Abstract
We investigated the effects of melatonin and its selected metabolites, i.e., N1-Acetyl-N2-formyl-5-methoxykynurenamine (AFMK) and 6-hydroxymelatonin (6(OH)Mel), on cultured human epidermal keratinocytes (HEKs) to assess their homeostatic activities with potential therapeutic implications. RNAseq analysis revealed a significant number of genes with distinct and overlapping patterns, resulting in common regulation of top diseases and disorders. Gene Set Enrichment Analysis (GSEA), Reactome FIViZ, and Ingenuity Pathway Analysis (IPA) showed overrepresentation of the p53-dependent G1 DNA damage response gene set, activation of p53 signaling, and NRF2-mediated antioxidative pathways. Additionally, GSEA exhibited an overrepresentation of circadian clock and antiaging signaling gene sets by melatonin derivatives and upregulation of extension of telomere signaling in HEKs, which was subsequently confirmed by increased telomerase activity in keratinocytes, indicating possible antiaging properties of metabolites of melatonin. Furthermore, Gene Ontology (GO) showed the activation of a keratinocyte differentiation program by melatonin, and GSEA indicated antitumor and antilipidemic potential of melatonin and its metabolites. IPA also indicated the role of Protein Kinase R (PKR) in interferon induction and antiviral response. In addition, the test compounds decreased lactate dehydrogenase A (LDHA) and lactate dehydrogenase C (LDHC) gene expression. These results were validated by qPCR and by Seahorse metabolic assay with significantly decreased glycolysis and lactate production under influence of AFMK or 6(OH)Mel in cells with a low oxygen consumption rate. In summary, melatonin and its metabolites affect keratinocytes’ functions via signaling pathways that overlap for each tested molecule with some distinctions.
Keywords: melatonin, metabolites of melatonin, RNA-sequencing, human keratinocytes, mitochondrial metabolism
1. Introduction
Melatonin, a tryptophan (Trp) metabolism product, is widely detected in many species, including animals, plants, unicellular eukaryotes, algae, bacteria and humans [1,2]. It regulates the circadian rhythm and exerts pleiotropic bioactivities mediated by interactions with high affinity G-protein coupled membrane bound melatonin receptors type 1 and type 2 (MT1 and MT2) or receptor-independent mechanisms [3,4]. Melatonin also acts as a broad-spectrum antioxidant, stimulator of antioxidative responses, and of DNA repair pathways at relatively high concentrations (>1 µM) [5,6]. It also has anticancer activities [7,8,9,10], which are shared by products of its metabolism [11,12,13,14,15,16].
Melatonin, mainly produced in the pineal gland, is also synthesized in peripheral organs, such as the skin [13,17,18,19,20,21,22]. This led to our proposal that the cutaneous melatoninergic system can defend this organ against environmental damage, with mitochondria playing an important role in this function [10,11,21,22,23], while being a part of a complex cutaneous neuroendocrine system that coordinates cutaneous responses to stress [4,24], including ultraviolet radiation (UVR) [23]. Extensive data from various laboratories [10,11,12,16,17,22,23,25,26,27,28] have documented that melatonin and/or its metabolites induce potent antioxidative actions, using DNA repair mechanisms against UVB-induced damages in human cutaneous cells including stimulation of NRF2 and p53-dependent pathways that are independent from MT1 and MT2 [12,16,28]. Importantly, melatonin and its metabolites accumulate in the human epidermis [29,30] and melatonin exerts anti-erythema and antiaging effects when applied topically to the skin [31,32].
Melatonin is also recognized for its complex immunomodulatory [33,34,35] and antiviral activities [36,37,38]. Specifically, it can regulate T-cell and macrophage functions, and possesses both immunostimulatory and anti-inflammatory properties that are context-dependent [33,35,39]. Thus, melatonin is a double-edged sword, being in some cases beneficial, while in others aggravating autoimmune responses [33,35,39,40,41]. Its anti-inflammatory action includes inhibition of NO synthases, TLR4 and NF-κB activation, as well as the upregulation of NRF2 [39]. Many of melatonin’s anti-inflammatory effects appear to be mediated by sirtuin-1 (SIRT1) [39,40]. Lastly, melatonin itself and its metabolites affect mitochondrial functions and energy yielding cellular metabolism, including in skin cells [1,9,14,16,22,42]. In addition, antiaging skin properties of melatonin are widely recognized [9,31,39,43].
To provide insight into the mechanisms of the above antioxidative, protective, metabolic, prodifferentiation, antiaging, anti-inflammatory, and antitumor mechanisms in the epidermis, we conducted RNAseq analyses using human epidermal keratinocytes treated with melatonin and its indolic (6-hydroxymelatonin) and kynuric N1-Acetyl-N2-formyl-5-methoxykynurenamine (AFMK) metabolites. We found overlapping and differential effects that were further investigated using biochemical and molecular assays. By using the comprehensive Omics method of estimation of gene expression, we obtained an insight not only into the expression of particular genes involved in the above processes, but also the correlation between these genes and their functional clustering. Based on these analyses, the ability of melatonin’s metabolites to regulate different pathways affecting the physiological and pathological states of epidermis remains under discussion.
2. Materials and Methods
2.1. Reagents
2-Hydroxymelatonin (2(OH)Mel), 6-hydroxymelatonin (6(OH)Mel), bovine serum albumin (BSA), ethanol (EtOH), glucose, melatonin, and sodium pyruvate were purchased from Sigma (St. Louis, MO, USA). AFMK and GlutaMAX™ supplements were purchased from Cayman Chemical (Ann Arbor, MI, USA) and Thermo Fisher Scientific (Waltham, MA, USA), respectively. Other reagents were supplied as follows: EpiGRO™ Human Epidermal Keratinocyte Complete Culture Media Kit (Millipore Merck KGaA, Darmstadt, Germany); RNA Miniprep Kit (Agilent Technologies, Santa Clara, CA, USA), high capacity cDNA Reverse Transcription Kit (Applied Biosystems), DyNamo Flash SYBR Green qPCR Kit (Thermo Scientific, Waltham, MA, USA), KAPA SYBR® FAST qPCR Kit (Kapa Biosystems, Wilmington, MA, USA), the Cell Mito Stress Test media was supplemented with 2 mM GlutaMAX™ Supplement (L-alanyl-L-glutamine dipeptide in NaCl), Proteome Profiler Array kit (R&D Systems, Minneapolis, MN, USA), lactate colorimetric assay kit (Cell Biolabs, Inc. San Diego, CA, USA), TeloTAGGG Telomerase Elisa assay (Roche, Basel, Switzerland), and a set of human primers for real-time PCR (Eurofins Genomics, Ebersberg, Germany).
2.2. Cell Culture and Treatment
Neonatal Human Epidermal Keratinocytes (HEKn, primary cells) were isolated from foreskins collected at the Woman and Infant Hospital (UAB) and cultured on Petri dishes (TPP) in EpiGRO™ Human Epidermal Keratinocyte Complete Culture Media Kit (Millipore Merck KGaA, Darmstadt, Germany) as described previously [12]. Cells were seeded into 24-well plates, maintained until they reached 70% confluency, and stimulated for 24 h with melatonin, 6(OH)Mel or AFMK at the final concentration of 10−5 M or with solvent only (control, 0.1% EtOH). HaCaT cells were grown in Dulbecco’s Modified Eagle Medium (DMEM) media containing 5% charcoal stripped FBS followed by treatment with 10−4 or 10−5 M melatonin and 10−5 M 6(OH)Mel and 2-hydroxymelatonin (2(OH)Mel) at 37 °C under 5% CO2 for 6 and 24 h. We have used similar doses of ligands in our previous studies, which demonstrated the paracrine mechanism of action secondary to production and metabolism of melatonin in the epidermis [12,13,29,30]. After 6 and 24 h, cells were collected in lysis reagent and RNA isolated using Absolutely RNA Miniprep Kit (Agilent Technologies, Santa Clara, CA, USA).
2.3. Reverse Transcription Reaction and Real-Time PCR
To study gene expression in material acquired from HaCaT cells, cDNA was synthesized using 0.4 µg RNA with High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) following manufacturers’ protocols. KAPA SYBR® FAST qPCR Kit (Kapa Biosystems, Wilmington, MA, USA) were used for qRT-PCR with Quant Studio 6 Real-Time PCR Systems (Applied Biosystems, Foster City, CA, USA) in presence of the following primers’ sequences: lactate dehydrogenase A (LDHA) (Fwd: 5′-ACCCAGTTTCCACCATGATT, Rev: 5′-CCCAAAATGCAAGGAACACT), PGC-1 (Fwd: 5′-GTCACCACCCAAATCCTTAT, Rev: 5’-ATCTACTGCCTGGAGACCTT), SIRT-1 (Fwd: 5′-TCGCAACTATACCCAGAACATAGACA, Rev: 5′-CTGTTGCAAAGGAACCATGACA), and cyclophilin B (Fwd: 5′-TGTGGTGTTTGGCAAAGTTC, Rev: 5′-GTTTATCCCGGCTGTCTGTC) as the house-keeping gene. The following temperature profile was set using Quant Studio 6 real-time PCR Systems (Applied Biosystems): hold stage (predenaturation): 95 °C for 20 s, PCR stage (40 cycles): (denaturation): 95 °C for 1 s, (annealing): 60 °C for 20 s.
2.4. RNA Sequencing and Bioinformatics Analysis
At least 200 ng of RNA from each HEKn sample with purity of 2.0 (A260/280) was sent for RNA sequencing by Novogene (Sacramento, CA, USA). Prior to sending, the sample quality and concentration were determined using Cytation 5 Imaging Reader. After sequencing, the raw sequence FASTQ files were trimmed to remove primer adapters using Trim Galore version 0.6.2 (parameters used: --paired; --trim1). The trimmed FASTQ files were then aligned to the Gencode GRCh38 p7 Release 25 genome using STAR version 2.7.1a (parameters used: --outReadsUnmappedFastx; --outSAMtype BAM SortedByCoordinate; --outSAMattributes All). Transcript abundances were then calculated from the alignments using HTSeq count version 0.11.2 (parameters used: -m union; -r pos; -t exon; -igene_id; -a 10; -s no; -f bam). The raw counts were then loaded into DESeq2 using their default settings to normalize and perform pairwise differential expression. All raw data were deposited in NCBI GEO (GSE147588).
