure the distribution of gene expression in the absence of enhancer, we also amplified and cloned 20 non-repetitive elements with a similar length distribution that were predicted to be inactive into the same SIN HIV vector. All positive and negative DNA elements were transduced and tested for activity in both forward and reverse orientations as enhancers are thought to function in an orientation-independent manner. Following the same procedures, we performed functional testing in duplicate in HOS, TZM-bl, and A549 cell lines in addition to H1-hESCs.
Performance comparison to other computational methods
We compared the performance of the matched filter to the peak-based models of the different epigenetic marks (Fig. 2), we used the histone (or DHS) peaks that overlapped with at least 50% (10%) of the STARR-seq peak to rank that prediction. We used a smaller threshold for DHS peaks as they are much shorter than histone peaks. We achieved similar results with thresholds of 25% for both histone and DHS peaks. The p-value of the intersecting peak was used to rank the peak-based predictions. The modENCODE histone peaks and DHS peaks [42] were compared to the matched filter scores in Figure 2.
We compared with other published enhancer prediction tools, including ChromHMM, a multivariate hidden Markov model [52]; CSIANN, a neural network based approach [53]; DELTA, an ensemble model integrating different histone modifications [54]; RFECS, a random forest model based on histone modifications [36], and REPTILE, a more recent published method that integrates histone modifications and whole-genome bisulfite sequencing data [55]. We used the mouse experimental data published in REPTILE for the comparison, and assessed the performance of our method compared to the four published methods mentioned above for all four mouse tissues with available experimental data, ChIP-seq data, and DNase data.
Our integrated model outperformed ChromHMM in all four tissues, with an AUROC value of 0.76 in hindbrain (versus ChromHMM 0.69), and 0.81 in limb (versus ChromHMM 0.75), etc (Supplementary Fig. 13a). For comparison with supervised algorithms like CSIANN, DELTA and REPTILE, our method had the highest AUROC in three out of four tissues (hindbrain, limb and neural tube) as shown in Supplementary Fig. 13b. In midbrain, the AUROC for our prediction was slightly lower than REPTILE and RFECS, possibly because the DNase experiment performed in midbrain was very noisy; the DNase experiment of mouse e11.5 midbrain was marked as “low SPOT score” in ENCODE, where SPOT stands for Signal Portion of Tag. We found that while 75% to 81% of the genome regions had DNase signals in the other three tissues, only 52% of the genome regions showed DNase signal in the experiment in midbrain. Overall, the comparison shows that our model trained using the Drosophila STARR-seq data had better performance than the other methods that were trained directly using mouse experimental data.
For human, we did not have an extensive amount of validated enhancer data. For comparison, we first checked the overlap of our predicted enhancers with the enhancer predictions from ChromHMM [56], and Segway [57]. We observed that a majority of our predictions overlap with either of them (See Supplementary materials). In addition, we compared our cell type-specific enhancer predictions with the integrative annotation of ChromHMM and Segway using CAGE-defined enhancers from the FANTOM5 Atlas [58]. We found that the percentage of overlap for our predicted enhancers was more than three times higher than that of the combined ChromHMM and Segway enhancers in each of these cell lines. Despite the fact that our framework predicted a smaller number of enhancers, the number of overlaps was still higher for our predictions. We also compared the predicted promoters from our model with their promoter annotations using FANTOM5 promoter sets. Again, the promoters predicted in our model had a higher fraction of overlaps with the FANTOM promoters (Supplementary Fig. 14). In addition to the integrative ENCODE annotation, we again compared with other supervised enhancer predictions like CSI-ANN [53], DEEP [59] and RFECS [36], using the FANTOM5 enhancer dataset. We found that our predicted K562 enhancers had a similar fraction of overlap with FANTOM5 enhancers compared to that of CSI-ANN, but the fraction was more than twice as high as that of DEEP and RFECS (Supplementary Fig. 14).
