Abstract
Millions of experimental animals are widely used in the assessment of toxicological or biological effects of manufactured nanomaterials in medical technology. However, the animal consciousness has increased and become an issue for debate in recent years. Currently, the principle of the 3Rs (i.e., reduction, refinement, and replacement) is applied to ensure the more ethical application of humane animal research. In order to avoid unethical procedures, the strategy of alternatives to animal testing has been employed to overcome the drawbacks of animal experiments. This article provides current alternative strategies to replace or reduce the use of experimental animals in the assessment of nanotoxicity. The currently available alternative methods include in vitro and in silico approaches, which can be used as cost-effective approaches to meet the principle of the 3Rs. These methods are regarded as non-animal approaches and have been implemented in many countries for scientific purposes. The in vitro experiments related to nanotoxicity assays involve cell culture testing and tissue engineering, while the in silico methods refer to prediction using molecular docking, molecular dynamics simulations, and quantitative structure–activity relationship (QSAR) modeling. The commonly used novel cell-based methods and computational approaches have the potential to help minimize the use of experimental animals for nanomaterial toxicity assessments.
Keywords: alternative animal test, nanotoxicity, cell-based test, tissue engineering, computational approach
1. Introduction
Experimental animals are widely used as a tool to evaluate the toxicological or biological effects of potential drug candidates for the development of new treatments. To date, animal ethics and animal consciousness have been growing, resulting in these topics becoming important issues over the last decade. Unfortunately, millions of animals continue to be used in medical research for scientific purposes each year. Alternative methods of animal research have been suggested as a good strategy to avoid unethical animal procedures and make scientific experiments more humane. Therefore, the concept of the replacement of animal testing was proposed by Charles Hume and William Russell in 1957 [1]. In 1959, the 3Rs (i.e., reduction, refinement, and replacement) strategy was first discussed in the book “The Principles of Humane Experimental Technique”, which was published by two English researchers, W. M. S. Russell (1925–2006) and R. L. Burch (1926–1996) [2,3]. The principle of Russell and Burch’s 3Rs, including reduction, refinement, and replacement, aimed to reduce the number of animals used and minimize the stress to such experimental animals in medical and biological research. At present, the principle of the 3Rs is applied in laboratories where animals are used to perform more humane animal research. Implementation of the 3Rs principle has the potential to overcome the drawbacks of traditional animal tests, such as the time-consuming nature, expensive feeding requirements, and associated ethical problems [4,5]. Nowadays, alternative methods are available to replace animal tests in the majority of commonly studied biomedical fields [6,7], including cell-based cytotoxic testing, genotoxicity, and biochemical assay. Non-animal approaches, which can replace animal tests, can provide quicker, more effective, and cheaper chemical safety assessments that function as substitutes for traditional animal experiments. Alternative methods to animal tests include chemical-based tests, in vitro cell culture systems, in silico computational biomodeling, and ex vivo tests using tissue from dead animals [8]. Compared to ex vivo tests, tissue engineering is a more ethical approach to the potential replacement of animal models. Tissue is taken from dead humans or animals to perform ex vivo tests, while artificial tissue utilizes cell and material methods in tissue engineering without any possible ethical problems. In this review article, we discuss the recent applications of alternative non-animal systems, including cell-based tests, computer-based modeling, and tissue engineering, for nanotoxicity assessments in scientific experiments.
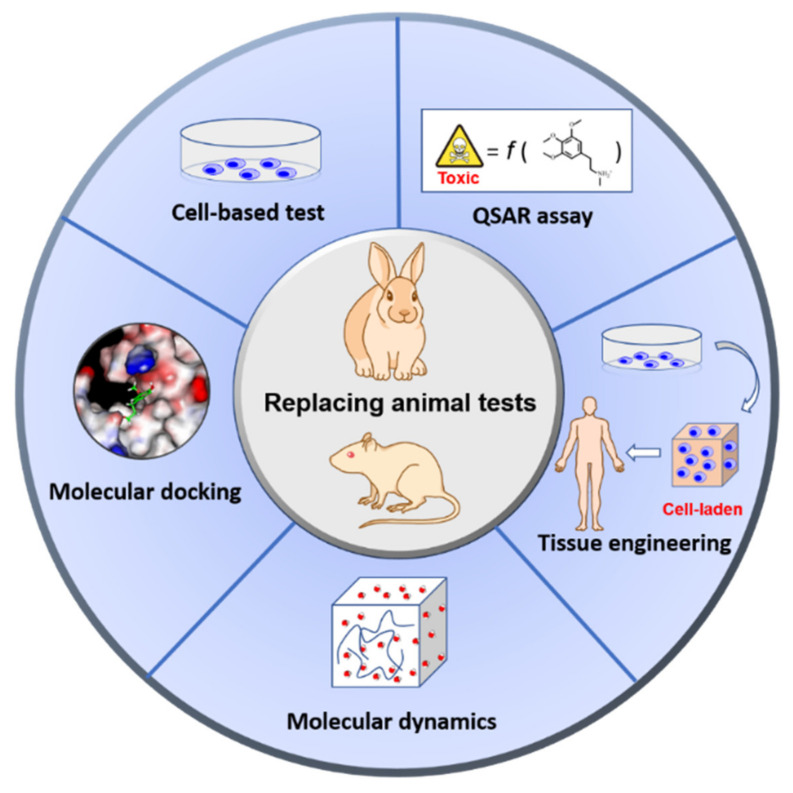
The rapid growth of nanotechnology has contributed to the urgent requirement of safety assessments for manufactured nanomaterials. Therefore, analytical methods of nanotoxicity have received a great deal of attention due to the application of nanomaterials in diverse research areas, such as agriculture [9], food industries [10], medicine [11], and biotechnology [12]. Upon the development of a novel nanomaterial, an assessment of its toxic and biological impacts must be performed to understand the potential risk factors before the material can finally be applied for medical use. Nevertheless, the number of experimental animals used in nanoparticle (NP) toxicity tests has increased each year [13]. In order to reduce the use of animal testing, humane experimental techniques have been employed to investigate biological and pathological effects as alternative forms of nanomaterial toxicity assessments. In this review, we introduce that in vitro cell-based models and in silico computational models are the most commonly used alternative testing tools to evaluate the safety and toxicity of chemical substances in the field of nanotechnology (Figure 1) [14,15,16].
Figure 1.
An overview of the current alternatives to the use of animals, including cell-based tests, tissue engineering, and computer-based techniques for nanotoxicity assessment. Cell-based tests and tissue engineering are in vitro approaches. In silico techniques include the application of prediction methods in nanotoxicity studies, such as molecular docking; quantitative structure–activity relationship (QSAR) assays; molecular dynamics simulations. Both in vitro and in silico approaches can be used to overcome ethical problems in medical research and nanotechnology.
For in vitro experiments, cell-based tests and tissue models are widely used alternative models to determine chemical hazards. In general, in vitro studies can be applied to investigate the minimum toxic effects in a specific cellular environment via cell viability protocols [17], while in vitro methods do not represent a complete replacement for in vivo assessments. Hence, in silico methods, in the replacement of animal testing, are a useful strategy to explore the relationship between chemical and biological systems in medical product development and safety assessments. Via the pharmacokinetic information database [18], numerous in silico program tools can predict absorption, distribution, metabolism, and excretion/toxicity (ADME/T) properties, derived entirely from the structures of the chemicals of interest [19,20,21].
2. Category of In Vitro Models
Cell-based tests, known as in vitro techniques, are commonly used to assess the safety and toxicity of drugs and chemicals in the replacement of animal testing [22]. In vitro studies are capable of providing faster, inexpensive, and valuable information that helps researchers to assess the potential risks or possible toxicity of newly developed nanomaterials before their final application [17]. Herein, we describe the commonly used novel in vitro methods that comply with the 3Rs principle and avoid the use of animal experimentation.