2.5. Bioinformatics
Pathway analysis was performed using Reactome and Ingenuity Pathway Analysis software (Ingenuity® Systems). Gene set enrichment analysis (GSEA) and diagram analysis with overlapping gene scores into pathways were made using Reactome FIViZ software. Data analyzed using IPA software were processed with the following steps: for generating networks, a data set containing gene identifiers and corresponding expression values were uploaded into the application. Each identifier was mapped to its corresponding object in Ingenuity’s Knowledge Base. A fold change cutoff of +/−2 was set to identify molecules whose expressions were significantly differentially regulated. These molecules, called Network Eligible molecules, were overlaid onto a global molecular network developed from information contained in Ingenuity’s Knowledge Base. Networks of Network Eligible Molecules were then algorithmically generated based on their connectivity. The functional analysis identified the biological functions and/or diseases that were most significant to the entire data set. Molecules from the data set that met the fold change cutoff of +/−2 and were associated with biological functions and/or diseases in Ingenuity’s Knowledge Base were considered for the analysis. Right-tailed Fisher’s exact test was used to calculate the p value determining the probability that each biological function and/or disease assigned to that data set is due to chance alone.
2.6. Analysis of Metabolic Function
Primary human keratinocytes were isolated from skin biopsies of healthy controls as previously described [44]. Keratinocytes were seeded at 0.25 × 105 cells/well overnight on Seahorse 96-well XFe96 microplates in culture medium. The next day, cells were treated with melatonin, AFMK and 6(OH)Mel for 24 h, all at a final concentration of 10−5 M. Just before the start of the experiment, cells were depleted of glucose for 1 h in a 37 °C non-CO2 incubator. Oxygen consumption rate (OCR or mitochondrial respiration) and extracellular acidification rate (ECAR or glycolytic function) in live cells in real-time were measured using the XF Cell Mito Stress Test and Agilent Seahorse XF96 Extracellular Flux Analyzer (Seahorse Bioscience, North Billerica, MA, USA). The Cell Mito Stress Test media was supplemented with 2 mM GlutaMAX™ Supplement, 1 mM sodium pyruvate, and 25 mM glucose and injected according to the Mito Stress Test. Results are automatically generated from Wave data that was exported to Excel.
2.7. Proteome Profiler and L-Lactate Functional Assays
To measure cytokine release, conditioned media from HEKn cells collected after treatment with melatonin, AFMK or 6(OH)Mel for 24 h were submitted for human cytokine array assay using Proteome Profiler Array kit (R&D Systems). To evaluate L-lactate production in HEKn cell media, the Lactate Assay kit (Colorimetric, Cell Biolabs) was used and the values were recorded using Cytation 5 Imaging Reader. Telomerase activity was measured using the TeloTAGGG Telomerase Elisa assay (Roche) in the cell pellets obtained after 24 h treatment of HaCaT keratinocytes with AFMK, melatonin or 6(OH)Mel at a concentration of 10−4 or 10−5 M. The data were collected with Cytation 5 Imaging Reader. All assays followed procedures provided by manufacturers.
2.8. Statistical Analysis
Experiments were performed at least three times, with results expressed in each case as the mean + standard deviation (SD). Significant differences between results were determined by both by Mann–Whitney U test and Student t-test, and an appropriate post hoc analysis using GraphPad Prism 7.05 software (La Jolla, CA, USA). p value of less than 0.05 was considered statistically significant.
3. Results and Discussion
3.1. RNAseq Results
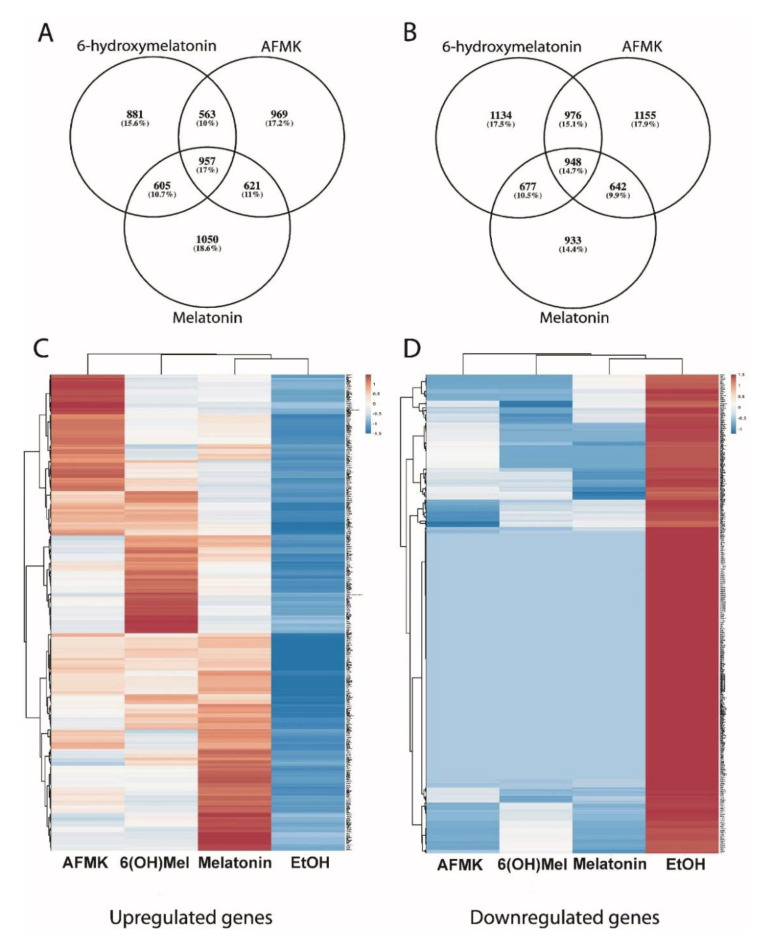
The analysis of RNAseq data with a fold change cutoff of +/−2 showed changes in the expressions of 6741 genes, 6831 genes and 6433 genes after respective treatments of keratinocytes with 6(OH)Mel, AFMK and melatonin. 6(OH)Mel, AFMK and melatonin enhanced expressions of 3735, 3721 and 3200 genes, and inhibited 3006, 3110 and 3233 genes, respectively (Figure 1A–D, Supplementary Materials Figure S1). An overlapping gene expression pattern included upregulation of 948 and downregulation of 957 common genes for all tested compounds. The Venn diagram also shows a distinct pattern of genes specific for each compound tested with 933 (14.4%), 1134 (17.5%) and 1155 (17.9%) genes upregulated in keratinocytes by melatonin, 6(OH)Mel and AFMK, respectively. Similarly, numbers of genes downregulated only by melatonin, 6(OH)Mel and AFMK, respectively, were 1050 (18.6%), 881 (15.6%) and 969 (17.2%).
Figure 1.
Analyses of differentially expressed genes in human keratinocytes treated with melatonin, 6-hydroxymelatonin and AFMK. Venn diagrams are in (A) for up- and in (B) for downregulated genes. Heatmaps for all upregulated genes are in (C) and for downregulated genes in (D), which are common for all tested compounds. Keratinocytes were treated with 10−5 M of melatonin, its metabolites or vehicle for 24 h and submitted for RNA-sequence analysis.
3.2. Bioinformatics Analysis
IPA demonstrated common regulation of top three diseases and disorders including cancer, dermatological diseases and conditions and organismal injuries and abnormalities by all compounds, with two additional ones having different sequence orders: reproductive system disease and endocrine system disorders for melatonin, reproductive system disease and psychological disorders for 6(OH)Mel, and endocrine system disorders and gastrointestinal diseases for AFMK (Table 1, Supplementary Materials Table S1). There were also similarities with some differences in regulation of molecular and cellular functions that included cell-to-cell signaling and interactions, gene expression, cell signaling, molecular transport and vitamin and mineral metabolisms for melatonin; molecular transport, cell-to-cell signaling and interaction, cell signaling, vitamin and mineral metabolisms and cell morphology for 6(OH)Mel; molecular transport, cellular function and maintenance, cell signaling, vitamin and mineral metabolisms and cellular movement for AFMK (Table 1).
Table 1.
Summary of Ingenuity Pathway Analysis (IPA) for top common classes of diseases and biofunctions regulated by melatonin and its metabolites based on the data obtained from primary human epidermal keratinocytes.
