Abstract
The nucleotide-binding site–leucine-rich repeat (NBS–LRR) gene family is the largest group of plant disease resistance (R) genes widespread in response to viruses, bacteria, and fungi usually involved in effector triggered immunity (ETI). Forty members of the Chinese cabbage CC type NBS–LRR family were investigated in this study. Gene and protein characteristics, such as distributed locations on chromosomes and gene structures, were explored through comprehensive analysis. CC–NBS–LRR proteins were classified according to their conserved domains, and the phylogenetic relationships of CC–NBS–LRR proteins in Brassica rapa, Arabidopsis thaliana, and Oryza sativa were compared. Moreover, the roles of BrCC–NBS–LRR genes involved in pathogenesis-related defense were studied and analyzed. First, the expression profiles of BrCC–NBS–LRR genes were detected by inoculating with downy mildew and black rot pathogens. Second, sensitive and resistant Chinese cabbage inbred lines were screened by downy mildew and black rot. Finally, the differential expression levels of BrCC–NBS–LRR genes were monitored at 0, 1, 3, 6, 12 and 24 h for short and 0, 3, 5, 7, 10 and 14 days for long inoculation time. Our study provides information on BrCC–NBS–LRR genes for the investigation of the functions and mechanisms of CC-NBS-LRR genes in Chinese cabbage.
Keywords: Chinese cabbage, CC–NBS–LRR, downy mildew, black rot
1. Introduction
Plants have developed specific mechanisms to protect themselves from abiotic or biotic stress [1,2]. Plant pathogen responses are rapidly activated when plants are attacked by pathogens, such as bacteria, fungi, oomycetes, viruses, nematodes, and insects, protecting plants from further harm [3]. Plant disease resistance (R) genes are involved in defense against pathogens, are triggered by pathogen signaling, and can target specific pathogens [4]. Most R genes encode a kind of nucleotide-binding site–leucine-rich repeat (NBS–LRR) protein. The NBS domain belongs to the NB-ARC domain and contains three conserved motifs, namely, P-loop, kinase-2, which coordinates divalent metal ions, and kinase-3a-binding nucleotide, which is a critical region for nucleotide binding in ATP/GTPs and plays a role in defense signal transmission [5]. Most LRR domains with 20–30 amino acid residues consist of two segments: a highly conserved segment (HCS) and variable segment (VS). However, the domains are usually 23–25 residues long, and a specific VS part in a plant LRR domain is involved in plant pathogen immune reaction and protein-to-protein interactions [6]. The NBS–LRR gene family is the largest class of R genes, which play multiple roles in direct or indirect host–pathogen recognition and downstream signaling transduction [7,8].
NBS–LRR proteins are divided into two types according to their conservative functional domains. One contains a toll/interleukin 1 receptor (TIR) domain at the N terminal and the other is the non-TIR(CC–NBS–LRR type), which assembles a coiled-coil (CC) domain at the N terminal instead of a TIR domain. Other domains in the N-terminal of CC–NBS–LRR replace or coexist with CC domains, particularly zinc fingers or RPW8 domains [9,10]. CC–NBS–LRR genes are found in monocotyledons and dicotyledons, and toll/interleukin-1 receptor-NBS-LRR (TNL) genes are rare in monocotyledons [9], revealing that CC–NBS–LRR is widely present in plants. Furthermore, approximately 80% of reported R genes encode the NBS–LRR domain, and more than 50 NBS genes have been proved in response to disease resistance [11]. The R gene Pi-ta belonging to the NBS-LRR type played a direct role in Magnaporthe grisea by interaction with the effector AVR-Pita in rice [12]. Additionally, the RRS1 protein of Arabidopsis thaliana interacted directly with PopP2, which was the pathogen protein of bacterial wilt [13]. RPS2 and RPM1 resistance genes from Arabidopsis were in response to Pseudomonas syringae by indirect interaction with AvrRpm1 and AvrB [14,15]. The ectopic overexpression of Arabidopsis RPW8 enhanced the resistant ability to powdery mildew in grapevine [16].
Chinese cabbage is a popular vegetable worldwide, especially in Asia. The leaf heading, which forms at the late heading stage, is the main edible part and commodious organ and is directly associated with the yield and quality of Chinese cabbage [17]. Downy mildew (DM) and black rot (BR) are the two common diseases affecting the heading leaves of Chinese cabbage in China and decreasing quality and yield [18]. Caused by Hyaloperonospora parasitica, DM is one of the three major diseases of Chinese cabbage and even the entire cruciferous family. BR is a bacterial disease caused by Xanthomonas campestris pv. campestris dowson, which infects many Brassica species, such as cabbage (Brassica oleracea var), Chinese cabbage (Brassica pekinensis), and oil seed rape (Brassica campestris) [19,20]. Thus, screening germplasm and identifying tolerant lines of Chinese cabbage will be helpful.
A total of 92 NBS-encoding resistance genes of Brassica rapa were identified without a reference genome [21], and the expression levels of 90 TIR–NBS–LRR genes against TuMV have been analyzed in Brassica rapa ssp. Pekinensis [22]. CC–NBS–LRR proteins in Chinese cabbage have not been explored, especially their roles in defense against bacterial and fungal infection. In our study, the 40 CC–NBS–LRR genes of Chinese cabbage were identified and analyzed in a genome-wide range. The basic information of the BrCC–NBS–LRR family was listed, including Genome position, exon No., the length of CDS and Protein, etc. A phylogenetic tree of CC–NBS–LRR genes in Brassica rapa, Arabidopsis thaliana, and Oryza sativa was built to elucidate the evolution. The expression profiles of BrCC–NBS–LRR genes in different tissues and in response to diseases were detected and analyzed, showing that BrCC–NBS–LRR genes were enriched in leaf, and their responses were quite different due to the different types of pathogen and time. Furthermore, the sensitive and insensitive inbred lines were obtained and inoculated with Hyaloperonospora parasitica and Xanthomonas campestris to explore the predicting functions of BrCC–NBS–LRR genes. These results suggest that they play an opposite or consistent function to diseases. Our study provides information on BrCC–NBS–LRR genes. This information is useful to further investigate the gene functions and mechanisms of CC–NBS–LRR genes in Chinese cabbage.
2. Results
2.1. Identifcation of the CC–NBS–LRR Family in Brassica rapa
The CC–NBS–LRR family usually encodes two typical domains, including the CC domain at the C terminal and the NBS–LRR domain at the N terminal. All CC–NBS–LRR family genes were searched using the Brassica rapa genome on Brassica Database (BRAD). A total of 40 CC–NBS–LRR family candidates were found, and their protein sequences were verified using online tools for protein functional domains. The tools were PfamScan (https://www.ebi.ac.uk/Tools/pfa/pfamscan/, accessed on 27 February 2021. EMBL-EBI 2013), InterPro (a protein sequence analysis and classifications website, http://www.ebi.ac.uk/interpro/, accessed on 27 February 2021. 2021), and ScanProsite (https://prosite.expasy.org/scanprosite/, accessed on 27 February 2021. 2018). Finally, 40 genes were screened, all of which have intact CC, NBS, and LRR domains. The CC–NBS–LRR family genes of Chinese cabbage were named according to their genes that were ID-searched on BRAD. Blast searches for the homologous genes of BrCC–NBS–LRR in Arabidopsis thaliana were conducted using TAIR (https://www.arabidopsis.org/, 31 March 2005), and 20 genes were homologous to CC–NBS–LRR members shown in Table S1. In our results, the main genetic characteristics of CC–NBS–LRR genes in Brassica rapa are summarized and shown in Table 1, including the gene ID chromosome location, the number of exons, protein length, molecular weights, and isoelectric point (PI). In all BrCC–NBS–LRR genes, exon length was between 1992 and 4842 bp, and the amount varied from 1 to 12. As for the proteins, the length ranged from 666 aa to 1613 aa. The isoelectric points between 5.42 and 8.98 are shown in Table 1.
Table 1.