2.1. Stem Cell Technology
Currently, cell-based assessments are utilized for the treatment of various prominent disorders—including cardiovascular, neurological, ophthalmologic, skeletal, and autoimmune disorders—and for evaluating the toxicity associated with such treatments [23]. To date, various stem cell sources exist that can be used for toxicity testing, such as fibromatosis-derived stem cells (FSCs) [24], mesenchymal stem cells (MSCs) [25], cardiac stem cells (CDCs) [26], and embryonic stem cells (ESCs) [27,28]. Human ESC research was first reported in 1998 [29], in which cells obtained from pre-implantation embryos were shown to be pluripotent in nature, with the ability to differentiate into various cell types. Thus, human ESCs may be suitable as a source of cells to be used in the development of tissue engineering [30]. Currently, several ethical and religious concerns remain within the context of using human ESCs for toxicological studies or experimentation purposes [31,32]. Human ESC research is considered an ethical problem due to the destruction of human embryos [33]. In this context, alternative stem cell sources have received a great deal of attention in order to avoid ethical quandaries for investigators [34].
Induced pluripotent stem cells (iPSCs) are multipotent stem cells that are directly obtained from skin or blood cells, which can overcome the ethical considerations related to research and publication [35]. According to the seminal report from Shinya Yamanaka’s lab in 2006 [36], the first iPSC technique used four encoding transcription factors (named Oct4, Sox2, Klf4, and C-myc) to convert reprogrammed adult mouse fibroblasts into pluripotent stem cells. The above four factors, also called the “Yamanaka factors”, have the ability to reprogram mouse or human somatic cells and differentiate them into iPSCs. The iPSCs have self-renewing properties and can differentiate into all cell types, demonstrating specific functions. Therefore, iPSCs have a great capacity for tissue repair and safety assessment applications, without the associated ethical concerns [37,38]. In the development of toxicity testing by using iPSCs, some studies have successfully incorporated iPSCs with genetic diversity into cardiotoxicity testing [39]. In a recent study, iPSCs as a cell-based in vitro model were regarded as a new approach in nanotoxicity assessment to evaluate the safety of engineered NPs [40]. For instance, hepatocyte-like cells (HLCs) induced from iPSCs have been suggested as an alternative in vitro hepatotoxicity model to study NP toxicity. The iPSC-derived HLCs can be used to assess the hepatotoxicity of silver NPs (AgNPs) [41]. Similarly, the toxicity of ZnO NPs can also be investigated by human iPSC-derived cardiomyocytes (hiPSC-CMs) for cardiac safety evaluation [42]. Thus, iPSCs not only play a role in preclinical drug toxicology studies but are also used as testing platforms for NP toxicity assessments.
2.2. Tissue Engineering
Tissue engineering is a multidisciplinary field that applies disciplines related to engineering, materials, medical, and biological sciences [43]. At present, tissue engineering could be used to customize the biochemical and physicochemical factors of different types of biological tissues. Scaffold-based tissue engineering is a commonly used technique in the formation of viable tissues for clinical or biological applications. For instance, chitosan-based materials represent one of the most well-known strategies for mimicking the physicochemical properties of the extracellular matrix (ECM). The advantage of using three-dimensional (3D) scaffolds is to provide a proper environment for cell adhesion and growth, which can facilitate cell differentiation associated with the replacement of biological tissues. In order to design 3D scaffolds for customization, the shape of the 3D scaffold can be established by the unique properties of smart material and 3D printing technologies. The customized shapes of 3D scaffolds provide a suitable environment for mimicking tissue [44].
The aim of this technique is to successfully produce ideal biopolymer-based 3D organ structures that can be used for NP toxicity assessments, with the eventual goal of replacing animal testing. The 3D organ structure models require the integration of synthetic or natural biological materials and stem cells for cell growth and differentiation, which provide cell-to-cell interactions and appropriate cell signaling pathways and facilitate growth in all directions during the process of cell culture. The thermally responsive properties, shape memory, and self-healing mechanisms of synthetic materials are regarded as crucial factors for approaching the design of cell-based screening systems in 3D models. Over the past decade, several studies have revealed that the synthetic materials used in tissue engineering are able to modulate stem cell differentiation via external stimulation to change their properties in 3D culture environments [45,46].
Tissue engineering has been employed to develop 3D culture environments as an alternative to animal testing toxicity assessment methods [47,48]. Traditional two-dimensional (2D) cell cultures are most commonly employed to study biological processes, mechanisms of diseases, drug development, and toxicity assays [49]. Nevertheless, 2D cultures present many limitations to mimicking in vivo conditions, such as the lack of tissue–tissue interfaces [50], presence of natural barriers, hypoxic gradients, and tight cell–cell junctions that reduce the efficiency of drug diffusion [51]. Cells grown in a 3D condition are able to mimic the natural conditions of tissues, thereby overcoming the limitations of 2D culture systems. Three-dimensional culture models can be classified into three typical types of preparation methods that are used in research studies: (i) scaffold-based techniques; (ii) cell spheroids grown on gel-like substances, and (iii) scaffold-free cell cultures in suspension [49]. Table 1 displays a comparison of the advantages and disadvantages of 2D and 3D cell culture methods. Notably, 2D cultures allow for easier cell observation and measurements, while they are also less expensive than 3D cultures.
Table 1.
The advantages and disadvantages of 2D and 3D cell culture models.
| Characteristics | 2D Culture | 3D Culture | Reference |
|---|---|---|---|
| Cell growth rate | The growth rate faster than in vivo test | The growth rate depends on the cell type | [52] |
| Quality of cell growth | Long-term and easy to culture | Long-term and easy to culture | [53] |
| Sub-culturing time | About 1 week | Up to 4 weeks | [54] |
| Cell–cell interactions | Lack of space for cell–cell or cell–ECM interactions | More available space to provide proper cell–cell or cell–ECM interactions | [55] |
| Cost of preparation for cell culture | Low-cost maintenance | More expensive and time-consuming | [56] |
| In vivo mimics | Limitation of imitating the natural organs | Natural structures are 3D | [57] |
| Preparation of cell culture | A few hours | From hours to many days | [58] |
For most fields of research, 2D cell cultures are still used in the majority of cell research studies and toxicity assessments. However, obviously, the available space in 2D cell cultures only exists in two dimensions. The limitations of 2D culture models include adhesion dependence, reduced biological environment size, and insufficient amount of cell–cell interactions. Therefore, 2D cell culture systems are not a good strategy for understanding how cells grow and function in an animal’s body. Several studies have demonstrated that cell cultures in 3D environments are more similar to in vivo cells compared to 2D cell cultures [59,60,61]. Co-culture systems can be carried out for 2D cultures as well as 3D models. These allow two or more different types of cells to be cultured on the same dish in order to study cellular interactions. However, 2D co-culture systems grow cells on a flat surface and have limited access for cell–cell contact. In contrast, 3D co-cultures can recapitulate the native cellular microenvironment. Thus, 3D co-culture system conditions are more capable of facilitating the evaluation of cell communication, cell–cell signaling, and interactions between different types of cultured cells. Three-dimensional cell culture systems can be successfully employed as an alternative to animal tests due to these systems being cost-effective and time-saving methods of tissue growth [62]. Additionally, 3D culture systems have the ability to reduce the time periods necessary for drug or toxic testing.
3. Category of In Silico Models
In silico modeling is a relatively new subject that combines experimental approaches, providing a powerful technique to help in understanding mechanisms at the atomic level [63,64]. Over the past decade, in the field of NP research, in silico modeling has been employed to investigate safer nanomaterials, in the replacement of animal experiments [65,66]. Since 2006, the Organization for Economic Co-operation and Development (OECD) has hosted the Working Party on Manufactured Nanomaterials (WPMN), aimed at developing appropriate strategies to ensure the safe use of nanomaterials and prevent the potential risks of nanotoxicity [67]. However, the assessment of a huge variety of different NPs is an inefficient and expensive method. Hence, in silico methods, such as bioinformatics and computational approaches, have become a comprehensive tool that allows one to evaluate the possible risks of NPs. Currently, nano-specific databases can be used to study NP risk assessments, such as the Online Chemical Modeling Environment (OCHEM), NanoDatabank, NanoHub, NanoMILE, and ModNanoTox [68]. To study NPs as new pharmaceutical drugs aimed at the prevention and treatment of diseases, the physico-chemical and structural properties of the nanomaterial must be investigated via in silico approaches. Molecular docking, quantitative structure–activity relationship (QSAR) studies, and molecular dynamics (MD) simulations are three typical categories of computational approaches that represent alternatives to animal testing in nanotoxicology studies. In this section, we summarize the various types of molecular modeling and their application in nanotoxicity assessments.