| Top Diseases and Biofunctions | Melatonin | AFMK | 6(OH)Mel | |||
|---|---|---|---|---|---|---|
|
p value range | Molecules | p value range | Molecules | p value range | Molecules |
| Cancer | 1.39 × 10−3 ÷ 4.30 × 10−21 | 1313 | 3.11 × 10−3 ÷ 2.06 × 10−23 | 1419 | 5.59 × 10−3 ÷ 8.62 × 10−21 | 1211 |
| Dermatological diseases and conditions | 2.99 × 10−4 ÷ 4.30 × 10−21 | 1073 | 1.60 × 10−3 ÷ 2.06 × 10−23 | 1127 | 5.22 × 10−3 ÷ 8.62 × 10−21 | 1065 |
| Organismal injury and abnormalities | 1.48 × 10−3 ÷ 4.30 × 10−21 | 1454 | 3.14 × 10−3 ÷ 2.06 × 10−23 | 1528 | 5.94 × 10−3 ÷ 8.62 × 10−21 | 1330 |
| Reproductive system disease | 1.48 × 10−3 ÷ 3.49 × 10−13 | 104 | 1.13 × 10−10 | 19 | 5.78 × 10−3 ÷ 6.37 × 10−9 | 47 |
| Endocrine system disorders | 1.47 × 10−3 ÷ 9.57 × 10−13 | 598 | 3.14 × 10−03 ÷ 1.56 × 10−12 | 252 | 6.1 × 10−8 | 194 |
| Gastrointestinal disease | 6.37 × 10−8 | 26 | 3.11 × 10−3 ÷ 1.56 × 10−12 | 404 | 1.6 × 10−7 | 86 |
| Psychological disorders | 1.7 × 10−7 | 18 | 1.35 × 10−5 | 31 | 5.93 × 10−3 ÷ 4.25 × 10−8 | 170 |
|
p value range | Molecules | p value range | Molecules | p value range | Molecules |
| Cell to cell signaling and interaction | 1.47 × 10−3 ÷ 2.19 × 10−9 | 415 | 1.2 × 10−5 | 86 | 5.96 × 10−3 ÷ 1.71 × 10−9 | 416 |
| Gene expression | 5.80 × 10−8 ÷ 5.80 × 10−8 | 25 | 1.96 × 10−6 | 23 | 5.15 × 10−5 | 20 |
| Cell signaling | 1.39 × 10−3 ÷ 7.55 × 10−7 | 172 | 3.11 × 10−3 ÷ 2.71 × 10−7 | 191 | 5.96 × 10−3 ÷ 1.23 × 10−8 | 205 |
| Molecular transport | 1.43 × 10−3 ÷ 7.55 × 10−7 | 267 | 3.12 × 10x−3 ÷ 7.72 × 10−10 | 375 | 5.96 × 10−3 ÷ 3.08 × 10−10 | 349 |
| Vitamin and mineral metabolism | 7.29 × 10−4 ÷ 7.55 × 10−7 | 138 | 2.43 × 10−3 ÷ 2.71 × 10−7 | 150 | 5.96 × 10−3 ÷ 1.23 × 10−8 | 169 |
| Cellular function and maintenance | 1.49 × 10−6 | 69 | 3.12 × 10−3 ÷ 1.06 × 10−9 | 157 | 2.51 × 10−7 | 18 |
| Cellular movement | 3.95 × 10−6 | 37 | 2.95 × 10−3 ÷ 4.56 × 10−7 | 208 | 7.3 × 10−6 | 62 |
| Cell morphology | 9.84 × 10−4 | 7 | 5.36 × 10−5 | 15 | 5.34 × 10−3 ÷ 2.51 × 10−7 | 97 |
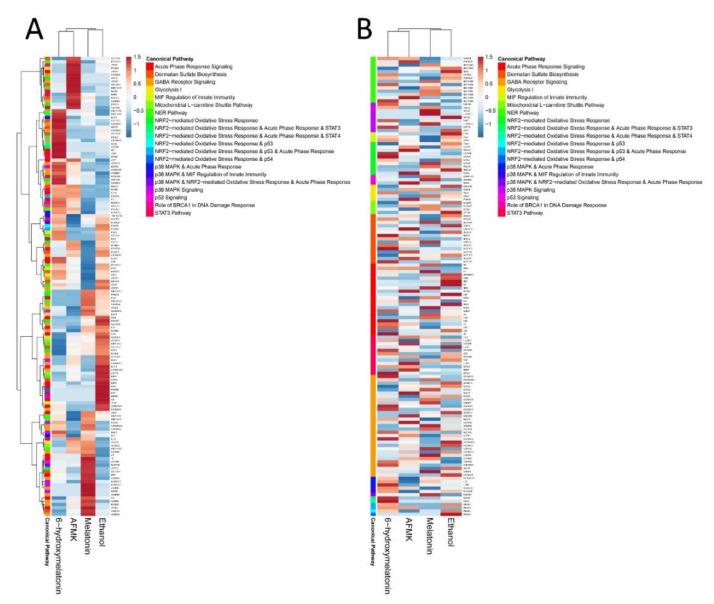
GSEA and Reactome FIViZ revealed upregulation of gene sets connected with responses to oxidative stress and DNA repair (Table 2). IPA was consistent with this analysis by showing stimulation of the p53 pathway and p38 MAPK signaling, NER pathway, NRF2-mediated oxidative stress response by melatonin, AFMK and 6(OH)Mel and the role of BRCA1 in DNA damage response only by melatonin and AFMK (Figure 2; Figure 3).
Table 2.
Gene Set Enrichment Analysis (GSEA) results for oxidative stress and DNA damage response gene sets affected by melatonin, AFMK and 6-hydroxymelatonin (6(OH)Mel) in human epidermal keratinocytes; NES—Normalized Enriched Score; (×)—the effect is absent.
| Reactome Pathway | GSEA for Melatonin | GSEA for AFMK |
GSEA for 6(OH)Mel |
|||
|---|---|---|---|---|---|---|
| NES | Direction | NES | Direction | NES | Direction | |
| Detoxification of reactive oxygen species | 1.09 | ↑ | 1.10 | ↑ | 1.10 | ↑ |
| DNA double-strand break response | 1.03 | ↑ | 1.04 | ↑ | 1.03 | ↑ |
| Base excision repair | 0.99 | ↑ | × | × | 0.99 | ↑ |
| p53-dependent G1/S DNA damage checkpoint | 1.11 | ↑ | 1.09 | ↑ | × | × |
| DNA double-strand break repair | 0.99 | ↑ | 1.00 | ↑ | 0.99 | ↑ |
| p53-independent G1/S DNA damage checkpoint | 1.10 | ↑ | 1.12 | ↑ | 1.11 | ↑ |
| TP53 regulates transcription of DNA repair genes | 1.01 | ↑ | 1.002 | ↑ | 0.99 | ↑ |
| Regulation of TP53 activity through methylation | 1.07 | ↑ | 1.10 | ↑ | 1.08 | ↑ |
| Regulation of TP53 activity through phosphorylation | 1.00 | ↑ | × | × | × | × |
| Recognition of DNA damage by PCNA-containing replication complex | 1.05 | ↑ | × | × | 1.06 | ↑ |
| Dual incision in GG-NER | 1.05 | ↑ | × | × | × | × |
| Gap-filling DNA repair synthesis and ligation in GG-NER | 1.05 | ↑ | × | × | 1.05 | ↑ |
| PCNA-dependent long patch base excision repair | 1.03 | ↑ | × | × | × | × |
| DNA damage recognition in GG-NER | × | × | 1.08 | ↑ | 1.07 | ↑ |
| Gap-filling DNA repair synthesis and ligation in TC-NER | × | × | 1.04 | ↑ | 1.04 | ↑ |
| G2/M DNA damage checkpoints | × | × | 1.02 | ↑ | × | × |
| G1/S DNA damage checkpoints | × | × | 1.09 | ↑ | 1.10 | ↑ |
| Stabilization of p53 | × | × | 1.12 | ↑ | × | × |
| DNA damage bypass | × | × | 1.05 | ↑ | × | × |
| Nucleotide excision repair | × | × | × | × | 1.04 | ↑ |
Figure 2.
Heatmaps created from canonical pathways results of IPA with clustering (A) or without clustering (B) of the sets of the genes involved in enriched pathways.
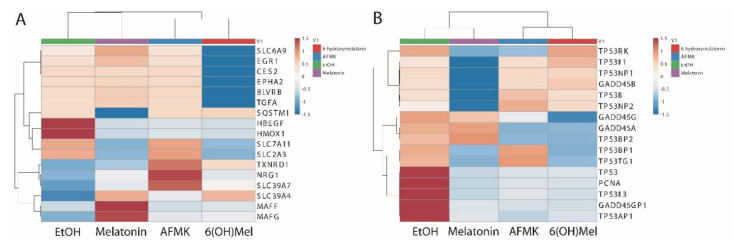
Figure 3.
Heatmaps for NRF2 (A) and TP53 (B) signaling pathways after treatment of human epidermal keratinocytes with melatonin and its derivatives.
The profile and Z-scores of expressions of the genes involved in NRF2 downstream signaling affected by tested compounds are shown in Figure 3A. Expression of solute carrier family 6 member 9 (SLC6A9) was stimulated by melatonin treatment, inhibited by 6(OH)Mel and did not change after AFMK treatment. Expression of early growth response protein 1 (EGR1) was upregulated by melatonin, downregulated by 6(OH)Mel and showed no change for AFMK, carboxylesterase 2-phase 1 protein (CES2), EPH receptor A2 (EPHA2, which belongs to ephrin receptor subfamily), biliverdin reductase B (BLVRB), and transforming growth factor α (TGFA) and had similar expression levels to the control after treatment with AFMK or melatonin and was downregulated by 6(OH)Mel. SQSTM1 (sequestosome 1) had a similar Z-score for AFMK and 6(OH)Mel and the control and a negative Z-score value (-1.5) for melatonin. Both heparin binding EGF-like growth factor (HBEGF) and heme oxygenase 1 (HMOX1) genes were inhibited by the tested compounds. Expression of solute carrier family 7 member 11 (SLC7A11) and solute carrier family 2 member 3 (SLC2A3) was inhibited by melatonin and 6(OH)Mel, but not by AFMK. Thioredoxin reductase 1 (TXNRD1) was strongly upregulated by AFMK and to a lower degree by 6(OH)Mel and slightly by melatonin. This observation is consistent with reported role of AFMK in protection against oxidative stress [10,26,44]. Expression of neuregulin 1 (NRG1) was only stimulated by AFMK. Transporter solute carrier family 39 member 7 (SLC39A7) only had a positive Z-score for AFMK, and solute carrier family 39 member 4 (SLC39A4) only had a positive Z-score for 6(OH)Mel and melatonin. MAF bZIP transcription factor F (MAFF) and v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G (MAFG), crucial regulators of mammalian gene expression, were strongly upregulated only by melatonin with weak stimulation by AFMK and 6(OH)Mel for MAFG only. In summary, these analyses are consistent with previous experimental data showing that melatonin and its metabolites differentially regulate the Nrf2 signaling pathway in keratinocytes [12,23,27,36] and such regulation would depend on melatonin metabolism [22].
The profile and Z-scores of expressions of the genes involved in TP53 signaling are presented in Figure 3B. TP53BP1 and TP53TG1 were upregulated by AFMK. The expression levels of growth arrest and DNA damage-inducible protein gamma (GADD45G) and GADD45A increased after melatonin treatment, and TP53I3 and TP53INP2 were upregulated by AFMK and 6(OH)Mel. The increased GADD45G level is consistent with melatonin involvement in cellular stress responses and DNA repair processes, as well as tumor suppression [1,10,12,21,26,27].
Increased GADD45A expression by melatonin was associated with DNA damage-induced transcription of this gene mediated by both p53-dependent and p53-independent mechanisms and responses to environmental stress by mediating activation of the p38/JNK pathway via MTK1/MEKK4 kinase. P53I11 and TP53INP1 were also upregulated by AFMK and 6(OH)Mel. All compounds increased the expression of GADD45B, indicating their involvement in the response to environmental stress by activation of p38/JNK signaling [45].
