Abstract
Latent HIV-1 proviruses are capable of reactivating productive lytic infection, but the precise molecular mechanisms underlying emergence from latency are poorly understood. In this study, we determined the contribution of the transcription factors NF-κB, NFAT, and AP-1 in the reactivation of latent HIV following T-cell receptor (TCR) activation using Jurkat T-cell clones harboring single latent HIV proviruses. Our findings demonstrate that during reactivation from latency, NF-κB enhances HIV transcription while NFAT inhibits it by competing with NF-κB for overlapping binding sites on the HIV long terminal repeat (LTR). We have also demonstrated for the first time the molecular contribution of AP-1 in the reactivation of HIV from latency, whereby AP-1 synergizes with NF-κB to regulate HIV transcriptional elongation following TCR activation.
Keywords: HIV Latency, HIV transcriptional elongation, TCR activation
Human immunodeficiency virus (HIV) establishes latent infections in memory CD4 + T cells despite prolonged intensive drug therapy. The ability to latently infect T cells allows HIV to escape strong humoral and cellular immune responses against the viral proteins, with the latent provirus pools capable of reactivating productive lytic infections following cessation or disruption of drug therapy. Current highly active antiretroviral therapy (HAART) mainly utilizes drugs that target viral proteins such as integrase, protease, and reverse transcriptase enzymes that are essential for HIV-1 replication [1]. Unfortunately, the latent provirus pools in memory T cells present stable reservoirs of viral variants that are prone to the development of drug resistance [2-5]. Transcription from the HIV long terminal repeat (LTR) is regulated at the level of initiation and elongation by both cellular transcription factors and viral Tat protein acting in concert [6-9]. Several studies have demonstrated that NF-κB is the major transcription factor required for proviral activation in T cells [10-12]. In unactivated T cells, NF-κB is sequestered in the cytoplasm by binding to an inhibitor of NF-κB (IκB-α) [10,13]. When T cells become activated by exposure to proinflammatory cytokines such as TNF-α, antigens, or mitogens, IκB-α is phosphorylated by IκB-α kinase-β (IKK-β) to undergo ubiquitination and subsequent proteasomal degradation to release NF-κB [14,15]. Activated NF-κB translocates into the nucleus to activate transcription from a wide variety of promoters including HIV LTR, cellular cytokine, and chemokine genes [16]. The HIV LTR core promoter region contains two NF-κB binding sites near the transcription start site which are overlapped by NFAT-binding sequences and both NF-κB and NFAT can bind to this same site. During HIV transcription, NF-κB and Sp-1 bind cooperatively to the promoter to enhance proviral activation [17]. Efficient induction of HIV transcription occurs following interaction of NF-κB-Sp-1 transcription complex with the preinitiation complex TFIID, a multiprotein complex comprising of TATA-box-binding protein (TBP) and TBP-associated factors (TAF) [18,19]. Transcriptional elongation of latent HIV proviruses is regulated by viral transactivator protein Tat [7,20]. In the absence of Tat, the vast majority of RNA polymerase (RNAP) II that initiates transcription from the HIV LTR is less processive and results in abortive transcription near the promoter region [21-23].
However, in the presence of viral Tat protein, the human cyclin T1 subunit of positive transcription elongation factor-b (P-TEFb) interacts with Tat and cooperatively binds to TAR, an RNA stem-loop structure encoded by the first 59 nucleotides of the nascent RNA transcript. Cooperative binding of Tat and cyclin T1 to TAR element activates the kinase subunit of P-TEFb called cyclin-dependent kinase-9 (CDK9) which hyperphosphorylates the C-terminal domain (CTD) of RNAP II within the elongation complex to form highly phosphorylated and processive form of RNAP II [9,24-26]. Due to the quiescent nature of the latently infected memory T cells, very low levels of viral Tat are generated. Before new Tat is synthesized to regulate proviral transcriptional elongation, promoter clearance is mediated by TFIIH which comprises of cyclin H and CDK7 subunits through phosphorylation of the CTD of RNAPs [27,28]. However, Kim and colleagues demonstrated that following activation of the TCR, P-TEFb, which is a cellular co-factor for viral Tat protein, is mobilized through an ERK-dependent pathway to enhance HIV transcriptional elongation before new Tat synthesis [29]. To further extend, besides ERK-dependent pathways, we have shown the crucial role of DNA-PK in facilitating both the initiation and elongation phases of HIV transcription [30-32]. Other cellular factors such as the phosphorylated-Spt5 subunit of the 5, 6-dichloro-1-b-D-ribo-furanosylbenzimidazole (DRB) sensitivity inducing factor (DSIF) stabilize the elongating transcription complexes by preventing premature RNAP II disengagement from DNA templates [6,26]. It is conceivable that these transcription regulation mechanisms allow the initial synthesis of new Tat proteins in latently infected T cells to subsequently regulate HIV transcriptional elongation.
Although NF-κB on its own can activate HIV LTR transcription, several studies have demonstrated that AP-1 and NF-κB form functional transcription ternary complexes [33,34] with a potentiated biological activity which enhances HIV LTR activation [35]. On the other hand, AP-1 is also known to synergize with NFAT to regulate the expression of a variety of cytokine and chemokine genes involved in immune functions and regulation [36,37]. Interestingly, activation of the T-cell receptor (TCR) induces multiple transcription factors that have been implicated in HIV transcription regulation, including NF-κB [10-12,38,39], NFAT [38,40-42], and AP-1 [35,43-45]. Despite the well-established sequence of molecular events that lead to proviral activation by NF-κB, the molecular contribution of NFAT and AP-1 in the regulation of latent HIV provirus transcription following activation of the TCR remains unknown. Therefore, in the current study, we aim to investigate the molecular contribution of the transcription factors NFAT and AP-1 in HIV proviral transcription upon TCR activation.
Materials and methods
Plasmid constructs
pHR’p-d2EGFP was derived by inserting the EcoRI and XhoI fragment of HIV-1 pNL4-3 into the pHR’ plasmid [46]. The short-lived version of green fluorescent protein (d2EGFP) replaces nef position at the MluI and XhoI sites. Site-directed mutagenesis was performed to replace histidine at position 13 with leucine (H13L) (CAT to TTA) within the HIV Tat gene. The H13L Tat was used over the WT Tat because unlike WT Tat, nearly 100% of the clones harboring H13L Tat are reactivatable following TNF-α stimulation [47]. The pHR’p-d2EGFP constructs contained either wild-type (WT) LTR or mutations (GGG to CTC) were introduced at the 3’ end of each of the two NF-κB binding sites to form mutant NF-κB (mKF-κB)-binding sites within the LTR [11]. Wild-type LTR and LTR mutants derived from firefly LTR-luciferase reporter constructs were subcloned into the pHR’ p-d2EGFP vector at BamH1 and XhoI sites.
Infection and isolation of Jurkat T-cell clones
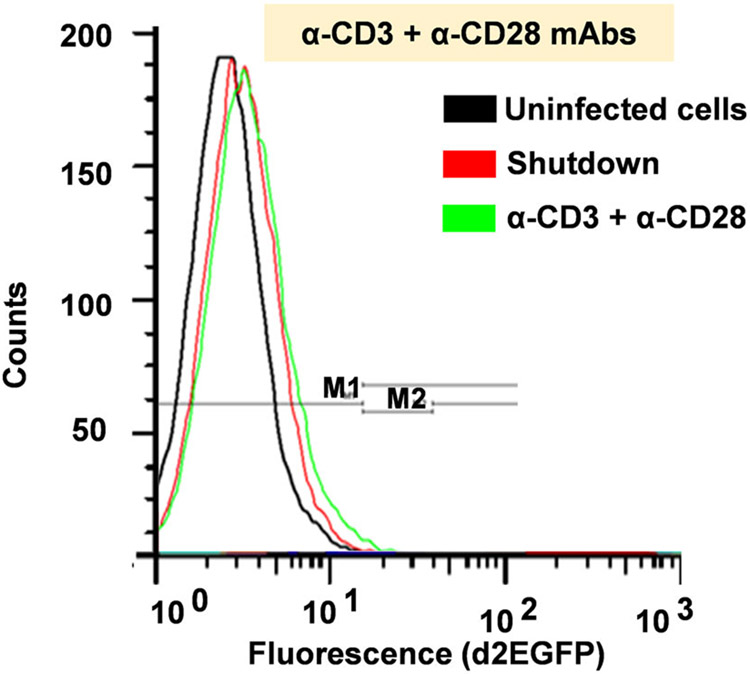
Infection of Jurkat T cells with lentiviral vectors and isolation of clones 2D10 (WT LTR and H13L Tat) and 2B5 (mNF-κB and H13L Tat) cells was previously described by Pearson et al. [47]. Vesicular stomatitis virus protein G (VSV-G)-pseudotyped HIV particles were produced by triple transfection of 293T cells using Lipofectamine 2000 reagent as described previously [27]. Virus titers were determined by infection of 2 × 106 Jurkat T cells with a serial dilution of concentrated virus preparation (harvested medium supernatant). Six hours postinfection, cells were washed with phosphate-buffered saline (PBS) and RPMI 1640 medium replaced. Expression of d2EGFP was assessed by fluorescently activated cell sorting analysis (FACS Calibur) 72 h postinfection, and d2EGFP expression subsequently analyzed every week until cells was fully shutdown without detectable d2EGFP expression before reactivation experiments.
Cell culture and reagents
Clone 2D10 and 2B5 cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), penicillin (100 IU·mL−1), streptavidin (100 IU·mL−1), and 25 mm HEPES at 37 °C in 5% CO2. Fresh medium was added to cells every 2–3 days, and cell density was maintained between 1–2 × 106 cells·mL−1. All the cell culture media, serum was procured from Gibco, USA, while the antibiotics (penicillin/streptomycin) were procured from the Thermo Fisher Scientific (USA). Antibodies for NF-κB, NFAT c1/c2, AP-1, and SPT-5 were also procured from Santa Cruz Biotechnology. Specific MAPK inhibitor PD98059 (PD) and cyclosporine A (CsA) inhibitor were procured from the Cell Signaling (Danvers, MA, USA) and Alfa Aesar (Haverhill, MA, USA), respectively.