TFs binding patterns at enhancers
To measure the differences in TF binding and co-binding patterns at promoters and enhancers, we overlapped the ChIP-seq peaks from ENCODE with our predicted enhancers and promoters using intersectBed. The two regions were considered to be overlapping if at least 25% of the ChIP-seq peak overlapped with the predicted enhancer or promoter.
To check if the STARR-seq-based enhancer predictions have different TF binding patterns, we referred to the fraction of TF occupancy of predicted enhancer from other methods. The comparison demonstrated in Supplementary Fig. 15 shows that the TF binding pattern of our prediction is very similar to previous literature report [36].
Code and data availability
We have implemented our methods in Python. The source code and the output annotations referenced in the paper are available at the website http://matchedfilter.gersteinlab.org. A dockerized image is also provided at this site.
A detailed description of the datasets used in this study is in the supplement. Specifically, the Drosophila epigenetics datasets used in this study were generated by the modENCODE consortium, available online (http://data.modencode.org). The mouse epigenetics datasets were generated by the ENCODE and Roadmap Epigenomics consortium, available online (https://www.encodeproject.org). We downloaded the Drosophila STARR-seq data [25] and the mouse FIREWACh data [30] from previous studies. The mouse transgenic enhancer assay results were generated by the Pennacchio lab at LBNL. Experiment results are summarized in supplementary tables 4–9, with the mouse images and additional details available on the VISTA Enhancer Browser (www.enhancer.lbl.gov). The human cell line enhancer reporter assay results were generated by the Sutton lab at Yale University. Experiment results are summarized in supplementary table 10. More detailed results for each cell line are available in the extended data table.
Supplementary Material
Acknowledgement:
M.Gerstein. were supported by the NIH grant HG009446–01. A.Visel. and L.A.Pennacchio. were supported by NHLBI grant R24HL123879, and NHGRI grants R01HG003988, and U54HG006997, and UM1HG009421 where research was conducted at the E.O. Lawrence Berkeley National Laboratory and performed under Department of Energy Contract DE-AC02–05CH11231, University of California. We thank A. Paccanaro, D. Galeano, M. Torres, and ENCODE consortium members for their helpful discussion about this work.
Footnotes
Competing Interest
The authors declare no competing interests.
References:
- 1.Banerji J, Rusconi S, and Schaffner W, Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell, 1981. 27(2 Pt 1): p. 299–308. [DOI] [PubMed] [Google Scholar]
- 2.Slattery M, et al. , Absence of a simple code: how transcription factors read the genome. Trends Biochem Sci, 2014. 39(9): p. 381–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levo M, et al. , Unraveling determinants of transcription factor binding outside the core binding site. Genome Res, 2015. 25(7): p. 1018–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wray GA, The evolutionary significance of cis-regulatory mutations. Nat Rev Genet, 2007. 8(3): p. 206–16. [DOI] [PubMed] [Google Scholar]
- 5.Corradin O. and Scacheri PC, Enhancer variants: evaluating functions in common disease. Genome Med, 2014. 6(10): p. 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gusev A, et al. , Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am J Hum Genet, 2014. 95(5): p. 535–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pennacchio LA, et al. , Enhancers: five essential questions. Nat Rev Genet, 2013. 14(4): p. 288–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erwin GD, et al. , Integrating diverse datasets improves developmental enhancer prediction. PLoS Comput Biol, 2014. 10(6): p. e1003677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pennacchio LA, et al. , In vivo enhancer analysis of human conserved non-coding sequences. Nature, 2006. 444(7118): p. 499–502. [DOI] [PubMed] [Google Scholar]