3.1. Molecular Docking
Molecular docking studies are a promising approach for computational simulations and the evaluation of chemical–biomolecule interactions based on 3D structural knowledge. Briefly, docking studies are simulation techniques that predict how small molecules (e.g., drugs or NPs) interact with large biomolecules (e.g., proteins or enzymes). In the first step of a docking study, all possible conformations and orientations of each ligand are generated according to the shape of the defined binding site in the protein structure. The second step involves carrying out scoring functions to approximately predict favorable interactions between the protein and the docked ligand, along with the possible orientations. After the docking study procedure, the docking scores, calculated by the scoring functions, are used to rank each properly fitted ligand in its binding site, which can then be used to define the best affinity ligand for the target protein. A good docking score means that the molecule has favorable intermolecular interactions, including hydrogen bonds, electrostatics, and hydrophobic interactions, for a given ligand and demonstrates the best affinity as a potent binder. The docking process places the rigid chemical compounds into the active site of the protein crystal structure that is obtained from the RCSB protein databank. Generally, the accuracy and speed of the docking conformation is strongly related to the types of search algorithms used. Each docking application is based on a conformational search algorithm, such as the genetic algorithm (GA) [69], Monte Carlo (MC) [70], and incremental construction (IC) [71].
A target protein interacts with the docked ligand and further generates a protein–ligand complex which may enhance or inhibit the biological function in the experimental test. In the past few years, the molecular docking technique has been applied to predict the binding affinity of bioactive compounds at the specific site of a target protein for studying the protein–ligand interactions. Recently, some studies have employed docking methods to analyze the binding conformations of ligands for toxicity assessments as alternatives to using animal models. Hence, the docking strategy can be applied to study the chemical interactions of NPs with target enzymes. This has provided insights into the possible mechanism from which the geometric structures of the protein–NP complex are formed.
Thus far, computational modeling has utilized the protein crystal structure to investigate the cytotoxic effects and potential risks associated with functionalized NPs in many fields of nanotechnology [72,73,74,75]. For instance, cytochrome P450 (CYP) is an important enzyme system for drug metabolism. Hence, molecular docking approaches can be applied to NP and CYP enzymes in order to estimate the molecular interactions of the protein–ligand complex on the basis of its 3D structure [76]. Inhibition of the CYP enzyme by co-administered drugs is clinically significant in terms of drug−drug interactions, which may reduce drug efficacy or increase toxicity [77,78,79]. In silico simulations provide insights into the potential mechanism of the inhibitory effects on the CYP enzyme after interaction with NPs. Additionally, these models can be used in collaboration with in vitro experiments for estimating toxicological properties in the development of novel nanomaterials. As another example, molecular docking has also been used to determine the potential toxicity of different types of NPs with biological macromolecules, including CuO, TiO2, Fe3O4, Au, Ag, ZnO, Mn2O3, and Fe3O4 [80]. Hence, docking analysis between NPs and biological molecules has gained increased attention in the field of nanotoxicology as an alternative method that is able to predict potential toxicity [81].
3.2. QSAR Assay
The QSAR assay—a computer-based technique—is a promising tool that can be used as an alternative method to replace animal tests in toxicity prediction. Regarding the QSAR model, biological activity or toxic effects are displayed as a function that is related to various types of molecular descriptors, as follows:
| Biological activity/toxicity = f (molecular descriptors) | (1) |
The computational technique helps to reduce the costs associated with labor, resources, and time in nanotoxicity assessments [82]. QSAR modeling is an efficient method of predicting the biological activity or toxicity of chemical substances on the basis of mathematical statistics and knowledge of machine learning (Figure 2). The concept of QSAR was proposed a century ago, and Crum-Brown and Fraser demonstrated that the composition of chemicals is associated with their physiological activity [83]. Following this, simple chemical compounds in response to the free energy relationship model were first proposed by Corwin Hansch in 1962 [84]. The basic aim of the QSAR model is to define an appropriate function that has a reasonable relationship between the chemical structure and biological activity. This has the potential to further summarize the physio-chemical and biological information in order to predict toxicity effects or develop ideal nanomaterials.
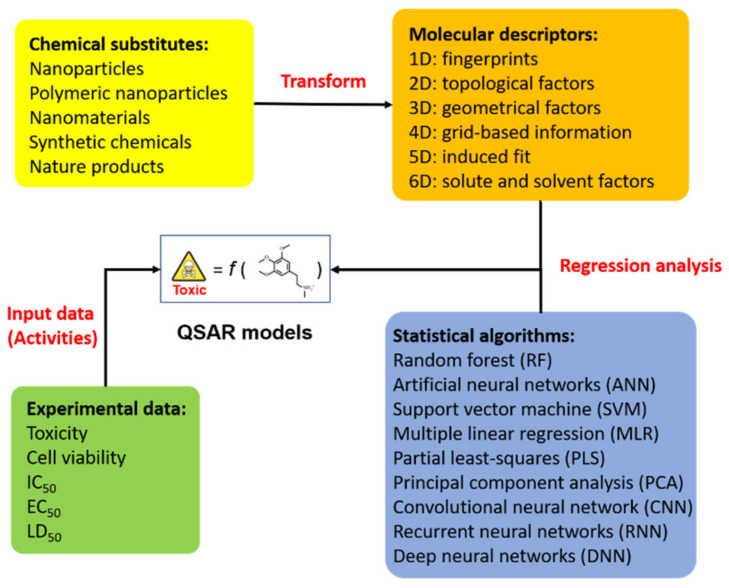
Figure 2.
A schematic overview of QSAR model generation in the assessment of chemical substitutes and prediction of their toxicity. The schematic diagram illustrates a typical method based on different statistical algorithms and specific molecular descriptors for building a predictive regression model. QSAR modeling could be employed to combine experimental measurements for in silico prediction in drug design or nanotoxicology research.
According to the dimensions of molecular descriptors used for model generation, QSAR methods can be classified into several classes of modeling, such as 1D, 2D, 3D, 4D, 5D, and so forth [85]. In most cases, 2D- and 3D-QSAR studies are commonly used to evaluate the series of chemical compounds [86,87,88]. The 1D-QSAR approach allows for the determination of correlations for 1D descriptors (pKa, log P, structural fragments, and fingerprints) with biological activity [89]. To date, the 2D-QSAR method has been widely explored in many studies for the study of toxic or medicinal chemistry, which was first proposed in the early 1970s [90]. In general, the physio-chemical properties of 2D-QSAR consider various parameters, including geometric parameters, the polar surface area, topological indices, and molecular fingerprints. In the process of model generation, 2D-QSAR correlates the above physio-chemical properties with biological activity, which can be applied to identify the predictive toxicology for organic compounds [91]. However, the lack of steric properties is a disadvantage of 2D-QSAR. To solve this problem, the 3D-QSAR method employs the 3D properties (steric and electrostatic fields) surrounding the interesting molecule and uses chemometric methods to build a relationship between the structural properties and the molecule’s activity [92].
As for the statistical methods used for QSAR models, the correlation technique is used to construct relationship models between structural information and biological activity/toxicity. According to the different types of machine learning algorithms used, QSAR generation can be classified into two major categories: linear and non-linear methods. The linear regression methods contain principal component analysis (PCA), multiple linear regression (MLR), and partial least-squares (PLS) methods. The non-linear regression algorithms consist of random forest (RF), support vector machine (SVM), and artificial neural networks (ANNs). For instance, 3D-QSAR employs statistical methods such as the ANN algorithm, PLS analysis, PCA methods, and cluster analysis for model generation. Therefore, the predictive power of 3D-QSAR makes it more suitable than 2D-QSAR [93]. Recently, artificial intelligence (AI) has been used as a statistic algorithm to search for potential regression models during the process of QSAR model generation. For example, the deep-learning-based QSAR model employs a convolutional neural network (CNN), recurrent neural networks (RNNs), and deep neural networks (DNNs) to optimize the quality of predictive models.