CC–NB–LRR genes in Chinese cabbage.
| Gene ID | Genome Position | CDS Length(bp) | Exon | Protein Length(aa) | Molecular Weight (kDa) | Isoelectric Point |
|---|---|---|---|---|---|---|
| Bra011432 | A01:2,191,374...2,196,276 | 2424 | 5 | 807 | 91.25906 | 5.96 |
| Bra013947 | A01:8,643,759...8,646,482 | 2724 | 1 | 907 | 103.78083 | 5.79 |
| Bra026368 | A01:9,720,079...9,723,012 | 2934 | 1 | 977 | 110.51354 | 6.56 |
| Bra031482 | A01:17,037,929...17,040,208 | 2145 | 2 | 714 | 81.85303 | 8.46 |
| Bra035424 | A01:16,404,803...16,407,820 | 2703 | 3 | 900 | 104.13992 | 6.56 |
| Bra029405 | A02:25352667...25357391 | 3489 | 4 | 1162 | 135.16142 | 6.22 |
| Bra013134 | A03:20,277,197...20,280,164 | 2736 | 3 | 911 | 104.87017 | 8.30 |
| Bra013213 | A03:19,840,894...19,844,167 | 2916 | 3 | 971 | 111.04147 | 7.81 |
| Bra019063 | A03:26,487,398...26,490,427 | 3030 | 1 | 1009 | 114.15502 | 6.30 |
| Bra036995 | A03:29,029,474...29,032,056 | 2583 | 1 | 860 | 97.34071 | 6.87 |
| Bra018245 | A05:6,909,119...6,911,701 | 2583 | 1 | 860 | 98.19411 | 6.50 |
| Bra027332 | A05:20,364,432...20,367,560 | 3129 | 1 | 1042 | 119.33079 | 8.42 |
| Bra009882 | A06:17,846,911...17,849,759 | 2466 | 5 | 821 | 93.52592 | 6.33 |
| Bra018834 | A06:1,698,860...1,701,415 | 2556 | 1 | 851 | 96.87193 | 5.85 |
| Bra018835 | A06:1,694,120...1,696,651 | 2532 | 1 | 843 | 95.76674 | 6.08 |
| Bra018863 | A06:1,554,378...1,556,936 | 2559 | 1 | 852 | 96.60849 | 5.99 |
| Bra019752 | A06:4,608,614...4,611,267 | 2583 | 2 | 860 | 98.23598 | 8.12 |
| Bra019754 | A06:4,599,425...4,602,097 | 2673 | 1 | 890 | 100.98419 | 6.63 |
| Bra019755 | A06:4,595,072...4,597,754 | 2586 | 2 | 861 | 98.09356 | 6.05 |
| Bra026094 | A06:6,170,240...6,176,552 | 4842 | 12 | 1613 | 182.73632 | 6.22 |
| Bra016781 | A08:19,944,556...19,947,218 | 2589 | 2 | 862 | 98.25993 | 5.64 |
| Bra016782 | A08:19,949,435...19,952,182 | 2748 | 1 | 915 | 104.28085 | 5.44 |
| Bra016785 | A08:19,969,925...19,972,603 | 2679 | 1 | 892 | 101.87473 | 6.10 |
| Bra030778 | A08:20,402,328...20,404,925 | 2598 | 1 | 865 | 98.73556 | 5.14 |
| Bra030779 | A08:20,398,567...20,401,178 | 2517 | 2 | 838 | 95.04727 | 5.82 |
| Bra034631 | A08:12,381,249...12,384,097 | 2655 | 3 | 884 | 99.63007 | 6.46 |
| Bra017572 | A09:16,599,373...16,602,112 | 2001 | 5 | 666 | 76.24774 | 5.48 |
| Bra026682 | A09:33,197,405...33,200,004 | 2514 | 2 | 837 | 95.01493 | 6.02 |
| Bra026923 | A09:34,338,460...34,341,119 | 2559 | 2 | 852 | 97.16891 | 6.09 |
| Bra026924 | A09:34,342,578...34,345,222 | 2544 | 2 | 847 | 96.53494 | 5.73 |
| Bra026977 | A09:34,580,053...34,582,713 | 2661 | 1 | 886 | 101.12124 | 5.72 |
| Bra026978 | A09:34,584,346...34,587,018 | 2673 | 1 | 890 | 101.55526 | 6.14 |
| Bra026979 | A09:34,589,539...34,592,283 | 2745 | 1 | 914 | 103.82301 | 6.74 |
| Bra027097 | A09:8,445,238...8,447,974 | 1992 | 3 | 663 | 75.57687 | 6.33 |
| Bra027866 | A09:9,368,958...9,371,944 | 2790 | 3 | 929 | 106.98517 | 6.51 |
| Bra036845 | A09:25,444,116...25,448,953 | 3876 | 5 | 1291 | 146.91249 | 6.22 |
| Bra037123 | A09:4,366,953...4,369,453 | 2415 | 2 | 804 | 93.69916 | 8.98 |
| Bra037139 | A09:4,297,244...4,300,267 | 2454 | 4 | 817 | 93.03288 | 5.42 |
| Bra002495 | A10:9,233,853...9,236,492 | 2640 | 1 | 879 | 99.47706 | 8.46 |
| Bra015597 | A10:747,361...750,005 | 2571 | 2 | 856 | 97.89404 | 7.43 |
2.2. Location and Distribution of CC–NBS–LRR Family Genes on Brassica rapa Chromosome
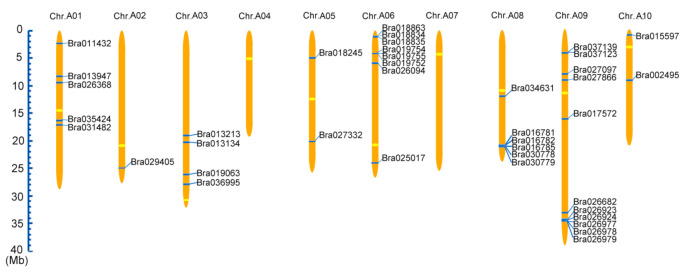
The physical chromosomal locations of BrCC–NBS–LRR family genes can be found on BRAD (http://brassicadb.org/brad/index.php, accessed on 27 February 2021), and are drawn and shown in Figure 1. Apart from A04 and A07, the BrCC–NBS–LRR family members showed asymmetric distribution on each chromosome of Brassica rapa from A1 to A10. The majority of BrCC–NBS–LRR genes were located on A09 with 12 members (30%), followed by A06 with 8 (15%) BrCC–NBS–LRR members. However, only one member was located on A02. Two members were located on A05 and A10, and four, five, and six members were located on A3, A01, and A08, respectively. A total of 24 CC–NBS–LRR family genes were found in Arabidopsis thaliana, 16 (67%) of which were located on Chr.1 and eight were located on Chr.5 (Table S2). However, 40 members of BrCC–NBS–LRR have uneven distribution on the eight chromosomes of Chinese cabbage because of chromosome expansion. Duplication analysis showed that only four pairs of genes had two copies, and two genes had three copies, which were distributed on different chromosomes. This result suggested that duplicate events in Chinese cabbage were caused by segment duplications without tandem.
Figure 1.
The chromosomal location of CC–NB–LRR genes from Chinese cabbage. The scale represents 40 Mb chromosomal distance. The chromosome numbers are labeled on the top of them. The duplicate genes are connected with red line.
2.3. Phylogenetic Analysis of the CC–NBS–LRR Family in Brassica rapa, Arabidopsis thaliana, and Oryza sativa
To further analyze the phylogenetic relationship of CC–NBS–LRR family genes among Chinese cabbage, Arabidopsis thaliana, and rice, we aligned a series of multiple protein sequences and built a phylogenetic tree with the neighbor-joining method. MEGA7.0 was used, and 40 CC–NBS–LRR members of Brassica rapa, 24 AtCC–NBS–LRR genes, and eight candidates belonging to rice were included (Figure 2, Table S3). The CC–NBS–LRR proteins were divided into nine subgroups according to their protein sequence features, namely, Groups A to G. The largest subgroup was Group A with 14 members, including 12 BrCC–NBS–LRR and two AtCC–NBS–LRR proteins. Groups E and H had four proteins each and thus had the smallest subgroups in terms of number. All the proteins in Group E were Chinese cabbage CC–NBS–LRR proteins, and two proteins in Group H were Chinese cabbage proteins and two were Arabidopsis thaliana, showing that Bra013134 shared high homology with AT1G58400 and Bra035424 shared high homology with At1G59620. Two types of CC–NBS–LRR proteins were found in Group B, including four AtCC–NBS–LRR members and seven BrCC–NBS–LRR members, with a total of 11 members. Similar to Group B, Group C contained four AtCC–NBS–LRR and five BrCC–NBS–LRR proteins. Seven CC–NBS–LRR proteins, including two Brassica rapa and five Oryza sativa members, were found. Groups F and G had three types of CC–NBS–LRR proteins, such as Arabidopsis thaliana, Oryza sativa, and Brassica rapa proteins. In Group F, two AtCC–NBS–LRR, two OsCC–NBS–LRR, and four BrCC–NBS–LRR proteins were found, whereas Group G had one Oryza sativa, two Arabidopsis thaliana, and three Brassica rapa members. These results showed that the phylogenetic tree of all the CC–NBS–LRR members is inclined to species differences because of the loose functional domain structures of the CC domain, NBS, and LRRs.
Figure 2.