Western blot analysis of NF-κB, NFAT, and AP-1 nuclear induction kinetics
Jurkat T-cell clones 2D10 and 2B5 were activated through the TCR using 0.125 μg·mL−1 of anti-CD3 monoclonal antibodies plus 1.0 μg·mL−1 of anti-CD28 monoclonal antibodies or 10 ng·mL−1 of TNF-α. For each time point consisting of 5 × 106 cells in a 6-h activation time course, samples were collected every 15 min until 1.25 h following activation; thereafter, samples were collected every 30 min until 6 h. Activated cells were washed with 500 μL of ice-cold PBS and allowed to swell in 500 μL of CE buffer (1 mm Hepes-KOH pH 7.9, 60 mm KCl, 1 mm EDTA, 0.5% NP-40, 1 mm DTT, 1 mm PMSF) and cells were vortexed for lysis. Nuclei were pelleted at 4K for 5 min. Cytoplasmic lysates were transferred to new Eppendorf tubes. Nuclei were washed with 500 μL of CE buffer and pelleted at 13K for 1 min. Nuclei were resuspended in 60 μL of NE buffer (250 mm Tris pH 7.8, 60 mm HCl, 1 mm EDTA, 1 mm DTT, 1 mm PMSF) and lysed by 4 freeze–thaw cycles in liquid nitrogen. The nuclear lysate was cleared by centrifugation at 13K for 1 min and supernatant transferred into a new microfuge tube. Total nuclear protein concentration in the samples was normalized using standard Bradford assay as described previously [48]. For each activation time point, 8.0 μL of total nuclear samples was loaded on to NuPAGE 10% Bis–Tris gels (Invitrogen, Carlsbad, CA, USA) for electrophoresis. Proteins on gels were transferred on to nitrocellulose membranes and detected using the ECL protein detection reagent (GE Healthcare, Chicago, IL, USA). The following mAbs specific for p65, p50 (NF-κB), NFATc1, NFATc2 (NFAT), c-Fos, Fra1, c-Jun, JunD (AP-1), and Spt5 were used in immunoblots. Densitometry analysis was used to determine relative protein levels in western blots using QuantityOne software (Bio-Rad, Hercules, CA, USA).
Electrophoretic Mobility Gel Shift Assay (EMSA)
Extraction of nuclear samples for Gel binding assay was performed using the protocol for western blot analysis. Ten percent of Glycerol was immediately added to nuclear lysate and samples frozen at −80 °C until ready for use. Two micrograms of nuclear lysate was reacted with 0.1 pmoles of 32P-labeled 30bp double-stranded oligonucleotides derived from the HIV-1 LTR containing the NF-κB binding sites or oligonucleotides derived from the IL-2 promoter containing the distal NF-AT binding sites in 6μl of binding buffer (10 mm Tris/HCl pH 7.5, 50 mm NaCl, 10% glycerol, 1% NP-40, 1 mm EDTA, 0.1 μg poly dI/dC) and incubated for 15 min at RT. The protein–oligonucleotide complexes were resolved on a nondenaturing 12.5% acrylamide (19:1) and 1X TGE buffer (24.8 mm Tris/HCl, 190 mm glycine, 1 mm EDTA) and visualized by standard autoradiography.
Preparation of double-stranded oligonucleotides
Two microliters of 12.5 μm single-stranded oligonucleotides was added to 5 μL of 32P-labeled γ-ATP, 2 μL of T4 polynucleotide kinase (PNK), 5 μL of 10X PNK buffer in 36 μL reaction volume and incubated at 37 °C for 1 h. Two microliters of 12.5 μm reverse-strand oligonucleotides was added to the reaction and boiled at 95 °C for 5 min; samples were spanned down and placed back on the heating block and allowed to cool down slowly to 30 °C. Double-stranded oligonucleotides were purified using nondenaturing 12.5% acrylamide (19 : 1) gel before use in binding assays.
Chromatin Immunoprecipitation (ChIP) assay
For each activation time point, 5 × 107 cells were fixed using 0.5% formaldehyde for 10 min at room temperature (RT). Cells were washed twice with 20 mL of ice-cold PBS and allowed to swell in 5 mL of CE buffer. Nuclei were pelleted at 2K for 10 min at +4 °C and resuspended in 1 mL of SDS-lysis buffer (1% SDS, 10 mm 191 EDTA, 50 mm Tris/HCl pH 8.1, 1 mm PMSF, 1 μ·mL−1 aprotinin, 1 μg·mL−1 pepstatin A). Genomic DNA was shredded to lengths less than 800 bp by sonication (Misonex 3000) under the following sonication conditions; output 2.5 for 20 s, 8 times. For each time point, 200 μL of sonicated samples was added to 800 μL of ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris/HCl pH 8.1, 167 mm NaCl). Samples were incubated with specific antibodies at + 4 °C overnight. One hundred microliters of 25% protein A-sepharose was used in DNA–protein immunoprecipitation. Antibody–DNA–protein complexes were washed with mL of each wash buffer in the order given below; low salt immune complex wash buffer (0.1% SDS, 1% Triton X-100, mm EDTA, 20 mm Tris/HCl pH 8.1, 150 mm NaCl), high salt immune complex wash buffer (0.1% SDS, 1% Triton X-v100, 2 mm EDTA, 20 mm Tris/HCl pH 8.1, 500 mm NaCl), RIPA buffer (20 mm Tris/HCl pH 7.5, 150 mm NaCl, 0.5% Triton X-100, 0.5% Na-deoxycholate, 0.1% SDS, 5 mm EDTA), and TE buffer (10 mm Tris/HCl pH 8.0, 1 mm EDTA pH 8.0) twice. Protein DNA complexes were eluted from protein A-sepharose twice using 250 μL of freshly prepared elution buffer (1% SDS, 0.1 mm NaHCO3). Twenty microliters of 5 m NaCl was added to total eluate and protein-DNA complexes reversed-cross-linked at 65 °C overnight. Ten microliters of 0.5 m EDTA, 10 μL of 2 m Tris/HCl pH 6.5, and 2 μL of 10 mg·mL−1 proteinase-K was added and samples incubated at 50 °C for 2hrs followed by phenol extraction and ethanol precipitation. ChIP was performed using the following antibodies, RNAP II (#39097, Active Motif) and c-Fos (K-25, Santa Cruz).
Real-time PCR analysis of eluted DNA
Precipitated DNA samples were dissolved in 250 μL of distilled water and 5 μL of the sample used in real-time PCR using the SYBR green PCR master mix (Bio-Rad) as described previously by Kim et al. [29]. No-antibody control values were subtracted from each sample value to remove the nonspecific background signal. The following HIV-1 primer sets were used in real-time PCR amplification. HIV Nuc-1 region (forward primer + 30) 5’ CTGGGA GCT CTC TGG CTA ACT-3’ (reverse primer + 134) 5’-TTA CCA GAG TCA CAC AAC AGA CG-3’. Downstream primers (HIV envelope region) (forward primers + 2593) 5’-TGA GGG ACT ATT GGA GAA 221 GTG A-3’ (reverse primer + 2691) 5’-TCT GCA CCA CTC TTC TCT TTG C-3’. Cellular gene primers used were EGR2 (forward primer + 35) 5’-CGA GGG GAC TCA CTG ACT GTT A-3’ (reverse primer + 126) 5’-TTG CCA CTG ACT CTC TCC TGT C-3’.
Statistical analysis
Data were analyzed using Microsoft Excel or Graphpad Prism 5.0 (GraphPad Software, San Diego, CA, USA). For paired samples, statistical analyses were performed using Student’s t-test. One-way analysis of variance (ANOVA) was performed for multiple data point comparisons. Experimental data are presented as the mean ± SD of at least three independent experiments. P < 0.05 was considered significant: P values were defined as * P < 0.05 and **P < 0.01.
Results
NF-κB is the major transcription factor induced by TNF-α stimulation
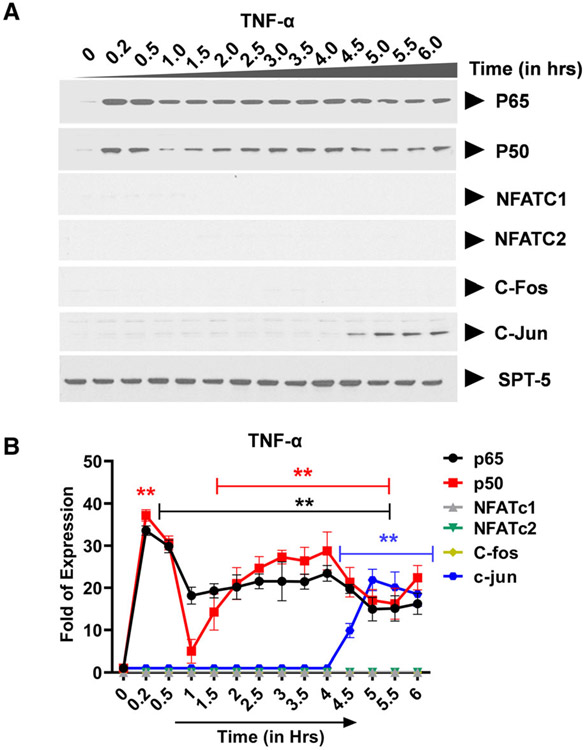
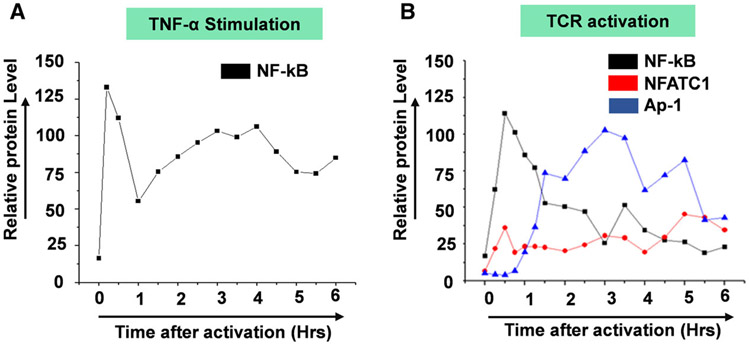
Earlier, most studies that have attempted to address the role of AP-1 transcription factors in the regulation of HIV transcription relied on ex vivo systems where cells are co-transfected with plasmid constructs that express high levels of exogenous proteins and the LTR-Luc reporters. The effects of exogenously expressed AP-1 transcription factors on HIV LTR activation are then assayed by measurement of the reporter protein expression usually 72 h post-transfection. Whereas this approach enabled determination of whether or not AP-1 activates HIV transcription, it was limited by the fact that it is insufficient to enable detailed kinetic studies of endogenously expressed AP-1 transcription factors and how they modulate the duration and magnitude of HIV transcriptional response in time-course experiments. In the experiments described here, we have chosen to study the nuclear induction kinetics of endogenously expressed transcription factors mobilized following TCR activation and how they modify the duration and magnitude of HIV provirus transcription. We utilized the well-established Jurkat T-cell line model of HIV latency; Clone 2D10 cells. The Clone 2D10 cells harbor a single latently infected HIV provirus. The latent HIV provirus in the Clone 2D10 cells contains WT LTR and H13L Tat in which nearly 100% of the latently infected Clone 2D10 cells are reactivatable following TNF-α stimulation. The use of immunoblots allowed us to determine transcription factors that become induced following TNF-α stimulation or TCR activation and their nuclear induction kinetics over a six-hour activation time course. Nuclear entry of NF-κB following TNF-α stimulation occurs in two distinct phases over a six-hour activation time course. Upon TNF-α stimulation, NF-κB is immediately induced into the nucleus reaching maximal nuclear levels by 30 min during the first induction phase lasting until 1.5 h (Fig. 1A,B). During the second activation phase, NF-κB nuclear levels peak between 3 and 4hrs. Albeit AP-1 c-Jun subunit is induced with a much-delayed kinetics after 4.5 h of TNF-α stimulation, AP-1 c-Fos, NFATc1, and NFATc2 are not induced by TNF-α stimulation (Fig. 1A,B).