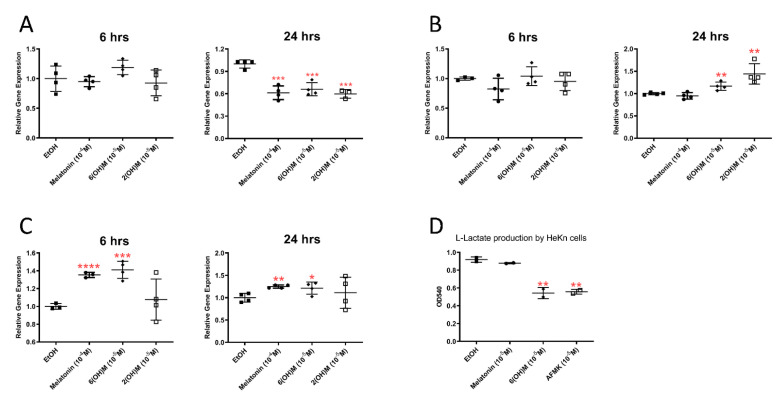
The expression profile of genes involved in a mitochondrial metabolism was similar for all tested compounds (Table 3). However, minor differences were seen in the regulation of pathway of glycogen synthesis, which was upregulated by melatonin and AFMK, but not by 6(OH)Mel. The mitochondrial fatty acid beta-oxidation signaling was over-represented only after melatonin treatment (Table 3). Selective investigation of genes from the tested pathways showed a decrease in LDHA expression after 6(OH)Mel (fold change: −1.02) AFMK (fold change: −1.03) melatonin (fold change: −1.05) treatments. Similarly, a decrease in lactate dehydrogenase C (LDHC) gene expression was seen (fold change: 6(OH)Mel: −3.22, AFMK: −2.07, melatonin: −1.19).
Table 3.
GSEA for mitochondrial metabolism/signaling in primary human epidermal keratinocytes treated with melatonin, 6(OH)Mel or AFMK; NES—Normalized Enriched Score; FDR—False Discovery Rate; (×)—the effect is absent.
| Influence of Melatonin Metabolites on Mitochondrial Metabolism | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Reactome Pathway | GSEA for Melatonin | GSEA for 6(OH)Mel | GSEA for AFMK | ||||||
| NES | FDR | Direction | NES | FDR | Direction | NES | FDR | Direction | |
| Citric acid cycle | 1.06 | 0.50 | ↑ | 1.07 | 0.49 | ↑ | 1.06 | 0.52 | ↑ |
| Glucose metabolism | 1.06 | 0.50 | ↑ | 1.05 | 0.51 | ↑ | 1.05 | 0.51 | ↑ |
| Mitochondrial translation | 1.07 | 0.50 | ↑ | 1.07 | 0.49 | ↑ | 1.07 | 0.50 | ↑ |
| Cristae formation | 1.07 | 0.50 | ↑ | 1.07 | 0.49 | ↑ | 1.09 | 0.54 | ↑ |
| Pyruvate metabolism and citric acid cycle | 1.05 | 0.50 | ↑ | 1.05 | 0.51 | ↑ | 1.05 | 0.51 | ↑ |
| Mitochondrial translation elongation | 1.08 | 0.51 | ↑ | 1.09 | 0.57 | ↑ | 1.09 | 0.53 | ↑ |
| Pyruvate metabolism | 1.05 | 0.52 | ↑ | 1.03 | 0.53 | ↑ | 1.06 | 0.50 | ↑ |
| Mitochondrial protein import | 1.04 | 0.53 | ↑ | 1.05 | 0.51 | ↑ | 1.03 | 0.54 | ↑ |
| Mitochondrial biogenesis | 1.04 | 0.53 | ↑ | 1.05 | 0.51 | ↑ | 1.03 | 0.54 | ↑ |
| Mitochondrial translation initiation | 1.09 | 0.54 | ↑ | 1.07 | 0.48 | ↑ | 1.08 | 0.51 | ↑ |
| Mitochondrial fatty acid β-oxidation |
1.03 | 0.56 | ↑ | × | × | × | × | × | × |
| The citric acid (TCA) cycle and respiratory electron transport | 1.09 | 0.57 | ↑ | 1.09 | 0.56 | ↑ | 1.08 | 0.52 | ↑ |
| Gluconeogenesis | 1.12 | 0.85 | ↑ | 1.13 | 0.77 | ↑ | 1.12 | 0.86 | ↑ |
| Glycogen metabolism | 1.11 | 0.76 | ↑ | 1.11 | 0.72 | ↑ | 1.11 | 0.79 | ↑ |
| Mitochondrial tRNA aminoacylation | 1.00 | 0.61 | ↑ | 1.00 | 0.59 | ↑ | 1.00 | 0.60 | ↑ |
| Respiratory electron transport | 1.10 | × | × | 1.10 | 0.63 | ↑ | 1.09 | 0.57 | ↑ |
| Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins | × | × | × | 1.10 | 0.68 | ↑ | 1.11 | 0.78 | ↑ |
| Glycogen synthesis | 1.10 | 0.67 | ↑ | × | × | × | 1.09 | 0.57 | ↑ |
Therapeutic implications for melatonin and its metabolites based on the GSEA are shown in Table 4. GSEA shows an increase in extension of telomeres, telomere maintenance, telomere C-strand (lagging strand) synthesis and packaging of telomere end signaling, which indicates antiaging properties of melatonin, AFMK and 6(OH)Mel in the skin. These analyses are consistent with the recognized antiaging properties of melatonin [46], supported further by the functional data presented in Section 3.4. In addition, the GSEA also suggests an antiviral activity of melatonin and its metabolites in keratinocytes with upregulation of innate immunity. This is consistent with IPA, indicating a role of RIG1-like receptors in the antiviral innate immunity pathway for 6(OH)Mel as well as the role of PKR in the interferon induction and antiviral responses. The role of RIG1-like receptors in antiviral innate immunity and the role of PKR in interferon induction and antiviral response signaling were also seen after AFMK treatment. The same set of pathways was found in IPA for melatonin. These data are consistent with previous reports on antiviral properties of melatonin [38].
Table 4.
Therapeutic implications for melatonin, AFMK and 6(OH)Mel based on GSEA. NES—normalized enriched score; FDR—false discovery rate; (×)—the effect is absent.
| Therapeutic Effects of Melatonin and Its Metabolites | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Reactome Pathway | GSEA for Melatonin | GSEA for 6(OH)Mel | GSEA for AFMK | ||||||
| NES | FDR | Direction | NES | FDR | Direction | NES | FDR | Direction | |
| Antiviral mechanism by IFN-stimulated genes | × | × | × | 1.03 | 0.57 | ↑ | 1.03 | 0.55 | ↑ |
| ISG15 antiviral mechanism | 1.03 | 0.55 | ↑ | × | × | × | 1.03 | 0.55 | ↑ |
| Defensins | 0.94 | 0.89 | ↑ | 0.94 | 0.90 | ↑ | 0.94 | 0.91 | ↑ |
| Beta defensins | 0.93 | 0.93 | ↑ | 0.92 | 0.95 | ↑ | 0.91 | 0.96 | ↑ |
| Tat-mediated HIV elongation arrest and recovery | 1.06 | 0.50 | ↑ | 1.05 | 0.51 | ↑ | 1.07 | 0.49 | ↑ |
| HIV elongation arrest and recovery | 1.07 | 0.51 | ↑ | 1.06 | 0.50 | ↑ | 1.08 | 0.49 | ↑ |
| Abortive elongation of HIV-1 transcript in the absence of Tat | 1.06 | 0.49 | ↑ | 1.06 | 0.50 | ↑ | × | × | × |
| Pausing and recovery of HIV elongation | 1.05 | 0.49 | ↑ | 1.06 | 0.51 | ↑ | × | × | × |
| Telomere maintenance | 0.98 | 0.69 | ↑ | 0.99 | 0.66 | ↑ | 0.99 | 0.64 | ↑ |
| Packaging of telomere ends | 1.01 | 0.60 | ↑ | 1.02 | 0.58 | ↑ | 1.03 | 0.55 | ↑ |
| Extension of telomeres | 0.96 | 0.83 | ↑ | 0.97 | 0.75 | ↑ | 0.97 | 0.76 | ↑ |
| Telomere C-strand (Lagging Strand) synthesis | 0.97 | 0.74 | ↑ | 0.96 | 0.83 | ↑ | 0.95 | 0.86 | ↑ |
| Interleukin-10 signaling | 1.06 | 0.49 | ↑ | 1.06 | 0.50 | ↑ | 1.07 | 0.49 | ↑ |
AFMK significantly downregulated IL-6 signaling pathway activation (Z-score = −2.121) in IPA. 6(OH)Mel also downregulated IL6 gene expression (fold change = −7.56), decreased the expression of IL6 signal transducer (Gp130) (fold change = −1.16) and decreased the expression of IL6R (fold change = −1.11). Similarly, AFMK decreased IL6 (fold change = −1.54), IL6ST (fold change = −1.06) and IL6R (fold change = −1.16) gene expressions. Melatonin also decreased expression of both IL6R (fold change = −1.29) and IL6ST (fold change = −1.28) genes. In GSEA, melatonin and its derivatives upregulated IL-10 signaling, which indicates their ability to suppress inflammation and immune responses (Table 4).
Melatonin and its metabolites (AFMK and 6(OH)Mel) protect primary keratinocytes against apoptosis by stimulation of expression of nuclear factor interleukin 3 (NFIL3) (RNAseq data) (Figure 1), indicating protection from programmed cell death (fold change = 1.28, = 1.11 for melatonin and AFMK, respectively) and by inhibition of apoptotic activator BCL2L11 (fold change = −1.35 and −1.08, for AFMK and 6(OH)Mel, respectively). IPA has shown that AFMK decreased signaling of calcium-induced T lymphocyte apoptosis (Z score = −1.67) and apoptosis signaling (Z score = −0.82). These analyses are consistent with anti-apoptotic and prodifferentiation effects of melatonin and/or its metabolites on human keratinocytes [7,10,13,17,23,28,44,47].