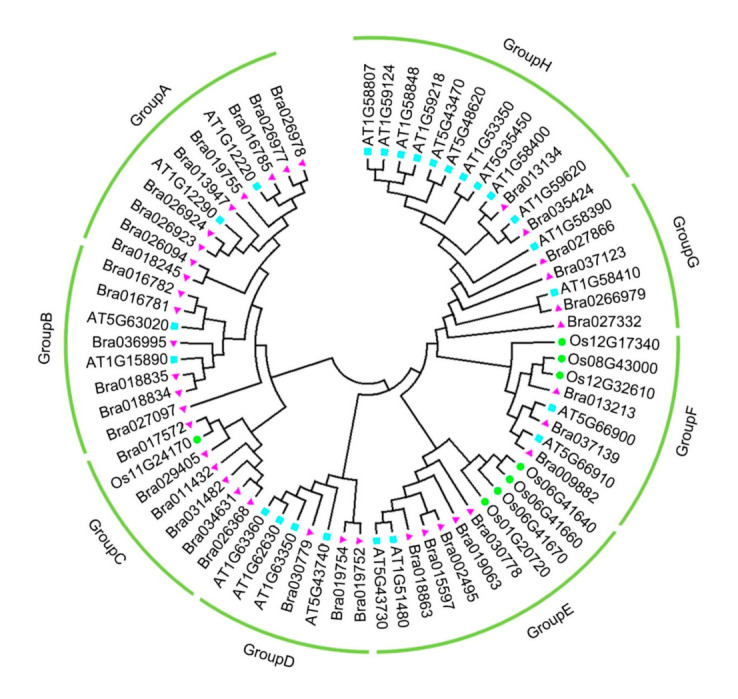
Phylogenetic relationships of CC–NB–LRR protein among Brassica rapa, Arabidopsis thaliana and Oryza. sativa. The unroot phylogenetic tree was constructed by MEGA 7.0 by neighbor-joining method with 1000 bootstrap replicates. CC–NB–LRRs of different plants are indicated with different colors and shapes. Pink triangles represent Brassica rapa, green circles represent Oryza. sativa and blue squares represent Arabidopsis thaliana.
2.4. Gene Structures and Protein Function Domains of CC–NBS–LRR Family in Chinese Cabbage
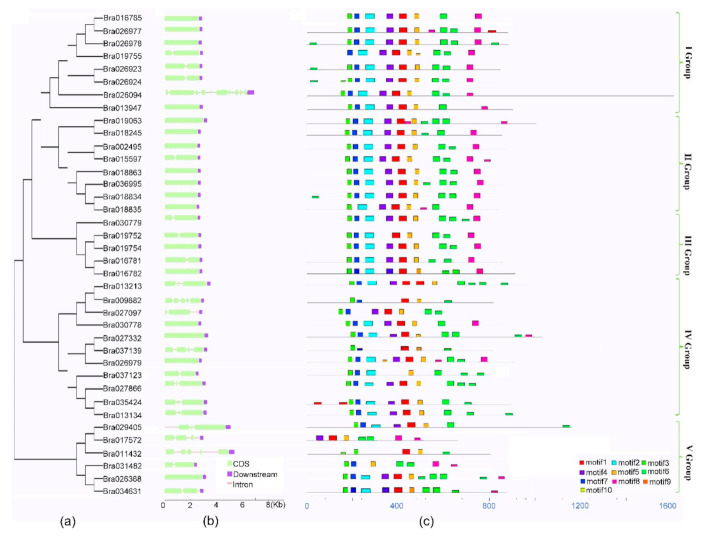
The gene structures and protein function domains of the BrCC–NBS–LRR family were analyzed using a phylogenetic tree built by aligning all the protein sequences of BrCC–NBS–LRR proteins through the neighbor-joining method, and the phylogenetic relationships among them were explored. This family was divided into five subgroups, namely, Groups I, II, III, IV, and V, as shown in Figure 3A. The IV Group had 11 members, which was the largest group. Group III was the smallest, with only four BrCC–NBS–LRR proteins. Groups I, II, and V had eight, eight, and six members, respectively (Figure 3A).
Figure 3.
The Phylogenetic tree, gene structure and multiple motifs of CC–NBS–LRR family in Chinese cabbage. (a) Phylogenetic tree was constructed based on the protein sequence of Chinese cabbage by MAGE7.0; (b) The gene structure of Chinese cabbage CC–NBS–LRRs. Exon and intron are represented by green boxes and lines; (c) The multiple conserved motifs of Chinese cabbage CC–NBS–LRR proteins. Different colored boxes represent function motifs, identified by an online tool MEME. Motifs 2, 3, 4, 7, 10 were NB-ARC function domains.
The BrCC–NBS–LRR gene structures were analyzed on the basis of the alignments between whole cDNA and genomic DNA sequences, which were downloaded from BRAD and blasted in the National Center for Biotechnology Information (NCBI). Then, the structures were drawn with Gene Structure Display Server, as shown in Figure 3B. Nearly half of all BrCC–NBS–LRR genes had only one exon (17 of 39), and nine genes had two exons and one intron. However, Bra026094 had most exons up to 12. The rest of the CC–NBS–LRR genes contained more than three exons. Specifically, six members had three exons, two had four, and four had five (Figure 3B). In terms of structure performance, most BrCC–NBS–LRR genes have similar exons and introns in the same phylogenetic subfamily.
The conserved protein motifs of BrCC–NBS–LRR were searched on the online tool MEME, which was set to monitor 10 putative protein domains (shown in Figure 3c) with the protein sequences listed in Table S3 and sequence logos of motifs in Figure S1. All the CC–NBS–LRR proteins of Chinese cabbage had the motif 6, which encodes LRR domains. However, the encoding proteins of motif 2, 3, 4, 7, and 10 belong to NB–ARC function domains, which were identified as central nucleotide-binding domains of resistance (R) proteins in plants. Moreover, the motif 3 was found in each BrCC–NBS–LRR protein, and it may be an essential motif for NBSs of Chinese cabbage. Additionally, most BrCC–NBS–LRR proteins contained motif 2 (85%) or motif 4 (82%). All of the 40 BrCC–NBS–LRR proteins contained one or more NB–ARC domain motifs and some other motifs, but Br011432 had the lowest number of motifs. In our searched results, the functions of motifs 1, 5, 8, and 9 were undefined, although motifs 1 and 5 were highly conserved in the CC–NBS–LRR family of Chinese cabbage. Similar motifs were found in the same BrCC–NBS–LRR superfamily, and Groups I, II, and III showed higher degrees of conservation than Groups IV and V.
2.5. Cis-Acting Elements Analysis of the CC–NBS–LRR Family Promoter in Brassica rapa
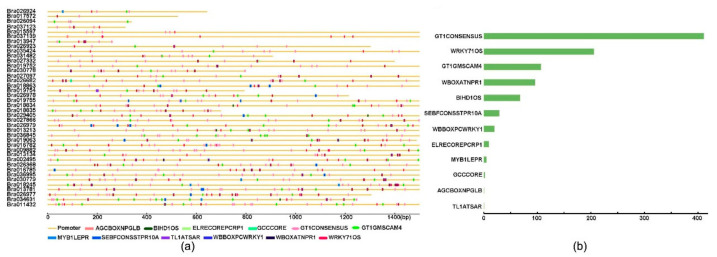
The promoter sequences of BrCC–NBS–LRR genes obtained from BRAD were approximately 265–1500 bp upstream of transcriptional start sites (Table S5). They were scanned with the online tool New PLACE for the prediction of putative cis-acting regulatory DNA elements. The disease-resistant related cis-elements of BrCC–NB–LRR promoters were further analyzed and drawn in Figure 4. A total of 12 types of disease-resistant related cis-elements were found, namely, AGCBOXNPGLB, BIHD1OS, ELRECOREPCRP1, GCCCORE, GT1CONSENSUS, GT1GMSCAM4, MYB1LEPR, SEBFCONSSTPR10A, TL1ATSAR, WBOXATNPR1, WBBOXPCWRKY1, and WRKY71OS (Figure 4). They were distributed randomly in their promoters with different items (Figures S2 and S3). The cis-elements of GT1CONSENSU were detected in each BrCC–NBS–LRR promoter. Especially, in GT1CONSENSU, the GT-1-like factors bind the PR-1a promoter which influences the expression level of SA-inducible genes [23]. GT1CONSENSU had the largest number of cis-elements among the CC–NBS–LRR members of Chinese cabbage (up to 411). BrCC–NBS–LRR proteins contained different GT1CONSENSU elements with numbers ranging from 3 to 24. WRKY71OS was the second largest number cis-element of in BrCC–NBS–LRR with a total of 206, which are related to the WRKY proteins that bind specifically to TGAC-containing W box elements within the pathogenesis-related class 10 (PR-10) genes [24]. Most CC–NBS–LRR members contained WRKY71OS elements, except Bra019754. A total of 38 BrCC–NBS–LRR members contained GT1GMSCAM4 in their promoters, the expression of which is induced by pathogens and mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. WBOXATNPR1 and WBBOXPCWRKY1, a salicylic acid (SA)-induced WRKY DNA binding protein and a “W box” WRKY protein that binds specifically to DNA sequence motifs [25], appeared in 31 and 25 BrCC–NBS–LRR gene promoters with a total of 96 and 20 items (Figure 4B). A total of 68 BIHD1OS elements were present in 32 BrCC–NBS–LRR genes; these elements are the binding sites of OsBIHD1 in disease resistance responses [26]. SEBFCONSSTPR10A, a binding site of the potato silencing element binding factor (SEBF) gene found in the promoter of the pathogenesis-related gene PR-10a, was present in 18 BrCC–NBS–LRR gene promoters with a total of 29. Nearly 10 elements were found in other BrCC–NBS–LRR gene promoters, such as ELRECOREPCRP1, which contained the consensus sequence of the elements W1 and W2 of PR1-1/PR1-2 promoters, and the WRKY1 protein binding site had 10 items in the 10 promoters of BrCC–NBS–LRRs. Five MYB1LEPR cis-elements were found in five BrCC–NBS–LRR genes and were related to Pti4(ERF), which regulates defense-related gene expression assisted by GCC box and non-GCC box cis elements [27]. Two GCC-box cores were present in many pathogen-responsive genes on the Bra026682 promoter, and only one TL1ATSAR was detected in the promoter regions of 13 NPR1-responsive ER-resident genes on the Bra019754 promoter. Only one AGCBOXNPGLB was conserved in most PR-protein genes on the Bra037139 promoter. Furthermore, 40 cis-elements were present on Bra011432 and Bra034631, and only five cis-elements were present on Bra026924 gene promoters listed in Figures S2 and S3. These results suggested that the expression of homology BrCC-NBS-LRR genes might be regulated by different mechanisms, and these genes play different roles in disease resistance in Chinese cabbage.