Fig. 1.
Transcription factor: NF-κB exhibits a unique nuclear induction kinetics following T-cell activation by TNF-α. (A) Latently infected Clone 2D10 cells were activated by TNF-α in a six-hour time-course experiment. Nuclear fraction for each activation time point was used in western blot analysis to determine the nuclear levels and induction kinetics of NF-κB (p65 and p50), NFAT (c1 and c2), and AP-1 (c-Fos and c-Jun) transcription factors. The Spt5 subunit of the DSIF nuclear protein complex is constitutively expressed, and it was used as the loading control. Note that the results presented here are a representation of immunoblot experiments performed in triplicates. (B) Densitometry analysis of relative nuclear NF-κB (p65 and p50), NFAT (c1 and c2), and AP-1 (c-Fos and c-Jun) protein levels in (A) above. Error bars represent the mean ± SD of three independent and separate experiments. The P value of statistical significance was set as; P < 0.05 (*) or 0.01 (**).
T-cell receptor (TCR) activation induces NF-κB, NFAT, and AP-1 with unique kinetics
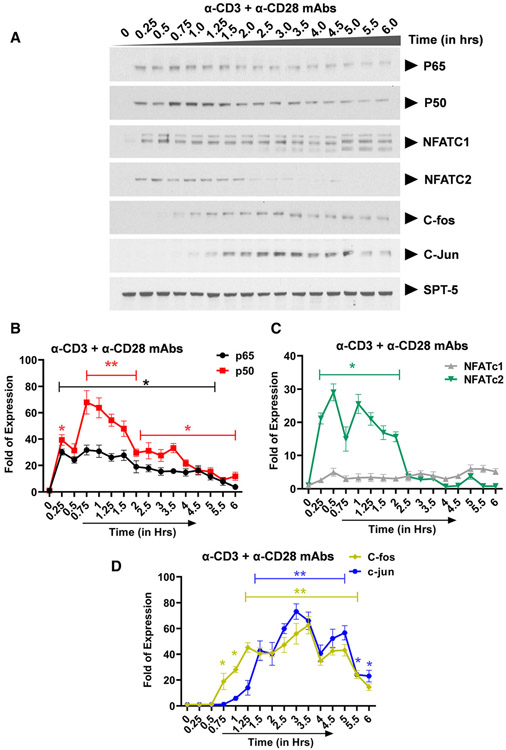
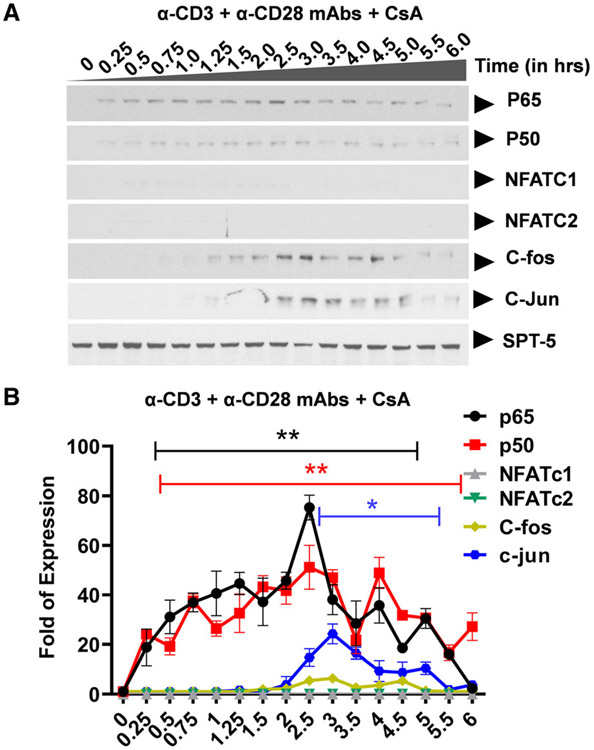
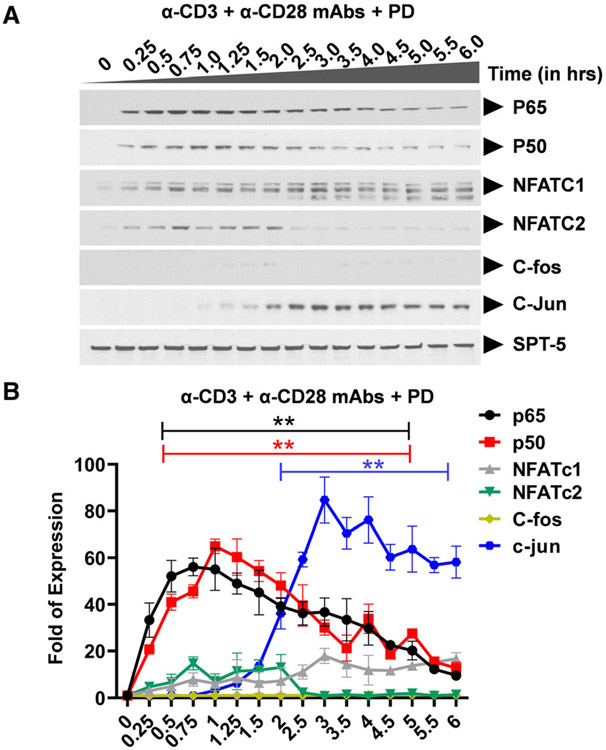
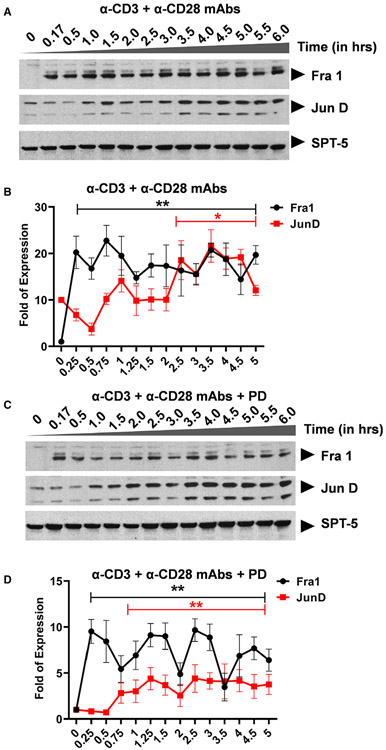
Ligation of the TCR results in activation of three distinct major signal transduction pathways, namely, (a) the MAPK pathway which leads to AP-1 induction, (b) the Ca2+-calcineurin pathway which results in NFAT induction, and (c) the protein kinase C (PKC) pathway which induces NF-κB via the CARMA1, Bcl10, and MALT1 (CBM) protein complex present upstream of the IκB kinase (IKK) complex. Activation of the TCR/CD3 complex using anti-CD3 and anti-CD28 mAbs enhances PKC-mediated NF-κB induction through activation of the Akt kinase (protein kinase B) which synergizes with the PKC activation signals. In contrast to TNF-α stimulation which induces NF-κB as the major factor, activation of the TCR using anti-CD3 and anti-CD28 mAbs is more complex and results in the induction of multiple transcription factors including NF-κB p65 and p50, NFATc1 and NFATc2, AP-1 c-Fos and c-Jun with varied induction kinetics. Western blot analysis using p65- and p50-specific antibodies demonstrated that basal levels of nuclear NF-κB are present in latently infected Jurkat T-cell clones, 2D10 cells (Fig. 2A,B and Fig. S1). SPT-5 was used as a nuclear protein loading control (Fig. S2). Soon after TCR activation, NF-κB is immediately mobilized into the nucleus reaching maximal nuclear levels by 30 min and between 3 to 4hrs during the first and second induction phases, respectively (Fig. 2A,B and Figure S1). The most notable differences in NF-κB induction following TNF-α or TCR activation occur during the second nuclear induction phase. Whereas the induction kinetics of NF-κB following TNF-α or TCR activation are comparable, there is significantly more nuclear NF-κB during the second activation phase following TNF-α stimulation than during TCR activation. The gel retardation assays further validated these findings, as we found a time-dependent binding of nuclear NF-κB to the HIV LTR, in a time-course experiment following TCR activation, with peak at around 30 min (Figure S1). This observation is not limited to the HIV LTR which harbors double NF-κB binding sites but it also applies to the canonical single NF-κB binding site derived from the IL-2 promoter. Like NF-κB, NFATc1 and NFATc2 are induced soon after activation. Whereas the induction levels of NFATc1 increase significantly during latter activation time points, NFATc2 is efficiently induced immediately following TCR activation but its induction is limited to early activation time points lasting until 2hrs (Fig. 2A-C). However, treatment of Clone 2D10 cells with cyclosporine A for 1 h before TCR activation resulted in inhibition of nuclear induction of both NFATc1 and NFATc2 (Fig. 3A,B). Unlike NF-κB and NFAT which are induced soon following TCR activation, nuclear induction of AP-1 c-Fos and c-Jun subunits is so unique and delayed until 45 min reaching maximal nuclear levels by 3 h (Fig. 2A,D). However, MAPK inhibitor PD98059 selectively and specifically inhibited AP-1 c-Fos but not c-Jun induction (Fig. 4A,B). Pretreatment of clone 2D10 cells with both PD98059 and cyclosporine A for 1hr before TCR activation inhibited both AP-1 c-Fos and NFAT induction (Fig. 5A,B). We also determined whether or not TCR activation induces other AP-1 subunits in addition to c-Fos and c-Jun. Interestingly, TCR activation also induced AP-1 Fra-1 and JunD, which was not inhibited by MAPK inhibitor PD98059 suggesting that unlike c-Fos, these AP-1 subunits are induced via a different MAPK signal pathway similar to c-Jun which is not affected by MAPK inhibitor PD98059 (Fig. 6A-D). The use of Clone 2D10 model system of HIV latency allowed us to study the detailed nuclear induction kinetics of the endogenous transcription factors that are mobilized following TCR activation. In this case, TCR activation, unlike TNF-α stimulation, induces multiple transcription factors with distinct nuclear induction kinetics over a six-hour time course.