We also found an influence of melatonin on an expression of genes involved in the regulation of molecular clock in the skin through GSEA. We noticed an overrepresentation of circadian clock genes set. Differential expression analysis of RNAseq data indicated changes in expression of particular genes associated with circadian clock under influence of melatonin. There were upregulations of the NPAS2 (fold change = 2.3) and PPARγ (fold change = 2.7) genes as well as of the HDAC4 gene (fold change = 2.1) (Figure 1A). The activation of PPARγ leads to strong anti-inflammatory effects [48]. Other genes affected by melatonin included ARNTL (fold change = −1.1), PER1 (fold change = 1.05), PER3 (fold change = 1.38), CRY1 (fold change = 1.01) and CRY2 (fold change = 1.48), CLOCK (fold change = −1.01), SIRT1 and RORA (fold change = 1.52). Genes typical of the circadian network in the liver that have been stimulated by melatonin in keratinocytes include AURKB (fold change = −1.03), BIRC5 and CENPA (fold change = −1.01), CHEK1, CKAP5 and CSNK2A1 (fold change = −1.02), HLAB, INCENP, KIF2C and MAD2L1 (fold change = −1.3), MCM3 and MCM4 (fold change = −1.1), MCM6 (fold change = −1.2), RFC4 (fold change = −1.2), SFN, SPC24, SRC and UBD (fold change = −1.11), and ZWINT (fold change = −1.1). Therefore, we suggest that melatonin can also affect the circadian molecular clock in keratinocytes in a similar manner as in other organs [49].
The epidermal lipids produced in keratinocytes play an essential role in the skin’s barrier formation and function [50]. These lipids constitute a barrier against the loss of water and electrolytes and a barrier against microbial and viral invasion. However, elevated sebum excretion with lipid content is a key factor involved in the acne pathology [50,51]. The GSEA also suggested hypolipidemic and antiatherogenic potential for melatonin and its metabolites (Table 5).
Table 5.
Implications for cardiovascular system based on the GSEA of gene expression pattern affected by melatonin, AFMK and 6(OH)Mel in primary human epidermal keratinocytes. NES—normalized enriched score; (×)—the effect is absent.
| Reactome Pathway | GSEA for Melatonin | GSEA for 6(OH)Mel |
GSEA for AFMK |
|||
|---|---|---|---|---|---|---|
| NES | Direction | NES | Direction | NES | Direction | |
| Plasma lipoprotein assembly, remodeling, and clearance | 1.08 | ↑ | 1.08 | ↑ | 1.08 | ↑ |
| ABC-family proteins-mediated transport | 1.08 | ↑ | 1.08 | ↑ | × | × |
| Triglyceride catabolism | 1.04 | ↑ | × | × | × | × |
| LDL clearance | 1.09 | ↑ | 1.09 | ↑ | 1.08 | ↑ |
| PPARα activated gene expression | 1.01 | ↑ | × | × | 1.00 | ↑ |
| Assembly of active LPL and LIPC lipase complexes | 1.00 | ↑ | × | × | 1.01 | ↑ |
| Platelet homeostasis | 1.00 | ↑ | 1.00 | ↑ | × | × |
| Plasma lipoprotein clearance | × | × | × | × | 1.07 | ↑ |
| Prostacyclin signaling through prostacyclin receptor | × | × | 1.10 | ↑ | 1.11 | ↑ |
| Platelet calcium homeostasis | × | × | × | × | 0.98 | ↑ |
| Regulation of lipid metabolism by PPARα | × | × | × | × | 0.97 | ↑ |
| Nitric oxide stimulated guanylate cyclase | × | × | × | × | 0.86 | ↑ |
| Vasopressin regulates renal water homeostasis via aquaporins | × | × | 1.04 | ↑ | × | × |
| Regulation of insulin secretion | × | × | 1.04 | ↑ | × | × |
| Triglyceride catabolism | × | × | 1.03 | ↑ | × | × |
| ABC transporters in lipid homeostasis | × | × | 0.89 | ↑ | × | × |
Interestingly, melatonin and 6(OH)Mel increased gamma-aminobutyric acid (GABA) receptor signaling in canonical IPA (Figure 2). The GABA receptors respond to GABA, the main inhibitory neurotransmitter in the mature vertebrate central nervous system (CNS). Therefore, we speculate that melatonin, AFMK and 6(OH)Mel might act similarly to GABA receptor agonists, which are the class of drugs that typically possesses sedative action with anticonvulsant, anxiolytic and muscle relaxant activities [52].
Finally, the analysis indicates the anticancer potential of melatonin, AFMK and 6(OH)Mel by inhibition of pathways involved in process of carcinogenesis or by activation of anticancer immune signaling (Table 6). Antitumor properties of the tested compound were confirmed by an additional pathway analysis of differential expressed genes using the reactome platform. This showed negative regulation of Notch 4 signaling (with entities ratio = 0.008, p value = 0.04, FDR = 0.451; reactions ratio = 4.92). These analyses indicate complex action of melatonin and its metabolites against multiple types of tumors and are consistent with well-established anticancer activity of melatonin [1,8]. Interestingly, 6(OH)Mel and AFMK show inhibition of the osteoarthritis pathway in IPA with a Z-score = −0.535 and ratio = 0.102 and Z-score = −0.688 and ratio = 0.124, respectively. This analysis is consistent with the reported protective role caused by application of melatonin in experimental models of osteoarthritis [53,54,55,56].
Table 6.
Indications for protective effects of melatonin, AFMK and 6(OH)Mel against common human tumors based on Canonical Ingenuity Pathway Analysis (IPA) of data obtained from primary human epidermal keratinocytes. (×)—the effect is absent.
| IPAs | IPA for Melatonin |
IPA for AFMK |
IPA for 6(OH)Mel |
|||
|---|---|---|---|---|---|---|
| Z-Score | Ratio | Z-Score | Ratio | Z-Score | Ratio | |
| PD-1, PD-L1 cancer immunotherapy pathway | 0.33 | 0.11 | 1.51 | 0.15 | × | × |
| Renal cell carcinoma signaling pathway | −2 | 0.06 | −1.34 | 0.07 | × | × |
| Basal cell carcinoma signaling | × | 0.08 | × | × | −2.24 | 0.14 |
| Glioma signaling | −1.63 | 0.06 | −0.70 | 0.08 | × | × |
| Acute myeloid leukemia signaling | −1.34 | 0.08 | −1.41 | 0.11 | −1.34 | 0.08 |
| SPINK1 general cancer pathway | × | × | −0.70 | 0.15 | × | × |
| Estrogen-dependent breast cancer signaling | × | × | −1 | 0.12 | × | × |
| SPINK1 pancreatic cancer pathway | × | × | −0.44 | 0.12 | × | × |
| Small cell lung cancer signaling | × | × | −1 | 0.07 | × | × |
| Endometrial cancer signaling | × | × | −1 | 0.08 | × | × |
| Non-small cell Lung cancer signaling | × | × | −1.34 | 0.09 | × | × |
| Pancreatic adenocarcinoma signaling | × | × | −1.63 | 0.05 | × | × |
| Glioma Invasiveness signaling | × | × | −1.13 | 0.12 | × | × |
3.3. Cytokine Array Analysis
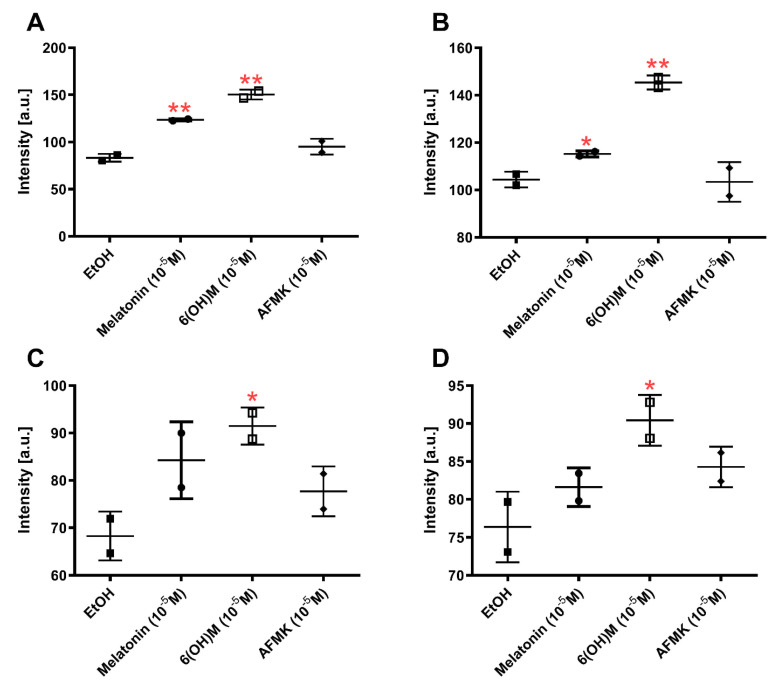
Keratinocytes are the main source of cytokines in the epidermis and secrete CSF, TNFα, IL-1, IL-6, IL-3, IL-8, TGFα, TGFβ, and PDGF [57]. They can act as a complex regulatory network in the epidermis influenced by physiological or pathological signals [58,59]. The cytokines’ activity is mediated by interaction with corresponding high affinity receptors [57]. The lower level of the expression of macrophage migration inhibitory factor (MIF) was noticed in DESeq2 analysis of RNAseq data for all tested compounds (fold change: 6(OH)Mel = −1.08, melatonin = −1.06, AFMK = −1.14). Tests using the cytokine profiler array have confirmed these results (Figure 4A). We also noticed elevated interleukin 1 receptor antagonist (IL1ra) protein expression after treatment with melatonin and with 6(OH)Mel (Figure 4B).
Figure 4.
Profile of cytokine expression for melatonin, AFMK and 6-hydroxymelatonin after 24 h incubation in HEKn as results of Proteome Profiler Cytokine Array: (A) MIF, (B) IL1ra, (C) SERPINE1, and (D) CXCL1 protein expression. The plots represent all values with calculated means and significance was evaluated by Mann–Whitney U test with * p < 0.05 and ** p < 0.01.