Figure 4.
The disease resistant related cis-elements on CC–NB–LRR promoters of Chinese cabbage. (a) The distribution of disease resistant related cis-elements on BrCC–NB–LRR promoters; (b) The total number of each disease resistant related cis-element on Chinese cabbage CC–NB–LRR promoters.
2.6. Tissue Expression Patterns of CC-NBS-LRR Genes in Chinese Cabbage
To explore the expression patterns of BrCC–NBS–LRR genes in the roots, stems, leaves, flowers, and siliques, the total mRNA of the different tissues of the Chinese cabbage cultivar A160 were isolated and reversed-transcribed into cDNAs, which were amplified with specific primers (Table S6) and normalized with BrActin. Expression levels were detected through real-time PCR, and the expression characteristics were analyzed, as shown in Figure S4. The leaves had the highest number of BrCC–NBS–LRR genes that exhibited significantly high expression levels. The expression levels of Bra027097, Bra027866, and Bra036845 in the leaves were 10-fold those in the other tissues. However, Bra026094 showed high expression levels in all tissues, especially in the roots, stems, flowers, and siliques. In the siliques, only three BrCC–NBS–LRR genes showed prominently enhanced expression levels. These results showed that the BrCC–NBS–LRR expression levels in the four tissues varied, indicating that they play different roles in the development of Chinese cabbage.
2.7. Expression Profiles of BrCC–NBS–LRR Genes in Response to Phytopathogens
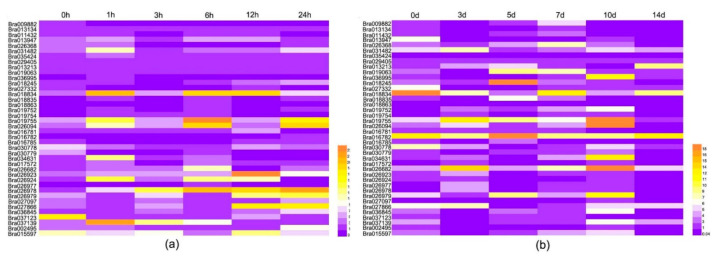
To further analyze the potential functions of BrCC–NBS–LRR genes in response to fungal diseases, the inbred A160 lines of Chinese cabbage in the five-leaf stage were inoculated with Hyaloperonospora parasitica (DM) or distilled water as a control. They were harvested after 1, 3, 6, 12, and 24 h for short inoculations and 3, 5, 7, 10, and 14 days for long inoculations for the detection of the expression levels of BrCC–NBS–LRR genes through real-time PCR. The expression characteristics were analyzed, as shown in Figure 5A,B. After the short inoculations, we found that most (63%) of the CC–NBS–LRR family genes were up-regulated after 1 h of inoculation. This result indicated that they quickly respond to DM. For example, Bra034631 expression was induced and increased 11-fold within 1 h. As for Bra031482, the expression was up-regulated 10-fold. The expression level of Bra026978 increased with inoculation time, showing that Bra026978 was continuously induced by DM. The expression levels of numerous BrCC–NBS–LRRs dramatically changed after 12 h of inoculation. However, the expression levels of BrCC–NBS–LRRs were relatively evenly up-regulated 3, 5, 7, and 10 days after inoculation (Figure 5B). In addition, the expression levels of Bra016782 and Bra026682 were consistently high at all inoculation periods. However, most BrCC–NBS–LRR genes (88%) showed decreased expression 14 days after the inoculation of DM relative to the expression levels detected at day 0 of inoculation (Figure 5B). Prominent diseased plaques were observed in Chinese cabbage leaves after 14 days of inoculation. The expression of Bra019755 was up-regulated 18-fold in 3, 5, 7, and 10 days after inoculation and then declined rapidly to 1.
Figure 5.
Expression patterns of BrCC–NBS–LRRs in response to DM. (a) The expression levels of BrCC–NBS–LRR genes in response to DM with short inoculation time for 0, 1, 3, 6, 12 and 24 h; (b) the expression levels of BrCC–NBS–LRR genes in response to DM with long inoculation time for 0, 3, 5, 7, 10 and 14 days.
Expression levels after exposure to Xanthomonas campestris, a kind of bacterial BR, were further detected through qPCR. The results are shown in Figure 6A,B. Different BrCC–NBS–LRR members were induced at different inoculation times (0, 1, 3, 6, 12, and 24 h (short inoculations) and 3, 5, 7, 10, and 14 days (long inoculations)). Seven of the 40 BrCC–NBS–LRR genes (18%) were up-regulated more than twofold after 1 h of inoculation relative to the expression at 0 h, and the expression levels of Br018245, Bra030778, and Bra017572 increased 3.5-, 7.3-, and 2.2-fold after 3 h of BR inoculations. Six hours after inoculation, only Bra016785 showed an increase in expression level. The expression of the others showed no change or was down-regulated. However, more BrCC–NBS–LRRs (15 of 40) were up-regulated after 12 h of inoculation. The expression levels of CC–NBS–LRRs were smooth and steady compared with those after the short inoculations. Bra030778 was up-regulated in the long inoculations (3, 5, 7, 10, and 14 days), and the expression levels of Bra029405, Bra019063, Bra017572, and Bra015597 increased remarkably 3 days after inoculation. The expression levels of Bra030779 and Bra015597 remarkably increased 5 days after inoculation. After 7 days of BR inoculation, 17 CC–NBS–LRR genes showed up-regulated expression, but 93% of the BrCC–NBS–LRR genes showed down-regulated expression 14 days after inoculation. Most of the Chinese cabbages showed numerous disease spots. Compared to long time inoculation, these dramatically differential expressions of BrCC-NBS-LRR genes in the 24 h short inoculation suggested that they respond quickly to BR.
Figure 6.
Expression patterns of BrCC–NBS–LRRs in response to BR. (a) The expression levels of BrCC–NBS–LRR genes in response to BR with short inoculation time for 0, 1, 3, 6, 12 and 24 h; (b) the expression levels of BrCC–NBS–LRR genes in response to BR with long inoculation time for 0, 3, 5, 7, 10 and 14 days.
2.8. Associated Expression of BrCC-NBS-LRR in Different Inbred Lines of Chinese Cabbage in Response to Phytopathogen
To further analyze the expression characteristics of BrCC-NBS-LRR in response to fungal and bacterial disease, 24 inbred lines of Chinese cabbage were applied to screen the sensitive and resistant lines to DM and BR. On the basis of statistics (Table S6), inbred lines A95 and A167 showed the most sensitivity compared to the other 23 lines to DM and BR, respectively, and, in contrast, A24 and A96 were resistant.