Fig. 2.
Transcription factors: NF-κB, NFAT, and AP-1 exhibit unique nuclear induction kinetics following T-cell activation via anti-CD3 and anti-CD28 mAbs. (A) Latently infected Clone 2D10 cells were activated by using anti-CD3 and anti-CD28 mAbs in a six-hour time-course experiment. Nuclear fraction for each activation time point was used in western blot analysis to determine the nuclear levels and induction kinetics of NF-κB (p65 and p50), NFAT (c1 and c2), and AP-1 (c-Fos and c-Jun) transcription factors. The Spt5 subunit of the DSIF nuclear protein complex is constitutively expressed, and it was used as the loading control. Note that the results presented here is a representation of immunoblot experiments performed in triplicates. Densitometry analysis of relative nuclear (B) NF-κB (p65 and p50), (C) NFAT (c1 and c2), and (D) AP-1 (c-Fos and c-Jun) protein levels in (A) above. Error bars represent the mean ± SD of three independent and separate experiments. The P value of statistical significance was set as P < 0.05 (*) or 0.01 (**).
Fig. 3.
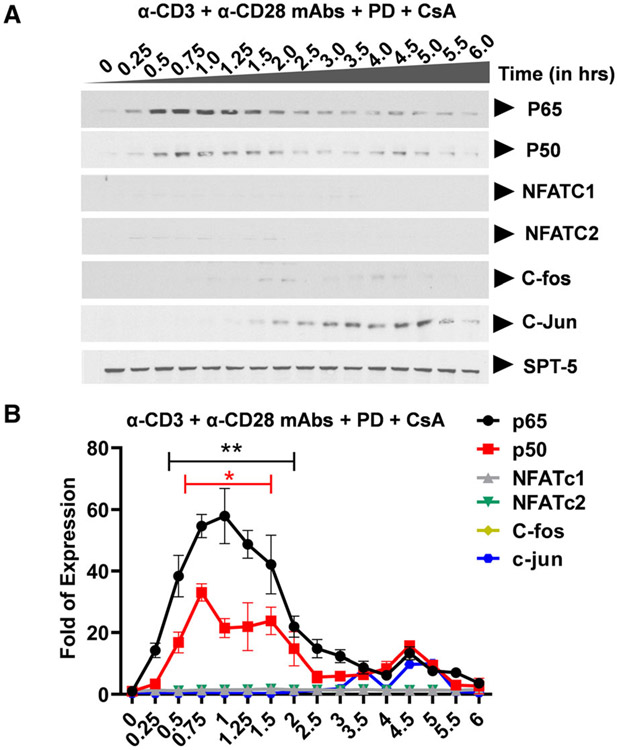
Western blot analysis showing specific inhibition of nuclear NFAT induction following TCR activation. (A) Clone 2D10 cells were pretreated with calcium-calcineurin inhibitor cyclosporine A (CsA) for one hour prior to the time-course activation experiment. Nuclear fractions were used to determine specific inhibition of NFAT, AP-1, and both AP-1 and NFAT nuclear induction using western blot analysis. Spt5 was used as the loading control. Note that the results presented here are a representation of immunoblot experiments performed in triplicates. (B) Densitometry analysis of relative nuclear NF-κB (p65 and p50), NFAT (c1 and c2), and AP-1 (c-Fos and c-Jun) protein levels in (A) above. Error bars represent the mean ± SD of three independent and separate experiments. The P value of statistical significance was set as P < 0.05 (*) or 0.01 (**).
Fig. 4.
Western blot analysis showing specific inhibition of nuclear AP-1 induction following TCR activation. (A) Clone 2D10 cells were pretreated with specific MAPK inhibitor PD98059 (PD) for one hour prior to the time-course activation experiment. Nuclear fractions were used to determine specific inhibition of NFAT, AP-1, and both AP-1 and NFAT nuclear induction using western blot analysis. Spt5 was used as the loading control. Note that the results presented here are a representation of immunoblot experiments performed in triplicates. (B) Densitometry analysis of relative nuclear NF-κB (p65 and p50), NFAT (c1 and c2), and AP-1 (c-Fos and c-Jun) protein levels in (A) above. Error bars represent the mean ± SD of three independent and separate experiments. The P value of statistical significance was set as P < 0.05 (*) or 0.01 (**).
Fig. 5.
Western blot analysis showing specific inhibition of nuclear AP-1 and NFAT induction following TCR activation. (A) Clone 2D10 cells were pretreated with combination of calcium-calcineurin inhibitor cyclosporine A (CsA) and specific MAPK inhibitor PD98059 (PD) for one hour prior to the time-course activation experiment. Nuclear fractions were used to determine specific inhibition of both AP-1 and NFAT nuclear induction using western blot analysis. Spt5 was used as the loading control. Note that the results presented here are a representation of immunoblot experiments performed in triplicates. (B) Densitometry analysis of relative nuclear NF-κB (p65 and p50), NFAT (c1 and c2), and AP-1 (c-Fos and c-Jun) protein levels in (A) above. Error bars represent the mean ± SD of three independent and separate experiments. The P value of statistical significance was set as; P < 0.05 (*) or 0.01 (**).
Fig. 6.
Activation of the TCR results in the induction of other AP-1 subunits. Western blot analysis using a nuclear fraction to determine additional AP-1 subunits other than c-Fos and c-Jun that become induced following TCR activation. (A) AP-1 Fra-1 and JunD subunits are induced following activation of the TCR using anti-CD3 and anti-CD28 mAbs. Spt5 nuclear protein was used as the loading control. (B) Densitometry analysis of relative nuclear Fra-1 and JunD protein levels in (A) above. Error bars represent the mean ± SD of three independent and separate experiments. The P value of statistical significance was set as P < 0.05 (*) or 0.01 (**). (C) Clone 2D10 cells were pretreated for 1hr with MAPK inhibitor PD prior to the TCR activation and nuclear fractions analyzed by western blot to determine whether or not PD inhibits Fra-1 or JunD induction. Spt5 nuclear protein was used as the loading control. (D) Densitometry analysis of relative nuclear Fra-1 and JunD protein levels in (C) above. Error bars represent the mean ± SD of three independent and separate experiments. The P value of statistical significance was set as; P < 0.05 (*) or 0.01 (**).
AP-1 contributes to latent HIV provirus transcription following TCR activation
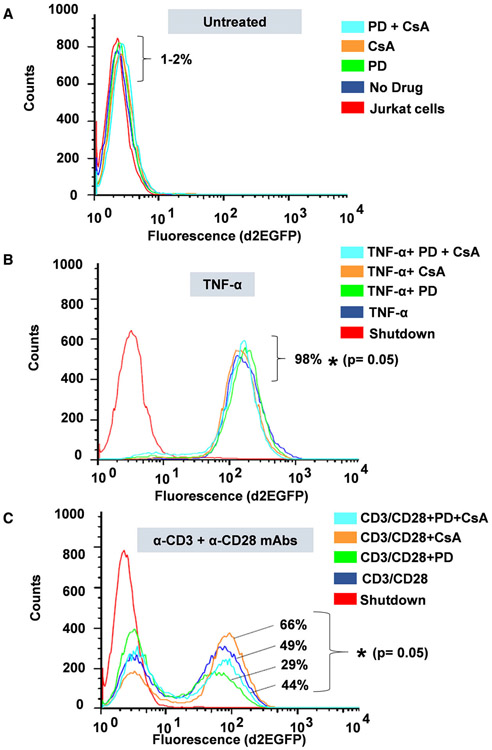
Using Clone 2D10 cells harboring single latently infected HIV proviruses, we determined the individual contribution of AP-1 c-Fos, NFAT, and NF-κB on d2EGFP expression as a measure of proviral transcription using FACS analysis following 16 h of TCR activation or TNF-α stimulation. Prior to FACS analysis after 16 h of cellular activation, Clone 2D10 cells were either untreated or pretreated for 1 h with MAPK inhibitor PD98059 to specifically inhibit c-Fos induction, CsA to inhibit NFATc1 and NFATc2 or PD/CsA drug combination to inhibit AP-1 c-Fos, NFATc1, and NFATc2 and cells activated using TNF-α or anti-CD3 and anti-CD28 mAbs. Drug treatments without activation were included to control for the effects of each inhibitor on provirus activation and were found to exhibit no activation effects on d2EGFP expression as expected (Fig. 7A). TNF-α stimulation of Clone 2D10 cells resulted in 98% d2EGFP expression demonstrating that activation of latent HIV provirus transcription in Jurkat T-cell clones is strictly dependent on NF-κB which is the major transcription factor induced following TNF-α stimulation (Fig. 7B). PD, CsA, or PD/CsA combination does not inhibit d2EGFP expression following TNF-α stimulation (Fig. 7B). The observation that TNF-α induces only NF-κB as the major factor which regulates HIV transcription and the fact that it does not induce AP-1 and NFAT allowed us to utilize TNF-α stimulation to control for TCR activation experiments which induce multiple factors via distinct signal pathways. In contrast to TNF-α stimulation, when clone 2D10 cells were activated through the TCR under similar drug conditions, we observed a robust 49% d2EGFP expression (Fig. 7C). However, PD which specifically inhibits AP-1 c-Fos induction significantly decreased d2EGFP from 49% to 29% (a significant 20% reduction) demonstrating that AP-1 c-Fos contributes to HIV provirus transcription following TCR activation (Fig. 7C). Surprisingly, unlike MAPK inhibitor PD98059 which inhibited HIV transcription, CsA treatment resulted in a strong increase in d2EGFP expression of 66% up from 49% (a significant 17% increase) demonstrating that NFAT inhibits HIV transcription in Jurkat T-cell cones following TCR activation such that blocking NFAT results in increased HIV LTR transcriptional activation after 16 h of activation (Fig. 7C). Inhibition of HIV provirus transcription by NFAT following TCR activation is because DNA binding sequences of NFAT overlaps the two NF-κB binding sites within the HIV LTR with the result that when NFAT and NF-κB are both induced and are available in the nucleus at the same time such as during TCR activation, binding of NFAT which has distinct binding sequence requirements directly blocks NF-κB recruitment to the HIV LTR. Interestingly in samples treated with PD/CsA drug combination, enhancement of d2EGFP expression by CsA was offset by the inhibitory effects of MAPK inhibitor PD98059 following TCR activation, thereby restoring d2EGFP expression (44%) to levels similar to TCR activation (49%) without drug inhibitors. These observations demonstrate the complexity and multifaceted effects of TCR activation and signaling in the modulation of HIV provirus transcription.