QSAR methods have been applied in many scientific disciplines. For instance, the QSAR approach is frequently used in risk assessments, toxicology, regulatory decision making, and chemical regulation. Thus, in silico techniques have the potential to replace animal tests on the basis of their ability to statistically and reliably predict the likelihood of a chemical substance being hazardous. Such in silico techniques replicate similar conditions to those existing in human biology. Table 2 shows various different QSAR modeling studies that are used to understand the nanotoxicity of chemical substitutes, such as NPs, metal oxide, and fullerenes. In some countries, governments utilize QSAR tools for toxic hazard prediction to avoid animal testing [94]. In recent years, researchers have applied QSAR models to analyze potential toxic effects involved in the manufacturing of nanomaterials [93].
Table 2.
Main approaches of QSAR models for nanotoxicity evaluation.
| Statistical Algorithm | Chemical Substitute | Statistical Software | Measurement | Reference |
|---|---|---|---|---|
| Random forest | Metal oxide | R | Cell viability | [95] |
| Artificial neural network | Carbon nanotubes, fullerenes, and silica NPs | CORAL | Cytotoxicity | [96] |
| Support vector machine | Q-dots and FeOx NPs | R and Python | Cellular uptake | [97] |
| Genetic algorithm and multiple linear regression | Thiol-gold NPs | TREOR | Cell viability | [98] |
| Partial least-squares | SiO2, TiO2, CeO2, AlOOH, ZnO, Ni(OH)2 | R | Cytotoxicity | [99] |
| Deep neural network and k-nearest neighbor | Q-dots and FeOx NPs | R | Cellular uptake | [100] |
| Bayesian networks | NPs | Python | Cytotoxicity | [101] |
3.3. MD Simulation
MD simulation is a mature method and is widely used to investigate chemical and physical properties in the modern era of computational nanotoxicology. MD simulations can be regarded as a complementary technique to examine the atomic motion of 3D structures that are obtained from experimental data, such as X-ray crystallography and nuclear magnetic resonance (NMR) spectrometry. Modern MD simulations not only assist with understanding the time-dependent behavior of the physical movements of atoms and molecules but also provide the thermodynamic and kinetic properties of the material systems at the atomic level. Therefore, detailed information on the conformational changes of macromolecules and NPs can be observed from MD simulations.
Computational methods, by using MD simulations, can be employed to generate toxicity prediction models that can be used in NP design and development. Toxicity assessments using traditional cell-based or animal tests have ethical considerations, financial problems, and are time-consuming. Hence, computational toxicology is widely applied in biomedical research to estimate toxic effects on various biological systems. Hence, this method is an alternative strategy for studying the toxicity of chemical compounds. In a recent study, MD simulations were applied to test various hypotheses of toxicity mechanisms in the field of computational toxicology and nanotoxicology. For instance, the aggregation and agglomeration of NPs are responsible for increased toxicity during the process of preparation or in the polymer matrix [102]. Consequently, this can induce apoptosis and generate intracellular reactive oxygen species (ROS) [103]. For example, agglomeration of titanium dioxide (TiO2) NPs was shown to trigger toxicity responses in cell-based and animal experiments. Meanwhile, large agglomerates of TiO2 NPs facilitated stronger biological effects, such as DNA damage, GSH depletion, and inflammation, compared to small agglomerates [104]. Therefore, studying the aggregation of multiple NPs is an important strategy to investigate their toxic effects.
4. Challenges of Alternative Testing Strategy
The major drawback of in vitro methods is the failure to mimic in vivo situations. For instance, in vitro models have no intrinsic circulation associated with the drug delivery system [105]. The toxicity effects of chemicals were not represented in the affected tissues. Additionally, chronic effects cannot be tested via in vitro studies due to difficulties in the consequences of long-term simulation [106]. Actually, in vitro test systems have limitations in establishing in vivo-relevant systems. In some cases, as the isolated primary cells have difficulty corresponding to the relevant cell type in biological organs, it may not easy to construct reasonable in vitro models corresponding to in vivo testing models. In addition, there are still problems in estimating the in vitro concentrations with regard to in vivo doses. In most research, in vitro alternative testing is hard to extrapolate from cellular pathways to in vivo toxicological outcomes [107].
For in silico computational approaches, assessing the accuracy of in silico prediction is still a major challenge for nanoparticles. For instance, NPs have different particle sizes, surface properties, aggregate sizes, and solubility influencing their toxicity [108]. Hence, there are still restrictions to identify the accurate physicochemical properties of NPs to predict reasonable hazards. However, the use of NPs in cytotoxicity assays presents drawbacks in common toxicity methods, such as water-soluble tetrazolium salts (WSTs), propidium iodide, and oxidative stress detection assays, due to optical interference. During the process of optical detection, the NPs may have the same spectral range as absorbing light and, thus, lead to incorrect results [108].
Therefore, the development of in vitro and in silico tests involves technical challenges to set up reliable culture conditions with in vivo-like levels to perform hazard testing of NPs. The use of non-mammalian alternative models may be an ideal strategy to overcome the ethical concerns related to the traditional animal models in the safety assessment of NPs. To assess the hazards of engineered NPs, non-mammalian models such as [109] Caenorhabditis elegans (C. elegans) [110], Drosophila (Drosophila melanogaster) [111], African clawed frog (Xenopus laevis) [112], chicken chorioallantoic membrane (Gallus gallus) [113], and zebrafish (Danio rerio) [114] could be considered as reliable approaches to improve the reliability of assessments of the toxicity of NPs.
5. Conclusions
Toxicity assessment allows us to explore the toxic effects of developed chemical substitutes, such as NPs and nanomaterials. However, animal consciousness and ethics are important issues to be considered during the process of manufacturing nanomaterials. Implementation of the 3Rs principle has facilitated the development of various alternative methods that have been suggested in the replacement of traditional animal tests. The use of analytical techniques, including in vitro cell-based assays, tissue engineering, in silico structure-based techniques, and QSAR studies, can lead to the avoidance of unethical experiments. The alternative methods can not only be used to study nanotoxicity but can also be extensively used in drug design and applied in clinics. In addition, in silico modeling could also combine in vitro experimental approaches for providing a powerful technique to investigate the mechanism for newly developed products. It is important to establish the relationship between the physicochemical characteristics of NPs and nanotoxicity. Nevertheless, achieving accurate toxicological results of NPs is dependent on factors such as particle size, surface properties, aggregate size, and solubility. Therefore, the prediction of possible nanotoxicity requires systemic examination to understand the molecular and cellular mechanisms involved. Nanotoxicity assays should utilize multidisciplinary approaches, including in vitro and in silico experimental models, to avoid contradictory results. These current strategies for alternatives to animal testing are regarded as important approaches that are necessary to meet the 3Rs principle, which will result in reducing the number of experimental animals used in the future.