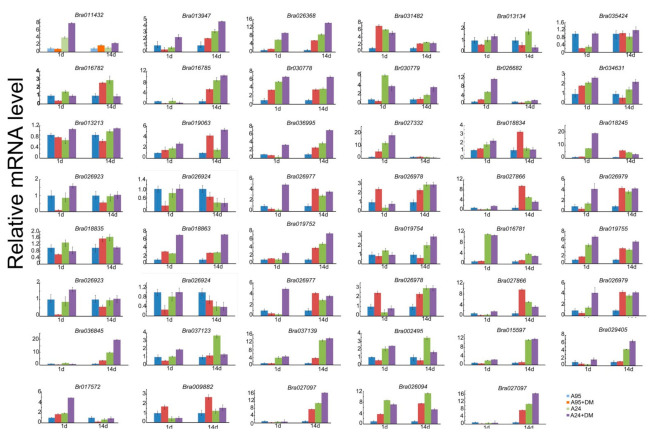
Then, the expression levels of inbred lines A95 and A24 were detected when they were exposed to DM for 1 day and 14 days, as shown in Figure 7. The expression levels of most BrCC–NBS–LRR genes in the sensitive inbred line A95 and resistant line A24 were up-regulated in response to DM. A total of 11 BrCC–NBS–LRR genes showed similar expression characteristics. The expression levels in the two lines were induced by DM to a higher degree than by the control. Moreover, the expression levels in A24 were even remarkably higher than those in A95, not only at 1 day of DM inoculation but also after the control inoculation, particularly those of Bra011432, Bra026368, Bra030778, Bra026682, Bra034631, Bra017572, Bra027332, Bra018245, Bra018863, Bra019755, and Bra037139. Meanwhile, 13 CC–NBS–LRR family members of Chinese cabbage had the same trend of expression as above in the long inoculations (14 days), particularly Bra013947, Bra026368, Bra029405, Bra030778, Bra030779, Bra036995, Bra027097, Bra018863, Bra019752, Bra019755, Bra036845, Bra037139, and Bra015597. The trends of Bra026368, Bra030778, Bra018863, Bra019755, and Bra015597 in the 1 day short inoculation were consistent with those in the 14 day long inoculation. By contrast, Bra009882 and Bra026978 expression in A24 and A95 decreased obviously after 1 day of DM inoculation.
Figure 7.
The BrCC–NBS–LRR expression levels of inbred line A95 and A24 with DM inoculation. The CC–NBS–LRR genes’ expression levels were detected in A95 and A24 for both 24 h (short inoculation) and 14 days (long inoculation) of DM inoculation with water as control. They represent results performed in triplicate. Error bars represent ± SE.
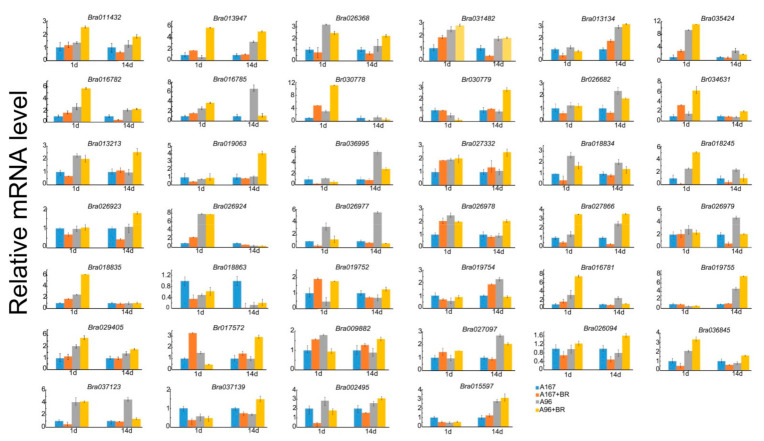
As for disease stress due to bacterial BR, the sensitive line A167 and resistant line A96 were also inoculated with Xanthomonas campestris, and the expression levels of the BrCC–NBS–LRR family were detected through qPCR 1 and 14 days after inoculation, as shown in Figure 8. A total of 14 BrCC–NBS–LRR genes were induced by BR after 1 day of inoculation in A167 and A96, and six BrCC–NBS–LRR genes were induced after 14 days of inoculation. In addition, the expression levels in A96 were even higher than those in A167 after inoculation. Especially, the expression levels of Bra035424 and Bra030778 in A167 were up-regulated more than 11-fold. Only one member showed the same expression trend as above (Bra027866). However, the expression of Bra030779 was down-regulated after 1 day of BR inoculation. These results revealed that the CC–NBS–LRR gene family of Chinese cabbage may play different roles in response to a disease and the type of role depends on inoculation time and disease types.
Figure 8.
The BrCC–NBS–LRR expression levels of inbred line A95 and A24 with BR inoculation. The CC–NBS–LRR genes’ expression levels were detected in A95 and A24 for both 24 h (short inoculation) and 14 days (long inoculation) of BR inoculation with water as control. They represent results performed in triplicate. Error bars represent ± SE.
3. Discussion
Plant disease resistance genes (R genes) have mechanisms for recognizing and providing direct or indirect protection against pathogens in plants and play a critical role in effector triggered immunity (ETI) [28]. Of all five classes of R genes, the NBS–LRR class encodes the largest protein family with one or more NBSs and an LRR domain at the C terminal [29]. The NBS–LRR protein family usually contains two major subfamily TNL genes and non-toll/interleukin-1 receptor-NBS-LRR (nTNL) genes, which is composed CC–NBS–LRR [30]. In our study, 40 CC–NBS–LRR members of Chinese cabbage were searched and identified on the basis of the entire genome of Chinese cabbage. Although 47.1% NBS encodings of Brassica rapa appeared to have undergone tandem duplication and to be distributed in tandem arrays [31], all BrCC–NBS–LRRs were unevenly distributed on A01–10, with five pairs of two copies (Figure 1) and two pairs of three copies. No BrCC–NBS–LRR members were found on A04 and A07; however, A09 had the most members on it with 12 items. Brassica rapa and Arabidopsis thaliana belong to Brassicaceae plants and shared high homology with each other. A total of 21 AtCC–NBS–LRR genes were found on the whole genome, 17 members were found on Chr.1, and only one was found on A05 (Table S2), which showed that the distribution was more extreme. Furthermore, CC–NBS–LRRs of Chinese cabbage were expanded 1.9-fold compared to Arabidopsis thaliana.
Although Brassica rapa shared high homology with model monocotyledonous plant Arabidopsis thaliana, based on the protein sequence the phylogenetic tree of the CC–NBS–LRR family was built to explore the homology among Arabidopsis thaliana, Brassica rapa and monocotyledon rice. To our surprise, not all these BrCC–NBS–LRRs were highly clustered in the same subgroup with their At orthologs; moreover, CC–NBS–LRRs of the same species were clustering together. Even two BrCC–NBS–LRRs shared high homology with rice instead of Arabidopsis, such as Bra013213 and Bra017572, as shown in Figure 1.
The protein structures of BrCC–NBS–LRRs were usually composed of three parts: one or more NBSs, an LRR domain at the C terminal, and a CC domain at the N terminal. NBS-encoding domains are necessary for the recognition of diverse pathogens in plants [32]. Five kinds of NBS-encoding motifs were found in BrCC–NBS–LRRs, such as motifs 2, 3, 4, 7, and 10 (Figure 3), and motif 3 was the most conserved in the BrCC–NBS–LRRs. LRR may be a core function domain to the recruitment of protein [33]. LRRs in general are irregular and variably responsible for interactions between resistance proteins on account of a hydrophobic backbone for three-dimensional integrity and many free solvent-exposed residues of the LRR domain [34,35]. The CC domains were detected in all the CC–NBS–LRR members of Brassica rapa on IntrePro. The CC motif was analogous to the LRR motif in the critical core of the three-dimensional structure composed of hydrophobic chains and free solvent-exposed residues. The CC domain of a CC–NBS–LRR gene of LR10 in wheat directly recognizes pathogens; this result indicated that the CC domain function of CC–NBS–LRRs needs to be further explored and excavated [36].