- 10.Visel A, et al. , Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat Genet, 2008. 40(2): p. 158–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nord AS, et al. , Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell, 2013. 155(7): p. 1521–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Visel A, et al. , ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature, 2009. 457(7231): p. 854–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andersson R, et al. , An atlas of active enhancers across human cell types and tissues. Nature, 2014. 507(7493): p. 455–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Narlikar L, et al. , Genome-wide discovery of human heart enhancers. Genome Res, 2010. 20(3): p. 381–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yip KY, et al. , Classification of human genomic regions based on experimentally determined binding sites of more than 100 transcription-related factors. Genome Biol, 2012. 13(9): p. R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghandi M, et al. , Enhanced regulatory sequence prediction using gapped k-mer features. PLoS Comput Biol, 2014. 10(7): p. e1003711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arnold CD, et al. , Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science, 2013. 339(6123): p. 1074–7. [DOI] [PubMed] [Google Scholar]
- 18.Yanez-Cuna JO, et al. , Dissection of thousands of cell type-specific enhancers identifies dinucleotide repeat motifs as general enhancer features. Genome Res, 2014. 24(7): p. 1147–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shlyueva D, Stampfel G, and Stark A, Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet, 2014. 15(4): p. 272–86. [DOI] [PubMed] [Google Scholar]
- 20.Maston GA, et al. , Characterization of enhancer function from genome-wide analyses. Annu Rev Genomics Hum Genet, 2012. 13: p. 29–57. [DOI] [PubMed] [Google Scholar]
- 21.Creyghton MP, et al. , Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A, 2010. 107(50): p. 21931–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heintzman ND, et al. , Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet, 2007. 39(3): p. 311–8. [DOI] [PubMed] [Google Scholar]
- 23.Yue F, et al. , A comparative encyclopedia of DNA elements in the mouse genome. Nature, 2014. 515(7527): p. 355–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerstein MB, et al. , Comparative analysis of the transcriptome across distant species. Nature, 2014. 512(7515): p. 445–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong X, et al. , Modeling gene expression using chromatin features in various cellular contexts. Genome Biol, 2012. 13(9): p. R53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng C. and Gerstein M, Modeling the relative relationship of transcription factor binding and histone modifications to gene expression levels in mouse embryonic stem cells. Nucleic Acids Res, 2012. 40(2): p. 553–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng Y, et al. , Principles of regulatory information conservation between mouse and human. Nature, 2014. 515(7527): p. 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyle AP, et al. , Comparative analysis of regulatory information and circuits across distant species. Nature, 2014. 512(7515): p. 453–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gjoneska E, et al. , Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer’s disease. Nature, 2015. 518(7539): p. 365–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zabidi MA, et al. , Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature, 2015. 518(7540): p. 556–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cotney J, et al. , Chromatin state signatures associated with tissue-specific gene expression and enhancer activity in the embryonic limb. Genome Res, 2012. 22(6): p. 1069–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ernst J, et al. , Mapping and analysis of chromatin state dynamics in nine human cell types. Nature, 2011. 473(7345): p. 43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burges CJC, A Tutorial on Support Vector Machines for Pattern Recognition. Data Mining and Knowledge Discovery, 1998. 2: p. 121–-167. [Google Scholar]
- 34.Roadmap Epigenomics C, et al. , Integrative analysis of 111 reference human epigenomes. Nature, 2015. 518(7539): p. 317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Consortium EP, An integrated encyclopedia of DNA elements in the human genome. Nature, 2012. 489(7414): p. 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajagopal N, et al. , RFECS: a random-forest based algorithm for enhancer identification from chromatin state. PLoS Comput Biol, 2013. 9(3): p. e1002968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koch CM, et al. , The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res, 2007. 17(6): p. 691–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murtha M, et al. , FIREWACh: high-throughput functional detection of transcriptional regulatory modules in mammalian cells. Nat Methods, 2014. 11(5): p. 559–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bailey SD, et al. , ZNF143 provides sequence specificity to secure chromatin interactions at gene promoters. Nat Commun, 2015. 2: p. 6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muerdter F, et al. , Resolving systematic errors in widely used enhancer activity assays in human cells. Nat Methods, 2018. 15(2): p. 141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar BVKV, Mahalanobis A, and Juday RD, Correlation pattern recognition. 2005, Cambridge, UK; New York: Cambridge University Press. xii, 390 p. [Google Scholar]