Author Contributions
Conceptualization, H.-J.H., Y.-F.L. and H.-W.C.; writing—original draft preparation, H.-J.H. and H.-W.C.; writing—review and editing, H.-J.H., Y.-H.L., Y.-H.H., C.-T.L., Y.-F.L. and H.-W.C.; supervision, Y.-F.L. and H.-W.C. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Ministry of Science and Technology, Taiwan (MOST 108-2314-B-039-061-MY3, MOST 109-2314-B-038-078-MY3, MOST 109-2314-B-038-089 and MOST 109-2314-B-038-109).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Doke S.K., Dhawale S.C. Alternatives to animal testing: A review. Saudi Pharm. J. 2015;23:223–229. doi: 10.1016/j.jsps.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balls M. It’s Time to Reconsider The Principles of Humane Experimental Technique. Altern. Lab. Anim. 2020;48:40–46. doi: 10.1177/0261192920911339. [DOI] [PubMed] [Google Scholar]
- 3.Swaminathan S., Kumar V., Kaul R. Need for alternatives to animals in experimentation: An Indian perspective. Indian J. Med. Res. 2019;149:584. doi: 10.4103/ijmr.IJMR_2047_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Norman G.A.V. Limitations of Animal Studies for Predicting Toxicity in Clinical Trials. JACC Basic Transl. Sci. 2019;4:845–854. doi: 10.1016/j.jacbts.2019.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y., Zhao Y., Song F. The Ethical Issues of Animal Testing in Cosmetics Industry. Humanit. Soc. Sci. 2020;8:112. doi: 10.11648/j.hss.20200804.12. [DOI] [Google Scholar]
- 6.Avonto C., Chittiboyina A.G., Khan S.I., Dale O.R., Parcher J.F., Wang M., Khan I.A. Are atranols the only skin sensitizers in oakmoss? A systematic investigation using non-animal methods. Toxicol. Vitr. 2021;70:105053. doi: 10.1016/j.tiv.2020.105053. [DOI] [PubMed] [Google Scholar]
- 7.Kimber I. The activity of methacrylate esters in skin sensitisation test methods II. A review of complementary and additional analyses. Regul. Toxicol. Pharmacol. 2021;119:104821. doi: 10.1016/j.yrtph.2020.104821. [DOI] [PubMed] [Google Scholar]
- 8.Taylor K. Animal Experimentation: Working Towards a Paradigm Change. Brill; Leiden, The Netherlands: 2019. Recent developments in alternatives to animal testing; pp. 585–609. [Google Scholar]
- 9.Usman M., Farooq M., Wakeel A., Nawaz A., Cheema S.A., Rehman H.U., Ashraf I., Sanaullah M. Nanotechnology in agriculture: Current status, challenges and future opportunities. Sci. Total Environ. 2020;721:137778. doi: 10.1016/j.scitotenv.2020.137778. [DOI] [PubMed] [Google Scholar]
- 10.Nile S.H., Baskar V., Selvaraj D., Nile A., Xiao J., Kai G. Nanotechnologies in Food Science: Applications, Recent Trends, and Future Perspectives. Nano Micro Lett. 2020;12:45. doi: 10.1007/s40820-020-0383-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joshi M., Prabhakar B. Nanotoxicity Assessment: A Necessity. Nanosci. Nanotechnol. Asia. 2020;10:248–265. doi: 10.2174/2210681209666190228142315. [DOI] [Google Scholar]
- 12.Singh A.V., Laux P., Luch A., Sudrik C., Wiehr S., Wild A.-M., Santomauro G., Bill J., Sitti M. Review of emerging concepts in nanotoxicology: Opportunities and challenges for safer nanomaterial design. Toxicol. Mech. Methods. 2019;29:378–387. doi: 10.1080/15376516.2019.1566425. [DOI] [PubMed] [Google Scholar]
- 13.Kar S., Leszczynski J. Exploration of Computational Approaches to Predict the Toxicity of Chemical Mixtures. Toxics. 2019;7:15. doi: 10.3390/toxics7010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poli D., Mattei G., Ucciferri N., Ahluwalia A. An Integrated In Vitro–In Silico Approach for Silver Nanoparticle Dosimetry in Cell Cultures. Ann. Biomed. Eng. 2020;48:1271–1280. doi: 10.1007/s10439-020-02449-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furxhi I., Murphy F., Mullins M., Arvanitis A., Poland C.A. Nanotoxicology data for in silico tools: A literature review. Nanotoxicology. 2020;14:612–637. doi: 10.1080/17435390.2020.1729439. [DOI] [PubMed] [Google Scholar]
- 16.Selvaraj K., Murugesan S., Banoth K., Pavadai P., Ewa B., Piotr M., Eliza G.-M., Sankarganesh A., Sureshbabu Ram Kumar P., Vigneshwaran R., et al. Capsaicin-loaded solid lipid nanoparticles: Design, biodistribution, in silico modeling and in vitro cytotoxicity evaluation. Nanotechnology. 2020;32:095101. doi: 10.1088/1361-6528/abc57e. [DOI] [PubMed] [Google Scholar]
- 17.Savage D.T., Hilt J.Z., Dziubla T.D. Nanotoxicity. Springer; Berlin/Heidelberg, Germany: 2019. In vitro methods for assessing nanoparticle toxicity; pp. 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sayre R.R., Wambaugh J.F., Grulke C.M. Database of pharmacokinetic time-series data and parameters for 144 environmental chemicals. Sci. Data. 2020;7:122. doi: 10.1038/s41597-020-0455-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durán-Iturbide N.A., Díaz-Eufracio B.I., Medina-Franco J.L. In silico ADME/Tox profiling of natural products: A focus on Biofacquim. ACS Omega. 2020;5:16076–16084. doi: 10.1021/acsomega.0c01581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falcón-Cano G., Molina C., Cabrera-Pérez M.Á. ADME Prediction with KNIME: Development and Validation of a Publicly Available Workflow for the Prediction of Human Oral Bioavailability. J. Chem. Inf. Model. 2020;60:2660–2667. doi: 10.1021/acs.jcim.0c00019. [DOI] [PubMed] [Google Scholar]
- 21.Wu F., Zhou Y., Li L., Shen X., Chen G., Wang X., Liang X., Tan M., Huang Z. Computational Approaches in Preclinical Studies on Drug Discovery and Development. Front. Chem. 2020:8. doi: 10.3389/fchem.2020.00726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lauschke K., Rosenmai A.K., Meiser I., Neubauer J.C., Schmidt K., Rasmussen M.A., Holst B., Taxvig C., Emnéus J.K., Vinggaard A.M. A novel human pluripotent stem cell-based assay to predict developmental toxicity. Arch. Toxicol. 2020;94:3831–3846. doi: 10.1007/s00204-020-02856-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buzhor E., Leshansky L., Blumenthal J., Barash H., Warshawsky D., Mazor Y., Shtrichman R. Cell-based therapy approaches: The hope for incurable diseases. Regen. Med. 2014;9:649–672. doi: 10.2217/rme.14.35. [DOI] [PubMed] [Google Scholar]
- 24.Wang J.P., Yu H.M., Chiang E.R., Wang J.Y., Chou P.H., Hung S.C. Corticosteroid inhibits differentiation of palmar fibromatosis-derived stem cells (FSCs) through downregulation of transforming growth factor-beta1 (TGF-beta1) PLoS ONE. 2018;13:e0198326. doi: 10.1371/journal.pone.0198326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kouroupis D., Sanjurjo-Rodriguez C., Jones E., Correa D. Mesenchymal Stem Cell Functionalization for Enhanced Therapeutic Applications. Tissue Eng. Part B Rev. 2019;25:55–77. doi: 10.1089/ten.teb.2018.0118. [DOI] [PubMed] [Google Scholar]
- 26.Pang L. Toxicity testing in the era of induced pluripotent stem cells: A perspective regarding the use of patient-specific induced pluripotent stem cell–derived cardiomyocytes for cardiac safety evaluation. Curr. Opin. Toxicol. 2020;23–24:50–55. doi: 10.1016/j.cotox.2020.04.001. [DOI] [Google Scholar]
- 27.Horie S., Masterson C., Devaney J., Laffey J.G. Stem cell therapy for acute respiratory distress syndrome: A promising future? Curr. Opin. Crit. Care. 2016;22:14–20. doi: 10.1097/MCC.0000000000000276. [DOI] [PubMed] [Google Scholar]
- 28.Rikhtegar R., Pezeshkian M., Dolati S., Safaie N., Afrasiabi Rad A., Mahdipour M., Nouri M., Jodati A.R., Yousefi M. Stem cells as therapy for heart disease: iPSCs, ESCs, CSCs, and skeletal myoblasts. Biomed. Pharmacother. 2019;109:304–313. doi: 10.1016/j.biopha.2018.10.065. [DOI] [PubMed] [Google Scholar]