The cis-acting elements of gene promoters were essential for transcriptional expression, and the types of cis-acting elements indicated the potential functions of genes, particularly in response to pathogens [37]. All the BrCC–NBS–LRR promoters were downloaded from BRAD and analyzed with an online tool New PLACE for the searching of cis-acting regulatory elements. A total of 12 disease-resistance-related cis-acting elements were found in the promoters of BrCC–NBS–LRR. The GT1CONSENSU (consensus GT-1) elements dominated as investors and appeared on the promoters in tandem. It was first reported as the binding sites of many light-regulated genes in many species, for example, ribulose bisphosphate carboxylase small chain, phytase gene [38,39]. The binding of GT-1-like factors to the PR-1a promoter can reduce TMV infection and SA treatment and influence the expression level of SA-inducible genes [23]. The function of GT1GMSCAM4 was analogous to that of GT1CONSENSU. WRKY71OS, WBOXATNPR1, and WBBOXPCWRKY1 are typical disease-related elements found in BrCC–NBS–LRRs. WRKY71OS is not only involved in plant defense signaling by binding specifically to W box elements of the PR-10, but also plays a role in GA and ABA signaling pathways [25,40]. “W-box” sequences were found in the promoters of WBOXATNPR1 and WBBOXPCWRKY1 to bind with NPR1 and PR1, participating in plant defense response [41,42]. A total of 68 BIHD1OS elements were found in BrCC–NBS–LRR genes, that is, the binding site of the BELL-type transcription factor of OsBIHD1 associated with resistance and response in rice [26]. Some minor cis-acting elements are associated with disease resistance, such as SEBFCONSSTPR10A, ELRECOREPCRP1, MYB1LEPR, TL1ATSAR, GCCCORE, and AGCBOXNPGLB.
In recent years, a large number of plant genomes have been sequenced, and an increasing number of NBS-encoding families of different species have been analyzed and surveyed [36]. In addition, five Brassicaceae NBS–LRR genes were comprehensively analyzed and compared on the basis of established cross-species phylogenetic and syntenic relationships of NBS genes for the study of the R gene in Brassicaceae, particularly in Arabidopsis lyrata, Arabidopsis thaliana, Brassica rapa, Capsella rubella (127), and Thellungiella salsuginea [43]. NBS-LRR genes exert critical effects in response to multiple pathogens, including viruses, bacteria, and fungi [44]. CC–NBS–LRR, as a main subfamily of NBS–LRR, is involved in disease resistance in plants, such as Arabidopsis thaliana, rice, and wheat. However, they are rarely reported in Chinese cabbage in terms of response to DM and BR. In our study, the A160 inbred lines of Chinese cabbage were inoculated with Hyaloperonospora parasitica and Xanthomonas campestris, and short and long inoculations were performed for exploring the expression profile. In the short inoculations, heatmaps were drawn after 1, 3, 6, 12, and 24 h of inoculation (Figure 5 and Figure 6). We found that most BrCC-NBS-LRR genes were induced drastically 1 h after inoculation, and a peak in expression level occurred 12 h after inoculation with Hyaloperonospora parasitica and Xanthomonas campestris. Bra026978 was continuously induced by Hyaloperonospora parasitica. The expression levels of Bra018834, Bra019755, and Bra026094 were up-regulated by Hyaloperonospora parasitica at each inoculation time in the short inoculations, which showed that these genes have a relationship with DM defense. Bra016785 and Bra027332 had high expression levels during BR inoculation. For long inoculations, the expressed BrCC-NBSL-RRs after inoculation with DM were more abundant than those observed after BR inoculation. Moreover, the differentially expressed genes responding to each pathogen slightly varied. Bra030778 was induced markedly at each inoculation of BR, but no change in response to DM was observed, revealing that Bra030778 preferred involvement in BR defense.
An increasing number of CC–NBS–LRRs have been reported, such as Lr10, which is involved in (CC–NBS–LRR)-mediated resistance [45]; Yr10 (CC–NBS–LRR), which plays a key role in resistance to Pst [46]; Lr34 (CC–NBS–LRR), which enhances resistance during adult stages; TaRPM1, which is involved in resistance to Pst [47]; and Pm21 from the wild species, which shows high resistance to Bgt [48]. Additionally, in rice Pb1 shows resistance to rice blast only during adult stages [49]. To further identify the function of BrCC-NBS-LRRs, the expression characteristics were carried out on the sensitive inbred lines A95 and A167 and resistant lines A24 and A96 to DM and BR for 1 day and 14 day inoculation, as shown in Figure 7 and Figure 8. There were 11 BrCC-NBS-LRR genes showing similar expression profiles induced by DM rather than the control; moreover, A24 was even higher than A95 DM for 1 day inoculation and for 13 for the long inoculation (14 days). The expression levels of BrCC–NBS–LRR genes after 1 day of inoculation with DM and BR were compared. Bra030778 and Bra034631 were induced and had the same expression level. In the long inoculation of DM and BR, three BrCC–NBS–LRRs (Bra013947, Bra019755, and Bra015597) showed higher expression levels. These results revealed that BrCC–NBS–LRR family genes play various roles in response to phytopathogens, and the roles depend on disease type and inoculation time.
4. Materials and Methods
4.1. Plant Materials and Inoculation
A total of 24 inbred lines of Chinese cabbage from Northeast Agricultural University were cultivated in a greenhouse subjected to day (28 °C)/night (18 °C) and 16 h light/8 h dark cycles. For the expression profiles of DM and BR, the Brassica rapa inbred line A160 of the fifth leaf was sprayed with pathogens and harvested 3, 5, 7, 10, and 14 days after infection, and non-infected Chinese cabbage was used as the control. Additionally, for screening the sensitive and insensitive plants, the fourth leaves of 24 inbred lines were infected by both Hyaloperonospora parasitica and Xanthomonas campestris for 14 days, respectively, until invasion of the two diseases with water as the control. The whole leaf and lesion areas of Chinese cabbage were surveyed, and three replications of biological duplication were carried out. The lesion areas of leaves were measured, and according to statistics, the inbred lines with a lesion area taking up greater than 75% of the whole leaves were sensitive lines, and at the same time less than 20% was used as the resistant line. Inbred lines A95 and A167 were the most sensitive to DM and BR, respectively, whereas A24 and A96 were resistant. The leaves of the sensitive and resistant plants were harvested and frozen in liquid nitrogen and then stored at −80 °C for RNA isolation.
The inbred line A160 of Chinese cabbage was grown in a greenhouse until four leaves were moved to light incubator with 6°C for 25 days of vernalization, and then they were grown under conventional environment until flowering. In total, the tissues of root, stem, leaf, flower and silique were harvested, respectively, for RNA isolation.
4.2. Identification and Analysis of CC–NBS–LRR in Chinese Cabbage
All the CC–NBS–LRR members of Chinese cabbage were obtained from BRAD (http://brassicadb.org/brad/, accessed on 27 February 2021), with the B. Rapa genome (version 1.5). They were identified with the NB–ARC domain according to the Hidden Markov Model of pfam00931 and the LRR domain of PLN03210 at the C terminal with the BLAST program of the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 14 April 2021). The CC domain of BrCC–NBS–LRRs was confirmed using MPI Bioinformatics Toolkit (https://toolkit.tuebingen.mpg.de/#/tools/quick2d, accessed on 27 February 2021. 2008–2021) [50]. The other CC–NBS–LRR sequence of Arabidopsis thaliana, with A thaliana genome: Araport11 and Oryza sativa were downloaded from TAIR (http://www.arabidopsis.org/, accessed on 27 February 2021. 2005) and Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/, 6 February 2013), which were confirmed using the Basic Local Alignment Search Tool of NCBI. InterPro (http://www.ebi.ac.uk/interpro/scan.html, accessed on 27 February 2021. 2021) and Pfam (http://pfam.xfam.org/search#tabview=tab1, accessed on 27 February 2021. 2018) were used for further confirmation [51].
4.3. The Cis-Acting Elements in Promoters, Chromosomal Locations, and Gene Structures
The promoter sequences of BrCC–NBS–LRR genes were searched on BRAD (http://brassicadb.org/brad/, accessed on 27 February 2021), and all of them were analyzed using New PLACE (https://sogo.dna.affrc.go.jp/cgi-bin/sogo.cgi?lang=en&pj=640&action=page&page=newplace, accessed on 27 February 2021. 2011–2021), which is a database containing plants used in the analysis of cis-acting regulatory elements [52]. Information on chromosomal location was obtained from BRAD and assessed by MapInspect. The genome and CDS sequences of CC–NBS–LRR were downloaded from BRAD, which were placed in the Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/, accessed on 27 February 2021. 2021) for the reconstruction of the gene structure [53].
4.4. Phylogenetics and Conserved Motif Analysis of CC–NBS–LRR Protein
The CC–NBS–LRR protein sequences of Brassica rapa and Arabidopsis thaliana and Oryza sativa were downloaded from BRAD, TAIR, and Rice Genome Annotation Project. First, multiple sequences were aligned with MEGA6, then the phylogenetic trees were calculated and constructed using the neighbor-joining method within 1000 replicates [54].
The protein sequences were analyzed using the online tool MEME Suite with the following settings: site distribution, any number of repetitions; number of motifs, 10.