Fig. 7.
Fluorescently activated cell sorting (FACS) analysis to determine d2EGEP expression. Prior to 16hrs of activation, Clone 2D10 cells were either untreated or pretreated for 1hr with PD, CsA, or PD/CsA drug inhibitors to specifically block induction of AP-1, NFAT, or both AP-1/NFAT transcription factors, respectively. (A) Drug controls (B) FACS analysis of untreated and drug-treated samples following TNF-α stimulation and (C) FACS analysis of untreated and drug-treated samples following TCR activation using anti-CD3 and anti-CD28 mAbs. Note that the results presented here represent experiments that were performed three times and each time samples were analyzed in duplicates. Error bars represent the mean ± SD of three independent and separate experiments. The P value of statistical significance was set as P < 0.05 (*).
AP-1 c-Fos modulates HIV provirus transcription by enhancing elongation
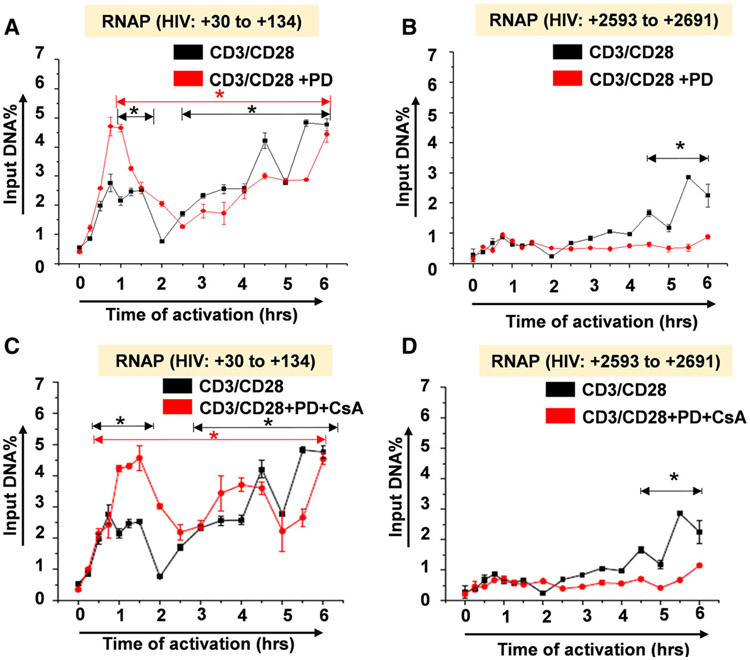
Given the specificity of MAPK inhibitor PD98059 in the inhibition of c-Fos induction which resulted in a significant decrease in d2EGFP expression by FACS analysis, we determined the precise molecular mechanism through which AP-1 c-Fos contributes to reactivation of latent HIV proviruses. Using ChIP analysis to measure RNAP levels along the provirus in kinetic experiments, we observed that PD98059 does not block the recruitment of RNAP to the HIV LTR (+30 to +134) indicating that initiation rates are constant with or without PD98059 treatment (Fig. 8A). However, PD98059 treatment induced a significant reduction in RNAP levels downstream of the HIV transcription start site within the envelope region (+2593 to +2691) following the initial round of transcription (Fig. 8B). This demonstrates that AP-1 c-Fos specifically contributes to HIV transcription by enhancing elongation such that inhibition of c-Fos nuclear mobilization results in a dramatic reduction in processive RNAP complexes following TCR activation. PD98059 in the PD/CsA inhibitor combination similarly inhibited elongation of latent HIV provirus transcription but not initiation (Fig. 8C,D). Therefore, AP-1 c-Fos contributes to the reactivation of latent HIV proviruses by enhancing transcriptional elongation.
Fig. 8.
Chromatin immunoprecipitation (ChIP) analysis measuring RNAP II recruitment to the HIV provirus following TCR activation. Analysis of HIV transcriptional response by measuring transcribing RNAP II levels along the provirus (A) analysis of transcribing RNAP II levels at Nuc-1 position in PD treated and untreated samples (B) analysis of transcribing RNAP II levels downstream from transcription start site in PD treated and untreated samples in (A). (C) Measurement of transcribing RNAP II levels at Nuc-1 position in PD/CsA combination-treated samples and (D) measurement of transcribing RNAP II levels downstream from transcription start site in PD/CsA combination-treated and untreated samples in (C). Note that the ChIP experiments were separately performed twice. During the RT-PCR analysis, samples for each time point were run in triplicates and the mean values used in the plots. Error bars represent the mean ± SD of three independent and separate experiments. The P value of statistical significance was set as; P < 0.05 (*).
NF-κB and NFAT induced by TCR activation regulate the expression of cellular genes
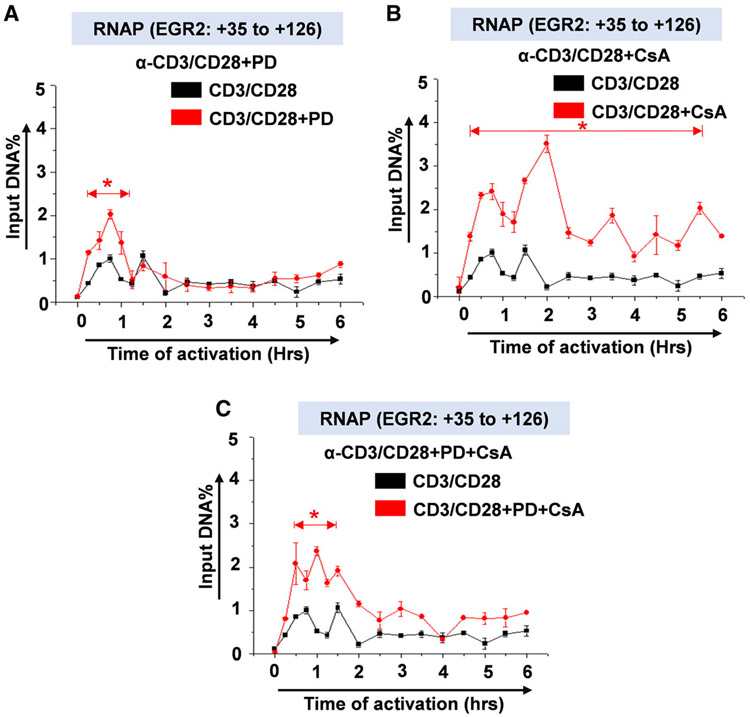
T-cell functions are regulated by a milieu of cytokines and chemokines, the majority of which expression are regulated either by NF-κB or NFAT or both. We took advantage of this observation and analyzed transcription of selected cellular genes including early growth response-2 (EGR2) to control for transcriptional activity of the TCR-mobilized transcription factors in our ChIP analysis using the same ChIP samples as shown in Fig. 8 and also to provide an internal control for the effects of drug inhibitors. EGR2 is a cellular transcription factor that regulates expression of Fas molecules known to modulate apoptosis or programmed cell death and EGR2 gene transcription itself is regulated by both NF-κB and NFAT transcription factors. Interestingly, the NF-κB binding sequence on the EGR2 promoter (GGGACTT) has homology to the NF-κB LTR binding sequence (GGGACTTTCC)x2 which binds not only NF-κB but also NFAT, suggesting that the EGR2 promoter is capable of binding both NF-κB and NFAT. It is conceivable that in the absence of NF-κB, NFAT can regulate EGR2 gene expression or inhibit NF-κB activation of EGR2 gene transcription through competitive binding to overlapping or similar binding sequences. Following activation of the TCR, NF-κB but not NFAT regulates EGR2 gene transcription during the first nuclear entry cycle of NF-κB and it was not affected by MAPK inhibitor PD98059 (Fig. 9A). However, blocking NFAT induction using CsA resulted in a significant increase in EGR2 gene transcription as measured by RNAP recruitment to the EGR2 promoter indicating that NFAT competitively inhibits NF-κB activation of EGR2 gene such that blocking NFAT mobilization using CsA significantly increases EGR2 transcription following TCR activation (Fig. 9B,C). ChIP analysis of cellular genes provided internal controls for the regulation of HIV LTR transcription by TCR-mobilized transcription factors which demonstrates that NF-κB, NFAT, and AP-1 induced upon TCR activation are functional and regulate not only HIV gene transcription but also cellular gene expression (Fig. 9).
Fig. 9.
Analysis of RNAP II levels to measure transcriptional response by cellular genes following TCR activation. ChIP assay measuring transcribing RNAP II levels along early growth response-2 (EGR2) cellular gene in (A) PD treated and untreated samples (B) CsA treated and untreated samples and (C) PD/CsA combination-treated and untreated samples in a 6-hr activation time-course experiment. Error bars represent the mean ± SD of three independent and separate experiments. The P value of statistical significance was set as; P < 0.05 (*).