IL1ra is a competitive inhibitor of IL1, which participates in the process of silencing of inflammatory responses [60]. This suggests that melatonin and its derivatives can attenuate the proinflammatory conditions by increasing expression of ILra protein. Serpine family E member 1 (SERPINE1)’s protein expression was significantly elevated by 6(OH)Mel only (Figure 4C). SERPINE1 is a protein required for stimulation of keratinocyte migration during cutaneous injury repair [61]. We also noticed that 6(OH)Mel can enhance the C-X-C motif chemokine ligand 1 (CXCL1) level (Figure 4D). CXCL1 is an antimicrobial protein, which also plays a role in the antiviral innate immune response [62]. These findings are consistent with the above bioinformatic analyses and well-documented immunomodulatory and protective properties of melatonin [63].
3.4. Results of Telomerase Assay
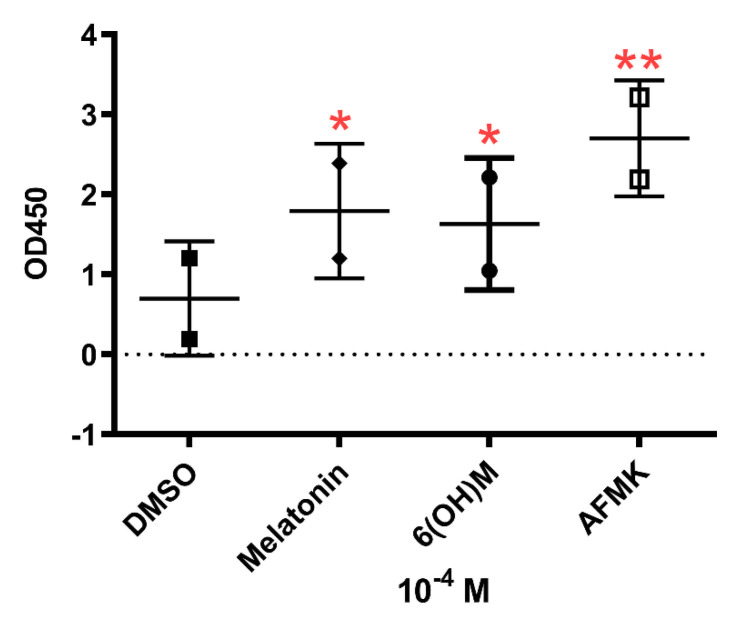
The RNAseq data and pathway analysis (Table 4) were confirmed by measurement of telomerase activity by TRAP assay in HaCaT keratinocytes treated with melatonin and its metabolites for 24 h at a concentration of 10−4 M (Figure 5). The use of an immortalized keratinocyte line was dictated by its phenotypic stability during passaging. The influence of the melatonin on telomerase activity has also been indicated by GO analysis of RNAseq data after stimulation with melatonin, which included overrepresentation of Positive-Regulation-of-Telomerase-Activity gene set enrichment with the parameter of Rank at Max equal 4064. These data and bioinformatics analyses support previous studies showing antiaging activity of melatonin in the human skin [31,43].
Figure 5.
Changes in telomerase activity in epidermal HaCaT keratinocytes under influence of melatonin, 6-hydroxymelatonin or AFMK at a concentration of 10−4 M. The plots represent all values with calculated means and significance was evaluated by paired Student t-Test with * p < 0.05; ** p < 0.01.
3.5. Energy Yielding Metabolism Assays
The bioinformatics analyses presented in Section 3.1 and Section 3.2 that indicated an effect on energy yielding metabolism were substantiated by qPCR analyses. Specifically, melatonin and its hydroxymetabolites stimulated sirtuin 1 (SIRT-1) and PGC1α, while inhibiting LDHA gene expressions in HaCaT keratinocytes (Figure 6). The latter finding has been confirmed by inhibition of lactate production in cultured cells treated with 6(OH)Mel and AFMK (Figure 6D).
Figure 6.
The effect of melatonin and its metabolites on the expressions of LDHA (A), PGC-1 (B) and SIRT-1 (C), and lactate production (D). Human epidermal (HaCaT) keratinocytes were cultured in presence of absence of the listed concentrations of the compounds and qPCR was performed as described in Materials and Methods with cyclophilin B as housekeeping gene. Each panel shows fold changes after calculated using delta-delta Ct normalized with housekeeping gene. Lactate accumulation in culture media is shown (D). The plots represent all values with calculated means and significance was evaluated both by Mann–Whitney U test and Student t-Test, which showed the same p values: * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001.
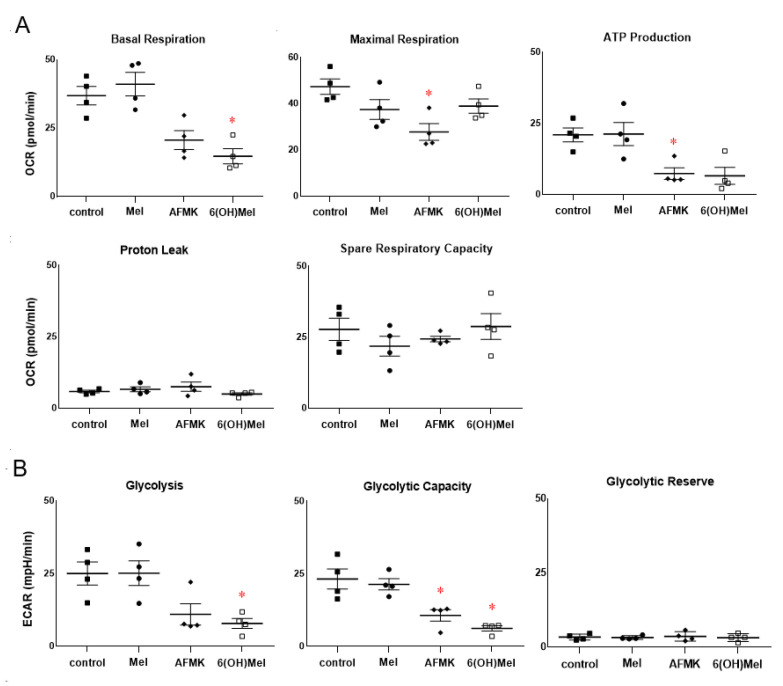
Next, we performed a comprehensive metabolic analysis using the Seahorse metabolic analyzer platform. The obtained data on the oxygen consumption rate (mitochondrial respiration, OCR, Figure 6A) and extracellular acidification rate (glycolytic function, ECAR, Figure 6B) showed consistency with the RNAseq, bioinformatics and functional analyses presented above. We noticed decreased basal respiration and ATP production 24 h following AFMK or 6(OH)Mel compared to control or melatonin stimulated conditions (Figure 7A). Maximal respiration, proton leak and spare respiratory capacity were unaffected by melatonin or its metabolites. Both AFMK and 6(OH)Mel, but not melatonin itself, significantly decreased glycolysis and glycolytic production in HEKn cells with a low oxygen consumption rate (Figure 7B), which is consistent with inhibition of lactate production by the compounds presented in Figure 5E. In general, these studies are in line with recognized abilities of melatonin and its metabolites to affect mitochondrial functions and their intramitochondrial metabolism [2,9,14,15,16,22,27,42,64].
Figure 7.
The Seahorse metabolic assay analysis performed in primary human epidermal keratinocytes treated with melatonin and its metabolites at a concentration of 10−5 M for 24 h. (A): OCR and (B): ECAR parameters are presented along the procedure as described in Materials and Methods. The plots represent all values with calculated means and significance was evaluated both by Mann–Whitney U test and Student t-Test, which showed the same p values: * p < 0.05. Passages 2 and 3 of primary epidermal keratinocytes were used for the experiments.
While previous studies on metabolic functions were predominantly performed on isolated mitochondria, the Seahorse metabolic platform utilizes the intact cells. The reported inhibition of basal respiration and ATP production and of glycolysis by melatonin metabolites in keratinocytes is consistent with reported previously inhibition of cell proliferation by melatonin and its downstream metabolites [13,21,30,44]. However, the lack of an effect of melatonin on the above metabolic parameters requires further investigation since melatonin is metabolized rapidly within mitochondria to 6(OH)Mel and AFMK and the presented cellular effect may represent an average of both melatonin effect and that of its metabolites. This issue will require additional studies that are beyond the scope of this manuscript.
4. Conclusions
Comprehensive analysis of RNAsequence data has shown significant changes in gene expression induced by melatonin and its two indolic (6(OH)Mel) and kynuric (AFMK) metabolites with an overlapping and differential pattern. This resulted in common regulation of top diseases and disorders and of molecular and cellular functions with some additional specificity for each of the tested molecules. This is common for melatonin, 6(OH)Mel and AFMF signaling pathways, with some signaling pathways being specific for each tested molecule. These may include common and separate nuclear receptors or regulatory proteins on which these molecules will act. GSEA, Reactome FIViZ and IPA indicated activation of p53 signaling and NRF2-mediated antioxidative pathways, which is consistent with our previous studies showing protective and antioxidative functions of melatonin and its metabolites. An overrepresentation of circadian clock and antiaging signaling by melatonin derivatives is consistent with antiaging properties of the melatonin. The GO showing an activation of the keratinocyte differentiation program by melatonin and the GSEA indicating antitumor activity are consistent with functional data reported previously demonstrating antiproliferative properties of melatonin and its metabolites in skin cells. Interestingly, the GSEA and IPA indicated, respectively, antilipidemic potential and antiviral responses controlled by melatonin and its metabolites. In addition, melatonin and metabolites had an effect on cellular metabolism and mitochondria functions in keratinocytes.
It is expected that the above changes will affect keratinocyte proliferation and differentiation programs to improve epidermal barrier formation, leading to increased protection against environmental insults and different pathogens. In addition, these analyses suggest possible applications of melatonin, AFMK and 6(OH)Mel in translational medicine.
Abbreviations
| BRCA1 | breast cancer gene 1 |
| DMSO | dimethyl sulfoxide |
| NRF2 | NF-E2–related factor 2 |
| p53 | tumor protein P53 |
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/antiox10040618/s1, Table S1: Summary of Ingenuity Pathway Analysis (IPA) for top biological functions regulated by melatonin and its metabolites based on the data obtained from primary human epidermal keratinocytes. Figure S1: Volcano Plot diagram of differentially expressed genes for melatonin vs EtOH (A), 6-hydroxymelatonin vs EtOH (B), and AFMK vs EtOH (C).
