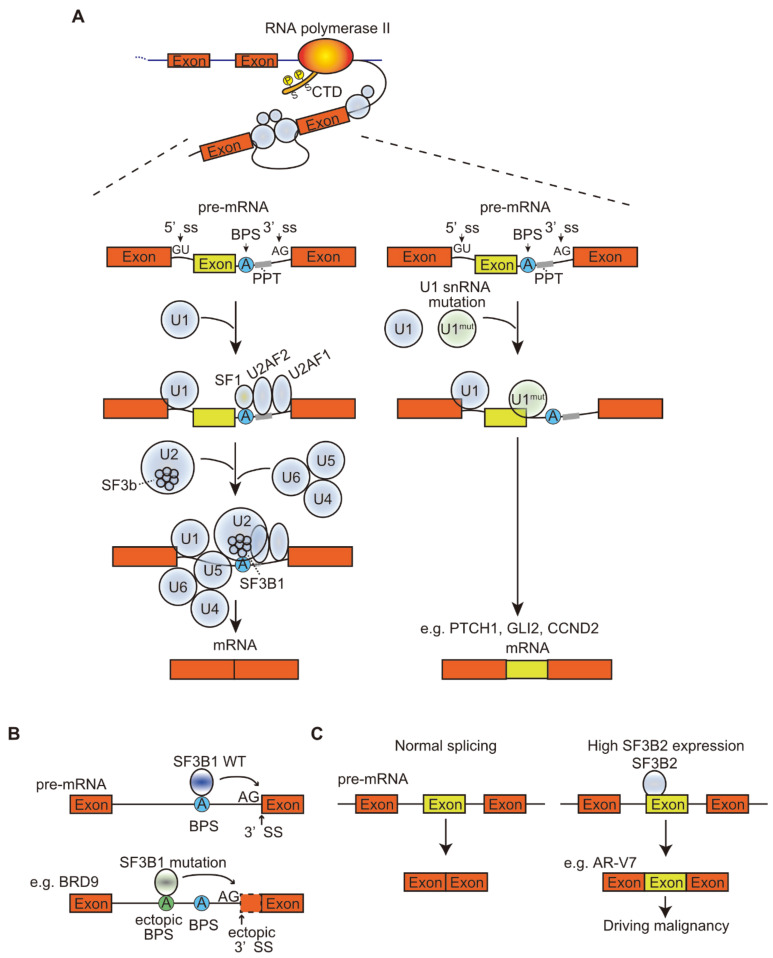
Figure 1.
Splicing is dysregulated by mutations in snRNA and splicing factors and expression levels of splicing factors. Splicing is a catalytical process that removes introns from precursor mRNA (pre-mRNA) to generate mature mRNA and is caused by dynamic changes in large protein-RNA complexes called spliceosomes. pre-mRNA is processed by the spliceosome in conjunction with transcription regulating factors. (A) The pre-mRNA is transcribed by RNA polymerase II (RNAPII) and spliced by spliceosomes. In the major spliceosomes, U1 and U2 small nuclear ribonucleoproteins (snRNPs) have a pivotal role to recognize 5′ and 3′ splice sites (ss). These complexes recognize functional intron sequences such as 5′ and 3′ ss, the branch point site (BPS), and the polypyrimidine tract (PPT). The U1 snRNP binds to the 5′ ss on the pre-mRNA and the U2 snRNP binds to the BPS with the aid of the U2 auxiliary factor (U2AF). This splicing machinery in association with other proteins removes the intron with a lariat structure from pre-mRNA to generate the mature mRNA. The mutation in U1 snRNA modifies the RNA recognition sequence of U1 snRNP, allowing the mutated U1 snRNP to bind to the 5′ ss of cryptic exon, which results in the inclusion of the cryptic exon. (B) SF3B1 is a critical factor to bind the SF3b complex to the BPS sequence in the intron. A mutation in SF3B1 modifies the RNA recognition sequence, allowing mutated SF3B1-binding to an ectopic BPS. The mis-binding shifts the position of the 3′ ss upstream, resulting in ectopic 3′ ss. (C) Expression levels of splicing factors are also involved in the regulation of splicing. SF3B2 is a component of the SF3b complex that is ubiquitously expressed. High SF3B2 expression promotes splicing in the target exons and introns, resulting in increased cancer malignancy-driving splicing variants, such as AR-V7.