Enhancement of HIV transcriptional elongation by AP-1 c-Fos is mediated via NF-κB binding sites
Some studies have demonstrated that AP-1 can form functional transcription complexes either with NF-κB or NFAT. On the other hand, the HIV LTR upstream sequences other that the core promoter region is known to contain AP-1 binding sites while other intragenic AP-1 binding sequences within the pol gene are reported to be critical for virus infectivity. Results of our studies with MAPK inhibitor PD98059 demonstrate that AP-1 c-Fos specifically contributes to HIV provirus transcription by enhancing elongation. However, whether AP-1 c-Fos fulfills this function by acting individually or through the formation of transcription ternary complexes with NF-κB is unknown. To test this hypothesis, we utilized Jurkat T-cell clone 2B5 cells which harbor mNF-κB LTR. This mutation blocks binding of both NF-κB and NFAT to the HIV LTR to activate transcription. Utilization of clone 2B5 cells therefore provided an excellent condition to determine whether or not, AP-1 alone can activate an HIV promoter without the requirement for NF-κB or NFAT binding. Analysis of d2EGFP expression as a measure of LTR activation in clone 2B5 cells following 16hrs of TCR activation showed no d2EGFP expression demonstrating that AP-1 on its own is unable to activate HIV transcription following TCR activation of Jurkat T-cell clones and that AP-1 c-Fos requires NF-κB to synergistically modulate HIV transcriptional elongation (Fig. 10). It is imperative to point out that FACS analysis of d2EGFP expression presented in Fig. 10 utilized a different gating system from those presented in Fig. 7. We further sought to determine whether or not synergistic interaction between AP-1 c-Fos and NF-κB to regulate HIV transcriptional elongation may require direct binding of AP-1 to the HIV promoter. By analyzing LTR structures in different HIV-1 subtypes, we observed that unlike HIV-1 subtypes; A, C, and E, subtypes B and D HIV LTRs harbor no AP-1 binding sites within its core promoter region (result not shown). Interestingly, the HIV molecular clone pNL4-3 utilized in our lentiviral pHR’ p-d2EGFP construct is derived from subtype B virus suggesting that this molecular clone contains no AP-1 binding sites within its core promoter region. Indeed, analysis of the LTR sequence of this molecular clone revealed no substantial AP-1 binding site which, therefore, demonstrates that synergy between AP-1 c-Fos and NF-κB to modulate HIV transcriptional elongation is devoid of direct binding of c-Fos to the LTR but rather requires NF-κB binding through which c-Fos is co-recruited to the HIV promoter to modulate HIV transcriptional elongation.
Fig. 10.
Fluorescently activated cell sorting (FACS) analysis measuring d2EGFP expression in clone 2B5 cells following 16hrs of TCR activation using anti-CD3 and anti-CD28 mAbs. Clone 2B5 cells harbor single latently infected proviruses with mNF-κB LTR which blocks binding of both NF-κB and NFAT to the HIV promoter. Clone 2B5 cells were activated through the TCR using anti-CD3 and anti-CD28 mAbs for 16 hrs and analyzed for d2EGFP expression. The mutated NF-κB binding sites are required for the activation of HIV transcription by NF-κB and NFAT.
Discussion
Physical and functional interplay between AP-1 and NF-κB, both of which belong to different families of transcription factors, was first reported by Stein et al. [34] who demonstrated that the bZIP domain of AP-1 c-Fos and c-Jun physically interacts with the Rel-homology domain (RHD) of NF-κB p65 forming a transcription complex with enhanced DNA binding and potentiated biological function including activation of the HIV LTR transcription. By utilizing the HIV-1 latently infected U1 monocytic cell lines, Yang and colleagues demonstrated that AP-1 and NF-κB transcription factors synergize to activate HIV LTR following cytokine stimulation and that this synergy was mediated through the NF-κB binding sites [35]. Functional interaction between AP-1 and NF-κB is demonstrated to regulate expression of cytokine genes and bimolecular fluorescence complementation (BiFC)-based techniques such as BiFC-FRET have allowed direct visualization of AP-1-NF-κB ternary complex formation in living cells [33]. Whereas these studies have demonstrated unique molecular interaction and mechanism of transcription regulation through functional complementation between different transcription factors, they are limited by the fact that it was not possible to determine the contribution of each transcription partner in the transcription complex. In this study, we directly determined the individual contribution of AP-1 in the AP-1-NF-κB transcription complex in kinetic studies of the reactivation of latently infected HIV proviruses following TCR activation.
Unlike TNF-α stimulation which induces NF-κB as the major and only transcription factor, activation of the TCR induces multiple distinct signal transduction pathways leading to induction of multiple transcription factors including NF-κB, NFAT, and AP-1 with distinct nuclear levels and entry phases. Uniquely, NF-κB p65 and p50 induction occur in parallel, and both exhibit two nuclear induction phases over a 6-h activation time course. The biphasic NF-κB nuclear entry was first reported by Saccani and colleagues [49] who demonstrated that certain NF-κB-dependent target genes were transcribed either during the first or second waves of NF-κB nuclear mobilization. Like NF-κB, induction of NFATc1 and NFATc2 is immediate following TCR activation but with varied kinetics. While NFATc2 induction lasts until 2hrs, NFATc1 is induced in three distinct isoforms; NFATc1/C (upper band) and NFATc1/B (middle band) induction increases during latter activation time points while isoform NFATc1/A (lower band) induction is delayed until 2.5 h after activation and the nuclear level increases during subsequent activation time points (Figs 2 and 11). The delayed nuclear induction of NFATc1/A isoform is because the synthesis and induction of this isoform are activation-induced such that there are time requirements for message translation followed by nuclear induction of the newly synthesized NFATc1/A proteins [50,51]. Unlike NF-κB and NFAT which are immediately induced upon TCR activation, induction of AP-1 c-Fos and c-Jun is delayed until 45 min, and once induced, its nuclear levels increase during subsequent time points (Figs 2 and 11). AP-1 is an early-expressed gene. Low levels of inducible residual AP-1 are present in the cytoplasm; however, following activation of the TCR, expression of AP-1 genes is activated resulting in more AP-1 induction during latter activation time points following message translation [52].
Fig. 11.
Densitometry analysis of western blot in figure 1 and figure 2 comparing nuclear induction kinetics of NF-κB, NFAT, and AP-1 transcription factors. (A) Induction kinetics of NF-κB (p65) following TNF-α stimulation and (B) Nuclear induction kinetics of NF-κB (p65), NFAT (c1), and AP-1 (c-Fos) following TCR activation.
Latent HIV provirus transcription in Jurkat T cells is dependent on NF-κB which is the major transcription factor induced by proinflammatory cytokine TNF-α stimulation. Consistent with the findings of Williams et al. [12], that sustained NF-κB induction is a prerequisite for efficient expression of latent HIV proviruses, we observed that sustained NF-κB nuclear mobilization results in enhanced HIV transcription following TNF-α stimulation of clone 2D10 cells. ChIP analysis demonstrated that RNAP II levels along the provirus during HIV transcriptional initiation and elongation closely mirrored NF-κB nuclear mobilization kinetics. In contrast to TNF-α stimulation, we observed that the kinetics of RNAP II recruitment to the LTR following TCR activation occurs in parallel to nuclear NF-κB levels particularly during the first phase of NF-κB nuclear mobilization when HIV transcription is strictly NF-κB-dependent. Cyclosporine A treatment enhanced RNAP II recruitment to the HIV LTR during early activation time points corresponding to the first phase of NF-κB nuclear induction by blocking NFAT which competitively inhibits NF-κB binding to the HIV promoter to activate HIV transcription (Fig. 8C). Because AP-1 also synergizes with NFAT other than NF-κB, it is conceivable that besides competitive binding with NF-κB to the HIV LTR, NFAT also competes with NF-κB for the limited availability of nuclear AP-1 c-Fos as another mechanism of NFAT inhibition of HIV transcription following TCR activation. During latter activation time points when AP-1 becomes available in the nucleus, activation time points corresponding to the second phase of NF-κB nuclear mobilization, HIV transcription occurs in parallel to both NF-κB and AP-1 nuclear induction kinetics during which AP-1 C-Fos specifically synergizes with NF-κB to enhance HIV transcriptional elongation. Blocking c-Fos specifically by PD98059 strongly inhibited HIV transcriptional elongation (Fig. 8B,D). Our observation that a transcription complex consisting of AP-1 c-Fos and NF-κB ternary complex modulates HIV transcriptional elongation is consistent with the observation by West et al. [11] that NF-κB p65 stimulates HIV transcriptional elongation in addition to stimulating initiation. Stein et al. [34] demonstrated that physical interaction between AP-1 and NF-κB is mediated via the bZIP region of AP-1 and RHD of NF-κB and that functional ternary complex formation required active TAD of both AP-1 and NF-κB transcription factors. Consistent with this observation, we measured NF-κB recruitment to the HIV promoter region using the Gel binding assay upon TCR activation (Fig. S1). We observed that NF-κB is recruited to the HIV LTR in parallel with its nuclear levels during the first phase of its nuclear induction with maximal binding occurring at 30 min. On the other hand, ChIP analysis revealed that both NFAT and NF-κB are recruited to the HIV LTR following TCR activation. However, treatment of cells with CsA significantly inhibited NFAT binding to the HIV LTR mean while binding of NF-κB to the LTR was enhanced (Fig. S3). Given that AP-1 and NF-κB are transcription partners and that AP-1 alone is unable to activate transcription from the HIV LTR, it is clearly demonstrable that AP-1 c-Fos and NF-κB synergize to form transcriptionally active ternary complexes which is co-recruited to the HIV promoter to modulate HIV transcriptional elongation following TCR activation.