- 29.Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 30.Khademhosseini A., Ashammakhi N., Karp J.M., Gerecht S., Ferreira L., Annabi N., Darabi M.A., Sirabella D., Vunjak-Novakovic G., Langer R. Chapter 27–Embryonic stem cells as a cell source for tissue engineering. In: Lanza R., Langer R., Vacanti J.P., Atala A., editors. Principles of Tissue Engineering. 5th ed. Academic Press; Cambridge, MA, USA: 2020. pp. 467–490. [DOI] [Google Scholar]
- 31.Volarevic V., Markovic B.S., Gazdic M., Volarevic A., Jovicic N., Arsenijevic N., Armstrong L., Djonov V., Lako M., Stojkovic M. Ethical and Safety Issues of Stem Cell-Based Therapy. Int. J. Med. Sci. 2018;15:36–45. doi: 10.7150/ijms.21666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kugler J., Huhse B., Tralau T., Luch A. Embryonic stem cells and the next generation of developmental toxicity testing. Expert Opin. Drug Metab. Toxicol. 2017;13:833–841. doi: 10.1080/17425255.2017.1351548. [DOI] [PubMed] [Google Scholar]
- 33.Niemiec E., Howard H.C. Ethical issues related to research on genome editing in human embryos. Comput. Struct. Biotechnol. J. 2020;18:887–896. doi: 10.1016/j.csbj.2020.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Afshar L., Aghayan H.-R., Sadighi J., Arjmand B., Hashemi S.-M., Basiri M., Samani R.O., Ashtiani M.K., Azin S.-A., Hajizadeh-Saffar E. Ethics of research on stem cells and regenerative medicine: Ethical guidelines in the Islamic Republic of Iran. Stem Cell Res. Ther. 2020;11:1–5. doi: 10.1186/s13287-020-01916-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zakrzewski W., Dobrzyński M., Szymonowicz M., Rybak Z. Stem cells: Past, present, and future. Stem Cell Res. Ther. 2019;10:68. doi: 10.1186/s13287-019-1165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 37.Peng B.Y., Dubey N.K., Mishra V.K., Tsai F.C., Dubey R., Deng W.P., Wei H.J. Addressing Stem Cell Therapeutic Approaches in Pathobiology of Diabetes and Its Complications. J. Diabetes Res. 2018;2018:7806435. doi: 10.1155/2018/7806435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Labusca L., Herea D.D., Mashayekhi K. Stem cells as delivery vehicles for regenerative medicine-challenges and perspectives. World J. Stem Cells. 2018;10:43–56. doi: 10.4252/wjsc.v10.i5.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luz A.L., Tokar E. Pluripotent Stem Cells in Developmental Toxicity Testing: A Review of Methodological Advances. Toxicol. Sci. Off. J. Soc. Toxicol. 2018;165:31–39. doi: 10.1093/toxsci/kfy174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Handral H.K., Tong H.J., Islam I., Sriram G., Rosa V., Cao T. Pluripotent stem cells: An in vitro model for nanotoxicity assessments. J. Appl. Toxicol. 2016;36:1250–1258. doi: 10.1002/jat.3347. [DOI] [PubMed] [Google Scholar]
- 41.Gao X., Li R., Sprando R.L., Yourick J.J. Concentration-dependent toxicogenomic changes of silver nanoparticles in hepatocyte-like cells derived from human induced pluripotent stem cells. Cell Biol. Toxicol. 2021;37:245–259. doi: 10.1007/s10565-020-09529-1. [DOI] [PubMed] [Google Scholar]
- 42.Li Y., Li F., Zhang L., Zhang C., Peng H., Lan F., Peng S., Liu C., Guo J. Zinc Oxide Nanoparticles Induce Mitochondrial Biogenesis Impairment and Cardiac Dysfunction in Human iPSC-Derived Cardiomyocytes. Int. J. Nanomed. 2020;15:2669. doi: 10.2147/IJN.S249912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garrod M., San Chau D.Y. An overview of tissue engineering as an alternative for toxicity assessment. J. Pharm. Pharm. Sci. 2016;19:31–71. doi: 10.18433/J35P6P. [DOI] [PubMed] [Google Scholar]
- 44.Bezek L.B., Cauchi M.P., De Vita R., Foerst J.R., Williams C.B. 3D printing tissue-mimicking materials for realistic transseptal puncture models. J. Mech. Behav. Biomed. Mater. 2020;110:103971. doi: 10.1016/j.jmbbm.2020.103971. [DOI] [PubMed] [Google Scholar]
- 45.Dawson E., Mapili G., Erickson K., Taqvi S., Roy K. Biomaterials for stem cell differentiation. Adv. Drug Deliv. Rev. 2008;60:215–228. doi: 10.1016/j.addr.2007.08.037. [DOI] [PubMed] [Google Scholar]
- 46.Singh A., Elisseeff J. Biomaterials for stem cell differentiation. J. Mater. Chem. 2010;20:8832–8847. doi: 10.1039/c0jm01613f. [DOI] [Google Scholar]
- 47.Movia D., Bruni-Favier S., Prina-Mello A. In vitro Alternatives to Acute Inhalation Toxicity Studies in Animal Models—A Perspective. Front. Bioeng. Biotechnol. 2020;8:549. doi: 10.3389/fbioe.2020.00549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmidt K., Berg J., Roehrs V., Kurreck J., Al-Zeer M.A. 3D-bioprinted HepaRG cultures as a model for testing long term aflatoxin B1 toxicity in vitro. Toxicol. Rep. 2020;7:1578–1587. doi: 10.1016/j.toxrep.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kapałczyńska M., Kolenda T., Przybyła W., Zajączkowska M., Teresiak A., Filas V., Ibbs M., Bliźniak R., Łuczewski Ł., Lamperska K. 2D and 3D cell cultures–a comparison of different types of cancer cell cultures. Arch. Med. Sci. AMS. 2018;14:910–919. doi: 10.5114/aoms.2016.63743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jensen C., Teng Y. Is It Time to Start Transitioning From 2D to 3D Cell Culture? Front. Mol. Biosci. 2020;7:33. doi: 10.3389/fmolb.2020.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koti P., Nath S., Blell J., Boyer C., Redwan I.N. Comparing Drug Response in 2D Cultures and 3D Bioprinted Tumoroids. CELLINK LLC; Boston, MA, USA: 2020. [Google Scholar]
- 52.Melissaridou S., Wiechec E., Magan M., Jain M.V., Chung M.K., Farnebo L., Roberg K. The effect of 2D and 3D cell cultures on treatment response, EMT profile and stem cell features in head and neck cancer. Cancer Cell Int. 2019;19:16. doi: 10.1186/s12935-019-0733-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lagies S., Schlimpert M., Neumann S., Wäldin A., Kammerer B., Borner C., Peintner L. Cells grown in three-dimensional spheroids mirror in vivo metabolic response of epithelial cells. Commun. Biol. 2020;3:246. doi: 10.1038/s42003-020-0973-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prabha M.S., Divakar K., Priya J.D.A., Selvam G.P., Balasubramanian N., Gautam P. Statistical analysis of production of protease and esterase by a newly isolated Lysinibacillus fusiformis AU01: Purification and application of protease in sub-culturing cell lines. Ann. Microbiol. 2015;65:33–46. doi: 10.1007/s13213-014-0833-z. [DOI] [Google Scholar]
- 55.Fontoura J.C., Viezzer C., Dos Santos F.G., Ligabue R.A., Weinlich R., Puga R.D., Antonow D., Severino P., Bonorino C. Comparison of 2D and 3D cell culture models for cell growth, gene expression and drug resistance. Mater. Sci. Eng. C. 2020;107:110264. doi: 10.1016/j.msec.2019.110264. [DOI] [PubMed] [Google Scholar]
- 56.Fernando E.H., Dicay M., Stahl M., Gordon M.H., Vegso A., Baggio C., Alston L., Lopes F., Baker K., Hirota S. A simple, cost-effective method for generating murine colonic 3D enteroids and 2D monolayers for studies of primary epithelial cell function. Am. J. Physiol. Gastrointest. Liver Physiol. 2017;313:G467–G475. doi: 10.1152/ajpgi.00152.2017. [DOI] [PubMed] [Google Scholar]
- 57.Nunes A.S., Barros A.S., Costa E.C., Moreira A.F., Correia I.J. 3D tumor spheroids as in vitro models to mimic in vivo human solid tumors resistance to therapeutic drugs. Biotechnol. Bioeng. 2019;116:206–226. doi: 10.1002/bit.26845. [DOI] [PubMed] [Google Scholar]