4.5. Total RNA Extraction and Real-Time PCR Analysis
The plants described in the Plant Materials section were harvested for RNA extraction with TRIzol™ reagent (Invitroge, Thermo Fisher, Waltham, MA, USA), and 2 mg of RNA was reversed-transcribed to cDNA with TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix kits (TransGen, Beijing, China). Approximately 15 L of total volume was used for real-time PCR, containing 1 L of specific primers, 3 L of cDNA samples diluted 30–50-fold and 7.5 L of SYBR Green real-time PCR master mix (TOYOBO, Osaka, Japan). The reactions were amplified using a qTower3G (Analytik Jena, Jena, Germany) real-time PCR detection system for 40 cycles with unimodal dissolution curve and tubulin as an internal reference [55]. The primers are listed in Table S7.
Acknowledgments
This work was supported by the Natural Science Foundation of Heilongjiang Province of China (LH2019C016), China Postdoctoral Science Foundation (2018M641799), “Young Talents” Project of Northeast Agricultural University of China (18QC07).
Abbreviation
| BR | black rot |
| CC–NBS–LRR | coil-coil nucleotide-binding site leucine-rich repeat |
| DM | Downy mildew |
| ETI | Effector triggered immunity |
| NB-ARC | nucleotide-binding adaptor shared by APAF-1, R proteins and CED-4 |
| PI | isoelectric point |
| SA | salicin |
| TIR | toll/interleukin 1 receptor. |
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms22084266/s1, Figure S1: Sequence logos of motifs in BrCC–NBS–LRR, Figure S1: Sequence logos of motifs in BrCC–NBS–LRR, Figure S2: The number of different type cis-elements on each BrCC–NBS–LRR promoter, Figure S3: The total disease resistant related cis-element number on each Chinese cabbage CC–NB–LRR promoter, Figure S4: Heatmap of BrCC–NBS–LRRs in root, stem, leaf, flower and silique, Table S1: The homologous gene of BrCC–NBS–LRRs in Arabidopsis thaliana and their positive function, Table S2: The chromosome locations of CC–NBS–LRR in Arabidopsis thaliana and Chinese cabbage, Table S3: CC–NBS–LRR genes in Arabidopsis thaliana, rice and Chinese cabbage, Table S4: Motif sequences detected by MEME, Table S5: The promoter cis-elements of CC–NB–LRR genes in Chinese cabbage, Table S6: Primers of qPCR.
Author Contributions
Y.L., A.W. and Y.Z. planned and designed the study and wrote the manuscript. Y.L., D.L. and N.Y. performed the experiments. X.Z. implemented the software. K.H. and R.G. provided supplementary experimental results. J.B. collected all of the plant materials in this work. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Natural Science Foundation of Heilongjiang Province of China, grant number LH2019C016, China Postdoctoral Science Foundation, grant number 2018M641799, “Young Talents” Project of Northeast Agricultural University of China, grant number 18QC07.
Institutional Review Board Statement
This study did not involve humans or animals.
Informed Consent Statement
This study did not involve humans.
Data Availability Statement
This statement if the study did not report any data.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ausubel F.M. Are innate immune signaling pathways in plants and animals conserved? Nat. Immunol. 2005;6:973–979. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]
- 2.Haak D.C., Fukao T., Grene R., Hua Z.H., Ivanov R., Perrella G., Li S. Multilevel Regulation of Abiotic Stress Responses in Plants. Front. Plant Sci. 2017;8:1564. doi: 10.3389/fpls.2017.01564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker C.M., Chitrakar R., Obulareddy N., Panchal S., Williams P., Melotto M. Molecular battles between plant and pathogenic bacteria in the phyllosphere. Braz. J. Med. Biol. Res. 2010;43:698–704. doi: 10.1590/S0100-879X2010007500060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belkhadir Y., Subramaniam R., Dangl J.L. Plant disease resistance protein signaling: NBS-LRR proteins and their partners. Curr. Opin. Plant Biol. 2004;7:391–399. doi: 10.1016/j.pbi.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 5.Tameling W.I., Elzinga S.D., Darmin P.S., Vossen J.H., Takken F.L., Haring M.A., Cornelissen B.J. The tomato R gene products I-2 and MI-1 are functional ATP binding proteins with ATPase activity. Plant Cell. 2002;14:2929–2939. doi: 10.1105/tpc.005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsushima N., Miyashita H. Leucine-Rich Repeat (LRR) Domains Containing Intervening Motifs in Plants. Biomolecules. 2012;2:288–311. doi: 10.3390/biom2020288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wan H., Yuan W., Ye Q., Wang R., Ruan M., Li Z., Zhou G., Yao Z., Zhao J., Liu S., et al. Analysis of TIR- and non-TIR-NBS-LRR disease resistance gene analogous in pepper: Characterization, genetic variation, functional divergence and expression patterns. BMC Genom. 2012;13:502. doi: 10.1186/1471-2164-13-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams M., Tummala N.R., Aziz S.G., Risko C., Bredas J.L. Influence of Molecular Shape on Solid-State Packing in Disordered PC61BM and PC71BM Fullerenes. J. Phys. Chem. Lett. 2014;5:3427–3433. doi: 10.1021/jz501559q. [DOI] [PubMed] [Google Scholar]
- 9.Tarr D.E., Alexander H.M. TIR-NBS-LRR genes are rare in monocots: Evidence from diverse monocot orders. BMC Res. Notes. 2009;2:197. doi: 10.1186/1756-0500-2-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sukarta O.C.A., Slootweg E.J., Goverse A. Structure-informed insights for NLR functioning in plant immunity. Semin. Cell Dev. Biol. 2016;56:134–149. doi: 10.1016/j.semcdb.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 11.Song W., Wang B., Li X., Wei J., Chen L., Zhang D., Zhang W., Li R. Identification of Immune Related LRR-Containing Genes in Maize (Zea mays L.) by Genome-Wide Sequence Analysis. Int. J. Genom. 2015;2015:231358. doi: 10.1155/2015/231358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jia Y., McAdams S.A., Bryan G.T., Hershey H.P., Valent B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 2000;19:4004–4014. doi: 10.1093/emboj/19.15.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deslandes L., Olivier J., Peeters N., Feng D.X., Khounlotham M., Boucher C., Somssich I., Genin S., Marco Y. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. USA. 2003;100:8024–8029. doi: 10.1073/pnas.1230660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeYoung B.J., Innes R.W. Plant NBS-LRR proteins in pathogen sensing and host defense. Nat. Immunol. 2006;7:1243–1249. doi: 10.1038/ni1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gururani M.A., Venkatesh J., Upadhyaya C.P., Nookaraju A., Pandey S.K., Park S.W. Plant disease resistance genes: Current status and future directions. Physiol. Mol. Plant P. 2012;78:51–65. doi: 10.1016/j.pmpp.2012.01.002. [DOI] [Google Scholar]
- 16.Hu Y., Li Y., Hou F., Wan D., Cheng Y., Han Y., Gao Y., Liu J., Guo Y., Xiao S., et al. Ectopic expression of Arabidopsis broad-spectrum resistance gene RPW8.2 improves the resistance to powdery mildew in grapevine (Vitis vinifera) Plant Sci. 2018;267:20–31. doi: 10.1016/j.plantsci.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y., Guan X.Y., Liu S.N., Yang M., Ren J.H., Guo M., Huang Z.H., Zhang Y.W. Genome-Wide Identification and Analysis of TCP Transcription Factors Involved in the Formation of Leafy Head in Chinese cabbage. Int. J. Mol. Sci. 2018;19:847. doi: 10.3390/ijms19030847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hashiba T., Narisawa K. The development and endophytic nature of the fungus Heteroconium chaetospira. Fems Microbiol Lett. 2005;252:191–196. doi: 10.1016/j.femsle.2005.08.039. [DOI] [PubMed] [Google Scholar]
- 19.Zeilmaker T., Ludwig N.R., Elberse J., Seidl M.F., Berke L., Van Doorn A., Schuurink R.C., Snel B., Van den Ackerveken G. DOWNY MILDEW RESISTANT 6 and DMR6-LIKE OXYGENASE 1 are partially redundant but distinct suppressors of immunity in Arabidopsis. Plant J. 2015;81:210–222. doi: 10.1111/tpj.12719. [DOI] [PubMed] [Google Scholar]