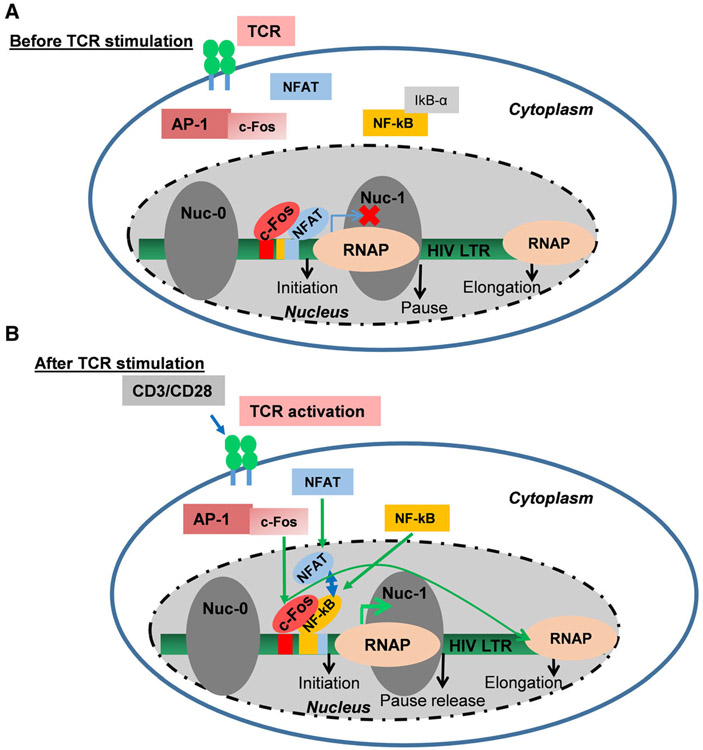
In addition to the two NF-κB binding sequences located with the core HIV promoter, multiple AP-1 binding sites have been identified within the HIV LTR [53,54] albeit not all HIV subtype-specific promoters harbor AP-1 binding sequences. The intragenic AP-1 binding sites within the pol gene have been shown to enhance virus infectivity [43,55]. We examined whether or not AP-1 can activate HIV LTR transcription in the absence of NF-κB by binding to its DNA binding site. Using clone 2B5 cells harboring mNF-κB LTR which blocks both NF-κB and NFAT bonding, we demonstrated that AP-1 alone without NF-κB is unable to activate HIV transcription. Because functional interaction between two different transcription factors usually requires binding of each factor to composite DNA binding sites, we next examined whether or not synergistic interaction between AP-1 c-Fos and NF-κB to regulate HIV transcriptional elongation occurs as a result of direct AP-1 binding to the LTR. Analysis of the LTR structures in different HIV-1 subtypes demonstrated that the pNL4-3 molecular clone utilized in the pHR’p-d2EGFP proviral construct lacks AP-1 binding sites at the LTR demonstrating that functional synergy between AP-1 c-Fos and NF-κB to regulate HIV transcriptional elongation following TCR activation is mediated via NF-κB DNA binding sequences such that when NF-κB binding is blocked, AP-1 alone is unable to activate d2EGFP expression in clone 2B5 cells. Regulation of HIV proviral transcription is a process tightly regulated by multiple but complementary T-cell signal pathways. AP-1 and NF-κB synergize to enhance HIV transcriptional elongation mediated via NF-κB LTR binding sites following TCR activation (Fig. 12A,B). In this investigation, we have utilized a well-established T-cell line-based model system of HIV latency. The current study is the extension of our previous findings that we obtained using a primary T-cell-based latency model system [56]. Based on our findings, we, for the first time, demonstrated that AP-1 synergizes with NF-κB to enhance HIV transcriptional elongation and eventually fully reactivate latent HIV proviruses, following TCR activation.
Fig. 12.
Basic model showing that AP-1 c-Fos subunit synergizes with NF-κB to enhance transcriptional elongation of latent HIV proviruses following TCR activation.
Conclusions
Persistence of latent HIV provirus pools in memory CD4 + T cells despite prolonged intensive drug therapy remains the greatest obstacle to successful HIV treatment and cure. T-cell receptor (TCR) signaling is the only physiological process known to reactivate latent HIV proviruses in vivo. Ligation of the TCR induces multiple transcription factors including NF-κB, NFAT, and AP-1, which have previously been reported to regulate HIV transcription via distinct signaling pathways. By utilizing MAPK inhibitor PD98059 and cyclosporine A to block AP-1 and NFAT, respectively, we analyzed the molecular contribution of each factor in the reactivation of latent HIV proviruses in Jurkat T-cell clones by monitoring the distribution of RNAP II along the latent HIV provirus using the high throughput chromatin immunoprecipitation assay following TCR activation. Upon TCR ligation, NF-κB and NFAT translocate into the nucleus within 15 min; however, AP-1 nuclear mobilization is delayed until 45 min. NF-κB exhibits two distinct nuclear entry phases and the kinetics of proviral activation following TNF-α treatment is strictly NF-κB-dependent during both nuclear entry phases. In contrast, following TCR activation, RNA polymerase II recruitment to the HIV LTR is NF-κB-dependent only during the first phase of NF-κB nuclear mobilization. However, 45 min following activation, AP-1 is induced and both AP-1 c-Fos subunit and NF-κB co-assemble on the promoter to regulate HIV transcriptional elongation during the second phase of NF-κB nuclear mobilization. Treatment of Jurkat T-cell clones with MAPK inhibitor PD98059 specifically blocked AP-1 c-Fos nuclear mobilization and recruitment to the HIV promoter, which strongly inhibited HIV transcriptional elongation as demonstrated by the remarkable reduction in RNAP II levels downstream of the provirus. Studies using Cyclosporine A demonstrated that NFAT inhibits HIV LTR activation through competitive binding with NF-κB to the HIV promoter. Other than P-TEFb, a cellular co-factor for viral Tat protein, this paper for the first time identified the involvement of AP-1 in the regulation of HIV transcriptional elongation through synergistic interaction with NF-κB following TCR activation.
Supplementary Material
Fig. S1. Electrophoretic mobility gel shift assay using nuclear fraction to determine binding of NF-κB to its cis regulatory elements using oligos derived from the HIV LTR (upper panel) and the Interleukin 2 promoter (lowerpanel).
Fig. S2. Immunoblot showing that SPT-5 is abundantly present in the nuclear extract.
Fig. S3. Binding of NF-κB and NFAT at HIV LTR in the absence and presence of Cyclosporin A.
Acknowledgements
We thank Drs. Jonathan Karn, Carlos Subauste, Eric Arts, David McDonald, and Koh Fijinaga, for helpful discussions. We also thank the AIDS Research and Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, US National Institutes of Health. Moreover, we would also like to thank the Center for Translational Medicine, Thomas Jefferson University, including all staff members for technical support and assistance for the experiment of this study. The research in the Tyagi laboratory is partially funded by the National Institute on Drug Abuse (NIDA), NIH Grants, 1R01DA041746-01 to MT. This research work was also supported by a doctoral training fellowship to J.H. from the Fogarty International Center (FIC) at the National Institutes of Health (NIH) through the Fogarty- AIDS International Training and Research Program (AITRP) (Training Grant: NIH 5D43 TW00011). The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the U.S. National Institutes of Health. The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- FBS
fetalbovine serum
- HAART
highly active antiretroviral therapy
- HIV
human immunodeficiency virus
- LTR
long terminal repeat
- PBS
phosphate-buffered saline
- PNK
polynucleotide kinase
- P-TEFb
positive transcription elongation factor-b
- RNAP
RNA polymerase
- TAF
TBP-associated factors
- TBP
TATA-box-binding protein
- VSV-G
vesicular stomatitis virus protein G
Footnotes
Supporting information
Additional supporting information may be found online in the Supporting Information section at the end of the article.
References
- 1.Arts EJ and Hazuda DJ (2012) HIV-1 antiretroviral drug therapy. Cold Spring Harb Perspect Med 2 (4), a007161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Condra JH, Schleif WA, Blahy OM, Gabryelski LJ, Graham DJ, Quintero JC, Rhodes A, Robbins HL, Roth E, Shivaprakash M et al. (1995) In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature 374, 569–571. [DOI] [PubMed] [Google Scholar]
- 3.Hare S, Vos AM, Clayton RF, Thuring JW, Cummings MD and Cherepanov P (2010) Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance. Proc Natl Acad Sci USA 107, 20057–20062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goethals O, Clayton R, Van Ginderen M, Vereycken I, Wagemans E, Geluykens P, Dockx K, Strijbos R, Smits V, Vos A et al. (2008) Resistance mutations in human immunodeficiency virus type 1 integrase selected with elvitegravir confer reduced susceptibility to a wide range of integrase inhibitors. J Virol 82, 10366–10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larder BA and Kemp SD (1989) Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT). Science 246 (4934), 1155–1158. [DOI] [PubMed] [Google Scholar]
- 6.Bourgeois CF, Kim YK, Churcher MJ, West MJ and Karn J (2002) Spt5 cooperates with human immunodeficiency virus type 1 Tat by preventing premature RNA release at terminator sequences. Mol Cell Biol 22, 1079–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Isel C and Karn J (1999) Direct evidence that HIV-1 Tat stimulates RNA polymerase II carboxyl-terminal domain hyperphosphorylation during transcriptional elongation. J Mol Biol 290 (5), 929–941. [DOI] [PubMed] [Google Scholar]
- 8.Karn J (1999) Tackling Tat. J Mol Biol 293 (2), 235–254. [DOI] [PubMed] [Google Scholar]
- 9.Kim YK, Bourgeois CF, Isel C, Churcher MJ and Karn J (2002) Phosphorylation of the RNA polymerase II carboxyl-terminal domain by CDK9 is directly responsible for human immunodeficiency virus type 1 Tat-activated transcriptional elongation. Mol Cell Biol 22, 4622–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nabel G and Baltimore D (1987) An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature 326 (6114), 711–713. [DOI] [PubMed] [Google Scholar]
- 11.West MJ, Lowe AD and Karn J (2001) Activation of human immunodeficiency virus transcription in T cells revisited: NF-kappaB p65 stimulates transcriptional elongation. J Virol 75 (18), 8524–8537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams SA, Kwon H, Chen LF and Greene WC (2007) Sustained induction of NF-kappa B is required for efficient expression of latent human immunodeficiency virus type 1. J Virol 81, 6043–6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganchi PA, Sun SC, Greene WC and Ballard DW (1992) I kappa B/MAD-3 masks the nuclear localization signal of NF-kappa B p65 and requires the transactivation domain to inhibit NF-kappa B p65 DNA binding. Mol Biol Cell 3, 1339–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayden MS and Ghosh S (2004) Signaling to NF-kappaB. Genes Dev 18 (18), 2195–2224. [DOI] [PubMed] [Google Scholar]
- 15.Karin M (1999) How NF-kappaB is activated: the role of the IkappaB kinase (IKK) complex. Oncogene 18 (49), 6867–6874. [DOI] [PubMed] [Google Scholar]
- 16.Chen FE and Ghosh G (1999) Regulation of DNA binding by Rel/NF-kappaB transcription factors: structural views. Oncogene 18 (49), 6845–6852. [DOI] [PubMed] [Google Scholar]