- 42.mod EC, et al. , Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science, 2010. 330(6012): p. 1787–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blanchard G, Bousquet O, and Massaer P, Statistical performance of support vector machines. Ann. Statist, 2008. 36: p. 489–531. [Google Scholar]
- 44.Hoerl AE and Kennard RW, Ridge Regression: Biased Estimation for Nonorthogonal Problems. Technometrics, 1970. 12(1): p. 55−−67. [Google Scholar]
- 45.Breiman L, Random Forests. Machine Learning, 2001. 45(1): p. 5–-32. [Google Scholar]
- 46.Stuart R. and Norvig P, Artificial Intelligence: A Modern Approach. 2nd ed. 2003. [Google Scholar]
- 47.Pedregosa F, et al. , Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research, 2011. 12: p. 2825–-2830. [Google Scholar]
- 48.Diao Y, et al. , A tiling-deletion-based genetic screen for cis-regulatory element identification in mammalian cells. Nat Methods, 2017. 14(6): p. 629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Villar D, et al. , Enhancer evolution across 20 mammalian species. Cell, 2015. 160(3): p. 554–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harrow J, et al. , GENCODE: The reference human genome annotation for The ENCODE Project. Genome Research, 2012. 22(9): p. 1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kothary R, et al. , Inducible Expression of an Hsp68-Lacz Hybrid Gene in Transgenic Mice. Development, 1989. 105(4): p. 707-&. [DOI] [PubMed] [Google Scholar]
- 52.Ernst J. and Kellis M, ChromHMM: automating chromatin-state discovery and characterization. Nat Methods, 2012. 9(3): p. 215–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Firpi HA, Ucar D, and Tan K, Discover regulatory DNA elements using chromatin signatures and artificial neural network. Bioinformatics, 2010. 26(13): p. 1579–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu YM, et al. , DELTA: A Distal Enhancer Locating Tool Based on AdaBoost Algorithm and Shape Features of Chromatin Modifications. Plos One, 2015. 10(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He YP, et al. , Improved regulatory element prediction based on tissue-specific local epigenomic signatures. Proceedings of the National Academy of Sciences of the United States of America, 2017. 114(9): p. E1633–E1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ernst J. and Kellis M, ChromHMM: automating chromatin-state discovery and characterization. Nature Methods, 2012. 9(3): p. 215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoffman MM, et al. , Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat Methods, 2012. 9(5): p. 473–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arner E, et al. , Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science, 2015. 347(6225): p. 1010–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kleftogiannis D, Kalnis P, and Bajic VB, DEEP: a general computational framework for predicting enhancers. Nucleic Acids Research, 2015. 43(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We have implemented our methods in Python. The source code and the output annotations referenced in the paper are available at the website http://matchedfilter.gersteinlab.org. A dockerized image is also provided at this site.
A detailed description of the datasets used in this study is in the supplement. Specifically, the Drosophila epigenetics datasets used in this study were generated by the modENCODE consortium, available online (http://data.modencode.org). The mouse epigenetics datasets were generated by the ENCODE and Roadmap Epigenomics consortium, available online (https://www.encodeproject.org). We downloaded the Drosophila STARR-seq data [25] and the mouse FIREWACh data [30] from previous studies. The mouse transgenic enhancer assay results were generated by the Pennacchio lab at LBNL. Experiment results are summarized in supplementary tables 4–9, with the mouse images and additional details available on the VISTA Enhancer Browser (www.enhancer.lbl.gov). The human cell line enhancer reporter assay results were generated by the Sutton lab at Yale University. Experiment results are summarized in supplementary table 10. More detailed results for each cell line are available in the extended data table.