Author Contributions
J.S. designed and performed the experiments, analyzed obtained data, performed bioinformatics analysis, and wrote the first draft of the manuscript. T.-K.K. conducted the data analysis and prepared the layout of the graphs. D.K.C. performed bioinformatics analyses, deposited RNAseq. K.K., F.S. and M.K.T. analyzed metabolic function and evaluated the obtained results. A.T.S. conceived and designed the experiments, analyzed obtained data, and, together with K.K., obtained funding. Finally, A.T.S. together with K.K., T.-K.K., F.S., Z.J., R.M.S., K.S., J.Z., R.J.R., and M.K.T. drafted and approved the final version of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the grants: 1R01AR056666-01A2, 1R01AR073004-01A1, 1RO1AR071189-01A1, and R21AI149267-01A1 from NIH, by a VA merit grant (no. 1I01BX004293-01A1) (A.T.S.), and the German Research Foundation (Deutsche Forschungsgemeinschaft (DFG)); grant number: KL2900/2-1 (K.K.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All raw RNAseq data were deposited in NCBI GEO (GSE147588).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Reiter R.J., Rosales-Corral S.A., Tan D.X., Acuna-Castroviejo D., Qin L., Yang S.F., Xu K. Melatonin, a full service anti-cancer agent: Inhibition of initiation, progression and metastasis. Int. J. Mol. Sci. 2017;18:843. doi: 10.3390/ijms18040843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan D.X., Reiter R.J. Mitochondria: The birth place, battle ground and the site of melatonin metabolism in cells. Melatonin Res. 2019;2:44–66. doi: 10.32794/mr11250011. [DOI] [Google Scholar]
- 3.Cipolla-Neto J., Gaspar do Amaral F. Melatonin as a hormone: New physiological and clinical insights. Endocr. Rev. 2018;39:990–1028. doi: 10.1210/er.2018-00084. [DOI] [PubMed] [Google Scholar]
- 4.Slominski R.M., Reiter R.J., Schlabritz-Loutsevitch N., Ostrom R.S., Slominski A.T. Melatonin membrane receptors in peripheral tissues: Distribution and functions. Mol. Cell. Endocrinol. 2012;351:152–166. doi: 10.1016/j.mce.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reiter R.J., Tan D.X., Rosales-Corral S., Manchester L.C. The universal nature, unequal distribution and antioxidant functions of melatonin and its derivatives. Mini Rev. Med. Chem. 2013;13:373–384. doi: 10.2174/1389557511313030006. [DOI] [PubMed] [Google Scholar]
- 6.Slominski A., Tobin D.J., Zmijewski M.A., Wortsman J., Paus R. Melatonin in the skin: Synthesis, metabolism and functions. Trends Endocrinol. Metab. 2008;19:17–24. doi: 10.1016/j.tem.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 7.Fischer T.W., Zmijewski M.A., Wortsman J., Slominski A. Melatonin maintains mitochondrial membrane potential and attenuates activation of initiator (casp-9) and effector caspases (casp-3/casp-7) and PARP in UVR-exposed HaCaT keratinocytes. J. Pineal Res. 2008;44:397–407. doi: 10.1111/j.1600-079X.2007.00542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jung B., Ahmad N. Melatonin in cancer management: Progress and promise. Cancer Res. 2006;66:9789–9793. doi: 10.1158/0008-5472.CAN-06-1776. [DOI] [PubMed] [Google Scholar]
- 9.Reiter R.J., Rosales-Corral S., Tan D.X., Jou M.J., Galano A., Xu B. Melatonin as a mitochondria-targeted antioxidant: One of evolution’s best ideas. Cell. Mol. Life Sci. 2017;74:3863–3881. doi: 10.1007/s00018-017-2609-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slominski A.T., Kleszczyński K., Semak I., Janjetovic Z., Zmijewski M.A., Kim T.K., Slominski R.M., Reiter R.J., Fischer T.W. Local melatoninergic system as the protector of skin integrity. Int. J. Mol. Sci. 2014;15:17705–17732. doi: 10.3390/ijms151017705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer T.W., Sweatman T.W., Semak I., Sayre R.M., Wortsman J., Slominski A. Constitutive and UV-induced metabolism of melatonin in keratinocytes and cell-free systems. FASEB J. 2006;20:1564–1566. doi: 10.1096/fj.05-5227fje. [DOI] [PubMed] [Google Scholar]
- 12.Janjetovic Z., Stuart G., Jarrett S.G., Lee E.F., Duprey C., Reiter R.J., Slominski A.T. Melatonin and its metabolites protect human melanocytes against UVB-induced damage: Involvement of NRF2-mediated pathways. Sci. Rep. 2017;7:1274. doi: 10.1038/s41598-017-01305-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim T.K., Kleszczyński K., Janjetovic Z., Sweatman T., Lin Z., Li W., Reiter R.J., Fischer T.W., Slominski A.T. Metabolism of melatonin and biological activity of intermediates of melatoninergic pathway in human skin cells. FASEB J. 2013;27:2742–2755. doi: 10.1096/fj.12-224691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Semak I., Naumova M., Korik E., Terekhovich V., Wortsman J., Slominski A. A novel metabolic pathway of melatonin: Oxidation by cytochrome C. Biochemistry. 2005;44:9300–9307. doi: 10.1021/bi050202d. [DOI] [PubMed] [Google Scholar]
- 15.Semak I., Korik E., Antonova M., Wortsman J., Slominski A. Metabolism of melatonin by cytochrome P450s in rat liver mitochondria and microsomes. J. Pineal Res. 2008;45:515–523. doi: 10.1111/j.1600-079X.2008.00630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slominski A.T., Semak I., Fischer T.W., Kim T.K., Kleszczyński K., Hardeland R., Reiter R.J. Metabolism of melatonin in the skin: Why is it important? Exp. Dermatol. 2017;26:563–568. doi: 10.1111/exd.13208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer T.W., Zbytek B., Sayre R.M., Apostolov E.O., Basnakian A.G., Sweatman T.W., Wortsman J., Elsner P., Slominski A. Melatonin increases survival of HaCaT keratinocytes by suppressing UV-induced apoptosis. J. Pineal Res. 2006;40:18–26. doi: 10.1111/j.1600-079X.2005.00273.x. [DOI] [PubMed] [Google Scholar]
- 18.Slominski A., Baker J., Rosano T.G., Guisti L.W., Ermak G., Grande M., Gaudet S.J. Metabolism of serotonin to N-acetylserotonin, melatonin, and 5-methoxytryptamine in hamster skin culture. J. Biol. Chem. 1996;271:12281–12286. doi: 10.1074/jbc.271.21.12281. [DOI] [PubMed] [Google Scholar]
- 19.Slominski A.T., Kim T.K., Kleszczyński K., Semak I., Janjetovic Z., Sweatman T., Skobowiat C., Steketee J.D., Lin Z., Postlethwaite A., et al. Characterization of serotonin and N-acetylserotonin systems in the human epidermis and skin cells. J. Pineal Res. 2020;68:e12626. doi: 10.1111/jpi.12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slominski A., Pisarchik A., Semak I., Sweatman T., Wortsman J., Szczesniewski A., Slugocki G., McNulty J., Kauser S., Tobin D.J., et al. Serotoninergic and melatoninergic systems are fully expressed in human skin. FASEB J. 2002;16:896–898. doi: 10.1096/fj.01-0952fje. [DOI] [PubMed] [Google Scholar]
- 21.Slominski A., Wortsman J., Tobin D.J. The cutaneous serotoninergic/melatoninergic system: Securing a place under the sun. FASEB J. 2005;19:176–194. doi: 10.1096/fj.04-2079rev. [DOI] [PubMed] [Google Scholar]
- 22.Slominski A.T., Zmijewski M.A., Semak I., Kim T.K., Janjetovic Z., Slominski R., Zmijewski J.W. Melatonin, mitochondria, and the skin. Cell. Mol. Life. Sci. 2017;74:3913–3925. doi: 10.1007/s00018-017-2617-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slominski A.T., Zmijewski M.A., Plonka P.M., Szaflarski J.P., Paus R. How UV light touches the brain and endocrine system through skin, and why. Endocrinology. 2018;159:1992–2007. doi: 10.1210/en.2017-03230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slominski A., Wortsman J. Neuroendocrinology of the skin. Endocr. Rev. 2000;21:457–487. doi: 10.1210/er.21.5.457. [DOI] [PubMed] [Google Scholar]
- 25.Fischer T.W., Kleszczyński K., Hardkop L.H., Kruse N., Zillikens D. Melatonin enhances antioxidative enzyme gene expression (CAT, GPx, SOD), prevents their UVR-induced depletion, and protects against the formation of DNA damage (8-hydroxy-2’-deoxyguanosine) in ex vivo human skin. J. Pineal Res. 2013;54:303–312. doi: 10.1111/jpi.12018. [DOI] [PubMed] [Google Scholar]
- 26.Fischer T.W., Slominski A., Zmijewski M.A., Reiter R.J., Paus R. Melatonin as a major skin protectant: From free radical scavenging to DNA damage repair. Exp. Dermatol. 2008;17:713–730. doi: 10.1111/j.1600-0625.2008.00767.x. [DOI] [PubMed] [Google Scholar]
- 27.Kleszczyński K., Zillikens D., Fischer T.W. Melatonin enhances mitochondrial ATP synthesis, reduces reactive oxygen species formation, and mediates translocation of the nuclear erythroid 2-related factor 2 resulting in activation of phase-2 antioxidant enzymes (γ-GCS, HO-1, NQO1) in ultraviolet radiation-treated normal human epidermal keratinocytes (NHEK) J. Pineal Res. 2016;61:187–197. doi: 10.1111/jpi.12338. [DOI] [PubMed] [Google Scholar]