- 17.Perkins ND, Edwards NL, Duckett CS, Agranoff AB, Schmid RM and Nabel GJ (1993) A cooperative interaction between NF-kappa B and Sp1 is required for HIV-1 enhancer activation. EMBO J 12, 3551–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freed EO (2001) HIV-1 replication. Somat Cell Mol Genet 26 (1–6), 13–33. [DOI] [PubMed] [Google Scholar]
- 19.Kingsman SM and Kingsman AJ (1996) The regulation of human immunodeficiency virus type-1 gene expression. Eur J Biochem 240 (3), 491–507. [DOI] [PubMed] [Google Scholar]
- 20.Marciniak RA and Sharp PA (1991) HIV-1 Tat protein promotes formation of more-processive elongation complexes. EMBO J 10 (13), 4189–4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marzio G, Tyagi M, Gutierrez Mi and Giacca M (1998) HIV-1 tat transactivator recruits p300 and CREB-binding protein histone acetyltransferases to the viral promoter. Proc Natl Acad Sci U S A 95 (23), 13519–13524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benkirane M, Chun RF, Xiao H, Ogryzko VV, Howard BH, Nakatani Y and Jeang K-T (1998) Activation of integrated provirus requires histone acetyltransferase. p300 and P/CAF are coactivators for HIV-1 Tat. J Biol Chem 273 (38), 24898–24905. [DOI] [PubMed] [Google Scholar]
- 23.Tyagi M and Bukrinsky M (2012) Human immunodeficiency virus (HIV) latency: the major hurdle in HIV eradication. Mol Med 18, 1096–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bieniasz PD, Grdina TA, Bogerd HP and Cullen BR (1999) Recruitment of cyclin T1/P-TEFb to an HIV type 1 long terminal repeat promoter proximal RNA target is both necessary and sufficient for full activation of transcription. Proc Natl Acad Sci U S A 96 (14), 7791–7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mancebo HS, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D et al. (1997) P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev 11, 2633–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ping YH and Rana TM (2001) DSIF and NELF interact with RNA polymerase II elongation complex and HIV-1 Tat stimulates P-TEFb-mediated phosphorylation of RNA polymerase II and DSIF during transcription elongation. J Biol Chem 276 (16), 12951–12958. [DOI] [PubMed] [Google Scholar]
- 27.Kim YK, Bourgeois CF, Pearson R, Tyagi M, West MJ, Wong J, Wu S-Y, Chiang C-M and Karn J (2006) Recruitment of TFIIH to the HIV LTR is a rate-limiting step in the emergence of HIV from latency. EMBO J 25 (15), 3596–3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar KP, Akoulitchev S and Reinberg D (1998) Promoter-proximal stalling results from the inability to recruit transcription factor IIH to the transcription complex and is a regulated event. Proc Natl Acad Sci U S A 95 (17), 9767–9772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim YK, Mbonye U, Hokello J and Karn J (2011) T-cell receptor signaling enhances transcriptional elongation from latent HIV proviruses by activating P-TEFb through an ERK-dependent pathway. J Mol Biol 410 (5), 896–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sahu G, Farley K, El-Hage N, Aiamkitsumrit B, Fassnacht R, Kashanchi F, Ochem A, Simon GL, Karn J, Hauser KF et al. (2015) Cocaine promotes both initiation and elongation phase of HIV-1 transcription by activating NF-kappaB and MSK1 and inducing selective epigenetic modifications at HIV-1 LTR. Virology 483, 185–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zicari S, Sharma AL, Sahu G, Dubrovsky L, Sun L, Yue H, Jada T, Ochem A, Simon G, Bukrinsky M et al. (2020) DNA dependent protein kinase (DNA-PK) enhances HIV transcription by promoting RNA polymerase II activity and recruitment of transcription machinery at HIV LTR. Oncotarget 11, 699–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tyagi S, Ochem A and Tyagi M (2011) DNA-dependent protein kinase interacts functionally with the RNA polymerase II complex recruited at the human immunodeficiency virus (HIV) long terminal repeat and plays an important role in HIV gene expression. J Gen Virol 92 (Pt 7), 1710–1720. [DOI] [PubMed] [Google Scholar]
- 33.Shyu YJ, Suarez CD and Hu CD (2008) Visualization of AP-1 NF-kappaB ternary complexes in living cells by using a BiFC-based FRET. Proc Natl Acad Sci U S A 105 (1), 151–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stein B, Baldwin AS Jr, Ballard DW, Greene WC, Angel P and Herrlich P (1993) Cross-coupling of the NF-kappa B p65 and Fos/Jun transcription factors produces potentiated biological function. EMBO J 12, 3879–3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang X, Chen Y and Gabuzda D (1999) ERK MAP kinase links cytokine signals to activation of latent HIV-1 infection by stimulating a cooperative interaction of AP-1 and NF-kappaB. J Biol Chem 274 (39), 27981–27988. [DOI] [PubMed] [Google Scholar]
- 36.Macian F, Garcia-Rodriguez C and Rao A (2000) Gene expression elicited by NFAT in the presence or absence of cooperative recruitment of Fos and Jun. EMBO J 19 (17), 4783–4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramirez-Carrozzi V and Kerppola T (2003) Asymmetric recognition of nonconsensus AP-1 sites by Fos-Jun and Jun-Jun influences transcriptional cooperativity with NFAT1. Mol Cell Biol 23 (5), 1737–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brooks DG, Arlen PA, Gao L, Kitchen CM and Zack JA (2003) Identification of T cell-signaling pathways that stimulate latent HIV in primary cells. Proc Natl Acad Sci USA 100, 12955–12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams SA and Greene WC (2007) Regulation of HIV-1 latency by T-cell activation. Cytokine 39 (1), 63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bosque A and Planelles V (2009) Induction of HIV-1 latency and reactivation in primary memory CD4+ T cells. Blood 113 (1), 58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cron RQ, Bartz SR, Clausell A, Bort SJ, Klebanoff SJ and Lewis DB (2000) NFAT1 enhances HIV-1 gene expression in primary human CD4 T cells. Clin Immunol 94, 179–191. [DOI] [PubMed] [Google Scholar]
- 42.Kinoshita S, Su L, Amano M, Timmerman LA, Kaneshima H and Nolan GP (1997) The T cell activation factor NF-ATc positively regulates HIV-1 replication and gene expression in T cells. Immunity 6, 235–244. [DOI] [PubMed] [Google Scholar]
- 43.Colin L, Vandenhoudt N, de Walque S, Van Driessche B, Bergamaschi A, Martinelli V, Cherrier T, Vanhulle C, Guiguen A, David A et al. (2011) The AP-1 binding sites located in the pol gene intragenic regulatory region of HIV-1 are important for viral replication. PLoS One 6, e19084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duverger A, Wolschendorf F, Zhang M, Wagner F, Hatcher B, Jones J, Cron RQ, van der Sluis RM, Jeeninga RE, Berkhout B et al. (2013) An AP-1 binding site in the enhancer/core element of the HIV-1 promoter controls the ability of HIV-1 to establish latent infection. J Virol 87, 2264–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perdomo MF, Hosia W, Jejcic A, Corthals GL and Vahlne A (2012) Human serum protein enhances HIV-1 replication and up-regulates the transcription factor AP-1. Proc Natl Acad Sci USA 109, 17639–17644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D and Naldini L (1998) A third-generation lentivirus vector with a conditional packaging system. J Virol 72, 8463–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pearson R, Kim YK, Hokello J, Lassen K, Friedman J, Tyagi M and Karn J (2008) Epigenetic silencing of human immunodeficiency virus (HIV) transcription by formation of restrictive chromatin structures at the viral long terminal repeat drives the progressive entry of HIV into latency. J Virol 82, 12291–12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simonian MH (2002) Spectrophotometric determination of protein concentration. Curr Protoc Cell Biol Appendix 3, p. Appendix 3B. [DOI] [PubMed] [Google Scholar]
- 49.Saccani S, Pantano S and Natoli G (2001) Two waves of nuclear factor kappaB recruitment to target promoters. J Exp Med 193 (12), 1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chuvpilo S, Avots A, Berberich-Siebelt F, Glockner J, Fischer C, Kerstan A, Escher C, Inashkina I, Hlubek F, Jankevics E et al. (1999) Multiple NF-ATc isoforms with individual transcriptional properties are synthesized in T lymphocytes. J Immunol 162, 7294–7301. [PubMed] [Google Scholar]
- 51.Chuvpilo S, Zimmer M, Kerstan A, Glockner J, Avots A, Escher C, Fischer C, Inashkina I, Jankevics E, Berberich-Siebelt F et al. (1999) Alternative polyadenylation events contribute to the induction of NF-ATc in effector T cells. Immunity 10, 261–269. [DOI] [PubMed] [Google Scholar]
- 52.Jain J, McCaffrey PG, Valge-Archer VE and Rao A (1992) Nuclear factor of activated T cells contains Fos and Jun. Nature 356 (6372), 801–804. [DOI] [PubMed] [Google Scholar]
- 53.Franza BR Jr, Rauscher F, Josephs S and Curran T (1988) The Fos complex and Fos-related antigens recognize sequence elements that contain AP-1 binding sites. Science 239 (4844), 1150–1153. [DOI] [PubMed] [Google Scholar]
- 54.Roebuck KA, Brenner DA and Kagnoff MF (1993) Identification of c-fos-responsive elements downstream of TAR in the long terminal repeat of human immunodeficiency virus type-1. J Clin Invest 92 (3), 1336–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Lint C, Amella CA, Emiliani S, John M, Jie T and Verdin E (1997) Transcription factor binding sites downstream of the human immunodeficiency virus type 1 transcription start site are important for virus infectivity. J Virol 71 (8), 6113–6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tyagi M, Pearson RJ and Karn J (2010) Establishment of HIV latency in primary CD4+ cells is due to epigenetic transcriptional silencing and P-TEFb restriction. J Virol 84 (13), 6425–6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Electrophoretic mobility gel shift assay using nuclear fraction to determine binding of NF-κB to its cis regulatory elements using oligos derived from the HIV LTR (upper panel) and the Interleukin 2 promoter (lowerpanel).
Fig. S2. Immunoblot showing that SPT-5 is abundantly present in the nuclear extract.
Fig. S3. Binding of NF-κB and NFAT at HIV LTR in the absence and presence of Cyclosporin A.