- 20.Zhang B., Li P., Su T.B., Li P.R., Xin X.Y., Wang W.H., Zhao X.Y., Yu Y.J., Zhang D.S., Yu S.C., et al. BrRLP48, Encoding a Receptor-Like Protein, Involved in Downy Mildew Resistance in Brassica rapa. Front. Plant Sci. 2018;9:1708. doi: 10.3389/fpls.2018.01708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mun J.H., Yu H.J., Park S., Park B.S. Genome-wide identification of NBS-encoding resistance genes in Brassica rapa. Mol. Genet. Genom. 2009;282:617–631. doi: 10.1007/s00438-009-0492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lv S.W., Changwei Z., Tang J., Li Y.X., Wang Z., Jiang D.H., Hou X.L. Genome-wide analysis and identification of TIR-NBS-LRR genes in Chinese cabbage (Brassica rapa ssp pekinensis) reveal expression patterns to TuMV infection. Physiol Mol Plant P. 2015;90:89–97. doi: 10.1016/j.pmpp.2015.04.001. [DOI] [Google Scholar]
- 23.Buchel A.S., Brederode F.T., Bol J.F., Linthorst H.J. Mutation of GT-1 binding sites in the Pr-1A promoter influences the level of inducible gene expression in vivo. Plant Mol. Biol. 1999;40:387–396. doi: 10.1023/A:1006144505121. [DOI] [PubMed] [Google Scholar]
- 24.Xie Z., Zhang Z.L., Zou X., Huang J., Ruas P., Thompson D., Shen Q.J. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 2005;137:176–189. doi: 10.1104/pp.104.054312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eulgem T., Rushton P.J., Robatzek S., Somssich I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 26.Luo H., Song F., Goodman R.M., Zheng Z. Up-regulation of OsBIHD1, a rice gene encoding BELL homeodomain transcriptional factor, in disease resistance responses. Plant Biol. 2005;7:459–468. doi: 10.1055/s-2005-865851. [DOI] [PubMed] [Google Scholar]
- 27.Chakravarthy S., Tuori R.P., D’Ascenzo M.D., Fobert P.R., Despres C., Martin G.B. The tomato transcription factor Pti4 regulates defense-related gene expression via GCC box and non-GCC box cis elements. Plant Cell. 2003;15:3033–3050. doi: 10.1105/tpc.017574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang X.P., Wang J.P. Genome-Wide Analysis of NBS-LRR Genes in Sorghum Genome Revealed Several Events Contributing to NBS-LRR Gene Evolution in Grass Species. Evol. Bioinform. 2016;12:9–21. doi: 10.4137/EBO.S36433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyers B.C., Kozik A., Griego A., Kuang H.H., Michelmore R.W. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15:809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shao Z.Q., Zhang Y.M., Hang Y.Y., Xue J.Y., Zhou G.C., Wu P., Wu X.Y., Wu X.Z., Wang Q., Wang B., et al. Long-term evolution of nucleotide-binding site-leucine-rich repeat genes: Understanding gained from and beyond the legume family. Plant Physiol. 2014;166:217–234. doi: 10.1104/pp.114.243626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu J.Y., Tehrim S., Zhang F.Q., Tong C.B., Huang J.Y., Cheng X.H., Dong C.H., Zhou Y.Q., Qin R., Hua W., et al. Genome-wide comparative analysis of NBS-encoding genes between Brassica species and Arabidopsis thaliana. BMC Genomics. 2014;15:1–18. doi: 10.1186/1471-2164-15-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sekhwal M.K., Li P.C., Lam I., Wang X.E., Cloutier S., You F.M. Disease Resistance Gene Analogs (RGAs) in Plants. Int. J. Mol. Sci. 2015;16:19248–19290. doi: 10.3390/ijms160819248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song W., Forderer A., Yu D., Chai J. Structural biology of plant defence. New Phytol. 2021;229:692–711. doi: 10.1111/nph.16906. [DOI] [PubMed] [Google Scholar]
- 34.Mackey D., Holt B.F., 3rd, Wiig A., Dangl J.L. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 2002;108:743–754. doi: 10.1016/S0092-8674(02)00661-X. [DOI] [PubMed] [Google Scholar]
- 35.Ade J., DeYoung B.J., Golstein C., Innes R.W. Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc. Natl. Acad. Sci. USA. 2007;104:2531–2536. doi: 10.1073/pnas.0608779104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu J., Zhu J.F., Wang L.F., Wang S.M. Genome-Wide Association Study Identifies NBS-LRR-Encoding Genes Related with Anthracnose and Common Bacterial Blight in the Common Bean. Front. Plant Sci. 2017;8:1398. doi: 10.3389/fpls.2017.01398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rushton P.J., Reinstadler A., Lipka V., Lippok B., Somssich I.E. Synthetic plant promoters containing defined regulatory elements provide novel insights into pathogen- and wound-induced signaling. Plant Cell. 2002;14:749–762. doi: 10.1105/tpc.010412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gilmartin P.M., Chua N.H. Spacing between GT-1 binding sites within a light-responsive element is critical for transcriptional activity. Plant Cell. 1990;2:447–455. doi: 10.1105/tpc.2.5.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Villain P., Mache R., Zhou D.X. The mechanism of GT element-mediated cell type-specific transcriptional control. J. Biol. Chem. 1996;271:32593–32598. doi: 10.1074/jbc.271.51.32593. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Z.L., Xie Z., Zou X.L., Casaretto J., Ho T.H.D., Shen Q.X.J. A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol. 2004;134:1500–1513. doi: 10.1104/pp.103.034967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu D., Chen C., Chen Z. Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell. 2001;13:1527–1540. doi: 10.1105/TPC.010115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rushton P.J., Torres J.T., Parniske M., Wernert P., Hahlbrock K., Somssich I.E. Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 1996;15:5690–5700. doi: 10.1002/j.1460-2075.1996.tb00953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y.M., Shao Z.Q., Wang Q., Hang Y.Y., Xue J.Y., Wang B., Chen J.Q. Uncovering the dynamic evolution of nucleotide-binding site-leucine-rich repeat (NBS-LRR) genes in Brassicaceae. J. Integr. Plant Biol. 2016;58:165–177. doi: 10.1111/jipb.12365. [DOI] [PubMed] [Google Scholar]
- 44.Dangl J.L., Horvath D.M., Staskawicz B.J. Pivoting the plant immune system from dissection to deployment. Science. 2013;341:746–751. doi: 10.1126/science.1236011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loutre C., Wicker T., Travella S., Galli P., Scofield S., Fahima T., Feuillet C., Keller B. Two different CC-NBS-LRR genes are required for Lr10-mediated leaf rust resistance in tetraploid and hexaploid wheat. Plant J. 2009;60:1043–1054. doi: 10.1111/j.1365-313X.2009.04024.x. [DOI] [PubMed] [Google Scholar]
- 46.Liu W., Frick M., Huel R., Nykiforuk C.L., Wang X.M., Gaudet D.A., Eudes F., Conner R.L., Kuzyk A., Chen Q., et al. The Stripe Rust Resistance Gene Yr10 Encodes an Evolutionary-Conserved and Unique CC-NBS-LRR Sequence in Wheat. Mol. Plant. 2014;7:1740–1755. doi: 10.1093/mp/ssu112. [DOI] [PubMed] [Google Scholar]
- 47.Wang J.H., Tian W., Tao F., Wang J.J., Shang H.S., Chen X.M., Xu X.M., Hu X.P. TaRPM1 Positively Regulates Wheat High-Temperature Seedling-Plant Resistance to Puccinia striiformis f. sp. tritici. Front. Plant Sci. 2020;10:1679. doi: 10.3389/fpls.2019.01679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xing L., Hu P., Liu J., Witek K., Zhou S., Xu J., Zhou W., Gao L., Huang Z., Zhang R., et al. Pm21 from Haynaldia villosa Encodes a CC-NBS-LRR Protein Conferring Powdery Mildew Resistance in Wheat. Mol. Plant. 2018;11:874–878. doi: 10.1016/j.molp.2018.02.013. [DOI] [PubMed] [Google Scholar]
- 49.Hayashi N., Inoue H., Kato T., Funao T., Shirota M., Shimizu T., Kanamori H., Yamane H., Hayano-Saito Y., Matsumoto T., et al. Durable panicle blast-resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication. Plant J. Cell Mol. Biol. 2010;64:498–510. doi: 10.1111/j.1365-313X.2010.04348.x. [DOI] [PubMed] [Google Scholar]
- 50.Zimmermann L., Stephens A., Nam S.Z., Rau D., Kubler J., Lozajic M., Gabler F., Soding J., Lupas A.N., Alva V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 2018;430:2237–2243. doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 51.Bailey T.L., Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers; Proceedings of the International Conference on Intelligent Systems for Molecular Biology; Stanford, CA, USA. 15–17 August 1994; Menlo Park, CA, USA: Association for the Advancement of Artificial Intelligence; 1994. pp. 28–36. [PubMed] [Google Scholar]
- 52.Higo K., Ugawa Y., Iwamoto M., Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu B., Jin J., Guo A.Y., Zhang H., Luo J., Gao G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saitou N., Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Y.W., Jin D., Xu C., Zhang L., Guo M.H., Fang Z.Y. Regulation of bolting and identification of the alpha-tubulin gene family in Brassica rapa L. ssp pekinensis. Genet. Mol. Res. 2016;15:1–13. doi: 10.4238/gmr.15017507. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This statement if the study did not report any data.