- 28.Skobowiat C., Brożyna A.A., Janjetovic Z., Jeayeng S., Oak A.S.W., Kim T.K., Panich U., Reiter R.J., Slominski A.T. Melatonin and its derivatives counteract the ultraviolet B radiation-induced damage in human and porcine skin ex vivo. J. Pineal Res. 2018;65:e12501. doi: 10.1111/jpi.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim T.K., Lin Z., Li W., Reiter R.J., Slominski A.T. N1-Acetyl-5-Methoxykynuramine (AMK) is produced in the human epidermis and shows antiproliferative effects. Endocrinology. 2015;156:1630–1636. doi: 10.1210/en.2014-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim T.K., Lin Z., Tidwell W.J., Li W., Slominski A.T. Melatonin and its metabolites accumulate in the human epidermis in vivo and inhibit proliferation and tyrosinase activity in epidermal melanocytes in vitro. Mol. Cell. Endocrinol. 2015;404:1–8. doi: 10.1016/j.mce.2014.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milani M., Sparavigna A. Antiaging efficacy of melatonin-based day and night creams: A randomized, split-face, assessor-blinded proof-of-concept trial. Clin. Cosmet. Investig. Dermatol. 2018;11:51–57. doi: 10.2147/CCID.S153905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheuer C., Pommergaard H.C., Rosenberg J., Gögenur I. Dose dependent sun protective effect of topical melatonin: A randomized, placebo-controlled, double-blind study. J. Dermatol. Sci. 2016;84:178–185. doi: 10.1016/j.jdermsci.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 33.Ren W., Liu G., Chen S., Yin J., Wang J., Tan B., Wu G., Bazer F.W., Peng Y., Li T., et al. Melatonin signaling in T cells: Functions and applications. J. Pineal Res. 2017;62:e12394. doi: 10.1111/jpi.12394. [DOI] [PubMed] [Google Scholar]
- 34.Tamtaji O.R., Mobini M., Reiter R.J., Azami A., Gholami M.S., Asemi Z. Melatonin, a toll-like receptor inhibitor: Current status and future perspectives. J. Cell. Physiol. 2019;234:7788–7795. doi: 10.1002/jcp.27698. [DOI] [PubMed] [Google Scholar]
- 35.Xia Y., Chen S., Zeng S., Zhao Y., Zhu C., Deng B., Zhu G., Yin Y., Wang W., Hardeland R., et al. Melatonin in macrophage biology: Current understanding and future perspectives. J. Pineal Res. 2019;66:e12547. doi: 10.1111/jpi.12547. [DOI] [PubMed] [Google Scholar]
- 36.Kleszczyński K., Slominski A.T., Steinbrink K., Reiter R.J. Clinical trials for use of melatonin to fight against COVID-19 are urgently needed. Nutrients. 2020;12:2561. doi: 10.3390/nu12092561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reiter R.J., Ma A., Sharma R. Treatment of ebola and other infectious diseases: Melatonin “goes viral”. Melatonin Res. 2020;3:43–57. doi: 10.32794/mr11250047. [DOI] [Google Scholar]
- 38.Tan D.X., Hardeland R. Potential utility of melatonin in deadly infectious diseases related to the overreaction of innate immune response and destructive inflammation: Focus on COVID-19. Melatonin Res. 2020;3:120–143. doi: 10.32794/mr11250052. [DOI] [Google Scholar]
- 39.Hardeland R. Melatonin and inflammation-Story of a double-edged blade. J. Pineal Res. 2018;65:e12525. doi: 10.1111/jpi.12525. [DOI] [PubMed] [Google Scholar]
- 40.Hardeland R. Aging, melatonin, and the pro- and anti-inflammatory networks. Int. J. Mol. Sci. 2019;20:1223. doi: 10.3390/ijms20051223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacDonald I.J., Huang C.C., Liu S.C., Tang C.H. Reconsidering the role of melatonin in rheumatoid arthritis. Int. J. Mol. Sci. 2020;21:2877. doi: 10.3390/ijms21082877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kleszczyński K., Bilska B., Stegemann A., Flis D.J., Ziolkowski W., Pyza E., Luger T.A., Reiter R.J., Böhm M., Slominski A.T. Melatonin and its metabolites ameliorate UVR-induced mitochondrial oxidative stress in human MNT-1 melanoma cells. Int. J. Mol. Sci. 2018;19:3786. doi: 10.3390/ijms19123786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bocheva G., Slominski R.M., Slominski A.T. Neuroendocrine aspects of skin aging. Int. J. Mol. Sci. 2019;20:2798. doi: 10.3390/ijms20112798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Janjetovic Z., Nahmias Z.P., Hanna S., Jarrett S.G., Kim T.K., Reiter R.J., Slominski A.T. Melatonin and its metabolites ameliorate ultraviolet B-induced damage in human epidermal keratinocytes. J. Pineal Res. 2014;57:90–102. doi: 10.1111/jpi.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin S., Hoffmann K., Gao C., Petrulionis M., Herr I., Schemmer P. Melatonin promotes sorafenib-induced apoptosis through synergistic activation of JNK/c-jun pathway in human hepatocellular carcinoma. J. Pineal Res. 2017;62:e12398. doi: 10.1111/jpi.12398. [DOI] [PubMed] [Google Scholar]
- 46.Kleszczyński K., Fischer T.W. Melatonin and human skin aging. Dermatoendocrinology. 2012;4:245–252. doi: 10.4161/derm.22344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kleszczyński K., Tukaj S., Kruse N., Zillikens D., Fischer T.W. Melatonin prevents ultraviolet radiation-induced alterations in plasma membrane potential and intracellular pH in human keratinocytes. J. Pineal Res. 2013;54:89–99. doi: 10.1111/j.1600-079X.2012.01028.x. [DOI] [PubMed] [Google Scholar]
- 48.Le P.H., Kuo C.J., Cheng H.T., Wu R.C., Chen T.H., Lin C.J., Chiang K.C., Hsu J.T. Melatonin attenuates acute pancreatitis-induced liver damage through Akt-dependent PPAR-γ pathway. J. Surg. Res. 2019;236:311–318. doi: 10.1016/j.jss.2018.11.056. [DOI] [PubMed] [Google Scholar]
- 49.Matsui M.S., Pelle E., Dong K., Pernodet N. Biological rhythms in the skin. Int. J. Mol. Sci. 2016;17:801. doi: 10.3390/ijms17060801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pappas A. Epidermal surface lipids. Dermatoendocrinology. 2009;1:72–76. doi: 10.4161/derm.1.2.7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zouboulis C.C. Acne and sebaceous gland function. Clin. Dermatol. 2004;22:360–366. doi: 10.1016/j.clindermatol.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 52.Rosenstein R.E., Cardinali D.P. Central gabaergic mechanisms as targets for melatonin activity in brain. Neurochem. Int. 1990;17:373–379. doi: 10.1016/0197-0186(90)90019-P. [DOI] [PubMed] [Google Scholar]
- 53.Guo J.Y., Li F., Wen Y.B., Cui H.X., Guo M.L., Zhang L., Zhang Y.F., Guo Y.J., Guo Y.X. Melatonin inhibits Sirt1-dependent NAMPT and NFAT5 signaling in chondrocytes to attenuate osteoarthritis. Oncotarget. 2017;8:55967–55983. doi: 10.18632/oncotarget.18356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hosseinzadeh A., Kamrava S.K., Joghataei M.T., Darabi R., Shakeri-Zadeh A., Shahriari M., Reiter R.J., Ghaznavi H., Mehrzadi S. Apoptosis signaling pathways in osteoarthritis and possible protective role of melatonin. J. Pineal Res. 2016;61:411–425. doi: 10.1111/jpi.12362. [DOI] [PubMed] [Google Scholar]
- 55.Lim H.D., Kim Y.S., Ko S.H., Yoon I.J., Cho S.G., Chun Y.H., Choi B.J., Kim E.C. Cytoprotective and anti-inflammatory effects of melatonin in hydrogen peroxide-stimulated CHON-001 human chondrocyte cell line and rabbit model of osteoarthritis via the SIRT1 pathway. J. Pineal Res. 2012;53:225–237. doi: 10.1111/j.1600-079X.2012.00991.x. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Y., Lin J., Zhou X., Chen X., Chen A.C., Pi B., Pan G., Pei M., Yang H., Liu T., et al. Melatonin prevents osteoarthritis-induced cartilage degradation via targeting microRNA-140. Oxid. Med. Cell. Longev. 2019;2019:9705929. doi: 10.1155/2019/9705929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ansel J., Perry P., Brown J., Damm D., Phan T., Hart C., Luger T., Hefeneider S. Cytokine modulation of keratinocyte cytokines. J. Investig. Dermatol. 1990;94:101S–107S. doi: 10.1111/1523-1747.ep12876053. [DOI] [PubMed] [Google Scholar]
- 58.Kupper T.S. Interleukin 1 and other human keratinocyte cytokines: Molecular and functional characterization. Adv. Dermatol. 1988;3:293–307. [PubMed] [Google Scholar]
- 59.Kupper T.S., Chua A.O., Flood P., McGuire J., Gubler U. Interleukin 1 gene expression in cultured human keratinocytes is augmented by ultraviolet irradiation. J. Clin. Investig. 1987;80:430–436. doi: 10.1172/JCI113090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gabay C., Lamacchia C., Palmer G. IL-1 pathways in inflammation and human diseases. Nat. Rev. Rheumatol. 2010;6:232–241. doi: 10.1038/nrrheum.2010.4. [DOI] [PubMed] [Google Scholar]
- 61.Providence K.M., Higgins S.P., Mullen A., Battista A., Samarakoon R., Higgins C.E., Wilkins-Port C.E., Higgins P.J. SERPINE1 (PAI-1) is deposited into keratinocyte migration “trails” and required for optimal monolayer wound repair. Arch. Dermatol. Res. 2008;300:303–310. doi: 10.1007/s00403-008-0845-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang Y., Gnanadurai C.W., Fu Z. Critical roles of chemokines and cytokines in antiviral innate immune responses during rabies virus infection. Front. Agr. Sci. Eng. 2017;4:260–267. doi: 10.15302/J-FASE-2016116. [DOI] [Google Scholar]
- 63.Juybari K.B., Pourhanifeh M.H., Hosseinzadeh A., Hemati K., Mehrzadi S. Melatonin potentials against viral infections including COVID-19: Current evidence and new findings. Virus Res. 2020;287:198108. doi: 10.1016/j.virusres.2020.198108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reiter R.J., Sharma R., Ma Q. Switching diseased cells from cytosolic aerobic glycolysis to mitochondrial oxidative phosphorylation: A metabolic rhythm regulated by melatonin? J. Pineal Res. 2021;70:e12677. doi: 10.1111/jpi.12677. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw RNAseq data were deposited in NCBI GEO (GSE147588).