- 58.De Hoogt R., Estrada M.F., Vidic S., Davies E.J., Osswald A., Barbier M., Santo V.E., Gjerde K., van Zoggel H.J.A.A., Blom S., et al. Protocols and characterization data for 2D, 3D, and slice-based tumor models from the PREDECT project. Sci. Data. 2017;4:170170. doi: 10.1038/sdata.2017.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kawai S., Yamazaki M., Shibuya K., Yamazaki M., Fujii E., Nakano K., Suzuki M. Three-dimensional culture models mimic colon cancer heterogeneity induced by different microenvironments. Sci. Rep. 2020;10:3156. doi: 10.1038/s41598-020-60145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chaicharoenaudomrung N., Kunhorm P., Noisa P. Three-dimensional cell culture systems as an in vitro platform for cancer and stem cell modeling. World J. Stem Cells. 2019;11:1065. doi: 10.4252/wjsc.v11.i12.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin S., Yang G., Jiang F., Zhou M., Yin S., Tang Y., Tang T., Zhang Z., Zhang W., Jiang X. A Magnesium-Enriched 3D Culture System that Mimics the Bone Development Microenvironment for Vascularized Bone Regeneration. Adv. Sci. 2019;6:1900209. doi: 10.1002/advs.201900209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scanarotti C., Rovida C., Penco S., Vernazza S., Tirendi S., Ciliberti R., Bassi A.M. Alternative approach to animal testing and cell cultures, according to European laws. Altex. 2017;34:441–442. doi: 10.14573/altex.1706231. [DOI] [PubMed] [Google Scholar]
- 63.Piñero J., Furlong L.I., Sanz F. In silico models in drug development: Where we are. Curr. Opin. Pharmacol. 2018;42:111–121. doi: 10.1016/j.coph.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 64.Kumaniaev I., Subbotina E., Galkin M.V., Srifa P., Monti S., Mongkolpichayarak I., Tungasmita D.N., Samec J.S.M. A combination of experimental and computational methods to study the reactions during a Lignin-First approach. Pure Appl. Chem. 2020;92:631–639. doi: 10.1515/pac-2019-1002. [DOI] [Google Scholar]
- 65.Shityakov S., Roewer N., Broscheit J.-A., Förster C. In silico models for nanotoxicity evaluation and prediction at the blood-brain barrier level: A mini-review. Comput. Toxicol. 2017;2:20–27. doi: 10.1016/j.comtox.2017.02.003. [DOI] [Google Scholar]
- 66.Furxhi I., Murphy F., Mullins M., Arvanitis A., Poland C.A. Practices and Trends of Machine Learning Application in Nanotoxicology. Nanomaterials. 2020;10:116. doi: 10.3390/nano10010116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rasmussen K., Rauscher H., Kearns P., González M., Riego Sintes J. Developing OECD test guidelines for regulatory testing of nanomaterials to ensure mutual acceptance of test data. Regul Toxicol. Pharm. 2019;104:74–83. doi: 10.1016/j.yrtph.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pikula K., Zakharenko A., Chaika V., Kirichenko K., Tsatsakis A., Golokhvast K. Risk assessments in nanotoxicology: Bioinformatics and computational approaches. Curr. Opin. Toxicol. 2020;19:1–6. doi: 10.1016/j.cotox.2019.08.006. [DOI] [Google Scholar]
- 69.Spiegel J.O., Durrant J.D. AutoGrow4: An open-source genetic algorithm for de novo drug design and lead optimization. J. Cheminform. 2020;12:1–16. doi: 10.1186/s13321-020-00429-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang W., Bell E.W., Yin M., Zhang Y. EDock: Blind protein–ligand docking by replica-exchange monte carlo simulation. J. Cheminform. 2020;12:1–17. doi: 10.1186/s13321-020-00440-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rarey M., Kramer B., Lengauer T. The particle concept: Placing discrete water molecules during protein-ligand docking predictions. Proteins Struct. Funct. Bioinform. 1999;34:17–28. doi: 10.1002/(SICI)1097-0134(19990101)34:1<17::AID-PROT3>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 72.Baimanov D., Cai R., Chen C. Understanding the Chemical Nature of Nanoparticle–Protein Interactions. Bioconjugate Chem. 2019;30:1923–1937. doi: 10.1021/acs.bioconjchem.9b00348. [DOI] [PubMed] [Google Scholar]
- 73.Chinnathambi S., Karthikeyan S., Hanagata N., Shirahata N. Molecular interaction of silicon quantum dot micelles with plasma proteins: Hemoglobin and thrombin. Rsc Adv. 2019;9:14928–14936. doi: 10.1039/C9RA02829C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ahmed L., Rasulev B., Kar S., Krupa P., Mozolewska M.A., Leszczynski J. Inhibitors or toxins? Large library target-specific screening of fullerene-based nanoparticles for drug design purpose. Nanoscale. 2017;9:10263–10276. doi: 10.1039/C7NR00770A. [DOI] [PubMed] [Google Scholar]
- 75.Singh K.P., Dhasmana A., Rahman Q. Elucidation the Toxicity Mechanism of Zinc Oxide Nanoparticle Using Molecular Docking Approach with Proteins. Asian J. Pharm. Clin. Res. 2018;11:441–446. doi: 10.22159/ajpcr.2018.v11i3.23384. [DOI] [Google Scholar]
- 76.Wasukan N., Kuno M., Maniratanachote R. Molecular Docking as a Promising Predictive Model for Silver Nanoparticle-Mediated Inhibition of Cytochrome P450 Enzymes. J. Chem. Inf. Model. 2019;59:5126–5134. doi: 10.1021/acs.jcim.9b00572. [DOI] [PubMed] [Google Scholar]
- 77.Hakkola J., Hukkanen J., Turpeinen M., Pelkonen O. Inhibition and induction of CYP enzymes in humans: An update. Arch. Toxicol. 2020;94:3671–3722. doi: 10.1007/s00204-020-02936-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guengerich F.P. A history of the roles of cytochrome P450 enzymes in the toxicity of drugs. Toxicol. Res. 2021;37:1–23. doi: 10.1007/s43188-020-00056-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fu Y., Yi Y., Fan Y., Shang R. Cytochrome P450 inhibition potential and initial genotoxic evaluation of 14-O-[(4,6-diaminopyrimidine-2-yl)thioacetyl] mutilin. Sci. Rep. 2020;10:13474. doi: 10.1038/s41598-020-70400-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Abdelsattar A.S., Dawoud A., Helal M.A. Interaction of nanoparticles with biological macromolecules: A review of molecular docking studies. Nanotoxicology. 2020;15:66–95. doi: 10.1080/17435390.2020.1842537. [DOI] [PubMed] [Google Scholar]
- 81.Chibber S., Ahmad I. Molecular docking, a tool to determine interaction of CuO and TiO2 nanoparticles with human serum albumin. Biochem. Biophys. Rep. 2016;6:63–67. doi: 10.1016/j.bbrep.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Buglak A.A., Zherdev A.V., Dzantiev B.B. Nano-(Q)SAR for Cytotoxicity Prediction of Engineered Nanomaterials. Molecules. 2019;24:4537. doi: 10.3390/molecules24244537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brown A.C., Fraser T.R. On the Connection between Chemical Constitution and Physiological Action; with special reference to the Physiological Action of the Salts of the Ammonium Bases derived from Strychnia, Brucia, Thebaia, Codeia, Morphia, and Nicotia. J. Anat. Physiol. 1868;2:224–242. [PMC free article] [PubMed] [Google Scholar]
- 84.Hansch C., Maloney P.P., Fujita T., Muir R.M. Correlation of Biological Activity of Phenoxyacetic Acids with Hammett Substituent Constants and Partition Coefficients. Nature. 1962;194:178–180. doi: 10.1038/194178b0. [DOI] [Google Scholar]
- 85.Peter S.C., Dhanja J.K., Malik V., Radhakrishnan N., Jayakanthan M., Sundar D. Encyclopedia of Bioinformatics and Computational Biology. Elsevier; Amsterdam, The Netherlands: 2019. Quantitative Structure-Activity Relationship (QSAR): Modeling Approaches to Biological Applications; pp. 661–676. [DOI] [Google Scholar]
- 86.Baviskar B.A., Deore S.L., Jadhav A.I. 2D and 3D QSAR Studies of Saponin Analogues as Antifungal Agents against Candida albicans. J. Young Pharm. 2020;12:48. doi: 10.5530/jyp.2020.12.10. [DOI] [Google Scholar]
- 87.Shukla A., Tyagi R., Meena S., Datta D., Srivastava S.K., Khan F. 2D-and 3D-QSAR modelling, molecular docking and in vitro evaluation studies on 18β-glycyrrhetinic acid derivatives against triple-negative breast cancer cell line. J. Biomol. Struct. Dyn. 2020;38:168–185. doi: 10.1080/07391102.2019.1570868. [DOI] [PubMed] [Google Scholar]
- 88.El Aissouq A., Toufik H., Stitou M., Ouammou A., Lamchouri F. In silico design of novel tetra-substituted pyridinylimidazoles derivatives as c-jun N-terminal kinase-3 inhibitors, using 2D/3D-QSAR studies, molecular docking and ADMET prediction. Int. J. Pept. Res. Ther. 2020;26:1335–1351. doi: 10.1007/s10989-019-09939-8. [DOI] [Google Scholar]
- 89.Mansouri K., Cariello N.F., Korotcov A., Tkachenko V., Grulke C.M., Sprankle C.S., Allen D., Casey W.M., Kleinstreuer N.C., Williams A.J. Open-source QSAR models for pKa prediction using multiple machine learning approaches. J. Cheminform. 2019;11:1–20. doi: 10.1186/s13321-019-0384-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.De Carvalho P.O.M., Ferreira M.M.C. 2D, 3D and Hybrid QSAR Studies of Nostoclide Analogues as Inhibitors of the Photosystem II. J. Braz. Chem. Soc. 2019;30:265–278. doi: 10.21577/0103-5053.20180175. [DOI] [Google Scholar]
- 91.Hansch C., Hoekman D., Leo A., Zhang L., Li P. The expanding role of quantitative structure-activity relationships (QSAR) in toxicology. Toxicol. Lett. 1995;79:45–53. doi: 10.1016/0378-4274(95)03356-P. [DOI] [PubMed] [Google Scholar]
- 92.Verma J., Khedkar V.M., Coutinho E.C. 3D-QSAR in drug design-a review. Curr. Top. Med. Chem. 2010;10:95–115. doi: 10.2174/156802610790232260. [DOI] [PubMed] [Google Scholar]
- 93.Cao J., Pan Y., Jiang Y., Qi R., Yuan B., Jia Z., Jiang J., Wang Q. Computer-aided nanotoxicology: Risk assessment of metal oxide nanoparticles via nano-QSAR. Green Chem. 2020;22:3512–3521. doi: 10.1039/D0GC00933D. [DOI] [Google Scholar]
- 94.Madden J.C., Enoch S.J., Paini A., Cronin M.T.D. A Review of In Silico Tools as Alternatives to Animal Testing: Principles, Resources and Applications. Altern. Lab. Anim. 2020;48:146–172. doi: 10.1177/0261192920965977. [DOI] [PubMed] [Google Scholar]
- 95.Ha M.K., Trinh T.X., Choi J.S., Maulina D., Byun H.G., Yoon T.H. Toxicity Classification of Oxide Nanomaterials: Effects of Data Gap Filling and PChem Score-based Screening Approaches. Sci. Rep. 2018;8:3141. doi: 10.1038/s41598-018-21431-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Singh A.V., Ansari M.H.D., Rosenkranz D., Maharjan R.S., Kriegel F.L., Gandhi K., Kanase A., Singh R., Laux P., Luch A. Artificial Intelligence and Machine Learning in Computational Nanotoxicology: Unlocking and Empowering Nanomedicine. Adv. Healthc. Mater. 2020;9:1901862. doi: 10.1002/adhm.201901862. [DOI] [PubMed] [Google Scholar]
- 97.Fourches D., Pu D., Tassa C., Weissleder R., Shaw S.Y., Mumper R.J., Tropsha A. Quantitative nanostructure− activity relationship modeling. ACS Nano. 2010;4:5703–5712. doi: 10.1021/nn1013484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schmidt J., Marques M.R.G., Botti S., Marques M.A.L. Recent advances and applications of machine learning in solid-state materials science. NPJ Comput. Mater. 2019;5:83. doi: 10.1038/s41524-019-0221-0. [DOI] [Google Scholar]
- 99.Forest V., Hochepied J.-F., Leclerc L., Trouvé A., Abdelkebir K., Sarry G., Augusto V., Pourchez J. Towards an alternative to nano-QSAR for nanoparticle toxicity ranking in case of small datasets. J. Nanoparticle Res. 2019;21:1–14. doi: 10.1007/s11051-019-4541-2. [DOI] [Google Scholar]
- 100.Yan X., Sedykh A., Wang W., Yan B., Zhu H. Construction of a web-based nanomaterial database by big data curation and modeling friendly nanostructure annotations. Nat. Commun. 2020;11:2519. doi: 10.1038/s41467-020-16413-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Furxhi I., Murphy F., Poland C.A., Sheehan B., Mullins M., Mantecca P. Application of Bayesian networks in determining nanoparticle-induced cellular outcomes using transcriptomics. Nanotoxicology. 2019;13:827–848. doi: 10.1080/17435390.2019.1595206. [DOI] [PubMed] [Google Scholar]
- 102.Zare Y. Study of nanoparticles aggregation/agglomeration in polymer particulate nanocomposites by mechanical properties. Compos. Part A Appl. Sci. Manuf. 2016;84:158–164. doi: 10.1016/j.compositesa.2016.01.020. [DOI] [Google Scholar]
- 103.Fu P.P., Xia Q., Hwang H.-M., Ray P.C., Yu H. Mechanisms of nanotoxicity: Generation of reactive oxygen species. J. Food Drug Anal. 2014;22:64–75. doi: 10.1016/j.jfda.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Murugadoss S., Brassinne F., Sebaihi N., Petry J., Cokic S.M., Van Landuyt K.L., Godderis L., Mast J., Lison D., Hoet P.H., et al. Agglomeration of titanium dioxide nanoparticles increases toxicological responses in vitro and in vivo. Part. Fibre Toxicol. 2020;17:10. doi: 10.1186/s12989-020-00341-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.D’Souza S. A review of in vitro drug release test methods for nano-sized dosage forms. Adv. Pharm. 2014;2014:304757. doi: 10.1155/2014/304757. [DOI] [Google Scholar]
- 106.Omidi M., Fatehinya A., Farahani M., Akbari Z., Shahmoradi S., Yazdian F., Tahriri M., Moharamzadeh K., Tayebi L., Vashaee D. 7–Characterization of biomaterials. In: Tayebi L., Moharamzadeh K., editors. Biomaterials for Oral and Dental Tissue Engineering. Woodhead Publishing; Cambridge, UK: 2017. pp. 97–115. [DOI] [Google Scholar]
- 107.Tice R.R., Austin C.P., Kavlock R.J., Bucher J.R. Improving the human hazard characterization of chemicals: A Tox21 update. Environ. Health Perspect. 2013;121:756–765. doi: 10.1289/ehp.1205784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ribeiro A.R., Leite P.E., Falagan-Lotsch P., Benetti F., Micheletti C., Budtz H.C., Jacobsen N.R., Lisboa-Filho P.N., Rocha L.A., Kühnel D., et al. Challenges on the toxicological predictions of engineered nanoparticles. NanoImpact. 2017;8:59–72. doi: 10.1016/j.impact.2017.07.006. [DOI] [Google Scholar]
- 109.Kucinska M., Murias M., Nowak-Sliwinska P. Beyond mouse cancer models: Three-dimensional human-relevant in vitro and non-mammalian in vivo models for photodynamic therapy. Mutat. Res. Rev. Mutat. Res. 2017;773:242–262. doi: 10.1016/j.mrrev.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 110.Oskouian B., Saba J.D. Death and taxis: What non-mammalian models tell us about sphingosine-1-phosphate. Semin. Cell Dev. Biol. 2004;15:529–540. doi: 10.1016/j.semcdb.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 111.López Hernández Y., Yero D., Pinos-Rodríguez J.M., Gibert I. Animals devoid of pulmonary system as infection models in the study of lung bacterial pathogens. Front. Microbiol. 2015;6:38. doi: 10.3389/fmicb.2015.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Little A.G., Pamenter M.E., Sitaraman D., Templeman N.M., Willmore W.G., Hedrick M.S., Moyes C.D. Utilizing comparative models in biomedical research. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2021;255:110593. doi: 10.1016/j.cbpb.2021.110593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Eckrich J., Kugler P., Buhr C.R., Ernst B.P., Mendler S., Baumgart J., Brieger J., Wiesmann N. Monitoring of tumor growth and vascularization with repetitive ultrasonography in the chicken chorioallantoic-membrane-assay. Sci. Rep. 2020;10:18585. doi: 10.1038/s41598-020-75660-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Couderq S., Leemans M., Fini J.-B. Testing for thyroid hormone disruptors, a review of non-mammalian in vivo models. Mol. Cell. Endocrinol. 2020;508:110779. doi: 10.1016/j.mce.2020.110779. [DOI] [PubMed] [Google Scholar]